Abstract
The nematode Caenorhabditis elegans produces a broad family of pheromones, known as the ascarosides, that are modified with a variety of groups derived from primary metabolism. These modifications are essential for the diverse activities of the ascarosides in development and various behaviors, including attraction, aggregation, avoidance, and foraging. The mechanism by which these different groups are added to the ascarosides is poorly understood. Here, we identify a family of over thirty enzymes, which are homologous to mammalian carboxylic ester hydrolase (CES) enzymes, and show that a number of these enzymes are responsible for the selective addition of specific modifications to the ascarosides. Through stable isotope feeding experiments, we demonstrate the in vivo activity of the CES-like enzymes and provide direct evidence that the acyl-CoA synthetase ACS-7, which was previously implicated in the attachment of certain modifications to the ascarosides in C. elegans, instead activates the side chain of certain ascarosides for shortening through β-oxidation. Our data provide a key to the combinatorial logic that gives rise to different modified ascarosides, which should greatly facilitate the exploration of the specific biological functions of these pheromones in the worm.
C. elegans uses the ascaroside family of pheromones to induce development of the stress-resistant dauer larval stage and to communicate with other worms in order to coordinate various behaviors.1 All of the ascarosides contain a core structure of an ascarylose sugar attached to a fatty acid-derived side chain that can be of various lengths and that often terminates with a carboxylic acid (Fig. 1A).2–4 The ascarosides can be decorated with a large number of additional groups on the sugar (“head groups”) or on the side chain (“terminus groups”). These modifications come from a variety of primary metabolic pathways, including amino acid, carbohydrate, and nucleoside pathways, and have dramatic consequences for the specific biological activities of the ascarosides.5–8 For example, ascarosides modified on the 4′-position with an indole-3-carbonyl (IC) group induce aggregation and can promote dauer development, while ascarosides modified with an (E)-2-methyl-2-butenoyl (MB) group or octopamine succinyl (OS) group induce avoidance (Fig. 1A).5–6, 9–10 The ascaroside pheromones are biosynthesized from ascarosides with long side chains that are shortened through β-oxidation cycles (center of Fig. 1B).11–15 However, the integration of β-oxidation cycles with the attachment of various modifications, such that only ascarosides with certain side-chain lengths are modified with certain modifications, is not well understood.
Figure 1.
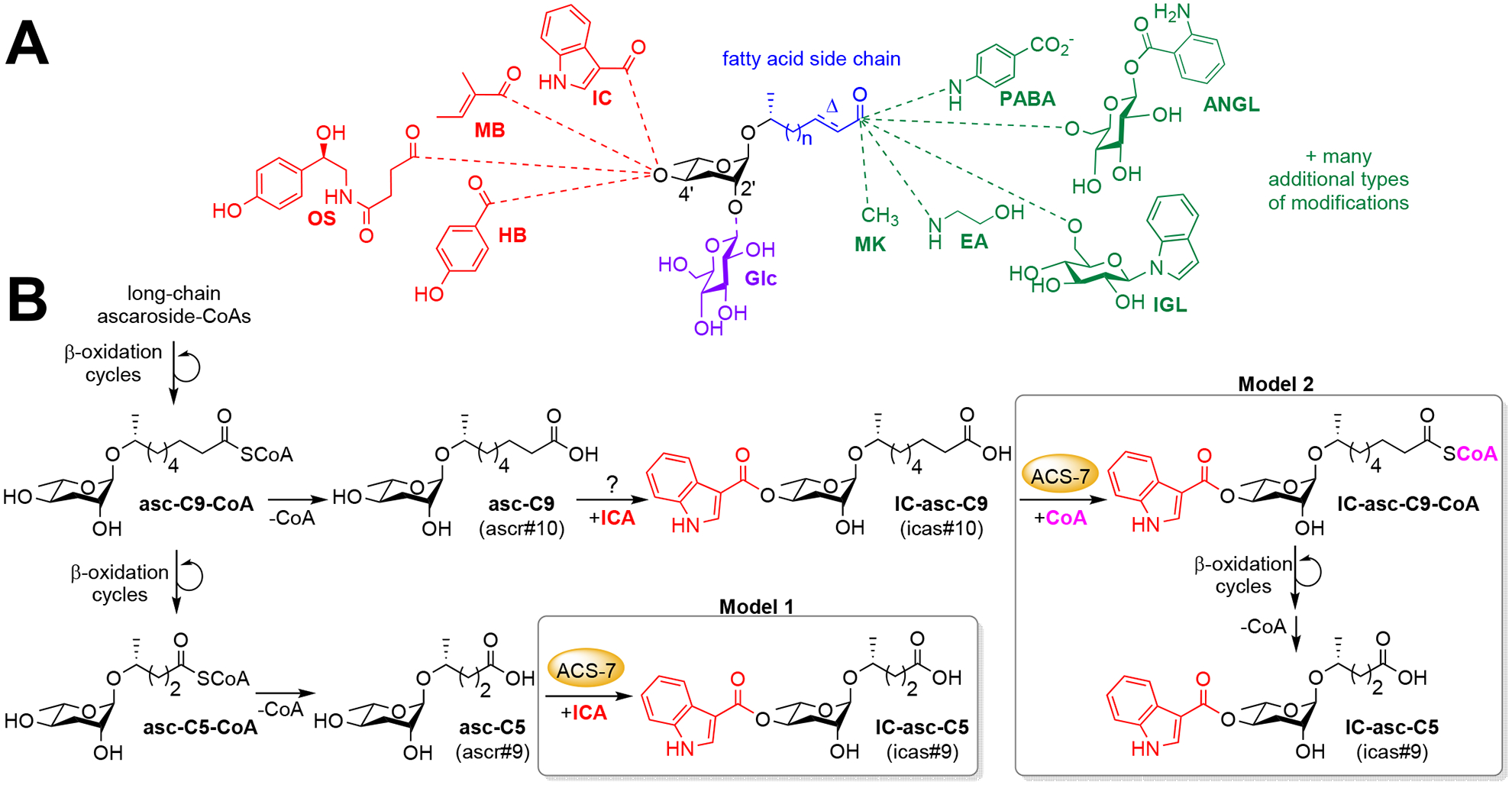
