Abstract
Anthropogenic fecal pollution in urban waterbodies can promote the spread of waterborne disease. The objective of this study was to test crAssphage, a novel viral human fecal marker not previously applied for fecal source tracking in Latin America, as a fecal pollution marker in an urban river in Chile. Human fecal markers crAssphage CPQ_064 and Bacteroides HF183, the human pathogen norovirus GII, and culturable fecal indicator bacteria (FIB) were quantified at six locations spanning reaches of the Mapocho River from upstream to downstream of Santiago, as well as in repeated sub-daily frequency samples at two urban locations. Norovirus showed positive correlation trends with crAssphage (τ = 0.57, p = 0.06) and HF183 (τ = 0.64, p = 0.03) in river water, but not with E. coli or enterococci. CrAssphage and HF183 concentrations were strongly linearly related (slope = 0.97, p < 0.001). Chlorinated wastewater effluent was an important source of norovirus GII genes to the Mapocho. Precipitation showed non-significant positive relationships with human and general fecal indicators. Concentrations of crAssphage and HF183 in untreated sewage were 8.35 and 8.07 log10 copy/100 ml, respectively. Preliminary specificity testing did not detect crAssphage or HF183 in bird or dog feces, which are predominant non-human fecal sources in the urban Mapocho watershed. This study is the first to test crAssphage for microbial source tracking in Latin America, provides insight into fecal pollution dynamics in a highly engineered natural system, and indicates river reaches where exposure to human fecal pollution may pose a public health risk.
Keywords: Norovirus, Microbial source tracking, Fecal pollution, crAssphage, Water quality, Urban river
Graphical abstract
Highlights
-
•
First application of crAssphage for microbial source tracking in Latin America.
-
•
Treated wastewater was an important source of norovirus genes to the Mapocho river.
-
•
Norovirus was tracked by human fecal markers rather than fecal indicator bacteria.
-
•
Fecal indicator bacteria were poor indicators of viral waterborne illness risk.
1. Introduction
Rivers are the primary source of renewable freshwater for humans and ecosystems globally, yet they face many anthropogenic threats, including extraction and pollution (Vörösmarty et al., 2010). In particular, urbanization around rivers tends to degrade water quality, increasing potential for the spread of waterborne disease (Meybeck, 2003). Waterborne disease risks are most commonly assessed by measuring concentrations of fecal indicator bacteria (FIB), such as E. coli or enterococci, which are recommended for monitoring fecal pollution globally (World Health Organization, 2011). While monitoring FIB has many benefits, FIB are markers of general fecal pollution and do not indicate specific animal sources of feces, which differ in their human health risks (Soller et al., 2010b). As bacteria, FIB may also have limited utility for indicating the presence of viruses (Harwood et al., 2013), which are thought to be predominant etiologies of recreational waterborne illness (Soller et al., 2010a, Soller et al., 2017).
To improve detection of health-relevant microbial pollution, methods to assess human fecal pollution have been developed, primarily using genetic microbial source tracking (MST) markers. Among the highest performing MST markers are genetic targets from human-associated organisms, which are quantified by quantitative polymerase chain reaction (qPCR) (Ahmed et al., 2016a, Ahmed et al., 2016b; Boehm et al., 2013; Mayer et al., 2018). Recently, MST methods were developed (Stachler et al., 2017) for a highly abundant human-gut-associated bacteriophage called crAssphage (Dutilh et al., 2014). As a virus, crAssphage could serve as a superior indicator of human viral pollution compared to bacterial indicators, though little data on the co-occurrence of crAssphage and human viral pathogens currently exists (Farkas et al., 2018). CrAssphage MST markers have been tested in the USA (Ahmed et al., 2018a; Stachler et al., 2018), Nepal (Malla et al., 2019), Thailand (Kongprajug et al., 2019), Australia (Ahmed et al., 2018b), the UK (Farkas et al., 2019), Spain (García-Aljaro et al., 2017), and Japan (Malla et al., 2019), but not in any Latin American countries. Although crAssphage and high-performing human bacterial markers, such as Bacteroides HF183, Bacteroidales BacHum, and Firmicutes Lachno2, have been shown to be quite specific to human feces in most geographic contexts tested thus far, cross-reaction with non-human fecal sources has been reported (Ahmed et al., 2018a; Ahmed et al., 2016a, Ahmed et al., 2016b; Boehm et al., 2013; Kildare et al., 2007; Malla et al., 2019; Mayer et al., 2018; Stachler et al., 2017). Thus, cross-reaction with locally relevant fecal sources should be examined prior to application in new regions (Stachler et al., 2017). CrAssphage MST marker abundance in sewage should also be examined to assess its local sensitivity for detecting sewage pollution because although crAssphage sequences in wastewater metagenomes have been found to be globally ubiquitous, there is also tremendous geographic sequence diversity (Edwards et al., 2019), and evidence of strong geographic dependence of crAssphage abundance in sewage (Stachler and Bibby, 2014).
While measuring MST markers is useful for assessing fecal contamination, directly measuring human pathogens informs the understanding of specific health risks posed by water contact or consumption. Epidemiological and microbial risk assessment modeling indicate that human norovirus is a predominant etiology of waterborne illness. Norovirus is a leading cause of gastroenteritis outbreaks globally, and primary outbreak cases are often linked to food or water exposure (Patel et al., 2009). Available data indicate that norovirus has been a leading cause of acute gastroenteritis in Chile since at least 2000. A study conducted in Santiago between 2000 and 2003 found that 45% of gastroenteritis outbreaks investigated were caused by human caliciviruses, a family of viruses to which norovirus belongs, with the most common etiologic agent being norovirus GII (Vidal et al., 2005). A 2008 study among hospitalized children with diarrhea in Concepción, Chile found that norovirus prevalence was higher (25.5%) than that of other tested viruses, rotavirus and adenovirus (Montenegro et al., 2014). In 2010, an outbreak led to 31,036 reported cases of norovirus GII infection in the Antofagasta region. Primary cases in this outbreak were attributed to consumption of raw vegetables irrigated with treated wastewater containing low levels of residual free chlorine (Díaz et al., 2012). From 2013 to 2017, norovirus GII was the most prevalent enteric virus in outbreak, diarrhea monitoring, and shellfish samples collected by the Chilean epidemiologic monitoring network (Instituto de Salud Pública de Chile, 2018). Quantitative microbial risk assessment (QMRA) models of US waters also consistently indicate that norovirus drives health risk in recreational waters polluted with sewage (Boehm et al., 2015; Wyn-Jones et al., 2011) and stormwater runoff (Soller et al., 2017). Thus, accurately assessing norovirus contamination is key to understanding risks to people who contact urban environmental waters.
The objective of this study was to test crAssphage for assessing fecal pollution in an urban river in Chile. This is the first study to apply human Bacteroides HF183 and crAssphage MST markers in Chile, and it adds new data on the co-occurrence of crAssphage and norovirus. Overall, this research extends knowledge of the utility of crAssphage in a new geographic context, provides insight into fecal pollution dynamics in a highly engineered natural system, and indicates river reaches where exposure to human fecal pollution may pose a public health risk.
2. Methods
2.1. Research questions
The specific research questions (RQ) investigated were: (RQ1) Do crAssphage and HF183 indicate human fecal pollution in the Mapocho River? Low concentrations of these markers in untreated wastewater or detection in non-human feces could suggest that these markers are poor indicators of human fecal contamination. (RQ2) Is crAssphage correlated with norovirus in Mapocho River water samples? To contextualize this correlation, correlations of norovirus with HF183 and culturable E. coli and enterococci were calculated as well. Norovirus is expected to be most correlated with crAssphage, since both are PCR-based measurements of viruses. (RQ3) Are crAssphage and HF183 correlated in Mapocho River water samples? Lack of correlation could indicate differential sources or environmental fate and transport, possibly caused by differences in viral and bacterial physiology. Finally, (RQ4) how do concentrations of fecal indicators vary with as a function of water turbidity, precipitation, and time of day? Correlation with these factors can provide insight into human and environmental processes governing fecal pollution in the Mapocho River.
2.2. Site description and sampling campaigns
Samples were collected from the Mapocho River in the greater metropolitan area of Santiago, Chile (33.440°W, 70.649°S), which has an estimated population of 7.1 million inhabitants (Instituto Nacional de Estadísticas de Chile, 2017). The Mapocho flows from its headwaters in the Andes Mountains east of Santiago, through the city, and then joins the Maipo River west of Santiago before discharging into the Pacific Ocean. Upstream of Santiago, the Mapocho’s catchments comprise 854 km2 of mountainous, undeveloped land, classified as 23% grassland, 23% shrubland, 51% barren, and <1% each cropland and impervious (Figure S1) (Alvarez-Garreton et al., 2018). The Mapocho system within and to the west of Santiago, however, is highly engineered: its urban reaches are channelized, large fractions of its flow are extracted for irrigation, and it receives large volumes of wastewater treatment plant effluent downstream of the city. It also receives input from the Maipo River via the San Carlos canal in central Santiago (indicated in Fig. 1), although this canal was closed for maintenance from May 18 through June 2, 2019. Thus, the Mapocho did not receive input from the San Carlos canal during the spatial sampling campaign described below. Santiago has separate municipal stormwater and sewage collection systems, and stormwater is discharged to the Mapocho River as well.
