Abstract
The actinomycete strain Streptomyces coelicolor LY001 was purified from the sponge Callyspongia siphonella. Fractionation of the antimicrobial extract of the culture of the actinomycete afforded three new natural chlorinated derivatives of 3-phenylpropanoic acid, 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid (1), 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid methyl ester (2), and 3-(3-chloro-4-hydroxyphenyl)propanoic acid (3), together with 3-phenylpropanoic acid (4), E-cinnamic acid (5), and the diketopiperazine alkaloids cyclo(l-Phe-trans-4-OH-l-Pro) (6) and cyclo(l-Phe-cis-4-OH-d-Pro) (7) were isolated. Interpretation of nuclear magnetic resonance (NMR) and high-resolution electrospray ionization mass spectrometry (HRESIMS) data of 1–7 supported their assignments. Compounds 1–3 are first candidates of the natural chlorinated phenylpropanoic acid derivatives. The production of the chlorinated derivatives of 3-phenylpropionic acid (1–3) by S. coelicolor provides insight into the biosynthetic capabilities of the marine-derived actinomycetes. Compounds 1–3 demonstrated significant and selective activities towards Escherichia. coli and Staphylococcus aureus, while Candida albicans displayed more sensitivity towards compounds 6 and 7, suggesting a selectivity effect of these compounds against C. albicans.
Keywords: Red Sea sponges, marine actinomycetes, Streptomycescoelicolor LY001, halogenated 3-phenylpropanoic acid derivatives, diketopiperazine alkaloids, structural determinations, antimicrobial activities
1. Introduction
The marine actinomycetes represent a vital source of biologically active secondary metabolites and a promising future source for drug discovery. It is well known that marine tunicates and sponges are highly associated with symbiotic microbes [1,2,3]. There are very few reports about investigation of Red Sea actinomycetes for their chemical diversity and biomedical importance [4].
Streptomycetes represent a group within the actinomycetes with an economical importance and represent a vigorous source of different bioactive secondary metabolites [5]. More than 75% of the marketed antibiotics, commercially available compounds and several agrochemicals are synthesized by streptomycetes [6,7,8]. In 1990, the genus Streptomyces alone represented the main source of almost 60% of the antibiotics and most of the agrochemicals [9]. Streptomycetes produce an array of diverse and bioactive compounds with antimicrobial [5,9,10,11], anticancer [5,12,13], insecticidal and antiparasitic [14], anti-inflammatory [15], anti-fouling [16], antiviral and anti-infective [17,18] properties. Moreover, the genus Streptomyces is considered as a candidate of industrial importance [19,20,21], and a producer of secondary metabolites with herbicidal activity and which promote plant growth [22], vitamins [23], ribonucleases [11,13,24,25,26,27], and enzyme inhibitors [13]. These features make these microorganisms an ideal and favorite research project for academia and industry [9]. Recent advances in marine microbiology, including their purification and identification of bacteria that produce rich arrays of bioactive natural products, provide a strong motivation to explore aggressively their potential as sources of novel pharmaceutical agents [28,29,30]. Likewise, new methodology for rapid and affordable sequencing of microbial chromosomes, bioinformatic and metabolic profiling enables information on the predicted secondary metabolic diversity contained within a bacterial genome to be rapidly evaluated and manipulated [31,32,33].
Due to the fast growing number of pathogenic microbes, viral infections and cancer cells that have resistance towards current therapies, drug leads, and new chemical entities for the enhancement of new drugs are in high demand. Tuberculosis, malaria, and Staphylococcus aureus infections are among just a few diseases that have become difficult to treat with antibiotics [34]. There is, therefore, a pressing need for the development of new methodologies to provide new drugs/drug leads for the future. Marine microorganisms, such as actinomycetes that exist in association with marine invertebrates and algae, produce novel chemical entities with potential pharmaceutical significance. The importance of natural products, however, extends far beyond that of drug discovery to that of addressing fundamental biological questions, taking full advantage of their structural complexity and functional diversity to probe biological function.
The decreasing number of FDA approved therapeutics, and the low number of drugs in the pipeline that have arisen from synthetic combinatorial libraries, has spurred renewed interest in natural products research with the aim of identifying new structural scaffolds. Marine-derived actinomycetes represent a promising and vigorous source of ubiquitous and diverse and bioactive secondary metabolites that can be obtained in large-scale cultures and developed as drug leads. Overall, there are relatively few reports of marine microbial cultivation from the highly biodiverse Red Sea. The results from this study confirm that Red Sea marine actinomycetes represent a promising source of new drug leads with antibiotic potential.
As a part of our endeavor to purify and characterize bioactive marine microbial candidates [35,36], the antimicrobial fractions of extract of the culture of Streptomyces coelicolor LY001 was investigated. Three chlorinated derivatives of 3-phenylpropanoic acid, including 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid (1), 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid methyl ester (2), and 3-(3-chloro-4-hydroxyphenyl)propanoic acid (3), along with 3-phenylpropanoic acid (4) [37], E-cinnamic acid (5) and the diketopiperazine alkaloids cyclo(l-Phe-trans-4-OH-l-Pro) (6) [38,39], and cyclo(l-Phe-cis-4-OH-d-Pro) (7) [40,41] were isolated and identified. Compounds 1–7 were determined by assignments of their NMR and HRESIMS data. Herein, the isolation, structure assignments, and the antimicrobial activities of 1–7 are presented.
2. Results and Discussion
2.1. Isolation of the Actinomycete, Streptomyces coelicolor LY001
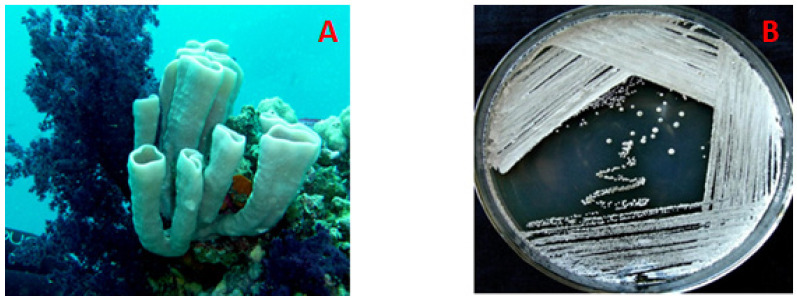
The marine-derived actinomycete LY001 (Figure 1) was isolated from the internal tissues of the sponge Callyspongia siphonella (Figure 1). The resulting sequence of the actinomycete strain was searched for homology with Basic Local Alignment Search Tool (BLAST) in the GenBank. The alignment with reported sequences in the GenBank showed that the LY001 isolate belongs to the genus Streptomyces and displayed 100% similarity with the strain Streptomyces coelicolor AB588124.