General structure of the modified ascarosides and proposed biosynthetic models. (A) Ascarosides can be modified with head groups on the 4′-position, including IC, MB, OS, and HB (4-hydroxybenzoyl), and 2′-position, including Glc (glucosyl), as well as terminus groups at the end of the side chain, including MK (methyl ketone), EA (ethanolamide), IGL (indole glucosyl), ANGL (anthranilic acid glucosyl), and PABA (para-aminobenzoic acid). Ascarosides are named using a structure-based nomenclature, (head group)-asc-(ω)(Δ)C#-(terminus group). (B) Two alternative models for the attachment of the IC group to ascarosides and the role of ACS-7 have been proposed. Analogous models have been proposed for the attachment of the OS group to the ascarosides.
The acyl-CoA synthetase ACS-7 was shown to be required for the production of IC- and OS-modified ascarosides with short (5-carbon) side chains (e.g., IC-asc-C5), but not those with longer side chains (e.g., IC-asc-C9).16 Based on these data, a model was proposed in which ACS-7 activates indole-3-carboxylic acid (ICA) and octopamine succinic acid as the corresponding CoA thioesters for attachment to ascarosides with short side chains, but not those with longer side chains (Model 1 in Fig. 1B).16 However, biochemical studies were not able to confirm this activity.16 Furthermore, later biochemical studies demonstrated that ACS-7’s preferred substrates are the side chains of ascarosides, specifically IC-modified ascarosides with longer side chains (e.g., IC-asc-C9).17 Additionally, β-oxidation was shown to influence the balance between IC-ascarosides with 9-carbon side chains and those with 5-carbon side chains, suggesting that ACS-7’s role is to activate IC-ascarosides with medium-length side chains for side-chain shortening through β-oxidation (Model 2 in Fig. 1B), thereby generating an unfavorable, dauer-inducing signal (IC-asc-C5).5, 17
More recently, in the nematode Pristionchus pacificus, a completely different type of enzyme was shown to be required for the attachment of an 2-ureidoisobutyryl head group to the 4′-position of the ascarylose ring of ascarosides, irrespective of the length of the ascaroside side chain.18 This enzyme (Ppa-UAR-1) is homologous to the CES enzymes, which are primarily known for their ability to hydrolyze a broad range of xenobiotics and endogenous lipid esters.19 However, CES enzymes, as well as other types of esterases, have also been shown in certain cases to both hydrolyze and form esters.20–21 Although Ppa-UAR-1 has not yet been investigated biochemically, it has been hypothesized to take a CoA-activated substrate and attach it to the 4′-position of the ascarylose sugar.18
