Abstract
Herbivorous insects use plant volatile compounds to find their host plants for feeding and egg deposition. The monophagous beetle Agasicles hygrophila uses a volatile (E)-4,8-dimethyl-1,3,7-nonanetriene (DMNT) to recognize its host plant Alternanthera philoxeroides. Alternanthera philoxeroides releases DMNT in response to A. hygrophila attack and nerolidol synthase (NES) is a key enzyme in DMNT biosynthesis; however, the effect of A. hygrophila on NES expression remains unclear. In this study, the A. philoxeroides transcriptome was sequenced and six putative NES genes belonging to the terpene synthase-g family were characterized. The expression of these NES genes was assayed at different times following A. hygrophila contact, feeding or mechanical wounding. Results showed that A. hygrophila contact and feeding induced NES expression more rapidly and more intensely than mechanical wounding alone. This may account for a large release of DMNT following A. hygrophila feeding in a previous study and subsequently facilitate A. hygrophila to find host plants. Our research provides a powerful genetic platform for studying invasive plants and lays the foundation for further elucidating the molecular mechanisms of the interaction between A. philoxeroides and its specialist A. hygrophila.
Subject terms: Herbivory, Wounding, Entomology
Introduction
Alligator weed, Alternanthera philoxeroides (Mart.) Griseb. (Caryophyllales: Amaranthaceae), is an invasive weed that has spread across 30 countries, including the USA, New Zealand, and China1. It not only causes serious economic losses by hindering river transportation and crop production, but also has a detrimental impact on native wildlife, causing many ecological problems in the invaded areas2–4. The most successful method of effectively controlling A. philoxeroides is the use of the alligator weed flea beetle Agasicles hygrophila (Selman and Vogt) (Coleoptera: Chrysomelidae), which is stably monophagous and has a high food requirement1. In China, the safety of using A. hygrophila near local crops was evaluated, revealing that the specific diet of this beetle did not change even many years after its introduction5,6. This stable and specific feeding relationship means that A. hygrophila and its host plant A. philoxeroides can be an useful model to study the interaction between plants and their host-specific herbivorous insects; however, few reports have been published on this particular relationship.
Research into the specific interaction between A. hygrophila and A. philoxeroides has largely focused on surface phenomena, such as the biological performance, life history, and host selection of A. hygrophila2,5,6. Recently, this plant–insect specificity was further elucidated in a report that host specificity is related to the response of A. hygrophila to specialized volatiles released by the plant, which can be detected at a distance7. This laboratory-based study found that under undamaged conditions the main volatile of A. philoxeroides, homoterpene (E)-4,8-dimethyl-1,3,7-nonanetriene (DMNT), was present in larger quantities in this species than in other plants tested. Agasicles hygrophila prefers the volatile emitted from plants fed by conspecific insects for 24 h than those from healthy plants, because insect injured plants released more DMNT. DMNT is a significant attractant of A. hygrophila, allowing A. hygrophila to discriminate host from non-host plants. DMNT is therefore considered to be an important volatile cue for A. hygrophila to orient, locate, and feed on A. philoxeroides.
DMNT is an important plant terpenoid volatile compound, emitted by the flowers of night-scented plants and plant leaves damaged by herbivores8,9, and is thought to attract pollinators or take part in indirect defense to reduce feeding damage and enhance plant fitness10,11. Undamaged individuals of most plant species do not release DMNT, or only release a small amount, however, healthy A. philoxeroides plants also emit DMNT7, which is a derivative of nerolidol. The terpene synthase enzyme nerolidol synthase (NES) was previously reported in lima beans (Phaseolus lunatus L.), maize (Zea mays L.), and cucumber (Cucumis sativus L.), and was shown to catalyze the biosynthesis of the DMNT precursor nerolidol from farnesyl pyrophosphate (FPP)12–14. After generating nerolidol, the P450 enzyme takes part in the further cleavage reaction to produce homoterpene DMNT15. During this process, terpene synthases determine the type and the quantity of terpenoids produced16. Here, nerolidol synthase belongs to the terpene synthase g (TPS-g) subfamily, of which proteins use FPP, geranyl pyrophosphate (GPP) or geranylgeranyl pyrophosphate (GGPP) as substrates to catalyze the production of acyclic monoterpenes16.
The expression of terpene synthase genes is involved in the interactions between plants and biotic environmental factors. For example, the linalool produced by transgenic Arabidopsis thaliana expressing FaNES1 from cultivated strawberry (Fragaria × ananassa Duch.) has a repellent effect on the green peach aphid (Myzus persicae (Sulzer)), while the volatiles produced by these plants ((3, S)-(E)-nerolidol and DMNT) attract predatory mites such as Phytoseiulus persimilis Athias-Henriot17,18. Likewise, transgenic chrysanthemum (Chrysanthemum morifolium Ramat.) expressing FaNES1 releases linalool and DMNT, rapidly attracting western flower thrips (Frankliniella occidentalis (Pergande))19. Transgenic rice (Oryza sativa L.) expressing a DMNT biosynthesis gene Pltps4 from P. lunatus enhanced the attraction of Cotesia chilonis (Matsumura), a parasitoid of trunk borers20. However, there is no report on the expression of NES genes in A. philoxeroides and the associated effect on A. hygrophila.
To elucidate the specific interaction between A. philoxeroides and A. hygrophila, we explored the role of the A. philoxeroides NES genes in this relationship. When we began this study, we lacked available A. philoxeroides gene data; therefore, we performed transcriptome sequencing on mixed A. philoxeroides tissues and screened the putative NES genes. In a previous study, A. hygrophila adults were placed on A. philoxeroides for 24 h to obtain herbivore-damaged plants7. However, in general, the biosynthesis of organic volatiles from damaged plants often takes several minutes to several hours, with the release of terpenoids being typically slower than that of the green leaf volatiles (GLVs)21–23. It is possible that this long period of wounding could mask changes in the expression levels of the NES genes in these plants. Therefore, in the present study, we examined the behavior of A. hygrophila adults on the leaves in the first two hours. We then compared the expression of NES genes in leaves subjected to contact, feeding by A. hygrophila, and mechanical wounding, revealing that A. hygrophila contact upregulates NES genes in A. philoxeroides before feeding, and A. hygrophila feeding significantly increases NES gene expression. Chemicals secreted by A. hygrophila likely induce the release of large amounts of DMNT from bitten A. philoxeroides.
Materials and methods
Plant and insect
Alternanthera philoxeroides was collected from a naturalized population in Yuhuan County, Zhejiang, China, then grown in the glasshouse of Shanxi Agricultural University, Taigu, Shanxi, China. Shoots from a single individual were separately cultured in pots at a constant temperature of 25 ± 1 °C, with 14 h light and 10 h dark.
Agasicles hygrophila was obtained from South China Agricultural University, Guangzhou, China, fed with fresh A. philoxeroides leaves, and maintained in an insectary at Shanxi Agricultural University for several generations under controlled conditions (25 ± 1 °C, 80 ± 5% relative humidity, and with a 14-h light: 10-h dark photoperiod).