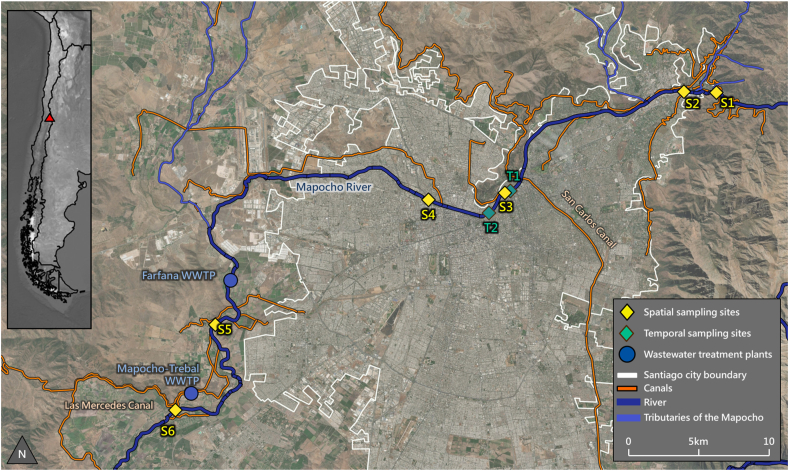
Fig. 1.
Map of sampling locations. S1 to S6 are spatial campaign sampling locations. T1 and T2 are temporal campaign sampling locations. Both wastewater treatment plants (WWTP) discharge into the Mapocho. The San Carlos Canal discharges a variable portion of its flow to the Mapocho; all other canals indicated extract water from the Mapocho. River geometry is from Digital Chart of the World (Digital Chart of the World, n.d.). Channel geometry is from the Chilean Ministry of Agriculture (Chilean Ministry of Agriculture, n.d.). City boundary geometry is from the Chilean Ministry of National Goods (Chilean Ministry of National Goods, n.d.). The basemap is used courtesy of ESRI.
Three sampling campaigns were undertaken to assess spatial and temporal water quality variation. The spatial campaign comprised 6 sampling sites along the length of the Mapocho River (Fig. 1), referred to from upstream to downstream as sites S1–S6 (detailed description in Table 1). Spatial sampling was conducted on 4 days during Fall 2019: May 22, 23, 25, and 26. S1 is located upstream of the city, in an unchannelized reach surrounded by low-density rural housing. S2–S4 are located in urban areas where the Mapocho is channelized. Much of the river’s flow is extracted for irrigation prior to reaching S2, and the San Carlos Canal replenishes the Mapocho’s flow by transferring water from the Maipo River to the Mapocho downstream of S2 but upstream of S3. S5 and S6 are located in agricultural areas on the western edge of Santiago, where the Mapocho is unchanellized. S5 receives chlorinated WWTP effluent from the Farfana WWTP, which performs activated sludge treatment and clarification before chlorination and discharge (Suez, 2020). S6 receives WWTP effluent from the Mapocho-Trebal WWTP, which also performs activated sludge treatment and secondary clarification prior to chlorination and discharge (Suez, 2020). The Farfana and Mapocho-Trebal plants each have annual median treatment capacity of 8.8 m3/s of raw sewage, a population equivalent of 3.7 million people (Suez, 2020, Suez, 2020).
Table 1.
Sampling site description. The column Inputs indicates inputs that are upstream of the corresponding site and downstream of the preceding site.
| Sampling campaign | Site name | Landmark | Coordinates (lat, long) | Description | Channelized | Inputs |
|---|---|---|---|---|---|---|
| Spatial | S1 | La Mancha School | −33.363904, −70.470417 | Upstream of city, residential | No | Headwaters |
| S2 | Las Condes Avenue | −33.362732, −70.496007 | Urban | Yes | Tributary | |
| S3 | Suecia Bridge | −33.418796, −70.610130 | Urban | Yes | San Carlos Canal, tributary | |
| S4 | Bulnes Bridge | −33.426565, −70.671327 | Urban | Yes | NA | |
| S5 | Rinconada-Maipu Bridge | −33.498992, −70.819940 | Downstream of city, agricultural | No | WWTP La Farfana, tributary | |
| S6 | Esperanza Bridge | −33.548990, −70.848974 | Downstream of city, agricultural | No | WWTP El Mapocho-Trebal | |
| Temporal (hourly & sub-daily) | T1 | Concepción Bridge | −33.422460, −70.618211 | Urban | Yes | San Carlos Canal, tributary |
| T2 | Los Candados Bridge | −33.434058, −70.629419 | Urban | Yes | NA |
Temporal sampling was split between two campaigns: a sub-daily campaign and an hourly campaign. The sub-daily campaign comprised two sampling sites, T1 and T2 from upstream to downstream, both located just downstream of S3. These sites were sampled at 7:30 and 13:30 h daily for 15 days in 2019 between May 22 and June 5. The hourly campaign was conducted at site T2 on a single day; hourly samples were collected from 7:30 to 16:30 on June 6, 2019.
2.3. Sample collection and processing
500 mL samples were collected in 1 L polycarbonate bottles, which were cleaned with isopropyl alcohol before sampling. Sample bottles were triple rinsed with sample water before transferring a final sample to a sterile, disposable Whirl-Pak bag. Samples were collected mid-river at the surface. Samples processed for FIB were stored in the dark at 4 °C for <6 h until lab processing. Samples from the spatial campaign were stored at 4 °C for up to 9 h prior to processing for molecular analysis. River temperature was measured in situ using a handheld probe (YSI-30, YSI, Xylem, Rye Brook, NY). Turbidity was measured using a bench top turbidity meter (HF Scientific DRT-15CE, Fort Myers, FL). E. coli and enterococci were quantified by defined substrate fluorescence assay (Colilert-18 and Enterolert, respectively, IDEXX, Westbrook, ME), per the manufacturer’s instructions, in sample water diluted 10-fold with deionized water. The lower and upper limits of detection for the IDEXX assays were 10 and 24,192 most probable number (MPN) per 100 ml of water sample.
For molecular analysis, virus and bacteria from 100 mL of each sample were concentrated by filtering water through a 47 mm, 0.45 μm-pore size mixed cellulose ester membrane filter (MilliporeSigma, Burlington, MA) using sterile disposable filtration funnels (Nalge Nunc International, Rochester, NY). To aid virus adsorption, prior to filtration samples were augmented with MgCl2 to a final concentration of 100 mM (Lukasik et al., 2000). Following filtration, 0.5 ml of RNA stabilization solution (RNAlater, Invitrogen, Waltham, MA) were added to the filter and incubated for 5 min before being aspirated through the filter. Process blanks tested the effectiveness of the sampling bottle and filter forceps cleaning procedure; a cleaned sampling bottle was used to collect deionized water, which was then processed along with river samples. Filters were stored in cryotubes overnight at 4 °C, and then transferred to −20 °C and stored for up to 1 month. Filters were then shipped to Stanford, CA, USA and stored at −80 °C for 4 months until nucleic acid extraction. This storage time is short compared to that of other studies which stored samples as long as 12–18 months (Li et al., 2019; Shanks et al., 2010). DNA and RNA were extracted simultaneously and directly from filters using the AllPrep PowerViral DNA/RNA kit (Qiagen, Hilden, Germany), which includes bead beating and β-mercaptoethanol denaturation of RNases. According to the manufacturer, this kit is designed for both bacterial and viral nucleic acid extraction. One extraction blank was included with every 15 samples to test for contamination in reagents or during the extraction process. Extracts were divided into 30 μl aliquots, stored at −80 °C, and then thawed only once for qPCR within 2 months. Microbial recovery was not quantified in this study. However, previous work indicated that extraction recovery efficiency measured by qPCR following direct extraction of filtered MgCl2-treated surface water was approximately 7% for non-enveloped virus (MS2 coliphage) and 17% for the bacterium E. faecium (Viau et al., 2011). Recent work also extracting nucleic acids directly from MgCl2-treated samples found whole process RNA recovery, including concentration, of murine hepatitis virus (an enveloped virus) of approximately 66% (Ahmed et al., 2020).
An untreated sewage influent sample was collected from the Farfana WWTP. Sewage was allowed to settle, and 25 ml were filtered following the filtration protocol above. Wastewater nucleic acid extracts were quantified fluorometrically (Qubit 2.0, Life Technologies, Thermo Fisher Scientific, Waltham, MA) and diluted to 1 ng/μl of dsDNA. Fresh bird (n = 5) and dog (n = 5) feces were collected aseptically from concrete surfaces near the Mapocho River. Fecal samples were transported to the lab and stored at −20 °C within 1 h of collection. Nucleic acids were extracted from each fecal sample in an extraction run containing no water samples, diluted to 1 ng dsDNA/μl, and then composited in equal mass ratios to create one sample per feces type at 1 ng dsDNA/μl. Compositing or pooling samples for MST assay sensitivity and specificity is well-established in the literature (Boehm et al., 2016; Shanks et al., 2009, 2008).
