Abstract
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related morbidity and mortality globally. Despite the remarkable improvements in comprehensive HCC treatment, the underlying mechanistic details of HCC remain elusive. We screened HCC patients for differentially expressed genes (DEGs) using the Gene Expression Omnibus (GSE113850) and The Cancer Genome Atlas (TCGA) datasets. LINC01554 expression in 40 paired samples was determined by quantitative reverse transcription polymerase chain reaction (RT-qPCR), and its clinical significance was assessed. LINC01554 was found to have a gain-of-function role in HCC in vitro. Additionally, the bioinformatics analysis of the genes co-expressed with LINC01554 was performed using the Co-LncRNA website, and potential molecular mechanisms were investigated using the Gene Ontology and Kyoto Encyclopaedia of Genes and Genomes resources and validated by in vitro experiments. A total of 229 DEGs were identified from the GSE113850 dataset. Among the identified DEGs, three long non-coding RNAs (lncRNAs) (DIO3OS, LINC01554, and LINC01093) with |logFC| ≥2 and P<0.05 were screened. A total of 148 lncRNAs with |logFC| ≥1 and P<0.05 were identified from TCGA dataset. Low LINC01554 expression levels were significantly correlated with overall survival, pathological stage, hepatitis B infection, tumour size, portal vein tumour thrombus, and TNM stage. Using gain-of-function assays, we further showed that LINC01554 inhibited the proliferation, migration, and invasion of the HCCLM9 and SK-Hep1 cells and promoted G0/G1 arrest, but it did not significantly affect apoptosis. Western blotting revealed that LINC01554 overexpression resulted in increased ZO-1 and E-cadherin expression levels, but decreased N-cadherin and vimentin expression levels. Moreover, LINC01554 overexpression inhibited Akt, p-Akt, β-catenin, and p-Gsk3β expression. Our results showed that LINC01554 repressed HCC cell invasiveness and epithelial-to-mesenchymal transition partly by inhibiting Wnt and PI3K-Akt signalling in vitro. Taken together, our findings provide new insights into the molecular mechanisms underlying HCC tumourigenesis and implicate LINC01554 as a potential target for HCC therapy.
Keywords: hepatocellular carcinoma, LINC01554, bioinformatics analysis, Wnt signalling, PI3K-Akt signalling
Introduction
Primary liver cancer is responsible for a large proportion of cancer-related deaths globally. More than half of all hepatocellular carcinoma (HCC) patients are in China, and Chinese HCC mortality rates are the highest in the world (1,2). Despite the advances in clinical and experimental research on HCC, the survival rate of HCC patients remains low (3). Emerging studies have reported that carcinogenesis is a complex process that involves multiple molecules (4). To this end, HCC carcinogenesis must be more extensively elucidated, and it is crucial to develop new approaches to improve HCC treatment and increase the long-term survival rate of HCC patients.
mRNAs that encode proteins account for <2% of the human transcriptome (5). Increasing evidence indicates that even though many transcripts are not translated into proteins, they function at the transcriptional level. Long non-coding RNAs (lncRNAs) are a new class of transcripts that consist of >200 nucleotides but are not translated into proteins (6). Some lncRNAs have been reported to be abnormally expressed in cancer, while others have been found to be related to cancer occurrence and development (7). LINC01554 has been reported as a significant lncRNA involved in the pathogenesis of non-alcoholic fatty liver disease and oesophageal cancer (8,9). In the present study, we found that LINC01554 was significantly downregulated in HCC tissues, and its expression levels were closely associated with the prognosis of HCC patients. Moreover, LINC01554 inhibited HCC proliferation and metastasis in vitro. Further experiments were performed to identify the mechanism of LINC01554 in HCC and determine its potential suitability as a diagnostic biomarker for early HCC detection and a potential target for HCC therapy.
Materials and methods
Gene expression profiles
We downloaded gene expression profiling data from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/gds/). The GSE113850 dataset, which consists of 39 tissue samples including 32 HCC and 7 normal liver tissues, was used (10). We also downloaded gene expression profiling data from The Cancer Genome Atlas database (TCGA, http://portal.gdc.cancer.gov/). This dataset consisted of 424 tissues, including 371 primary HCC, 3 recurrent HCC, and 50 normal liver tissues.
Screening of HCC patients for differentially expressed genes (DEGs)
We used the GEO2R statistical analysis tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) and Student's t-test to identify DEGs in the GSE113850 dataset. The adjusted P-value was set at <0.05, and the logFC value was set at >2 (upregulated genes) or <-2 (downregulated genes). Data from the TCGA database were analysed using the R packages, DESeq and edgeR (https://www.bioconductor.org/), and Student's t-test to identify DEGs. Meanwhile, heat maps and volcano plots were created by a hierarchical clustering method.
Comparison of the expression levels of three lncRNAs and the clinicopathological characteristics of the HCC patients
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) is an online tool used for analysing RNA sequencing (RNA-Seq) data from TCGA and GTEx databases. It includes RNA sequencing expression data of 9,736 tumors and 8,587 normal samples and related clinical datasets of TCGA and the GTEx projects. It provides key interactive and customizable functions including differential expression analysis, profiling plotting, correlation analysis, patient survival analysis, similar gene detection and dimensionality reduction analysis (11). Using GEPIA, we compared the gene expression levels of HCC and normal liver tissues. We used log-rank test to analyse the correlation between the levels of gene expression and the duration of patient survival. Moreover, we analysed the gene expression levels according to the tumour stage of the HCC patients. The adjusted P-value was set at <0.05, and the datasets were set at LIHC (liver hepatocellular carcinoma).
Clinical specimens and cell culture
Forty patients (age range, 20–76 years; mean age, 54.78±10.64 years; males, 33; females, 7), who attended the Zhongnan Hospital of Wuhan University from April 2018 to April 2019, were enrolled in this study. All patients were diagnosed with HCC by histological examination, and they provided written informed consent for their tissues to be used for medical research. Ethical approval for this study was obtained from the Zhongnan Hospital of Wuhan University. Liver cancer cell lines (SK-Hep1, HCCLM9 and HepG2) were obtained from the Cell Bank of the Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). Cells were cultured in Dulbecco's modified Eagles medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.), supplemented with 10% foetal bovine serum (FBS) and 1% penicillin-streptomycin, at 37°C in a 5% CO2 humidified atmosphere.
