Abstract
Renal cell carcinoma (RCC) is the most common type of renal cancer. Long non-coding RNA (lncRNA) has been reported to play a vital role in the development and progression of various types of cancer type. However, the underlying molecular mechanisms of PLK1S1 in regulating RCC progression remain unclear. In the present study, PLK1S1 was upregulated in RCC tissues and cells, and PLK1S1 expression was also significantly elevated in stage IV RCC tissues. Kaplan-Meier analysis showed that patients with high PLK1S1 expression had a shorter overall survival time compared with those with low PLK1S1 expression. Moreover, bioinformatics analysis and luciferase reporter assay demonstrated that PLK1S1 inhibited microRNA (miR)-653 expression by direct interaction. Functional analyses demonstrated that a miR-653 inhibitor promoted short hairpin PLK1S1-attenuated cell proliferation, invasion and sorafenib resistance of RCC cells. In addition, C-X-C motif chemokine receptors 5 (CXCR5) was identified as an effector of PLK1S1/miR-653-mediated tumorigenesis and drug resistance in RCC cells. Lastly, xenograft experiments demonstrated that PLK1S1 knockdown inhibited tumor growth in vivo. Reverse transcription-quantitative PCR and western blot analysis revealed that PLK1S1 knockdown upregulated the expression level of miR-653, whilst downregulating the expression level of CXCR5. In conclusion, the present study revealed that PLK1S1 promoted tumor progression and sorafenib resistance in RCC through regulation of the miR-653/CXCR5 axis, which may offer a novel treatment strategy for patients with RCC.
Keywords: renal cell carcinoma, PLK1S1, miR-653, CXCR5, chemoresistance
Introduction
Renal cell carcinoma (RCC) is one of the most lethal urological neoplasms, which contributed to ~5% of all malignant carcinomas, worldwide (1). Chemotherapy (such as sorafenib) is one of the primary treatment strategies for RCC (2). However, chemoresistance remains a major cause of cancer recurrence and cancer-associated mortality (3). Therefore, it is important to improve the understanding of RCC pathogenesis and identify novel targets for the development of diagnostic and therapeutic approaches.
Long non-coding RNAs (lncRNAs) are transcripts >200 nucleotides in length, which are not protein coding (4). Accumulating evidence indicates that lncRNAs are involved in the physiological and pathological processes in most types of cancer, such as hepatocellular carcinoma, colorectal cancer and prostate cancer (5,6). In previous reports, lncRNAs could function as both oncogenes and tumor suppressors in RCC. For example, lncRNA SARCC inhibited RCC progression by regulating androgen receptor/miR-143 signals (7). On the other hand, lncRNA DUXAP8 promoted RCC tumorigenesis by downregulating miR-126 (8). Moreover, it has been reported that lncRNAs have been associated with chemoresistance in cancer, including RCC. For example, Xu et al (9) demonstrated that the knockdown of lncRNA SRLR reduced chemoresistance to sorafenib in RCC cells. PLK1S1 has been reported to be upregulated in tamoxifen-resistant MCF7 cells, lung adenocarcinoma and bone metastasis (10–12). However, the exact mechanisms of PLK1S1 in RCC remains unclear.
MicroRNAs (miRNAs/miRs) are another type of endogenous non-coding RNAs with a length of 22–25 nucleotides, which regulate gene expression at the post-transcriptional level (13). miRNAs have been reported to play vital roles in cell proliferation, apoptosis and metastasis in human cancer (14). For example, miR-296 inhibited cell invasion and migration in esophageal squamous cell carcinoma by targeting STAT3 (15). Recently, miR-653 was found to suppress non-small cell lung cancer and breast cancer tumorigenesis (16,17). Nonetheless, whether miR-653 is involved in RCC remains to be further clarified.
C-X-C chemokine receptors (CXCRs) are a family of cellular G-protein coupled receptors (18). Among the CXCRs, the chemokine receptor CXCR5, which is primarily expressed in B cells and CD4+ T cells, has been associated with tumorigenesis and progression of various types of cancer, such as breast, prostate and colon cancer (19–21). Recently, Zheng et al (22) reported that CXCR5 was associated with clear cell renal cell carcinoma (ccRCC) progression and predicted poor prognosis, which led to the hypothesis that CXCR5 might be a vital modulator of RCC tumorigenesis. The findings of the present study provide a further understanding into the development of RCC, which may facilitate the development of a novel therapeutic targets in the treatment of RCC.
Materials and methods
Clinical specimens
In total, 33 pairs of RCC tissues and adjacent normal tissues were obtained from patients (19 males and 14 females), who had undergone surgical resection, with a median age of 56 years (range, 31–78 years) between February 2016 and August 2018 at the Shanghai JiaoTong University School of Medicine. Written informed consent was provided from all participators prior to the start of the study. All tissues were instantly frozen in liquid nitrogen, and then stored at −80°C for further analysis. The clinicopathological data was obtained from the medical records at the Shanghai Jiaotong University School of Medicine. All patients were classified according to Union International Cancer Control and the American Joint Committee on Cancer (23,24). The study was approved by the Ethics Committee of Shanghai Jiaotong University School of Medicine.
The Cancer Genome Atlas (TCGA) analysis
The expression of PLK1S1 and TCGA kidney renal clear cell carcinoma (TCGA-KIRC) clinical data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Patients with corresponding gene expression were included in the present study, while those with missing overall survival data were excluded.
Cell culture
The human RCC cells (ACHN, Caki1, A498 and 786-O), papillary renal cell carcinoma cells (Caki2) and the immortalized renal proximal tubules epithelial cells (RPTEC) were purchased from American Type Culture Collection. The cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.), and supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere containing 5% CO2. To generate ACHN and 786-O resistant cell lines (ACHN-R and 786-O-R, respectively), ACHN and 786-O cells were incubated with increasing concentrations of sorafenib (1 to 20 µM) for >6 months.