Figure 1.
The Red Sea sponge Callyspongia siphonella (A) and the actinomycete Streptomyces coelicolor LY001 (B).
2.2. Structure Elucidation of the Compounds
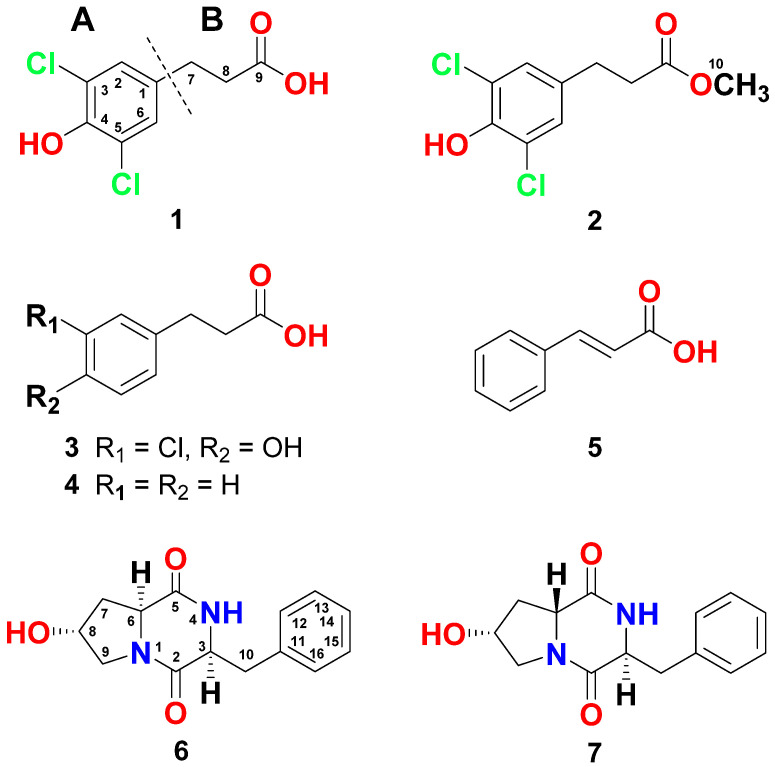
The structural assignment of compound 1 (Figure 2) was supported by interpretation of its NMR spectra (Figures S1–S5). The molecular formula of C9H8Cl2O3 was suggested for 1 as supported by HRESIMS (234.9932, C9H9Cl2O3, [M + H]+) (Figure S6), suggesting five degrees of unsaturation. Its 13C NMR spectrum along with the heteronuclear single-quantum correlation spectroscopy (HSQC) experiment showed seven signals equivalent for nine carbons including the equivalent methines (C-2 and C-6), two methylenes (C-7 and C-8) and five quaternary carbons, including two chemically equivalent carbons (C-3 and C-5) (Table 1). Its 1H NMR demonstrated a 2-proton singlet at δH 7.12 for the chemically equivalent protons H-2 and H-6 (Table 1). This two-proton singlet at δH 7.12 was correlated to the signal at δC 128.1 (CH, C-2/C-6) in the HSQC experiment. In the 1H-1H correlation spectroscopy (COSY) spectrum, vicinal couplings between the methylene protons at δH 2.86 (2H, H2-7) and 2.64 (2H, H2-8) was observed. Further, the protons of the methylenes at δH 2.86 (H2-7) and 2.64 (H2-8) were correlated, in the HSQC experiment, to the 13C NMR signals at δC 39.3 (CH2, C-7) and 34.5 (CH2, C-8), respectively. In addition, the existence of the quaternary carbons at δC 133.6 (qC, C-1), 122.1 (qC, C-3, C-5), and 146.0 (qC, C-4) together with δC 128.1 (2 × CH, C-2/C-6) suggested a 1,3,4,5-tetrasubstituted benzene moiety with OH at C-4 and symmetrical substitutions with chlorine atoms at C-3 and C-5.
Figure 2.
Structures of 1–7.
Table 1.
NMR data of 1–3 (CDCl3, 1H at 850 MHz, 13C at 213 Hz).
| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC (mult.) a | δH (mult., J in Hz) | δC (mult.) a | δH (mult., J in Hz) | δC (mult.) a | δH (mult., J in Hz) | |
| 1 | 133.6, qC | 133.8, qC | 133.5, qC | |||
| 2 | 128.1, CH | 7.12 (s) | 128.1, CH | 7.10 (s) | 128.6, CH | 7.17 (d, 2.5) |
| 3 | 122.1, qC | 121.0, qC | 120.8, qC | |||
| 4 | 146.9, qC | 146.6, qC | 151.7, qC | |||
| 5 | 122.1, qC | 121.0, qC | 116.1, CH | 6.93 (d, 8.5) | ||
| 6 | 128.1, CH | 7.12 (s) | 128.1, CH | 7.10 (s) | 128.4, CH | 7.02 (dd, 8.5, 2.5) |
| 7 | 29.3, CH2 | 2.86 (t, 7.6) | 29.8, CH2 | 2.84 (t, 7.6) | 29.5, CH2 | 2.86 (t, 7.6) |
| 8 | 34.5, CH2 | 2.64 (t, 7.6) | 35.3, CH2 | 2.59 (t, 7.6) | 34.8, CH2 | 2.65 (t, 7.6) |
| 9 | 173.8, qC | 173.3, qC | 173.9, qC | |||
| 10 | 51.9, CH3 | 3.67 (s) | ||||
a Multiplicities of the 13C NMR signals were assigned from HSQC experiment.
