Abstract
Accumulating evidence has emerged revealing that noncoding RNAs (ncRNAs) play essential roles in the occurrence and development of hepatocellular carcinoma (HCC). However, the complicated regulatory interactions among various ncRNAs in the development of HCC are not entirely understood. The newly discovered mechanism of competing endogenous RNAs (ceRNAs) uncovered regulatory interactions among different varieties of RNAs. In recent years, a growing number of studies have suggested that ncRNAs, including long ncRNAs, circular RNAs and pseudogenes, play major roles in the biological functions of the ceRNA network in HCC. These ncRNAs can share microRNA response elements to affect microRNA affinity with target RNAs, thus regulating gene expression at the transcriptional level and both physiological and pathological processes. The ncRNAs that function as ceRNAs are involved in diverse biological processes in HCC cells, such as tumor cell proliferation, epithelial-mesenchymal transition, invasion, metastasis and chemoresistance. Based on these findings, ncRNAs that act as ceRNAs may be promising candidates for clinical diagnosis and treatments. In this review, we discuss the mechanisms and research methods of ceRNA networks. We also reviewed the recent advances in studying the roles of ncRNAs as ceRNAs in HCC and highlight possible directions and possibilities of ceRNAs as diagnostic biomarkers or therapeutic targets. Finally, the limitations, gaps in knowledge and opportunities for future research are also discussed.
Keywords: Hepatocellular carcinoma, Noncoding RNA, Competing endogenous RNA
Background
Hepatocellular carcinoma (HCC), the most prevalent subtype of hepatic malignancy, is one of the leading causes of cancer-associated mortality worldwide [1]. There are an estimated 62 million new cases of HCC annually, 85% of which are diagnosed in developing countries [2]. Owing to the insidious symptoms and early metastases, most HCC patients are diagnosed at an advanced stage, leading to limited efficacy and even ineffectiveness of therapeutic approaches [3]. Despite continuous therapeutic advances, such as in surgical resection, liver transplantation and radiofrequency ablation, the 5-year survival rate for HCC patients is still under 20% [4, 5]. Therefore, it is important to identify the underlying mechanism as well as precise diagnostic and prognostic biomarkers for early diagnosis and risk assessment. Furthermore, identifying effective therapeutic targets for HCC is critical.
Carcinogenesis is a complicated multi-step, multi-stage process involving both genetic and epigenetic alterations [3]. However, the molecular pathogenesis of HCC remains unclear. Furthermore, the majority of previous studies that examined the molecular mechanisms of hepatic tumorigenesis have focused on protein-coding genes and not yet provided comprehensive and detailed mechanisms of HCC. High-throughput sequencing technology has revealed numerous large and small non-coding RNAs (ncRNAs) that are critically involved in carcinogenesis [6]. NcRNAs, which universally exist in a broad range of organisms, are a group of RNAs without protein-coding functions and include long non-coding RNA (lncRNA), microRNA (miRNA), pseudogene, circular RNA (circRNA), small interfering RNA, small nucleolar RNA, ribosomal RNA and transfer RNA (Fig. 1). LncRNA, miRNA, pseudogene and circRNA are primarily involved in post-transcriptional regulation [7]. Recently, accumulating evidence has demonstrated that these ncRNAs are potential diagnostic and prognostic biomarkers for HCC [8–10]. Nevertheless, the specific functions and mechanisms of most ncRNAs in HCC remain unclear.
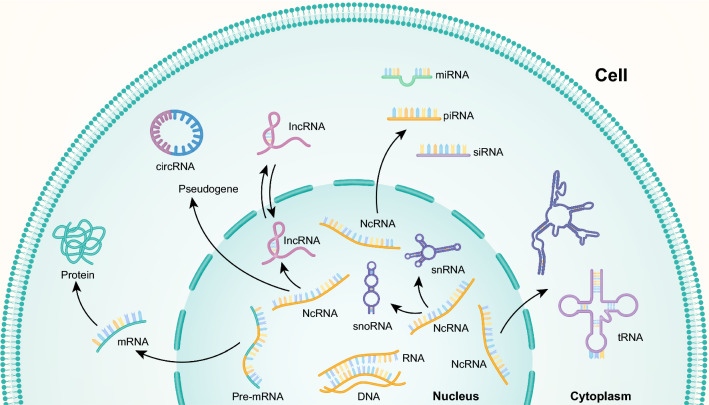
Fig. 1.
The classification of coding and noncoding RNA. Eukaryotic mRNA molecules are usually composed of small segments of the original gene and are generated by a process of cleavage and rejoining from an original precursor RNA (pre-mRNA) molecule, which is an exact copy of the gene. Noncoding RNA (ncRNA) mainly include long non-coding RNA (lncRNA), microRNA (miRNA), pseudogene, circular RNA (circRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), ribosomal RNA (rRNA) and transfer RNA (tRNA)
Recent studies have reported competing endogenous RNA (ceRNA) regulatory networks (ceRNET), in which ceRNAs modulate each other at the post-transcriptional level via competition of shared miRNAs. Functionally, ceRNA networks are considered the bridge to connect the functions of protein-coding mRNAs with the functions of ncRNAs, including lncRNAs, circRNAs, miRNAs and pseudogenes. According to the ceRNA hypothesis, ceRNAs sharing miRNA response elements (MREs) may affect miRNA affinity with mRNAs, thereby triggering gene silencing [11]. Because all transcripts with MREs could theoretically serve as ceRNAs, these transcripts may be considered as universal regulators of post-transcriptional events under physiological and pathological conditions. Aberrantly expressed ceRNAs may cause dysregulation of ceRNA networks, leading to human diseases, including cancer [12–14].
Several reports have demonstrated the roles of ncRNAs as ceRNAs in multiple processes of pathogenesis in HCC (Fig. 2). Based on the demonstrated function of ncRNAs as ceRNAs in HCC, ncRNAs may serve as potential biomarkers and therapeutic targets. In this review, we will discuss ncRNAs that function as ceRNAs in HCC and their relevance to current clinical practice. We first summarize the mechanisms and research methods of ceRNA and ceRNETs and provide some examples of the far-reaching roles for these molecules in affecting HCC processes. We then discuss how the basic science insights into the function of ncRNAs as ceRNAs are being applied to develop next-generation biomarkers and therapies in HCC. As the so-called “dark matter” of the genome continues to be brought into the light, it is evident that targeting ncRNAs that function as ceRNAs and ceRNETs has great potential to impact HCC patient care.
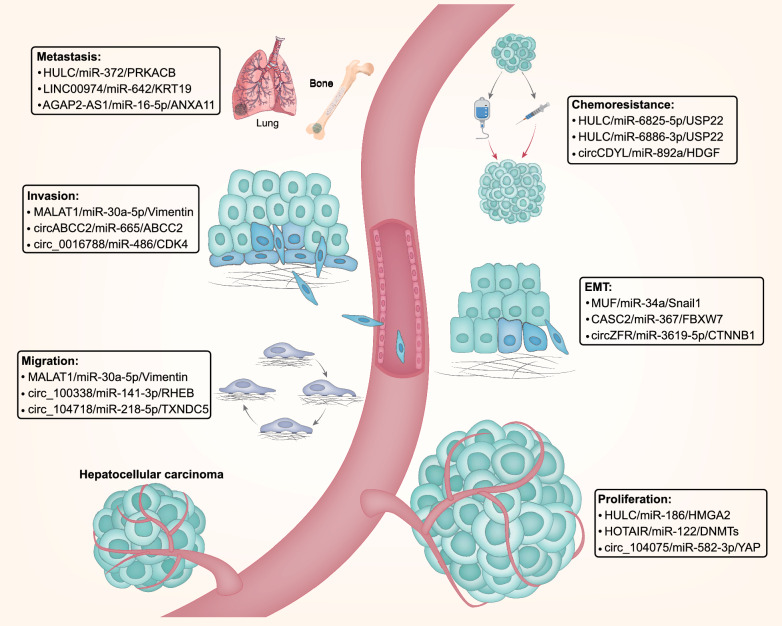
Fig. 2.