Cell transfection
The short hairpin (sh)RNA specific to PLK1S1 (shPLK1S1; 5′-UCAGCUGCUGUCGUAUUCAUGAG-3′) and its negative control (shNC; 5′-AAUUCUCCGAACGUGUCACGU-3′), miR-653 mimic (5′-GUGUUGAAACAAUCUCUACUG-3′) and its negative control (NC mimic; 5′-GACAACUUACAAUCUCUACUG-3′), and the miR-653 inhibitor (5′-AGCCUUGAUCGAGGUCGGGAU-3′) and its negative control (NC inhibitor; 5′-CAGUAGAGAUUGUAAGUUGUC-3′), were synthesized by Shanghai GenePharma Co., Ltd.. For the overexpression of CXCR5, the CXCR5 cDNA was cloned into the pCDNA3.1 vector (Shanghai GenePharma Co., Ltd). Transfection of the cells with vectors, shPLK1S1or shNC and miR-653 mimic or NC mimic, miR-653 inhibitor or NC inhibitor, and co-transfection with shPLK1S1 and miR-653 inhibitor (all at 10 nM) were conducted with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturers instructions. All functional experiments were performed 48 h post-transfection.
Cell Counting Kit-8 (CCK-8) assay
To determine the IC50 value, ACHN and 786-O cells (1×104 cells/well) were seeded into 96-well plates and then treated with varying concentrations of sorafenib (2.5, 5, 10, 20, 40 and 80 µg/ml) for 48 h. For the detection of cell viability, transfected ACHN and 786-O cells were treated with sorafenib (2 µg/ml) for 48 h. Next, cell viability was determined using the CCK-8 assay kit (Dojindo Molecular Technologies, Inc.). Briefly, 10 µl CCK-8 solution was added to each well and incubated for 3 h, and then the absorbance at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Inc.). IC50 were determined as the concentration of the drug at which sorafenib produced 50% growth inhibition, with higher IC50 values indicated higher chemoresistance potential.
Colony formation assay
Transfected RCC cells were seeded in 6-well plates at a density of 200 cells/well. Following culturing for two weeks, PBS (Sigma-Aldrich; Merck KGaA) was used to rinse each well. Subsequently, RCC cells were fixed in 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA), and then stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) both at room temperature for 10 min. The number of colonies were then counted using a light microscope (magnification, ×200). Groups of >50 cells were considered a clone.
Matrigel assay
The invasion abilities of the RCC cells were assessed using Transwell chambers (8.0-µm pore size; EMD Millipore) pre-coated with Matrigel for 1 h (Corning Inc.) at room temperature. Transfected cells (8×104 cells) were added to the upper chamber containing 150 µl RPMI-1640 without FBS. In addition, 550 µl RPMI-1640 medium was added to the lower chamber. After 24 h, cells in the upper chamber were removed, and cells in the lower membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) both for 20 min at room temperature. Invaded cells were counted in 3 randomly selected visual fields using a light microscope (magnification, ×200; Zeiss GmBH).
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
The TUNEL apoptosis kit (Roche Diagnostics GmbH) was used to assess cell apoptosis. In brief, cells were washed with PBS for 5 min, three times and fixed in 4% paraformaldehyde (cat. no. AR1069; Wuhan Boster Biological Technology, Ltd.) at 4°C for 20 min. The cells were then incubated with the TUNEL enzyme for 60 min at 3°C. Finally, the fluorescent reaction was counterstained with DAPI (1:1,000 in PBS; Sigma-Aldrich; Merck KGaA) to stain the nucleus for 10 min at room temperature. Antifade mounting medium (cat. no. P0126; Beyotime Institute of Biotechnology) was used. Images from 4 fields of view were used to obtain images with a fluorescent microscope (magnification, ×20; Olympus Corporation).
Western blot analysis
Proteins were extracted from transfected RCC cells using RIPA buffer (Thermo Fisher Scientific, Inc.). Protein concentration was measured using the bicinchoninic acid assay (Beyotime Institute of Biotechnology). A total of 10 µg protein/lane were separated using 10% SDS-PAGE (EMD Millipore), and then transferred to PVDF membranes (Bio-Rad Laboratories, Inc.). After blocking with 5% skimmed milk, membranes were probed with primary antibodies against CXCR5 (1:1,000; cat. no. ab133706; Abcam) and anti-GAPDH (1:1,000; cat. no. ab8245; Abcam) overnight at 4°C. Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies, goat anti-mouse IgG, (cat. no. ab205719) and goat anti-rabbit IgG, (cat. no. ab205718) (both 1,1000; both from Abcam) at room temperature for 2 h. The protein bands were visualized using an enhanced chemiluminescence kit (Bio-Rad Laboratories, Inc.). GAPDH served as the loading control.
Reverse transcription-quantitative PCR (RT-qPCR)
At 48-h post-transfection, total RNA was isolated from tissues and cell lines using TRIzol® (Thermo Fisher Scientific, Inc.). The RNA was reverse transcribed into cDNA using a reverse transcriptase kit (Takara Bio, Inc.) or the TaqMan® miRNA reverse transcription kit (Thermo Fisher Scientific, Inc.) at 37°C for 15 min. RT-qPCR was performed on the ABI 7900 Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR-Green PCR Master Mix kit (Takara Bio, Inc). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 3 min; 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The expression levels of genes were calculated using the 2−ΔΔCq method (25). U6 and GAPDH were set as the internal control. The sequences of the primers were as follows: PLK1S1 forward, 5′-CCCACATTCACACCGACAGA-3′ and reverse, 5′-ACTCTTGCCATGACGTGTGT-3′; miR-653 forward, 5′-ACCAGCTTCAAACAAGTTCACTG-3′ and reverse, 5′-GCTTCCATCTTATCATTCTTGCA-3′; CXCR5 forward, 5′-CCCTCATGGCCTCCTTCAAG-3′ and reverse, 5′-AGGGCAAGATGAAGACCAGC-3′; GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′; and reverse, 5′-GCCTTCTCCATGGTGGTGAA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACATATACTA-3′ and reverse, 5′-ACGAATTTGCGTGTCATCCTTGCG-3′.
Luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn) online tool was used to predict the potential miRNAs that could bind to PLK1S1, while TargetScan (http://www.targetscan.org) was used to predict the potential downstream target of miR-653. Mutants within the miR-653 binding site were created using the QuikChange II Site Directed Mutagenesis kit (Agilent Technologies, Inc.). The wild-type (WT) and mutant (Mut) PLK1S1 or CXCR5 was sub-cloned into pmirGLO dual-luciferase vector (Shanghai GenePharma Co., Ltd.) to construct PLK1S1-WT/Mut or CXCR5-WT/Mut vectors. Subsequently, PLK1S1-WT/Mut or CXCR5-WT/Mut vectors were co-transfected with NC mimic, miR-653 mimic and miR-653 inhibitor into 293T cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was evaluated using the Dual-Luciferase Reporter Analysis system (Promega Corporation), following incubation for 48 h. Firefly luciferase activity was normalized to Renilla (Promega Corporation) luciferase gene activity.
Xenograft experiment
A total of 10 male BALB/c-nu nude mice (age, 5 to 6-weeks-old; weight, 18–20 g) were randomly divided into two groups and maintained under 26°C, 50% relative humidity, with a 12-h light/dark cycle, with ad libitum access to food and water. All in vivo experimental procedures were approved by the Ethics Committee of Shanghai Jiaotong University School of Medicine. The ACHN cells transfected with shNC or shPLK1S1 were subcutaneously injected into the mice. Tumors were examined every 7 days. Mice were euthanized according to the following criteria: i) the weight of the mouse was excessively reduced or increased; ii) abnormal behavior; iii) tumor diameter was >1.5 cm; iv) the tumor was ulcerated. No mouse died or had significant weight loss during the experiment. On 28th day, the mice were sacrificed by cervical dislocation following anesthesia with an intraperitoneal injection of sodium pentobarbital (60 mg/kg). Animal death was confirmed by cardiac and respiratory arrest, muscle relaxation and lack of reflection. The tumors were photographed, and the tumor weights were measured. Tumor volume was calculated using the following formula: Volume = (length × width2)/2. The maximum tumor volume was 709 mm3 and the maximum diameter of tumor was 16 mm.
Statistical analysis
Statistical analysis was performed using SPSS v16.0 (SPSS, Inc.). The experiments were performed three times and the data are presented as mean ± standard deviation. Comparisons among multiple groups were performed using one-way analysis of variance, followed by Tukey's post hoc test. Comparison between RCC and adjacent normal tissue samples from patients with RCC was performed using a paired Student's t-test, while comparison between the experimental and control groups was performed using an unpaired Student's t-test. Kaplan-Meier analysis and the log-rank test were used to analyze survival curves. Cut-off values were determined using the mean expression level of PLK1S1. P<0.05 was considered to indicate a statistically significant difference.
Results
PLK1S1 is upregulated in RCC cells and tissues and is associated with poor prognosis
The mRNA expression level of PLK1S1 was investigated in RCC cells, and the results indicated that expression in the RCC cells (Caki1, ACHN, A498 and 786-O) and the Caki2 papillary RCC cell line was significantly upregulated compared with that in the RPTEC cell line (Fig. 1A). Furthermore, the PLK1S1 mRNA expression level was also significantly increased in clinical tissues, (Fig. 1B). In addition, it was found that PLK1S1 expression was associated with histological grade, tumor stage, lymph node metastasis and distant metastasis, while there was no association with age or sex (Table I).
Figure 1.
PLK1S1 is upregulated in RCC cells and tissues and associated with poor prognosis. Reverse transcription-quantitative PCR was used to determine the relative mRNA expression level of PLK1S1 in (A) RCC cells (ACHN, Caki1, A498 and 786-O), papillary RCC cells (Caki2) and immortalized epithelial cells of renal proximal tubules (RPTEC) and (B) in clinical RCC tissues and adjacent normal tissues (n=33). (C) TCGA datasets were used to investigate the mRNA expression levels of PLK1S1 in normal tissues and RCC tissues. (D) Kaplan-Meier analysis was used to identify an association between PLK1S1 mRNA expression levels and overall survival of patients with RCC from TCGA datasets. (E) Violin plot showed that high mRNA expression levels of PLK1S1 was associated with advanced stage in patients with RCC. F, 3.18, adj. P=0.0299. All data are represented as the mean ± SD. *P<0.05. Adj., adjusted.
Table I.
