Abstract
Double-stranded RNA-binding protein Staufen homolog 1 (STAU1) is a highly conserved multifunctional double-stranded RNA-binding protein, and is a key factor in neuronal differentiation. RNA sequencing was used to analyze the overall transcriptional levels of the upregulated cells by STAU1 and control cells, and select alternative splicing (AS). It was determined that the high expression of STAU1 led to changes in the expression levels of a variety of inflammatory and immune response genes, including IFIT2, IFIT3, OASL, and CCL2. Furthermore, STAU1 was revealed to exert a significant regulatory effect on the AS of genes related to the ‘nerve growth factor receptor signaling pathway’. This is of significant importance for neuronal survival, differentiation, growth, post-damage repair, and regeneration. In conclusion, overexpression of STAU1 was associated with immune response and regulated AS of pathways related to neuronal growth and repair. In the present study, the whole transcriptome of STAU1 expression was first analyzed, which laid a foundation for further understanding the key functions of STAU1.
Keywords: double-stranded RNA-binding protein Staufen homolog 1, RNA-binding protein, RNA-sequencing, alternative splicing, nerve growth factor receptor signaling pathway
Introduction
RNA binding proteins (RBPs) are an important family of proteins related to RNA metabolism. They dynamically bind RNA to form a variety of complexes, including ribonucleoprotein particles (RNPs) (1). RBPs play a significant role in various cellular processes, including RNA splicing, polyadenylation, RNA editing, RNA transport, maintenance of RNA stability and degradation, intracellular localization, and translation control (2). Recent studies have revealed that RBPs are closely associated with muscular dystrophy, neurological disease, cancer, and Mendelian disease (3,4).
Staufen is a member of the double-stranded RNA-binding protein family, which is involved in mRNA transport and localization to different subcellular compartments (5). Staufen proteins contain several double-stranded RNA-binding domains (6). Double-stranded RNA-binding protein Staufen homolog 1 (STAU1) plays a crucial role in mRNA output, re-localization, translation, and STAU1-mediated mRNA decay (SMD) (7,8). SMD is also involved in the developmental process, such as myogenesis and adipogenesis, and possibly in angiogenesis (9). STAU1, by binding to the inverted repeat Alu elements (IRAlus) on the 3′UTR of mRNA, inhibits retention of IRAlus-containing mRNA in the nuclei, thereby enhancing its nuclear export. Moreover, STAU1 binding to the IRAlus on the 3′UTR of mRNA can promote translation of these mRNAs by inhibiting the binding of protein kinase R (PKR) (10). In addition, STAU1 is also an important mRNA transport factor in neurons. After binding to the 3′UTR of mRNA, STAU1 can induce translation-dependent mRNA degradation through direct interaction with UPF1. An upregulation of transcripts due to depletion of STAU1 and UPF1 may play a pivotal role in the differentiation process (11). STAU1 expression can be used to regulate neuronal differentiation (12). Studies have revealed that STAU1 plays a substantial role in the immunity against the influenza virus and human immunodeficiency virus type I (HIV-1) (13–15).
In order to study the possible biological functions of STAU1, a STAU1-regulated transcriptome in HeLa cells was obtained. To determine the gene expression profile and the alternative splicing (AS) events in the genome, which are regulated by STAU1, high-throughput RNA sequencing (RNA-seq) was employed. In addition, the related results were validated in HeLa cells. A previous research demonstrated that PTB knockdown converted highly transformed HeLa cells to neuronal-like cells. They extended this analysis to multiple cell types of diverse origin, including human embryonic carcinoma stem cells (NT2), mouse neural progenitor cells (N2A), human retinal epithelial cells (ARPE19), and primary mouse embryonic fibroblasts (MEFs). Upon PTB knockdown, all of these cells exhibited a neuronal-like morphology (16). Comparative transcriptome analysis revealed that STAU1 can selectively regulate the expression of inflammatory genes, including interferon-induced protein with tetratricopeptide repeats 2 (IFIT2), 2′-5′-oligoadenylate synthetase-like protein (OASL), and interferon-induced protein with tetratricopeptide repeats 3 (IFIT3). Furthermore, it was revealed that AS of genes widespread in the ‘nerve growth factor receptor signaling pathway’, including PLEKHG2, ARHGEF11, NR4A1, PDGFB, FGFR4, and RALGDS, was regulated by STAU1. Overexpression of STAU1 was closely associated with inflammation. Collectively, the present research defined a potential regulatory pattern in which AS of inflammatory and immune response genes was regulated by STAU1. It indicated that STAU1 may be involved in the proliferation and survival of neurons (17), differentiation, cell growth and apoptosis (18), post-damage repair and regeneration, neurite outgrowth and retraction (19), and myelination (20).
Materials and methods
Cloning and construction of plasmids
CE Design 1.04 software (Vazyme Biotech Co., Ltd.). was employed to design primers for Hot Fusion (21). There was a gene-specific sequence and a pIRES-hrGFP-1a vector (part no. 240031; Agilent Technologies, Inc.) sequence (17–30 bp) in each primer. The designed primers contained two parts of a sequence, the sequence before ‘ATG’ in the forward (F)-primer or ‘AGC’ in the reverse (R)-primer matches of the pIRES-hrGFP-1a vector and the other part of the sequence belonged to STAU1 gene sequence, presented as follows: F-primer: AgcccgggcggatccgaattcATGAAACTTGGAAAAAAACCAATGT and R-primer: GtcatccttgtagtcctcgagAGCACCTCCCACACACAGACA.
EcoRI and XhoI (NEB) were used to digest pIRES-hrGFP-1a vectors for ~2–3 h at 37°C, which were then subjected to agarose (1.0%) gel electrophoresis and purification using Qiagen spin-column-based kit (Qiagen GmbH) according to the manufacturer's instructions. TRIzol reagent (Ambion) was used to extract total RNA from HeLa cells, which was used to synthesize cDNA by oligo dT primer. The cDNA is the template to amplify insert fragments of the STAU1 gene using the primers by polymerase chain reaction (PCR). The vectors and insert fragments were added to a microtube and ligated by ClonExpress II One Step Cloning kit (Vazyme Biotech Co., Ltd.). The ligand product was transformed into E. coli DH5α, which was then added to a Luria-Bertani (LB) agar plate, containing 1 µl/ml ampicillin (Sigma-Aldrich; Merck KGaA), followed by overnight incubation at 37°C. The template used for PCR was DNA extracted from DH5α cells, using DNA polymerase 2X Green Taq Mix (Vazyme Biotech Co., Ltd.). The sequences of the Universal primers were as follows: Forward (F)-primer: AATTAACCCTCACTAAAGGG and Reverse (R)-primer: GTCCTTATCATCGTCGTCTT.
PCR procedures were carried out as follows: The samples were first denatured at 95°C for 5 min, denaturation at 95°C for 3 min, followed by 28 cycles of annealing at 55°C for 30 sec and extension at 72°C for 1 min. Universal primers were employed to screen colonies. Sanger sequencing (22) was used to verify the insert sequence.
Cell culture and transfections
Human cervical carcinoma cell line, HeLa (CCTCC no. GDC0009) was obtained from the China Center for Type Culture Collection. HeLa cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; GE Healthcare), 100 µg/ml streptomycin (Hyclone), and 100 U/ml penicillin (Hyclone). Plasmid (400 ng) (part no. 240031; Agilent Technologies, Inc.) was transfected into HeLa cells using Lipofectamine™ 2000 Transfection Reagent (cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Transfected cells were harvested after 48 h for reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis.
Gene overexpression
STAU1 overexpression was determined using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the control. cDNA was synthesized according to the standard instructions followed by real-time quantitative PCR. RT-qPCR was conducted on the Bestar SYBR Green RT-PCR Master Mix (DBI Bioscience). The primers are presented in Table SI. The concentrations of the transcripts were then normalized to GAPDH mRNA levels using the 2−ΔΔCq method (23). The data were analyzed by Student's t-test using GraphPad Prism software (version 8.0; GraphPad Software, Inc.).