Summary of ncRNAs act as ceRNAs mediated functions. NcRNAs can regulate hepatocellular carcinoma progression, such as cell proliferation, migration, epithelial–mesenchymal transition (EMT), invasion, distant metastasis and chemotherapy resistance
Mechanisms and research methods of ceRNA and ceRNETs
Our expanded understanding of the transcriptome has led to the identification of diverse MREs in various RNA transcripts. The ceRNA hypothesis, which was initially established in 2011 [15], proposes that RNAs that share MREs can function as ceRNAs and lead to an impairment of miRNA activity (Fig. 3) [11, 16]. The upregulated expression of one particular transcript could sequestrate more miRNA copies from the miRNA pool and subsequently derepress other transcripts and vice versa. Moreover, the existence of MREs in lncRNAs, circRNAs and pseudogenes has suggested a modulation and control among these ncRNAs through a complicated ceRNET [17].
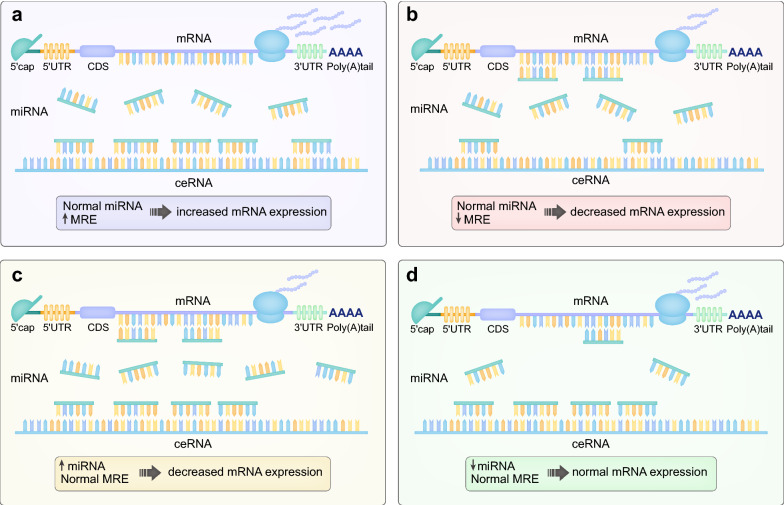
Fig. 3.
The mechanisms of ceRNA regulatory networks. CeRNAs and mRNAs share a pool of miRNAs, and the competition for miRNAs leads to a dynamic regulation of the expression level of mRNAs. a ceRNA with up-regulated miRNA response elements (MREs) can “absorb” more miRNAs, thus the mRNA expression is increased. b When the MRE is down-regulated, miRNAs tend to bind with and silence mRNAs, leading to a decrease in mRNA expression. c miRNA up-regulation exceeds the “sponge effect” of ceRNAs, and mRNA expression is inhibited by excessive miRNAs. d miRNA level is decreased, and mRNA can be expressed normally. In addition, the subtle regulation of one certain mRNA can be achieved by the interactions of multiple miRNAs and ceRNAs
The in-depth studies on ceRNA interactions have revealed several potential factors for optimal ceRNA activity. First, the interaction can be affected by the abundance and relative concentration of ceRNAs and interacting miRNAs. Studies have reported that ceRNA activity reaches an optimal state when the levels of miRNA and ceRNA are nearly equimolar [18, 19]. CeRNAs have limited sequestration ability on highly abundant miRNAs; however, lowly expressed miRNAs exhibit minimal interaction with active ceRNAs due to their restricted number. Second, the location of ceRNA components is another potential factor. The expression of miRNAs is cell- and tissue-specific, and thus some ceRNAs only function in specific cells with shared accessible miRNAs [20]. Moreover, the subcellular localization of ceRNAs may also impact the availability of ceRNAs to shared miRNAs, thereby affecting ceRNA activity [21]. Third, since each miRNA can competitively bind to multiple transcripts, the binding affinities of these transcripts to miRNAs can impact the competing effects; in other words, higher affinity indicates more potent competition [22]. Of note, the binding affinity between target RNAs and miRNAs is primarily affected by the binding of MREs (located in target RNA) with seed regions (located in miRNA). Binding is additionally influenced by single-nucleotide polymorphisms, RNA editing and alternative splicing [23–26], as these factors can alter the seed regions and the target spectrum, which further lead to the production, degradation or alteration of miRNA binding sites. Fourth, the abundance of argonaute (Ago), which is the catalytic component of the RNA-induced silencing complex, is likely to be a rate-limiting factor for ceRNA crosstalk. The seed region in the RNA-induced silencing complex forms base pairs with the MREs located within the 3′-untranslated region of target RNA to modulate target RNA [27]. Finally, RNA transcripts are also likely to compete for shared RNA binding proteins, which are involved in RNA degradation, stability and splicing [28], in addition to shared miRNAs. This could hinder miRNA-target binding via MRE occupancy or inversely promote miRNA-target binding via the recruitment of miRNAs to target RNA, implicating their involvement in the ceRNET [29, 30].
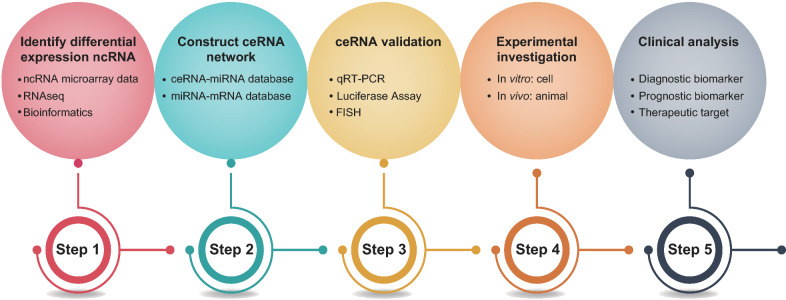
The prediction, construction and validation of ceRNETs have become possible from the transcriptome data derived from various malignancies and the development of bioinformatics and computational methods. The construction of ceRNETs can effectively predict the function and underlying mechanisms of related ceRNAs. CeRNA crosstalk, a posttranscriptional event, is mediated by miRNAs and dependent on MREs located within each transcript [11]. Therefore, MRE identification in the related transcript of interest plays a decisive role in predicting ceRNA crosstalk. Silicon-based or high-throughput databases have been developed to facilitate the identification of ceRNETs (Table 1). The research methods and processes involving ceRNETs in HCC to date are presented in Fig. 4.
Table 1.