Lentiviral construction and cell transfections
The sequence of lncRNA LINC01554 was sub-cloned into the lentiviral expression vector Lv105 (GeneCopoeia, Guangzhou, China). 293T cells were plated into 6-well plates. After 293T cell confluence reached 80%, cells were co-transfected with LINC01554-Lv105 (or control vector EX-EGFP-Lv105) and Lenti-Pac CMV Expression Packaging vectors (gag-pol-RRE vector, REV vector and VSV-G vector) (GeneCopoeia). The mass of each vector in each well is 1.0 µg lentiviral plasmid, 1.2 µg gag-pol-RRE vector, 1.0 µg REV vector, and 1.0 µg VSV-G vector. The transfection was performed by using LipoFectMax™ Transfection Reagent (ABP Biosciences, USA). After incubation for 48 h, the lentiviral particles were harvested from the cell medium supernatant by centrifuging at 500 × g for 10 min. The SK-Hep1 and HCCLM9 cells were then transduced with Lv-LINC01554 lentivirus controlled with the Lv-NC lentivirus.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the clinical specimens and cells using TRIzol reagent (Yeasen Biotech). cDNA was generated using a Hieff First Strand cDNA Synthesis Super Mix for RT-qPCR kit (Yeasen Biotech). RT-qPCR analysis was performed using the Hieff qPCR SYBR Green Master Mix kit (Yeasen Biotech) and a StepOnePlus instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). Relative lncRNA expression was analysed using the 2−ΔΔCq method (12), with normalisation to β-actin expression. Primer sequences used for RT-qPCR were as follows: LINC01554 forward, 5′-TGTGGCAAACGCAAACGA-3′ and reverse, 5′-GCCCAGAGTAAAGGGAAATGTA-3′; β-actin forward, 5′-TGGCACCCAGCACAATGAA-3′ and reverse, 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
Western blot analysis
Total protein was isolated from the HCCLM9 and SK-Hep1 cells using RIPA lysis buffer (Wuhan Servicebio Biotechnology). Protein concentrations were determined using the bicinchoninic acid assay (Wuhan Servicebio Biotechnology). The mass of protein loaded per lane was 25 µg. Proteins were separated using 10% SDS-PAGE gels and electro-transferred to PVDF membranes. The membranes were then blocked by TBST (with 5% skim milk powder) for 2 h at room temperature and incubated overnight, respectively, with primary antibodies at 4°C. After washing with TBST three times, the membranes were incubated, respectively, with goat anti-rabbit secondary antibody at 37°C for 2 h. The dilutions and suppliers of the primary antibodies and secondary antibodies are documented in Table I. Protein blots were detected using a chemiluminescence kit (NCM Biotech) and Tanon 4500 Immunodetection System (Tanon). The protein bands were visualised with chemiluminescence ECL reagent and recorded with X-ray film. The signal intensity was analysed by ImageJ v1.42q software (National Institutes of Health). GAPDH content was used to standardise sample loading.
Table I.
Diluted concentrations and supplier information for the antibodies used for western blotting.
| Antibodies | Catalog no. | Company/Supplier | Dilution |
|---|---|---|---|
| ZO-1 | 21773-1-AP | Wuhan Proteintech Group | 1:1,000 |
| E-cadherin | AF0131 | Affinity Biosciences | 1:1,000 |
| N-cadherin | GB11135 | Wuhan Servicebio Biotechnology | 1:1,000 |
| Vimentin | GB11192 | Wuhan Servicebio Biotechnology | 1:1,000 |
| AKT | 10176-2-AP | Wuhan Proteintech Group | 1:1,000 |
| p-AKT | AF0908 | Affinity Biosciences | 1:1,000 |
| GSK3β | GB11099 | Wuhan Servicebio Biotechnology | 1:1,000 |
| p-GSK3β | 5558 | Cell Signaling Technology | 1:1,000 |
| β-catenin | GB11015 | Wuhan Servicebio Biotechnology | 1:1,000 |
| GAPDH | GB12002 | Wuhan Servicebio Biotechnology | 1:1,000 |
| Goat anti-rabbit IgG | GB23303 | Wuhan Servicebio Biotechnology | 1:3,000 |
| Goat anti-mouse IgG | GB23301 | Wuhan Servicebio Biotechnology | 1:3,000 |
Flow cytometric analysis
After transfection, the cells were collected and washed twice with phosphate-buffered saline (PBS). For cell cycle analysis, the cells were stained with a cell cycle staining kit (KeyGEN) for 15 min. Apoptosis was detected using an Annexin V-FITC and Propidium Iodide Apoptosis Detection Kit (KeyGEN). Finally, the proportion of cells in different phases of the cell cycle and the apoptosis rate were measured using a FACSCalibur flow cytometer (BD Biosciences).
Fluorescence in situ hybridisation (FISH)
Briefly, the SK-Hep1 and HCCLM9 cells were first fixed with 4% formaldehyde. They were then placed on slides, and hybridisation reactions were performed using LINC01554-, U6-, and 18S rRNA-specific probes. Cells were then counterstained with DAPI. Fluorescence images were captured using a confocal microscope. The FAM-labelled LINC01554, TAMRA-labelled 18S rRNA, and TAMRA-labelled U6 probe sequences were 5′-ACTTCCCGACTGCTCATCAACCGACCTCCCTGGGGCCGAGAGGCA-3′, 5′-CGCTGAGCCAGTCAGTGTAGCGCGCGTGCAGCCCCGGACATCTAAGGGCATCACAGACCTGTTATTGCTCAATCTCGGGTGGCTGAACGCCACTTGTCCCTCTAAGAAGTTGGGGGACGCCGACCGCTCGGGGGTCGCGTAACTAGTTAGCATGCCAGAGTCTCGTTCGTTATCGGAATTAACCAGACAAATCGCTCCACCAACTAAGAACGGCCAT-3′ and 5′-AAAAATATGGAACGCTTCACGAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAATCTTCTCTGTATCGTTCCAATTTTAGTATATGTGCTGCCGAAGCGAGCAC-3′, respectively. The probes were purchased from AxlBioCo., Ltd.
Cell Counting Kit-8 (CCK-8) assay
Cells in the logarithmic growth phase were digested with trypsin and seeded (2×103) in a 96-well plate. CCK-8 reagent (10 µl) (Dojindo Molecular Technologies) was added to each well at 0, 24, 48, and 72 h. After incubating the cells with the CCK-8 reagent for 4 h, we measured the optical density (OD) at 450 nm.
Scratch test assay
Cells were seeded in 6-well plates at 50% density of the cells. When they were approximately 90% confluent, the tip of a sterile 1,000-µl pipette was used to scratch a line wound. Cells that migrated to the wound region were observed under a microscope at 0 and 24 h.
Transwell assays
A 24-well Transwell chamber with Matrigel was used to evaluate the cell invasion rates. Cells (5×104) from each group were added to the upper Transwell chamber, and DMEM (600 µl), containing 10% FBS, was added to the lower Transwell chamber. Cells were then incubated at 37°C in 5% CO2 for 48 h. Finally, after fixation with 4% paraformaldehyde for 20 min and crystal violet staining for 15 min at room temperature, the number of cells passing through the membrane was observed under a light microscope. Cells were counted in five random fields, and the average number of cells per field was calculated.
Gene set enrichment analysis (GSEA)
Thirty-two HCC samples and 7 normal liver samples from GSE113850 were divided into two groups (HCC vs. NC). In order to investigate the further function of LINC01554, GSEA (http://software.broadinstitute.org/gsea/index.jsp) was carried out between the two groups. Annotated gene were selected as the reference gene sets. False discovery rate (FDR) <0.25, and gene size ≥100 were regarded as the cut-off criteria.