Cell proliferation assay
Cell proliferation was measured by the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. Briefly, 1×103 cells were seeded in 96-well plates in triplicate. After 48 h of transfection, 5 mg/ml MTT solution was added into the cells. After 4 h of incubation at 37°C, the MTT solution was removed. The insoluble MTT was dissolved in DMSO. Absorbance at 490 nm was measured using a Benchmark Plus microplate reader (Bio-Rad Laboratories, Inc.).
RNA isolation and high-throughput sequencing
Four groups of samples were prepared, namely, control cells and overexpression (OE)-STAU1 cells (two biological replicates). Prior to RNA isolation, HeLa cells were first harvested. TRIzol reagent (Ambion; Thermo Fisher Scientific, Inc.) was used to isolate total RNA, which was then purified by phenol-chloroform and treated with RQ1 DNase (Promega Corporation) to eliminate DNA. The quantity and quality of the RNA were verified by using SmartSpec Plus (Bio-Rad Laboratories, Inc.) to detect the absorbance at a wavelength of 260–280 nm. Agarose (1.5%) gel electrophoresis was employed to detect RNA integrity.
RNA-seq library was prepared using VAHTS Stranded mRNA-seq Library Prep kit (cat. no. NR602; Vazyme Biotech Co., Ltd.), and 1 µg total RNA was used for one sample. After polyadenylated mRNAs were purified, they were fragmented and double-strand cDNAs were produced. The double-strand cDNAs were subjected to end repair, and poly(A) tails were added. They were then ligated to VAHTS RNA Adapters (Vazyme Biotech Co., Ltd.) and digested by heat-labile uracil-DNA glycosylase (UDG). Before sequencing, amplification, purification, and quantification of the single-strand cDNA were performed, the quantity of the cDNA was redetermined by qPCR using Agilent 2100 (Agilent Technologies, Inc.), and was finally stored at −80°C.
The library was prepared according to the manufacturer's protocol, which was applied to 150-nt paired-end sequencing using Illumina HiSeq X Ten system (Illumina, Inc.). ABLife performed the sequencing using the sequencing kit provided from Illumina, Inc.
Data processing and alignment
Raw reads containing more than 2-N bases were first discarded. FASTX-Toolkit (Ver. 0.0.13) was used to trim the adaptors and bases of low quality from raw sequencing reads, and short reads less than 16-nt were removed, filtered for quality (fastq_quality_filter -q 25 -p 80) and against artifact sequences (fastx_artifacts_filter) and collapsed (fastx_collapse). Base quality Q30 was used to indicate the proportion of bases with a sequencing error rate <0.1%. RNA-seq data presented in this study have been deposited in the Gene Expression Omnibus of NCBI and are accessible through GEO Series accession number GSE136890. Using TopHat v.2.0.10 (24), clean reads were mapped to the GRch38 genome and four mismatches were allowed. For calculation of fragments per kilobase of transcript per million fragments mapped (FPKM) (25) and gene read counting, we applied specifically mapped reads, and by calculating the Pearson correlation coefficient (PCC) for cluster analysis.
Analysis of differentially expressed genes (DEGs)
The R Bioconductor package edgeR (26) was utilized to screen the DEGs. A false-discovery rate (FDR)<0.05 and fold-change (FC)>2 or <0.5 were set as the cut-off points for identifying DEGs. Next, a volcano map was drawn to reveal the number of DEGs.
AS analysis
By applying the ABLas algorithm (27,28), regulated differential splicing events (RDSEs) and differential splicing events (DSEs) were identified. Briefly, 10 types of DSEs were detected by ABLas on the basis of splice junctions directly extracted from mapping reads, including exon skipping (ES), alternative 5′splice site (A5SS), alternative 3′ splice site (A3SS), intron retention (IR), mutually exclusive exons (MXE), mutually exclusive 5′UTRs (5pMXE), mutually exclusive 3′UTRs (3pMXE), cassette exon, A3SS and ES, and A5SS and ES. Sashimi plot by IGV Tools was used for AS analysis.
To assess RBP-regulated DSEs, Student's t-test was performed to evaluate the significance of AS events. The events, which were significant at a P-value equal to 5%, were considered RBP-regulated DSEs.
We also analyzed the overlapping genes from the DEGs and regulated alternative splicing events (RASEs) between the samples were defined and quantified by using the ABLas pipeline; and a Venn diagram was drawn.
AS events and DEGs are validated by RT-qPCR
In order to validate RNA-seq data, RT-qPCR was performed. The primers used for RT-qPCR are presented in Table SI. RNA was reversely-transcribed into cDNA using an M-MLV Reverse Transcriptase (Vazyme Biotech Co., Ltd.). RT-qPCR was performed by StepOne Real-Time PCR System using the SYBR-Green PCR Reagent Kit (Yeasen Biotechnology Co., Ltd.). PCR procedures were carried out as follows: The samples were first denatured at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec, and then annealed and extended at 60°C for 1 min. The procedures were repeated three times for all samples. The RNA expression levels of all the DEGs were normalized against those of GAPDH.
In addition, an RT-qPCR assay was undertaken for DSE validation. The primers used for detecting DSEs are presented Table SI. To detect alternative isoforms, a boundary-spanning primer of constitutive and alternative exons was used, as well as an opposing primer in one constitutive exon. The boundary-spanning primer of the alternative exon was designed according to a ‘model exon’ to detect model splicing, or an ‘altered exon’ to detect altered splicing.
Functional enrichment analysis
To sort out functional categories of DEGs, KOBAS 2.0 server (29) was employed to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms. Enrichment was assessed using the hypergeometric test followed by Benjamini-Hochberg FDR adjustment for P-values. The heatmap was constructed by calculating the Pearson correlation coefficient (PCC) of the DEGs.
Results
Overexpression of FLAG-tagged STAU1 promotes cell proliferation
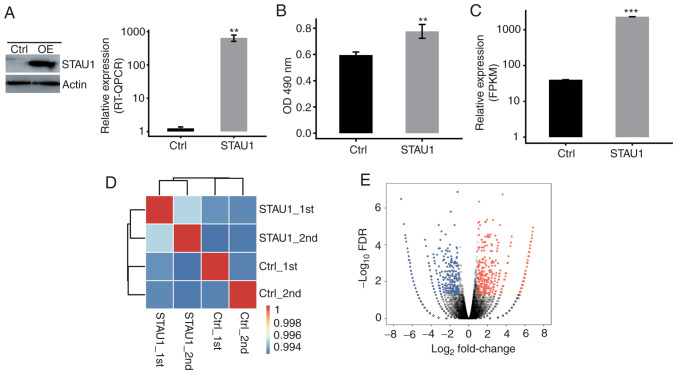
In order to analyze the expression of STAU1-expressing vectors and its influence on the proliferation of human HeLa cells, the STAU1-transfected cells with GFP label and flag label (fused with the target gene) were successfully constructed. In addition, cells transfected with blank controls were considered as the negative control. After 48 h of transfection, the expression level of GFP was detected to indicate whether the transfection was successful. The cells were harvested and detected by RT-qPCR and western blot assays. The results indicated a significant overexpression after cell transfection with STAU1-expressing vectors (Fig. 1A). Thereafter, proliferation of STAU1-transfected cells was detected by MTT assay. Compared with the control group, overexpression of STAU1 promoted cell proliferation (Fig. 1B).
Figure 1.
STAU1 overexpression promotes cell proliferation. (A) STAU1 expression was quantified by western blotting and RT-qPCR. (B) An MTT assay revealed that STAU1 overexpression promoted cell proliferation of HeLa cells. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01. (C) STAU1 expression quantified by RNA sequencing data. FPKM values were calculated as described in the Materials and methods. ***P<0.001. (D) Heatmap revealing the hierarchically clustered Pearson correlation matrix that resulted from comparing the transcript expression values of the control and STAU1-overexpression samples. (E) Identification of STAU1-regulated genes. Upregulated genes are labeled in red, whereas downregulated are labeled in blue in the volcano plot. STAU1, double-stranded RNA-binding protein Staufen homolog 1; RT-qPCR, reverse transcription-quantitative PCR; FPKM, fragments per kilobase of transcript per million fragments mapped; Ctrl, control; OE, overexpression.