Summary of computational approaches for identifying the ceRNA networks
| Databases | Brief description | Link | Ref |
|---|---|---|---|
| starBase v2.0 | Database supplies comprehensive interaction network of ncRNAs (lncRNAs, miRNAs, and ceRNAs), mRNA, and proteins in cancer cells and normal tissues based on 108 CLIP-Seq | http://starbase.sysu.edu.cn/ | [31] |
| NPInter v3.0 | Database supplies comprehensive interaction among ncRNAs (except tRNAs and rRNAs), lncRNAs and others. It provides different types of basic information about the interaction | http://www.bioinfo.org/NPInter/ | [32] |
| DIANA-LncBase v2 | Database supplies two different miRNA–lncRNA interaction modules. One module is experimentally supported, and the other is in silico predicted interactions | http://www.microrna.gr/LncBase/ | [33, 34] |
| circBase | Database provides scripts to identify known and novel circRNAs in sequencing data. | http://www.circbase.org/ | [35] |
| LNCediting | Database supplies information about RNA editing in lncRNAs and miRNA–lncRNA interactions | http://bioinfo.life.hust.edu.cn/LNCediting/ | [36] |
| CircInteractome | Database supplies information about circRNAs and their interaction with proteins or miRNAs | http://circinteractome.nia.nih.gov/ | [37] |
| Cancer-Specific-CirRNA-Database (CSCD) | Database for cancer-specific circRNAs | http://gb.whu.edu.cn/CSCD/ | [38] |
| spongeScan | Database supplies information about microRNA binding elements in lncRNA sequences | http://spongescan.rc.ufl.edu | [39] |
| SomamiR 2.0 | Database provides information and functional analysis of expected miRNA–ceRNA interaction | http://compbio.uthsc.edu/SomamiR | [40] |
| lnCeDB | Database provides information of human lncRNA that acts as ceRNAs | http://gyanxet-beta.com/lncedb/ | [41] |
| miRBase | Database provides miRNA gene hunters with unique names for novel miRNA genes prior to publication of results | http://www.mirbase.org/ | [42] |
| Targetscan | Database provides information of predicted microRNA targets | http://www.targetscan.org/vert_72/ | [43] |
| miRcode | Database provides “whole transcriptome” human microRNA target predictions based on the comprehensive GENCODE gene annotation. | http://www.mircode.org/ | [44] |
ncRNA: noncoding RNA; ceRNA: competing endogenous RNA; lncRNA: long noncoding RNA; mRNAs: messenger RNAs; miRNA: microRNA; circRNA: circularRNA; Ref: reference
Fig. 4.
The steps and research methods for studying competing endogenous RNA network in HCC. First, differentially expressed ncRNAs in HCC should be obtained using various resources and software. Second, computer algorithms and public databases may be used to predict interactions between different transcripts and to construct ceRNET. Third, ceRNA interactions within ceRNET should be validated experimentally. Fourth, the dysregulation of ceRNAs in HCC pathogenesis may be further investigated by functional studies involving cell and animal experiments. Finally, the hub genes within ceRNET should be translated into clinical manegement of HCC. ncRNA: Non-coding RNA; ceRNA: Competing endogenous RNA; ceRNET: ceRNA regulatory network; mRNA: Messenger RNA; miRNA: MicroRNA; qRT-PCR: Quantitative reverse transcription polymerase chain reaction; FISH: Fluorescence in situ hybridization
The types of ncRNAs acting as ceRNAs and their possible roles in HCC
LncRNAs as ceRNAs
LncRNAs, ncRNAs over 200 nt in length that originate from promoter-proximal, antisense and intergenic regions, carry out their regulatory function by interacting with genomic DNA, mRNAs and proteins as well as other ncRNAs [45, 46]. LncRNAs may act as sponges, scaffolds, decoys, signals and guides. LncRNAs can regulate gene expression at epigenetic, transcriptional, posttranscriptional and translational levels. Thus, these ncRNAs are considered to have pleiotropic impacts and considered “master regulators” of the genome [47, 48]. They exert regulatory effects in both physiological and pathological conditions, including in various cancers such as HCC [49–51]. Emerging evidence has revealed that a large family of lncRNAs function as ceRNAs for modulation of the expression and biological features of miRNAs in HCC [48] (Table 2).
Table 2.
Validated ceRNA networks shaped by the ceRNA function of lncRNAs in HCC
| ceRNA network type | ceRNA member | Shared miRNA(s) | Competing target (mRNA) | ceRNA role | Experiments related to mechanism | Related HCC function mechanisms | Ref |
|---|---|---|---|---|---|---|---|
| LncRNA–miRNA–mRNA | HULC | miR-372 | PRKACB | Oncogenic | Cellular | Invasion and metastasis | [12] |
| miR-6825-5p, miR-6845-5p, miR-6886-3p | USP22 | Oncogenic | shRNA knockdown in mouse xenografts | Chemosensitivity | [52] | ||
| miR-200a-3p | ZEB1 | Oncogenic | siRNA knockdown in mouse xenografts | Invasion and EMT | [53] | ||
| miR-186 | HMGA2 | Oncogenic | Overexpression, antimiRs in mouse xenografts | Proliferation | [54] | ||
| HOTAIR | miR-122 | DNMTs | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation | [55] | |
| miR-23b-3p | ZEB1 | Oncogenic | Overexpression in mouse xenografts | Invasion and EMT | [56] | ||
| MALAT1 | miR-30a-5p | Vimentin | Oncogenic | shRNA knockdown in mouse xenografts | Migration and invasion | [57] | |
| miR-204 | SIRT1 | Oncogenic | Cellular | Migration and invasion | [58] | ||
| miR-143-3p | ZEB1 | Oncogenic | Cellular | Proliferation and invasion | [59] | ||
| MIAT | miR-214 | EZH2 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and invasion | [60] | |
| LINC00974 | miR-642 | KRT19 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [61] | |
| CCAT1 | Let-7 | HMGA2, c-Myc | Oncogenic | Cellular | Proliferation and migration | [62] | |
| DANCR | miR-214, miR-320a, miR-199a | CTNNB1 | Oncogenic | siRNA knockdown in mouse xenografts | Invasion and metastasis | [63] | |
| miR-27a-3p | ROCK1, LIMK1, COFILIN1 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [64] | ||
| miR-216a-5p | KLF12 | Oncogenic | shRNA knockdown in mouse xenografts | Migration and invasion | [65] | ||
| HOTTIP | miR-125b | HOXA | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and migration | [66] | |
| ATB | miR-200 family | ZEB1, ZEB2 | Oncogenic | Overexpression, knockdown in mouse xenografts | Invasion and EMT | [67] | |
| UCA1 | miR-216b | FGFR1 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and metastasis | [68] | |
| miR-203 | Snail2 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and invasion | [69] | ||
| FAL1 | miR-1236 | AFP, ZEB1 | Oncogenic | Cellular | Proliferation and migration | [70] | |
| MUF | miR-34a | Snail1 | Oncogenic | overexpression in mouse xenografts | EMT | [71] | |
| HOXD-AS1 | miR-130a-3p | SOX4 | Oncogenic | shRNA knockdown in mouse xenografts | Migration and invasion | [72] | |
| SNHG8 | miR-149-5p | PPM1F | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [73] | |
| CDKN2B-AS1 | let-7c-5p | NAP1L1 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [74] | |
| FLVCR1-AS1 | miR-513c | MET | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [75] | |
| ZFAS1 | miR-150 | ZEB1, MMP14, MMP16 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation and metastasis | [76] | |
| NEAT1 | miR-485 | STAT3 | Oncogenic | Cellular | Migration and invasion | [77] | |
| TUG1 | miR-142-3p | ZEB1 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and EMT | [78] | |
| miR-144 | JAK2 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and migration | [79] | ||
