Abstract
The apoptotic nuclease EndoG is involved in mitochondrial DNA replication. Previous results suggested that, in addition to regulate cardiomyocyte hypertrophy, EndoG could be involved in cell proliferation. Here, by using in vivo and cell culture models, we investigated the role of EndoG in cell proliferation. Genetic deletion of Endog both in vivo and in cultured cells or Endog silencing in vitro induced a defect in rodent and human cell proliferation with a tendency of cells to accumulate in the G1 phase of cell cycle and increased reactive oxygen species (ROS) production. The defect in cell proliferation occurred with a decrease in the activity of the AKT/PKB-GSK-3β-Cyclin D axis and was reversed by addition of ROS scavengers. EndoG deficiency did not affect the expression of ROS detoxifying enzymes, nor the expression of the electron transport chain complexes and oxygen consumption rate. Addition of the micropeptide Humanin to EndoG-deficient cells restored AKT phosphorylation and proliferation without lowering ROS levels. Thus, our results show that EndoG is important for cell proliferation through the control of ROS and that Humanin can restore cell division in EndoG-deficient cells and counteracts the effects of ROS on AKT phosphorylation.
Keywords: EndoG, Cell proliferation, Mitochondria, Reactive oxygen species, Cell signaling, Humanin, Romo1
Highlights
-
•
Reduced expression of the mitochondrial nuclease EndoG induces ROS production.
-
•
EndoG deficiency hampers cell proliferation through ROS-dependent signaling.
-
•
Increased ROS in EndoG-deficient cells limits the Akt/Gsk3/cyclin axis activity.
-
•
Humanin sustains proliferation despite high ROS levels induced by Endog deficiency.
-
•
Romo-1 deficiency reduces cell proliferation independently of EndoG and ROS.
1. Introduction
Cell division or growth and cell's mitochondrial function are mutually regulated in order to adjust energy supply and molecular precursors to each cell function. Also the switch between one and another depends on nutrient availability [1,2]. Mitochondrial electron transport chain (METC) activity has been directly correlated with cell division and inversely correlated with cell growth [1]. Cyclin D1 regulates mitochondrial respiration in cycling epithelial cells [3] and, recently, phosphorylation of METC complex I by Cyclin B/cdk1 has been shown to improve respiration and shorten cell cycle time [4]. Reactive oxygen species (ROS) generated by the METC are directly related to the control of cell proliferation in Drosophila [5] and redox signaling influences diverse aspects of normal cell function, particularly in the heart, as reviewed in Ref. [6]. High ROS production due to the increase of environmental oxygen concentration after birth and ROS-induced sub-lethal DNA damage have been shown to trigger cardiomyocyte cell cycle arrest in neonatal mice [7]. However, the mediators and regulators naturally influencing ROS production and, hence, the effect of ROS on cell division and growth have not been identified.
EndoG is a nuclease that was first characterized in bovine heart mitochondrial extracts [8]. In addition to its implication in caspase-independent cell death [9], EndoG has been shown to influence mitochondrial DNA (mtDNA) replication in vitro [10] and DNA recombination [11]. However, the relevance of EndoG in vivo was unclear because a first characterization of two independently generated Endog-deficient mice showed no overt phenotype [12,13]. We previously identified Endog as a determinant of cardiac hypertrophy [14] and showed that spontaneously hypertensive rats, which naturally lack Endog expression, and Endog knockout mice, have increased ROS abundance in the heart and larger cardiomyocytes [14]. Our recent results show that EndoG is involved in the control of mtDNA replication and abundance, which influences mitochondrial ROS production, modulating cardiomyocyte size [15]. However the increase in cardiomyocyte size did not fully translate into increased heart mass in Endog-deficient mice [14].
Oxidative stress influences cardiomyocyte division at birth [7], ROS control cell division in the fly [5] and Endog deficiency induces increased ROS abundance [14,15]. Therefore, we wanted to assess whether, in addition to influence cell size, EndoG function influences cell division and, if so, whether the effect is mediated by the control of ROS generation. In the present work, we analyzed the influence of EndoG in cell proliferation and characterized the signaling mediators controlling it, using different approaches to reduce Endog/ENDOG gene expression in different cell types; we also quantified ROS abundance and assessed their impact on the EndoG-dependent changes in cell signaling and proliferation.
2. Materials and Methods
2.1. Bioethics statement for Endog knockout mouse line and rat neonates
The investigation with experimental animals was approved by the Experimental Animal Ethic Committee (CEEA) of the University of Lleida (codes CEEA06-01/10,07-01/10, 08-01/09 and 09-01/09), comply with the ARRIVE Guidelines and conforms to the Guide for the Care and Use of Laboratory Animals, 8th Edition, published in 2011 by the US National Institutes of Health. Endog mouse colony is derived from founders given by Dr. Michael Lieber, University of Southern California, LA, CA, USA (Irvine, R.A.; Adachi, N. 2005), have a C57BL/6J background and was housed in Tecniplast GM500 cages (391 × 199 × 160 mm) never exceeding 5 adults/cage. Rats used for neonatal cardiomyocyte culture had a Sprague-Dawley background and were housed in conventional rooms. All animals were housed at the Experimental Animal Housing Facility—University of Lleida, lights on from 7 a.m. to 7 p.m., temperature = 18–22 °C and 30–70% humidity. Enriched environment included autoclaved cellulose material. Animals were fed 2914 diet (Irradiated Teklad Global 14% Protein Rodent Maintenance Diet, Harlan) and sterilized tap water, both ad libitum. Wellbeing of animals is monitored daily by visual inspection and, for SPF housed mice, pathogen analysis is monitored from sentinel animals in periods of 8 weeks following the standards determined by the Federation of European Laboratory Animal Science Association (FELASA). Adult mice and neonatal rat pups were sacrificed following the Guidelines of our CEEA.
2.2. Heart cardiomyocyte cell counting, histological procedures and primary cell culture
Mouse hearts were dissected, weighted and cardiomyocytes were counted as previously described [16]. Briefly, hearts were dissected and formol-fixed at 4 °C followed by a treatment with 12.5 M KOH overnight at 4 °C. After a 10min vortex to dissociate the cells, they were passed through a 250 μm mesh, centrifuged, suspended in phosphate buffered saline (PBS) and cardiomyocytes were counted using a Neubauer chamber. Cardiomyocytes in the counting chamber were distinguished from fibroblasts and debris by cytoplasmic size and cell shape. Immunohistochemistry was performed in paraffin-included heart slices as previously described [16] using primary antibodies against Ki67 (MAB4190, Merck) and Cyclin D (SC-20044), biotinylated secondary antibodies (Jackson) and developed with deaminobenzidine (DAB, DAKO) contrasted with hematoxylin. Microscopic images were obtained using a Leica DMD108 microscope and 1000–2000 nuclei were assessed for Ki67 staining/mouse using ImageJ software (Wayne Rasband, NIH). Rat and mouse neonatal cardiomyocyte cultures were performed as previously described [17], using all available pups (usually 2–3 pups per genotype), and purity was assessed by fluorescent detection of α-sarcomeric actinin (A7811, SIGMA) visualized with an Olympus IX70 vertical epifluorescence phase-contrast microscope. Methods for neonatal mouse cardiomyocyte and fibroblast cultures were adapted from Bahi et al., 2006 [18].
2.3. Cell lines
Human embryonic kidney cell line HEK293 (CRL-1573) and Rat-1 (CRL-2210) rat fibroblasts were purchased from the American Type Culture Collection. All reagents for cell culture were from GIBCO. All cell lines were grown in Dulbecco's modified Eagle medium (#41965-039) supplemented with 10% fetal bovine serum and, except for Rat-1 cells, also supplemented with 1 mmol/L HEPES (#15630-056), 1 mmol/L sodium pyruvate (#11360-039), 2 mmol/L l-glutamine (#25030-024), 1 mmol/L MEM NEAA (#11140-035) and 100 U/ml of penicillin/100 μg/ml streptomycin (#15140-122) at 37 °C with saturating humidity and 5% CO2. All experiments were conducted with low passage cells from recently resuscitated frozen stocks.
2.4. Modification of ENDOG/Endog expression in cultured cells
To reduce EndoG expression in cultured cells, two approaches were used. Lentiviral vector pLVTHM (Dr. Trono's lab., Switzerland) containing small hairpin RNA interference sequences targeting different regions of the rat EndoG mRNA sequence were prepared as previously described and used to silence Endog expression in Rat-1 cells. Endog shRNA1 construct targets the sequence 5′-GGAACAACCTTGAGAAGTA-3′ and Endog shRNA2 is against the sequence 5′-GCAGCTTGACTCGAACTTA-3’. A lentiviral vector containing a scrambled sequence from shRNA1 was used as control [15]. ENDOG expression was disrupted in human HEK293 cells by CRISPR-Cas9 based gene scission and repair. In brief, we selected four sequences (5′-CCGGCCATGCGGGCGCTGCGGGC-3′, 5′-CCGGCCCGCAGCGCCCGCATGGC-3′, 5′-CCCGACGCCAGGGTCAGGCCGGC-3′ and 5′-CCGGGACTGCTGGGCCGGCTGCC-3′), spanning the first third of the ENDOG ORF within exon 1, for their use as single-guide RNAs (sgRNA1 to 4). Of note, ENDOG exons 2 and 3 overlap with the 5′UTR of SPOUT1 (UniProt Q5T280). Each sequence was subcloned in the BsbI site of the pSpCas9(BB)-2A-GFP vector (Addgene), which contains the rest of the sgRNA scaffold and also includes the Cas9 nuclease ORF [19]. In addition, we generated a PCR construct containing 1 kb-sequences at 5′ and 3′ of the Endog gene region containing the target sgRNA sequences, flanking the puromycin resistance ORF (PuroR). In the cells, this DNA fragment would disrupt the ENDOG gene after insertion of the PuroR sequence by homology-directed repair after Cas9-driven gene scission at the locus determined by the sgRNAs. This fragment was agarose gel-purified and co-transfected with the vector containing the sgRNA and Cas9 sequences in HEK293 cells growing in 96-well culture plates. Puromycin was added five days after transfection and wells containing single puromycin resistant cell clones were selected and expanded. Several HEK293 clones for each sgRNA (1–4) were PCR-tested for the recombination event and ENDOG expression was checked by qPCR and Western Blot. Cell proliferation and ROS production were compared between control cells and one positive clone per each sgRNA.