RNA-seq profiling of the transcriptional response to STAU1 overexpression
In order to assess STAU1-mediated transcriptional regulation in human HeLa cells, four groups of samples were prepared, namely, control cells and overexpression (OE)-STAU1 cells (two biological replicates). Total RNA extraction was carried out for the 4 groups of samples, and the cDNA library was prepared. Then, the library was subjected to paired-end sequencing on Illumina HiSeq X Ten to extract high-quality transcriptome data. Quality analysis of clean reads indicated that the mean Q30 quality score was 93.93% (30). Next, high-quality clean reads were aligned against GRCH38 human reference genome using TopHat2 software (Table I). RNA-Seq data were analyzed and the expression levels of STAU1 were quantified, which further demonstrated overexpression of STAU1 (Fig. 1C). Correlation analysis was undertaken to determine the variability in the gene expression level between each pair of the samples. Moreover, cluster analysis was performed between the samples (Fig. 1D). As revealed in Fig. 1D, there was a correlation between STAU1 OE cells and control cells; there was also a significant correlation between the biological replicates. With analysis of DEGs among samples, criteria for significant difference were set to FC≥2 or ≤0.5 and FDR<0.05. A volcano plot was drawn and the results revealed that 801 significant DEGs were related to STAU1 overexpression. Among them, 423 upregulated genes and 378 downregulated genes were identified (Fig. 1E). This indicated that STAU1 plays an extensive transcriptional regulatory role in HeLa cells.
Table I.
Mapping of the high-quality clean reads on the reference genome.
| Sample | STAU1_1st | STAU1_2nd | Ctrl_1st | Ctrl_2nd | Mean |
|---|---|---|---|---|---|
| Raw reads | 83246680 | 92553878 | 83563062 | 80069538 | 84858289.5 |
| Clean reads | 79647695 | 89540818 | 79864334 | 76548941 | 81400447 |
| Q30 (%) | 94.08 | 93.93 | 93.62 | 94.09 | 93.93 |
| Paired-end reads | 78141536 | 87841376 | 78169980 | 75050020 | 79800728 |
| Total mapped (%) | 71511926 (91.52) | 81141734 (92.37) | 70870280 (90.66) | 69467494 (92.56) | 73247858.5 (91.78) |
| Uniquely mapped (%) | 69078606 (96.6) | 78068680 (96.21) | 67729842 (95.57) | 67403229 (97.03) | 70570089.3 (96.35) |
| Splice reads (%) | 30027459 (43.47) | 33845649 (43.35) | 29491479 (43.54) | 28788551 (42.71) | 30538284.5 (43.27) |
STAU1, double-stranded RNA-binding protein Staufen homolog 1.
STAU1 regulates the expression of genes enriched in inflammatory and immune response
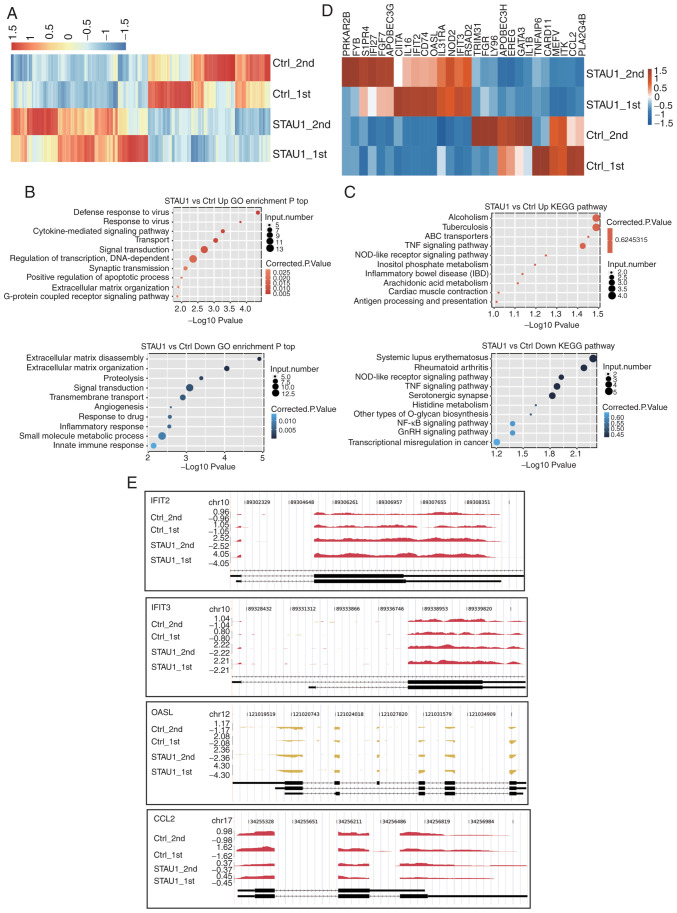
A heatmap of the expression levels of DEGs was plotted. The upregulated genes were represented in the red region of the experimental group, and downregulated ones in the red region of the control group. The results revealed a high level of consistency in STAU1-mediated transcription between the two groups of data (Fig. 2A). The functions and potential biological roles of these DEGs were further analyzed. GO enrichment and KEGG pathway analyses were conducted for the upregulated and downregulated genes, respectively, and the top 10 GO terms were presented. It is generally believed that a P<0.05 indicates significant difference, i.e., significant enrichment. Thus, this threshold was defined as the cut-off. GO functions were divided into three categories, namely, molecular function, biological process, and cellular component. As revealed in Fig. 2B, genes regulating STAU1 overexpression were primarily enriched in pathways related to inflammation and immune response. The upregulated genes in the OE-STAU1 were enriched in ‘defense response to virus’, ‘cytokine-mediated signaling pathways’, ‘transport’, ‘signal transduction’ and ‘synaptic transmission’; the downregulated genes were enriched in ‘signal transduction’, ‘transmembrane transport’, ‘inflammatory response’, and ‘innate immune response’. Studies have confirmed that STAU1 is positively correlated with the function and number of neuronal synapses, and can induce morphological changes of the dendritic spine (31–33). After overexpression of STAU1, upregulated genes associated with inflammatory and immune responses (IFIT2, OASL, IFIT3, CCL5, and CD74) were detected (GEO accession number GSE136890).
Figure 2.
RNA-seq analysis of STAU1-regulated transcriptome. (A) Hierarchical clustering of DEGs in the control and STAU1-overexpression samples. FPKM values were log2-transformed and then median-centered by each gene. (B) The top 10 representative GO biological processes of upregulated and downregulated genes. (C) The top 10 representative KEGG pathways of upregulated and downregulated genes. (D) Hierarchical clustering of DEGs enriched in cytokine-mediated signaling, inflammatory response, and immune response. (E) Reads coverage and distribution across representative DEGs involved in cytokine-mediated signaling and immune response. RNA-seq, RNA sequencing; STAU1, double-stranded RNA-binding protein Staufen homolog 1; DEGs, differentially expressed genes; FPKM, fragments per kilobase of transcript per million fragments mapped; Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology.
For KEGG pathway analysis, the cut-off was also set to P<0.05. The results revealed that the genes were enriched in a variety of pathways related to the immune system, inflammatory response, and nervous system (Fig. 2C); the upregulated genes were primarily enriched in ‘ABC transporters’ and ‘TNF signaling pathway’. The downregulated genes were mainly enriched in ‘Systemic lupus erythematosus’, ‘Rheumatoid arthritis’, ‘NOD-like receptor signaling pathway’, ‘TNF signaling pathway’, ‘Serotonergic synapse’, and ‘NF-κB signaling pathway’.