To search for enzymes that might attach modifications to the ascarosides, we used BLAST to identify homologs of the Ppa-UAR-1 enzyme from P. pacificus in C. elegans. This search revealed a family of 32 CES-like enzymes (Fig. 2A). These enzymes are characterized by a conserved Glu-His-Ser catalytic triad, including a GXSXG motif that contains the catalytic Ser and an HGGG/HGGA motif that serves as an oxyanion hole (Fig. S1). To test whether the CES-like enzymes are involved in the attachment of modifications to the ascarosides, we used LC-MS to analyze ascaroside production by worm strains containing mutations in 13 of the family members, focusing on those modified ascarosides that are the most abundant (i.e., those modified with IC, MB, and Glc groups) (Fig. 1A, Fig. S2). We also screened four additional genes by knocking them down using RNA interference (Fig. S3). Two of the mutant strains, cest-3 and cest-9.2, showed virtually no production of IC- and MB-modified ascarosides, respectively (Fig. S2). We confirmed this result by profiling a larger number of ascarosides using HR-LC-MS (Fig. 2B, Fig. S4, Fig. S5). Because the cest-3 strain contains a deletion of both cest-3 and a neighboring gene, we verified that deletion of cest-3 was responsible for the defect in IC-ascaroside production by rescuing production through transgenic expression of the cest-3 gene (Fig. S6). This rescue strain showed expression of CEST-3 in the intestine, which is a major site of ascaroside biosynthesis (Fig. S7). Unfortunately, our efforts to heterologously express cest-3 and cest-9.2 as either full-length or truncated proteins lacking their predicted transmembrane domains were unsuccessful, and thus, we were not able to biochemically characterize these enzymes (Supplementary Methods). To provide further evidence for CEST-3’s role in IC group attachment, we used CRISPR-Cas9 to generate a worm strain in which the Ser residue in the catalytic triad of CEST-3 was mutated to an Ala residue (Fig. S1). This strain was unable to produce the IC-ascarosides while producing normal amounts of other ascarosides (Fig. 2C, Fig. S8). Strikingly, this strain, unlike wild-type worms, does not localize to a lawn of bacterial food (Fig. S9). This result is consistent with the role of the IC-ascarosides in promoting attraction, promoting aggregation on food, and suppressing foraging.9, 22
Figure 2.
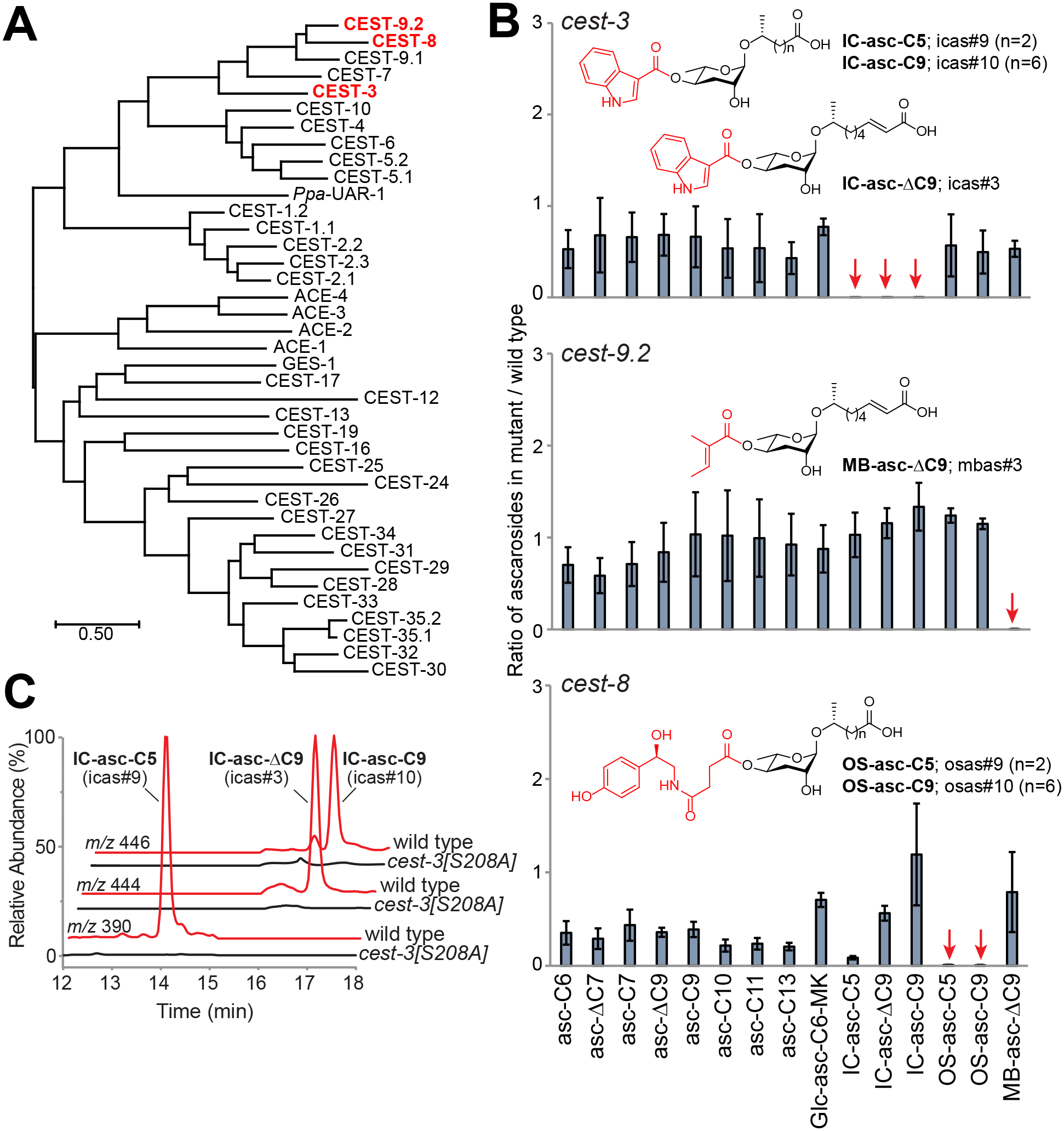
Role of CES-like enzymes in attaching different modifications to the ascarosides. (A) Phylogenetic tree of CES-like enzymes generated with MEGA X.24 Enzymes that have been shown here to attach specific modifications are highlighted in red. Acetylcholinesterases ACE-1 through −4 are included for comparison purposes. (B) Abundance of ascarosides produced by cest-3(tm8654), cest-9.2(gk952063), and cest-8(gk952062) worms relative to wild type, as determined by HR-LC-MS. Data represent the mean of three experiments ± standard deviation. (C) LC-MS extracted ion chromatogram of IC-ascarosides produced by wild-type and cest-3(reb30[S208A]) worms. Abundance of all ascarosides produced by the mutant relative to wild type is shown in Figure S8.