| ANRIL | miR-122-5p | NR | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [80] | |
| n335586 | miR-924 | CKMT1A | Oncogenic | Overexpression in mouse xenografts | Migration and invasion | [81] | |
| HOXA-AS2 | miR-520c-3p | GPC3 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and EMT | [82] | |
| PCAT-1 | miR-215 | CRKL | Oncogenic | shRNA knockdown, miR upregulation in mouse xenografts | Proliferation | [83] | |
| MCM3AP-AS1 | miR-194-5p | FOXA1 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation | [84] | |
| SNHG6-003 | miR-26a/b | TAK1 | Oncogenic | Overexpression in mouse xenografts | Proliferation | [85] | |
| TP73-AS1 | miR-200a | HMGB1, RAGE | Oncogenic | Cellular | Proliferation | [86] | |
| DSCR8 | miR-485-5p | FZD7 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation | [87] | |
| LINC00707 | miR-206 | CDK14 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [88] | |
| AGAP2-AS1 | miR-16-5p | ANXA11 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation and metastasis | [89] | |
| miat | miR-22-3p | sirt1 | Oncogenic | Knockdown in mouse xenografts | Proliferation | [90] | |
| DSCAM-AS1 | miR-338-3p | CyclinD1, SMO | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [91] | |
| XIST | miR-194-5p | MAPK1 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [92] | |
| miR-497-5p | PDCD4 | Tumor suppressive | Overexpression in mouse xenografts | Proliferation and metastasis | [93] | ||
| miR-92b | Smad7 | Tumor suppressive | Overexpression, knockdown in mouse xenografts | Proliferation and metastasis | [94] | ||
| LINC00657 | miR-106a-5p | PTEN | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [95] | |
| MEG3 | miR-9-5p | SOX11 | Tumor suppressive | Cellular | Proliferation | [96] | |
| SNHG16 | miR-93 | NR | Tumor suppressive | Overexpression in mouse xenografts | Cell proliferation and chemosensitivity | [97] | |
| CASC2 | miR-367 | FBXW7 | Tumor suppressive | Overexpression downregulation in mouse xenografts | EMT | [98] | |
| MIR31HG | miR-575 | ST7L | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation and metastasis | [99] | |
| FTX | miR-374a | WIF1, PTEN, WNT5A | Tumor suppressive | Overexpression in mouse xenografts | Invasion and EMT | [100] |
ceRNA: competing endogenous RNA; lncRNA: long noncoding RNA; mRNAs: messenger RNAs; miRNA: microRNA; HCC: hepatocellular carcinoma; EMT: epithelial–mesenchymal transition; Ref: reference
LncRNAs can exert their functions in the HCC pathophysiological process by participating in a certain pathway but through interactions with different miRNAs. The lncRNA metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) promotes HCC migration and invasion by three pathways [57–59]. First, MALAT1 competes with miR-30a-5p to regulate vimentin gene expression. Pan et al. [57] first demonstrated the sponge role of MALAT-1 for miR-30a-5p and described its functions in HCC as a ceRNA. The authors measured MALAT1 and vimentin expression levels in paired HCC and normal adjacent tissues by RT-PCR and predicted potential miR-30a-5p binding sites in MALAT1 and the target site of miR-30a-5p in vimentin using bioinformatic analyses. Co-transfection of plasmids and miR-30a-5p mimics into 293T cells was used to discover how miR-30a-5p regulates MALAT1 expression levels. Dual luciferase assay showed that miR-30a-5p significantly decreased vimentin expression level, and subsequent experiments showed that miR-30a-5p inhibits HCC cell migration and invasion. The role of MALAT1 in HCC cell growth was also tested using in vivo experiments, in which the authors injected HepG2 cells transfected with sh-MALAT1 into nude mice; the tumor volume and size in the shRNA knockdown group were smaller than that of the negative control group. These results provided in vivo evidence that MALAT1 promotes HCC growth. Second, MALAT1 competes with miR-204 to regulate SIRT1 gene expression. Hou et al. [58] used shRNA knockdown and loss-of-function strategies on MALAT1 in HepG2 cells to screen potential MALAT-1-interacting miRNA candidates, and identified miR-204 had shown the best correlation. The authors treated HepG2 cells with miR-204 inhibitors and mimics and found that MALAT-1 level was increased and the migration and invasion abilities of HepG2 cells were enhanced in miR-204 inhibitor group. The results demonstrated that MALAT1 derepresses SIRT1 by sponging and competitively binding to miR-204, causing enhanced migration, invasion and epithelial-mesenchymal transition (EMT) of HCC. Finally, MALAT1 also competes with miR-143-3p for the tumor suppressor ZEB1. Chen et al. [59] conducted a clinicopathological analysis in 56 paired HCC and non-tumor liver samples to explore the association between MALAT-1 expression level and clinical characteristics including age, sex, tumor size, tumor differentiation, TNM stage and distant metastasis. The authors also predicted possible interactions among MALAT1, miR-143-3p and ZEB1 based on a bioinformatics analysis. SiRNA knockdown of MALAT-1 was performed in several cell lines, and Matrigel invasion assays proved that si-MALAT-1 inhibited HCC cell invasion. Subsequent experiments confirmed the existence of the MALAT1/miR-143-3p/ZEB1 regulatory pathway and showed that high expression of MALAT1 in HCC led to increased ZEB1, thus promoting the proliferation and migration of HCC.
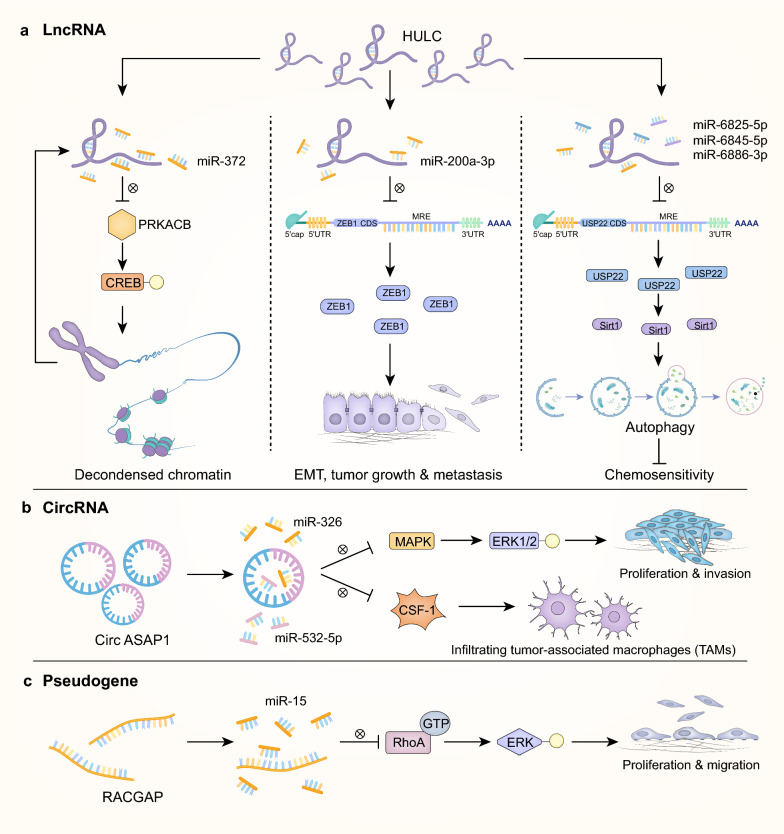
Several lncRNAs can influence HCC behavior by several distinct mechanisms through interactions with different miRNAs and their corresponding mRNAs. HULC is one of the most upregulated lncRNAs in HCC and was found to function in HCC by at least three mechanisms (Fig. 5a). Wang et al. [12] performed chromatin accessibility by real-time PCR (CHART-PCR) assays to evaluate the accessibility of genomic DNA to nuclease and analyze different regions of open chromatin around the HULC promoter. ChIP assays demonstrated that increased euchromatic histone modifications are correlated with transcription activation of HULC. Subsequent experiments revealed significantly increased miR-372 level after siRNA-mediated inhibition of HULC, indicating that HULC decreases miR-372 expression in HCC and enhances chromatin accessibility and transcription. Li et al. [53] observed that a higher expression level of HULC in HCC tissues is associated with many enhanced EMT features, including histological morphology, physiological behavior and EMT markers such as E-cadherin, N-cadherin, ZO-1, vimentin, β-catenin, Snail and ZEB1. The authors proposed that HULC exhibits a negative regulatory effect on miR-200a-3p and upregulates ZEB1, resulting in enhanced EMT and promoted growth and metastasis of HCC. A HULC/USP22/Sirt1 protective autophagy pathway was further identified that attenuates HCC cell sensitivity to chemotherapeutic agents [52]. In the study, autophagy-related proteins were markedly increased upon HULC overexpression, but were significantly reduced upon Sirt-1 silencing. These results revealed a HULC-Sirt1-autophagy pathway, which was further discovered to be able to weaken the chemosensitivity of HCC cells toward oxaliplatin. As another example, the lncRNA X inactive-specific transcript (XIST) inhibits the proliferation of HCC cells through the miR-497-5p/PDCD4 or miR-92b/Smad7 pathway, but promotes HCC progression by silencing miR-194-5p and derepressing MAPK1 [92–94].