Then, among all the DEGs, 28 genes related to cytokine-mediated signaling, inflammatory response, and immune response were selected and presented in the DEGs-based hierarchical clustering plot (Fig. 2D). Among them, there were 15 upregulated genes and 13 downregulated genes. Herein, 3 upregulated genes (IFIT2, OASL, and IFIT3) and 1 downregulated gene [chemokine (C-C motif) ligand 2 (CCL2)] were analyzed in details in terms of coverage and distribution of reads (Fig. 2E). Distribution of reads reflected the relative location of genes and the relative read abundance, which further demonstrated differential expression in control cells and STAU1 OE cells. Therefore, it could be concluded that STAU1 selectively regulated genes related to inflammation and immune responses.
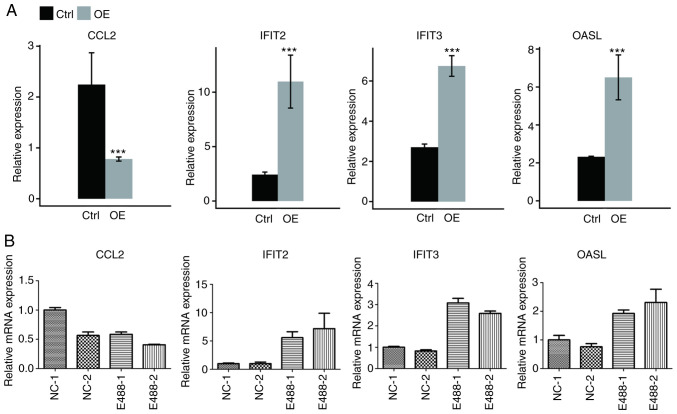
STAU1 positively regulates the expression of IFIT2, OASL, IFIT3, and negatively regulates the expression of CCL2 in HeLa cells
The expression levels of the aforementioned 4 genes are presented in Fig. 3A. These genes were enriched in the cytokine-mediated signaling pathway, inflammatory response or other biological processes in the GO database. To verify the reliability of RNA sequencing, qPCR was conducted for HeLa cells. It was revealed that IFIT2, OASL, and IFIT3 were significantly upregulated, while CCL2 was significantly downregulated (Fig. 3B), which was consistent with results of RNA-seq analysis. The results of qPCR of the 3 genes (IFI27, S1PR4, CCL5) in Fig. S1 were also consistent with RNA-seq analysis (data not shown), and the non-DEG CD44 was the control gene.
Figure 3.
Validation of the expression level of DEGs enriched in cytokine-mediated signaling and immune response. Gene expression quantified by (A) RNA seq data and (B) RT-qPCR. Error bars represent mean ± SEM. ***P<0.001. FPKM values were calculated as described in the Materials and methods. DEGs, differentially expressed genes; RNA seq, RNA sequencing; RT-qPCR, reverse transcription-quantitative PCR; FPKM, fragments per kilobase of transcript per million fragments mapped; CCL2, chemokine (C-C motif) ligand 2; IFIT2, interferon-induced protein with tetratricopeptide repeats 2; IFIT3, interferon-induced protein with tetratricopeptide repeats 3; OASL, 2′-5′-oligoadenylate synthetase-like protein; Ctrl, control; OE, overexpression.
STAU1 regulates the AS of genes enriched in the ‘nerve growth factor receptor signaling pathway’
Regulated AS events (RASEs) of STAU1 in human HeLa cells were further analyzed. Every sample in the RNA-seq data was aligned to unique mapped reads on the reference genome for RASE analysis. The results of exon detection in 4 samples are presented in Table II. A total of 237,791 detected exons were achieved, accounting for 64.74% of all annotated exons in the reference genome. Splice junctions of each sample were then analyzed by using TopHat2 software, and 160,308 known splice junctions (Known_Junction) and 163,225 novel splice junctions (Novel_Junction) were obtained (Table III).
Table II.
Exon detection results in RNA-seq data.
| Sample | Detected exon | Annotated exon | Ratio (%) |
|---|---|---|---|
| Ctrl_1st | 207169 | 367321 | 56.40 |
| Ctrl_2nd | 204690 | 367321 | 55.73 |
| STAU1_1st | 213908 | 367321 | 58.23 |
| STAU1_2nd | 219650 | 367321 | 59.80 |
| Total | 237791 | 367321 | 64.74 |
RNA-seq, RNA sequencing; STAU1, double-stranded RNA-binding protein Staufen homolog 1.
Table III.
Splicing junction analysis of samples from RNA-seq data.
| Sample | All_Junction | Novel_Junction | Known_Junction |
|---|---|---|---|
| Ctrl_1st | 195462 | 55232 | 140230 |
| Ctrl_2nd | 186313 | 48162 | 138151 |
| STAU1_1st | 216284 | 71098 | 145186 |
| STAU1_2nd | 234013 | 85203 | 148810 |
| Total | 323533 | 163225 | 160308 |
RNA-seq, RNA sequencing; STAU1, double-stranded RNA-binding protein Staufen homolog 1.
Various RASEs were statistically analyzed using ABLas, and the detection results in each sample are presented in Table IV. There were 76,259 detected RASEs, including 19,746 annotated RASEs in the genome (known AS) and 56,513 non-annotated novel RASEs (novel AS).
Table IV.
All AS events detected from all samples.
| Sample | KAS (%) | NAS (%) | All AS |
|---|---|---|---|
| Ctrl_1st | 12728 (36.58) | 22063 (63.42) | 34791 |
| Ctrl_2nd | 11916 (36.40) | 20818 (63.60) | 32734 |
| STAU1_1st | 14251 (37.53) | 23717 (62.47) | 37968 |
| STAU1_2nd | 15375 (37.63) | 25480 (62.37) | 40855 |
| Total | 19746 (25.89) | 56513 (74.11) | 76259 |
AS, alternative splicing; KAS, known AS; NAS, novel AS; STAU1, double-stranded RNA-binding protein Staufen homolog 1.
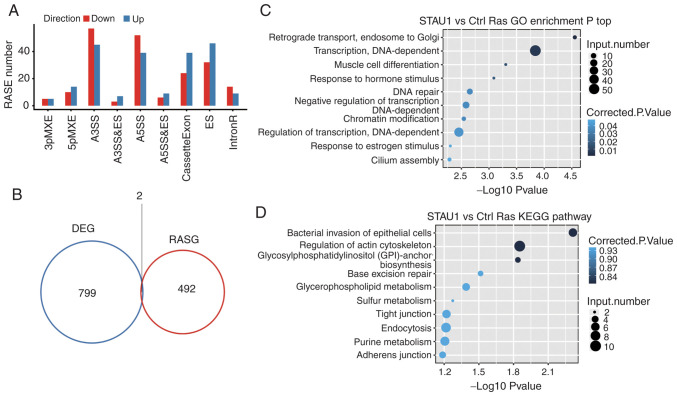
Due to the set-up of biological replicates, a Student's t-test was used to compare the variation of AS levels of each gene between two samples, and the criterion for significantly different AS was set to P≤0.05. As revealed in Fig. 4A, a total of 549 RASEs were detected, which are presented in Table V. Among them, the major types of RASEs included 102 A3SS, 91 A5SS, 78 ES, and 63 cassette exons. This indicated that STAU1 had a retaining and promoting effect on exons in the entire genome. Number and types of other differential RASEs were as follows: 24 5pMXE, 10 3pMXE, 10 A3SS and ES, 16 MXE, and 15 A5SS and ES. The aforementioned results indicated that STAU1 could regulate AS in the genome of HeLa cells.
Figure 4.
Identification and functional analysis of STAU1-regulated AS events. (A) Classification of STAU1 overexpression-regulated AS events. (B) Venn diagram revealing the overlap of STAU1-regulated DEGs and RASGs. (C) The top 10 enriched GO biological processes of the STAU1-regulated alternatively spliced genes. (D) The top 10 enriched KEGG pathways of the STAU1-regulated alternatively spliced genes. STAU1, double-stranded RNA-binding protein Staufen homolog 1; AS, alternative splicing; DEGs, differentially expressed genes; RASGs, regulated alternatively spliced genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table V.