Given that CEST-3 and CEST-9.2 are phylogenetically related, we hypothesized that the enzymes that attach other modifications to the 4′-position, such as those that attach the OS modification, might cluster in a similar region of the phylogenetic tree. OS-modified ascarosides are abundantly produced by starved, early larval stage worms.6 However, using HR-LC-MS we were able to detect the OS-modified ascarosides in mixed-stage cultures and screen the cest-3, cest-7, cest-8, cest-9.1, and cest-9.2 mutant strains for the ability to make these compounds. A cest-8 mutant strain was specifically unable to make the OS-modified ascarosides (Fig. 2B).
Previously, it was hypothesized that ACS-7 attaches the IC and OS groups to short-chain ascarosides (Model 1 in Fig. 1B),16 but our data would suggest CEST-3 and CEST-8 are required for attaching the IC- and OS-groups, respectively, to medium-chain ascarosides and that the side chains of those ascarosides are then activated by ACS-7 for β-oxidation to make the short-chain IC- and OS-ascarosides (Model 2 in Fig. 1B). Previously, we showed that daf-22 mutant worms, which are defective in the final step in the β-oxidation cycles that shorten the side chains of the ascarosides and thus do not make any short- or medium-chain ascarosides, will only attach the IC group to exogenously added ascarosides with medium-length (C8-C11) side chains.11, 17 To provide direct evidence for Model 2, we synthesized a deuterium-labeled version of an ascaroside with a 9-carbon side chain, d2-asc-C9 (Supplementary Methods; Fig. S10–S11) and then cultured wild-type worms with this ascaroside (Fig. 3A). By stable isotope labeling the ascaroside, we could follow how the ascaroside is processed inside the worm independently of endogenous ascarosides, and thus, we could perform these studies in wild-type worms, rather than daf-22 worms, such that the β-oxidation process is still intact (Fig. 3A). Importantly, large amounts of deuterium-labeled IC-ascarosides with both 9-carbon side chains (d2-IC-asc-C9) and 5-carbon side chains (d2-IC-asc-C5) were detected (Fig. 3B), consistent with our model (Model 2 in Fig. 1B). Addition of ICA to the cultures, which we had previously shown increases the production of IC-ascarosides, enhanced this effect.23 Meanwhile, the d2-asc-C9 itself is not activated as a CoA-thioester to a significant degree for shortening through β-oxidation because only minor amounts of non-IC-ascarosides with shorter side chains were detected (Fig. 3A,B). These results also agree with our previous biochemical experiments showing that ACS-7 prefers to activate the side chains of IC-ascarosides with medium-length side chains, not the side chains of unmodified (non-IC) ascarosides.17 Thus, our data strongly argue that medium-chain IC-ascarosides (e.g., IC-asc-C9) can be converted to short-chain IC-ascarosides (e.g., IC-asc-C5) through activation followed by β-oxidation.