2.5. Glutathione, N-Acetyl-Cysteine, MitoTEMPO™, ethidium bromide and humanin treatments
l-Glutathione reduced (GSH, Sigma-Aldrich; G4251) was stored at 163 mM and prepared at a concentration of 2 mM in culture medium at the time of the experiment, as previously described [15,20]. The Superoxide dismutase (SOD) mimetic MitoTEMPO™ (Sigma-Aldrich; SML0737) was stored at 20 mM, prepared in water and added to the culture at a final concentration of 25 μM during 24 h [15,21]. N-Acetyl-l-cysteine (NAC, Sigma-Aldrich; A7250), a ROS scavenger, was dissolved in complete DMEM media to produce a 612 mM stock solution. Following filter sterilization (0.2 μm pore-size filters), NAC was added to the cell media at a concentration of 0.2 mM during the time required according to the experimental design, as previously described [15]. In order to reduce the mitochondrial DNA (mtDNA) content, cells were cultured with Ethidium bromide solution (EtBr) 25 ng/ml (Sigma-Aldrich; E1510) during 1 week until the cells approached a state containing 10–20% of the normal amount of mtDNA as previously described [15]. Lyophilized Humanin peptide, sequence MAPRGFSCLLLLTGEIDLPVK (custom synthesized, purity>91%; GenScript, USA) was reconstituted in DMSO to obtain a 10 mM stock solution that was subsequently dissolved in culture media to obtain the final working concentration [15].
2.6. Silencing of Romo1 expression
Romo1 gene silencing was achieved in cultured mouse fibroblasts by lentiviral transduction of validated pLKO.1 vector-subcloned Romo1 shRNA sequences (MISSION® SHCLNG-NM_025946, SIGMA). Romo1 silencing vas verified at the time indicated in the figure legend by RT-qPCR using the TaqMan specific probe Mm01604571_g1 (Thermo-Fisher).
2.7. Cell proliferation analysis
Equal number of cells were seeded from low passage plates (lower than 20 in the case of cell lines and 2 passages from tissue dissection in the case of primary cultures). For gene silencing experiments, lentiviral transduced cells were incubated 3 days and after trypsinization, cells were seed at equal densities. In every experiment, cells from 2 plates were counted a few hours after seeding to confirm equal initial cell number. This was the time of drug/vehicle addition if required. Cultures were let to grow during 48–72 h. At the end of the period, plates were rinsed with PBS and trypsinized. After mild centrifugation, pellets were resuspended in PBS and counted in a Neubauer cell chamber under a phase contrast microscope. The exact number of independent experiments performed in duplicate is specified in the figure legends.
2.8. Quantification of ROS
ROS production was determined by using the fluorescent probe MitoSOX™ Red, a cationic derivative of DHE, which is rapidly and selectively targeted to the mitochondria where, as stated by the manufacturer, it is oxidized mainly by superoxide anions (O2.−) (Thermo Fisher Scientific; M36008). We have used previously MitoSOX™ Red to detect mitochondrially generated superoxide anions by flow cytometry [15]. In brief, cells were washed twice in PBS, trypsinized and counted using a Neubauer cell chamber. Equal amount of cells from each population were incubated in PBS containing 2,5 μM MitoSOX™ Red for 10 min. Fluorescence of the cell population is proportional to the levels of intracellular ROS generated and measured with a BD FACSCanto II cytometer (Becton Dickinson, Mountain View, CA, USA) using 488 nm laser excitation and detection with BP 585/42 filter. For some control experiments we also used DHE (Sigma-Aldrich; D7008), and Dihydrorhodamine-123 (SIGMA; D1054), a probe that, following the manufacturer's instructions, localizes in the mitochondria and emits green fluorescence when oxidized.
2.9. Cell cycle analysis
Cells were seeded in 60-mm petri dishes at 1.25–2.5 × 106 cells/dish, left to adhere and treated as indicated. Afterward, cells were trypsinized and washed in ice-cold PBS, fixed in 70% ethanol and stored at –20 °C until analysis. Fixed cells were suspended in 500 μl propidium iodide (PI)/RNase staining buffer (Becton Dickinson, Franklin Lakes, NJ, USA), incubated for 30 min at 37 °C and analyzed in a BD FACSCanto II cytometer (Becton Dickinson) as in previous works [20]. Data analysis was performed using ModFit LT software (Verity software house, Topsham, ME, USA).
2.10. Analysis of gene and protein expression
Total RNA extraction from cell cultures and reverse transcription were performed as described [16,18] and quantitative Real Time PCR was performed in a iCycler iQ PCR detection system and iQ v.3 and iQ v.5 software (BioRad), using the TaqMan Gene Expression Master Mix (Applied Biosystems Cat.N. 4369016) and specific Gene Expression Assays from Applied Biosystems to amplify the transcripts of human and/or rodent ENDOG/Endog, Romo1, Irf7 (Mm00516788_m1) and Isg15 (Mm01604571_g1). Gene expression calculations were carried out as described previously for other genes [16]. Protein extraction, SDS-PAGE in standard polyacrylamide gels and Western Blot were performed and analyzed as described elsewhere [16,18]. Antibodies were against human and rodent EndoG (ab76122, Abcam), pAkt (#4060, Cell Signaling), Akt (sc-1618, Santa Cruz Biotechnology), pGSK3 (9331, Cell Signaling), GSK3 (ab18893, Abcam), pCDK1 (ab18, Abcam), CDK1 (ab32384 Abcam), Cyclin D (SC-20044 Santa Cruz Biotechnology), Cyclin B (ab181593, Abcam), HDAC4 (SC-11418, Santa Cruz Biotechnology), GAPDH (ab8245, Abcam), Lamin A/C (ab8984, Abcam) and), Catalase (sc-271803, Santa Cruz Biotechnology), MnSOD (sc-133134, Santa Cruz Biotechnology), Complex I (NDUFS3; 459130, Invitrogen), Complex II (SDHα; a11142, Molecular Probes), Complex III (Core 2; a11143, Molecular Probes), Complex IV (Cox4; A21348, Thermo Fisher) and Complex V (F1F0; a21350, Molecular Probes).
2.11. Coenzyme Q quantification
Primary neonatal skin fibroblasts were seeded in 100-mm petri dishes and let to grow until 80–90% confluence. To collect the cells for the extraction, cells were scrapped with ice cold PBS, centrifuged at 1000 g for 5 min at 4 °C and the pellet was stored at −80 °C until the day of the extraction. For total CoQ determination, cells were disrupted by adding sodium dodecyl sulphate (1%) in PBS and immediately vortexing during 1 min. One hundred of CoQ6 was added as internal control to check extraction procedure. Ethanol: isopropanol (95:5) in a proportion 2:1 was added and mixed again during 1 min. Organic extraction was performed with 600 μl of hexane. Mixture was vortexed again during 1 min and centrifuged at 1000×g for 10 min 4 C. The upper organic phase was removed and stored. Organic extraction was repeated twice. All the organic phases were pulled and dried with speed-vac at 35 °C. Dried lipid extract was dissolved in 60 μl ethanol and injected in duplicate in a Beckman 166-126 HPLC system (Beckman Coulter, Brea, CA, USA) equipped with a 20 μl loop and a 15-cm Kromasil C-18 column (Sigma Aldrich, Barcelona, Spain) maintained at 40 °C. CoQ was separated in a flux of 1 ml/min of mobile phase of 65:35 methanol/2-propanol y 1.42 mM lithium perchlorate. Total levels of CoQ were detected by an electrochemical detector and expressed as pmol/mg protein. For redox ratio between CoQred/oxidized, Cells were homogenized in potassium-phosphate buffer. To 91.5 μl of sample, 3.5 μl mercaptoethanol and 5 μl internal standard were added to avoid oxidation and control extraction. Then, 300 μl propanol was added to the mixture and vortexed 30 s. After 3 min of incubation, mixtures were vortexed again and centrifuged at 12.000×g for 1 min. Upper phase was immediately injected into a 100 μl loop of HPLC as indicated previously [22]. Determination of the CoQH2/CoQ was performed by using the electrochemical detector. Data are from three independent experiments including 3 Endog+/+ and 3 Endog−/− mice.
2.12. Nucleoid analysis
For immunofluorescence detection of dsDNA (ab27156, from Abcam) and TOM20 (sc-11415, from Santa Cruz Biotechnology), cells were plated on 12 mm diameter coverslips in 24-well plates, 15.000 cells/well to obtain 50–70% confluence upon fixation next day. Cells were fixed with 4% paraformaldehyde (PFA) for 20 min and washed twice with PBS. Then they were permeabilized with buffer A (0.1% Triton X-100, 3% FBS in PBS) for 30 min, and coverslips were incubated for 10 min with buffer B (0.05% Saponin, 2% FBS in PBS) to allow moderate permeabilization and blocking of the fixed cells. Then, coverslips were incubated with primary antibody drops (prepared in Buffer B at a dilution of 1:400) in a dark wet chamber for 30 min, washed three times for 10 min with Buffer B, and incubated with secondary antibody drops (prepared in Buffer B at a dilution of 1:800) in a dark wet chamber for 30 min. Finally, cells were washed three times for 10 min with Buffer B, nuclei were stained with Hoechst for 10 min and washed twice with PBS. Nucleoid microscope images were obtained using ZEISS Elyra 7 super-resolution microscope at x64 magnification. Nucleoid number and size quantification was performed with the ImageJ plugin 3D Object Counter, while co-localization quantification between dsDNA-TOM20 was performed with JACoP plugin, establishing the same thresholds for each plugin in each image. All quantifications were done on stacks of images. Representative images are a maximum intensity projection of all stacks. Nucleoid number and size were quantified in 42 dermal fibroblasts per genotype from 6 independent animals.
2.13. Statistical analysis
Statistics were performed with GraphPad Prism (GraphPad Software, San Diego, CA, USA). The effect of Endog expression on the experimental values was assessed by the Student's t-test. The effect of Endog expression in gene expression in Fig. 7B was performed by the Mann-Whitney U test. The Kruskal-Wallis test was used to analyze changes in MitoSOX™ fluorescence due to Endog expression and NAC addition (Fig. 1 D, G, J) and gene expression in tissues from Endog+/+ and Endog−/− mice (Fig. 7C and D), followed by the Dunn's test for selected post hoc comparisons. Moreover, one-way analysis of variance (ANOVA) followed by the Bonferroni test was performed to determine the impact of Endog expression in cell cycle in different experimental groups, and two-way ANOVA was performed to determine the impact of Endog expression and gender or drug addition or gene silencing on the experimental values and for possible interaction. Experiments were performed at least three times in duplicates; the exact number is specified in each figure legend. All statistical tests were two-sided at a significance level of <0.05.
Fig. 7.