Classification of all RASE events between sample groups.
| Type | STAU1_vs_Ctrl Up | STAU1_vs_Ctrl Down |
|---|---|---|
| 3pMXE | 5 | 5 |
| 5pMXE | 14 | 10 |
| A3SS | 45 | 57 |
| A3SS and ES | 7 | 3 |
| A5SS | 39 | 52 |
| A5SS and ES | 9 | 6 |
| ES | 46 | 32 |
| IR | 62 | 79 |
| MXE | 9 | 6 |
| Cassette Exon | 39 | 24 |
| Total | 275 | 274 |
RASE, regulated alternative splicing events; STAU1, double-stranded RNA-binding protein Staufen homolog 1; 3pMXE, mutually exclusive 3′UTRs; 5pMXE, mutually exclusive 5′UTRs; A3SS, alternative 3′splice site; ES, exon skipping; A5SS, alternative 5′splice site.
An integrated analysis was performed for differentially regulated alternatively spliced genes (RASGs) and DEGs in different samples. There were 2 genes with significant difference in terms of both the expression level and AS level (Fig. 4B).
Similarly, GO enrichment and KEGG pathway analyses were undertaken on the differential RASGs, and the top 10 terms are presented in Fig. 4C and D. GO terms and KEGG pathways, in which RASGs were enriched are presented in Table SII and SIII, respectively.
It was revealed that the genes whose AS level was regulated by STAU1 were mainly enriched in ‘retrograde transport, endosome to Golgi’, ‘muscle cell differentiation’, and other reported STAU1-related pathways. The GO term ranking the 16th was enriched in ‘nerve growth factor receptor signaling pathway’ (~P=0.01).
STAU1-regulated AS of PLEKHG2, ARHGEF11, NR4A1, PDGFB, FGFR4, RALGDS in HeLa cells
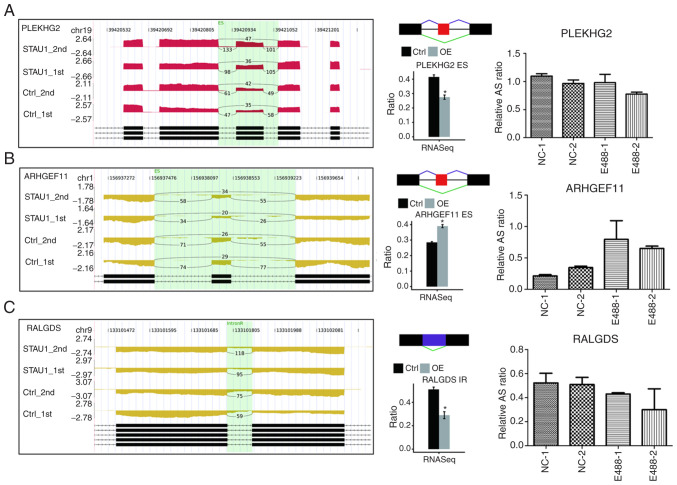
As revealed in Table SII, 6 key genes were selected for the detection of RASE, namely, ES, A5SS, 5pMXE, and IR. According to the results of RNA-seq (Fig. 5 and Fig. S2), the number of reads sequenced for each gene was over 10. Then, for each gene, the number of reads for AS between STAU1 OE cells and control cells was compared, and a significant difference in the AS levels was noted. To verify the reliability of the results, qPCR was performed in the HeLa cells. Primers for the qPCR verification are presented in Table SI. RASEs detected by qPCR were consistent with those by RNA-seq, which demonstrated that STAU1 may play a significant regulatory role in the AS of ‘nerve growth factor receptor signaling pathway’.
Figure 5.
Validation of STAU1-regulated AS events. IGV-Sashimi plot revealed two (A and B) two ES and (C) one IR AS events in three different genes. Reads distribution of each AS event was plotted in the left panel with the transcripts of each gene shown below. The schematic diagrams depict the structures of AS events, AS1 (purple line) and AS2 (green line). The exon sequences are denoted by black boxes, intron sequences by a horizontal line (right panel, top), while the retained intron by a purple box. RNA-seq quantification and RT-qPCR validation of ASEs are presented in the panels on the right. STAU1, double-stranded RNA-binding protein Staufen homolog 1; AS, alternative splicing; ES, exon skipping; IR, intron retention; RNA-seq, RNA sequencing; RT-qPCR, reverse-transcription quantitative PCR. Error bars represent mean ± SEM. *P<0.05.
Discussion
RNA-seq based on high-throughput sequencing is currently the most widely used transcriptome sequencing technology, which can promptly extract all the genetic information of the samples. RNA-seq, has become one of the most representative high-throughput sequence-based techniques due to its high-throughput, high accuracy, and cost-effectiveness. It can be used to study the structure and function of genes, identify changes in gene expression, and explore AS patterns that are regulated (34). In the field of life sciences, this method has been used to explore the pathogenesis of diseases, clinical diagnosis, and pharmacological research (35,36).
In the present study it was revealed that overexpression of STAU1 promoted the proliferation of HeLa cells, which are useful for the study of gene regulation in the central nervous system, while the proliferation of neurons and glial cells in the central nervous system plays an important role in neuropathic pain (37). Upregulation of STAU1 caused upregulation or downregulation of numerous genes, including IFIT2, IFIT3, OASL and CCL2. Through functional analysis, changes in the expression levels of these genes may affect signaling pathways, such as ‘defense response to virus’, ‘cytokine-mediated signaling pathway’, and ‘inflammatory response’, which are closely associated with inflammatory immune response. In addition, the AS of multiple genes was also regulated by STAU1, and the main enriched pathways not only include ‘retrograde transport’ and ‘muscle cell differentiation’, but also the ‘nerve growth factor receptor signaling pathway’.
Significant upregulation of IFIT2, IFIT3, and OASL genes was consistently indicated by RNA-seq and qPCR of HeLa cells. The IFIT family performs multiple functions, including antitumor effects and regulation of cell apoptosis and innate immune pathways (38,39). It can also inhibit replication of flavivirus and coronavirus (40). Siegfried et al stimulated wild-type bone marrow-derived macrophages (BMDMs), IFIT2−/−, and IFNAR−/− BMDMs with LPS, respectively. Results of enzyme-linked immunosorbent assay (ELISA) test indicated that the mutant BMDMs exhibited a significant reduction in the expression levels of TNF-α and interleukin-6 (IL-6) than the wild-type BMDMs. Furthermore, shRNA interference targeting IFIT2 was performed in RAW264.7 macrophages. It was revealed that TNF-α and IL-6 were also downregulated, suggesting a pro-inflammatory role of IFIT2 (41). Berchtold et al demonstrated that overexpression of IFIT2 decreased the secretion of TNF-α in the RAW264.7 cells (42). Niess et al compared highly metastatic L3.6pl pancreatic tumor cells and lowly metastatic COL0357FG pancreatic tumor cells. It was determined that upregulation of IFIT3 promoted synthesis and secretion of IL-6 (43). Liu et al revealed that the expression of exogenous IFIT3 enhanced the inducing effect of NF-κB on TNF-α, without influencing TNF-α-mediated activation of NF-κB (44). Furthermore, OASL is an interferon-stimulated gene (ISG), playing a significant role in the immune response to viruses (45). Activation of OASL can be induced by interferon (IFN). The expression of OASL can further stimulate the production of IFN, thereby forming a positive feedback (46). IFN-γ has a neuroprotective effect, and significantly promotes secretion of IL-6 in astrocytes (47). Inflammatory cytokines (TNF-α and IL-6) are important molecules, mediating enhancement of hyperalgesia via increasing glutamic acid-induced excitatory current, thereby promoting the development of pain (48). Excitatory synaptic transmission is mainly regulated by AMPA and NMDA receptors. Inflammatory factors enhance their degree of excitation, promote the release of excitatory mediators, such as glutamic acid and substance P, and participate in the regulation of various pain signaling pathways (49,50). Therefore, TNF-α and IL-6 play a vital role in NP.