Bioinformatics analysis of the co-expressed genes (CEGs)
Co-LncRNA (http://bio-bigdata.hrbmu.edu.cn/Co-LncRNA/) is a web-based computational tool that predicts the CEGs of single or multiple lncRNAs. We analysed the same RNA-Seq datasets to identify the CEGs. The regression/correlation method was set as Linear Regression. The corresponding coefficient threshold values were set at >2, and the adjusted P-value was set at <0.01.
Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses of CEGs
The GO resource (http://www.geneontology.org/) describes gene functions and groups them into three categories: Molecular function, cellular component, and biological process. KEGG (http://www.genome.jp/kegg/) contains a set of genomes and biological pathways that provide information on genome functions and the relationships among genes. We visualised all known GO functional annotations and KEGG pathways of the CEGs using DAVID (http://david.ncifcrf.gov). GO and KEGG enrichment plots were also created.
Statistical analysis
Statistical analysis was performed using the SPSS 22.0 software (IBM Corp.). All experiments were performed in triplicate, and all data are presented as means ± standard error of the mean. All data are presented as a comparison between the groups and analysed by Student's t-test or analysis of variance (ANOVA). Receiver operating characteristic (ROC) curves were used to determine the correlation between the true- and false-positive rates, and this was important for assessing the diagnostic value of the genes. Results were regarded as statistically significant at P<0.05.
Results
Identification of the DEGs
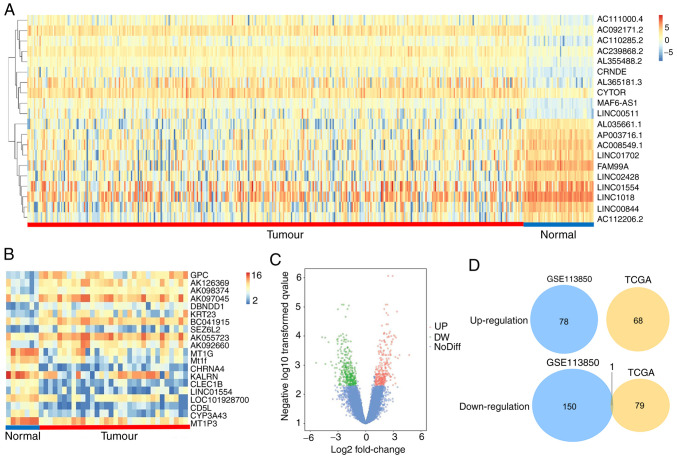
After analysing the GSE113850 dataset, we identified 229 DEGs, of which 151 were downregulated and 78 were upregulated, as shown in a volcano plot (Fig. 1C). Hierarchical clustering analysis of the top 10 DEGs with a fold change >2 is presented as a heat map (Fig. 1B). Three lncRNA genes (DIO3OS, LINC01554, and LINC01093) were among these DEGs. After analysing TCGA data, we identified 148 differentially expressed lncRNAs, and hierarchical clustering analysis of the top 10 lncRNAs with a fold change >1 is presented as a heat map (Fig. 1A). The Venn plot shows that LINC01554 was the only differentially expressed lncRNA in both the GSE113850 and TCGA datasets (Fig. 1D).
Figure 1.
(A) Hierarchical clustering analysis of the top 10 differentially expressed lncRNAs (|logFC| >1, P<0.05) from TCGA datasets. (B) Hierarchical clustering analysis of the top 10 differentially expressed genes (DEGs, |logFC| >2, P<0.05) from the GSE113850 dataset. (C) After analysing the GSE113850 dataset, we identified 229 DEGs, of which 151 were downregulated and 229 were upregulated, as indicated in a volcano plot (|logFC| >2; P<0.05). Red indicates upregulated DEGs; green indicates downregulated DEGs; blue indicates no difference. (D) Venn plot shows that LINC01554 is a common differentially expressed lncRNA in the GSE113850 and TCGA datasets. TCGA, Cancer Genome Atlas.
Relationship between the expression levels of three lncRNAs and the prognosis of HCC patients
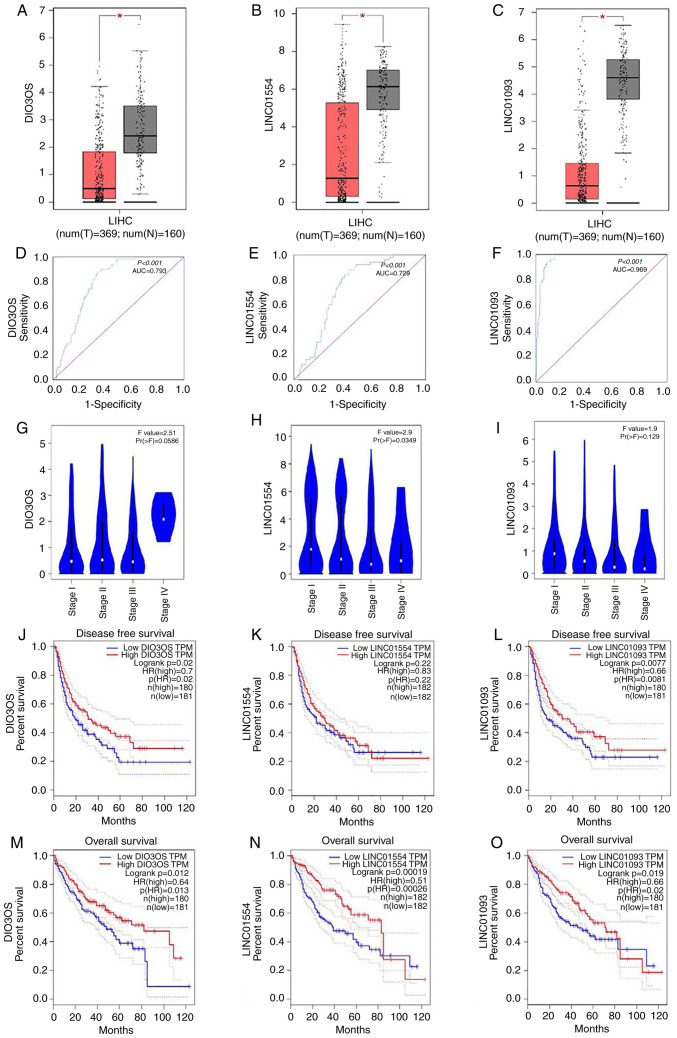
Using GEPIA, we compared the expression levels of the three lncRNAs, DIO3OS, LINC01554 and LINC01093, in the HCC and normal liver tissues. The results showed that the lncRNA expression levels were lower in HCC tissues than these in the normal liver tissues (Fig. 2A-C). The area under the curve (AUC) values for DIO3OS, LINC01554 and LINC01093 were 0.793, 0.729 and 0.969, respectively (Fig. 2D-F), indicating a moderate diagnostic value of their expression levels in HCC. These results were based on the original data from TCGA. We also analysed the lncRNA expression levels according to the tumour stage of HCC patients. Among the three lncRNAs, LINC01554 showed expression levels that significantly differed according to the HCC stage (Fig. 2G-I). We further explored the effects of these lncRNAs on the survival of HCC patients. These results showed that decreased DIO3OS LINC01554 and LINC01093 were significantly correlated with disease-free survival (DFS) of all HCC patients (Fig. 2J-L), and decreased levels of all three lncRNAs were significantly correlated with overall survival (OS) of all HCC patients (Fig. 2M-O). Meanwhile, we also explored the correlation between LINC01554 expression and the clinicopathological features in 40 HCC patients to evaluate the clinical role of LINC01554 in HCC (Table II). Lower LINC01554 expression levels were strongly linked to certain clinical characteristics, including HBV infection (P=0.0044), pathological stage (P=0.0079), tumour size (P=0.0252), portal vein tumour thrombus (P=0.0471), and TNM stage (P=0.0022).