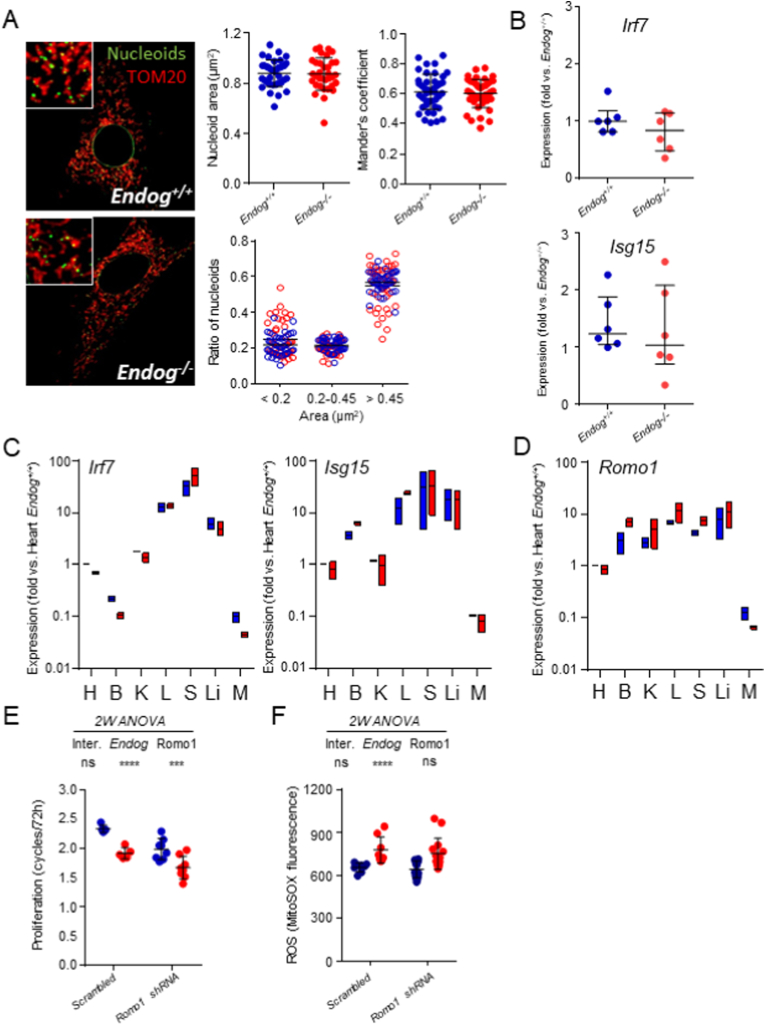
Analysis of mitochondrial DNA nucleoid morphology and expression of Interferon pathway's markers in EndoG deficient cells, and assessment of the role of Romo1 expression and function in cell proliferation and ROS production. A) Double strand DNA (nucleoids, green) and mitochondria (TOM20 staining, red) were imaged as described in the Materials and Methods section in primary cultures of skin fibroblast obtained from Endog+/+ and Endog−/− mice. Representative composite images of a single cell for each genotype are presented in the panels (insets are a magnification of the picture for detail). Nucleoid area, area group distribution and colocalization with mitochondria (Mander's coefficient) for Endog+/+ (blue) and Endog−/− (red) measured and calculated from 42 dermal fibroblasts per genotype from 6 independent animals are shown in the graphs plus mean ± SD. B) Expression of interferon pathway markers Irf7 and Isg15 was quantified in total RNA extracts obtained from cultured skin fibroblast of Endog+/+ (blue) and Endog−/− (red) mice. N = 6, dots are individual measurements performed in duplicate and corrected by Gapdh expression, median with interquartile range is indicated. Mann-Whitney U test was performed. C) Irf7 and Isg15 mRNA expression was quantified also in several tissues of Endog+/+ (blue) and Endog−/− (red) mice (N = 3/genotype). H: Heart, B: Brain, K: Kidney, L: Lung, S: Spleen, Li: Liver, M: Skeletal muscle. Bars represent min to max. values plus median of data referred to Heart. D) Romo1 mRNA expression was quantified in the same samples as in C. Bars represent min to max. values plus median of data referred to Heart. In C) and D) the Kruskal-Wallis test followed by Dunn's post hoc test was performed showing no influence of Endog expression in the expression of Irf7, Isg15 and Romo1. E) Endog+/+ and Endog−/− mouse-derived skin fibroblasts transduced with a scrambled shRNA or a mixture of 2 independent Romo1-specific shRNA and equal amounts of cells were seeded. At time zero, cell number in replicate plaques was counted to confirm equal cell number and the rest of plates were counted after 72 h. Data are expressed as the number of cell cycles completed in 72 h. All values from 5 independent experiments are plotted. F) MitoSOX™ fluorescence was quantified by flow cytometry in preparations of the same experimental groups as in (E). All values from six experiments are plotted. In E and F mean ± SD are also indicated. Two-way ANOVA was used to analyze the influence of Endog and Romo1 expression and the interaction between them in cell proliferation and ROS abundance. Ns: not significant; ***, p < 0.001; ****, p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 1.
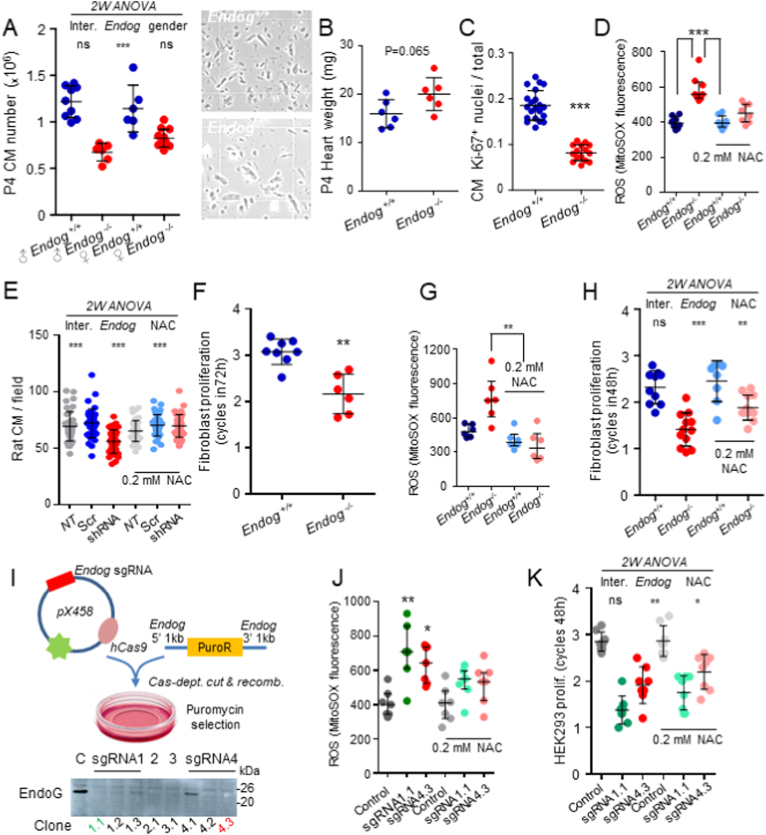
Analysis of the impact of Endog expression in cell proliferation and ROS abundance in diverse mouse, rat and human cell types. A) Cardiomyocyte (CM) number was counted from 5 to 9 hearts of Endog+/+ and Endog−/− of 4-days-old (P4) male and female mice (representative images shown at right), after digestion of the ventricular tissue that was previously weighted (B). C) Quantification of the expression of the proliferation-related Ki-67 nuclear antigen in histological ventricular tissue preparations from P4 Endog+/+ and Endog−/− mice. Ki-67+ nuclei were counted in 3 different slices from 3 hearts/genotype and data are expressed as Ki-67+ nuclei/total nuclei in the slice. All individual quantifications are plotted (1000–2000 nuclei/genotype). D) MitoSOX™ fluorescence was quantified by flow cytometry in preparations of ventricular cardiomyocytes from P4 Endog+/+ and Endog−/− mice cultured in the presence or absence of 0.2 mM N-Acetyl-Cysteine (NAC; n = 6). E) Neonatal rat ventricular cardiomyocyte cultures were left untreated (NT, not transduced), transduced with a scrambled sequence (Scr) or an Endog-specific silencing sequence (shRNA) and then treated or not with 0.2 mM NAC for 48 h. Cells were fixed and stained with muscle-specific α-actinin (cardiomyocytes) and Hoechst dye (nuclei). Cardiomyocytes were counted in 10 different microscopic fields/treatment in 4 independent experiments. All 40 values/treatment are plotted. F) Skin fibroblasts were obtained from P4 Endog+/+ and Endog−/− mice, plated and amplified. Equal number of cells were seeded in 2 plates/genotype and counted after 72 h. Data are expressed as the number of cell cycles completed in 72 h.Cells in replicate plates were counted at time zero to confirm equal initial cell number. All values from 4 independent experiments are plotted. G) MitoSOX™ fluorescence was quantified as in (D) in preparations of Endog+/+ and Endog−/− fibroblasts treated or not with NAC. H) Endog+/+ and Endog−/− fibroblasts were seeded in presence or absence of NAC and counted after 48 h. Conditions are as in F. I) Strategy for the CRISPR-Cas9-dependent ENDOG gene truncation by insertion of the puromycin resistance cassette (PuroR), using homologous recombination in 4 different selected sites within the ENDOG gene in HEK293 human cells. Expression of ENDOG was analyzed in control cells (C) and several clones from each of the four PuroR insertion sites (see Material and methods section). J) MitoSOX™ fluorescence was quantified as in (D) in preparations of control cells and cells from CRISPR-Cas9-treated clones 1.1 and 4.3 treated or not with 0.2 mM NAC (n = 7). K) HEK293 cells were counted in control and ENDOG-deficient cultures from clones 1.1 and 4.3 cultured for 48 h, treated or not with NAC. Proliferation was expressed as cell cycles completed in 48 h. Data plotted are from four experiments performed in duplicates. Graphs show experimental values plus mean ± SD, except D, G and J where median ± interquartile range is depicted. Analysis of the effect of Endog expression in the experimental values was performed with the Student's t-test (B, C and F). 2-way ANOVA was used to analyze the influence of Endog expression and gender (A) or NAC treatment (E, H and K) and the interaction between them in cell number. The Kruskal-Wallis test was used to compare the effects of Endog expression and NAC addition on MitoSOX™ fluorescence followed by the Dunn's test for selected post hoc comparisons (D, G and J). ns = not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
3. Results
3.1. EndoG deficiency restrains cell proliferation in the developing mouse heart
Previous results showed that Endog is a blood pressure-independent cardiac hypertrophy candidate gene, which regulates mitochondrial function and cardiomyocyte growth [14]. We also found that neonatal [15] and adult [14] Endog−/− mice have abnormally big cardiomyocytes but this was not fully translated into a bigger heart mass. This apparent paradox prompted us to further investigate the role of EndoG in the heart. We counted cardiomyocyte number and found that Endog deficiency was associated with a 33% decrease in cardiomyocyte number (Fig. 1A). Bigger cardiomyocyte size (Fig. 1A, right images), was not associated with a significant difference in heart weight in Endog−/− mice (Fig. 1B). This made us to suspect that hearts of Endog−/− mice were hypoplastic. A fraction of postnatal cardiomyocytes still proliferate after cell isolation and plating, therefore we analyzed the percentage of proliferating cells by immunodetection of Ki-67, which is broadly used as a proliferation marker. Our results showed a reduction in the number of cardiomyocyte nuclei expressing Ki-67 in Endog−/− cells (Fig. 1C). Consistent with our previous results in adult mice, our data showed increased ROS production in neonatal Endog−/− cardiomyocytes compared to wild type myocytes (Fig. 1D), which was diminished by the addition of the antioxidant compound N-Acetyl-l-Cysteine (NAC) (Fig. 1D). Endog-specific gene silencing in a culture of rat neonatal cardiomyocytes caused a reduction in cell number at 72 h, which was hampered by NAC addition (Fig. 1E). Taken together, these results showed that EndoG is involved in the regulation of cell division and growth in the developing rodent heart.