Transcriptome sequencing, assembly, and annotation
Alternanthera philoxeroides individuals with similar growth were selected for sampling. The fourth and fifth pairs of leaves, the third and fourth internodes from the top, flowers, and roots were harvested and frozen in liquid nitrogen for further processing. Total RNA was isolated from each of the four tissues (leaves, stems, flowers, and roots) using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) and RNA from each tissue was mixed in equal amounts. The RNA was then submitted to Biomarker Technologies (Beijing, China) for transcriptome sequencing on a HiSeq 2500 system (Illumina, San Diego, CA, USA). The sequencing adaptors, primer sequences, and low-quality data (reads that were< 50 bp and contained more N-bases) were removed from the original data, after which the clean reads were assembled using Trinity (Broad Institute, Cambridge, MA, USA)24 to obtain transcripts and unigenes. The unigenes were annotated by using them as a template for BLASTX-searches of multiple public repositories of known proteins, including RefSeq non-redundant proteins (NR), UniProtKB/Swiss-Prot25, Gene Ontology (GO; https://geneontology.org/)26, Clusters of Orthologous Genes (COG; https://www.ncbi.nlm.nih.gov/COG/)27, EuKaryotic Orthologous Groups (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/kegg/kegg1.html)28, Pfam (https://pfam.xfam.org/)29, and other databases. The results were parsed to calculate the total number of positive identities per query–subject pair and further filtered using an E-value ≤ 1e−05. The KEGG Orthology results of the unigenes were predicted using KOBAS 2.0 (https://kobas.cbi.pku.edu.cn).
Identification of terpene synthase genes
The identified nerolidol synthase FaNES1 was used to align to the unigene data from the A. philoxeroides transcriptome. The Unigenes in the results annotated as “terpene synthase” or “TPS” were then selected and aligned with the NCBI NR database using an online BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi)30. The open reading frames (ORFs) were searched with ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) and translated into amino acid sequences. DNAMan 7.0 (Lynnon Biosoft, San Ramon, CA, USA) was used to analyze the sequence identity of the A. philoxeroides TPS protein sequences. After removing incomplete sequences shorter than 500 bp and those shorter than 150 aa of translated protein sequences, we used the Maximum Likelihood method and JTT + G + I model to construct a phylogenetic tree of the TPS protein sequences in A. philoxeroides together with other plant species (Table S1) by MEGA6.0 (https://www.megasoftware.net/)31. The phylogenetic tree was verified using 1000 bootstrap replicates and was drawn by iTOL 5.6.1 (https://itol.embl.de/)32.
Investigation of the short-term feeding behavior in A. hygrophila adults
Newly emerged adults of A. hygrophila were placed in two glass bottles separately by their genders. The genders of adults were distinguished by individual size and abdominal morphological characteristics. Specifically, the female adult was larger than the male. The female adult has a ventral flat, and two abdominal segments at the end of its abdomen exposed outside the elytra. The abdomen of the male adult is covered with the elytra, and an oblong cavity that contains retractable external genitalia is in the 5th abdominal sternite. Ten newly emerged healthy A. hygrophila adults (five males and five females) were taken out from the bottles and starved for 12 h, then placed in a cylindrical box 5 cm in diameter and 4 cm in height. At the beginning of the light period, fresh leaves with stem were placed into the containers. The feeding duration of each beetle was observed and recorded for 2 h, and the area consumed by each adult was evaluated after 1 and 2 h using scanning pixel method. Specifically, we scanned a piece of square paper 10 cm in side length, the same leaf before and after insects feeding with a scanner for grayscale scanning. To calculate the gray part pixels, Photoshop CS6 was used. According to the paper size and the number of pixels, the area size of the insect feeding part was calculated as following: Consumed area = (pixel of leaf − pixel of leaf after consumed)·centimeter/pixel ratio. Centimeter/pixel ratio was calculated by Photoshop CS6.0 using a scanned picture contains a square paper 10 cm in side length. Each experiment was repeated three times.
Analyses of NES expression
Plant treatments
Plants with similar levels of growth were selected and cultured in cylindrical glass tubes (3.5 cm diameter, 30 cm height) containing 50 ml water until the roots were grown. One of the fourth pair of leaves was covered with a gauze cage (3 cm diameter, 6 cm height). Six newly emerged A. hygrophila adults (three males and three females) were starved for 12 h before they were placed in the cage. Starting from the time adults touched the leaves, a 5-min insect contact treatment was recorded as long as no feeding or excretion occurred. After 1 h of feeding, A. hygrophila was removed, and the leaves were collected at 0, 15, 30, 60, 180, 360, and 720 min after insect feeding. For each time point, different individual plants were used. Mechanical wounding was used to distinguish the effect caused by A. hygrophila secretion or mechanical damage by A. hygrophila feeding. The area and duration of mechanical wounding affect the plants’ volatile release22. Therefore, a 6-mm aperture puncher was used to mechanically injure the plant leaf margin for about 5 min until the area was similar to the insect feeding area, and the leaves were collected respectively from different individual plant at 0, 15, 30, 60, 180, 360, and 720 min after the mechanical wounding. At each point, a control was also performed. The leaves were frozen with liquid nitrogen for subsequent RNA extraction. Each treatment has five biological replicates, and each contained three leaves from different plant.
Sequence search and primer design
The sequences of the housekeeping and NES genes were identified in the transcriptome data based on their similarity with the sequences of these genes in other species stored in the NR protein database. The actin gene ACT, the α-tubulin gene TUA, the β-tubulin gene TUB, and the gene encoding ubiquitin-binding enzyme, UBC33, were selected as the reference genes. Real-time quantitative PCR (qPCR) primers (Table S2) were designed using Primer3 (https://primer3.ut.ee/) and synthesized by Sangon Biotech (Shanghai, China). To check the specificity of all primers, total RNA was reverse transcribed using the PrimeScript RT reagent kit with gDNA eraser (Takara Bio, Kusatsu, Japan). PCR was performed on the cDNA, and the PCR products were analyzed on a 1% agarose gel.
qPCR and data analysis
The total RNAs of the collected materials were extracted using the TRIzol method outlined above. The RNAs were quantified using a BioPhotometer Plus (Eppendorf, Hamburg, Germany), and their quality was verified using electrophoresis on an agarose gel. A 1-μg aliquot of total RNA was reverse transcribed at 37 °C for 15 min with an oligo-dT primer using the PrimeScript RT reagent kit with gDNA eraser (Takara Bio, Kusatsu, Japan). The gDNA eraser was used to remove genomic DNA. Two independent reactions were performed for each RNA sample.
The qPCR was performed using an Applied Biosystems 7500 Fast Real-Time Detection system (Thermo Fisher Scientific, Waltham, MA, USA) with the SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan). Each reaction was carried out according to the manufacturer’s protocol in a 20-µl volume, which contained 2 µl tenfold diluted reverse-transcribed cDNA as a template and 500 nM of each primer. In all qPCR experiments, the transcript levels of three independent biological replicates were measured, with three technical replicates performed for each sample. A no-template control was included in each run for each gene. The qPCR reaction was performed using the following conditions: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s. Finally, a melting curve was generated by increasing the temperature from 65 °C to 95 °C to determine the specificity of the reactions.
The stability of the four candidate reference genes UBC, ACT, TUA, and TUB were evaluated in the control group, A. hygrophila contact group, A. hygrophila feeding group, and mechanical wounding group using the reference screening software geNorm34, NormFinder35, and BestKeeper36. The most stable reference gene, UBC, was selected as the reference for the qPCR experiment.