Figure 2.
LINC01554 expression levels in HCC tissues and their association with clinicopathological characteristics of HCC patients based on the GEPIA and TCGA databases. DIO3OS (A) LINC01554 (B) and LINC01093 (C) lncRNA expression levels in HCC (T, tumour; n=369) and normal liver (N, non-tumour; n=160) tissue samples. ROC curves of DIO3OS (D) LINC01554 (E) and LINC01093 (F) lncRNAs in HCC patients based on the TCGA database. Association of DIO3OS (G) LINC01554 (H) and LINC01093 (I) lncRNAs with the pathological stage of HCC. Association of DIO3OS (J) LINC01554 (K) and LINC01093 (L) lncRNAs with the disease-free survival of HCC patients. Association of DIO3OS (M) LINC01554 (N) and LINC01093 (O) lncRNAs with the overall survival of HCC patients. HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; lncRNA, long non-coding RNA; ROC, receiver operating characteristic. *P<0.05, comparison with the Lv-NC group.
Table II.
Associations of LINC1554 expression with clinicopathological parameters in HCC.
| LINC01554 expression | ||||
|---|---|---|---|---|
| Variable | No. of cases | Low (n=20) | High (n=20) | P-value |
| Sex | 0.4075 | |||
| Male | 33 | 15 | 18 | |
| Female | 7 | 5 | 2 | |
| Age (years) | 0.2351 | |||
| ≤50 | 8 | 6 | 2 | |
| >50 | 32 | 14 | 18 | |
| HBV infection | 0.0044a | |||
| Negative | 19 | 5 | 14 | |
| Positive | 21 | 15 | 6 | |
| Serum AFP (ng/ml) | 0.1274 | |||
| ≤8.78 | 9 | 2 | 7 | |
| >8.78 | 31 | 18 | 13 | |
| Pathological stage | 0.0079a | |||
| I+II | 25 | 8 | 17 | |
| III+IV | 15 | 12 | 3 | |
| Cirrhosis | 0.7233 | |||
| No | 11 | 6 | 5 | |
| Yes | 29 | 14 | 15 | |
| Tumour number | 0.1274 | |||
| Singular | 31 | 18 | 13 | |
| Multifocal | 9 | 2 | 7 | |
| Tumour size (cm) | 0.0252a | |||
| ≤5 | 23 | 8 | 15 | |
| >5 | 17 | 12 | 5 | |
| PVTT | 0.0471a | |||
| No | 35 | 15 | 20 | |
| Yes | 5 | 5 | 0 | |
| TNM (AJCC) | 0.0022a | |||
| I+II | 26 | 8 | 18 | |
| III+IV | 14 | 12 | 2 | |
HCC, hepatocellular carcinoma; HBV, hepatitis B virus; AFT, α-fetoprotein; PVTT, portal vein tumour thrombus; AJCC, American Joint Committee on Cancer.
P<0.05, significant difference (in bold print).
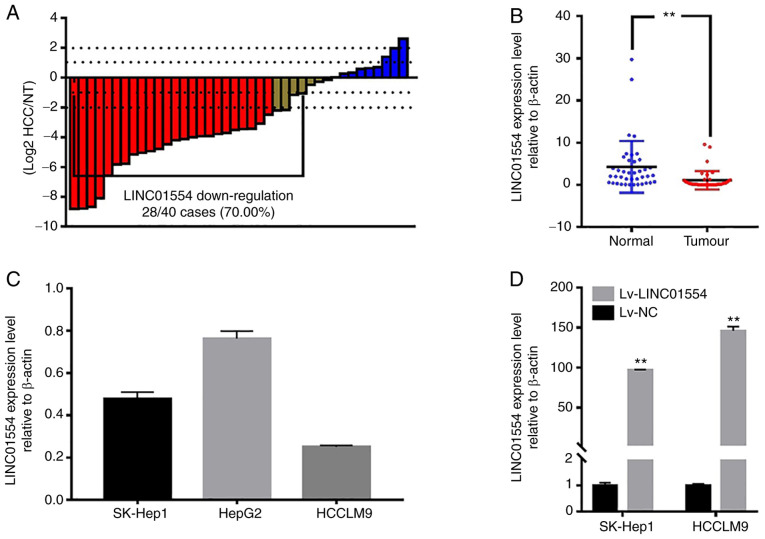
LINC01554 expression is downregulated in human HCC samples. We analysed LINC01554 expression in 40 paired HCC and normal tumour-adjacent tissue samples by RT-qPCR. The results showed that LINC01554 expression was significantly lower in the HCC samples (P<0.01, Fig. 3B), and downregulated LINC01554 expression was observed in 70.0% (28/40) of the HCC samples (Fig. 3A). We also investigated LINC01554 expression in the liver cancer cell lines (SK-Hep1, HepG2, and HCCLM9). RT-qPCR results showed that LINC01554 expression was relatively lower in the SK-Hep1 and HCCLM9 cells than in the HepG2 cells (Fig. 3C). Therefore, SK-Hep1 and HCCLM9 cells were used for subsequent experiments. To study LINC01554 functions in HCC cell lines, a recombinant lentivirus that carried the LINC01554 DNA sequence (Lv-LINC01554) and a recombinant lentivirus without the LINC01554 DNA sequence (Lv-NC), as a control, were transfected into HCC cells. Transfection efficiencies were confirmed by RT-qPCR (P<0.01; Fig. 3D).
Figure 3.
LINC01554 expression is downregulated in human HCC samples. (A) Downregulation of LINC01554 (>2-fold) is detected in 70.0% (28/40) of primary HCC tissues. (B) The relative expression of LINC01554 in HCC and adjacent non-tumour liver tissues (n=40) was analysed by RT-qPCR. (C) The relative expression of LINC01554 in liver cancer cell lines (HepG2, SK-Hep1 and HCCLM9) is analysed. (D) LINC01554 expression in the SK-Hep1 and HCCLM9 cells, after transfection with Lv-LINC01554, is measured by RT-qPCR. All results are from at least three independent experiments. β-actin was used as a control. **P<0.01, comparison with normal tissues or Lv-NC group. HCC, hepatocellular carcinoma.