3.2. EndoG deficiency limits proliferation of rodent and human dividing cells
Despite its high expression in striated muscle, Endog is also expressed in many other tissues [14,17]. We posited that the influence of EndoG in cell division was broad and affected other cell types. Primary dermal fibroblasts from Endog−/− neonates proliferated more slowly than those from wild type mice in vitro, and NAC addition restored normal ROS levels and proliferation (Fig. 1F–H). The mitochondrial origin of ROS generated in EndoG-deficient cells was also assessed using the mitochondria-specific ROS probe Dihydrorhodamine-123 (Rho123) (Suppl. Fig. 1A and B). ROS levels and cell proliferation were partially normalized in EndoG-deficient primary fibroblasts by Glutathione (GSH) and MitoTEMPO™, which induced ROS neutralization, in addition to NAC (Suppl. Fig. B, C), supporting the role of ROS in reducing cell proliferation. We then checked whether EndoG had a similar function in cycling human cells. ENDOG-specific lentiviral-driven gene silencing in the human cell line HEK293 only achieved a 50% reduction of ENDOG expression and, under these conditions, we did not detect changes neither in ROS abundance nor in cell division. Therefore, we designed a CRISPR-Cas9-based strategy to disrupt ENDOG in these cells (Fig. 1I). Complete lack of ENDOG induced accumulation of ROS (Fig. 1J) and reduced cell proliferation in two different ENDOG-deficient clones (Fig. 1K). The coinciding results of all these approaches _ in vivo and in vitro gene targeting and gene silencing_ discarded the possibility that reduced proliferation was due to the alteration of an overlapping gene [23] and gave support to a role of ENDOG in the control of cell proliferation. Addition of 0.2 mM NAC diminished ROS production and partially reversed the effect on cell proliferation pointing out to a mediator role of ROS in the effect of ENDOG silencing on HEK293 proliferation (Fig. 1K).
3.3. The effect of EndoG deficiency on cell proliferation depends on the mitochondrial DNA content
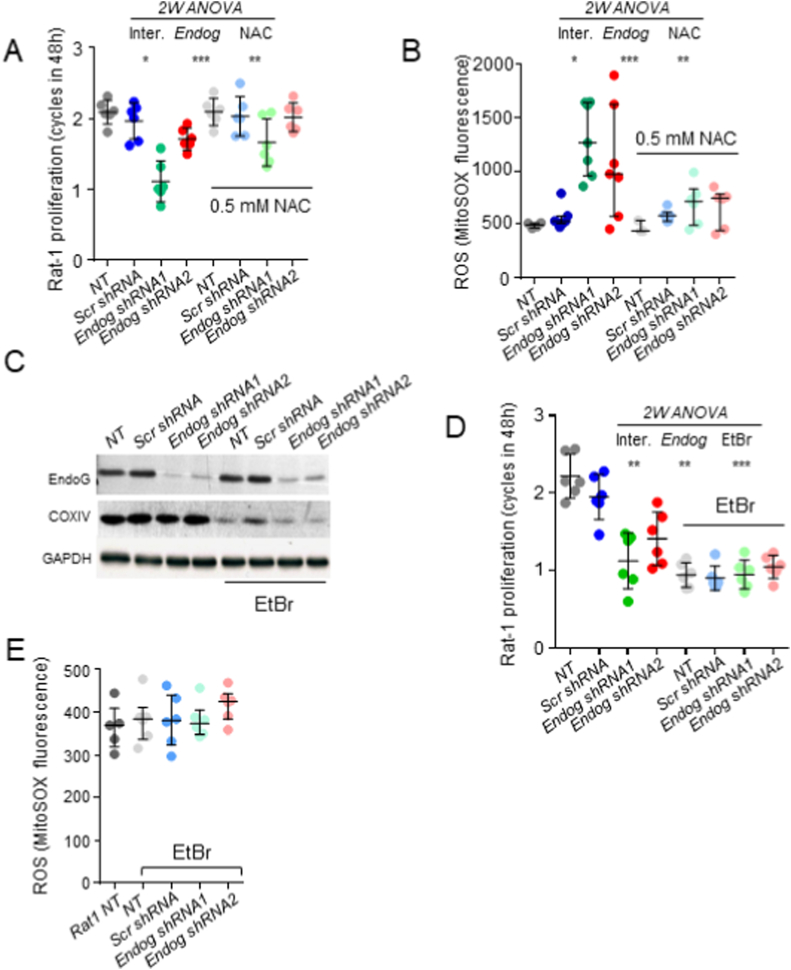
Our previous results showed that EndoG deficiency impairs mtDNA replication [15]. We wanted to determine whether the effects of EndoG deficiency on cell proliferation were dependent on the mtDNA content. To explore this, we used the rat fibroblast cell line Rat-1. EndoG-deficient Rat-1 cells reduced cell proliferation in a ROS-dependent manner as demonstrated by addition of the ROS scavenger NAC (Fig. 2A). Increased production of ROS triggered by Endog silencing in Rat-1 cells was confirmed by DHE (Suppl. Fig. 1D) and the mitochondria-specific probes MitoSOX™ (Fig. 2B), and Dihydrorhodamine-123 (Rho123) (Suppl. Fig. 1E). The increase in ROS production and the reduction in cell proliferation caused by EndoG deficiency was partially abolished by addition of the ROS scavengers NAC, GSH and MitoTEMPO™ (Suppl. Fig. 1E and F). We selected this cell line for the study of the impact of mtDNA on the effects of EndoG deficiency because primary mouse fibroblasts used previously did not maintain proliferation during the process to reduce their mtDNA content by exposure to ethidium bromide (EtBr) [15], and HEK293 cells viability was compromised when exposed to the same treatment. On the contrary, Rat-1 cells still proliferated at a rate of 0.5 duplications/day when their mtDNA was reduced enough as for reducing COXIV expression, a mitochondrially encoded protein (Fig. 2C and D). Silencing of Endog expression using two independent constructs had no effect on cell proliferation in EtBr-treated cells compared to scrambled-transduced cells (Fig. 2D). In parallel, no increase in ROS production was observed by EndoG silencing in mtDNA depleted cells (Fig. 2E). These data suggest that the capability of EndoG to influence cell proliferation could be indirect and dependent on its function in mtDNA replication.
Fig. 2.
Analysis of the influence of Endog expression in cell proliferation and ROS production in the Rat-1 cell line, and its dependence on the abundance of mitochondrial DNA. A) Equal number of Rat-1 control cells (NT, not transduced), scrambled-transduced (Scr) cells and cells transduced with either of two independent Endog silencing constructs were seeded in duplicates and the number of cells after 48 h was counted. Data are expressed as the number of cell cycles completed in 48 h in the presence or absence of the ROS scavenger NAC (0.5 mM). All values from 3 independent experiments are plotted plus mean ± SD. B) MitoSOX™ fluorescence was quantified by flow cytometry in preparations of the same experimental groups as in (A). C) Rat-1 cells were pre-treated with EtBr to reduce the mitochondrial DNA (mtDNA) content (see Materials and Methods section for details). Then, normal cells and EtBr-treated cells (EtBr) were transduced with scrambled (Scr) or two independent Endog-specific silencing constructs (Endog shRNA1, 2). Western Blot of EndoG assessing the efficacy of the silencing constructs and COXIV expression assessing the effect of EtBr treatment on mtDNA depletion, performed with samples of control and EtBr-treated cells not transduced (NT) or transduced with the different constructs. GAPDH was used as loading control. D) Equal number of control and EtBr-treated Rat-1 cells were seeded, left not transduced (NT) or transduced with scrambled (Scr) or Endog-directed silencing constructs (Endog shRNA1 and 2) in duplicate plates/treatment and counted after 48 h. Data are expressed as the number of cell cycles completed in 48 h. All values from 3 independent experiments in duplicates are plotted plus mean ± SD. E) MitoSOX™ fluorescence was quantified by flow cytometry in preparations of control not transduced cells (NT) or EtBr-treated cells not transduced (NT) or transduced with scrambled (Scr) or Endog-directed silencing constructs (Endog shRNA1 and 2). Values of four independent experiments are plotted plus mean ± SD. Two-way ANOVA was used to analyze the influence of Endog expression and NAC treatment (A, B) or EtBr treatment (D) and the interaction between them in cell proliferation and ROS production. Ns: not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
3.4. Proliferation slow down induced by EndoG deficiency associates with accumulation of cells in the G1 phase of the cell cycle
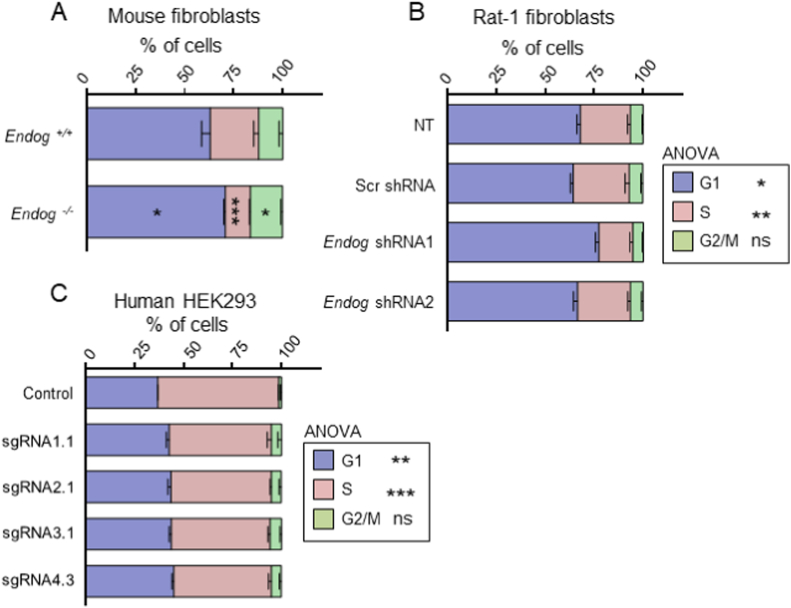
In order to characterize the intracellular events leading to cell cycle slow down induced by EndoG deficiency, we first analyzed the distribution of actively dividing cells throughout the main phases of cell cycle and the influence of Endog expression. Cells with reduced expression of Endog (Endog-specific shRNA-treated Rat-1 fibroblasts) or completely lacking its expression (primary Endog−/− fibroblasts, ENDOG-directed CRISPR-Cas9-treated HEK293 cells) tended to accumulate in the G1 phase compared to their respective controls (Fig. 3A–C).