The normalized expression values of the genes of interest were calculated using the 2–∆∆Ct method37. For calculating the basal gene expression in the control group, ApTPS19 was used as the control gene and the basal expression of ApTPS19 was normalized equal to 1. Other NES gene basal expression levels in the A. philoxeroides control group leaves were calculated and compared as the folds of ApTPS19 basal expression. The transcript levels of the NES genes in the control leaves and each of those subjected to contact, feeding by A. hygrophila, and mechanical wounding were transformed with logarithmic transformation to ensure normal distribution and compared using independent two-sample Student’s t-tests performed in IBM SPSS Statistics 22.0 (International Business Machines Corporation, Amonk, New York, USA). One-way ANOVA was used to analyze the basal expression of each NES gene. Two-way ANOVA was used to analyze the expression of each NES gene in each treatment group, with treatment methods and post-treatment time as the influential factors. Two-way ANOVAs were also used to analyze the expression of the NES genes in the four treatment groups at the same time point. Multiple comparisons were performed using Tukey’s post-hoc test. Student’s t-test was used to compare the expression levels of NES genes between two different treatments, after the homogeneity test.
Results
RNA sequencing, assembly, and annotation
To identify the terpene synthase genes involved in DMNT biosynthesis in A. philoxeroides, we performed transcriptome sequencing on the major tissues of this plant. A total of 9.91 Gb nucleotides and 40,984,113 raw reads were obtained from the A. philoxeroides sequencing, and the percentage of Q30 bases reached 91.14%. The original data were deposited in the NCBI SRA library under the accession number SRR10537333. In total, 95.97% of the clean reads were assembled into 4,904,951 contigs, 182,318 transcripts, and 83,179 unigenes (Table 1).
Table 1.
Summary of the RNA-seq data and the de novo assembly of the A. philoxeroides unigenes.
| Sequencing | |
|---|---|
| Raw reads | 40,984,113 |
| Clean reads | 39,332,453 |
| Q30 | 91.14% |
| N | 0.01% |
| N50 | 1103 |
| Unigene number | 83,179 |
| Unigene length (nt) | 67,987,255 |
| Mean length (nt) | 817.36 |
| Unigenes annotations based on homology with the NR protein database | 63,076 |
| Unigenes annotations based on homology with the KEGG database | 23,984 |
| Unigenes annotations based on homology with the COG database | 20,587 |
| Unigenes annotations based on homology with the GO database | 42,386 |
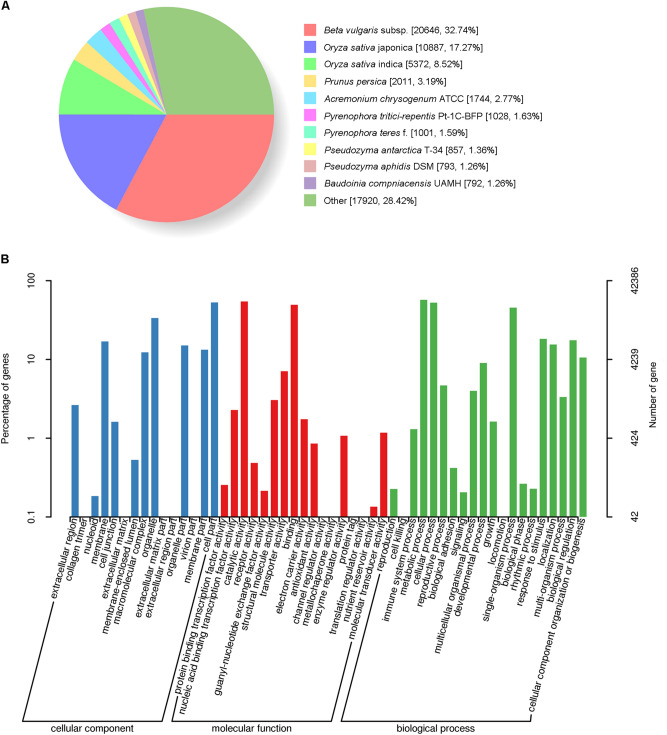
Homologs of the unigenes were identified by BLASTX; each publicly available protein database was searched using the transcriptome data, which resulted in the successful annotation of 63,536 (76.4%) of the unigenes. Among them, 63,076 genes were annotated based on their homology with sequences in the NR protein database, which accounted for 99.28% of the total number of annotations (Table 1). 32.74% of A. philoxeroides annotated sequences were aligned to beet (Beta vulgaris subsp. vulgaris L.) (Fig. 1A).
Figure 1.
Species with high levels of similarity in the NR database and GO classification statistics of A. philoxeroides annotated unigenes. (A) Species with relatively high levels of similarity to the A. philoxeroides unigenes in the NR protein database; (B) GO classification statistics of A. philoxeroides annotated unigenes.
We classified the predicted genes of A. philoxeroides using the GO, COG, KOG, KEGG, and other protein databases. A total of 42,386 unigenes were divided into three major categories and 52 functional groups (Fig. 1B); 23,984 unigenes were mapped to 277 KEGG pathways, including the glycolytic, gluconeogenesis, and pentose phosphate pathways, among which the numbers of unigenes associated with the carbon cycle (ko01200), amino acid synthesis (ko01230), and ribosome (ko03010) pathways were 1172, 1057, and 1016, respectively (Table 1; Fig. S1, S2).
Identification and analysis of the terpene synthase genes and NES genes in A. philoxeroides
In the A. philoxeroides transcriptome data, a total of 34 terpene synthase genes were identified (Table S3). Their encoded protein sequences were compared with those of other species. The sequences of terpene synthase genes in A. philoxeroides had the highest similarity to those of B. vulgaris.
The phylogenetic tree showed that the 12 terpene synthase proteins were divided into five subfamilies: TPS-a, TPS-b, TPS-c, TPS-e, and TPS-g (Fig. 2). The TPS-a, TPS-b, and TPS-g subfamilies contained the terpene synthases specific to angiosperm species, which were isolated from the other TPS subfamilies. These angiosperm-specific proteins were class-III terpene synthases, which was in accordance with the species classification of A. philoxeroides. ApTPS7, ApTPS9, ApTPS17, ApTPS18, ApTPS21, and ApTPS22 were classified into the TPS-a subfamily; ApTPS5 and ApTPS6 were classified into the TPS-b subfamily; and ApTPS16 and ApTPS19 were classified into TPS-g family. The TPS-e subfamily largely comprised kaurene synthase, which had a function similar to the annotated function of ApTPS23. The TPS-c subfamily mainly contained the monofunctional diterpene synthase, copalyl diphosphate synthase (CPS). ApTPS25, which was annotated as CPS, was also grouped into the TPS-c subfamily.
Figure 2.