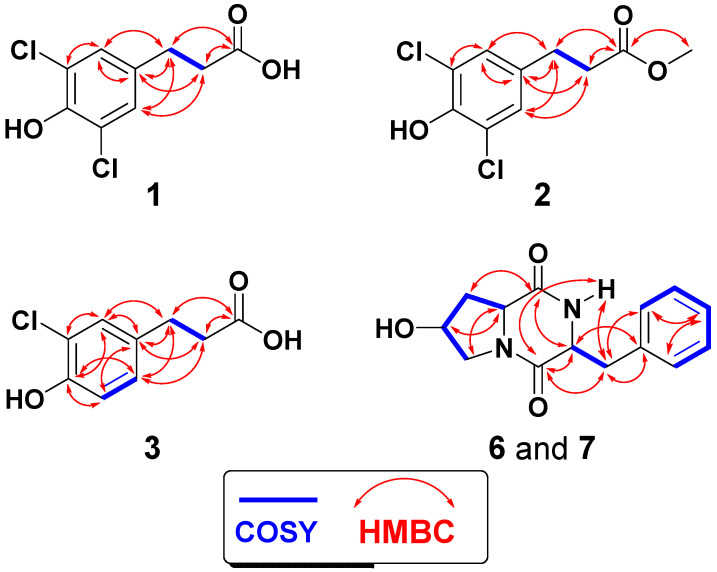
1H-13C Heteronuclear multiple bond correlation spectroscopy (HMBC) experiment supported the substitution on the phenyl moiety. HMBC of H-2,6/C-1 (qC, δC 133.6), H-2,6/C-3,5 (qC, δC 122.1), and H-2,6/C-4 (qC, δC 146.9) supported this substitution (Figure 3). Thus, 3,5-dichloro-4-hydroxyphenyl moiety was assigned as part A of the molecule (Figure 2). Furthermore, the 1H chemical shifts values of H-2 and H-6, and 13C values of C-1–C-6 in 1 are similar with those of (S)-2-amino-3-(3,5-dichloro-4-hydroxyphenyl)-propanoic acid [42]. The substituent at C-1 of the phenyl moiety was assigned as 3-substituted propanoic acid (part B) as supported from the signals at δH/C 2.86 (2H)/29.3 (H2-7/C-7), 2.64 (2H)/34.5 (H2-8/C-8), and δC 173.8 (qC, C-9) (Table 1). The HMBC from the protons at H2-7 and H2-8 to C-9 (δC 173.8) (Figure 3) supported this assignment. Finally, the connection between the two parts (A and B) of 1 was proved by cross-peaks in the HMBC of H2-7/C-1, H2-7/C-2,6, H2-8/C-1, and H-2,6/C-7 (Figure 3). Thus, compound 1 was assigned as 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid and this is the first report about its natural occurrence.
Figure 3.
1H-1H COSY and 1H-13C HMBC of 1-3, 6, and 7.
Interpretation of the one- and two-dimensional NMR data (Figures S7–S11) supported the structural determination of 2 (Figure 2). It possesses molecular formula C10H10Cl2O3 as supported by HRESIMS (249.0088, C10H11Cl2O3, [M + H]+) (Figure S12), being 15 mass units larger than 1, proving the existence of an additional CH3 in 2. The structure of 2 was assigned by interpretation of its NMR spectra. The NMR data of 2 are identical with those of 1 (Table 1), suggesting similar structures. Furthermore, the existence of an additional three-proton singlet at δH 3.67 correlated to the signal at δC 51.9 (CH3, C-10) in the HSQC, which supports the existence of a terminal methoxyl group in 2. The HMBC from H3-10 (δH 3.67) to C-9 (δC 173.3) supported the existence of a methyl ester group in 2 instead of a free carboxylic acid in 1.
Further, the assignment of the substituents on the aromatic moiety was supported by HMBC (Figure 3) as previously discussed under compound 1. Similarly, the 1H chemical shifts of H-2 and H-6, and 13C chemical shifts values of C-1–C-6 in 2 are similar to those of (S)-2-amino-3-(3,5-dichloro-4-hydroxyphenyl)-propanoic acid [42]. Accordingly, compound 2 was assigned as 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid methyl ester and this is its first natural occurrence.
The structure determination of compound 3 (Figure 2) was assigned by examination of its NMR spectroscopic data (Figures S13–S17). Compound 3 with molecular formula of C9H9ClO3 (201.0321, C9H10ClO3, [M + H]+) (Figure S18), being 34 units less than 1, suggests the presence of only one chlorine atom in 3. Its 13C NMR spectrum and HSQC exhibited nine signals for three aromatic methines (C-2, C-5, and C-6), two methylenes (C-7 and C-8), and four quaternary carbons (Table 1). Its 1H NMR spectrum showed two spin-spin coupling systems. The first one includes the aromatic ABX coupling system at δH 7.17 (H-2, d, J = 2.5 Hz), 6.93 (H-5, d, J = 8.5 Hz), and 7.02 (H-6, dd, J = 8.5, 2.5 Hz) (Table 1). These protons are correlated in the HSQC experiment to the signals at δC 128.6 (C-2), 116.1 (C-5), and 128.4 (C-6), respectively. This system supported the existence of a 1,3,4-trisubstituted aromatic moiety in 3. The second spin coupling system includes the vicinal coupling between the methylenes at δH 2.86 (H2-7, t, J = 7.6 Hz) and 2.65 (H2-8, t, J = 7.6 Hz). In the HMBC, H2-7 (δH 2.86) and H2-8 ((δH 2.65) displayed correlation to the signal at δC 173.9 (C-9) (Figure 3), supporting the existence of a 3-substituted propanoic acid moiety as in 1. Again, the placement of the side chain (3-substituted propanoic acid) at C-1 was secured from HMBC of H-2/C-7, H-6/C-7, H2-7/C-1, and H2-8/C-1 (Figure 3). Finally, HMBC of H-2/C-3 (qC, δC 120.8), H-2/C-4 (qC, δC 151.7), H-5/C-4, H-5/C-3, H-5/C-1 (qC, δC 133.5), H2-7/C-9 (qC, δC 173.9), and H2-8/C-9 (Figure 3) supported the substitution on the benzene ring in 3. Again, the chemical shifts of the 1H (H-2, H-5 and H-6) and 13C (C-1–C-6) NMR signals of the 3,5-dichloro-4-hydroxy phenyl moiety in compound 3 are similar with reported data for (S)-3-(3-chloro-4-hydroxyphenyl)-2-(dimethylamino)propanoic acid [42]. Thus, 3 was assigned as 3-(3-chloro-4-hydroxyphenyl)propanoic acid and this is the first report about its natural occurrence.
Interpretation of the NMR (Figures S19–S23) and MS spectra of 4 (Figure 2) supported its structure. The NMR are identical with those reported for 3-phenylpropanoic acid from a terrestrial Streptomyces strain [37]. Accordingly, 4 was assigned as 3-phenylpropanoic acid and this is its first occurrence from a marine Streptomyces.