Association between PLK1S1 mRNA expression levels and clinicopathological features in patients with renal cell carcinoma.
| Expression of PLK1S1 | ||||
|---|---|---|---|---|
| Clinicopathological features | Number | High, n (%) | Low, n (%) | P-value |
| Age, years | 0.374 | |||
| ≤60 | 15 | 7 (46.7) | 8 (53.3) | |
| >60 | 18 | 10 (55.6) | 8 (44.4) | |
| Sex | 0.462 | |||
| Male | 19 | 9 (47.4) | 10 (52.6) | |
| Female | 14 | 8 (57.1) | 6 (42.9) | |
| Histological grade | 0.011 | |||
| Well | 21 | 9 (42.9) | 12 (57.1) | |
| Moderate and poor | 12 | 9 (75.0) | 3 (25.0) | |
| Tumor stage | 0.028 | |||
| I and II | 20 | 8 (40.0) | 12 (60.0) | |
| III and IV | 13 | 9 (69.2) | 4 (30.8) | |
| Lymph node metastasis | 0.011 | |||
| Positive | 10 | 8 (80.0) | 2 (20.0) | |
| Negative | 23 | 11 (47.8) | 12 (52.2) | |
| Distant metastasis | 0.014 | |||
| Positive | 11 | 9 (81.8) | 2 (18.2) | |
| Negative | 22 | 10 (45.5) | 12 (54.5) | |
To investigate the mRNA expression levels further, TCGA datasets were analyzed to determine the involvement of PLK1S1 in RCC. The data revealed that PLK1S1 mRNA expression levels in RCC tissues was significantly increased compared with that in adjacent normal tissues (Fig. 1C). In addition, Kaplan-Meier analysis showed that patients with high PLK1S1 mRNA expression level had a shorter overall survival time compared with that in patients with low PLK1S1 expression (Fig. 1D). Notably, it was also found that the PLK1S1 mRNA expression level was significantly increased in patients with stage IV RCC (F, 3.18; adjusted P=0.0299) (Fig. 1E). Taken together, these data indicated that PLK1S1 might be an oncogene for RCC progression.
PLK1S1 interacts with miR-653
Using the StarBase bioinformatics analysis software, PLK1S1 was found to bind to miR-653 via complementary base pairing (Fig. 2A). The results from the luciferase reporter assay indicated that miR-653 mimic significantly reduced luciferase activity of pmirGLO-PLK1S1-WT vectors, whereas the miR-653 inhibitor significantly increased luciferase activity (Fig. 2B). In addition, PLK1S1 was significantly downregulated in shPLK1S1 transfected ACHN cells and significantly upregulated in PLK1S1 overexpressed ACHN cells. The downregulation of PLK1S1 significantly increased the expression of miR-653 whereas the upregulation of PLK1S1 significantly decreased the expression of miR-653 (Fig. 2C). Taken together, these results demonstrated that PLK1S1 inhibited miR-653 expression by direct interaction.
Figure 2.
PLK1S1 interacts with miR-653. The binding sequences between PLK1S1 and miR-653 were (A) predicted by StarBase and confirmed using (B) a luciferase reporter assay in 293T cells. (C) Reverse transcription-quantitative PCR was used to determine the mRNA expression levels of miR-653 and PLK1S1 in ACHN cells transfected with shNC, shPLK1S1 and pcDNA3.1-PLK1S1. All data are represented as the mean ± SD. *P<0.05; **P<0.01. miRNA, microRNA; sh, short hairpin; NC, negative control.
miR-653 inhibitor rescues PLK1S1 knockdown-attenuated tumorigenesis of RCC cells
To explore whether miR-653-mediated and PLK1S1-regulated RCC tumorigenesis and progression, stable PLK1S1-knockdown RCC cells (ACHN and 786-O) were generated (Fig. 3A). Subsequently, the miR-653 inhibitor was introduced into shPLK1S1-expressing ACHN and 786-O cells. RT-qPCR showed that the introduction of miR-653 inhibitor significantly reduced shPLK1S1-mediated miR-653 upregulation in RCC cells (Fig. 3B). Matrigel and colony formation assays revealed that the miR-653 inhibitor abrogated the inhibitory effects of PLK1S1 knockdown on the invasion and proliferation of RCC cells (Fig. 3C-F). Based on these results, it was confirmed that miR-653 was involved in PLK1S1-modulated tumorigenesis of RCC cells.
Figure 3.
miR-653 inhibitor rescues PLK1S1 knockdown-attenuated tumorigenesis of renal cell carcinoma cell lines. Reverse transcription-quantitative PCR was used to determine the mRNA levels of (A) PLK1S1 in ACHN and 786-O cells transfected with shNC and shPLK1S1 and (B) miR-653 in ACHN and 786-O cells transfected with shNC, shPLK1S1, shPLK1S1 plus miR-653 inhibitor. (C) Matrigel assay was used to investigate the invasion ability of ACHN and 786-O cells transfected with shNC, shPLK1S1, shPLK1S1 plus miR-653 inhibitor and the results were subsequently (D) quantified. (E) Colony formation assay was used to determine the colony number of ACHN and 786-O cells transfected with shNC, shPLK1S1, shPLK1S1 plus miR-653 inhibitor and the results were then (F) quantified. All data are represented as the mean ± SD. *P<0.05; **P<0.01. miRNA, microRNA; sh, short hairpin; NC, negative control.