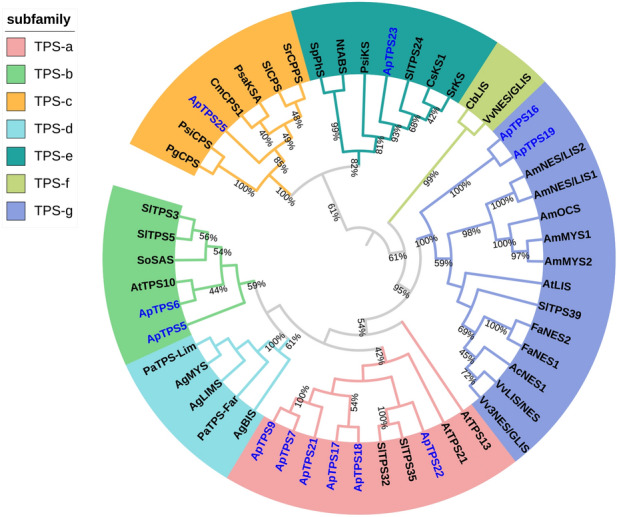
Phylogenetic tree of A. philoxeroides terpene synthase (TPSs) together with previously characterized plant TPSs.The tree was created in MEGA6 using the Maximum Likelihood method with JTT + G + I model, verified by bootstrap method, and drawn using iTOL (https://itol.embl.de/). The outermost circle layer indicates the species name and associated clade of each protein. Proteins labeled in blue font are terpene synthase proteins obtained from the A. philoxeroides transcriptome. The proteins and accession numbers used in the figure are shown in Table S3. The number on the branches indicate the supporting bootstrap value of proteins on the branches (between 40 and 100%). The closer the bootstrap value is to 1, the higher confident the branch is. The proteins on the same branch mean they are the most related proteins, which may share high homology and have similar function.
In total, six NES genes were obtained: ApTPS10, ApTPS12, ApTPS14, ApTPS15, ApTPS16, and ApTPS19 (Table 2). ApTPS19 was the longest of these genes (2161 bp), and was predicted to have an ORF of 1791 bp and to encode a protein of 596 aa in length. Four NES proteins were removed due to their shorter sequences, leaving ApTPS16 and ApTPS19, both of which were classified into the TPS-g subfamily. These proteins were annotated as NES, and shared about a 40% similarity with the other TPS enzymes in A. philoxeroides.
Table 2.
Nerolidol synthase genes in the A. philoxeroides transcriptome.
| Genes | Length (bp) | Identity (%) | e-value | Annotated function | Gene accession |
|---|---|---|---|---|---|
| ApTPS10 | 656 | 43 | 3e−10 | (3S)-linalool/(E)-nerolidol /(E,E)-geranyl linalool synthase [Vitis vinifera] | NP_001267990.1 |
| ApTPS12 | 415 | 42 | 3e–28 | nerolidol synthase [Tripterygium wilfordii] | AQA26342.1 |
| ApTPS14 | 336 | 50 | 7e–15 | (3S,6E)-nerolidol synthase 1 [Beta vulgaris subsp. vulgaris] | XP_010686920.1 |
| ApTPS15 | 830 | 61 | 3e–81 | terpene synthase 3 [Populus trichocarpa] | AEI52903.1 |
| ApTPS16 | 1495 | 46 | 1e–123 | (3S)-linalool/(E)-nerolidol /(E,E)-geranyl linalool synthase [Vitis vinifera] | ADR74213.1 |
| ApTPS19 | 2161 | 45 | 3e–147 | (3S)-linalool/(E)-nerolidol /(E,E)-geranyl linalool synthase [Vitis vinifera] | ADR74213.1 |
Short-term feeding behavior of A. hygrophila
In the present study, we examined the behavior of A. hygrophila adults on the leaves of A. philoxeroides within the first two hours. These actions include contacting, eating and resting. The time between the male and female adults touching the leaves and the initiation of feeding was 5.9 min and 6.2 min, respectively, after a 12-h starvation. The duration of a single feed was 19.6 min for males and 26.5 min for females. No statistical differences were observed between the feeding behaviors of male and female adults (Table 3). The interval between the first and second feeding periods varied from 1 to 90 min, during which the adults usually remained inactive. We, therefore, selected a 5-min period after placing the adults into the cage for use as the contact treatment in future experiments.
Table 3.
Early feeding behaviors of A. hygrophila adults.
| Gender | Time between touch and feeding (min) | Duration of single feeding behavior (min) | Leaf area consumed by a single adult within 1 h (mm2) |
|---|---|---|---|
| Female | 6.2 ± 1.27 | 26.5 ± 2.63 | 618.91 ± 59.67* |
| Male | 5.9 ± 1.35 | 19.6 ± 3.17 | 433.01 ± 28.44 |
The data in the table are presented as means ± SE. Asterisks (*) indicate a statistical difference between male and female adults, determined using a Student's t test (P < 0.05).
In addition, the leaf areas consumed by one adult in 1 h and the areas consumed by different numbers of adults in 1 h were also evaluated (Tables 3, S4). The feeding of male and female adults resulted in different leaf areas consumed; therefore, the use of six adults (three males and three females) was chosen as the insect feeding group for the subsequent 1-h feeding experiment. The leaf area consumed was about 20% of the entire leaf using this treatment method, which provided a basis for the mechanical wounding treatment of the A. philoxeroides leaves.
Selection of reference genes in leaves under A. hydrophila contact, feeding, or mechanical wounding
To ensure the accuracy of the quantitative analysis of NES expression under the different treatment conditions, the expression stabilities of the commonly used reference genes ACT, TUA, TUB, and UBC were screened. The results showed that the Ct values of the four candidate reference genes were 19–26 (Fig. S3), with the lowest Ct values observed for UBC (19–22) and the highest Ct values observed for TUB (23–26). The expression levels of each candidate reference gene were different when they were analyzed using GeNorm, NormFinder, and BestKeeper.
The GeNorm and NormFinder software analysis revealed that the M values of the four reference genes were all less than 1.5, and therefore these genes were considered to be relatively stable when expressed. Among them, UBC had the smallest M value and the best expression stability, whereas TUA had the largest M value and a relatively poorer stability. The BestKeeper software analysis revealed that the SD values of the Ct values for the four reference genes were < 1, indicating relatively stable gene expressions. Among them, UBC had the smallest SD value and the best expression stability, whereas TUA had the largest SD value and the worst stability (Table 4). Based on these results, the expression stabilities of the four candidate reference genes were ranked as follows: UBC > TUB > ACT > TUA. We therefore used UBC as the reference gene to investigate the expression of the NES genes in the leaves of A. philoxeroides under the different damage treatments.
Table 4.
Stable values of data analyses performed using GeNorm, NormFinder, and BestKeeper.
| GeNorm | NormFinder | BestKeeper | ||||
|---|---|---|---|---|---|---|
| Gene name | M | Gene name | M | Gene name | SD | CV |
| UBC | 0.776 | UBC | 0.319 | UBC | 0.53 | 2.60 |
| TUB | 0.797 | TUB | 0.319 | TUB | 0.59 | 2.38 |
| ACT | 0.843 | ACT | 0.431 | ACT | 0.85 | 3.92 |
| TUA | 0.968 | TUA | 0.573 | TUA | 0.86 | 3.67 |
M dictates the stability index: the larger the M value, the worse the expression stability of the gene. SD is standard deviation of the Ct value. CV is coefficient of variation of the Ct value. If SD < 1, the gene was considered to be relatively stable; therefore, the smaller the standard deviation and coefficient of variation, the more stable the gene expression.
Basal expression of various NES genes in healthy leaves
We investigated the basal expression level of the NES genes in the healthy leaves because of the large amount of DMNT from healthy A. philoxeroides plants. The results revealed statistical differences in the basal expression of the six NES genes in the leaves of the control plants (P < 0.05). Among them, ApTPS19 had the lowest basal expression, while ApTPS15 had the highest basal expression, which was 2058.9 times higher than that of ApTPS19 (Fig. 3).