Fig. 3.
Quantification of the Influence of Endog expression in the distribution of proliferating non-synchronized cells throughout the cell cycle phases. A) Skin fibroblast cultures were obtained from P3 Endog+/+ (n = 3) and Endog−/− (n = 6) mice and passaged two times. After 48 h in the plate, cells were detached, washed, fixed and stained with propidium iodide. The percentage of cells in each cell cycle phase was determined by flow cytometry. B) The same procedure was used for analyzing Rat-1 fibroblasts non transduced (NT) or transduced with scrambled (Scr) or Endog silencing constructs 1 and 2 (Endog shRNA) from 5 experiments; and C) for cultures of control HEK293 cells and HEK293 cell clones harboring a disruption of the ENDOG gene within exon 1 (4 experiments). Data depicted on stacked bar graphs are mean ± SEM. Statistical analysis was performed in A by the Student's t-test, and in C and D by one-way ANOVA followed by the Bonferroni test (ns: not significant; *. p < 0.05; **, p < 0.01; ***, p < 0.001).
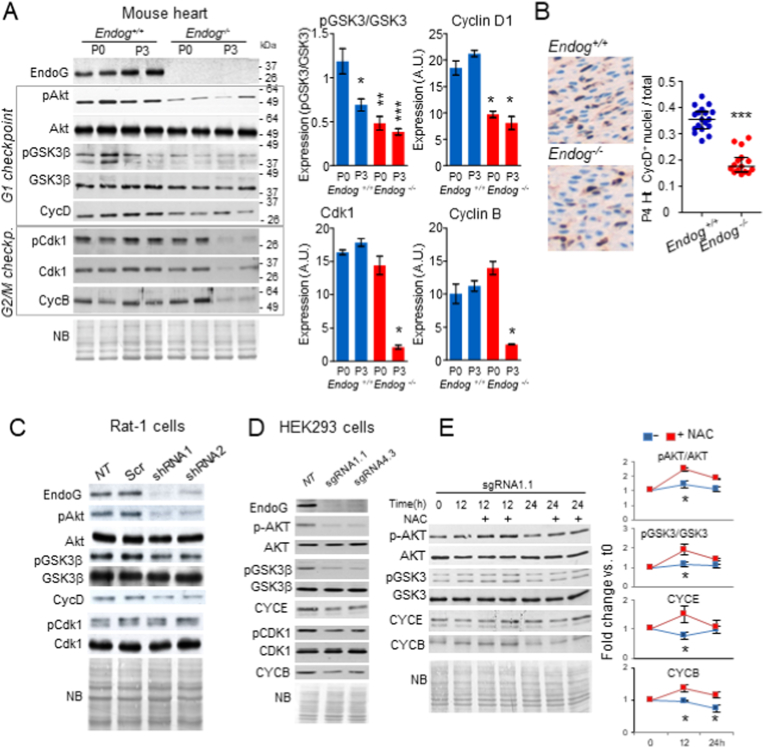
The above results lead us to analyze the expression of key regulators of cell cycle. Akt/PKB kinase regulates a plethora of intracellular events influencing cell metabolism, cell growth and cell division [24]. Akt transduces signals regulating cell division through the control of GSK-3β activity [25]. We assessed Akt phosphorylation at S473 (active Akt) and Akt-dependent GSK-3β phosphorylation at S9/S21 (inactive GSK-3β) in hearts of Endog+/+ and Endog−/− neonates at day 0 and day 3 postpartum (P0, P3), and we found lower amount of p-Akt and p- GSK-3β in EndoG-deficient hearts at both ages compared to wild type hearts (Fig. 4A). These events were associated with reduced Cyclin D expression (Fig. 4A), in accordance with the hypoplasty of EndoG-deficient myocardium. Interestingly, phosphorylation of CDK1 (cyclin-dependent kinase-1 or cdc2, cell division control protein 2 homolog) and expression of Cyclin B were only reduced in Endog−/− P3 hearts compared to controls but not at P0 (Fig. 4A). Cyclin D expression was also analyzed by immunostaining in P4 heart histological samples, corroborating a decrease in Cyclin D abundance in Endog−/− hearts compared to Endog+/+ samples (Fig. 4B). Expression analysis of these proteins in Rat-1 fibroblasts and HEK293 cells were in agreement with the results obtained in neonatal hearts, confirming the association of reduced Endog expression with lower Akt and GSK-3β phosphorylation and lower expression of Cyclin D (in Rat-1 cells) and Cyclin E (in HEK293 cells, in which Cyclin D was not detected) (Fig. 4C and D, respectively). These results showed that EndoG deficiency affects the progression through the G1 phase of cell cycle and suggest that this event involves a reduced activity of the Akt/GSK-3β axis, which influences the expression of G1 cyclins D and E.
Fig. 4.
Analysis of the influence of Endog depletion in the expression of signal transduction and cell cycle proteins. A) Expression of cell cycle regulators involved in the control of G1/S (Akt, GSK3, and their phosphorylated forms pAkt and pGSK3, Cyclin D1, Cyclin E), and G2/M (CDK1 and its phosphorylated form pCDK1 and Cyclin B in total ventricular extracts of P0 and P3 Endog+/+ and Endog−/− neonatal mice. Densitometry analysis of the Western Blot bands is represented for key genes in the bar-graphs at right. Values are means (n = 3 ± SEM). A.U., arbitrary units. The Kruskal-Wallis test was performed followed by Dunnett's test to compare all values vs. P0 Endog+/+. *, p < 0.005; **, p < 0.001; ***, p < 0.0001. B) Cyclin D immunostaining in ventricular histological preparations from Endog+/+ and Endog−/− neonatal mice. CyclinD+ nuclei were counted and referred to total nuclei in the graph at right. All 18 values from 6 hearts of each genotype are plotted. The Mann-Whitney U test was applied to check for significant differences. ***, p < 0.0001. C) The same cell cycle regulator proteins were analyzed in Rat-1 fibroblasts untreated (NT, not transduced), scrambled-transduced (Scr) and transduced with different Endog-specific silencing constructs (shRNA1 and 2) and D) in human HEK293 control cells and ENDOG-deficient clones using CRISPR-Cas9 technology to truncate ENDOG at two different regions (sgRNA1.1, sgRNA4.3; see Materials and Methods section for details). E) The effect of ROS scavenger N-Acetyl-l-Cysteine (NAC) on signaling proteins involved in cell cycle regulation was analyzed on protein extracts of ENDOG-deficient HEK293 clone sgRNA1.1 cells untreated or treated with 0.5 mM NAC for 12 and 24 h. Densitometry analysis of blots was performed and expression of key regulatory genes was referred to the values of untreated cells at time 0 in the graphs on the right. The Kruskal-Wallis test was performed followed by Dunn's test to compare control and NAC-treated values for each time point. Blots are representative of 3–5 independent experiments. NB: Naphthol blue staining of the membranes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In order to find out whether Akt-mediated signal transduction profile observed in EndoG-deficient cells was influenced by increased ROS abundance, we cultured a HEK293 clone lacking ENDOG expression (sgRNA1.1) in the presence or absence of the ROS scavenger NAC and analyzed the potential changes in cell signaling. We observed a transient recovery of pAkt/Akt, pGSK-3β/GSK-3β ratios and β-catenin and cyclin expression (Fig. 4E). Together, these results suggested an upstream role of ROS in the signal transduction linking EndoG deficiency with its biological effects on cell proliferation.
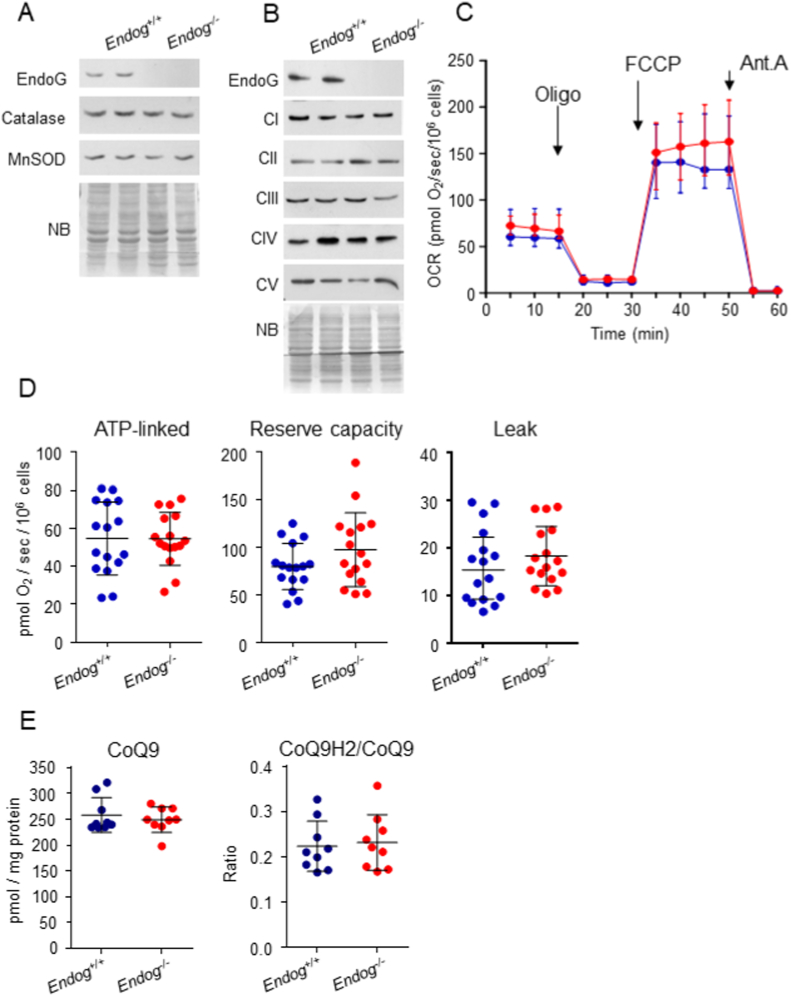
The relevant implication of ROS in the control of cell proliferation by EndoG prompted us to investigate the reason for their accumulation in EndoG-deficient cells. Despite the existence of ROS-dependent regulation of antioxidant enzymes [26,27], the major ROS metabolizing enzymes catalase and MnSOD were expressed at similar levels in fibroblasts from Endog+/+ and Endog−/− mice (Fig. 5A). Due to the fact that the METC activity is a major source of ROS and that EndoG regulates mtDNA replication and abundance [15,28], which contains genes codifying for several components of the METC, we checked the expression of several mitochondrial- and nuclear-encoded METC complex subunits in Endog+/+ and Endog−/− fibroblasts and observed no differences (Fig. 5B). Because METC activity regulation can proceed independently of METC complex abundance, we assessed O2 consumption of Endog+/+ and Endog−/− primary fibroblasts in vitro. We did not detect any genotype-dependent difference in basal, ATP-linked or maximal respiration (in presence of the uncoupler molecule FCCP), nor in the reserve respiratory capacity of these fibroblasts (Fig. 5C and D). Ubiquinone/Coenzyme Q, a lipophilic molecule which transfers electrons from either Complex I or Complex II to Complex III, is involved in the regulation of mitochondrial respiration and ROS production, and its abundance is affected by mtDNA content [29]. We analyzed the content of Coenzyme Q9, the most abundant form of Ubiquinone in mouse cells, as well as its redox state and found no differences in either parameter comparing fibroblasts from Endog+/+ and Endog−/− mice (Fig. 5E). Together, these results show that EndoG does not influence ETC function nor the expression of ROS detoxifying enzymes in living cells and, therefore, its influence in ROS abundance must involve subtler mechanisms.