PLK1S1 knockdown-mediated inhibitory effect on sorafenib resistance is reduced by the miR-653 inhibitor in sorafenib-resistant RCC cells
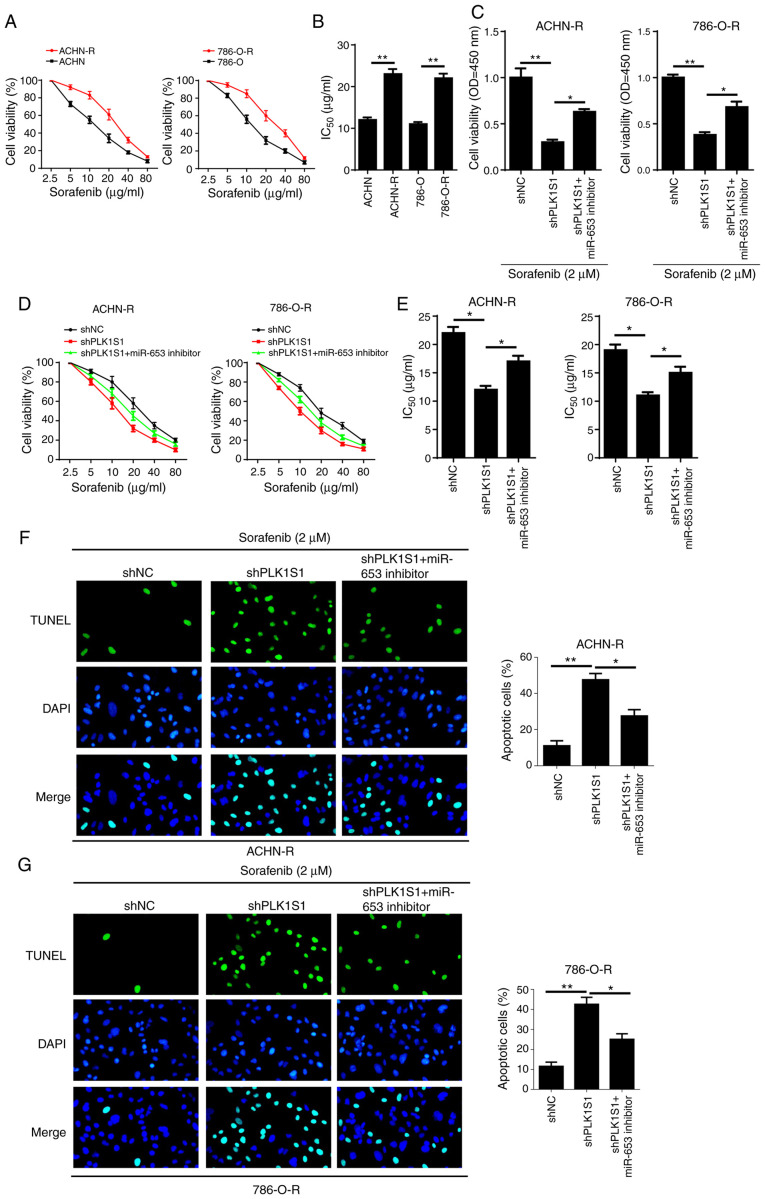
To investigate the potential role of PLK1S1 and miR-653 in drug resistance to sorafenib in RCC cells, sorafenib-resistant ACHN (ACHN-R) and 786-O cells (786-O-R) were established. As shown in Fig. 4A and B, the IC50 of sorafenib was significantly increased in ACHN-R and 786-O-R cells, indicating that sorafenib-resistant RCC cells were successfully generated. In addition, the miR-653 inhibitor partially reversed PLK1S1 knockdown-attenuated cell viability in sorafenib-resistant RCC cells under sorafenib treatment (Fig. 4C). Furthermore, PLK1S1 knockdown significantly decreased the IC50 of sorafenib in sorafenib-resistant RCC cells, while miR-653 inhibitor partially reversed shPLK1S1-induced decrease in IC50 values (Fig. 4D and E). The TUNEL assay revealed that the miR-653 inhibitor significantly decreased cell apoptosis in shPLK1S1 sorafenib-resistant RCC cells (Fig. 4F and G). In summary, the data suggested that PLK1S1 enhanced chemoresistance in RCC cells to sorafenib by regulating miR-653.
Figure 4.
PLK1S1 depletion-mediated inhibitory effect on sorafenib resistance is eliminated by the miR-653 inhibitor in sorafenib-resistant renal cell carcinoma cell lines. (A and B) The IC50 values of sorafenib in ACHN, ACHN-R, 786-O and 786-O-R cells was measured using the CCK-8 assay. CCK-8 was used to determine the (C) cell viability and (D and E) IC50 values of sorafenib of ACHN-R and 786-O-R cells transfected with shNC, shPLK1S1, shPLK1S1 plus miR-653 inhibitor treated with or without 2 µM sorafenib. TUNEL assay was used to investigate cell apoptosis in (F) ACHN-R and (G) 786-O-R cells transfected with shNC, shPLK1S1, shPLK1S1 plus miR-653 inhibitor treated with sorafenib. All data are represented as the mean ± SD. *P<0.05; **P<0.01. miRNA, microRNA; sh, short hairpin; NC, negative control; TUNEL, Terminal deoxynucleotidyl transferase dUTP nick end labeling; CCK-8; Cell Counting Kit-8; R, resistant.
CXCR5 is a target of miR-653
Using TargetScan (http://www.targetscan.org), CXCR5 was predicted as a downstream target of miR-653 (Fig. 5A). Furthermore, luciferase reporter assay demonstrated that miR-653 mimic reduced luciferase activity of WT CXCR5, while the opposite effect occurred in cells transfected with miR-653 inhibitor (Fig. 5B). In addition, RT-qPCR revealed that miR-653 was significantly upregulated in miR-653 mimic transfected ACHN cells and significantly downregulated in miR-653 inhibitor transfected ACHN cells. The upregulation of miR-653 significantly decreased the expression of CXCR5, while the downregulation of miR-653 significantly increased the expression of CXCR5 (Fig. 5C). Thus, these data indicate that miR-653 directly targets CXCR5.
Figure 5.
CXCR5 is a target of miR-653. The binding sequences between miR-653 and CXCR5 were (A) predicted using the TargetScan website and (B) confirmed using a luciferase reporter assay. (C) Reverse transcription-quantitative PCR was used to determine the mRNA expression levels of miR-653 and CXCR5 in ACHN cells transfected with NC mimic, miR-653 mimic, miR-653 inhibitor. All data are represented as the mean ± SD. *P<0.05; **P<0.01; ***P<0.001. miRNA, microRNA; sh, short hairpin; NC, negative control; CXCR5, C-X-C motif chemokine receptors 5.