2.4. qPCR assays
Primers, probes, and standard materials for all assays are given in Table S1. HF183/BacR287 (Green et al., 2014) and crAssphage CPQ_064 (Stachler et al., 2017) reactions were 25 μl, consisting of 1X Environmental Master Mix 2.0 (ABI, Thermo Fisher Scientific, Waltham, MA), 1 μM of each primer, 0.08 μM probe, 0.2 mg/ml BSA, 5 μl template, and the remainder water. Thermal cycling was performed in an ABI Step One Plus and comprised a 10-min hold at 95 °C, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 min. Standards were synthetic ssDNA ultramers (IDT, Coralville, IA). Norovirus GII ORF1-ORF2 (Loisy et al., 2005) reactions were 25 μl, consisting of 1X AgPath-ID 1-step RT-qPCR master mix (ABI, Thermo Fisher Scientific, Waltham, MA), 0.2 μM of each primer and probe, 5 μl template, and the remainder water. Thermal cycling conditions comprised 30-min at 50 °C for reverse transcription and a 10-min hold at 95 °C, followed by 45 cycles of 95 °C for 15 seconds and 60 °C for 1 min. Standard material was synthetic ssRNA spanning the ORF1-ORF2 junction (product no. VR-3235SD, ATCC, Manassas, VA).
Six-point triplicate standard curves and no-template controls (NTC) were quantified for crAssphage, HF183, and norovirus GII on 8, 7 and 3 instrument runs, respectively, including on each plate of samples. Mixed effects regression, with instrument run as a random intercept, was used to combine data across multiple runs while retaining plate-specific standard equation estimates (Sivaganesan et al., 2010). The limit of detection (LOD) was defined as the lowest concentration that can be reliably detected, i.e., at which at least 95% of standard reactions are expected to amplify. The lower limit of quantification (LLOQ) was defined as the lowest standard that can be quantified with a coefficient of variation less than 0.35 (Klymus et al., 2020). See SM for details. All samples were run in duplicate reactions. To test for PCR inhibition, for each assay at least 20% of extracts were run at 1:1 and 1:10 dilutions. Inhibition was defined as ΔCt <2.3 between 1:1 and 1:10 dilutions.
2.5. Precipitation, flow, and historical FIB data
Cumulative six-hour precipitation measurements were obtained from the Meteorological Directorate of Chile online database, station ID 330019 (Chilean Directorate of Meteorology, n.d.), which is located near sampling station S3. To compute cumulative precipitation in the 24-hr period prior to each water sample, each 6-hr cumulative measurement was assumed to comprise 6 1-hr intervals of equal precipitation. The preceding 24 of these 1-hr intervals was then summed. The river’s historical fecal coliform data were obtained courtesy of Aguas Andinas S.A. (internal monitoring program for river health). Enumeration of historical fecal coliform samples was performed by an external certified lab following Standard Method 9222 for membrane filtration (Rice et al., 2012).
2.6. Data analysis
This study’s data presented several statistical properties common in environmental monitoring that were accounted for to accurately assess patterns: non-detects, serial correlation, and spatial clustering. If the data subset used to investigate a research question contained >15% non-detects, statistical methods that explicitly account for censoring were used. If statistically significant serial correlation in residuals was found (applying the astsa::acf2() function (Shumway and Stoffer, 2017) in R to model residuals), appropriate models were used. All analysis was performed on log10-transformed microbial concentrations. Non-detect values were assigned half the limit of detection in log space. Spatial clustering by sampling location was assessed with the Kruskall-Wallis rank sum test, and further analyses accounted for spatial clustering in several ways. Details of these methods are described below.
For RQ2, to examine the correlation between the four indicators with norovirus, concentrations of all five microbes in all available field samples were used. In these data, norovirus was not detected in >15% of samples, and both serial correlation (autocorrelation coefficient significant at p < 0.05) and spatial clustering (by sampling site, p < 0.05) were evident. Suitable rank-based regression or correlation routines that can account for clustered data are not available in R as of February 2020. Thus, concentrations were averaged for each of the eight sampling sites, yielding eight values for each microbe, and Kendall’s tau-b was computed. Kendall’s tau-b was used for computing rank correlations because it accounts for rank ties, which are common in censored data, while Spearman’s rho does not (Agresti, 2010). Tau is expected to be about 0.15 units smaller than rho for the same degree of correlation (Helsel and Hirsch, 2002).
For RQ3, to test the correlation between crAssphage and HF183, all available crAssphage and HF183 concentrations from field samples were used. This included the temporal and spatial sampling campaigns, but not the hourly campaign since only FIB were measured. In these data, ≤15% of values were non-detects and spatial clustering (by sampling site, p < 0.05) was evident. Thus, a generalized estimating equation (GEE) with clustering by sampling site, an exchangeable correlation error structure, and robust standard errors was used. Serial correlation was not evident in model residuals.
For RQ4, to examine variation in indicator concentrations as a function of turbidity and precipitation, all concentrations of the four indicator organisms measured in the temporal campaign were used. In these data, ≤15% of values were non-detects, so ordinary least squares models were fit to indicator concentrations, with precipitation (cumulative over preceding 24 hours) and log10 turbidity as independent variables. The temporal sampling campaign comprised two proximate sampling sites, and model residuals were highly correlated between these two sites. Thus, to avoid pseudo-replication, concentrations were averaged across the two sites at each time point. Model residuals did not show serial correlation, but crAssphage and HF183 model residuals showed a temporal trend, so time was included as a continuous covariate for these two models.
Multi-collinearity was assessed by computing variance inflation factors (VIF) on models with multiple covariates. Bonferroni correction was applied to family-wise error rates, where a family of tests was defined as a group of analogous tests used to investigate an individual research question. Bonferroni was selected as a conservative correction because tests within a family were generally not independent (e.g., correlation of A with B and A with C). The pre-adjusted alpha value was 0.05. Data analysis was performed in R version 3.5.1 using Tidyverse packages (Wickham et al., 2019). Analyses can be reproduced from code available at https://github.com/wileyjennings/chile_mapocho_2019.
3. Results
3.1. qPCR and MST performance
Assay performance characteristics are given in Table S2. No processing blanks, extraction blanks, or NTCs amplified for any targets. Assays efficiencies were 95%, 92%, and 85% for HF183, crAssphage, and norovirus GII respectively. No inhibition was observed for crAssphage or HF183. For norovirus, three samples amplified in 1:10 dilutions, and two of those samples showed inhibition. Given that norovirus concentration estimates were generally low, concentration estimates from undiluted samples were used for analysis, recognizing that these estimates may be biased low due to RT-qPCR reaction inhibition.
Neither crAssphage nor HF183 amplified in composite dog or bird fecal samples. CrAssphage and HF183 concentrations in raw sewage were estimated at 8.35 (σ of qPCR replicates = 0.002) and 8.07 (0.02) log10 cp/100 ml, respectively.
3.2. Turbidity, temperature, and precipitation
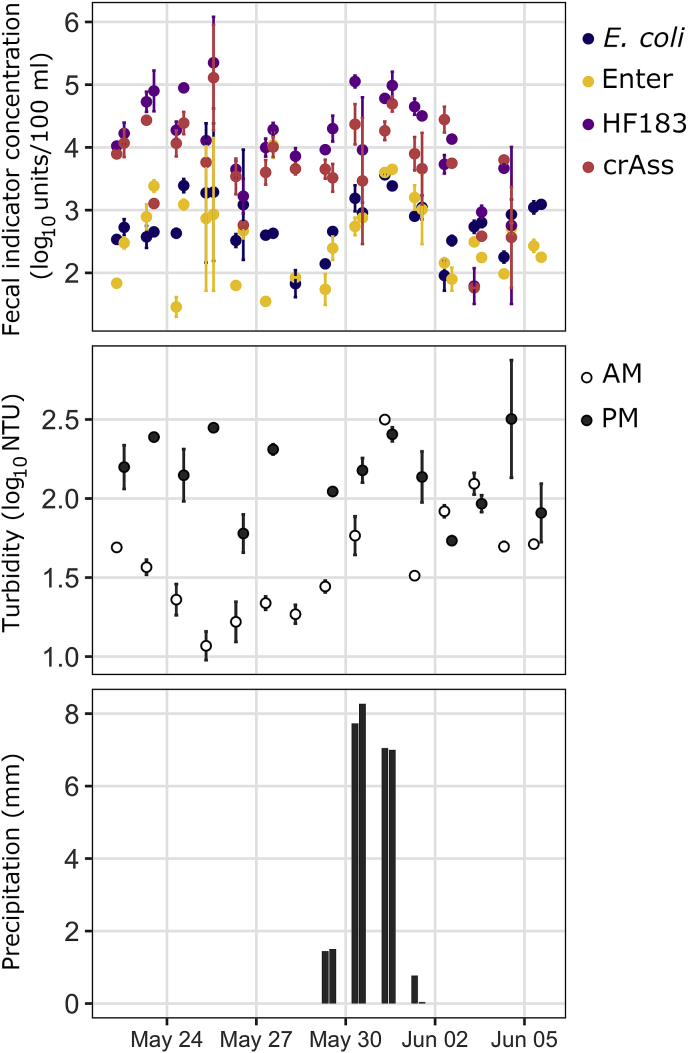
During the spatial campaign, turbidity (Table S3) was lowest at upstream sites S1 (mean = 0.91 log10 NTU, standard deviation = 0.11 log10 NTU) and S2 (1.44, 0.23 log10 NTU). Turbidity was highest at mid-city sites S3 (1.92, 0.81 log10 NTU) and S4 (2.35, 0.74 log10 NTU). Water temperature (Table S4) showed an increasing trend from upstream sites to downstream sites, with lowest temperature at S1 (mean = 6.5 °C, standard deviation = 2.3 °C) and highest temperature at S5 (16.4 °C, 1.3 °C). No precipitation occurred during or in the week prior to the spatial sampling campaign. During the temporal sampling campaign, turbidity was very similar at sites S1 (1.9, 0.5 log10 NTU) and S2 (1.5, 0.4 log10 NTU), and temperature was not measured. Mean precipitation in the 24 hours preceding samples was 1.2 mm and standard deviation was 2.6 mm (Table S4 shows data disaggregated by time of day). Linear regression of turbidity on time of day and precipitation indicated that turbidity was significantly associated with time of day (p < 0.001) but not 24-hr cumulative precipitation (p = 0.06). Controlling for precipitation, expected turbidity was 0.61 log10 NTU higher at 13:30 than 7:30.