Fig. 5.
Simplified examples of the roles that different classes of ceRNAs play in HCC oncogenesis. a LncRNA: HULC functions in HCC by three mechanisms: (1) Transcription activation of HULC decreases miR-372 level in HCC, derepresses PRKACB function and CREB phosphorylation, hence enhancing chromatin accessibility and transcription. (2) HULC exerts a negative regulatory effect on miR-200a-3p and subsequent upregulation of ZEB1, which facilitates EMT, tumor growth and metastasis. (3) HULC sponges miR-6825-5p, miR-6845-5p and miR-6886-3p, leading to increased expression of USP22 and Sirt1, which activates the protective autophagy pathway and attenuates HCC cell sensitivity to chemotherapeutic agents. b CircRNA: CircASAP1 decreases miR-326 and miR-532-5p level, thus promoting proliferation and invasion of HCC through miR-326/MAPK/ERK1/2 signaling, and raising TAMs infiltration through CSF-1 activation and secretion. c Pseudogene: RACGAP binds to miR-15, enhances the binding of GTP to RhoA and further ERK phosphorylation, promoting tumor cell proliferation and migration
Together, these studies on the associations between lncRNAs and HCC show that lncRNAs function in a multidimensional network to influence HCC, not only by interacting with a large group of miRNAs and affecting their correlated pathways, but also by participating in a number of distinct mechanisms. Notably, some lncRNAs show both tumor suppressive and oncogenic functions, which calls for additional studies to determine how these seemingly contrary functions are balanced and regulated.
CircRNAs as ceRNAs
CircRNAs are relatively more resistant to degradation compared with linear RNA because the 3′ and 5′ ends of circRNAs are covalently linked [101]. Apart from higher stability compared with linear RNAs, circRNAs are highly homologous to their linear counterparts and contain more MREs. Thus, circRNAs can act as effective miRNA sponges to disrupt miRNA-mediated target repression and regulate genes at both transcriptional and post-transcriptional levels as robust ceRNAs [102]. Emerging evidence has demonstrated that circRNAs may counteract miRNA-mediated repression of linear mRNAs by functioning as miRNA sponges (Table 3).
Table 3.
Validated ceRNA networks shaped by the ceRNA function of circRNAs in HCC
| ceRNA network type | ceRNA member | Shared miRNA(s) | Competing target (mRNA) | ceRNA role | Experiments related to mechanism | Related HCC function mechanisms | Ref |
|---|---|---|---|---|---|---|---|
| CircRNA–miRNA–mRNA | circASAP1 | miR-326, miR-532-5p | MAPK1, CSF-1 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation and invasion | [103] |
| circCdr1as | miR-1270 | AFP | Oncogenic | Cellular | Proliferation and migration | [104] | |
| circCdr1as | miR-7 | CCNE1, PIK3CD | Oncogenic | Cellular | Proliferation and invasion | [105] | |
| circABCC2 | miR-665 | ABCC2 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and invasion | [106] | |
| circ_104718 | miR-218-5p | TXNDC5 | Oncogenic | overexpression, siRNA knockdown in mouse xenografts, miR mimics, miR inhibitors | Proliferation, migration and invasion | [107] | |
| circ_104075 | miR-582-3p | YAP | Oncogenic | CRISPR/Cas9 knockout of upstream positive regulator in mouse models | Proliferation | [108] | |
| circFBLIM1 | miR-346 | FBLIM1 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and invasion | [109] | |
| circ_100338 | miR-141-3p | RHEB | Oncogenic | Cellular | Migration and invasion | [110] | |
| circ_0067934 | miR-1324 | FZD5 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [111] | |
| circ_0078710 | miR-31 | HDAC, CDK2 | Oncogenic | Overexpression in mouse xenografts | Proliferation, migration and invasion | [112] | |
| circ_ CDYL | miR-892a, miR-328-3p | HDGF, HIF1AN | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and chemoresistance | [113] | |
| circ_ DB | miR-34a | USP7 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation | [114] | |
| circ_101368 | miR-200a | HMGB1, RAGE | Oncogenic | Cellular | Migration and invasion | [115] | |
| circ_001569 | miR-411-5p, miR-432-5p | – | Oncogenic | shRNA knockdown in mouse xenografts | Migration and invasion | [116] | |
| circ_0015756 | miR-7 | FAK | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [117] | |
| circRBM23 | miR-138 | CCND3 | Oncogenic | Overexpression, siRNA knockdown in mouse xenografts | Proliferation and migration | [118] | |
| circ_0016788 | miR-486 | CDK4 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation and invasion | [119] | |
| circ_0000673 | miR-767-3p | SET | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and invasion | [120] | |
| circ_0005075 | miR-431 | ------- | Oncogenic | Cellular | Proliferation, migration and invasion | [121] | |
| miR-335 | MAPK1 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [122] | ||
| circ_BIRC6 | miR-3918 | Bcl2 | Oncogenic | Overexpression, shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [123] | |
| circ_ZNF652 | miR-203, miR-502-5p | Snail | Oncogenic | Knockdown in mouse xenografts | EMT | [124] | |
| circ ZFR | miR-3619-5p | CTNNB1 | Oncogenic | Cellular | Proliferation and EMT | [125] | |
| circ_0008450 | miR-214-3p | EZH2 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [126] | |
| circ_0000517 | miR‑326 | SMAD6 | Oncogenic | shRNA knockdown in mouse xenografts | Proliferation, colony formation, migration, and invasion | [127] | |
| circNFATC3 | miR-548I | NFATC3 | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation, migration and invasion | [128] | |
| circ_103809 | miR-620 | ------- | Tumor suppressive | Cellular | Proliferation, migration and invasion | [129] | |
| circC3P1 | miR-4641 | PCK1 | Tumor suppressive | Overexpression in mouse xenografts, miR mimics | Proliferation, migration and invasion | [130] | |
| circ_0091570 | miR-1307 | ISM1 | Tumor suppressive | Overexpression, siRNA knockdown in mouse xenografts | Proliferation and migration | [131] | |
| circ_0001649 | miR-127-5p, miR-612, miR-4688 | SHPRH | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation and migration | [132] | |
| circMTO1 | miR-9 | p21 | Tumor suppressive | siRNA knockdown in mouse xenografts | Proliferation and invasion | [133] | |
| cSMARCA5 | miR-17-3p, miR-181b-5p | TIMP3 | Tumor suppressive | Overexpression in mouse xenografts | Proliferation and migration | [134] | |
| circSETD3 | miR-421 | MAPK-14 | Tumor suppressive | Overexpression in mouse xenografts | Proliferation | [135] | |
| circTRIM33-12 | miR-191 | TET1 | Tumor suppressive | Overexpression, shRNA knockdown in mouse xenografts | Proliferation, migration, invasion and immune evasion | [136] | |
| circADAMTS13 | miR-484 | ADAMTS13 | Tumor suppressive | Cellular | Proliferation | [137] | |
| circADAMTS14 | miR-572 | RCAN1 | Tumor suppressive | Overexpression in mouse xenografts, miR mimics, miR inhibitors | Proliferation and invasion | [138] | |
| circSMAD2 | miR-629 | SMAD2 | Tumor suppressive | Cellular | Migration, invasion and EMT | [139] | |