In the present study it was also revealed that CCL2 was markedly downregulated. CCL2, also known as monocyte chemotactic protein-1 (MCP-1), can activate monocytes in the inflammatory state, induce leukocyte migration reaction, regulate T-cell function, and participate in inflammation and immune response (51). Recently, it has been revealed that CCL2 is highly expressed in DRG neurons and spinal dorsal horn surface neurons during peripheral nerve injury (52). CCL2 is released in an activity-dependent manner from the synaptic vesicles in the central nervous system into the spinal cord (52,53). The CCL2 expression in the spinal cord is not limited to neurons. After spinal nerve ligation, astrocytes can also upregulate CCL2. Additionally, the in vitro cultured astrocytes exhibited an upregulation of CCL2 by over 100-fold, which was rapidly released in a JNK-dependent manner (54). CCL2 secreted by astrocytes acts on CCR2 in the dorsal horn neurons. CCL2 can strengthen the release of glutamic acid from the injured neuronal presynaptic membrane and promote the function of glutamic acid receptors in the postsynaptic membrane. This inhibits GABA-induced inhibitory synaptic transmission, while causing rapid phosphorylation of eukaryotic protein kinase (EPK) and activation of NMDA receptors. As a result, central sensitization is induced in a direct, rapid and non-transcriptional manner (55). Another study shows that MCP-1 and its receptor CCR2 in primary sensory neurons are involved in maintaining paclitaxel induced peripheral neuropathy (56). Therefore, overexpression or depletion of CCL2 and CCR2 has a direct influence on NP.
The regulatory role of STAU1 overexpression on RASE of HeLa cells was further studied. A total of 549 significantly differential RASEs were identified, which verified our speculation that STAU1 can globally regulate the AS events in the genome in HeLa cells. Pathways in which the differential RASGs were enriched have been previously aforementioned. PLEKHG2 and ARHGEF11 are both Rho guanine nucleotide exchange factors and activators of Rho GTPases. A variety of biological effects can be regulated by Rho GTPases, such as transmembrane transport, cell migration, adhesion, and proliferation (57). Moreover, Rho GTPases can participate in the immune response by regulating the Rho/ROCK signaling pathway (58,59). Another study demonstrated that PLEKHG2/FLJ 00018 can regulate the morphology of Neuro-2a cells, thereby playing a significant role in nerve growth and cell proliferation (60). ARHGEF11 is involved in the regulation of axonal growth by regulating the activity of RhoA (61). RALGDS is one of the Ras effectors and functions as a guanine nucleotide exchange factor for the small G-protein, Ral, which regulates membrane trafficking and cytoskeletal remodeling (62). Notably, RALGDS has been revealed to promote neuronal differentiation (63) and exert a key regulatory effect on neuronal plasticity and memory formation (64). RALGDS has also been revealed to mediate cytoskeletal remodeling (65), promote cell proliferation (66), and facilitate oncogenic transformation (67). Rondaij et al revealed that RalGDS overexpression was conducive to promote the exocytosis of endothelial Weibel-Palade bodies (WPBs) (68). The proteins encoded by the FGFR1 and FGFR4 genes are all members of the fibroblast growth factor receptor (FGFR) family (69). They trigger the downstream cascade by binding to FGFRs, thereby playing a substantial role in promoting embryonic growth and development (69), development of the nervous system (70), and regulating the metabolism (71). Other biological functions of the proteins are manifested in promoting injury repair (72), bone formation (73), and vascular and neural regeneration (74,75). FGFR overexpression has also been revealed to be associated with tumor and bone diseases, as well as arthritis (76–78). All of the aforementioned genes were subjected to AS analysis, and the results were validated by qPCR, which indicated consistency with RNA-seq except for the qPCR result for the PDGFB gene that was inconsistent with that of RNA-seq.
It is already known that PDGFB plays a significant role in the growth and proliferation of vessels and nerves (79). Herein, we further discussed the AS events induced by PDGFB and the resultant alterations in gene functions as an example. Platelet-derived growth factor (PDGF) is an important factor promoting cell growth. It consists of five homotypic or heterotypic dimerized ligands (PDGF-AA, -AB, -BB, -CC, -CD), which are formed by polypeptide chains (PDGF-A, PDGF-B, PDGF-C, PDGF-D), encoded by four different genes, via the disulfide bonds (80). Both of its receptors PDGFR-α and PDGFR-β belong to the receptor tyrosine kinase (RTK) family (81). It has been demonstrated that PDGF can stimulate the division growth of fibroblasts (82), neuroglial cells (83) and smooth muscle cells (84). In particular, PDGF-BB has been revealed to promote neuronal development and differentiation (85), and to play a neurotrophic role as well (86,87). A number of scholars have demonstrated that PDGF-BB can regulate neuronal proliferation and differentiation by activating the PI3K/Akt and ERK pathways (88), restore the proliferation and differentiation of damaged neural precursor cells, and reverse neuronal excitotoxicity. PDGFB has been demonstrated to play an important role in neuropathic pain (89,90), and in the present study it was confirmed that STAUI regulated the alternative splicing of genes enriched in the ‘nerve growth factor receptor signaling pathway’ including PDGFB, and thus is also associated with neuropathic pain.
The present study confirmed that the PDGFB gene undergoes A5SS events. The normal secretion of PDGF-B protein into the extracellular domain to bind to the PDGFR receptor-α and -β subunits and fulfill the biological effect is consequently affected. The qPCR results were consistent with our theory that STAU1 promotes the retention of the PDGFB signal peptide, which mediates the neuroprotective mechanism and relieves neuropathic pain. STAU1 promotes the retention of PDGFB signal peptide, which mediates the neuroprotective mechanism and relieves the neuropathic pain. We surmised that an even more complex regulatory mechanism is herein involved, while further studies should be conducted to confirm our findings.
The present study revealed that overexpression of STAU1 had a regulatory effect on gene splicing and transcription in HeLa cells. STAU1 could positively regulate the transcription of genes related to inflammation and immune response. This regulatory effect also influenced the expression levels of pro-inflammatory factors and chemotactic factors. Moreover, AS of genes enriched in the ‘nerve growth factor receptor signaling pathway’ as well as ‘retrograde transport, endosome to Golgi’, and ‘muscle cell differentiation’ was regulated by STAU1. A recent study demonstrated that several RNA binding factors involved in local translation may play a crucial role in pain, including STAU1, a double-stranded dsRNA binding protein, which is expressed in peripheral sensory neurons and may play a role in axonal mRNA transport (91). Therefore, it can be concluded that STAU1 may be a novel potential therapeutic target for NP.
Supplementary Material
Acknowledgements
We deeply thank Dr Wen Chen for his significant technical and scientific comments.
Funding
The present study was financially supported by the Natural Science Foundation of Hubei Province, China (grant no. 2011CDC032). This study was partially supported by ABLife (experimental project no. ABL-7702121).
Availability of data and materials
The data discussed in this publication are available under GEO Series accession number GSE136890.