Figure 3.
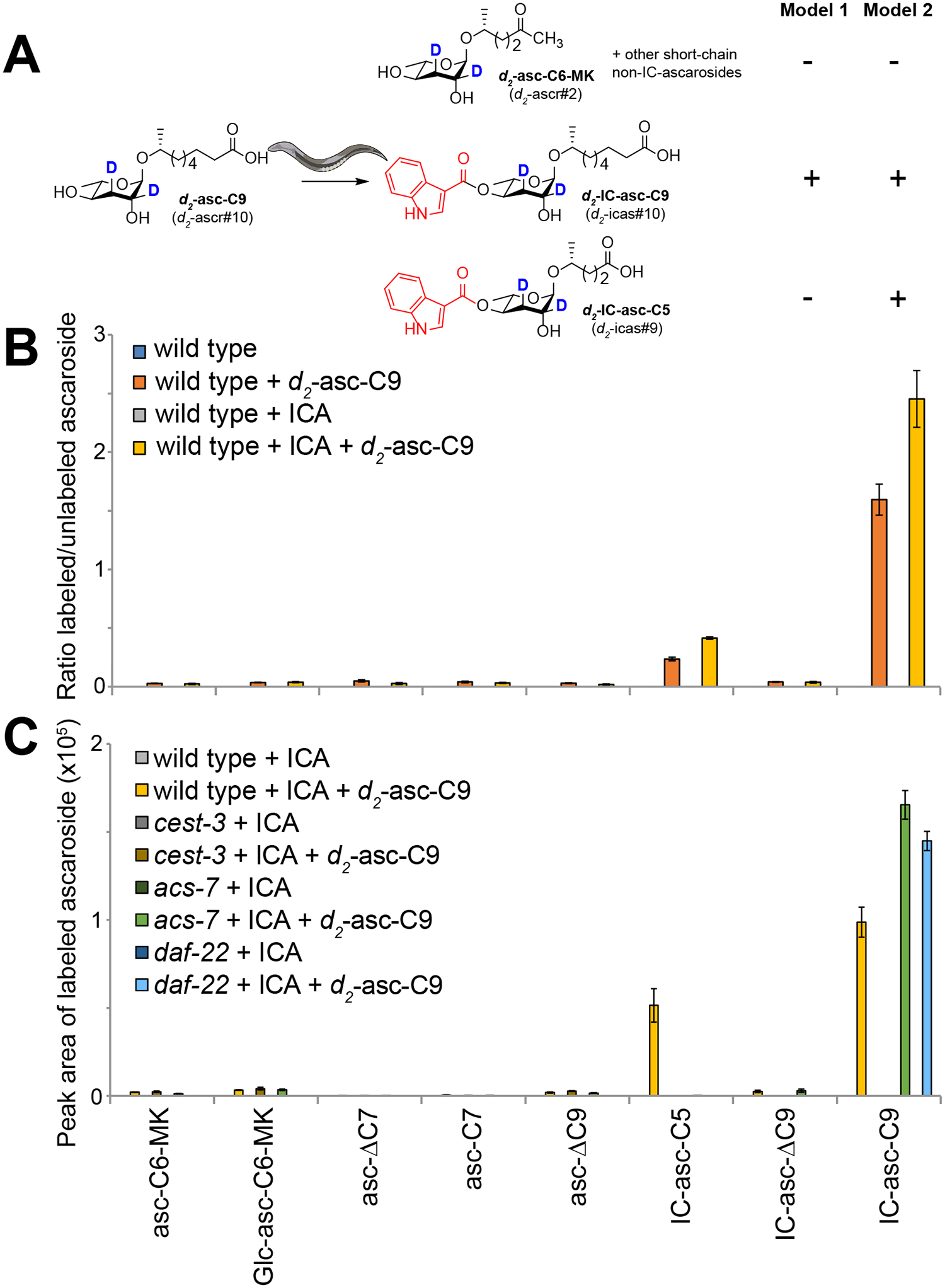
Processing of the deuterium-labeled ascaroside, d2-asc-C9, by wild-type and mutant worms. (A) The deuterium-labeled ascarosides that are expected to be produced when wild-type worms are fed d2-asc-C9, according to either Model 1 or Model 2. (B) Wild-type worms were grown with either vehicle or d2-asc-C9 in the absence or presence of ICA, and the amounts of deuterated and nondeuterated ascarosides in the culture medium were analyzed. (C) Wild-type, cest3(tm8654), acs-7(tm6781), and daf-22(m130) worms were grown with either vehicle or d2-asc-C9 in the presence of ICA, and the amounts of deuterated ascarosides in the culture medium were analyzed. Data represent the mean of three experiments ± standard deviation.
To demonstrate that CEST-3 is required for the production of the IC-ascarosides, we cultured cest-3 mutant worms in the presence of d2-asc-C9 and showed that these worms could not produce any deuterium-labeled IC-ascarosides, even though they were supplied with excess ICA (Fig. 3C). To confirm that ACS-7 and DAF-22 are required for the conversion of d2-IC-asc-C9 to d2-IC-asc-C5, we cultured acs-7 and daf-22 mutant worms in the presence of d2-asc-C9. In both cultures, the mutant worms could make d2-IC-asc-C9, but could not make d2-IC-asc-C5 (Fig. 3C). Thus, once the IC group is attached to an ascaroside with a 9-carbon side chain (to generate d2-IC-asc-C9), the side chain requires