| circLARP4 | miR-761 | RUNX3 | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation | [140] | |
| circLARP4 | miR-761 | RUNX3 | Tumor suppressive | shRNA knockdown in mouse xenografts | Proliferation | [140] | |
| circ_0005986 | miR-129-5p | Notch1 | Tumor suppressive | Cellular | Proliferation | [141] |
ceRNA: competing endogenous RNA; circRNA: circularRNA; mRNAs: messenger RNAs; miRNA: microRNA; HCC: hepatocellular carcinoma; EMT: epithelial–mesenchymal transition; Ref: reference
Similar to other ceRNAs, circRNAs competitively inhibit miRNAs via their MREs. CircRNAs function in tumor signaling pathways through their regulation of cell proliferation, apoptosis, tumor invasion and migration. Qin et al. [142] demonstrated significantly decreased expression of hsa_circ_0001649 expression in HCC tissues (P = 0.0014), which was related to tumor embolus (P = 0.017) as well as tumor size (P = 0.045). Yao et al. [143] reported the significantly decreased expression of circZKSCAN1 (hsa_circ_0001727) in HCC (P < 0.05), which was associated with vascular invasion (P = 0.002) and cirrhosis (P = 0.031) as well as tumor number (P < 0.01). Hu et al. reported increased levels of circASAP1 in HCC patients with postoperative metastasis or recurrence [103]. Transplanted HCC mouse models verified that circASAP1 promoted proliferation as well as invasion of HCC through miR-326/miR-532-5p-MAPK signaling (Fig. 5b). Xenografts of tumors with shRNA-mediated knockdown of circASAP1 showed decreased numbers of metastatic pulmonary nodules, while the xenograft model derived from circASAP1-overexpressing cells had a higher rate of lung metastasis. In addition, circASAP1 enhanced the infiltration of tumor associated macrophages in xenografts, but no alterations of infiltrating neutrophils or fibroblasts were observed. Zhang et al. observed that overexpression of circSMAD2 attenuated migration, invasion and EMT of HCC by sponging miR-629 [139]. Taken together, these results provide evidence that circRNAs have a dual role in regulating tumor signaling pathways by competing with miRNAs with contrary functions.
CircRNAs are resistant to exonucleases, resulting in relatively stable expression and intracellular accumulation. Exosomal excretion is thus an important route of in vivo elimination of circRNAs. In a study by Su et al., SMMC-7721 and HepG2 cell-derived exosomal circRNA Cdr1 was transfected into 293T cells [104]. Consistent with its intracellular function, exosomal Cdr1as increased the proliferation and migration of 293T cells, indicating that circRNAs function in the cytoplasm and exosomes. Zhang et al. reported a similar finding in the circ-DB/miR-34a/USP7/Cyclin A2 pathway [114].
The activating receptor natural-killer group 2 member D (NKG2D) is a tumor-associated immune response marker, and its expression indicates immune response mediated by CD8 + T cells and γδ + T cells as well as NK cells in tumors. Zhang et al. proposed the circTRIM22-12/miR-191/TET1 pathway and validated it in circTRIM33-12-overexpressing nude mice xenograft models. circTRIM33-12 expression level was positively correlated with NKG2D-positive cell numbers in HCC, suggesting that circTRIM33-12 sponged miR-191/TET1 and exerted an anti-tumor function by not only participating in the tumor proliferation–related signaling pathway, but also contributing to immune surveillance and T cell activation [136].
Considering the high stability of circRNAs in body fluids, including blood and saliva, circRNAs are potential superior biomarkers for HCC diagnosis. The miR-328-3p/HIF1AN-NOTCH2 and miR-892a/HDGF-NCL-PI3K-AKT pathways regulated by circCDYL facilitate Survivin and Myc expression in HCC, suggesting that a combination of circCDYL, HIF1AN and HDGF can be used for the diagnosis of Barcelona Clinic Liver Cancer stage 0 and A of HCC [144]. CircRNA_104075, another potential diagnostic biomarker, was discovered to upregulate the expression of YAP by inhibiting miR-582-3p. Zhang et al. used the CRISPR/Cas9 strategy to establish a knockdown mouse model of HNF4a. HNF4a was formerly proved to be an HCC-promoting transcription promoter. HNF4a knockout mice were smaller in size, with a low liver weight, and had low expression of circ_104075, which indicates that circ_104075 transcription is stimulated by HNF4a. Intriguingly, an m6A modification in the 3′-untranslated region of YAP mRNA facilitates the interaction between YAP, miR-582-3p and circRNA_104075, demonstrating how circRNA function can be influenced by methylation or other modifications and the role of epigenetics in circRNA function [108].
Similar to lncRNAs, circRNAs exert their diverse functions in HCC through sponging miRNAs. CircRNAs exhibit multiple roles in different pathways and participate in HCC pathogenesis through various pathophysiologic processes. In addition, circRNAs show enhanced stability, making them promising candidates for novel cancer biomarkers.
Pseudogenes as ceRNAs
Pseudogenes were previously described as non-functional, genomic junk DNAs sharing homology with the parental encoding sequence. Pseudogenes were characterized by a loss of protein-coding regions due to truncation or mutation and suggested to have no function. Transcriptomic and proteomic analyses have not only confirmed the ability of pseudogenes to produce transcripts and proteins, but also revealed novel miRNA/gene/pseudogene regulatory networks in cancer biology [145, 146]. Studies on the influence of pseudogenes in different types of tumors have revealed their roles in HCC (Table 4).
Table 4.
Validated ceRNA networks shaped by the ceRNA function of pseudogene in HCC
| ceRNA network type | ceRNA member | Shared miRNA(s) | Competing target (mRNA) | ceRNA role | Experiments related to mechanism | Related HCC function mechanisms | Ref |
|---|---|---|---|---|---|---|---|
| Pseudogene–miRNA–mRNA | OCT4-pg4 | miR-145 | OCT4 | Oncogenic | siRNA knockdown in mouse xenografts | Proliferation and colony formation | [10] |
| RACGAP1P | miR-15-5p | RACGAP1 | Oncogenic | Overexpression in mouse xenografts | Proliferation and migration | [147] | |
| INTS6P1 | miR-17-5p | INTS6 | Tumor suppressive | Overexpression in mouse xenografts | Apoptosis and cell growth | [148] |
ceRNA: competing endogenous RNA; mRNAs: messenger RNAs; miRNA: microRNA; HCC: hepatocellular carcinoma; Ref: reference
OCT4-pg4 is a pseudogene related to OCT4, a transcription factor that is involved in proliferation, pluripotency and self-renewal ability in embryonic stem cells and germ cells [149]. The expression of OCT4-pg4 is positively associated with OCT4, and both are increased in HCC. Wang et al. found that OCT4-pg4 serves as a molecular decoy towards miR-145 and prevents OCT4 inhibition, thus promoting HCC growth and tumorigenicity [10]. Survival analysis of 54 cases of HCC suggested a significant correlation between increased OCT4-pg4 expression levels and shortened overall survival as well as disease-free survival in HCC patients.