The structure of 5 (Figure 2) was assigned, from the NMR (Figures S24 and S25) and MS spectra, as E-cinnamic acid. Its NMR spectra exhibited signals for a monosubstituted benzene ring. The J value (J7,8 = 16.1 Hz) supported E configuration in 5.
The structures of 6 and 7 (Figure 2) were identified from interpretation of the corresponding NMR (Figures S26–S30 for 6 and Figures S31–S35 for 7) and MS spectra. Both compounds (Figure 2) displayed the same molecular formula of (C14H16N2O3). The diketopiperazine nature of 6 and 7 are obvious from the corresponding 1H and 13C resonating signals (Table 2). Those include the resonances of two amidic carbonyls (C-2 and C-5) as well as the resonances for the methine signals at C-3 and C-6 (Table 2). The remaining of the signals are characteristic for the presence of a hydroxylated proline and phenylalanine moieties in 6 and 7 [38,39,40,41]. The NMR signals at δH/C 4.60/68.4 (in 6) and δH/C 4.40/68.1 (in 7) are characteristic for the hydroxylated C-8 supporting the hydroxylation of the proline moieties in both compounds [38,39,40,41]. The NMR data of 6 and 7 (Table 2) are similar to those of cyclo(l-Phe-trans-4-OH-l-Pro) [38,39] and cyclo(l-Phe-cis-4-OH-d-Pro) [40,41], respectively. Furthermore, the 2D experiments (HSQC, COSY, HMBC) (Figure 3) supported the assignment of all signals of the compounds (Table 2 and Figure 3). Accordingly, compounds 6 and 7 were assigned as cyclo(l-Phe-trans-4-OH-l-Pro) and cyclo(l-Phe-cis-4-OH-d-Pro).
Table 2.
NMR data of 6 and 7 (CDCl3).
| Position | 6 (1H at 600 MHz, 13C at 150 MHz) | 7 (1H at 850 MHz, 13C at 213 Hz) | ||
|---|---|---|---|---|
| δC (mult.) a | δH (mult., J in Hz) | δC (mult.) a | δH (mult., J in Hz) | |
| 2 | 165.0, qC | 165.2, qC | ||
| 3 | 56.1, CH | 4.33 (dd, 10.8, 3.0) | 59.1, CH | 4.23 (tt, 4.2) |
| 4 (NH) | 5.60 (brs) | 5.80 (brs) | ||
| 5 | 169.5, qC | 168.4, qC | ||
| 6 | 57.3, CH | 4.46 (dd, 10.8, 6.0) | 55.8, CH | 3.17 (t, 8.5) |
| 7a | 37.8, CH2 | 2.37 (dd, 13.8, 6.0) | 38.0, CH2 | 2.37 (ddd,13.7, 8.5, 5.1) |
| 7b | 2.06 (ddd, 13.8, 11.4, 4.2) | 2.20 (m) | ||
| 8 | 68.4, CH | 4.60 (t, 4.2) | 68.1, CH | 4.40 (quin, 4.0) |
| 9a | 54.4, CH2 | 3.80 (dd, 13.2, 4.2) | 53.4, CH2 | 3.82 (dd, 12.7, 2.5) |
| 9b | 3.58 (d, 13.2) | 3.35 (dd, 12.7, 5.1) | ||
| 10a | 36.6, CH2 | 3.63 (dd, 14.5, 4.2) | 40.8, CH2 | 3.13 (dd, 14.0, 6.8) |
| 10b | 2.77 (dd, 15.5, 10.8) | 3.10 (dd, 14.0, 4.2) | ||
| 11 | 135.7, qC | 135.2, qC | ||
| 12 | 129.0, CH | 7.22 (d, 7.5) | 129.7, CH | 7.21 (d, 7.3) |
| 13 | 129.3, CH | 7.36 (t, 7.5) | 128.9, CH | 7.31 (t, 7.3) |
| 14 | 127.6, CH | 7.29 (t, 7.5) | 127.6, CH | 7.30 (t, 7.3) |
| 15 | 129.3, CH | 7.36 (t, 7.5) | 128.9, CH | 7.31 (t, 7.3) |
| 16 | 129.0, CH | 7.22 (d, 7.5) | 129.7, CH | 7.21 (d, 7.3) |
a Multiplicities of the 13C NMR signals were assigned from HSQC experiment.
The derivatives of 3-phenylpropanoic acid are rarely reported from microbial organisms. We believe that there is only one report about the occurrence of 3-phenylpropanoic acid from a terrestrial Streptomyces strain [37]. Further, it is worth pointing out that this is the first report about the natural existence of chlorinated derivatives of 3-phenylpropanoic acid. These results give an understanding about the biosynthetic potential and structural diversity of the cultured marine-derived microbes and the future application of these metabolites in drug discovery.
Phenylpropanoids area ubiquitous group of organic compounds including flavonoids, coumarins, phenolic acids, stilbenes, and lignins. They originate from phenylalanine and tyrosine [43]. The name phenylpropanoid is derived from a six-carbon forming a phenyl moiety connected to a three-carbon propene unit. The coumaric acid represents the key biosynthetic intermediate in the biosynthesis of all phenylpropanoids. Biosynthetically, a variety of natural products including flavonoids, phenylpropanoids, isoflavonoids, catechins, coumarins, stilbenes, aurones, and lignols originates from 4-coumaroyl-CoA [44], which is originated from cinnamic acid [44].
Three pathogens were used to determine the antimicrobial effects and the minimum inhibitory concentration (MIC) values of 1–7. Compound 1 showed the greatest inhibitory effects towards S. aureus and E. coli with inhibition zones of 17 and 23 mm (Table 3). In addition, compounds 2 and 3 were less active than 1 against these pathogens with inhibition zones of 12–20 mm (Table 3). Compound 4 displayed the lowest activity against these pathogens with inhibition zones of 15 and 11 mm. These findings suggest the importance of the substitution with a “3,5-dichloro-4-hydroxy” moiety on the phenyl moiety as well as the presence of a free terminal carboxylic acid moiety for a maximum antibacterial activity (as in 1). On the other hand, 1–4 showed modest effect towards C. albicans (ATCC 14053) with 6–9 mm inhibition zone (Table 3), suggesting selective antibacterial effects of these compounds against E. coli and S. aureus. Finally, the diketopiperazine alkaloids 6 and 7 displayed better activities towards C. albicans with 12–14 mm inhibition zones, while they were less active towards E. coli and S. aureus with 7–11 mm inhibition zones, suggesting their selective antifungal activity against C. albicans.