ACS-7 for activation as a CoA-thioester and DAF-22 for shortening through β-oxidation (to generate d2-IC-asc-C5). Interestingly, however, in wild-type worms, we did not see the production of the α-β−unsaturated d2-IC-asc-ΔC9 (Fig. 3B–C). This result suggests that the biosynthesis of the important aggregation pheromone IC-asc-ΔC9 (icas#3) occurs primarily by CEST-3 adding an IC group to the corresponding unmodified ascaroside, asc-ΔC9 (ascr#3) (as shown in Fig. 4), not by CEST-3 adding an IC group to asc-C9 (ascr#10), followed by activation of the side chain and α-β-unsaturation in the first step of β-oxidation.
Figure 4.
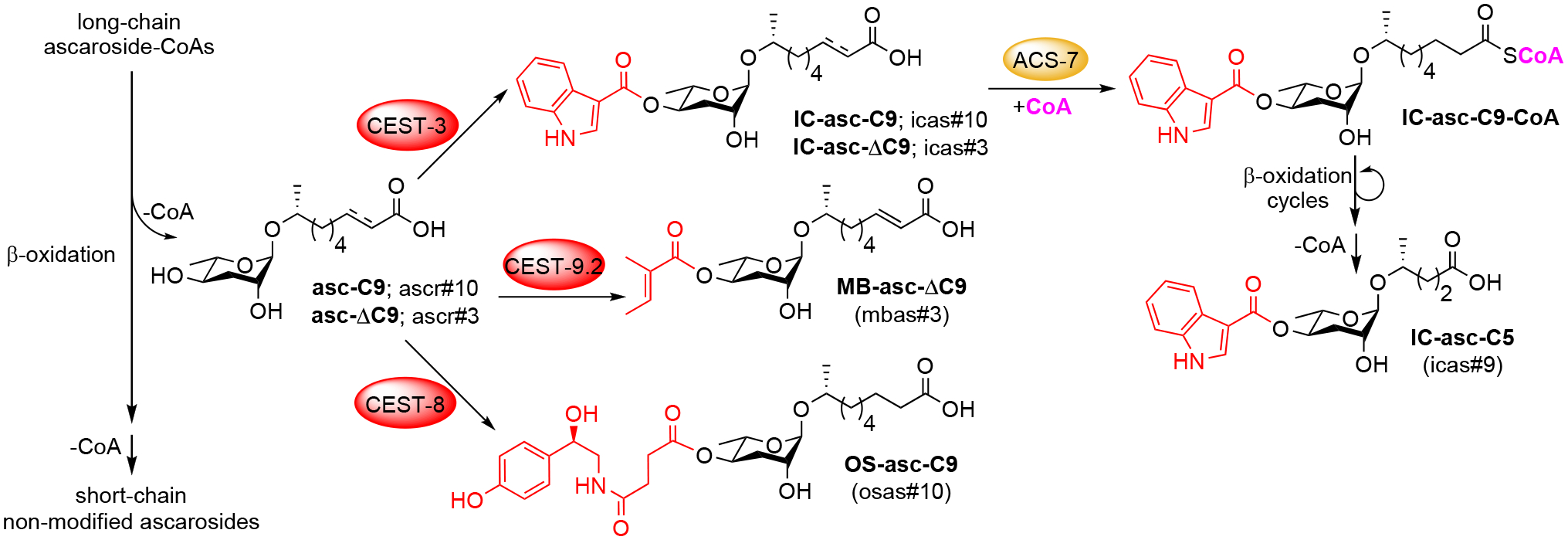
Revised model for the biosynthesis of modified ascarosides. CEST-3 likely prefers asc-C9 and asc-ΔC9 as substrates, CEST-9.2 prefers asc-ΔC9, and CEST-8 prefers asc-C9. In addition to activating the side chain of IC-asc-C9 for β-oxidation, ACS-7 may also activate the side chain of OS-asc-C9 for β-oxidation.
In summary, we identify a large family of CES-like enzymes in C. elegans and show that several of these enzymes are required for the biosynthesis of ascarosides modified with specific head groups (Fig. 4). Furthermore, we demonstrate using stable isotope labeling that ACS-7 is not involved in the attachment of modifications to ascarosides, but is instead required for activating the side chains of modified ascarosides for shortening through β-oxidation (Fig. 4). Although the