CXCR5 mediates PLK1S1/miR-653-regulated RCC tumorigenesis
To further determine whether PLK1S1/miR-653 promoted RCC progression through CXCR5, CXCR5 was introduced into shPLK1S1 and miR-653 mimic-expressing ACHN cells. Firstly, it was demonstrated that overexpression of CXCR5 significantly reversed the inhibitory effects of PLK1S1 knockdown and miR-653 overexpression on CXCR5 mRNA expression levels (Fig. 6A and B). Matrigel and colony formation assays demonstrated that the overexpression of CXCR5 significantly reversed the shPLK1S1- and miR-653 mimic-attenuated ACHN cell invasion and proliferation (Fig. 6C-H). Taken together, these data revealed that the PLK1S1/miR-653/CXCR5 axis promoted the development and progression of RCC.
Figure 6.
CXCR5 mediates PLK1S1/miR-653-regulated renal cell carcinoma tumorigenesis. Reverse transcription-quantitative PCR was used to determine the relative mRNA expression of CXCR5 in (A) ACHN cells transfected with shNC, shPLK1S1, shPLK1S1 plus CXCR5 and (B) ACHN cells transfected with NC mimic, miR-653 mimic, miR-653 mimic plus CXCR5. Matrigel assay was used to investigate cell invasion in ACHN cells transfected with (C) shNC, shPLK1S1, shPLK1S1 plus CXCR5 and (D) NC mimic, miR-653 mimic, miR-653 mimic plus CXCR5 and the results were subsequently (E) quantified. Colony formation assay was used to determine the colony number of ACHN cells transfected with (F) shNC, shPLK1S1, shPLK1S1 plus CXCR5 and (G) NC mimic, miR-653 mimic, miR-653 mimic plus CXCR5 and the results were subsequently (H) quantified. All data are represented as the mean ± SD. *P<0.05; **P<0.01. miRNA, microRNA; sh, short hairpin; NC, negative control; CXCR5, C-X-C motif chemokine receptors 5.
CXCR5 rescues PLK1S1 knockdown- or miR-653 mimic-attenuated chemoresistance of RCC cells
Next, the role of CXCR5 in PLK1S1 and miR-653-modulated drug resistance of RCC cells to sorafenib was assessed. As shown in Fig. 7A, CXCR5 increased the cell viability of sorafenib-resistant ACHN cells expressing shPLK1S1 or miR-653 mimic in the presence of sorafenib treatment. In addition, overexpression of CXCR5 reduced the inhibitory effect of shPLK1S1 or miR-653 mimic on IC50 values in sorafenib-resistant RCC cells (Fig. 7B and C). The TUNEL assay showed that overexpression of CXCR5 significantly reversed shPLK1S1 or miR-653 mimic-mediated promotion of cell apoptosis in ACHN-R cells following sorafenib treatment (Fig. 7D-G). Taken together, the data indicate that CXCR5 is a key effector of PLK1S1 and miR-653-regulated sorafenib resistance in RCC cells.
Figure 7.
CXCR5 rescues PLK1S1 knockdown- or miR-653 mimic-attenuated chemoresistance of renal cell carcinoma cell lines. Cell Counting Kit-8 assay was used to determine (A) cell viability and (B and C) IC50 values in ACHN-R cells transfected with shNC, shPLK1S1, shPLK1S1 plus CXCR5 and NC mimic, miR-653 mimic, miR-653 mimic plus CXCR5 treated with and without 2 µM sorafenib, respectively. (D-G) TUNEL assay was used to investigate cell apoptosis in ACHN-R cells transfected with (D) shNC, shPLK1S1, shPLK1S1 plus CXCR5 and the results were subsequently (E) quantified, and with (F) NC, miR-653 mimic, miR-653 mimic plus CXCR5, treated with 2 µM sorafenib and the results were (G) quantified. All data are represented as the mean ± SD. *P<0.05; **P<0.01. miRNA, microRNA; sh, short hairpin; NC, negative control; TUNEL, Terminal deoxynucleotidyl transferase dUTP nick end labeling; R, resistant; CXCR5, C-X-C motif chemokine receptors 5; OD, optical density.
PLK1S1 knockdown prevents tumor growth in vivo
To further determine whether PLK1S1 promotes RCC growth in vivo, the xenograft experiment was performed. The results demonstrated that PLK1S1 knockdown significantly reduced the growth of the tumor compared with that in the shNC group (Fig. 8A-C). RT-qPCR showed that the mRNA expression level of PLK1S1 was significantly decreased in the shPLK1S1 group compared with that in the shNC group, while the expression level of miR-653 was significantly increased in the shPLK1S1 group compared with that in the shNC group (Fig. 8D and E). Furthermore, western blot analysis demonstrated that knockdown of PLK1S1 reduced the protein expression level of CXCR5 (Fig. 8F). In summary, these data reveal that PLK1S1 promotes tumor growth of RCC via the miR-653/CXCR5 axis.
Figure 8.
PLK1S1 knockdown prevents tumor growth in vivo. (A) Images of the tumors were obtained, and (B) tumor volumes and (C) the weight of the tumors in mice injected with either shNC or shPLK1S1 were determined. n=5. (D) PLK1S1 and (E) miR-653 mRNA expression levels were determined using reverse transcription-quantitative PCR. (F) The protein expression level of CXCR5 was examined in tumors using western blot assay, with GAPDH as the loading control. All data are represented as the mean ± SD. *P<0.05; **P<0.01; ***P<0.001. miRNA, microRNA; sh, short hairpin; NC, negative control; CXCR5, C-X-C motif chemokine receptors 5.