Figure 3.
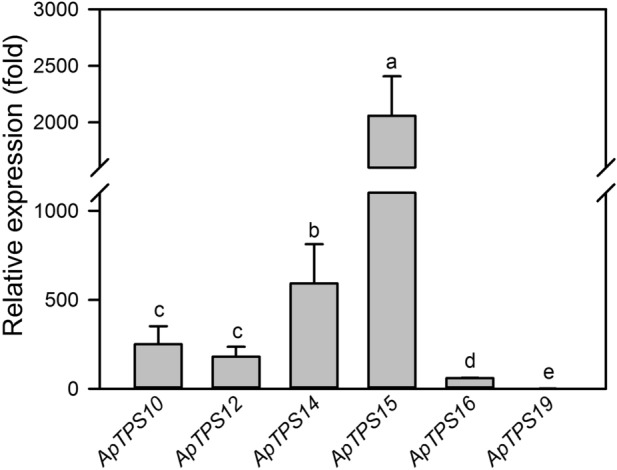
Relative transcription levels of the six nerolidol synthase genes in the plant of A. philoxeroides under undamaged control conditions. UBC was used as the reference gene to calculate the gene transcript level. The relative expression level of each NES is shown in fold relative to that of ApTPS19 which had the lowest transcription level that was taken as “1”. The values are means ± SD of three biological replications. Different letters indicate statistical differences between the genes, revealed using a one-way ANOVA and a Tukey’s post-hoc test (P < 0.05).
Induction of NES genes in A. philoxeroides prior to feeding by A. hygrophila
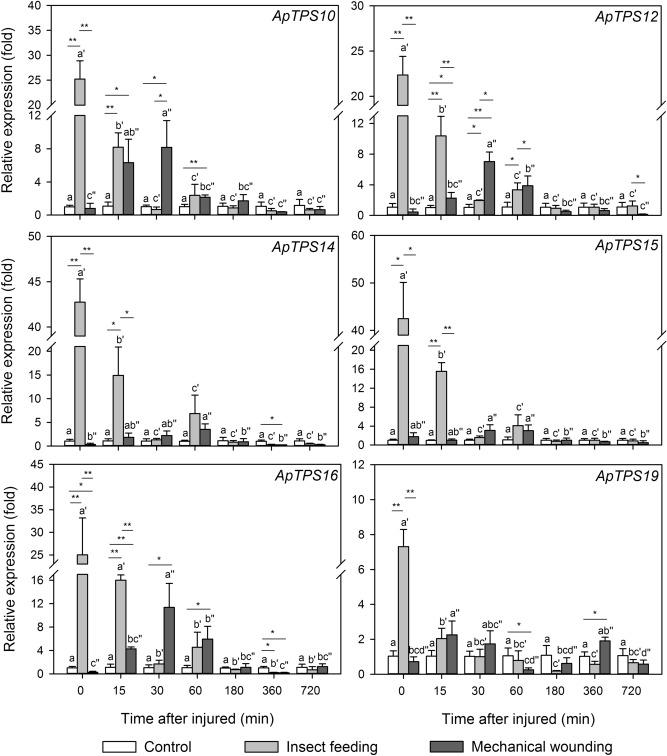
The early feeding behavior of A. hygrophila adults was investigated, revealing that the time between the flea beetles touching the leaves and the initiation of feeding was typically longer than 5 min. The changes in NES gene expression were evaluated in the leaves touched by A. hygrophila for 5 min. The expression levels of ApTPS10, ApTPS12, ApTPS16, and ApTPS19 increased significantly in A. philoxeroides leaves touched by A. hygrophila (P < 0.05), with the expression of ApTPS16 being 6.27 times higher than that of the control group. By contrast, ApTPS15 expression decreased significantly in the leaves touched by A. hygrophila (P < 0.05), whereas ApTPS14 expression was not statistically different between the control and A. hygrophila contact group (Fig. 4).
Figure 4.
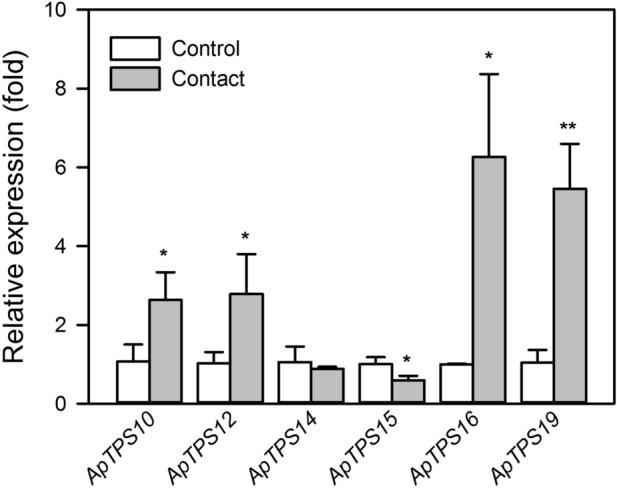
Nerolidol synthase gene expression levels in the control group and the A. hygrophila contact group of A. philoxeroides plants. UBC was selected as the reference gene. The values are means ± SD (normalized that the expression level in the control group equal to 1) of three biological replications. Asterisks indicate statistical differences between the treatments (*P < 0.05; **P < 0.01; Student’s t-test).
A. hygrophila feeding strongly induces NES gene expression in A. philoxeroides
The changes in the NES gene expression upon A. hygrophila feeding were also analyzed in a time series. All of the six NES genes in the leaves were significantly up-regulated immediately after 1 h insect-damaged (P < 0.05) (Fig. 5).
Figure 5.
Nerolidol synthase gene expression levels in the control group, A. hygrophila feeding group, mechanical wounding group of A. philoxeroides plants. In the A. hygrophila feeding group, the A. philoxeroides leaves were sampled at the indicated time points following a 1-h feeding experiment. In the mechanical wounding group, the A. philoxeroides leaves were sampled at the indicated time points following their mechanical damage. The values are means ± SD (normalized that the expression level in the control group equal to 1) of three biological replications. Different letters indicate statistical differences between different time treatments in control group, as determined using a one-way ANOVA and a Tukey’s post-hoc test (P < 0.05). Different letters with single quotation mark indicated statistical differences between different time treatments after insect feeding, as determined using a one-way ANOVA and a Tukey’s post-hoc test (P < 0.05). Different letters with double quotation marks indicate statistical differences between different time treatments after mechanical wounding, as determined using a one-way ANOVA and a Tukey’s post-hoc test (P < 0.05). Asterisks indicate statistical differences between every two treatments in the same time treatment (*P < 0.05; **P < 0.01; Student’s t-test).
ApTPS14 expression showed the greatest up-regulation, reaching a level 42.7 times higher than that of the control plants (P < 0.05). The expression of ApTPS19 showed the smallest up-regulation by insect feeding, reaching a level only 7.3 times higher than that of the control plants (P < 0.05). The four other NES genes showed several-fold up-regulation of their expression following A. hygrophila feeding, and were significantly different from the control plants (P < 0.05). Fifteen minutes after feeding ended, the expression of the six NES genes remained statistically higher than in the control plants (P < 0.05), but as time progressed their expression levels gradually decreased, returning to their normal levels within 15 to 30 min. ApTPS12, ApTPS14, ApTPS15, and ApTPS16 also showed a small peak in expression 1 h after feeding ended (P < 0.05) (Fig. 5).