Table 3.
Antimicrobial effects of 1–7.
| Compound | E. coli | S. aureus | C. albicans | |||
|---|---|---|---|---|---|---|
| Inhibition Zone (mm) | MIC (µg/mL) | Inhibition Zone (mm) | MIC (µg/mL) | Inhibition Zone (mm) | MIC (µg/mL) | |
| 1 | 23 | 16 | 17 | 32 | 9 | 125 |
| 2 | 20 | 32 | 12 | 64 | 7 | 250 |
| 3 | 18 | 32 | 15 | 32 | 7 | 250 |
| 4 | 15 | 64 | 11 | 125 | 6 | 250 |
| 5 | NT | NT | NT | NT | NT | NT |
| 6 | 7 | 250 | 9 | 250 | 12 | 32 |
| 7 | 11 | 125 | 7 | 250 | 14 | 32 |
| Ciprofloxacin a | 30 | 0.25 | 22 | 0.5 | NT | NT |
| Ketoconazole b | NT | NT | NT | NT | 30 | 0.5 |
a positive antibacterial drug; b positive antifungal drug.
To determine the MIC values of the compounds, a microdilution method was carried out (Table 3). Compound 1 displayed the highest activity towards E. coli with an MIC of 16 µg/mL, while compounds 6 and 7 displayed the highest antifungal activities with an MIC of 32 µg/mL towards C. albicans. On the other hand, compounds 2 and 3 displayed lower activities with MIC values of 32–64 µg/mL towards E. coli and S. aureus. Other compounds were weakly active with MIC values of 64–250 μg/mL.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations, ultraviolet (UV), infra-red (IR), NMR, and HRESIMS were acquired as previously reported [35,36]. Fractionation of the extracts and successive fractions were performed on SiO2 and Sephadex LH-20. The purification of the compounds was carried out on an analytical Shim-Pack C18 (250 × 4.6 mm, Shimadzu, Kyoto, Japan).
3.2. The Host Organism, C. siphonella
The marine sponge C. siphonella was harvested in May 2016 using scuba at a depth up to 20 m off Jizan at the Saudi Red Sea. The pink tubular sponge is dichotomously divided with a smooth thin-walled surface. It possesses a soft compressible consistency, which is difficult to tear. The voucher specimen measures up to 10 cm, while the branching tubes measure up to 5 cm in height and up to 2.5 cm in diameter. A comprehensive description of the sponge and the specimen’s codes are previously reported [45,46].
3.3. Isolation of the Actinomycete Streptomyces coelicolor LY001
After surface sterilization, about 1 cm3 of the internal tissue of the sponge was finely mixed in sterile seawater (10 mL), diluted, and spread on International Streptomyces Project-2 (ISP2) medium. The medium was amended with 3% NaCl. Afterwards, cultured plates were incubated at 30 °C and checked after actinomycetes growth regularly. The actinomycete LY001 was obtained in a pure state after several purification steps.
3.4. Characterization of the Actinomycete, Streptomyces coelicolor
The LY001 strain was identified by analysis of its 16S rRNA sequence. DNA preparation was used for 16S rRNA gene PCR amplification using 27f and 1492r primers. The reaction mixture composed of 50 µL included 1000 ng of gDNA, primers (each 20 pmol), and GoTaq Master Mixes (25 µL). The polymerase chain reaction (PCR) thermocycler initiated with denaturation at 95 °C (2 min), 30 cycles at 95 °C (30 s for denaturation), 30 s at 58 °C (for annealing), 60 s at 72 °C (for extension), and at 72 °C for 5 min for final completion of DNA extension. Purification of PCR products was accomplished on Agarose Gel DNA Purification Kit (Biocompare, South San Francisco, CA, USA) as supported by the supplier. 16S rRNA sequence displayed 100% similarity with Streptomyces coelicolor (Accession No. AB588124). The sequence of the Red Sea Streptomyces coelicolor LY001 was placed in the NCBI GenBank under the Accession Number MN883509 on 30 December 2019 (http://getentry.ddbj.nig.ac.jp/).
3.5. Large-Scale Culture of Streptomyces coelicolor
Spores of Streptomyces coelicolor were cultured in 2.0 L flasks, each containing ISP2 media (500 mL) [47] including 10 g of malt extract, 4.0 g of yeast extract, 4.0 g of dextrose, and 3% NaCl (w/v) in 1 L distilled water at pH of 7. Incubation of the culture was accomplished by shaking at 180 rpm at 28 °C for 14 days. The combined culture broth (10 L) was shaken against EtOAC three times (each 3 L). The resulted EtOAc extracts dried to give 1.3 g.
3.6. Purification of 1–7
The dried extract (1.3 g) was partitioned on C18 (ODS) silica (Sigma Aldrich, St. Louis, MO, USA) using vacuum liquid chromatography (VLC)column using H2O-MeOH gradients to give 10 fractions (Fractions A–J). The antimicrobial fractions C (180 mg) and D (118 mg) were separately fractionated on Sephadex LH-20 using MeOH-CH2Cl2 (1:1) mixture to give five subfractions, each. Subfraction C4 (65 mg) was purified on an ODS HPLC column using 30% CH3CN to afford compounds 5 (9.1 mg), 6 (3.7 mg), and 7 (2.6 mg). Subfraction D4 (54 mg) was purified on an ODS HPLC using 45% CH3CN to afford compounds 1 (6.2 mg), 2 (3.6 mg), 3 (3.1 mg), and 4 (4.2 mg).
Spectral Data of 1–7
3-(3,5-Dichloro-4-hydroxyphenyl)propanoic acid (1). Colorless solid; UV (MeOH) λmax (log ε): 227 (4.15), 312 (4.17) nm; IR (film) νmax 3340, 3029, 1700, 1301, 1219, 935 cm−1; HRESIMS m/z 234.9932 (calcd for C9H9Cl2O3, [M + H]+, 234.9929), NMR: Table 1.