Authors' contributions
YZ performed the experiments and data analysis and was the main author of the manuscript. ZH participated in the experimental design, carried out bioinformatics analysis, and partly contributed to the writing of the manuscript. JW and FD conducted the cell experiments and interpretation of the data. FL and YX designed the project, contributed to the analysis and interpretation of the data, and revision of the manuscript. All the authors read and approved the submitted final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 5.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with staufen1- and staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondy-Chorney E, Crawford Parks TE, Ravel-Chapuis A, Klinck R, Rocheleau L, Pelchat M, Chabot B, Jasmin BJ, Côté J. Staufen1 regulates multiple alternative splicing events either positively or negatively in DM1 Indicating its role as a disease modifier. PLoS Genet. 2016;12:e1005827. doi: 10.1371/journal.pgen.1005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10:1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Oh Y, Park J, Kim JI, Chang MY, Lee SH, Cho YH, Hwang J. Lin28B and miR-142-3p regulate neuronal differentiation by modulating staufen1 expression. Cell Death Differ. 2018;25:432–443. doi: 10.1038/cdd.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lucas S, Peredo J, Marion RM, Sanchez C, Ortin J. Human staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. J Virol. 2010;84:7603–7612. doi: 10.1128/JVI.00504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Oh JY, Pascua PN, Kim EG, Choi YK, Kim HK. Impairment of the staufen1-NS1 interaction reduces influenza viral replication. Biochem Biophys Res Commun. 2011;414:153–158. doi: 10.1016/j.bbrc.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 15.Rao S, Hassine S, Monette A, Amorim R, DesGroseillers L, Mouland AJ. HIV-1 requires staufen1 to dissociate stress granules and to produce infectious viral particles. RNA. 2019;25:727–736. doi: 10.1261/rna.069351.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentry JJ, Casaccia-Bonnefil P, Carter BD. Nerve growth factor activation of nuclear factor kappaB through its p75 receptor is an anti-apoptotic signal in RN22 schwannoma cells. J Biol Chem. 2000;275:7558–7565. doi: 10.1074/jbc.275.11.7558. [DOI] [PubMed] [Google Scholar]
- 18.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, et al. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 19.Rabizadeh S, Rabizadeh S, Ye X, Wang JJ, Bredesen DE. Neurotrophin dependence mediated by p75NTR: Contrast between rescue by BDNF and NGF. Cell Death Differ. 1999;6:1222–1227. doi: 10.1038/sj.cdd.4400614. [DOI] [PubMed] [Google Scholar]
- 20.Notterpek L. Neurotrophins in myelination: A new role for a puzzling receptor. Trends Neurosci. 2003;26:232–234. doi: 10.1016/S0166-2236(03)00099-7. [DOI] [PubMed] [Google Scholar]
- 21.Fu C, Donovan WP, Shikapwashya-Hasser O, Ye X, Cole RH. Hot fusion: An efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One. 2014;9:e115318. doi: 10.1371/journal.pone.0115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinn S, Gut IG. DNA sequencing-spanning the generations. N Biotechnol. 2013;30:366–372. doi: 10.1016/j.nbt.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin L, Li G, Yu D, Huang W, Cheng C, Liao S, Wu Q, Zhang Y. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus verticillium dahliae. BMC Genomics. 2017;18:130. doi: 10.1186/s12864-017-3507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech. 2017;1860:911–921. doi: 10.1016/j.bbagrm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39((Web Server issue)):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-cyclic: A self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebeau G, DesGroseillers L, Sossin W, Lacaille JC. mRNA binding protein staufen 1-dependent regulation of pyramidal cell spine morphology via NMDA receptor-mediated synaptic plasticity. Mol Brain. 2011;4:22. doi: 10.1186/1756-6606-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille JC. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol. 2008;28:2896–2907. doi: 10.1128/MCB.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, et al. A loss of function allele for murine staufen1 leads to impairment of dendritic staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci USA. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286:7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Liu X, Chen W, Chen L. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim Biophys Sin (Shanghai) 2013;45:867–874. doi: 10.1093/abbs/gmt088. [DOI] [PubMed] [Google Scholar]
- 41.Siegfried A, Berchtold S, Manncke B, Deuschle E, Reber J, Ott T, Weber M, Kalinke U, Hofer MJ, Hatesuer B, et al. IFIT2 is an effector protein of type I IFN-mediated amplification of lipopolysaccharide (LPS)-induced TNF-α secretion and LPS-induced endotoxin shock. J Immunol. 2013;191:3913–3921. doi: 10.4049/jimmunol.1203305. [DOI] [PubMed] [Google Scholar]
- 42.Berchtold S, Manncke B, Klenk J, Geisel J, Autenrieth IB, Bohn E. Forced IFIT-2 expression represses LPS induced TNF-alpha expression at posttranscriptional levels. BMC Immunol. 2008;9:75. doi: 10.1186/1471-2172-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niess H, Camaj P, Mair R, Renner A, Zhao Y, Jäckel C, Nelson PJ, Jauch KW, Bruns CJ. Overexpression of IFN-induced protein with tetratricopeptide repeats 3 (IFIT3) in pancreatic cancer: Cellular ‘pseudoinflammation’ contributing to an aggressive phenotype. Oncotarget. 2015;6:3306–3318. doi: 10.18632/oncotarget.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu XY, Chen W, Wei B, Shan YF, Wang C. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J Immunol. 2011;187:2559–2568. doi: 10.4049/jimmunol.1100963. [DOI] [PubMed] [Google Scholar]
- 45.Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR, Hartmann R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- 46.Zhu J, Ghosh A, Sarkar SN. OASL-a new player in controlling antiviral innate immunity. Curr Opin Virol. 2015;12:15–19. doi: 10.1016/j.coviro.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun L, Li Y, Jia X, Wang Q, Li Y, Hu M, Tian L, Yang J, Xing W, Zhang W, et al. Neuroprotection by IFN-ү via astrocyte-secreted IL-6 in acute neuroinflammation. Oncotarget. 2017;8:40065–40078. doi: 10.18632/oncotarget.16990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis A, Bennett DL. Neuroinflammation and the generation of neuropathic pain. Br J Anaesth. 2013;111:26–37. doi: 10.1093/bja/aet128. [DOI] [PubMed] [Google Scholar]
- 49.Kawasaki Y, Zhang L, Cheng JK, Ji RR. Cytokine mechanisms of central sensitization: Distinct and overlapping role of interleukin-1beta, interleukin-6, and tumor necrosis factor-alpha in regulating synaptic and neuronal activity in the superficial spinal cord. J Neurosci. 2008;28:5189–5194. doi: 10.1523/JNEUROSCI.3338-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung L, Cahill CM. TNF-alpha and neuropathic pain-a review. J Neuroinflammation. 2010;7:27. doi: 10.1186/1742-2094-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): An overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dansereau MA, Gosselin RD, Pohl M, Pommier B, Mechighel P, Mauborgne A, Rostene W, Kitabgi P, Beaudet N, Sarret P, Melik-Parsadaniantz S. Spinal CCL2 pronociceptive action is no longer effective in CCR2 receptor antagonist-treated rats. J Neurochem. 2008;106:757–769. doi: 10.1111/j.1471-4159.2008.05429.x. [DOI] [PubMed] [Google Scholar]