3.3. Variation of molecular indicators, FIB, and norovirus
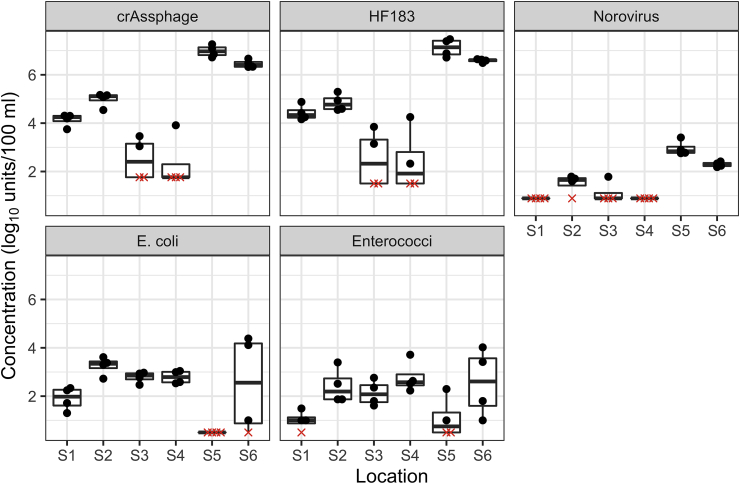
Seventy-eight field samples were tested for molecular and culturable targets. An additional 14 samples were tested only for culturable targets. Across all samples, the median (25th, 75th percentile) E. coli and enterococci concentrations were 2.70 (2.43, 3.04) and 2.29 (1.85, 2.80) log10 MPN/100 ml, respectively. Median crAssphage, HF183, and norovirus concentrations were 3.85 (3.38, 4.46), 4.21 (3.80, 4.79), and <1.78 (<1.78, <1.78) log10 cp/100 ml, respectively. Norovirus was detected in 14 of 78 samples. During the spatial sampling campaign, median concentrations of crAssphage and HF183 were highest (6.69 and 6.68 log10 cp/100 ml, respectively) at downstream sites S5 and S6 and lowest at mid-city sites S3 and S4 (<3.52 and 1.91 log10 cp/100 ml, respectively) (Fig. 2). Norovirus was detected in three of eight upstream (S1 and S2) samples, one of eight mid-city samples, and all eight downstream samples, where medians were approximately 2.6 log10 cp/100 ml. E. coli and enterococci did not significantly vary by site (p > 0.05). Median E. coli concentrations ranged from <1.00 (S5) to 3.34 (S2) log10 MPN/100 ml. Median enterococci concentrations ranged from <1.00 (S1, S5) to 2.61 (S6) log10 MPN/100 ml.
Fig. 2.
Microbial concentrations from the spatial sampling campaign. Crassphage, HF183, and norovirus are in units of log10 cp/100 ml. E. coli and enterococci are in units of log10 MPN/100 ml. Filled circles represent measured concentrations. Red Xs represent non-detects. Boxplot centerlines are medians, bottoms and tops of boxes are 25th and 75th percentiles, and whiskers extend to the furthest data point within a distance of 1.5 × interquartile range from the 25th and 75th percentiles, respectively. Individual points are displaced horizontally to aid visualization. N = 4 for each sampling site (S1 through S6). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Median log10 unit/100 ml concentrations during the temporal campaign across sites T1 and T2 were 3.82 for crAssphage, 4.13 for HF183, 2.69 for E. coli, and 2.49 for enterococci (Fig. 4). Norovirus was not detected at sites T1 or T2 during the temporal campaign. During the temporal campaign, median measurements at sites T1 and T2 differed by 7% or less for each indicator.
Fig. 4.
Indicator concentrations, turbidity, and precipitation during the temporal sampling campaign, averaged across sites. The legend indicates microbial targets shown in the top plot, which are E. coli, enterococci, HF183, and crAssphage. Norovirus is not depicted because it was detected in only two of 54 temporal campaign samples. Crassphage, HF183, and norovirus are in units of log10 cp/100 ml. E. coli and enterococci are in units of log10 MPN/100 ml. Points represent means of two samples (one taken at site T1 and one at T2), and error bars represent standard errors of those two samples. Precipitation is cumulative over the 24-hour period preceding a sample. Samples are not spaced evenly in time because samples were collected at 7:30 (AM) and 13:30 (PM) each day.
CrAssphage and HF183 concentrations in river water were strongly correlated. A 1 log10-unit increase in crAssphage concentration (per 100 ml) was associated with a 0.97 (95% CI 0.89, 1.05; p < 0.001) log10-unit increase in HF183 concentration. This estimate describes covariation of HF183 and crAssphage both within an individual sampling site and between sampling sites, while accounting for spatial clustering and temporal autocorrelation.
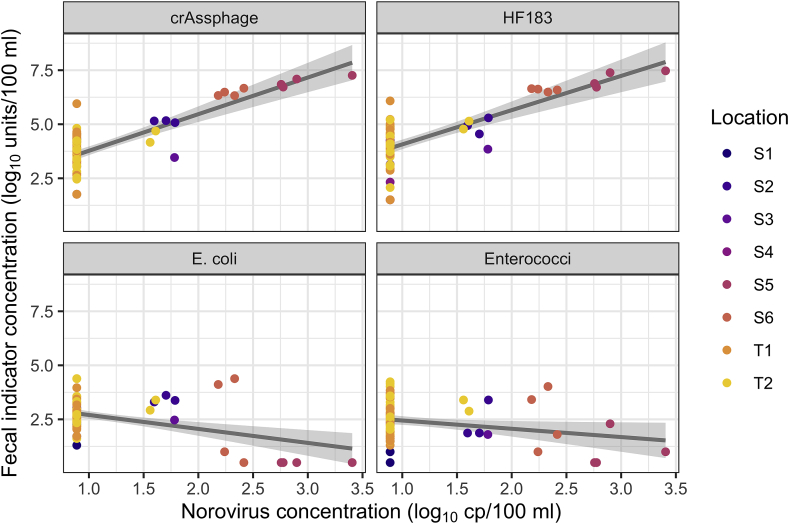
Norovirus was consistently detected only at wastewater effluent-impacted sites, where crAssphage and HF183 were highest. However, no indicator was significantly correlated with norovirus after adjusting for multiple comparisons. Scatter plots of norovirus against human fecal and general fecal indicator concentrations are shown in Fig. 3. Correlation coefficients between norovirus and indicator concentrations across spatial and temporal sampling campaigns were as follows: norovirus with crAssphage, 0.57 (Kendall’s tau-b, p = 0.06); with HF183, 0.64 (p = 0.03); with E. coli, −0.11 (p = 0.70); and with enterococci, −0.19 (p = 0.52). The alpha value used was 0.0125 (0.05 adjusted for four comparisons). Site-averaged values at the six spatial and two temporal sites were used for these estimates, as described in Methods.
Fig. 3.
Human fecal marker and general fecal indicator concentrations compared to norovirus concentrations. Crassphage, HF183, and norovirus are in units of log10 cp/100 ml. E. coli and enterococci are in units of log10 MPN/100 ml. Linear regression lines with 95% confidence bands about the mean are shown for illustrative purposes. These trend lines do not represent correlation estimates reported in Results because they are linear rather than rank relationships (so poorly handle the large number of non-detects) and do not account for statistical clustering by sampling location.