3-(3,5-Dichloro-4-hydroxyphenyl)propanoic acid methyl ester (2). Colorless solid; UV (MeOH) λmax (log ε): 229 (4.15), 315 (4.17) nm; IR (film) νmax 3335, 3030, 1665, 1305, 1219, 937 cm−1; HRESIMS m/z 249.0088 (calcd for C10H11Cl2O3, [M + H]+, 249.0085); NMR: Table 1.
3-(3-Chloro-4-hydroxyphenyl)propanoic acid (3). Colorless solid; UV (MeOH) λmax (log ε): 225 (3.95), 307 (4.00) nm; IR (film) νmax 3341, 3030, 1702, 1303, 1221, 936 cm−1; HRESIMS m/z 201.0321 (calcd for C9H10ClO3, [M + H]+, 201.0318), NMR: Table 1.
3-Phenylpropanoic acid (4). Colorless solid; ESIMS m/z 151.07 (C9H11O2, [M + H]+), NMR data: 1H NMR (850 MHz, CDCl3) δH: 7.22 (2H, m, H-2,6), 7.30 (2H, m, H-3,5), 7.21 (1H, m, H-4), 2.97 (t, J = 7.5 H2-7), 2.69 (t, J = 7.5 H2-8); 13C NMR (312 MHz, CDCl3) δC: 140.1 (qC, C-1), 128.5 (CH, C-2,6), 128.2 (CH, C-3,5), 126.4 (CH, C-4), 29.7 (CH2, C-7), 34.7 (CH2, C-8), 171.6 (qC, C-9).
E-Cinnamic acid (5). Colorless solid; ESIMS m/z 149.06 (C9H9O2, [M + H]+); NMR data: 1H NMR (850 MHz, CDCl3) δH: 7.55 (2H, m, H-2,6), 7.41 (2H, m, H-3,5), 7.40 (1H, m, H-4), 7.79 (1H, d, J = 16.1 Hz, H-7), 6.45 (1H, d, J = 16.1 Hz, H-8); 13C NMR (312 MHz, CDCl3) δC 134.4 (C-1), 128.1 (C-2,6), 128.9 (C-3,5), 130.8 (C-4), 147.2 (C-7), 117.0 (C-8), 171.6 (C-9).
Cyclo(l-Phe-trans-4-OH-l-Pro) (6). Colorless solid; [α]D −55° (c 0.10, MeOH); UV (MeOH) λmax (log ε): 230 (4.10), 305 (4.07) nm; IR (film) νmax 3451, 1663, 1629 cm−1; HRESIMS m/z 261.1241 (calcd for C14H17N2O3, [M + H]+, 261.1239), NMR: Table 2.
Cyclo(l-Phe-cis-4-OH-d-Pro) (7). Colorless solid; [α]D +38° (c 0.10, MeOH); UV (MeOH) λmax (log ε): 230 (4.10), 305 (4.07) nm; IR (film) νmax 3450, 1662, 1629 cm−1; HRESIMS m/z 261.1241 (calcd for C14H17N2O3, [M + H]+, 261.1239), NMR: Table 2.
3.7. Antimicrobial Evaluation of Compounds 1–7
3.7.1. Disc Diffusion Assay
Using the disc diffusion assay, the antimicrobial effects of 1–7 were evaluated at 100 μg/disc against several pathogenic microbes including E. coli (ATCC 25922), C. albicans (ATCC 14053), and S. aureus (ATCC 25923) and as previously described [48,49,50]. Ciprofloxacin (5.0 μg/disc) and ketoconazole (50 μg/disc) were used as positive antibiotics.
3.7.2. Determination of the MIC of 1–7
The MIC values of the compounds was evaluated using a macrodilution method [51]. Briefly, MeOH was used to dissolve the compounds at a final concentration of 2000 µg/mL, while distilled water was used to dissolve ciprofloxacin and ketoconazole at final concentrations of 100 µg/mL. All solutions were sterilized using syringe filters (0.2 µm). A two-fold serial dilution of the solutions was used in Mueller Hinton Broth (MHB) to afford concentrations between 1.0 and 1000 µg/mL for the compounds and between 0.125 and 64 µg/mL for ciprofloxacin and ketoconazole. From the 106 colony-forming units (CFU)/mL microbial suspensions, 500 μL were added in sterile tubes giving inoculua of 5 × 105 CFU/mL. Additional 100 μL of each stock solution of the compounds and antibiotics were added into the tubes. A control tube, which contains only the test microorganisms and methanol was prepared. The MeOH displayed no antimicrobial effect. Incubation of the tubes was accomplished at 37 °C for 48 h. The lowest concentrations of the compounds/antibiotics, which show no microbial growth were considered as MIC.
4. Conclusions
Purification of the antimicrobial fractions of the culture of the Streptomyces coelicolor LY001 afforded three chlorinated 3-phenylpropanoic acid derivatives including 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid (1), 3-(3,5-dichloro-4-hydroxyphenyl)propanoic acid methyl ester (2), and 3-(3-chloro-4-hydroxyphenyl)propanoic acid (3) along with 3-phenylpropanoic acid (4), E-cinnamic acid (5), and the dipeptides cyclo(l-Phe-trans-4-OH-l-Pro (6) and cyclo(l-Phe-cis-4-OH-d-Pro) (7). Structures of 1–7 were determined from interpretation of their NMR and HRESIMS spectroscopic data. The chlorinated 3-phenylpropanoic acid derivatives (1–3) showed selective and significant antimicrobial activities against S. aureus and E. coli and displayed modest effects towards C. albicans. On the other hand, the diketopiperazine alkaloids 6 and 7 were more active against and selective against C. albicans and less active than E. coli and S. aureus. Interestingly, this is the first report about natural occurrence of chlorinated 3-phenypropionic acid derivatives from a cultured marine-derived Streptomyces. These results suggest insight into the biosynthetic capacities of the cultured marine actinomycetes. Thus, compounds 1–3, 6, and 7 represent interesting compounds for the design of novel and effective antibiotic drugs.
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant No. (G-270-141-39). The authors, therefore, acknowledge with thanks the DSR for the technical and financial support. We would like to thank Rob van Soest for the identification of the sponge specimen.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/9/450/s1, Figure S1–S36: 1H NMR, 13NMR, 1H-1H COSY, multiplicity-edited HSQC, HMBC, and HRESIMS data of compounds 1-7.