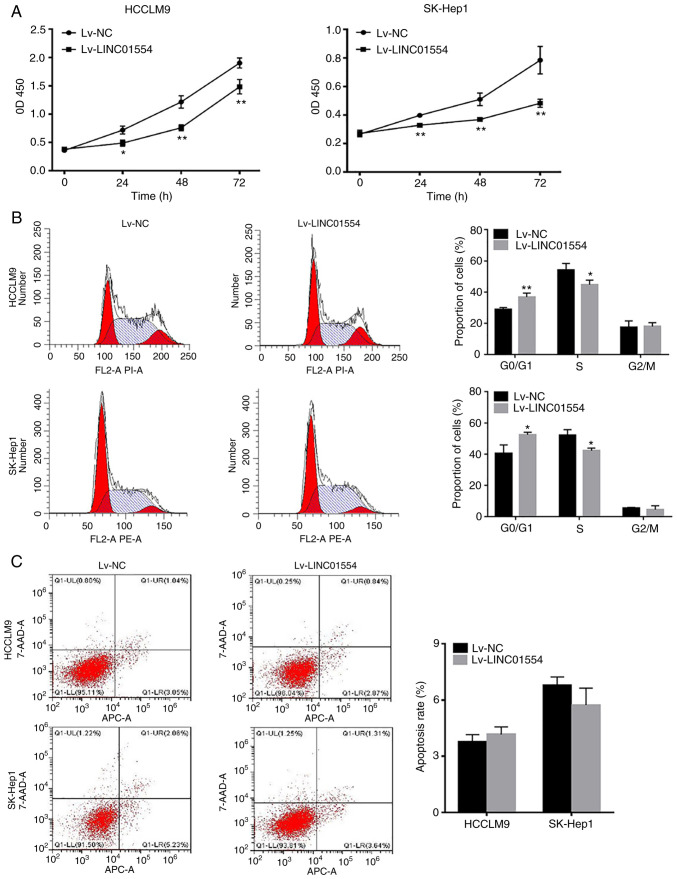
LINC01554 overexpression inhibits SK-Hep1 and HCCLM9 cell proliferation and promotes G0/G1 arrest, but it does not significantly affect apoptosis
According to the CCK-8 assay results, the OD450 values of the SK-Hep1 and HCCLM9 cell lines in the Lv-LINC01554 group at 0, 24, 48 and 72 h were significantly lower than those in the Lv-NC group (P<0.01; Fig. 4A). We examined the distribution of cells at different stages of the cell cycle using flow cytometry. The results revealed that LINC01554 overexpression promoted G0/G1 arrest (P<0.05; Fig. 4B), but it did not significantly affect apoptosis (Fig. 4C).
Figure 4.
LINC01554 overexpression inhibits SK-Hep1 and HCCLM9 cell proliferation and promotes G0/G1 arrest. (A) CCK-8 assay of LINC01554-overexpressing HCC SK-Hep1 and HCCLM9 cells. (B) Cell cycle analysis of LINC01554-overexpressing SK-Hep1 and HCCLM9 cells. (C) After transfection with an LINC01554-overexpression vector, the SK-Hep1 and HCCLM9 cells are double-stained and analysed by flow cytometry. Cells positive for Annexin V staining are counted as apoptotic cells. All results are from at least three independent experiments. *P<0.05, **P<0.01, comparison with the Lv-NC group. HCC, hepatocellular carcinoma.
LINC01554 inhibits epithelial-mesenchymal transition (EMT) and tumour invasion in the SK-Hep1 and HCCLM9 cells
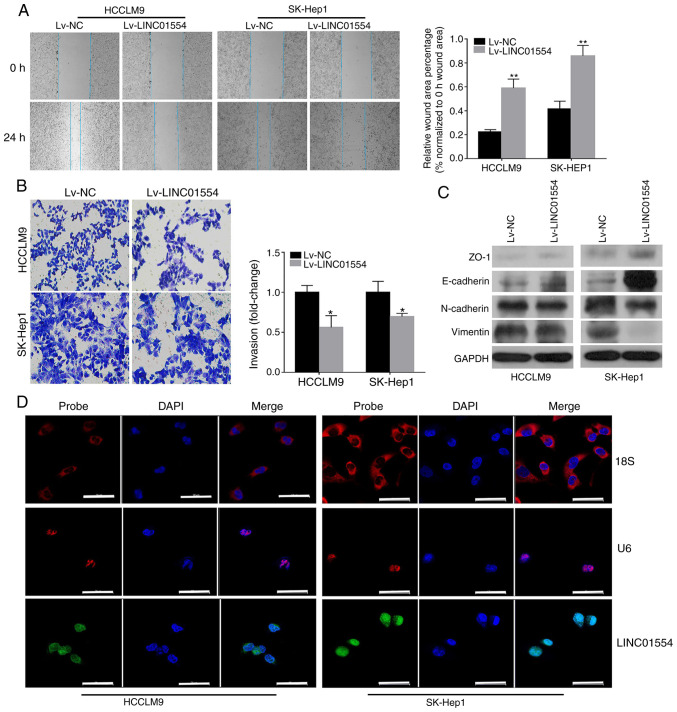
Wound-healing and Transwell invasion assays were used to investigate the role of LINC01554 in HCC cell migration and invasion. Scratch assays showed that the migration rate of the Lv-LINC01554 transfection group was significantly lower than that of the Lv-NC transfection group (P<0.01; Fig. 5A). Transwell experimental results showed that the number of migrated cells in the Lv-LINC01554 transfection group was significantly lower than that in the Lv-NC transfection group (P<0.01; Fig. 5B). Next, we assessed EMT-related gene expression in the HCCLM9 and SK-Hep1 cells. The results of the western blot analysis showed that LINC01554 overexpression led to increased ZO-1 and E-cadherin expression, but decreased N-cadherin and vimentin expression (Figs. 5C and S1). Moreover, FISH assays showed that LINC01554 was mainly located in the nuclei of the SK-Hep1 and HCCLM9 cells (Fig. 5D).
Figure 5.
LINC01554 overexpression suppresses migration, invasion, and EMT-related gene expression in HCC cells. (A) Wound-healing assay in LINC01554-overexpressing SK-Hep1 and HCCLM9 cells. (B) Invasion ability of LINC01554-overexpressing SK-Hep1 and HCCLM9 cells. (C) Expression levels of EMT markers (ZO-1, E-cadherin, N-cadherin and vimentin) in LINC01554-overexpressing HCCLM9 and SK-Hep1 cells are analysed by western blotting. (D) FISH analysis of LINC01554 in the SK-Hep1 and HCCLM9 cells. The U6 and 18S rRNA probes are labelled red, the LINC01554 RNA probe is labelled green, and nuclear DNA with DAPI staining is labelled blue. Scale bar, 50 µm. All results are from at least three independent experiments. *P<0.05, **P<0.01, comparison with the Lv-NC group. HCC, hepatocellular carcinoma; EMT, epithelial-mesenchymal transition.