metabolites that are used to modify the ascarosides, such as ICA and octopamine succinic acid, may need to be activated in some way (e.g., as the corresponding CoA-thioesters) in order for the CES-like enzymes to attach them to the ascarosides, ACS-7 is not involved in this process. Identification of the CES family of enzymes opens the door to uncovering the combinatorial biosynthetic pathway to different modified ascarosides, which should in turn greatly facilitate dissecting the specific biological functions of these ascarosides in the worm.
Supplementary Material
Acknowledgements
We thank Krishna Ghanta for advice on CRISPR-Cas9. HR-LC-MS/MS analysis was performed at the University of Florida Mass Spectrometry Research and Education Center, which is funded by the NIH (S10 OD021758-01A1). Some strains were provided by the National Bioresource Project (Japan) and by the Caenorhabditis Genetics Center, which is funded by NIH (P40 OD010440). This work was supported by the NIH (R01 GM118775) and the NSF (Career, 1555050).
Footnotes
Supporting Information.
All methods including synthesis methods; multiple sequence alignment of CES enzymes; ascaroside production in mutant and RNAi screens; representative chromatograms and MS-MS spectra used for ascaroside identification; cest-3 rescue experiment; imaging of cest-3 reporter strain; ascaroside production and behavior of cest-3(reb30[S208A]) strain; NMR spectra of d2-asc-C9; worm strain list.