Sub-daily sampling at two proximate sites revealed that only enterococci was associated with turbidity, and no indicators were associated with precipitation. A 1-log10 NTU increase in turbidity was associated (p < 0.001) with a 0.87 (95% CI 0.42, 1.32) log10 increase in enterococci concentration (Table 2), estimated by linear regression. No other coefficients for either precipitation or turbidity were statistically significant in models of crAssphage, HF183, E. coli, or enterococci (p-values > 0.0125, the alpha value adjusted for four comparisons). All variance inflation factors were near 1, indicating that multi-collinearity was not a problem.
Table 2.
Linear regression results describing variation of indicator concentrations with precipitation and turbidity. Indicator concentrations are log10 units/100 ml, as throughout this paper. The rationale for including time as a covariate in crAssphage and HF183 models is given in Methods.
| crAssphage |
HF183 |
E. coli |
Enterococci |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (std err) | p-value | Estimate (std err) | p-value | Estimate (std err) | p-value | Estimate (std err) | p-value | ||
| Parameter | Units | ||||||||
| Intercept | log10 concentration | 4.09 | <0.001 | 4.20 | <0.001 | 2.07 | <0.001 | 0.77 | 0.10 |
| (0.61) | (0.59) | (0.33) | (0.45) | ||||||
| Precipitation | mm | 0.09 | 0.09 | 0.12 | 0.03 | 0.05 | 0.08 | 0.05 | 0.21 |
| (0.05) | (0.05) | (0.03) | (0.04) | ||||||
| Turbidity | log10 NTU | 0.00 | 0.99 | 0.23 | 0.47 | 0.35 | 0.06 | 0.93 | <0.001 |
| (0.32) | (0.32) | (0.18) | (0.24) | ||||||
| Time | days | −0.07 | 0.04 | −0.10 | <0.01 | – | – | – | – |
| (0.03) | (0.03) | – | – | ||||||
| n | – | 27 | 27 | 29 | 29 | ||||
| Adj. R-squared | 0.13 | 0.30 | 0.23 | 0.41 | |||||
Hourly samples collected between 7:30 and 16:30 on a single day and measured for E. coli and enterococci revealed no significant linear trend over time. A plot of these data (Figure S4) did not suggest other diurnal patterns.
4. Discussion
4.1. CrAssphage is useful for indicating wastewater impact in Chile’s Mapocho River
Concentrations of crAssphage in untreated sewage using assay CPQ_064 were high, indicating that crAssphage has the potential to be a sensitive indicator of wastewater impact in Santiago. Concentrations measured in this study were comparable to or greater than concentrations measured in other countries. For example, crAssphage (CPQ_056) was estimated in untreated sewage in Florida, USA at 8.08–8.98 log10 cp/100 ml (Ahmed et al., 2018a); in Australia (CPQ_064) at 7.91 log10 cp/100 ml (Ahmed et al., 2018b); in the UK (CPQ_056) at 4–8 log10 cp/100 ml (Farkas et al., 2019); and in Thailand (CPQ_056) at 6.57 log10 cp/100 ml (Kongprajug et al., 2019). Although this study did not measure crAssphage concentrations in WWTP effluent samples, studies have reported 0- to 2-log10 (Thailand (Kongprajug et al., 2019) and UK (Farkas et al., 2019)) and 3-log10 (USA (Wu et al., 2020)) reductions of crAssphage concentrations through WWTPs with activated sludge units. More generally, this study’s measurements of crAssphage support the finding that crAssphage is a sensitive human-associated viral MST marker, with concentrations in sewage at least 2–3 orders of magnitude greater than concentrations typically measured of other human-associated viral tracking markers, including human adenovirus, human polyomavirus, and pepper mild mottle virus (Ahmed et al., 2016a, Ahmed et al., 2016b; Farkas et al., 2019; Hughes et al., 2017; Stachler et al., 2018).
CrAssphage was specific to human fecal waste among animal fecal samples tested. It did not amplify in bird or dog feces, two important non-human fecal sources in the Mapocho watershed. However, this study cannot rule out cross-reaction with other fecal sources, which could be important in agricultural areas. Most studies, with the exception of work done in Nepal (Malla et al., 2019), have reported high specificity of crAssphage to human feces, with limited cross-reaction in cat feces and cattle wastewater in Australia (Ahmed et al., 2018b), poultry litter in Florida, USA (Ahmed et al., 2018a), and swine feces in Thailand (Kongprajug et al., 2019).
crAssphage was highly correlated with the human-associated bacterial fecal marker HF183 in field samples in this study. This suggests that both markers derive from similar sources and have similar environmental fate on the time scale required for water to pass through the urban portion of the Mapocho River. No data are available that describe the degree to which covariance between CPQ_064 and HF183 gene targets may be driven by crAssphage genes being located inside Bacteroides cells or genomes. Other studies have reported mixed results regarding correlations between crAssphage and HF183 in environmental waters. Samples collected in wastewater-impacted urban river in the USA showed strong correlation between crAssphage CPQ_064 and HF183, and moderate correlation between crAssphage CPQ_056 and HF183 (Stachler et al., 2018). A study in Spain that used a different crAssphage assay found statistically significant linear correlations with HF183 in a creek that does not receive WWTP effluent, but not in a river that does receive WWTP effluent (Ballesté et al., 2019). Indeed, another study in the USA using CPQ_056 found that crAssphage was not correlated with HF183 in storm drain outfalls or a wetland contaminated with sewage in Tampa, Florida (Ahmed et al., 2018a). One possible explanation for the observed differences between HF183 and crAssphage is differential environmental persistence. Both crAssphage decay studies published to date have found evidence that crAssphage markers persist about 2–3 times longer than HF183 markers in freshwater mesocosms (Ahmed et al., 2019; Ballesté et al., 2019).
4.2. Norovirus patterns were more closely tracked by molecular MST markers than FIB
Correlation coefficients of norovirus with indicators were suggestive of positive relationships with crAssphage and HF183, and null relationships with E. coli and enterococci. It is important to note that these correlation analyses used site-averaged concentrations, and so they had low power. The only other study to date that measured crAssphage and norovirus simultaneously found statistically significant positive rank correlations between crAssphage and norovirus GII in wastewater influent and effluent from the UK (Farkas et al., 2019). No other study has examined co-occurrence of norovirus and crAssphage in an environmental water. The lack of correlation between FIB and human viruses in environmental waters is commonly reported (Korajkic et al., 2018) and may be attributable to differential environmental fate and transport (USEPA, 2015) or removal in wastewater treatment plant processes (Rose et al., 1996; Zhang and Farahbakhsh, 2007), as well as differential sources.
4.3. Although FIB pollution in the Mapocho River has declined dramatically since at least 2004, human fecal pollution and norovirus were elevated at some locations
In 2005, Santiago was treating only 4% of its sewage and discharging the rest untreated to the Mapocho River. With the construction of two large conventional treatment plants (activated sludge with chlorine disinfection), by 2016 Santiago was treating 100% of its wastewater (La Farfana WWTP site visit and UNFCCC website (UNFCCC, 2020, n.d.)). This increase in sewage treatment has resulted in a dramatic decline in fecal coliform concentrations in the Mapocho at locations coincident with sites sampled in this study (Figure S3). Compared to historical data, E. coli concentrations measured in this study generally corroborate the lowered levels of fecal coliform concentrations reported in recent years, considering methodological differences (Figure S3).
However, given the consistent detection of norovirus GII genes in the WWTP effluent-impacted waters at sites S5 and S6, these waters may present a health risk to those who contact it directly or who consume products recently irrigated with it. For example, the 2010 norovirus GII outbreak in the Antofagasta region of Chile originated from consumption of raw vegetables irrigated with treated wastewater containing low levels of residual free chlorine (Díaz T. et al., 2012). The national government and citizen groups, who have expressed growing interest in reclaiming the Mapocho for recreational use, should also consider the risk of waterborne illness potentially posed by viral pathogens in river extents receiving large loads of chlorinated effluent – a risk that may not be indicated by FIB. It is important to note that genome-based detection of norovirus does not indicate the infectivity of these viruses, and thus potential health risks cannot be stated unequivocally. However, a recent meta-analysis found that culturable male-specific coliphage often survive secondary treatment and chlorination, and that removal of qPCR-measured norovirus GII (2.7 log10 reduction) and culturable surrogate male-specific coliphage (2.9 log10 reduction) was similar (Pouillot et al., 2015). More generally, conventional biological treatment with chlorination is thought to be less effective at removing viruses than bacteria (Rose et al., 1996; Zhang and Farahbakhsh, 2007). These studies, along with the low infectious dose (ID) of human norovirus (ID50 estimated between 10 and 103 genome copies (Atmar et al., 2014; Teunis et al., 2008)), support the conjecture that the qPCR-measured norovirus concentrations at sites S5 and S6 may pose a human health risk.