Gene set enriched analysis
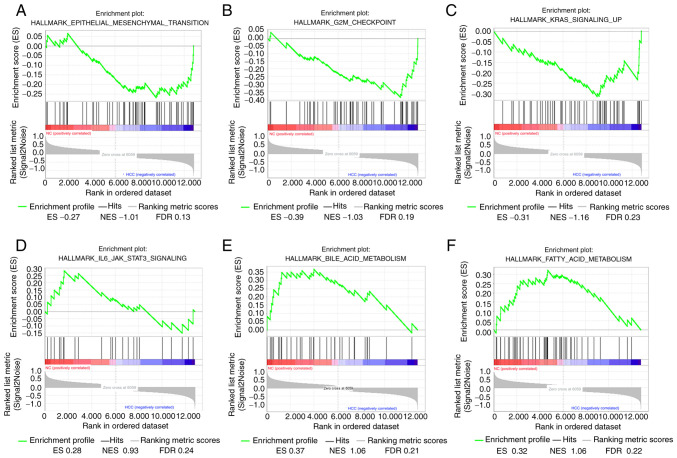
GSEA was used to map into 6 functional gene sets. A total of 6 functional gene sets were enriched, which were mainly ‘Epithelial mesenchymal transition’, ‘G2M checkpoint’, ‘Kras signaling up’, ‘IL6/JAK/STAT3 signaling’, ‘Bile acid metabolism’ and ‘Fatty acid metabolism’ (Fig. 6).
Figure 6.
Gene set enriched analysis (GSEA). (A-C) LINC01554 is positively correlated with Epithelial mesenchymal transition, G2M checkpoint and Kras signaling up. (D-F) LINC01554 is negatively correlated with IL6/JAK/STAT3 signaling, Bile acid metabolism and Fatty acid metabolism.
GO and KEGG pathway enrichment analysis
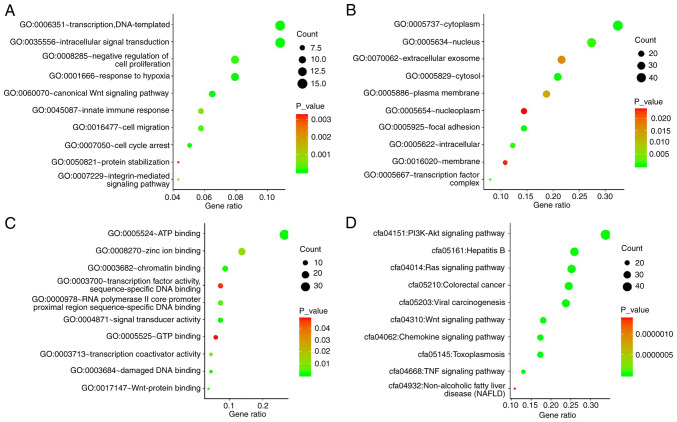
We analysed the GO functions and KEGG pathways of the genes co-expressed with LINC01554 using the DAVID online database. Only the top 10 pathways for each category are listed (Fig. 7A-D). GO analysis showed that the CEGs were mainly enriched in ‘intracellular signal transduction’, ‘transcription, DNA-templated’, ‘negative regulation of transcription by the RNA polymerase II promoter’, ‘negative regulation of cell proliferation’, ‘response to hypoxia’, ‘canonical Wnt signalling pathway’, ‘cell migration’, and ‘cell cycle arrest’. KEGG pathway analysis showed that these genes were related to PI3K-Akt, Ras, Wnt, chemokine, and tumour necrosis factor (TNF) signalling pathways; viral carcinogenesis; and toxoplasmosis, hepatitis B, and non-alcoholic fatty liver disease pathways.
Figure 7.
Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses of genes co-expressed with LINC01554. Three GO categories: Biological process (A) cellular component (B) and molecular function (C) (D) KEGG pathway analysis of genes co-expressed with LINC01554.
LINC01554 overexpression affected the expression of proteins related to Wnt and PI3K-Akt signalling in HCC cells
To examine whether LINC01554 exerts its function through Wnt and PI3K-Akt signalling in HCC cells, the expression levels of proteins related to these pathways (Akt, p-Akt, Gsk3β, p-Gsk3β and β-catenin) were measured in the LINC01554-overexpressing HCCLM9 and SK-Hep1 cells (Figs. 8A and B and S2). The results showed that LINC01554 overexpression inhibited Akt, p-Akt, β-catenin, and p-Gsk3β expression.
Figure 8.
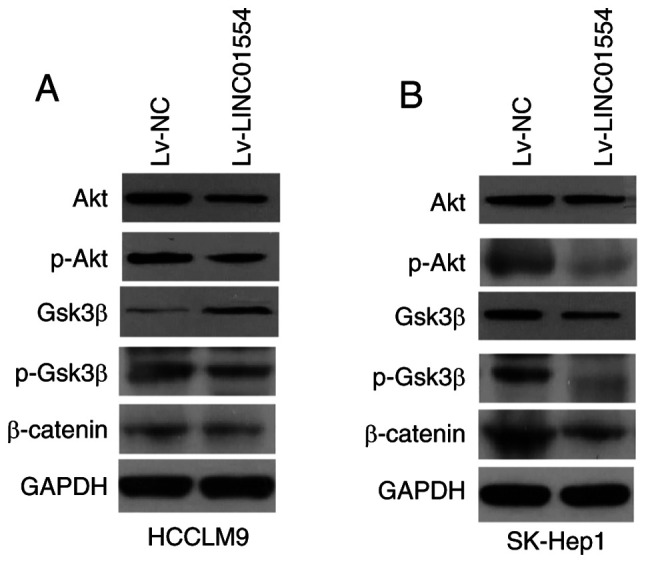
LINC01554 overexpression affects the expression of proteins related to Wnt and PI3K-Akt signalling in HCC cells. Expression levels of proteins related to Wnt and PI3K-Akt signalling (Akt, p-Akt, Gsk3β, p-Gsk3β, and β-catenin) in HCC cells are analysed by western blotting in the LINC01554-overexpressing HCCLM9 (A) and SK-Hep1 (B) cells. HCC, hepatocellular carcinoma; PI3K, phosphoinositide 3-kinase; Gsk3β, glycogen synthase kinase 3β; Akt, protein kinase B or (PKB); p-, phosphorylated.