RACGAP, a member of the GTPase activation family, is an oncoprotein that enhances the proliferation and migration of HCC by activating the RhoA/ERK signaling pathway. The RACGAP gene is also the first gene reported as an independent biomarker for HCC recurrence. Wang et al. discovered the significant up-regulation of the RACGAP1P pseudogene in HCC, which was related to shortened survival, larger tumor size, elevated AFP level and advanced clinical stage [147]. Luciferase assays and in vivo assays demonstrated that miR-15-5p was sequestered from its endogenous target RACGAP by RACGAP1P, causing increased RACGAP expression and contributing to the RACGAP oncogenic network (Fig. 5c).
In addition to oncogenic pseudogenes, tumor suppressive pseudogenes have also been identified. The INTS6 pseudogene inhibits the tumor growth in several types of human cancers through G1 cell cycle arrest. Peng et al. showed that INTS6 inhibited HCC cell growth, migration and survival [148]. Furthermore, INTS6P1 facilitated tumor suppression by competing with oncogenic miR-17-5p. In vitro and in vivo assays also showed that both INTS6 and INTS6P inhibited HCC cell growth, migration and survival.
These studies indicate that pseudogenes serve as ceRNAs and affect tumorigenesis through ceRNETs. With respect to their role in HCC carcinogenesis, pseudogenes may be used as prognostic indicators, stratification factors or therapeutic targets, and these functions may lead to the development of precise and individualized therapeutic strategies for HCC. It is likely that HCC-associated pseudogenes acting as ceRNAs are rare. Nevertheless, with the widespread application of whole-genome high-throughput sequencing technology, we believe that more HCC-associated pseudogenes will be uncovered and applied in disease management.
The clinical application of ncRNAs as ceRNAs in HCC
The current commonly used screening methods for HCC include the alpha-fetoprotein (AFP) diagnostic marker, ultrasonography, computed tomography, magnetic resonance imaging and liver biopsy [150]. Among these methods, imaging examination and serum AFP testing are the most common and basic screening approaches. However, even if a low-level cutoff is applied, the sensitivity value of AFP for diagnosing HCC is nearly 60% and the specificity is still inadequate [151]. Moreover, approximately 30% of patients with early-stage HCC cannot be detected using serum AFP as diagnostic biomarker [3]. According to the European Association for the Study of the Liver (EASL) and the American Association for the Study of Liver Diseases (AASLD) guidelines for HCC, serum AFP level can also be used to monitor HCC progression [152, 153]. Although the diagnostic rate of HCC has markedly increased from the adoption of serum AFP screening, the limited specificity of serum AFP has led to overdiagnosis and overtreatment [154]. In treating HCC patients with antineoplastic drugs, currently available regimens have shown disappointing results: both first-line therapy sorafenib and lenvatinib only prolong survival by nearly two months for patients with advanced-stage HCC [155]. Given the emerging evidence indicating that ncRNAs act as ceRNAs and suggesting their use as biomarkers and even therapeutic targets for HCC, further studies on ncRNAs may provide new insights and novel strategies for HCC diagnosis, surveillance and treatments.
Potential diagnostic biomarkers in HCC
Several ncRNAs that act as ceRNAs have been identified in the circulation and other body fluids in HCC and other cancers [108, 156–158]. These results suggested the potential that ncRNAs that act as ceRNAs and their associated ceRNETs may be useful for clinical applications as diagnostic biomarkers of HCC (Table 5).
Table 5.
Potential clinical application of ncRNAs function as ceRNAs in HCC
| ncRNA as ceRNA | Location | Clinical application | Description | Ref |
|---|---|---|---|---|
| circRNA_104075# | Serum | Diagnosis | AUC: 0.973, Se: 96.0%, Sp: 98.3% | [108] |
| BCYRN 1* | Serum | Diagnosis, prognosis, potential therapeutic target | Diagnosis: AUC: 0.7834, Se: 62.10%, Sp: 90.54%; Prognosis: overexpression associates with worse prognosis | [159, 160] |
| lncRNA-RP11‐513I15.6, miR‐1262, RAB11A† | Serum | Diagnosis | AUC: NA, Se: 100%, Sp: 76.7% | [156] |
| OCT4-pg4^ | Tissue | Prognosis, potential therapeutic target | Associates with prognosis | [10] |
| HOXD-AS1* | Tissue | Prognosis, potential therapeutic target | Associates with TNM stage and prognosis | [72] |
| SNHG6-003* | Tissue | Prognosis, potential therapeutic target | Associates with portal vein tumor thrombus, BCLC stage, distant metastasis and prognosis | [85] |
| AGAP2-AS1* | Tissue | Prognosis, potential therapeutic target | Associates with tumor size, histological grade, TNM stage, venous invasion and prognosis | [89] |
| CASC2* | Tissue | Prognosis, potential therapeutic target | Associates with venous infiltration, histological grade, TNM stage and prognosis | [98] |
| circRNA_104718# | Tissue | Prognosis, potential therapeutic target | Associates with vascular invasion and prognosis | [107] |
| circ_0000517# | Tissue | Prognosis, potential therapeutic target | Associated with tumor size, TNM stage, lymph node metastasis and prognosis | [127] |
| circNFATC3# | Tissue | Prognosis, potential therapeutic target | Associates with vascular invasion, histological grade and prognosis | [128] |
| MYCNOS, DLX6-AS1, LINC00221, CRNDE* | Tissue | Prognosis | Associates with prognosis | [161] |
| 13 lncRNAs prognositc model† | Tissue | Prognosis | Associates with prognosis | [162] |
| 3 lncRNAs and 6mRNAs prognostic model† | Tissue | Prognosis | Associates with prognosis | [163] |
ncRNA: non-coding RNA; ceRNA: competing endogenous RNA; HCC: hepatocellular carcinoma; AUC: Area under the curve; Se: Sensitivity; Sp: Specificity; NA: Not available; BCLC: Barcelona Clinic Liver Cancer; Ref: reference
* Long non-coding RNA; #circular RNA; ^ pseudogene; †ceRNA panel
Ming et al. demonstrated that the lncRNA BCYRN1 functions as a ceRNA to play a vital role in HCC [159]. The authors analyzed BCYRN1 expression in plasma from 124 HCC patients, 79 cirrhosis patients, 68 hepatitis B patients and 74 healthy controls. Plasma BCYRN1 expression of HCC patients was significantly higher than that of healthy controls (p < 0.001) or hepatitis B patients (p < 0.01) [159]. Furthermore, combined detection of BCYRN1 and AFP improved the diagnosis of HCC in their cohort. Their data also indicated that BCYRN1 in plasma performed well in HCC diagnosis.