References
- 1.Butcher RA, Decoding chemical communication in nematodes. Nat Prod Rep 2017, 34 (5), 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeong PY; Jung M; Yim YH; Kim H; Park M; Hong E; Lee W; Kim YH; Kim K; Paik YK, Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 2005, 433 (7025), 541–5. [DOI] [PubMed] [Google Scholar]
- 3.Butcher RA; Fujita M; Schroeder FC; Clardy J, Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol 2007, 3 (7), 420–2. [DOI] [PubMed] [Google Scholar]
- 4.Butcher RA; Ragains JR; Kim E; Clardy J, A potent dauer pheromone component in C. elegans that acts synergistically with other components. Proc Natl Acad Sci U S A 2008, 105, 14288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher RA; Ragains JR; Clardy J, An indole-containing dauer pheromone component with unusual dauer inhibitory activity at higher concentrations. Org Lett 2009, 11 (14), 3100–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artyukhin AB; Yim JJ; Srinivasan J; Izrayelit Y; Bose N; von Reuss SH; Jo Y; Jordan JM; Baugh LR; Cheong M; Sternberg PW; Avery L; Schroeder FC, Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J Biol Chem 2013, 288 (26), 18778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pungaliya C; Srinivasan J; Fox BW; Malik RU; Ludewig AH; Sternberg PW; Schroeder FC, A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2009, 106 (19), 7708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artyukhin AB; Zhang YK; Akagi AE; Panda O; Sternberg PW; Schroeder FC, Metabolomic “dark matter” dependent on peroxisomal β-oxidation in Caenorhabditis elegans. J Am Chem Soc 2018, 140 (8), 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan J; von Reuss SH; Bose N; Zaslaver A; Mahanti P; Ho MC; O’Doherty OG; Edison AS; Sternberg PW; Schroeder FC, A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol 2012, 10 (1), e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YK; Sanchez-Ayala MA; Sternberg PW; Srinivasan J; Schroeder FC, Improved synthesis for modular ascarosides uncovers biological activity. Org Lett 2017, 19 (11), 2837–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher RA; Ragains JR; Li W; Ruvkun G; Clardy J; Mak HY, Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A 2009, 106 (6), 1875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joo HJ; Kim KY; Yim YH; Jin YX; Kim H; Kim MY; Paik YK, Contribution of the peroxisomal acox gene to the dynamic balance of daumone production in Caenorhabditis elegans. J Biol Chem 2010, 285 (38), 29319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Reuss SH; Bose N; Srinivasan J; Yim JJ; Judkins JC; Sternberg PW; Schroeder FC, Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J Am Chem Soc 2012, 134 (3), 1817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X; Feng L; Chinta S; Singh P; Wang Y; Nunnery JK; Butcher RA, Acyl-CoA oxidase complexes control the chemical message produced by Caenorhabditis elegans. Proc Natl Acad Sci U S A 2015, 112 (13), 3955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X; Wang Y; Perez DH; Jones Lipinski RA; Butcher RA, Acyl-CoA oxidases fine-tune the production of ascaroside pheromones with specific side chain lengths. ACS Chem Biol 2018, 13 (4), 1048–1056. [DOI] [PubMed] [Google Scholar]
- 16.Panda O; Akagi AE; Artyukhin AB; Judkins JC; Le HH; Mahanti P; Cohen SM; Sternberg PW; Schroeder FC, Biosynthesis of modular ascarosides in C. elegans. Angew Chem Int Ed Engl 2017, 56 (17), 4729–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y; Wang Y; Zhang X; Bhar S; Jones Lipinski RA; Han J; Feng L; Butcher RA, Biosynthetic tailoring of existing ascaroside pheromones alters their biological function in C. elegans. Elife 2018, 7, 10.7554/eLife.33286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcke JM; Bose N; Artyukhin AB; Rodelsperger C; Markov GV; Yim JJ; Grimm D; Claassen MH; Panda O; Baccile JA; Zhang YK; Le HH; Jolic D; Schroeder FC; Sommer RJ, Linking genomic and metabolomic natural variation uncovers nematode pheromone biosynthesis. Cell Chem Biol 2018, 25 (6), 787–796 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian J; Nelson R; Lehner R, Carboxylesterases in lipid metabolism: from mouse to human. Protein Cell 2018, 9 (2), 178–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saerens SM; Verstrepen KJ; Van Laere SD; Voet AR; Van Dijck P; Delvaux FR; Thevelein JM, The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem 2006, 281 (7), 4446–56. [DOI] [PubMed] [Google Scholar]
- 21.Brzezinski MR; Abraham TL; Stone CL; Dean RA; Bosron WF, Purification and characterization of a human liver cocaine carboxylesterase that catalyzes the production of benzoylecgonine and the formation of cocaethylene from alcohol and cocaine. Biochem Pharmacol 1994, 48 (9), 1747–55. [DOI] [PubMed] [Google Scholar]
- 22.Greene JS; Brown M; Dobosiewicz M; Ishida IG; Macosko EZ; Zhang X; Butcher RA; Cline DJ; McGrath PT; Bargmann CI, Balancing selection shapes density-dependent foraging behaviour. Nature 2016, 539 (7628), 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y; Zhang X; Butcher RA, Tryptophan metabolism in Caenorhabditis elegans links aggregation behavior to nutritional status. ACS Chem Biol 2019, 14 (1), 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S; Stecher G; Li M; Knyaz C; Tamura K, MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35 (6), 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