Fig. 5.
Analysis of the effects of EndoG deficiency in the expression of ROS metabolizing enzymes and METC complexes, cellular respiration and CoQ redox status. A) Expression of Catalase and Mn-Superoxide Dismutase in Endog+/+ and Endog−/− skin fibroblasts. B) Expression of electron transport chain subunits I to IV and ATP-synthase (CV) in Endog+/+ and Endog−/− skin fibroblasts (NB: Naphthol Blue). C) Oxygen consumption rate (OCR) of Endog+/+ (blue) and Endog−/− (red) cultures in the presence of Oligomycin (ATP-synthase inhibitor), FCCP (METC-ATP synthase uncoupler), or Antimycin-A (Coenzyme-Q: Cytochrome-c Oxidoreductase, CIII, inhibitor). Data represent mean ± SD of 8 independent experiments comparing 2 Endog+/+ and 2 Endog−/− primary neonatal skin fibroblast cultures. D) ATP-linked respiration, reserve capacity and oxygen leak were measured in cultures of Endog+/+ (blue) and Endog−/− (red) skin fibroblast cultures from data presented in (C) following the methods described in the Materials and Methods section; individual experimental values are shown plus mean ± SD. E) Total coenzyme Q (CoQ9) and CoQ redox state (ratio of reduced CoQ vs. total CoQ) in Endog+/+ (blue) and Endog−/− (red) skin fibroblast cultures. Nine individual experimental measures are shown plus mean ± SD. Student's t-test did not find significant effects of Endog expression on the experimental values. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
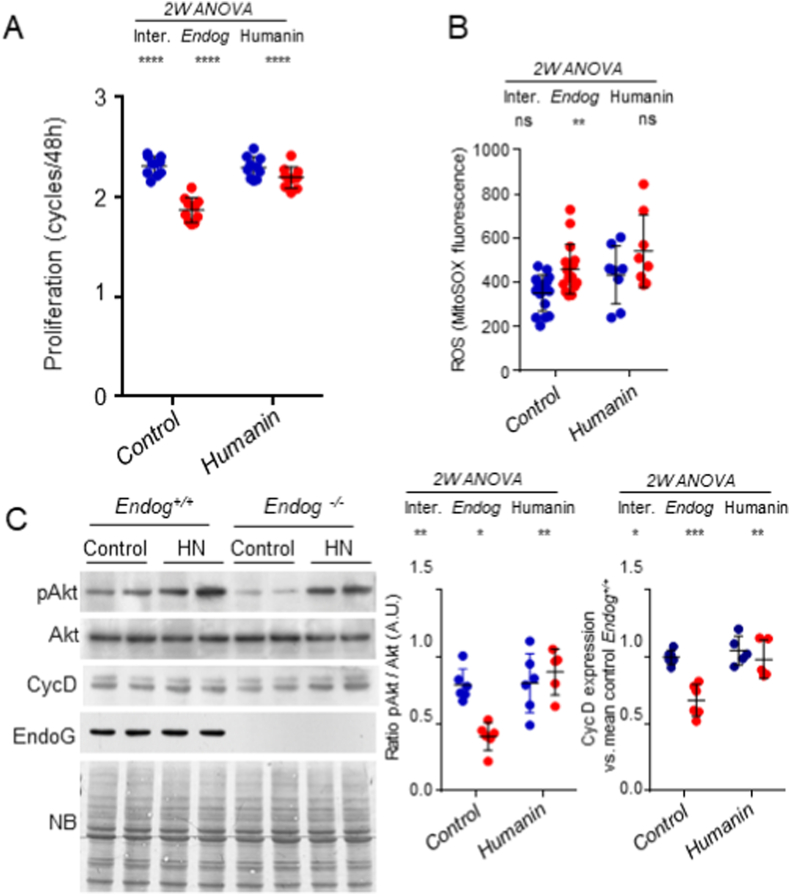
Humanin (HN) is a micropeptide that has been claimed to be encoded by an open reading frame within the 16S ribosomal RNA gene (MT-RNR2) in the mtDNA [30]. HN addition to the culture medium reduced ROS accumulation and improved metabolism in several cell models as reviewed in Ref. [30,31]. Our previous results showed that nanomolar HN concentrations can restore ROS levels and cell size in Endog-deficient cardiomyocytes [15]. Therefore, we treated cultures of primary skin fibroblasts from Endog+/+ and Endog−/− mice with HN and assessed their proliferation. Direct cell counting showed that 100 nM HN restored the proliferative capacity of Endog−/− fibroblasts (Fig. 6A) without modifying ROS production (Fig. 6B). HN also restored the normal pAkt/Akt ratio and Cyclin D expression (Fig. 6C). Together these results showed that HN can bypass the effects of ROS production, restoring Akt phosphorylation and cell proliferation in EndoG-deficient cells.
Fig. 6.
Effects of Humanin addition on the effects caused by EndoG deficiency in cell proliferation, ROS production and signal transduction. A) Equal number of Endog+/+ and Endog−/− mouse-derived skin fibroblasts were seeded. At time zero, cell number in replicate plaques was counted. DMSO (Control) or 100 nM Humanin (HN) diluted in DMSO were added in duplicate plates/treatment and cells were counted after 48 h. Data are expressed as the number of cell cycles completed in 48 h. All values from 5 independent experiments in duplicates are plotted. B) MitoSOX™ fluorescence was quantified by flow cytometry in preparations of the same experimental groups as in (A). Eight experiments for controls and four experiments for HN treatment in duplicates are plotted. C) Western Blot of pAkt, total Akt and EndoG was performed with cultures of primary skin fibroblasts of 6 Endog+/+ and 6 Endog−/− adult mice (NB: Naphthol blue staining of the membrane). After densitometric quantification of pAkt and Akt Western Blot bands, the ratio was calculated for each sample and plotted in the graph at right. Data in all the graphs represent individual values, mean ± SD (Endog+/+, blue dots; Endog−/−, red dots). Two-way ANOVA was used to analyze the influence of Endog expression and Humanin treatment and the interaction between them in cell proliferation, ROS production and Akt phosphorylation. Ns: not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Inflammation is frequently associated with increased ROS [32,33] and mitochondrial DNA (mtDNA) stress has been shown to induce changes in nucleoid morphology, mtDNA release and activation of inflammation leading to the induction of IFN signaling and Interferon-stimulated gene 15 (Isg15) expression [34,35]. EndoG deficiency induces a reduction in mtDNA content [15] and this event has been related to nucleoid morphology alteration and induction of Isg15 expression in other experimental models [35]. Therefore, we wanted to explore the possibility that inflammation was present in EndoG-deficient cells. We assessed nucleoid morphology and distribution in Endog+/+ and Endog−/− primary skin fibroblasts and did not find any genotype-dependent difference (Fig. 7A). The transcripts of Irf7, which is frequently required in the pathway [35], and Isg15 were expressed at similar levels in wild type and EndoG-deficient cells (Fig. 7B). No changes in Irf7 or Isg15 expression related to Endog absence were observed in any tissue analyzed in adult mice (Fig. 7C).
In search for alternative sources of ROS that could be influenced by EndoG, our interest was focused in Reactive Oxygen Species Modulator-1 (Romo1), a recently characterized inner mitochondrial membrane protein shown to be involved in ROS production and cell proliferation [36]. Although its function is still being characterized and its role in ROS production is debated [[37], [38], [39]], we decided to assess whether its expression could contribute to the changes in ROS production and cell proliferation observed in EndoG-deficient cells. Romo1 expression was similar in tissues of adult Endog+/+ and Endog−/− mice at the transcript level (Fig. 7D) and Romo1 gene silencing by shRNA-directed RNA interference reduced proliferation at similar levels in primary fibroblasts of both genotypes (Fig. 7E), without affecting ROS abundance (Fig. 7F). These results did not support a relevant contribution of Romo1 to the effects of EndoG in cell proliferation and ROS production, yet add novel information about this protein.
4. Discussion
The results presented here show that the mitochondrial nuclease EndoG is important for normal cell proliferation. A reduction of Endog expression slowed cell proliferation associated to an increase in ROS abundance, the accumulation of cells in the G1 phase of cell cycle and the reduction in the activity of the Akt-GSK-3β-Cyclin D axis. These effects were replicated in vivo in mouse cardiomyocytes, and in vitro in primary cell cultures, cell lines of rodent and human origin and by using several methods for downregulating Endog gene expression ranging from in vivo gene knock-out, to RNA interference using either small hairpin-RNA lentiviral vectors and CRISPR-Cas9-driven gene targeting by homologous recombination in cell cultures. Addition of ROS scavengers NAC, GSH or MitoTEMPO™ reduced ROS accumulation in EndoG-deficient cells and recovered cell proliferation, Akt phosphorylation and Cyclin D expression, showing that ROS contribute significantly to the reduction of EndoG-deficient cell proliferation. The results also show that changes in mtDNA influence the increase in ROS occurring in EndoG-deficient cells, and that ROS increase is not due to changes in mitochondrial respiration, expression of ROS detoxifying enzymes, nor to mtDNA-induced inflammation. Furthermore, our results also show that Romo1, which is located in the same compartment than EndoG and has a debated role in cell proliferation and ROS production, does not mediate the effects of EndoG deficiency in these events.