Human bacterial and viral fecal pollution concentrations were also high at upstream sites S1 and S2. Since these sites do not receive treated wastewater effluent, this pollution likely comes from untreated human feces, delivered by direct discharge of household-scale wastewater, leaking sewage infrastructure, or dry weather flows from irrigation or other anthropogenic washing activities. No precipitation occurred during or immediately prior to the spatial sampling campaign, which included sites S1 and S2. At S1 and S2, norovirus was detected in three of eight samples, and median crAssphage and HF183 concentrations exceeded QMRA-based risk thresholds for recreational exposure to sewage-polluted waters of 30 illnesses per 1000 bathers (3.67 log10 cp/100 ml for crAssphage (Crank et al., 2019) and 3.62 log10 cp/100 ml for HF183 (Boehm et al., 2015)).
Median human fecal marker concentrations declined substantially from S2 to S3. This decline may be attributable to dilution from tributary inflows between S2 and S3 (Fig. 1). If the decline were based on microbial decay alone, crAssphage would be expected to decay less than HF183 and culturable FIB given the cold water temperatures at sites S2 and S3 (Ballesté et al., 2019). This expectation is contradicted by observations that crAssphage declined by two to three orders of magnitude between S2 and S3, while HF183 declined by a similar amount and E. coli and enterococci declined by much less (zero to one orders of magnitude). For dilution to be the primary process governing these concentrations reduction, tributary inflows would need to contain similar concentrations of FIB and considerably lower concentrations of human markers compared to the Mapocho at S2. However, further work would be required to confirm that dilution was more important than microbial decay, and also to investigate the roll of sediment resuspension, in controlling concentrations between S2 and S3. Furthermore, while the San Carlos canal (Fig. 1), which delivers water from the Maipo River into the Mapocho River just upstream of S3, would typically influence microbial concentrations at S3, it did not contribute any flow during the spatial sampling campaign because it was closed for maintenance activities.
4.4. River channel maintenance activities may elevate FIB concentrations
The Mapocho River receives stormwater from Santiago’s stormwater system, which is separate from its sewage system, and from direct overland drainage. Human and general fecal pollution in stormwater flows have been extensively documented (Brownell et al., 2007; Parker et al., 2010; Sidhu et al., 2013; Tiefenthaler et al., 2011). Thus, concentrations of both human and general indicators of fecal pollution were expected to increase following precipitation events. While HF183, E. coli, and enterococci concentrations showed upward trends with increasing precipitation (p < 0.10), none of these associations were statistically significant after adjusting for multiple comparisons. Likely, the effect of precipitation cannot be distinguished in this data set (n = 27 for molecular markers, 29 for FIB) because unmeasured anthropogenic and environmental processes cause substantial variation in fecal pollution concentrations.
Turbidity, however, was positively associated with enterococci concentrations, and showed a positive trend with E. coli concentrations. Interestingly, in multiple linear regression, turbidity was strongly associated with time of day (7:30 vs 13:30), but not precipitation (data not shown). In the absence of precipitation driving turbidity levels via stormwater flows or sediment resuspension from elevated river flows, this diurnal turbidity pattern may be related to river channel bed maintenance activities that occurred throughout the sampling campaign. These activities included the use of heavy machinery alongside and occasionally in the river, and were conducted during business hours, implying the potential for greater sediment disturbance at 13:30 than 7:30. The association between turbidity and FIB concentrations is supported by laboratory and modeling studies showing that riverine sediment can harbor elevated levels of FIB, and that FIB concentrations in the water column may increase when sediment is resuspended (Droppo et al., 2011, 2009). Time of day was not included in linear regression models of microbial concentrations because time of day was strongly correlated with turbidity.
4.5. Limitations
This field study yielded insight into applying crAssphage and HF183 in a new geographic context, as well as fecal pollution patterns in the Mapocho River. It also has several limitations. First, more extensive characterization of crAssphage and HF183 concentrations in sewage would improve confidence in this study’s assessment of these markers as sensitive indicators of sewage pollution. Repeat sampling of influent sewage and effluent at both the Farfana and Mapocho-Trebal plants would aid interpretation of this study’s data downstream of these plants. Previous studies have reported no variation in crAssphage concentrations in sewage over the course of a year (García-Aljaro et al., 2017; Malla et al., 2019), so year-round characterization is likely unnecessary. Second, more extensive specificity evaluation of crAssphage and HF183 would be desirable. This would include assessment of these markers’ concentrations in other fecal sources potentially important in the Mapocho watershed. The approach we used of compositing the DNA from the fecal samples could potentially lead to false negatives if the target is diluted to below the assay detection limit when it is combined with the other DNA extracts; however this approach is established in the MST field (Boehm et al., 2016; Shanks et al., 2009, 2008). Third, PCR inhibition of the norovirus GII assay was observed, even though both the extraction protocol and the master mix were selected for their capacity to remove PCR inhibitors. Inhibition potentially yielded underestimates of norovirus concentrations. Dilution of extracts to reduce inhibitory effects was not feasible, given the low concentrations of norovirus present. Filtering larger sample volumes could potentially increase method sensitivity, but could also increase inhibition of PCR reactions. Similar to this study, other studies of urban environmental waters have found little or no inhibition of qPCR assays, yet substantial inhibition of RT-qPCR assays in the same samples (Steele et al., 2018).
5. Conclusions
-
•
CrAssphage and HF183 are useful indicators of human fecal pollution in Santiago, Chile.
-
•
Fecal indicator bacteria are inadequate indicators of viral pollution from chlorinated wastewater effluent.
-
•
Anthropogenic impacts on Mapocho River microbial water quality are extensive.
-
•
Viral pollution must be considered in efforts to reclaim the Mapocho River for recreational activities.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge Juan Antonio Garcés (Aguas Andinas S.A.) and Roberto Araya (Junta de Vigilancia del Río Mapocho) for providing background information on the Mapocho River system. We thank Mauricio Tapia for assistance with study logistics. This study was supported by a Stanford Bing Overseas Studies Program grant. WCJ was supported by a U.S. National Science Foundation Graduate Research Fellowship (award no. 2015202460).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.wroa.2020.100071.
Contributor Information
Wiley C. Jennings, Email: wileyjen@stanford.edu.
Elías Gálvez-Arango, Email: hegalvez@stanford.edu.
Ana L. Prieto, Email: ana.prieto@uchile.cl.
Alexandria B. Boehm, Email: aboehm@stanford.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agresti A. second ed. John Wiley & Sons; New York: 2010. Analysis of Ordinal Categorical Data. [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Warish, Hughes B., Harwood V.J. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water. 2016;8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- Ahmed W., Lobos A., Senkbeil J., Peraud J., Gallard J., Harwood V.J. Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res. 2018;131:142–150. doi: 10.1016/j.watres.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Besley C., Power K. Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Res. 2018 doi: 10.1016/j.watres.2018.08.049. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Sidhu J.P.S., Smith K., Beale D.J., Gyawali P., Toze S. Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters. Appl. Environ. Microbiol. 2016;82:1316–1323. doi: 10.1128/AEM.03765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Zhang Q., Kozak S., Beale D., Gyawali P., Sadowsky M.J., Simpson S. Comparative decay of sewage-associated marker genes in beach water and sediment in a subtropical region. Water Res. 2019;149:511–521. doi: 10.1016/j.watres.2018.10.088. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garreton C., Mendoza P.A., Boisier J.P., Addor N., Galleguillos M., Zambrano-Bigiarini M., Lara A., Puelma C., Cortes G., Garreaud R., McPhee J., Ayala A. The CAMELS-CL dataset: catchment attributes and meteorology for large sample studies – Chile dataset. Hydrol. Earth Syst. Sci. 2018;22:5817–5846. doi: 10.5194/hess-22-5817-2018. [DOI] [Google Scholar]
- Atmar R.L., Opekun A.R., Gilger M.A., Estes M.K., Crawford S.E., Neill F.H., Ramani S., Hill H., Ferreira J., Graham D.Y. Determination of the 50% human infectious dose for norwalk virus. J. Infect. Dis. 2014;209:1016–1022. doi: 10.1093/infdis/jit620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesté E., Pascual-Benito M., Martín-Díaz J., Blanch A.R., Lucena F., Muniesa M., Jofre J., García-Aljaro C. Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 2019;155:233–244. doi: 10.1016/j.watres.2019.02.042. [DOI] [PubMed] [Google Scholar]
- Boehm A.B., Soller J.A., Shanks O.C. Human-associated fecal quantitative polymerase chain reaction measurements and simulated risk of gastrointestinal illness in recreational waters contaminated with raw sewage. Environ. Sci. Technol. Lett. 2015;2:270–275. doi: 10.1021/acs.estlett.5b00219. [DOI] [Google Scholar]
- Boehm A.B., Van De Werfhorst L.C., Griffith J.F., Holden P.A., Jay J.A., Shanks O.C., Wang D., Weisberg S.B. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res., Microbial source track. 2013;47:6812–6828. doi: 10.1016/j.watres.2012.12.046. [DOI] [PubMed] [Google Scholar]
- Boehm A.B., Wang D., Ercumen A., Shea M., Harris A.R., Shanks O.C., Kelty C., Ahmed A., Mahmud Z.H., Arnold B.F., Chase C., Kullmann C., Colford J.M., Luby S.P., Pickering A.J. Occurrence of host-associated fecal markers on child hands, household soil, and drinking water in rural Bangladeshi households. Environ. Sci. Technol. Lett. 2016;3:393–398. doi: 10.1021/acs.estlett.6b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell M.J., Harwood V.J., Kurz R.C., McQuaig S.M., Lukasik J., Scott T.M. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res., Identifying Sources Fecal Pollut. 2007;41:3747–3757. doi: 10.1016/j.watres.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Chilean Directorate of Meteorology https://climatologia.meteochile.gob.cl/application/informacion/buscadorDeEstaciones/ n.d. Climatic services [WWW Document]. Stn. Lookup. URL. (accessed 2.27.20)
- Chilean Ministry of Agriculture http://ide.minagri.gob.cl/geoweb/ n.d. Spatial data infrastructure [WWW Document]. URL. (accessed 2.27.20)
- Chilean Ministry of National Goods http://www.ide.cl/descarga/capas/item/zonificacion-plan-regulador-metropolitano-de-santiago-prms.html n.d. Metropolitan regulatory zoning plan for Santiago [WWW Document]. URL. (accessed 2.27.20)
- Crank K., Petersen S., Bibby K. Quantitative microbial risk assessment of swimming in sewage impacted waters using CrAssphage and pepper mild mottle virus in a customizable model. Environ. Sci. Technol. Lett. 2019;6:571–577. doi: 10.1021/acs.estlett.9b00468. [DOI] [Google Scholar]
- Digital Chart of the World, n.d. Chile inland water data [WWW Document]. DIVA-GIS. URL diva-gis.org/gdata (accessed 2.27.20).