- 53.Thacker MA, Clark AK, Bishop T, Grist J, Yip PK, Moon LD, Thompson SW, Marchand F, McMahon SB. CCL2 is a key mediator of microglia activation in neuropathic pain states. Eur J Pain. 2009;13:263–272. doi: 10.1016/j.ejpain.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Gao YJ, Zhang L, Samad OA, Suter MR, Yasuhiko K, Xu ZZ, Park JY, Lind AL, Ma Q, Ji RR. JNK-induced MCP-1 production in spinal cord astrocytes contributes to central sensitization and neuropathic pain. J Neurosci. 2009;29:4096–4108. doi: 10.1523/JNEUROSCI.3623-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuang ZY, Kawasaki Y, Tan PH, Wen YR, Huang J, Ji RR. Role of the CX3CR1/p38 MAPK pathway in spinal microglia for the development of neuropathic pain following nerve injury-induced cleavage of fractalkine. Brain Behav Immun. 2007;21:642–651. doi: 10.1016/j.bbi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Boyette-Davis JA, Kosturakis AK, Li Y, Yoon SY, Walters ET, Dougherty PM. Induction of monocyte chemoattractant protein-1 (MCP-1) and its receptor CCR2 in primary sensory neurons contributes to paclitaxel-induced peripheral neuropathy. J Pain. 2013;14:1031–1044. doi: 10.1016/j.jpain.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rougerie P, Delon J. Rho GTPases: Masters of T lymphocyte migration and activation. Immunol Lett. 2012;142:1–13. doi: 10.1016/j.imlet.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Sato K, Sugiyama T, Nagase T, Kitade Y, Ueda H. Threonine 680 phosphorylation of FLJ00018/PLEKHG2, a Rho family-specific guanine nucleotide exchange factor, by epidermal growth factor receptor signaling regulates cell morphology of Neuro-2a cells. J Biol Chem. 2014;289:10045–10056. doi: 10.1074/jbc.M113.521880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu Y, Dobashi K, Iizuka K, Horie T, Suzuki K, Tukagoshi H, Nakazawa T, Nakazato Y, Mori M. Contribution of small GTPase Rho and its target protein rock in a murine model of lung fibrosis. Am J Respir Crit Care Med. 2001;163:210–217. doi: 10.1164/ajrccm.163.1.2001089. [DOI] [PubMed] [Google Scholar]
- 60.Sato K, Kimura M, Sugiyama K, Nishikawa M, Okano Y, Nagaoka H, Nagase T, Kitade Y, Ueda H. Four-and-a-half LIM domains 1 (FHL1) protein interacts with the rho guanine nucleotide exchange factor PLEKHG2/FLJ00018 and regulates cell morphogenesis. J Biol Chem. 2016;291:25227–25238. doi: 10.1074/jbc.M116.759571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X, Zhou Z, Fink DJ, Mata M. HspB1 silences translation of PDZ-RhoGEF by enhancing miR-20a and miR-128 expression to promote neurite extension. Mol Cell Neurosci. 2013;57:111–119. doi: 10.1016/j.mcn.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofer F, Fields S, Schneider C, Martin GS. Activated ras interacts with the ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci USA. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of ras by sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 64.Mitra S, Cheng KW, Mills GB. Rab GTPases implicated in inherited and acquired disorders. Semin Cell Dev Biol. 2011;22:57–68. doi: 10.1016/j.semcdb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharya M, Anborgh PH, Babwah AV, Dale LB, Dobransky T, Benovic JL, Feldman RD, Verdi JM, Rylett RJ, Ferguson SS. Beta-arrestins regulate a Ral-GDS Ral effector pathway that mediates cytoskeletal reorganization. Nat Cell Biol. 2002;4:547–555. doi: 10.1038/ncb821. [DOI] [PubMed] [Google Scholar]
- 66.Hao Y, Wong R, Feig LA. RalGDS couples growth factor signaling to Akt activation. Mol Cell Biol. 2008;28:2851–2859. doi: 10.1128/MCB.01917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, Kelly K. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rondaij MG, Bierings R, van Agtmaal EL, Gijzen KA, Sellink E, Kragt A, Ferguson SS, Mertens K, Hannah MJ, van Mourik JA, et al. Guanine exchange factor RalGDS mediates exocytosis of weibel-palade bodies from endothelial cells. Blood. 2008;112:56–63. doi: 10.1182/blood-2007-07-099309. [DOI] [PubMed] [Google Scholar]
- 69.Anteby EY, Natanson-Yaron S, Hamani Y, Sciaki Y, Goldman-Wohl D, Greenfield C, Ariel I, Yagel S. Fibroblast growth factor-10 and fibroblast growth factor receptors 1–4: Expression and peptide localization in human decidua and placenta. Eur J Obstet Gynecol Reprod Biol. 2005;119:27–35. doi: 10.1016/j.ejogrb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Kang K, Song MR. Diverse FGF receptor signaling controls astrocyte specification and proliferation. Biochem Biophys Res Commun. 2010;395:324–329. doi: 10.1016/j.bbrc.2010.03.174. [DOI] [PubMed] [Google Scholar]
- 71.Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaye M, Schlessinger J, Dionne CA. Fibroblast growth factor receptor tyrosine kinases: Molecular analysis and signal transduction. Biochim Biophys Acta. 1992;1135:185–199. doi: 10.1016/0167-4889(92)90136-Y. [DOI] [PubMed] [Google Scholar]
- 73.Barnes GL, Kostenuik PJ, Gerstenfeld LC, Einhorn TA. Growth factor regulation of fracture repair. J Bone Miner Res. 1999;14:1805–1815. doi: 10.1359/jbmr.1999.14.11.1805. [DOI] [PubMed] [Google Scholar]
- 74.Grothe C, Nikkhah G. The role of basic fibroblast growth factor in peripheral nerve regeneration. Anat Embryol (Berl) 2001;204:171–177. doi: 10.1007/s004290100205. [DOI] [PubMed] [Google Scholar]
- 75.Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, Richardson WD. Fgfr3 expression by astrocytes and their precursors: Evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130:93–102. doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- 76.Beenken A, Mohammadi M. The FGF family: Biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bono F, De Smet F, Herbert C, De Bock K, Georgiadou M, Fons P, Tjwa M, Alcouffe C, Ny A, Bianciotto M, et al. Inhibition of tumor angiogenesis and growth by a small-molecule multi-FGF receptor blocker with allosteric properties. Cancer Cell. 2013;23:477–488. doi: 10.1016/j.ccr.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 78.Murphy PR, Knee RS. Basic fibroblast growth factor binding and processing by human glioma cells. Mol Cell Endocrinol. 1995;114:193–203. doi: 10.1016/0303-7207(95)96800-W. [DOI] [PubMed] [Google Scholar]
- 79.Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, Franklin RA, McCubrey JA. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: Potential targeting for therapeutic intervention. Leukemia. 2003;17:1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- 80.Fredriksson L, Li H, Eriksson U. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 81.Reigstad LJ, Varhaug JE, Lillehaug JR. Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family. FEBS J. 2005;272:5723–5741. doi: 10.1111/j.1742-4658.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 82.Kohler N, Lipton A. Platelets as a source of fibroblast growth-promoting activity. Exp Cell Res. 1974;87:297–301. doi: 10.1016/0014-4827(74)90484-4. [DOI] [PubMed] [Google Scholar]
- 83.Westermark B, Wasteson A. A platelet factor stimulating human normal glial cells. Exp Cell Res. 1976;98:170–174. doi: 10.1016/0014-4827(76)90476-6. [DOI] [PubMed] [Google Scholar]
- 84.Ross R, Glomset J, Kariya B, Harker L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc Natl Acad Sci USA. 1974;71:1207–1210. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams BP, Park JK, Alberta JA, Muhlebach SG, Hwang GY, Roberts TM, Stiles CD. A PDGF-regulated immediate early gene response initiates neuronal differentiation in ventricular zone progenitor cells. Neuron. 1997;18:553–562. doi: 10.1016/S0896-6273(00)80297-4. [DOI] [PubMed] [Google Scholar]
- 86.Pietz K, Odin P, Funa K, Lindvall O. Protective effect of platelet-derived growth factor against 6-hydroxydopamine-induced lesion of rat dopaminergic neurons in culture. Neurosci Lett. 1996;204:101–104. doi: 10.1016/0304-3940(96)12326-0. [DOI] [PubMed] [Google Scholar]
- 87.Smits A, Kato M, Westermark B, Nister M, Heldin CH, Funa K. Neurotrophic activity of platelet-derived growth factor (PDGF): Rat neuronal cells possess functional PDGF beta-type receptors and respond to PDGF. Proc Natl Acad Sci USA. 1991;88:8159–8163. doi: 10.1073/pnas.88.18.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Erlandsson A, Enarsson M, Forsberg-Nilsson K. Immature neurons from CNS stem cells proliferate in response to platelet-derived growth factor. J Neurosci. 2001;21:3483–3491. doi: 10.1523/JNEUROSCI.21-10-03483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng F, Dhillon N, Callen S, Yao H, Bokhari S, Zhu X, Baydoun HH, Buch S. Platelet-derived growth factor protects neurons against gp120-mediated toxicity. J Neurovirol. 2008;14:62–72. doi: 10.1080/13550280701809084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao H, Duan M, Yang L, Buch S. Platelet-derived growth factor-BB restores human immunodeficiency virus Tat-cocaine-mediated impairment of neurogenesis: Role of TRPC1 channels. J Neurosci. 2012;32:9835–9847. doi: 10.1523/JNEUROSCI.0638-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de la Pena JB, Campbell ZT. RNA-binding proteins as targets for pain therapeutics. Neurobiol Pain. 2018;4:2–7. doi: 10.1016/j.ynpai.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication are available under GEO Series accession number GSE136890.