Author Contributions
Conceived and designed the experiments, L.A.S. and D.T.A.Y.; prepared a large-scale culture and extracted the culture, S.S.E.; performed the experiments and purified the compounds, D.T.A.Y. and L.A.S.; interpreted the NMR and MS data, L.A.S., T.A.A. and D.T.A.Y.; wrote and revised the manuscript, L.A.S. and D.T.A.Y. All authors have read and agreed to the published version of the manuscript.
Funding
Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant No. (G-270-141-39).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Faulkner D.J., Harper K.K., Haygood M.G., Salomon C.E., Schmidt E.W. Symbiotic bacteria in sponges: Sources of bioactive substances. In: Fusetani N., editor. Drugs from the Sea. Karger; Basel, Switzerland: 2000. pp. 107–119. [Google Scholar]
- 2.Tsukimoto M., Nagaoka M., Shishido Y., Fujimoto J., Nishisaka F., Matsumoto S., Haruari E., Imada C., Matsuzaki T. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J. Nat. Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann C.M., Marker A., Kurtböke D.I. An Overview on Marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity. 2017;9:40. doi: 10.3390/d9040040. [DOI] [Google Scholar]
- 4.Carroll A.R., Copp B.R., Davis R.A., Keyzers R.A., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2020;37:175–223. doi: 10.1039/C9NP00069K. [DOI] [PubMed] [Google Scholar]
- 5.Berdy J. Bioactive microbial metabolites. J. Antibiot. Tokyo. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 6.Newman D.J., Cragg G.M., Snader K.M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez-Esquilín A.E., Roane E.T.M. Antifungal activities of actinomycete strains associated with high-altitude sagebrush rhizosphere. J. Ind. Microbiol. Biotechnol. 2005;32:378–381. doi: 10.1007/s10295-005-0007-x. [DOI] [PubMed] [Google Scholar]
- 8.Olanrewaju O.S., Babalola O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019;103:1179–1188. doi: 10.1007/s00253-018-09577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramesh S., Rajesh M., Mathivanan N. Characterization of a thermostable alkaline protease produced by marine Streptomyces fungicidicus MML1614. Bioprocess. Biosyst. Eng. 2009;32:791–800. doi: 10.1007/s00449-009-0305-1. [DOI] [PubMed] [Google Scholar]
- 10.Prabavathy V.R., Mathivanan N., Murugesan K. Control of blast and sheath blight diseases of rice using antifungal metabolites produced by Streptomyces sp. PM5. Biol. Control. 2006;39:313–319. doi: 10.1016/j.biocontrol.2006.07.011. [DOI] [Google Scholar]
- 11.Prapagdee B., Kuekulvong C., Mongkolsuk S. Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. Int. J. Biol. Sci. 2008;4:330–337. doi: 10.7150/ijbs.4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006;9:245–251. doi: 10.1016/j.mib.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Hong K., Gao A.H., Xie Q.Y., Gao H., Zhuang L., Lin H.P., Yu H.P., Li J., Yao X.S., Goodfellow M., et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs. 2009;7:24–44. doi: 10.3390/md7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pimentel-Elardo S.M., Kozytska S., Bugni T.S., Ireland C.M., Moll H., Hentschel U. Antiparasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar. Drugs. 2010;8:373–380. doi: 10.3390/md8020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renner M.K., Shen Y.C., Cheng X.C., Jensen P.R., Frankmoelle W., Kauffman C.A., Fenical W., Lobkovsky E., Clardy J. Cyclomarins A-C, new anti-inflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.) J. Am. Chem. Soc. 1999;121:11273–11276. doi: 10.1021/ja992482o. [DOI] [Google Scholar]
- 16.Xu Y., He H., Schulz S., Liu X., Fusetani N., Xiong H., Xiao X., Qian P.Y. Potent antifouling compounds produced by marine Streptomyces. Bioresour. Technol. 2010;101:1331–1336. doi: 10.1016/j.biortech.2009.09.046. [DOI] [PubMed] [Google Scholar]
- 17.Sacramento D.R., Coelho R.R.R., Wigg M.D., Linhares L.F.T.L., Santos M.G.M., Semedo L.T.A.S., da Silva A.J.R. Antimicrobial and antiviral activities of an actinomycete (Streptomyces sp.) isolated from a Brazilian tropical forest soil. World J. Microbiol. Biotechnol. 2004;20:225–229. doi: 10.1023/B:WIBI.0000023824.20673.2f. [DOI] [Google Scholar]
- 18.Rahman H., Austin B., Mitchell W.J., Morris P.C., Jamieson D.J., Adams D.R., Spragg A.M., Schweizer M. Novel anti-infective compounds from marine bacteria. Mar. Drugs. 2010;8:498–518. doi: 10.3390/md8030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams S.T., Goodfellow M., Alderson G., Wellington E.M.H., Sneath P.H.A., Sackin M.J. A probability matrix for identification of some Streptomyces. J. Gen. Microbiol. 1983;129:1815–1830. doi: 10.1099/00221287-129-6-1815. [DOI] [PubMed] [Google Scholar]
- 20.Tamehiro N., Hosaka T., Xu J., Hu H., Otake N., Ochi K. Innovative approach for improvement of an antibiotic–overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol. 2003;69:6412–6417. doi: 10.1128/AEM.69.11.6412-6417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higginbotham S.J., Murphy C.D. Identification and characterisation of a Streptomyces sp. isolate exhibiting activity against methicillin-resistant Staphylococcus aureus. Microbiol. Res. 2010;165:82–86. doi: 10.1016/j.micres.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Sousa C.S., Soares A.C.F., Garrido M.S. Characterization of streptomycetes with potential to promote plant growth and biocontrol. Sci. Agric. Piracicaba Braz. 2008;65:50–55. doi: 10.1590/S0103-90162008000100007. [DOI] [Google Scholar]
- 23.Atta H.M. Production of vitamin B12 by Streptomyces fulvissimus. Egypt. J. Biomed. Sci. 2007;23:166–184. [Google Scholar]