Previously, we identified the Endog gene as a determinant of cardiac hypertrophy and showed that EndoG is involved in the regulation of cardiomyocyte growth through the control of mitochondrial ROS production [14]. However, cardiomyocyte hypertrophy was not fully translated into an overt increase of heart's weight in Endog−/− mice [15]. Increased cell death in the EndoG-deficient heart could account for the above discrepancy. However, the original reports on two independent Endog−/− mouse models found no differences in the induction of apoptosis compared to Endog+/+ cells [12,13]. In addition, our previous results showed that cardiomyocytes lacking Endog expression have less apoptotic nuclei, assessed both in vitro [17] and in vivo [40]. Therefore, we decided to further explore the cardiac phenotype of these mice and found a 30% reduction in cardiomyocyte number at birth, indicating that EndoG is important for normal cardiomyocyte proliferation during mouse's heart development. ENDOG gene silencing was previously suggested to reduce the proliferation of HEK293 cells [23]. However, in that report the possible impact of the silencing method and the transformed nature of the human cell line used questioned the actual implication of EndoG in cell proliferation in this cell model. Here, the effect of Endog deletion in primary skin fibroblast proliferation in vitro showed a cell-autonomous role of EndoG in the regulation of cell proliferation, which was confirmed using several alternative strategies to reduce Endog expression and was demonstrated in several cell types. Thus, our results unequivocally demonstrate a cell-autonomous role of EndoG in cell proliferation. Increased ROS generation and decreased cell proliferation rate in EndoG-deficient cells was influenced by mtDNA, because mtDNA-depleted Rat1 fibroblasts did not experiment changes in ROS abundance nor in cell proliferation after Endog gene silencing. These results are in agreement with our previous findings showing that ROS scavengers did not normalize mtDNA replication in Endog−/− mouse embryonic fibroblasts (MEF) [15]. The analysis of cell cycle consistently showed a tendency of EndoG-deficient cells to accumulate in the G1 phase, contrary to the G2 arrest reported elsewhere [23]. This was accompanied by a reduction in both Akt and GSK-3β phosphorylation and decreased Cyclin D expression, which preceded Cyclin B downregulation in postnatal cardiomyocytes. The AKT-dependent signaling pathway is involved in the control of the G1/S checkpoint and our results suggest that, by controlling ROS abundance, EndoG is important for proliferating cells to successfully overcome this step of the cell cycle.
ROS-dependent regulation of protein kinases and phosphatases involves the reversible oxidation of key Cys residues and S-glutathionylation [[41], [42], [43], [44]], with impact in many aspects of cell biology. In particular, hydrogen peroxide-induced formation of a disulfide bond in Akt has been shown to inhibit Akt phosphorylation and induce apoptosis [42]. Furthermore, a recent report has identified an evolutionarily conserved mechanism of redox regulation of Ser/Thr kinases involving a conserved Cys in the activation segment, which can control the enzyme's activity without affecting its phosphorylation [43]. The report shows that this redox sensitive regulation of protein kinase activity can affect, among many others, Akt and Aurora, which are important for the regulation of cell proliferation [43]. Increased ROS production in dividing EndoG-deficient cells shown here, and our previous results showing low amounts of free GSH in EndoG-deficient Rat-1 cells, which were restored by NAC addition [15] could be involved in the reduced phosphorylation of Akt and in the proliferation slow down due to EndoG deficiency through any of the mechanisms described by Murata and collaborators and Byrne and collaborators, and affect other key enzymes, which remain to be explored.
ROS are known to influence cell proliferation at many levels [45]. Due to the involvement of ROS in the effects of EndoG deficiency on cell proliferation, we investigated the possible alterations leading to an increase in the abundance of these molecules. The mitochondrial location of EndoG and previous findings suggesting changes in METC complex activity in isolated mitochondria of Endog−/− cells [14], despite a lack of changes in the in vitro activity of isolated METC complexes [15], prompted us to analyze potential alterations of METC function as the source of ROS in living cells. However, neither changes in the oxygen consumption rate, nor in any measured parameter related to the function and coupling of the METC were found in intact primary Endog−/− skin fibroblasts. Mitochondrial MnSOD and catalase are important ROS detoxifying enzymes [26,46], but their expression was unaffected by EndoG deficiency. Humanin is a 2.4 kDa micropeptide suggested to be codified by the mtDNA [30], and having antioxidative effects [47]. Our previous results showed that addition of HN normalized ROS abundance and growth in postnatal mouse Endog−/− and Endog-specific shRNA treated rat cardiomyocytes [15], but in proliferative cells, the results presented here show that HN restores the proliferation rate, Akt signaling pathway's activity and cyclin expression without affecting ROS abundance. Indeed, HN was shown previously to induce Akt phosphorylation through the interaction with GP130/IL6ST receptor complex [30]. Taken together, these results suggest that HN could disrupt the effect of ROS on Akt signaling, allowing normal proliferation in EndoG-deficient cells.
Oxidative stress is associated with inflammation, and mtDNA stress has been shown to induce inflammation and the expression of IFN-dependent genes [34,35]. In particular, Tfam deficiency was shown to trigger a 50% reduction of the mtDNA content inducing changes in nucleoid morphology, the release of mtDNA to the cytosol, and the activation of the IFN signaling pathway leading to increased expression of Isg15 in MEFs [35]. However, EndoG deficiency, which causes a 30% reduction in mtDNA content [14], did not affect nucleoid morphology and the expression of Irf7 and Isg15 neither in cultured skin fibroblasts, nor in adult mouse tissues, to a detectable extent. These results suggest that in EndoG-deficient cells either the defect in mtDNA is not sufficient to induce changes in the morphology of nucleoids and Isg15 expression, or that the effects depend on the cell type. Recently, Endog−/− MEFs have been used as a model to show induction of the IFN-dependent signaling allegedly in the absence of alterations in the mtDNA content and cell proliferation [48]. Unfortunately, the authors did not contrast their results with those previously published by us showing that EndoG deficiency does lead to a reduction in the mtDNA content [14], and induces a defect in mtDNA replication in cardiomyocytes and primary MEFs [15]. In addition, the results shown in the present work discard any change in Isg15 expression related to Endog expression in postnatal fibroblasts and adult tissues in vivo. One possible explanation for the discrepancies on the mtDNA content, proliferation rate and Isg15 expression between the work published by Kim and co-workers and our previous and present results could be the use of immortalized Endog−/− MEFs by these authors. However, contrary to the identification of other MEF lines used in their article, the source of Endog-/- MEFs was not specified. In any case, our data robustly demonstrate that EndoG deficiency does induce changes in mtDNA content and in cell proliferation without affecting nucleoid morphology and Isg15 gene expression. In fact, another report showed that a 50% reduction of mtDNA abundance induced by Opa-1 gene deletion triggered changes in nucleoid morphology but neither induced mtDNA release, nor activation of Isg15 expression [49]. Together with previous data published elsewhere, our results demonstrate that important mtDNA stress is not always directly related to the activation of the IFN pathway.
Reactive oxygen species modulator-1, Romo1/MTGM, a nuclearly encoded inner mitochondrial membrane protein, has been shown to be required for or to inhibit ROS production [37,50] and to stimulate [36,39] or to hamper [51] cell proliferation. Our results show that Romo1 silencing induces a similar reduction in cell proliferation in Endog+/+ and Endog−/− skin fibroblasts without significant effects on ROS abundance, suggesting that Romo1 is not involved in the role of EndoG in the regulation of ROS and cell proliferation. Thus, our data contribute to the understanding of Romo1 biological roles by showing that Romo1 is important for cell proliferation and dispensable for ROS biology, in agreement with Richter's group results [39].
In conclusion, our results show that the mitochondrial nuclease EndoG is required for normal cell proliferation through the control of ROS-dependent signaling. They also suggest that the increase of ROS abundance in EndoG-deficient cells hamper Akt phosphorylation and Akt-dependent progression through the G1/S cell cycle checkpoint. The micropeptide Humanin can overcome the effect of ROS and recover normal proliferation rate in the absence of EndoG. Our results also show that low mtDNA due to EndoG deficiency does not trigger IFN-dependent inflammation, and that the mitochondrial protein Romo1 is required for normal cell division independently of EndoG. Therefore, in addition to point to EndoG as an important gene for the control of cell proliferation through the control of ROS production, our data contribute to improve the knowledge on the biological roles of Humanin and Romo1.
Author contributions
Conceptualization, D.S.; methodology, G. L-LL., A.Z., X.D., M.L. and D.S.; investigation, N.B.; A.B., G.B., C.G. R.N., A.I., G. L-LL.; validation, all authors; formal analysis, N.B., G.B., A.B., M.L. and D.S.; resources, G. L-LL., A.Z., X.D. and D.S.; writing—original draft preparation, M.L. and D.S.; writing—review and editing, D.S. with the contributions of all authors; supervision, D.S.; project administration, D.S.; funding acquisition, D.S.
Funding
This research was funded by Ministerio de Ciencia e Innovación, Gobierno de España, grant numbers SAF2013-44942-R and PID2019-104509RB-I00 to D.S. and SAF2016-80157-R to X.D.; Fundació La Marató, Catalunya, grant number 20153810 to D.S.; AGAUR, Generalitat de Catalunya, Catalunya, grant number 2014-SGR-1609 to D.S. G.B. holds a contract from the University of Lleida; A.B. contract has been funded by Fundació La Marató TV3 and Diputació de Lleida/IRBLleida.