Mechanical wounding induces delayed and low levels of NES gene expression
Upon mechanical wounding, all of six NES gene expression showed no statistical difference from that in the leaves of the control group (Fig. 5).
After the mechanical wounding, the expression levels of the differentially expressed NES genes first increased to a peak at around 30–60 min, then gradually decreased to a level similar to that of the control plants after 1–3 h. After 30 min, ApTPS10, ApTPS12, ApTPS15, and ApTPS16 were significantly differentially expressed in the leaves subjected to mechanical wounding in comparison with the control plants (P < 0.05). After 1 h, ApTPS14 expression remained significantly different in the two groups (P < 0.05). However, at several time points, ApTPS19 expression was similar between the two groups (Fig. 5).
Discussion
Here, we showed that the presence of A. hygrophila affects NES gene expression in A. philoxeroides even before feeding and short-term feeding (1 h), and by A. hygrophila strongly and rapidly induces the up-regulation of the NES genes. This up-regulation of NES expression was likely the main reason why chewed leaves released more DMNT, in turn attracting more A. hygrophila to feed on these plants.
Initially, second-generation transcriptome sequencing was performed on the roots, stems, leaves, and flowers of A. philoxeroides to provide genetic and protein data that were lacking for this species. Compared with the previously reported transcriptomes of A. philoxeroides roots38 and A. philoxeroides individuals growing in ponds and uplands33, our sequencing provided novel genetic data for the flowers, which are an important organ for the release of volatile compounds39,40. Also, the annotation of the A. philoxeroides genes and proteins was improved. Along with the full-length transcriptome sequencing of A. hygrophila41, we provided valuable data for the comprehensive study of the molecular mechanisms underlying the specificity between A. philoxeroides and A. hygrophila. Few studies have reported sequencing data for other species in the Alternanthera genus; the genetic and protein data obtained from sequencing other related species in this family are limited to several edible and medicinal crops, such as Amaranthus caudatus L.42, Amaranthus hypochondriacus L.43–45, and Achyranthes bidentata Blume46. The transcriptome data of A. philoxeroides has therefore enriched the genetic data available for the Amaranthaceae, especially the Alternanthera genus. It also enriched the genetic data available for invasive plants, providing powerful genetic support for exploring their invasive properties, and their interactions with herbivorous insects.
In total, 34 TPS genes were predicted in the A. philoxeroides transcriptome (Table S3). In plants with published genomes, the number of TPS genes ranges from 19 to 15216; for example, there are 40 TPS genes in Arabidopsis thaliana, of which 32 have known or predicted functions47. Based on the materials and methods used for this transcriptome sequencing, we speculate that the A. philoxeroides genome may contain more TPS genes, which should be further explored using genome sequencing. The discovery of these TPS genes will enhance our understanding of the TPS genes in the Amaranthaceae as a whole.
Feeding by the specialist insect A. hygrophila affects A. philoxeroides, causing the release of DMNT7. The NES genes, which belong to TPS-g subfamily, are related to DMNT biosynthesis. Six NES genes were screened in this transcriptome. Following the contact or feeding by A. hygrophila, the expression pattern of A. philoxeroides NES genes was further studied. The NES genes were up-regulated before feeding by A. hygrophila. In the 5 min before feeding, when adults merely touched the leaves of A. philoxeroides, the initial contact caused significant up-regulation of most of the NES genes. After injury, plants usually activated a transcription cascade, leading to the expression of genes involved in the release of volatile organic compounds; however, it generally takes a few hours to a day (24 h) for the compounds to be released48–52. Compared with the Lilium ‘Siberia’ terpene synthase gene LoTPS1, which is activated in response to wounding, NES genes in A. philoxeroides responded to damage much faster48.
The rapid up-regulation of these genes following contact with A. hygrophila may have two explanations. In the first case, when the beetle touches the leaf surface, it might cause slight mechanical damage to the leaf, which in turn up-regulates NES expression. The tarsus of an insect typically has claws, which can cause fine scratches on leaf blades and break their trichomes as the insect crawls along the leaf surface. This has been confirmed in tobacco (Nicotiana tabacum L.), soybean (Glycine max (L.) Merr.)53,54, and tomato (Solanum lycopersicum L.)55. Also, the minor mechanical damage causes the release of plant terpenoid volatiles, which has been documented in a touch study of potato (Solanum tuberosum L.)56. However, the expression level of NES in the A. hygrophila contact group was significantly higher than that in the mechanical wounding group. The other possibility is the chemical substances secreted by insects may generate a chemical “footprint” on the leaf and up-regulate the expression of the NES genes. In other species, insect chemical footprints have been shown to mediate the transmission of the insect’s intraspecific and interspecific information. For example, some pollinators has been showed to transmit flower information to the same species by chemical “footprint”57,58, some parasitoids have located host insects by recognition of the chemical traces that host insects deposited after crawling59. It is not yet clear whether these secretions can trigger plant responses60, a hypothesis that will need to be tested in future studies.
When A. hygrophila was in contact with the plant for prolonged periods of time, the beetles began to feed. Short-term feeding (1 h) by A. hygrophila caused rapid and intense up-regulation of NES genes, which is consistent with the previous study showing release of large amounts of DMNT after feeding by A. hygrophila7. Plants responded quickly to insects’ damage, including the rapid biosynthesis of monoterpenes and sesquiterpenes61. For example, the feeding behavior of insect herbivores such as Spodoptera littoralis (Boisduval)62, Lymantria dispar L.63, and Hylobius abietis (L.)64 also induced the expression of plant terpene synthase genes and the release of terpene volatiles. For insects with chewing mouthparts, such as A. hygrophila, the feeding behavior not only causes mechanical wounding to the leaves but also deposes oral secretions, such as saliva, in the leaf incisions. Both of these can induce plant volatile production through special transcriptional pathways65. In this study, we used a puncher simulating similar feeding areas to rule out the effect of mechanical wounding on gene expression changes. However, NES expression in the A. hygrophila feeding group was still significantly greater than that in the mechanical wounding group. In our previously work, we measured the DMNT released from A. philoxeroides following mechanical wounding and feeding by A. hygrophila. The DMNT released due to insect feeding was 240 units ng·h−1·10 g−1·FW−1; by contrast, the DMNT release by mechanical wounding was only 13 units ng·h−1·10 g−1·FW−17. The magnitudes of NES expression caused by insect feeding and mechanical wounding in this study reflected the magnitudes of DMNT in response to these two treatments, respectively7. In Li7, the DMNT was collected over a 12 h period which covered the time frame used in this study. If there would be any DMNT released by mechanical wounding, it would be very minimal compared to the large quantity of DMNT caused by insect feeding. This suggests that the chemicals secreted by A. hygrophila might trigger the up-regulation of NES, which leads to the release of a large amount of DMNT after A. hygrophila feeding7.