- 24.Cal S., Aparicio J.F., De Los Reyes-Gavilan C.G., Nicieza J., Sanchez A. A novel exocytoplasmic endonuclease from Streptomyces antibioticus. J. Biochem. 1995;306:93–100. doi: 10.1042/bj3060093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunakova Z., Godnay A., Timko J. An extracellular endodeoxyribonuclease from Sterptomyces aureofaciens. Biochim. Biophys. Acta. 2004;1721:116–123. doi: 10.1016/j.bbagen.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Ramesh S., Mathivanan N. Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J. Microbiol. Biotechnol. 2009;25:2103–2111. doi: 10.1007/s11274-009-0113-4. [DOI] [Google Scholar]
- 27.Nicieza R.G., Huergo J., Connolly B.A., Sanchex J. Purification, characterization and role of nucleases and serine proteases in Streptomyces differentiation. J. Biol. Chem. 1999;274:20366–20375. doi: 10.1074/jbc.274.29.20366. [DOI] [PubMed] [Google Scholar]
- 28.Magarvey N.A., Keller J.M., Bernan V., Dworkin M., Sherman D.H. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl. Environ. Microbiol. 2004;70:7520–7529. doi: 10.1128/AEM.70.12.7520-7529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenical W., Jensen P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 30.Jensen P.R., Williams P.G., Oh D.C., Zeigler L., Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zazopoulos E., Huang K., Staffa A., Liu W., Bachmann B.O., Nonaka K., Ahlert J., Thorson J.S., Shen B., Farnet C.M. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat. Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 32.McAlpine J.B., Bachmann B.O., Piraee M., Tremblay S., Alarco A.M., Zazopoulos E., Farnet C.M. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J. Nat. Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 33.Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W., Jensen P.R., Moore B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakata T., Winzeler E.A. Genomics, systems biology and drug development for infectious diseases. Mol. Biosyst. 2007;3:841–848. doi: 10.1039/b703924g. [DOI] [PubMed] [Google Scholar]
- 35.Shaala L.A., Youssef D.T.A., Badr J.M., Harakeh S.M. Bioactive 2(1H)-pyrazinones and diketopiperazine alkaloids from a tunicate-derived actinomycete Streptomyces sp. Molecules. 2016;21:1116. doi: 10.3390/molecules21091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaala L.A., Youssef D.T.A., Badr J.M., Harakeh S.M., Genta-Jouve G. Bioactive diketopiperazines and nucleoside derivatives from a sponge-derived Streptomyces species. Mar. Drugs. 2019;17:584. doi: 10.3390/md17100584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayana K.J.P., Prabhakar P., Vijayalakshm M., Venkateswarlu Y., Krishna P.S.J. Biological activity of phenylpropionic acid isolated from a terrestrial Streptomycetes. Pol. J. Microbiol. 2007;56:191–197. [PubMed] [Google Scholar]
- 38.Adamczeski M., Quinoa E., Crews P. Novel sponge-derived amino acids. 5. Structures, stereochemistry, and synthesis of several new heterocycles. J. Am. Chem. Soc. 1989;111:647–654. doi: 10.1021/ja00184a037. [DOI] [Google Scholar]
- 39.Li X.-Y., Wang Y.-H., Yang J., Cui W.-Y., He P.-J., Munir S., He P.-F., Wu Y.-X., He Y.-Q. Acaricidal activity of cyclodipeptides from Bacillus amyloliquefaciens W1 against Tetranychus urticae. J. Agric. Food Chem. 2018;66:10163–10168. doi: 10.1021/acs.jafc.8b03806. [DOI] [PubMed] [Google Scholar]
- 40.Shigemori H., Tenma M., Shimazaki K., Kobayashi J. Three new metabolites from the marine yeast Aureobasidium pullulans. J. Nat. Prod. 1998;61:696–698. doi: 10.1021/np980011u. [DOI] [PubMed] [Google Scholar]
- 41.De Rosa S., Mitova M., Tommonaro G. Marine bacteria associated with sponge as source of cyclic peptides. Biomol. Eng. 2003;20:311–316. doi: 10.1016/S1389-0344(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 42.Restrepo M.P., Jaramillo E.G., Martínez A.M., Arango A.M., Restrepo S.R. Anti-parasite and cytotoxic activities of chloro and bromo L-tyrosine derivatives. J. Braz. Chem. Soc. 2018;29:2569–2579. doi: 10.21577/0103-5053.20180136. [DOI] [Google Scholar]
- 43.Barros J., Serrani-Yarce J.C., Chen F., Baxter D., Venables B.J., Dixon R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants. 2016;2:16050. doi: 10.1038/nplants.2016.50. [DOI] [PubMed] [Google Scholar]
- 44.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 45.Jain S., Laphookhieo S., Shi Z., Fu L., Akiyama S., Chen Z., Youssef D.T.A., van Soest R.W.M., El Sayed K.A. Reversal of P-glycoprotein-mediated multidrug resistance by sipholane triterpenoids. J. Nat. Prod. 2007;70:928–931. doi: 10.1021/np0605889. [DOI] [PubMed] [Google Scholar]
- 46.Youssef D.T.A., Ibrahim A.K., Khalifa S.I., Mesbah M.K., Mayer A.M.S., van Soest R.W.M. New anti-inflammatory sterols from the Red Sea sponges Scalarispongia aqabaensis and Callyspongia siphonella. Nat. Prod. Commun. 2010;5:27–31. [PubMed] [Google Scholar]
- 47.Küster E. Outline of a comparative study of criteria used in characterization of the actinomycetes. Int. Bull. Bacteriol. Nomencl. Taxon. 1959;9:97–104. doi: 10.1099/0096266X-9-2-97. [DOI] [Google Scholar]
- 48.Acar J.F. The disc susceptibility test. In: Lorian V., editor. Antibiotics in Laboratory Medicine. Williams and Wilkins; Baltimore, MD, USA: 1980. pp. 24–54. [Google Scholar]
- 49.Kiehlbauch J.A., Hannett G.E., Salfinger M., Archinal W., Monserrat C., Carlyn C. Use of the National Committee for Clinical Laboratory Standards Guidelines for Disk Diffusion Susceptibility Testing in New York State Laboratories. J. Clin. Microbiol. 2000;38:3341–3348. doi: 10.1128/JCM.38.9.3341-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaala L.A., Khalifa S.I., Mesbah M.K., van Soest R.W.M., Youssef D.T.A. Subereaphenol A, a new cytotoxic and antimicrobial dibrominated phenol from the Red Sea sponge Suberea mollis. Nat. Prod. Commun. 2008;3:219–222. doi: 10.1177/1934578X0800300222. [DOI] [Google Scholar]
- 51.CLSI, Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk Susceptibility Tests. 9th ed. West Valley Road; Wayne, PA, USA: 2007. CLSI Documents M07-A9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.