Declaration of competing interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101736.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lee I.H., Finkel T. Metabolic regulation of the cell cycle. Curr. Opin. Cell Biol. 2013;25:724–729. doi: 10.1016/j.ceb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miettinen T.P., Pessa H.K., Caldez M.J., Fuhrer T., Diril M.K., Sauer U., Kaldis P., Björklund M. Identification of transcriptional and metabolic programs related to mammalian cell size. Curr. Biol. 2014;24:598–608. doi: 10.1016/j.cub.2014.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakamaki T., Casimiro M.C., Ju X., Quong A.A., Katiyar S., Liu M., Jiao X., Li A., Zhang X., Lu Y., Wang C., Byers S., Nicholson R., Link T., Shemluck M., Yang J., Fricke S.T., Novikoff P.M., Papanikolaou A., Arnold A., Albanese C., Pestell R. Cyclin D1 determines mitochondrial function in vivo. Mol. Cell Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z., Fan M., Candas D., Zhang T.Q., Qin L., Eldridge A., Wachsmann-Hogiu S., Ahmed K.M., Chromy B.A., Nantajit D., Duru N., He F., Chen M., Finkel T., Weinstein L.S., Li J.J. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev. Cell. 2014;29:217–232. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owusu-Ansah E., Yavari A., Mandal S., Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 6.Kimura W., Muralidhar S., Canseco D.C., Puente B., Zhang C.C., Xiao F., Abderrahman Y.H., Sadek H.A. Redox signaling in cardiac renewal. Antioxidants Redox Signal. 2014;21:1660–1673. doi: 10.1089/ars.2014.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puente B.N., Kimura W., Muralidhar S.A., Moon J., Amatruda J.F., Phelps K.L., Grinsfelder D., Rothermel B.A., Chen R., Garcia J.A., Santos C.X., Thet S., Mori E., Kinter M.T., Rindler P.M., Zacchigna S., Mukherjee S., Chen D.J., Mahmoud A.I., Giacca M., Rabinovitch P.S., Aroumougame A., Shah A.M., Szweda L.I., Sadek H.A. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings O.W., King T.C., Holden J.A., Low R.L. Purification and characterization of the potent endonuclease in extracts of bovine heart mitochondria. J. Biol. Chem. 1987;262:2005–2015. http://www.ncbi.nlm.nih.gov/pubmed/3818585 [PubMed] [Google Scholar]
- 9.Li L.Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 10.Côté J., Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261(80):765–769. doi: 10.1126/science.7688144. http://www.ncbi.nlm.nih.gov/pubmed/7688144 [DOI] [PubMed] [Google Scholar]
- 11.Robertson A.B., Robertson J., Fusser M., Klungland A. Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination. Nucleic Acids Res. 2014;42:13280–13293. doi: 10.1093/nar/gku1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irvine R.A., Adachi N., Shibata D.K., Cassell G.D., Yu K., Karanjawala Z.E., Hsieh C.L., Lieber M.R. Generation and characterization of endonuclease G null mice. Mol. Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David K.K., Sasaki M., Yu S.W., Dawson T.M., Dawson V.L. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- 14.McDermott-Roe C., Ye J., Ahmed R., Sun X.-M., Serafin A., Ware J., Bottolo L., Muckett P., Canas X., Zhang J., Rowe G.C., Buchan R., Lu H., Braithwaite A., Mancini M., Hauton D., Marti R., Garcia-Arumi E., Hubner N., Jacob H., Serikawa T., Zidek V., Papousek F., Kolar F., Cardona M., Ruiz-Meana M., Garcia-Dorado D., Comella J.X., Felkin L.E., Barton P.J.R., Arany Z., Pravenec M., Petretto E., Sanchis D., Cook S.A. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasco N., Cámara Y., Núñez E., Beà A., Barés G., Forné C., Ruíz-Meana M., Girón C., Barba I., García-Arumí E., García-Dorado D., Vázquez J., Martí R., Llovera M., Sanchis D. Cardiomyocyte hypertrophy induced by Endonuclease G deficiency requires reactive oxygen radicals accumulation and is inhibitable by the micropeptide humanin. Redox Biol. 2018;16:146–156. doi: 10.1016/j.redox.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardona M., Antonio Lopez J., Serafin A., Rongvaux A., Inserte J., Garcia-Dorado D., Flavell R., Llovera M., Canas X., Vazquez J., Sanchis D. Executioner caspase-3 and 7 deficiency reduces myocyte number in the developing mouse heart. PloS One. 2015;10 doi: 10.1371/journal.pone.0131411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J.S., Ye J.M., Altafaj A., Cardona M., Bahi N., Llovera M., Canas X., Cook S.A., Comella J.X., Sanchis D. EndoG links Bnip3-induced mitochondrial damage and caspase-independent DNA fragmentation in ischemic cardiomyocytes. PloS One. 2011;6 doi: 10.1371/journal.pone.0017998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahi N., Zhang J.S., Llovera M., Ballester M., Comella J.X., Sanchis D. Switch from caspase-dependent to caspase-independent death during heart development - essential role of endonuclease G in ischemia-induced DNA processing of differentiated cardiomyocytes. J. Biol. Chem. 2006;281:22943–22952. doi: 10.1074/jbc.M601025200. [DOI] [PubMed] [Google Scholar]
- 19.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornago M., Garcia-Alberich C., Blasco-Angulo N., Vall-Llaura N., Nager M., Herreros J., Comella J.X., Sanchis D., Llovera M. Histone deacetylase inhibitors promote glioma cell death by G2 checkpoint abrogation leading to mitotic catastrophe. Cell Death Dis. 2014;5:e1435. doi: 10.1038/cddis.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z., Kirton H.M., Al-Owais M., Thireau J., Richard S., Peers C., Steele D.S. Epac2-Rap1 signaling regulates reactive oxygen species production and susceptibility to cardiac arrhythmias. Antioxidants Redox Signal. 2017;27:117–132. doi: 10.1089/ars.2015.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodríguez-Aguilera J., Cortés A., Fernández-Ayala D., Navas P. Biochemical assessment of coenzyme Q10 deficiency. J. Clin. Med. 2017;6:27. doi: 10.3390/jcm6030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang K.J., Ku C.C., Lehman I.R., Endonuclease G. A role for the enzyme in recombination and cellular proliferation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawlor M.A., Alessi D.R. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. https://www.ncbi.nlm.nih.gov/pubmed/11686294 [DOI] [PubMed] [Google Scholar]
- 25.Lal H., Ahmad F., Woodgett J., Force T. The GSK-3 family as therapeutic target for myocardial diseases. Circ. Res. 2015;116:138–149. doi: 10.1161/CIRCRESAHA.116.303613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glorieux C., Zamocky M., Sandoval J.M., Verrax J., Calderon P.B. Regulation of catalase expression in healthy and cancerous cells. Free Radic. Biol. Med. 2015;87:84–97. doi: 10.1016/j.freeradbiomed.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Miao L., St Clair D.K. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med. 2009;47:344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiehe R.S., Gole B., Chatre L., Walther P., Calzia E., Ricchetti M., Wiesmüller L. Endonuclease G promotes mitochondrial genome cleavage and replication. Oncotarget. 2018;9:18309–18326. doi: 10.18632/oncotarget.24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Hekimi S. Understanding Ubiquinone. Trends Cell Biol. 2016;26:367–378. doi: 10.1016/j.tcb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Kim S.J., Xiao J., Wan J., Cohen P., Yen K. Mitochondrially derived peptides as novel regulators of metabolism. J. Physiol. 2017 doi: 10.1113/JP274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C., Yen K., Cohen P. Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metabol. 2013;24:222–228. doi: 10.1016/j.tem.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B., Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J. Clin. Invest. 2018;128:3716–3726. doi: 10.1172/JCI120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harijith A., Ebenezer D.L., Natarajan V. Reactive oxygen species at the crossroads of inflammasome and inflammation. Front. Physiol. 2014;5:1–11. doi: 10.3389/fphys.2014.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rongvaux A., Jackson R., Harman C.C., Li T., West A.P., de Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.Y., Taniguchi T., Shadel G.S., Chen Z.J., Iwasaki A., Flavell R.A. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., Kaech S.M., Smiley J.R., Means R.E., Iwasaki A., Shadel G.S. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Na A.R., Chung Y.M., Lee S.B., Park S.H., Lee M.S., Do Yoo Y. A critical role for Romo1-derived ROS in cell proliferation. Biochem. Biophys. Res. Commun. 2008;369:672–678. doi: 10.1016/j.bbrc.2008.02.098. [DOI] [PubMed] [Google Scholar]
- 37.Norton M., Ng A.C.H., Baird S., Dumoulin A., Shutt T., Mah N., Andrade-Navarro M.A., McBride H.M., Screaton R.A. ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2004374. [DOI] [PubMed] [Google Scholar]
- 38.Lee G.Y., You D.G., Lee H.R., Hwang S.W., Lee C.J., Yoo Y.D. Romo1 is a mitochondrial nonselective cation channel with viroporin-like characteristics. J. Cell Biol. 2018;217:2059–2071. doi: 10.1083/jcb.201709001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richter F., Dennerlein S., Nikolov M., Jans D.C., Naumenko N., Aich A., MacVicar T., Linden A., Jakobs S., Urlaub H., Langer T., Rehling P. ROMO1 is a constituent of the human presequence translocase required for YME1L protease import. J. Cell Biol. 2019;218:598–614. doi: 10.1083/jcb.201806093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inserte J., Cardona M., Poncelas-Nozal M., Hernando V., Vilardosa U., Aluja D., Parra V.M., Sanchis D., Garcia-Dorado D. Studies on the role of apoptosis after transient myocardial ischemia: genetic deletion of the executioner caspases-3 and-7 does not limit infarct size and ventricular remodeling. Basic Res. Cardiol. 2016;111 doi: 10.1007/s00395-016-0537-6. [DOI] [PubMed] [Google Scholar]
- 41.Kemble D.J., Sun G. Direct and specific inactivation of protein tyrosine kinases in the Src and FGFR families by reversible cysteine oxidation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5070–5075. doi: 10.1073/pnas.0806117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murata H., Ihara Y., Nakamura H., Yodoi J., Sumikawa K., Kondo T. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J. Biol. Chem. 2003;278:50226–50233. doi: 10.1074/jbc.M310171200. [DOI] [PubMed] [Google Scholar]
- 43.Byrne D.P., Shrestha S., Galler M., Cao M., Daly L.A., Campbell A.E., Eyers C.E., Veal E.A., Kannan N., Eyers P.A. Aurora A regulation by reversible cysteine oxidation reveals evolutionarily conserved redox control of Ser/Thr protein kinase activity. Sci. Signal. 2020;13:1–22. doi: 10.1126/scisignal.aax2713. [DOI] [PubMed] [Google Scholar]
- 44.Corcoran A., Cotter T.G. Redox regulation of protein kinases. FEBS J. 2013;280:1944–1965. doi: 10.1111/febs.12224. [DOI] [PubMed] [Google Scholar]
- 45.Diebold L., Chandel N.S. Mitochondrial ROS regulation of proliferating cells. Free Radic. Biol. Med. 2016;100:86–93. doi: 10.1016/j.freeradbiomed.2016.04.198. [DOI] [PubMed] [Google Scholar]
- 46.Fridovich I. Related matters *. J. Biol. Chem. 1997:18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 47.Yen K., Lee C., Mehta H., Cohen P. The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J. Mol. Endocrinol. 2013;50:R11–R19. doi: 10.1530/JME-12-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., Gupta R., Blanco L.P., Yang S., Shteinfer-Kuzmine A., Wang K., Zhu J., Yoon H.E., Wang X., Kerkhofs M., Kang H., Brown A.L., Park S.J., Xu X., van Rilland E.Z., Kim M.K., Cohen J.I., Kaplan M.J., Shoshan-Barmatz V., Chung J.H. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366(80):1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodríguez‐Nuevo A., Díaz‐Ramos A., Noguera E., Díaz‐Sáez F., Duran X., Muñoz J.P., Romero M., Plana N., Sebastián D., Tezze C., Romanello V., Ribas F., Seco J., Planet E., Doctrow S.R., González J., Borràs M., Liesa M., Palacín M., Vendrell J., Villarroya F., Sandri M., Shirihai O., Zorzano A. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 2018;37:1–18. doi: 10.15252/embj.201796553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung Y.M., Kim J.S., Yoo Y.D. A novel protein, Romo1, induces ROS production in the mitochondria. Biochem. Biophys. Res. Commun. 2006;347:649–655. doi: 10.1016/j.bbrc.2006.06.140. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J., Liu T., Jin S.B., Tomilin N., Castro J., Shupliakov O., Lendahl U., Nistér M. The novel conserved mitochondrial inner-membrane protein MTGM regulates mitochondrial morphology and cell proliferation. J. Cell Sci. 2009;122:2252–2262. doi: 10.1242/jcs.038513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.