- Díaz T. J., Solari G. V., Cáceres C. O., Mena A. J., Baeza P. S., Muñoz U. X., O’Ryan G. M., Galeno A. H., Maldonado B. A., Mamani M. N. Brote de gastroenteritis aguda en la Región de Antofagasta,Chile: 2010. Rev. Chil. Infectol. 2012;29:19–25. doi: 10.4067/S0716-10182012000100003. [DOI] [PubMed] [Google Scholar]
- Droppo I.G., Krishnappan B.G., Liss S.N., Marvin C., Biberhofer J. Modelling sediment-microbial dynamics in the South Nation River, Ontario, Canada: towards the prediction of aquatic and human health risk. Water Res. 2011;45:3797–3809. doi: 10.1016/j.watres.2011.04.032. [DOI] [PubMed] [Google Scholar]
- Droppo I.G., Liss S.N., Williams D., Nelson T., Jaskot C., Trapp B. Dynamic existence of waterborne pathogens within river sediment compartments. Implications for water quality regulatory affairs. Environ. Sci. Technol. 2009;43:1737–1743. doi: 10.1021/es802321w. [DOI] [PubMed] [Google Scholar]
- Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G.Z., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K., Felts B., Dinsdale E.A., Mokili J.L., Edwards R.A. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.A., Vega A.A., Norman H.M., Ohaeri M., Levi K., Dinsdale E.A., Cinek O., Aziz R.K., McNair K., Barr J.J., Bibby K., Brouns S.J., Cazares A., Jonge P.A. de, Desnues C., Muñoz S.L.D., Fineran P.C., Kurilshikov A., Lavigne R., Mazankova K., McCarthy D.T., Nobrega F.L., Muñoz A.R., Tapia G., Trefault N., Tyakht A.V., Vinuesa P., Wagemans J., Zhernakova A., Aarestrup F.M., Ahmadov G., Alassaf A., Anton J., Asangba A., Billings E., Cantu V.A., Carlton J.M., Cazares D., Cho G.-S., Condeff T., Cortés P., Cranfield M., Cuevas D.A., Iglesia R.D. la, Decewicz P., Doane M.P., Dominy N.J., Dziewit L., Elwasila B.M., Eren A.M., Franz C., Fu J., Garcia-Aljaro C., Ghedin E., Gulino K.M., Haggerty J.M., Head S.R., Hendriksen R.S., Hill C., Hyöty H., Ilina E.N., Irwin M.T., Jeffries T., Torroella J.J., Junge R.E., Kelley S.T., Kowalewski M., Kumaresan D., Leigh S., Lisitsyna E.S., Llagostera M., Maritz J.M., Marr L.C., McCann A., Mirzaei M.K., Molshanski-Mor S., Monteiro S., Moreira-Grez B., Morris M., Mugisha L., Muniesa M., Neve H., Nguyen N., Nigro O.D., Nilsson A.S., O’Connell T., Odeh R., Oliver A., Piuri M., Prussin A.J., Qimron U., Quan Z.-X., Rainetova P., Rojas A.A.R., Raya R., Rice G.A.O., Rossi A., Santos R., Shimashita J., Stachler E.N., Stene L.C., Strain R., Stumpf R., Torres P.J., Twaddle A., Ibekwe M.U., Villagra N., Wandro S., White B., Whitely A., Whiteson K.L., Wijmenga C., Zambrano M.M., Zschach H., Dutilh B.E. 2019. Global Phylogeography and Ancient Evolution of the Widespread Human Gut Virus crAssphage. bioRxiv 527796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Adriaenssens E.M., Walker D.I., McDonald J.E., Malham S.K., Jones D.L. Critical evaluation of CrAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ. Virol. 2019;11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- García-Aljaro C., Ballesté E., Muniesa M., Jofre J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017;10:1775–1780. doi: 10.1111/1751-7915.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H.C., Haugland R.A., Varma M., Millen H.T., Borchardt M.A., Field K.G., Walters W.A., Knight R., Sivaganesan M., Kelty C.A., Shanks O.C. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol. 2014;80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood V.J., Boehm A.B., Sassoubre L.M., Vijayavel K., Stewart J.R., Fong T.-T., Caprais M.-P., Converse R.R., Diston D., Ebdon J., Fuhrman J.A., Gourmelon M., Gentry-Shields J., Griffith J.F., Kashian D.R., Noble R.T., Taylor H., Wicki M. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res., Microbial source tracking. 2013;47:6929–6943. doi: 10.1016/j.watres.2013.04.064. [DOI] [PubMed] [Google Scholar]
- Helsel D.R., Hirsch R.M. U.S. Geological Survey, Techniques of Water-Resources Investigations Book 4. U.S. Geological Survey; 2002. Statistical methods in water resources. Chapter A3. [Google Scholar]
- Hughes B., Beale D.J., Dennis P.G., Cook S., Ahmed W. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00028-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto de Salud Pública de Chile . Vol. 8. Instituto de Salud Pública de Chile; 2018. (Resultados de Laboratorio de Determinación de Norovirus. Chile, 2013-2017). No. 4) [Google Scholar]
- Instituto Nacional de Estadísticas de Chile . Instituto Nacional de Estadísticas de Chile; 2017. Resultados Definitivos Censo 2017. [Google Scholar]
- Kildare B.J., Leutenegger C.M., McSwain B.S., Bambic D.G., Rajal V.B., Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J., Goldberg C.S., Helbing C.C., Hunter M.E., Jackson C.A., Lance R.F., Mangan A.M., Monroe E.M., Piaggio A.J., Stokdyk J.P., Wilson C.C., Richter C.A. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA. 2020;2:271–282. doi: 10.1002/edn3.29. [DOI] [Google Scholar]
- Kongprajug A., Mongkolsuk S., Sirikanchana K. CrAssphage as a potential human sewage marker for microbial source tracking in southeast asia. Environ. Sci. Technol. Lett. 2019;6:159–164. doi: 10.1021/acs.estlett.9b00041. [DOI] [Google Scholar]
- Korajkic A., McMinn B.R., Harwood V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Publ. Health. 2018;15 doi: 10.3390/ijerph15122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Sivaganesan M., Kelty C.A., Zimmer-Faust A., Clinton P., Reichman J.R., Johnson Y., Matthews W., Bailey S., Shanks O.C. Large-scale implementation of standardized quantitative real-time PCR fecal source identification procedures in the Tillamook Bay Watershed. PLoS One. 2019;14 doi: 10.1371/journal.pone.0216827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisy F., Atmar R.L., Guillon P., Le Cann P., Pommepuy M., Le Guyader F.S. Real-time RT-PCR for norovirus screening in shellfish. J. Virol. Method. 2005;123:1–7. doi: 10.1016/j.jviromet.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lukasik J., Scott T.M., Andryshak D., Farrah S.R. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 2000;66:2914–2920. doi: 10.1128/aem.66.7.2914-2920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla B., Ghaju Shrestha R., Tandukar S., Sherchand J.B., Haramoto E. Performance evaluation of human-specific viral markers and application of pepper mild mottle virus and CrAssphage to environmental water samples as fecal pollution markers in the kathmandu valley, Nepal. Food Environ. Virol. 2019;11:274–287. doi: 10.1007/s12560-019-09389-x. [DOI] [PubMed] [Google Scholar]
- Mayer R.E., Reischer G.H., Ixenmaier S.K., Derx J., Blaschke A.P., Ebdon J.E., Linke R., Egle L., Ahmed W., Blanch A.R., Byamukama D., Savill M., Mushi D., Cristóbal H.A., Edge T.A., Schade M.A., Aslan A., Brooks Y.M., Sommer R., Masago Y., Sato M.I., Taylor H.D., Rose J.B., Wuertz S., Shanks O.C., Piringer H., Mach R.L., Savio D., Zessner M., Farnleitner A.H. Global distribution of human-associated fecal genetic markers in reference samples from six continents. Environ. Sci. Technol. 2018;52:5076–5084. doi: 10.1021/acs.est.7b04438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meybeck M. Global analysis of river systems: from Earth system controls to Anthropocene syndromes. Philos. Trans. R. Soc. B Biol. Sci. 2003;358:1935–1955. doi: 10.1098/rstb.2003.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro S., Pineda S., Enríquez I., Enríquez N., Rivera N., Delgado C. Detección de norovirus en niños con diarrea adquirida en la comunidad o nosocomial en el Hospital Guillermo Grant Benavente de Concepción, Chile. Rev. Chil. Infectol. 2014;31:298–304. doi: 10.4067/S0716-10182014000300008. [DOI] [PubMed] [Google Scholar]