Discussion
As key regulators of tumour pathogenesis, long non-coding RNAs (lncRNAs) are involved in the multilevel regulation of gene expression in cancer (13). However, the lncRNAs with aberrant expression in HCC remain unknown. In the present study, we screened hepatocellular carcinoma (HCC) patients for differentially expressed genes (DEGs) from the Gene Expression Omnibus (GEO) dataset, GSE113850, and found that DIO3OS, LINC01554, and LINC01093 were in the top 30 DEGs. Based on a literature review, we chose DIO3OS, LINC01554, and LINC01093 for further investigation as they showed relevant biological functions. For example, previous studies have shown that DIO3OS is highly expressed in pancreatic cancer and DIO3OS was found to regulate pancreatic cancer cell proliferation and metastasis by affecting the miR-122/ALDOA axis (14). LINC01093 is a liver-specific lncRNA that was found to be significantly downregulated in HCC samples and could inhibit HCC cell growth and metastasis (15). LINC01554 downregulation was found to promote high aerobic glycolysis in cancer cells and cell growth by regulating the PKM2 and Akt/mTOR signalling pathways (16). We used GEPIA datasets to verify the differences in DIO3OS, LINC01554, and LINC01093 expression levels between the HCC and normal liver tissues. We further explored the effects of DIO3OS, LINC01554, and LINC01093 expression levels on overall survival (OS) and disease-free survival (DFS). These results showed that decreased DIO3OS, LINC01554, and LINC01093 levels were significantly correlated with OS and decreased DIO3OS levels were significantly associated with DFS. The AUC values of DIO3OS, LINC01554, and LINC01093 indicated that their expression levels were of moderate diagnostic value in HCC. Therefore, we suggested that these three lncRNAs could be used as tumour biomarkers of HCC. Meanwhile, we also screened the patients for differentially expressed lncRNAs using TCGA datasets. Venn plots showed that LINC01554 was a common differentially expressed lncRNA in the GSE113850 and TCGA datasets. Moreover, we found that lower LINC01554 expression levels were positively correlated with pathological stage, hepatitis B infection, tumour size, portal vein tumour thrombus, and TNM stage in 40 HCC patients. Therefore, we chose LINC01554 as the focus of our subsequent experiments.
A search of the NCBI databases (https://www.ncbi.nlm.nih.gov/) revealed that LINC01554 is a 1,931-bp lncRNA, with three exons, encoded by a gene on chromosome 5q15. Using RT-qPCR, we confirmed that LINC01554 expression levels were significantly lower in the HCC tissues than these levels in the paired normal tumour-adjacent tissues obtained from 40 HCC patients. This finding was consistently observed in the GSE113850 and TCGA datasets. A gain-of-function assay further showed that LINC01554 inhibited HCCLM9 and SK-Hep1 cell proliferation, migration, and invasion and promoted G0/G1 arrest, but it did not significantly affect apoptosis. Cancer cell proliferation and metastasis are known to play significant roles in cancer development, and the adhesion of cancer cells in the microcirculation is related to cancer metastasis through the vessels (17,18). Epithelial-mesenchymal transition (EMT) is an important step in liver cancer metastasis, and extracellular matrix (ECM)-receptor interactions lead to the direct or indirect control of cellular adhesion and migration (19,20). Moreover, EMT is an early event that is critical for cancer invasion and metastasis (21). In our study, LINC01554 affected the expression levels of EMT markers in the HCCLM9 and SK-Hep1 cells. LINC01554 overexpression promoted the expression of epithelial markers (E-cadherin and ZO-1) but inhibited the expression of mesenchymal markers (N-cadherin and vimentin) in the HCCLM9 and SK-Hep1 cells. These findings strongly implicate the downregulation of LINC01554 in the metastatic growth of HCC.
The mechanism of action of lncRNAs is mainly associated with their subcellular localisation. Nuclear lncRNAs mostly show effects at the transcriptional levels, while cytoplasmic lncRNAs mostly function as endogenous sponges for miRNAs or show effects at post-transcriptional levels (22,23). Furthermore, FISH assays showed that LINC01554 is mainly located in the nuclei of HCC cells, indicating its function in transcriptional regulation. To gain a better understanding of the potential molecular mechanism of LINC01554 in HCC, we performed LINC01554 co-expression analysis using the Co-LncRNA website. lncRNA-mRNA co-expression analyses are commonly used to identify potential target genes of lncRNAs and research the molecular mechanisms of lncRNAs further (24). We identified 139 genes as potential LINC01554 targets using this analysis. Meanwhile, we performed GO and KEGG pathway analyses of these 139 genes using the DAVID online database. The analyses identified multiple signalling pathways that involved these genes. The pathways included PI3K-Akt, Ras, Wnt, chemokine, and TNF signalling pathways; viral carcinogenesis, and hepatitis B disease pathway. Importantly, the abnormal activation of the Wnt and PI3K/Akt pathways has been widely reported in tumour development. The Wnt signalling pathway is commonly affected in HCC of different aetiologies, which most likely involve common pathogenetic mechanisms. Furthermore, the Wnt signalling pathway is reported to have a close association with EMT (25,26). Emerging evidence indicates that some lncRNAs participate in tumour invasion and metastasis by regulating the Wnt/β-catenin signalling pathway and inhibiting E-cadherin expression. The mitogen-activated protein kinase (MAPK) and RAS/RAF/MEK/ERK pathways, which regulate cell growth and involve a number of receptors such as epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene homologue (KRAS), also play key roles in HCC development (27–30). OTUD6B was found to inhibit clear cell renal cell carcinoma by regulating the Wnt/β-catenin pathway and suppressing EMT-related protein expression (31). Moreover, it has been reported that the PI3K/Akt signalling pathway mediates EMT to increase tumour aggressiveness (32,33). PI3K/Akt promotes non-small-cell lung carcinoma metastasis by regulating EMT (34). Additionally, emerging studies have reported that some lncRNAs promote cancer progression by regulating the PI3K/Akt pathway (35). One study demonstrated that knockdown of the lncRNA NR027113 inhibited the activity of the PI3K/Akt signalling pathway and restrained EMT. Furthermore, this study reported that PTEN silencing promoted Akt protein phosphorylation and HCC cell function (36). These data suggest that the Wnt and PI3K/Akt pathways are tightly linked to each other and both promote EMT. In the present study, the Wnt and PI3K/Akt pathways were repressed by LINC01554 overexpression in in vitro investigations. However, the mechanism by which LINC01554 regulates EMT through the Wnt and PI3K/Akt pathways warrants further exploration. KEGG pathway analysis also showed that genes co-expressed with LINC01554 were associated with precancerous diseases, such as hepatitis B virus infection and viral carcinogenesis, which are factors critical for triggering liver carcinogenesis (37). These studies consistently suggest that low LINC01554 levels may play a role in tumour pathogenesis.
The present study has some limitations. Firstly, the functions and potential mechanisms of LINC01554 in HCC were not confirmed in vivo. We did perform inhibition experiments of LINC01554. Regrettably, the efficiencies of interference were extremely low. The possible reason may be the location of LINC01554 in the nucleus of HCC cells. Secondly, the experimental validation of LINC01554 expression was only performed in 40 pairs of HCC and matched normal tissues. Therefore, investigations and corresponding experiments with a larger clinical sample size will be the focus of our future research.