Discussion
The present study revealed that PLK1S1 promoted RCC tumorigenesis and enhanced sorafenib resistance of RCC through the miR-653/CXCR5 axis (Fig. 9). The present study not only discovered a novel regulatory mechanism in RCC, but also identified a potential novel therapy for patients with RCC.
Figure 9.

Schematic diagram shows the mechanism by which PLK1S1 contributes to renal cell carcinoma tumorigenesis and chemoresistance.
Over the last decade, lncRNAs have attracted increasing attention and demonstrated their pivotal roles in various types of cancer, including RCC. For example, Dong et al (26) reported that lncRNA SNHG7 promoted proliferation and inhibited apoptosis of RCC cells by inhibiting the protein expression level of CDKN1A. Yue et al (27) revealed that the knockdown of lncRNA DLEU1 inhibited RCC progression by regulating the Akt and EMT signaling pathway. In the present study, it was demonstrated that PLK1S1 was upregulated in RCC tissues and cells, and that high expression of PLK1S1 was associated with an advanced TNM stage and poor prognosis in patients with RCC.
The competing endogenous RNA (ceRNA) network exhibits its regulatory function in human cancer, including RCC. For example, Yang et al (28) found that the lncRNA TUG1 acted as a ceRNA of miR-196a to accelerate the proliferation, migration and invasion of RCC. Shi et al (29) demonstrated that the lncRNA ROR sponged miR-206 to promote RCC progression through increasing the protein expression level of VEGF. Furthermore, Xie et al (17) demonstrated that hsa_circ_0004771 facilitated the proliferation and inhibited apoptosis in breast cancer by functioning as a ceRNA of miR-653 to regulate the ZEB2 signaling pathway. In the present study, it was confirmed that miR-653 had the ability to bind to PLK1S1. To the best of our knowledge this is the first time that the sponging effect of PLK1S1 on miR-653 in RCC has been discovered. In addition, the inhibitory effects of shPLK1S1 on RCC progression and chemoresistance of RCC cells to sorafenib were reduced by the miR-653 inhibitor. Therefore, to the best of our knowledge it has been demonstrated for the first time that PLK1S1 acted as a ceRNA to sponge miR-653 to promote tumorigenesis and enhance the chemoresistance of RCC.
CXCRs, comprising of CXCR 1 to 7, are not only involved in the immune system but also in tumorigenesis and cancer development (30). For example, Saintigny et al (31) found that CXCR2 was associated with a low 5-year survival rate and promoted the invasion and metastasis of lung adenocarcinoma. Furthermore, Sun et al (32) reported that the knockdown of CXCR2 and CXCR3 suppressed the migration, invasion, colony formation and sphere-forming abilities of RCC cells. CXCR5 has been reported to be upregulated in ccRCC, and CXCR5 knockdown reduced the promoting effect of CXCL13 on the proliferation and migration of ccRCC cells (22). However, there are no previous studies that investigated the involvement of CXCR5 in RCC. In the present study, it was found that CXCR5 was a target of miR-653. In addition, the overexpression of CXCR5 rescued PLK1S1 knockdown- or miR-653 mimic-attenuated progression and chemoresistance of RCC cells, suggesting that CXCR5 was crucial for PLK1S1/miR-653-regulated progression of RCC.
In conclusion, the present study reported the potential molecular mechanisms of PLK1S1 in the tumorigenesis and chemoresistance of RCC. To the best of our knowledge it has been demonstrated for the first time that PLK1S1 contributed to RCC progression and enhanced the chemosensitivity of RCC cells via the miR-653/CXCR5 pathway. The results provide a further understanding in the treatment of RCC using a PLK1S1-targeted approach. However, further investigation is still required. Firstly, other lncRNAs may exist and serve as ceRNAs to regulate crucial gene expression in RCC. Secondly, PLK1S1 can bind to a number of miRNAs, of which, other miRNAs can also affect the development of RCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Science and Technology Committee of Changning District of Shanghai (grant no. CNKW2018Y03).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
WL, DL and ML designed the present study. DY and YZ and SZ performed all the experiments. WL and ML analyzed the data and prepared the figures. WL and DL drafted the initial manuscript. ML reviewed and revised the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
All in vivo experimental procedures were approved by the Ethics Committee of Shanghai Jiaotong University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, Gore JL, Sun M, Wood C, Russo P. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 3.Brugarolas J. Renal-cell carcinoma-molecular pathways and therapies. N Engl J Med. 2007;356:185–187. doi: 10.1056/NEJMe068263. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 6.Li M, Wang Y, Cheng L, Niu W, Zhao G, Raju JK, Huo J, Wu B, Yin B, Song Y, Bu R. Long non-coding RNAs in renal cell carcinoma: A systematic review and clinical implications. Oncotarget. 2017;8:48424–48435. doi: 10.18632/oncotarget.17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng J, Lin W, Huang Q, Li G, Zheng J, Chang C. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ. 2017;24:1502–1517. doi: 10.1038/cdd.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang T, Wang X, Yang X, Ji J, Wang Q, Yue X, Dong Z. Long non-coding RNA DUXAP8 enhances renal cell carcinoma progression via downregulating miR-126. Med Sci Monit. 2018;24:7340–7347. doi: 10.12659/MSM.910054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Yang F, Wei D, Liu B, Chen C, Bao Y, Wu Z, Wu D, Tan H, Li J, et al. Long noncoding RNA-SRLR elicits intrinsic sorafenib resistance via evoking IL-6/STAT3 axis in renal cell carcinoma. Oncogene. 2017;36:1965–1977. doi: 10.1038/onc.2016.356. [DOI] [PubMed] [Google Scholar]