- Parker J.K., McIntyre D., Noble R.T. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, USA. Water Res. 2010;44:4186–4194. doi: 10.1016/j.watres.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Patel M.M., Hall A.J., Vinjé J., Parashar U.D. Noroviruses: a comprehensive review. J. Clin. Virol. 2009;44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Pouillot R., Doren J.M.V., Woods J., Plante D., Smith M., Goblick G., Roberts C., Locas A., Hajen W., Stobo J., White J., Holtzman J., Buenaventura E., Burkhardt W., Catford A., Edwards R., DePaola A., Calci K.R. Meta-analysis of the reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants. Appl. Environ. Microbiol. 2015;81:4669–4681. doi: 10.1128/AEM.00509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice E.W., Baird R.B., Eaton A.D., Clesceri L.S. twenty-second ed. APHA; AWWA, WEF: 2012. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Rose J.B., Dickson L.J., Farrah S.R., Carnahan R.P. Removal of pathogenic and indicator microorganisms by a full-scale water reclamation facility. Water Res. 1996;30:2785–2797. doi: 10.1016/S0043-1354(96)00188-1. [DOI] [Google Scholar]
- Shanks O.C., Atikovic E., Blackwood A.D., Lu J., Noble R.T., Domingo J.S., Seifring S., Sivaganesan M., Haugland R.A. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 2008;74:745–752. doi: 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks O.C., Kelty C.A., Sivaganesan M., Varma M., Haugland R.A. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 2009;75:5507–5513. doi: 10.1128/AEM.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks O.C., White K., Kelty C.A., Sivaganesan M., Blannon J., Meckes M., Varma M., Haugland R.A. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ. Sci. Technol. 2010;44:6281–6288. doi: 10.1021/es100311n. [DOI] [PubMed] [Google Scholar]
- Shumway R.H., Stoffer D.S. fourth ed. Springer; 2017. Time Series Analysis and its Application with R Examples. [Google Scholar]
- Sidhu J.P.S., Ahmed W., Gernjak W., Aryal R., McCarthy D., Palmer A., Kolotelo P., Toze S. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci. Total Environ. 2013;463–464:488–496. doi: 10.1016/j.scitotenv.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M., Haugland R.A., Chern E.C., Shanks O.C. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Res. 2010;44:4726–4735. doi: 10.1016/j.watres.2010.07.066. [DOI] [PubMed] [Google Scholar]
- Soller J.A., Bartrand T., Ashbolt N.J., Ravenscroft J., Wade T.J. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res., Shifting paradigms in the assessment of recreational water quality. 2010;44:4736–4747. doi: 10.1016/j.watres.2010.07.064. [DOI] [PubMed] [Google Scholar]
- Soller J.A., Schoen M., Steele J.A., Griffith J.F., Schiff K.C. Incidence of gastrointestinal illness following wet weather recreational exposures: harmonization of quantitative microbial risk assessment with an epidemiologic investigation of surfers. Water Res. 2017;121:280–289. doi: 10.1016/j.watres.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Soller J.A., Schoen M.E., Bartrand T., Ravenscroft J.E., Ashbolt N.J. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res., Shifting paradigms in the assessment of recreational water quality. 2010;44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- Stachler E., Akyon B., Carvalho N.A. de, Ference C., Bibby K. Correlation of crAssphage qPCR markers with culturable and molecular indicators of human fecal pollution in an impacted urban watershed. Environ. Sci. Technol. 2018 doi: 10.1021/acs.est.8b00638. [DOI] [PubMed] [Google Scholar]
- Stachler E., Bibby K. Metagenomic evaluation of the highly abundant human gut bacteriophage CrAssphage for source tracking of human fecal pollution. Environ. Sci. Technol. Lett. 2014;1:405–409. doi: 10.1021/ez500266s. [DOI] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J.A., Blackwood A.D., Griffith J.F., Noble R.T., Schiff K.C. Quantification of pathogens and markers of fecal contamination during storm events along popular surfing beaches in San Diego, California. Water Res. 2018;136:137–149. doi: 10.1016/j.watres.2018.01.056. [DOI] [PubMed] [Google Scholar]
- Suez, 2020. El Trebal Mapocho Wastewater Treatment Plants [WWW Document]. URL https://www.suezwaterhandbook.com/case-studies/wastewater-treatment/El-Trebal-Mapocho-wastewater-treatment-plant-Chile.
- Suez, 2020. La Farfana Wastewater Treatment Plant [WWW Document]. URL https://www.suezwaterhandbook.com/case-studies/wastewater-treatment/La-Farfana-wastewater-treatment-plant-Chile.
- Teunis P.F.M., Moe C.L., Liu P.E., Miller S., Lindesmith L., Baric R.S., Le Pendu J., Calderon R.L. Norwalk virus: how infectious is it? J. Med. Virol. 2008;80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- Tiefenthaler L., Stein E.D., Schiff K.C. Levels and patterns of fecal indicator bacteria in stormwater runoff from homogenous land use sites and urban watersheds. J. Water Health. 2011;9:279–290. doi: 10.2166/wh.2010.056. [DOI] [PubMed] [Google Scholar]
- UNFCCC . 2020. https://unfccc.int/climate-action/momentum-for-change/planetary-health/santiago-biofactory-chile n.d. Santiago Biofactory, Chile [WWW Document]. URL. (accessed 3.31.20) [Google Scholar]
- USEPA . USEPA; 2015. Review of Coliphages as Possible Indicators of Fecal Contamination for Ambient Water Quality (No. 820- R-15–098) [Google Scholar]
- Viau E.J., Lee D., Boehm A.B. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ. Sci. Technol. 2011;45:7158–7165. doi: 10.1021/es200984b. [DOI] [PubMed] [Google Scholar]
- Vidal R., Solari V., Mamani N., Jiang X., Vollaire J., Roessler P., Prado V., Matson D.O., O’Ryan M.L. Caliciviruses and foodborne gastroenteritis, Chile. Emerg. Infect. Dis. 2005;11:1134–1137. doi: 10.3201/eid1107.041062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vörösmarty C.J., McIntyre P.B., Gessner M.O., Dudgeon D., Prusevich A., Green P., Glidden S., Bunn S.E., Sullivan C.A., Liermann C.R., Davies P.M. Global threats to human water security and river biodiversity. Nature. 2010;467:555–561. doi: 10.1038/nature09440. [DOI] [PubMed] [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., Kuhn M., Pedersen T., Miller E., Bache S., Müller K., Ooms J., Robinson D., Seidel D., Spinu V., Takahashi K., Vaughan D., Wilke C., Woo K., Yutani H. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- World Health Organization . WHO; Geneva: 2011. Guidelines for Drinking-Water Quality (No. 4) [Google Scholar]
- Wu Z., Greaves J., Arp L., Stone D., Bibby K. Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environ. Int. 2020;136 doi: 10.1016/j.envint.2019.105452. [DOI] [PubMed] [Google Scholar]
- Wyn-Jones A.P., Carducci A., Cook N., D’Agostino M., Divizia M., Fleischer J., Gantzer C., Gawler A., Girones R., Höller C., de Roda Husman A.M., Kay D., Kozyra I., López-Pila J., Muscillo M., José Nascimento M.S., Papageorgiou G., Rutjes S., Sellwood J., Szewzyk R., Wyer M. Surveillance of adenoviruses and noroviruses in European recreational waters. Water Res. 2011;45:1025–1038. doi: 10.1016/j.watres.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Farahbakhsh K. Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: implications to water reuse. Water Res. 2007;41:2816–2824. doi: 10.1016/j.watres.2007.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.