In damaged A. philoxeroides leaves, high expression of NES genes causes a large amount of DMNT to be released, which attracts more specialist A. hygrophila7. The release of plant volatiles has a significant attraction to insects (herbivore and their parasitoids) and may provide signals for conspecifics to find host plants or increase aggregation. For example, the root-feeding larvae of a specialized insect Bikasha collaris (Baly) can induce its host plant Triadica sebifera (L.) Small to produce volatiles, attracting aboveground conspecific adults66. Also, Spodoptera littoralis caterpillars could be attracted by volatiles emitted by maize (Zea mays) damaged by conspecific larvae, and thereby find a less suitable but easily detectable host plant67. We speculated that the specialist A. hygrophila might use the chemicals it secretes to affect the release of volatiles from the host plants and facilitate population development by making it easier for conspecifics to locate their host plants A. phyloxeroides. These questions will need to be explored in future research.
Conclusions
In this study, we identified six NES genes from the A. philoxeroides transcriptome. When the leaves were contact by A. hygrophila, NES gene expression increased rapidly, and was strongly up-regulated after A. hygrophila feeding. Our results suggested that the rapid and strong increase of NES transcript levels promotes the large-scale release of DMNT. The released DMNT helps conspecific species of A. hygrophila locate host plants. Our results also suggested that chemicals secreted by A. hygrophila could be responsible for the rapid and strong increase of NES transcript levels. Our results represent an initial explanation of the molecular mechanisms underlying the interaction between A. philoxeroides and its natural enemy A. hygrophila and provide a molecular basis for understanding the relationships between obligate herbivorous insects and their host plants.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No.31570436), Applied Basic Research Program of Shanxi Province (No. 201901D211354), and Shanxi Province Graduate Education Innovation Project (No. 2019BY072). We are very grateful to Professor Thomas James Ridsdill-Smith (University of Western Australia, Perth) for his suggestion to consider the effects of insect-touch. We thank Biomarker Technologies for help in sequencing and analyzing the transcriptome data.
Author contributions
Y.W. designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft. Y.L. analyzed the data, authored or reviewed drafts of the paper, and approved the final draft. X.W., D.J. and J.H. designed the experiments, authored or reviewed drafts of the paper, and approved the final draft. L.L.G. directed the design of the experiments, helped the outline of this manuscript and co-write the draft of this paper, and approved the final draft. R.M. is the principal investigator of this grant, directed and supervised this research work, co-authored and reviewed the drafts of the paper, and approved the final draft.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73130-z.
References
- 1.Julien MH, Skarratt B, Maywald GF. Potential geographical distribution of Alligator Weed and its biological control by Agasicles hygrophila. J.Aquat. Plant Manag. 1995;33:55–60. [Google Scholar]
- 2.Coulson, J. R. Biological Control of Alligatorweed, 1959–s1972: A Review and Evaluation. (Department of Agriculture, Agricultural Research Service, 1977).
- 3.Vogt, G. B., Quimby, P. C. & Kay, S. H. Effects of Weather on the Biological Control of Alligator Weed in the Lower Mississippi Valley Region, 1973–83. (Technical bulletin/United States Department of Agriculture, USA, 1992).
- 4.Van Driesche, R., Blossey, B., Hoddle, M., Lyon, S. & Reardon, R. Biological Control of iInvasive Plants in the Eastern United States. (Forest Health Technology Enterprise Team, USDA Forest Service, 2002).
- 5.Lu J, et al. Non-target plant testing of the flea beetle Agasicles hygrophila, a biological control agent for Alternanthera philoxeroides (alligatorweed) in China. J.Aquat. Plant Manag. 2015;53:88–94. doi: 10.1080/09583157.2012.708019. [DOI] [Google Scholar]
- 6.Zhao L, Lu J, Hu S, Li N, Ma R. The fitness of Agasicles hygrophila on several non-target plant species. Acta Phytophyl. Sin. 2013;40:350–354. [Google Scholar]
- 7.Li N, et al. Manipulating two olfactory cues causes a biological control beetle to shift to non-target plant species. J. Ecol. 2017;105:1534–1546. doi: 10.1111/1365-2745.12778. [DOI] [Google Scholar]
- 8.Boland W, Feng Z, Donath J, Gäbler A. Are acyclic C11 and C16 homoterpenes plant volatiles indicating herbivory? Naturwissenschaften. 1992;79:368–371. doi: 10.1007/BF01140183. [DOI] [Google Scholar]
- 9.Kaiser R. Trapping, investigation and reconstitution of fower scents. Perfumes: Art. Sci.Technol. 1994 doi: 10.1007/978-94-011-3826-0_7. [DOI] [Google Scholar]
- 10.Turlings TC, Tumlinson JH, Lewis WJ. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science. 1990;250:1251–1253. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- 11.Dicke M, Sabelis MW, Takabayashi J, Bruin J, Posthumus MA. Plant strategies of manipulating predatorprey interactions through allelochemicals: Prospects for application in pest control. J. Chem. Ecol. 1990;16:3091–3118. doi: 10.1007/BF00979614. [DOI] [PubMed] [Google Scholar]
- 12.Boland W, Hopke J, Donath J, Nüske J, Bublitz F. Jasmonic acid and coronatin induce odor production in plants. Angew. Chem. Int. Ed. 1995;34:1600–1602. doi: 10.1002/anie.199516001. [DOI] [Google Scholar]
- 13.Bouwmeester HJ, Verstappen FWA, Posthumus MA, Dicke M. Spider mite-induced (3S)-(E)-nerolidol synthase activity in cucumber and lima bean. The first dedicated step in acyclic C11-homoterpene biosynthesis. Plant Physiol. 1999;121:173–180. doi: 10.1104/pp.121.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopke J, Donath J, Blechert S, Boland W. Herbivore-induced volatiles: the emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a beta-glucosidase and jasmonic acid. FEBS Lett. 1994;352:146–150. doi: 10.1016/0014-5793(94)00948-1. [DOI] [PubMed] [Google Scholar]
- 15.Tholl D, Sohrabi R, Huh J-H, Lee S. The biochemistry of homoterpenes–common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry. 2011;72:1635–1646. doi: 10.1016/j.phytochem.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 17.Aharoni A, et al. Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell. 2003;15:2866–2884. doi: 10.1105/tpc.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kappers IF, et al. Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science. 2005;309:2070–2072. doi: 10.1126/science.1116232. [DOI] [PubMed] [Google Scholar]
- 19.Yang T, Stoopen G, Thoen M, Wiegers G, Jongsma MA. Chrysanthemum expressing a linalool synthase gene 'smells good', but 'tastes bad' to western flower thrips. Plant Biotechnol. J. 2013;11:875–882. doi: 10.1111/pbi.12080. [DOI] [PubMed] [Google Scholar]
- 20.Li F, et al. Expression of lima bean terpene synthases in rice enhances recruitment of a beneficial enemy of a major rice pest. Plant Cell Environ. 2018;41:111–120. doi: 10.1111/pce.12959. [DOI] [PubMed] [Google Scholar]