- 10.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng F, Wang R, Zhang Y, Zhao Z, Zhou W, Chang Z, Liang H, Zhao W, Qi L, Guo Z, Gu Y. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer. 2017;16:98. doi: 10.1186/s12943-017-0666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dat Le T, Matsuo T, Yoshimaru T, Kakiuchi S, Goto H, Hanibuchi M, Kuramoto T, Nishioka Y, Sone S, Katagiri T. Identification of genes potentially involved in bone metastasis by genome-wide gene expression profile analysis of non-small cell lung cancer in mice. Int J Oncol. 2012;40:1455–1469. doi: 10.3892/ijo.2012.1348. [DOI] [PubMed] [Google Scholar]
- 13.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang RT, Xu M, Xu CX, Song ZG, Jin H. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin Cancer Res. 2014;20:4705–4516. doi: 10.1158/1078-0432.CCR-14-0517. [DOI] [PubMed] [Google Scholar]
- 15.Wang ZZ, Luo YR, Du J, Yu Y, Yang XZ, Cui YJ, Jin XF. MiR-296-5p inhibits cell invasion and migration of esophageal squamous cell carcinoma by downregulating STAT3 signaling. Eur Rev Med Pharmacol Sci. 2019;23:5206–5214. doi: 10.26355/eurrev_201906_18185. [DOI] [PubMed] [Google Scholar]
- 16.Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510:462–466. doi: 10.1016/j.bbrc.2019.01.131. [DOI] [PubMed] [Google Scholar]
- 17.Xie R, Tang J, Zhu X, Jiang H. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019;39:BSR20181919. doi: 10.1042/BSR20181919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CY, Chen YM, Hsu HH, Shiu CT, Kuo HC, Chen TY. Grouper (epinephelus coioides) CXCR4 is expressed in response to pathogens infection and early stage of development. Dev Comp Immunol. 2012;36:112–120. doi: 10.1016/j.dci.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhao K, Yao Y, Luo X, Lin B, Huang Y, Zhou Y, Li Z, Guo Q, Lu N. LYG-202 inhibits activation of endothelial cells and angiogenesis through CXCL12/CXCR7 pathway in breast cancer. Carcinogenesis. 2018;39:588–600. doi: 10.1093/carcin/bgy007. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Tang H, Huang J, An H. Upregulation of CXCR7 is associated with poor prognosis of prostate cancer. Med Sci Monit. 2018;24:5185–5191. doi: 10.12659/MSM.906180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates RC, DeLeo MJ, III, Mercurio AM. The epithelial-mesenchymal transition of colon carcinoma involves expression of IL-8 and CXCR-1-mediated chemotaxis. Exp Cell Res. 2004;299:315–324. doi: 10.1016/j.yexcr.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Z, Cai Y, Chen H, Chen Z, Zhu D, Zhong Q, Xie W. CXCL13/CXCR5 axis predicts poor prognosis and promotes progression through PI3K/AKT/mTOR pathway in clear cell renal cell carcinoma. Front Oncol. 2018;8:682. doi: 10.3389/fonc.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guinan P, Sobin LH, Algaba F, Badellino F, Kameyama S, MacLennan G, Novick A. TNM staging of renal cell carcinoma: Workgroup no. 3. Union international contre le cancer (UICC) and the American joint committee on cancer (AJCC) Cancer. 1997;80:992–993. doi: 10.1002/(SICI)1097-0142(19970901)80:5<992::AID-CNCR26>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 24.Sobin LH, Fleming ID. TNM classification of malignant tumors, fifth edition (1997). Union internationale contre le cancer and the American joint committee on cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(SICI)1097-0142(19971101)80:9<1803::AID-CNCR16>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Dong JS, Wu B, Jiang B. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of renal cell cancer cells by downregulating CDKN1A. Eur Rev Med Pharmacol Sci. 2019;23:10241–10247. doi: 10.26355/eurrev_201912_19661. [DOI] [PubMed] [Google Scholar]
- 27.Yue G, Chen C, Bai L, Wang G, Huang Y, Wang Y, Cui H, Xiao Y. Knockdown of long noncoding RNA DLEU1 suppresses the progression of renal cell carcinoma by downregulating the Akt pathway. Mol Med Rep. 2019;20:4551–4557. doi: 10.3892/mmr.2019.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Sun DM, Yu JF, Zhang M, Yi C, Yang R, Dan BH, Li AJ. Long noncoding RNA TUG1 promotes renal cell carcinoma cell proliferation, migration and invasion by downregulating microRNA196a. Mol Med Rep. 2018;18:5791–5798. doi: 10.3892/mmr.2018.9608. [DOI] [PubMed] [Google Scholar]
- 29.Shi J, Zhang D, Zhong Z, Zhang W. lncRNA ROR promotes the progression of renal cell carcinoma through the miR-206/VEGF axis. Mol Med Rep. 2019;20:3782–3792. doi: 10.3892/mmr.2019.10636. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yu C, Zhang Y. Characterization of the prognostic values of CXCR family in gastric cancer. Cytokine. 2019;123:154785. doi: 10.1016/j.cyto.2019.154785. [DOI] [PubMed] [Google Scholar]
- 31.Saintigny P, Massarelli E, Lin S, Ahn YH, Chen Y, Goswami S, Erez B, O'Reilly MS, Liu D, Lee JJ, et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73:571–582. doi: 10.1158/0008-5472.CAN-12-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun KH, Sun GH, Wu YC, Ko BJ, Hsu HT, Wu ST. TNF-α augments CXCR2 and CXCR3 to promote progression of renal cell carcinoma. J Cell Mol Med. 2016;20:2020–2028. doi: 10.1111/jcmm.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.