In conclusion, we report the novel finding that LINC01554 inhibited HCC cell proliferation, migration, and invasion and promoted G0/G1 arrest. Taken together, our findings provide new insights into the molecular mechanisms underlying HCC tumourigenesis and implicated LINC01554 as a potential target for HCC therapy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This project was supported by the Health Commission of the Hubei Province Scientific Research Project (grant no. WJ2019H074), The Medical Science Advancement Program (Clinical Medicine) of Wuhan University (grant no. TFLC2018003) and National Key Laboratory of Virology (grant no. 2018KF005).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
QY and GP designed the research and revised the manuscript. LL, KH and ZL performed the experiments and wrote the draft manuscript. LL, KH, ZL, HZ and HL analyzed the experimental results. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided written informed consent for their tissues to be used for medical research. Ethical approval (2019073) for this study was obtained from the Zhongnan Hospital of Wuhan University (Wuhan, Hubei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.De Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanism of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 4.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumour-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 5.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 6.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep. 2012;45:604–611. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryaboshapkina M, Hammar M. Human hepatic gene expression signature of non-alcoholic fatty liver disease progression, a meta-analysis. Sci Rep. 2017;7:12361. doi: 10.1038/s41598-017-10930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Q, Liu B. Identification of a RNA-Seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163–5172. doi: 10.12659/MSM.902615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klingenberg M, Groß M, Goyal A, Polycarpou-Schwarz M, Miersch T, Ernst A, Leupold J, Patil N, Warnken U, Allgayer H, et al. The long noncoding RNA cancer susceptibility 9 and RNA binding protein heterogeneous nuclear ribonucleoprotein L form a complex and coregulate genes linked to AKT signaling. Hepatology. 2018;68:1817–1832. doi: 10.1002/hep.30102. [DOI] [PubMed] [Google Scholar]
- 11.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi: 10.1158/0008-5472.CAN-19-0255. [DOI] [PubMed] [Google Scholar]
- 14.Cui K, Jin S, Du Y, Yu J, Feng H, Fan Q, Ma W. Long noncoding RNA DIO3OS interacts with miR-122 to promote proliferation and invasion of pancreatic cancer cells through upregulating ALDOA. Cancer Cell Int. 2019;19:202. doi: 10.1186/s12935-019-0922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z, Deng X, Yang C, Ruan H, Yu C, et al. A noval, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019;450:98–109. doi: 10.1016/j.canlet.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, Li MQ, He JZ, Zeng TT, Ban XJ, et al. LIN01554-mediated glucose metabolism reprogramming suppresses tumourigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9:796–810. doi: 10.7150/thno.28992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labernadie A, Kato T, Brugues A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, Gonzalez-Tarrago V, Elosegui-Artola A, Albertazzi L, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164:257–264. doi: 10.1093/jb/mvy047. [DOI] [PubMed] [Google Scholar]
- 19.Cheng N, Li Y, Han ZG. Argonaute2 promotes tumour metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology. 2013;57:1906–1918. doi: 10.1002/hep.26202. [DOI] [PubMed] [Google Scholar]
- 20.Liu YA, Liang BY, Guan Y, You J, Zhu L, Chen XP, Huang ZY. Loss of N-cadherin is associated with loss of E-cadherin expression and poor outcomes of liver resection in hepatocellular carcinoma. J Surg Res. 2015;194:167–176. doi: 10.1016/j.jss.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Cho ES, Kang HE, Kim NH, Yook JI. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT) Arch Pharm Res. 2019;42:14–24. doi: 10.1007/s12272-018-01108-7. [DOI] [PubMed] [Google Scholar]
- 22.Yu S, Yang D, Ye Y, Liu P, Chen Z, Lei T, Pu J, Liu L, Wang Z. Long noncoding RNA actin filament-associated protein 1 antisense RNA 1 promotes malignant phenotype through binging with lysine-specific demethylase 1 and repressing HMG box-containing protein 1 in non-small-cell lung cancer. Cancer Sci. 2019;110:2211–2225. doi: 10.1111/cas.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang YH, He GL, Huang SZ, Zhong KB, Liao H, Cai L, Gao Y, Peng ZW, Fu SJ. The long noncoding RNA AK002107 negatively modulates miR-140-5p and targets TGFBR1 to induce epithelial-mesenchymal transition in hepatocellular carcinoma. Mol Oncol. 2019;13:1296–1310. doi: 10.1002/1878-0261.12487. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Zhang J, Le TD, Liu L, Li J. Inferring and analysing module-specific lncRNA-mRNA causal regulatory networks in human cancer. Brief Bioinform. 2019;20:1403–1419. doi: 10.1093/bib/bby008. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BKP, Ooi LL, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinomain association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Cai H, Sun L, Zhan P, Chen M, Zhang F, Ran Y, Wan J. LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J Exp Clin Cancer Res. 2018;37:225. doi: 10.1186/s13046-018-0864-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y, Zhang F, Lu Y, Zheng L, Zhang W, Li X, Li X. Long non-coding RNA CASC11 interacts with hnRNA-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376:62–73. doi: 10.1016/j.canlet.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Guo P, He Y, Chen Z, Chen L, Luo Y, Qi L, Liu Y, Wu Q, Cui Y, et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signaling pathway. Cell Death Dis. 2018;9:513. doi: 10.1038/s41419-018-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding D, Huang H, Jiang W, Yu W, Zhu H, Liu J, Saiyin H, Wu J, Huang H, Jiang S, Yu L. Reticulocalbin-2 enhances hepatocellular carcinoma proliferation via modulating the EGFR-ERK pathway. Oncogene. 2017;36:6691–6700. doi: 10.1038/onc.2017.230. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD, Meng YY, Yuan C, Li HM, Yu YP, et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/β-catenin signaling pathway. Mol Cancer. 2019;18:15. doi: 10.1186/s12943-019-0942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng J, Zhang XT, Liu XL, Fan L, Li C, Sun Y, Liang XH, Wang JB, Mei QB, Zhang F, Zhang T. WSTF promotes proliferation and invasion of lung cancer cells by inducing EMT via PI3K/Akt and IL-6/STAT3 signaling pathways. Cell Signal. 2016;28:1673–1682. doi: 10.1016/j.cellsig.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Pan Q, Fuhler GM, Smits R, Peppelenbosch MP. Action and function of Wnt/β-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol. 2017;52:419–431. doi: 10.1007/s00535-016-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Yu Z, Chen F, Xu H, Shen S, Chen W, Chen L, Su Q, Zhang L, Bi J, et al. miR-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed Pharmacother. 2018;103:1632–1642. doi: 10.1016/j.biopha.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Wu Q, Liu Z, Luo X, Fan Y, Liu Y, Zhang Y, Hua S, Fu Q, Zhao M, et al. Downregulation of FAP suppresses cell proliferation and metastasis through PTEN/PI3K/AKT and Ras-ERK signaling in oral squamous cell carcinoma. Cell Death Dis. 2014;5:e1155. doi: 10.1038/cddis.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Zhou ZY, He CC, Zhang JL, Wang J, Xiao ZY. Down-regulation of LncRNA NR027113 inhibits cells proliferration and metastasis via PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:7222–7232. doi: 10.26355/eurrev_201811_16256. [DOI] [PubMed] [Google Scholar]
- 37.Chaturvedi VK, Singh A, Dudey SK, Hetta HF, John J, Singh MP. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb Pathog. 2019;128:184–194. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.