- 21.Loughrin JH, Manukian A, Heath RR, Turlings TC, Tumlinson JH. Diurnal cycle of emission of induced volatile terpenoids by herbivore-injured cotton plant. Proc. Natl. Acad. Sci. USA. 1994;91:11836–11840. doi: 10.1073/pnas.91.25.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mithöfer A, Wanner G, Boland W. Effects of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiol. 2005;137:1160–1168. doi: 10.1104/pp.104.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turlings TCJ, Erb M. Tritrophic interactions mediated by herbivore-induced plant volatiles: mechanisms, ecological relevance, and application potential. Annu. Rev. Entomol. 2018;63:433–452. doi: 10.1146/annurev-ento-020117-043507. [DOI] [PubMed] [Google Scholar]
- 24.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apweiler R, et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2013;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L, Geng Y, Yang H, Hu Y, Yang J. Gene expression reaction norms unravel the molecular and cellular processes underpinning the plastic phenotypes of Alternanthera Philoxeroides in contrasting hydrological conditions. Front. Plant Sci. 2015;6:991–991. doi: 10.3389/fpls.2015.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Xu L, Wang X, Pan G, Lu LD. novo characterization of the alligator weed (Alternanthera philoxeroides) transcriptome illuminates gene expression under potassium deprivation. J. Genet. 2015;94:95–104. doi: 10.1007/s12041-015-0493-1. [DOI] [PubMed] [Google Scholar]
- 39.Dudareva N, Negre F, Nagegowda DA, Orlova I. Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 2006;25:417–440. doi: 10.1080/07352680600899973. [DOI] [Google Scholar]
- 40.Sohrabi R, et al. In planta variation of volatile biosynthesis: An alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell. 2015;27:874–890. doi: 10.1105/tpc.114.132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia D, et al. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt) Sci. Rep. 2018;8:2197–2197. doi: 10.1038/s41598-018-20181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetter MG, Schmid KJ. Analysis of phylogenetic relationships and genome size evolution of the Amaranthus genus using GBS indicates the ancestors of an ancient crop. Mol. Phylogenet. Evol. 2017;109:80–92. doi: 10.1016/j.ympev.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 43.Délano-Frier JP, et al. Transcriptomic analysis of grain amaranth (Amaranthus hypochondriacus) using 454 pyrosequencing: Comparison with A. tuberculatus, expression profiling in stems and in response to biotic and abiotic stress. BMC Genomics. 2011;12:363. doi: 10.1186/1471-2164-12-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunil M, et al. The draft genome and transcriptome of Amaranthus hypochondriacus: A C4 dicot producing high-lysine edible pseudo-cereal. DNA Res. 2014;21:585–602. doi: 10.1093/dnares/dsu021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunil M, Hariharan N, Dixit S, Choudhary B, Srinivasan S. Differential genomic arrangements in Caryophyllales through deep transcriptome sequencing of A. hypochondriacus. PLoS ONE. 2017;12:e0180528. doi: 10.1371/journal.pone.0180528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang YH, et al. The root transcriptome of Achyranthes bidentata and the identification of the genes involved in the replanting benefit. Plant Cell Rep. 2018;37:611–625. doi: 10.1007/s00299-018-2255-z. [DOI] [PubMed] [Google Scholar]
- 47.Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 48.Abbas F, Ke Y, Yu R, Fan Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium 'Siberia'. Planta. 2019;249:71–93. doi: 10.1007/s00425-018-3006-7. [DOI] [PubMed] [Google Scholar]
- 49.Erb M, Reymond P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- 50.Huang X-Z, et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018;41:261–274. doi: 10.1111/pce.13088. [DOI] [PubMed] [Google Scholar]
- 51.Rauwane ME, Odeny DA, Millar I, Rey C, Rees J. The early transcriptome response of cassava (Manihot esculenta Crantz) to mealybug (Phenacoccus manihoti) feeding. PLoS ONE. 2018;13:e0202541–e0202541. doi: 10.1371/journal.pone.0202541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamiru A, et al. A maize landrace that emits defense volatiles in response to herbivore eggs possesses a strongly inducible terpene synthase gene. Ecol. Evol. 2017;7:2835–2845. doi: 10.1002/ece3.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bown AW, Hall DE, MacGregor KB. Insect footsteps on leaves stimulate the accumulation of 4-aminobutyrate and can be visualized through increased chlorophyll fluorescence and superoxide production. Plant Physiol. 2002;129:1430–1434. doi: 10.1104/pp.006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall DE, MacGregor KB, Nijsse J, Bown AW. Footsteps from insect larvae damage leaf surfaces and initiate rapid responses. Eur. J. Plant Pathol. 2004;110:441–447. doi: 10.1023/B:EJPP.0000021072.89968.de. [DOI] [Google Scholar]
- 55.Peiffer M, Tooker JF, Luthe DS, Felton GW. Plants on early alert: Glandular trichomes as sensors for insect herbivores. New Phytol. 2009;184:644–656. doi: 10.1111/j.1469-8137.2009.03002.x. [DOI] [PubMed] [Google Scholar]
- 56.Markovic D, Nikolic N, Glinwood R, Seisenbaeva G, Ninkovic V. Plant responses to brief touching: A mechanism for early neighbour detection? PLoS ONE. 2016;11:e0165742–e0165742. doi: 10.1371/journal.pone.0165742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eltz T. Tracing pollinator footprints on natural flowers. J. Chem. Ecol. 2006;32:907–915. doi: 10.1007/s10886-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 58.Witjes S, Witsch K, Eltz T. Reconstructing the pollinator community and predicting seed set from hydrocarbon footprints on flowers. Oecologia. 2011;166:161–174. doi: 10.1007/s00442-010-1824-9. [DOI] [PubMed] [Google Scholar]
- 59.Rostás M, Wölfling M. Caterpillar footprints as host location kairomones for Cotesia marginiventris: persistence and chemical nature. J. Chem. Ecol. 2009;35:20–27. doi: 10.1007/s10886-009-9590-z. [DOI] [PubMed] [Google Scholar]
- 60.Hilker M, Meiners T. How do plants "notice" attack by herbivorous arthropods? Biol. Rev. Camb. Philos. Soc. 2010;85:267–280. doi: 10.1111/j.1469-185X.2009.00100.x. [DOI] [PubMed] [Google Scholar]
- 61.Holopainen JK. Multiple functions of inducible plant volatiles. Trends Plant Sci. 2004;9:529–533. doi: 10.1016/j.tplants.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Degenhardt J, Gershenzon J. Demonstration and characterization of (E)-nerolidol synthase from maize: A herbivore-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta. 2000;210:815–822. doi: 10.1007/s004250050684. [DOI] [PubMed] [Google Scholar]
- 63.Irmisch S, Jiang Y, Chen F, Gershenzon J, Köllner TG. Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa) BMC Plant Biol. 2014;14:270–270. doi: 10.1186/s12870-014-0270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovalchuk A, et al. Activation of defence pathways in Scots pine bark after feeding by pine weevil (Hylobius abietis) BMC Genomics. 2015;16:352–352. doi: 10.1186/s12864-015-1546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felton GW, Tumlinson JH. Plant-insect dialogs: Complex interactions at the plant-insect interface. Curr. Opin. Plant Biol. 2008;11:457–463. doi: 10.1016/j.pbi.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 66.Sun, X. et al. Root feeding larvae increase their performance by inducing leaf volatiles that attract aboveground conspecific adults. J.Ecol. 107, 2713–2723. 10.1111/1365-2745.13196 (2019).
- 67.von Mérey GE, Veyrat N, D’ssAlessandro M, Turlings TCJ. Herbivore-induced maize leaf volatiles affect attraction and feeding behavior of Spodoptera littoralis caterpillars. Front Plant Sci. 2013;4:209–209. doi: 10.3389/fpls.2013.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.