Zhang et al. found that circ_104075 acts as ceRNA to sponge miR-582-3p to stimulate tumorigenesis via YAP [108]. Circ_104075 exhibited a sensitivity of 96.0% and a specificity of 98.3% in HCC diagnosis. Moreover, some recent studies revealed that panels consisting of ncRNAs and their related ceRNETs can also be used as diagnostic biomarkers for HCC. Asmaa et al. [156] constructed an lncRNA-associated ceRNET (lncRNA-RP11-513I15.6-miR1262-RAB11A mRNA) according to an in silico analysis, followed by validation in serum specimens and additional clinical and experimental assays. Receiver operating characteristic curve analysis revealed that RAB11A mRNA, lncRNA-RP11-513I15.6 and miR-1262 in the ceRNET were effective biomarkers to differentiate HCC subjects from healthy controls (area under curve: 0.963, 0.847 and 0.822, respectively). Moreover, the combination of serum exosomal miR-1262, lncRNARP11-513I15.6 and AFP measurements improved the diagnostic accuracy to detect HCC at the early stage to nearly 100% sensitivity and 76.7% accuracy [156]. With the development of specific algorithms for ncRNA detection and quantification, we believe that more and more ncRNAs that function as ceRNAs will be identified as diagnostic biomarkers for HCC.
Potential prognostic biomarkers and therapeutic targets in HCC
Several studies have explored the clinical application of ncRNAs that function as ceRNA as prognostic biomarkers and potential therapeutic targets for HCC (Table 5). Cao et al. found that the lncRNA SNHG6-003 functions as a ceRNA to promote cell proliferation and induce drug resistance in HCC [85]. High expression of SNHG6-003 closely correlated with tumor progression and poor survival in HCC patients. The results suggested that targeting the ceRNA network involving SNHG6-003 may be a treatment strategy against HCC. Besides the tremendous diagnostic value in HCC, the prognostic value of BCYRN1 was also proven in a recent study. Ding et al. demonstrated that overexpression of BCYRN1 significantly expedited HCC cell growth, clone formation and movement abilities, while downregulation of BCYRN1 had the opposite effects [160]. The authors also found that BCYRN1 was overexpressed in HCC samples, which was associated with unfavorable prognosis in patients with HCC. The study findings indicated that BCYRN1, miR-490-3p, and POU3F2 formed a ceRNA mechanism to modulate the occurrence and progression of HCC, suggesting these can be served as potential target molecules for the management of HCC [160]. Recently, Lin et al. established a circRNA-miRNA-mRNA regulatory network by integrating the analysis of differentially expressed circRNAs, miRNAs and mRNAs in HCC [9]. The functional enrichment analysis of differentially expressed circRNA-related mRNAs, screened from the constructed HCC-associated ceRNETs, revealed that differentially expressed cirRNAs were significantly associated with cell proliferation, cell adhesion and cell migration. The differentially expressed circRNAs are involved in various processes including cell cycle, peroxisome proliferator-activated receptor signaling, chemical carcinogenesis and p53 signaling, playing vital roles in HCC progression. The refined circRNA-miRNA-mRNA regulatory modules associated with HCC carcinogenesis further uncovered three key circRNAs (hsa_circ_0004913, hsa_circ_0007456 and hsa_circ_0078279) that may play important roles in carcinogenesis and progression of HCC [9]. These circRNAs may competitively bind to miR-182 and miR-346 to affect their respective regulatory networks. Functionalization of previously uncharacterized transcripts could be achieved, partially by identifying the ceRNA interactors, to present a framework for predicting and validating ceRNA interactions; this strategy may be universally applied to any transcript. In a scale-free network, links between nodes follow a power-law distribution, which suggests that most nodes have only a few links, while a few nodes can possess a large number of links [164]. These post-transcriptional ceRNETs can be considered as scale-free networks, because their structure is constructed upon smaller interconnected subnetworks, in which the nodes (ceRNAs) are linked by a large number of connections (miRNAs) [11]. The few highly connected nodes, also known as target hubs, were found to be critically involved in biological processes [164]. Therefore, these critical nodes (three circRNAs and two miRNAs), identified by the constructed circRNA-associated ceRNETs, may represent ideal therapeutic targets for HCC.
As promising biomarkers in the screening, diagnosis and prognosis for HCC, ncRNAs as ceRNAs are also potential therapeutic targets. However, there are still significant gaps in our current understanding of ncRNA functions as ceRNAs and we are still far from being able to incorporate ncRNAs into clinical practice. Large-scale studies and clinical trials are required to validate the role of ncRNAs as ceRNAs in the clinical application for HCC.
Conclusions
In this review, we briefly discussed ceRNAs as well as the principles and influencing factors of the interactions between ceRNAs, concentrating on the roles and molecular mechanisms of ncRNAs that function as ceRNAs in HCC. We also summarized the ideas and methods to study ceRNAs and ceRNETs in HCC and some commonly used databases.
Recently, significant progress has been made in the studies of ceRNAs in HCC. To date, studies on ncRNAs that act as ceRNAs in HCC have primarily involved overexpression and knockout assays in cells and animals. However, ceRNA activity is affected by other factors, including subcellular location and ceRNA component abundance, interactions with RNA binding proteins, RNA editing and ceRNA affinity in the endogenous cellular context. Whether the results shown in overexpression assays truly reflect the spontaneous ceRNA crosstalk during carcinogenesis in patients with HCC remains unknown. Therefore, more animal experiments and clinical trials should be performed to validate these results.
Additionally, the majority of identified ceRNA interactions reflect single binding partners, although emerging evidence indicates ceRNA crosstalk in large interconnected networks. Aside from direct interactions via shared miRNAs, secondary and indirect interactions might also exert significant effects on ceRNA modulation. Thus, further investigations of ceRNAs should not only concentrate on identifying binary ceRNA interactions but enroll network analyses of potential complex miRNA and ceRNA networks. Moreover, the scale-free network property of ceRNA regulation also poses a challenge in selecting HCC-related molecular therapeutic targets. Targeting nonessential nodes within regulatory networks could cause ineffective therapeutic responses, as cancer cells may overcome the resulting damage through alternative signaling pathways. Therefore, the selection of therapeutic targets situated in a hub position of a ceRNET should be considered in future screening studies for HCC therapeutic targets.
In summary, the recently developed research techniques and computational approaches as well as the continued uncovering of ceRNET components will facilitate more detailed studies of ncRNAs that act as ceRNAs in HCC. These findings will not only provide a more comprehensive understanding of the underlying mechanism of HCC pathogenesis and progression but also establish a novel direction for future strategies for the diagnosis, treatment and prevention of HCC.
Acknowledgements
We gratefully acknowledge all of the other researchers and work in this field that we were unable to cite due to space restrictions. We thank Sitong Liu and Yunzhu Li for assistance with graphic design. We also thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China, for editing the English text of a draft of this manuscript.
Abbreviations
- HCC
Hepatocellular carcinoma
- ceRNA
Competing endogenous RNA
- ncRNA
Non-coding RNA
- miRNA
MicroRNA
- lncRNA
Long non-coding RNA
- circRNA
Circular RNA
- mRNA
Messenger RNA
- MRE
MicroRNA response element
- ceRNET
Competing endogenous RNA regulatory network
- Ago
Argonaute
- EMT
Epithelial-mesenchymal transition
- AFP
Alpha-fetoprotein
- AUC
Area under the curve
Authors’ contributions
GX and WYX designed the study. GX, WYX, YX and BJ were mainly responsible for data collection and drafted the manuscript. YLM, SDD and ZTZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Technologies R&D Program (Grant Number: 2015BAI13B09), the National Natural Science Foundation of China (Grant Number: 81972698) and Peking Union Medical College Graduate Student Innovation Fund (Grant Number: 2019-1002-45).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gang Xu, Wei-Yu Xu and Yao Xiao contributed equally to this article.
Contributor Information
Shun-Da Du, Email: dushd@pumch.cn.
Zhong-Tao Zhang, Email: zhangzht@ccmu.edu.cn.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A, Minguez B, Forner A, et al. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–28. doi: 10.1146/annurev.med.080608.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28(5):753–70. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M. Systemic therapy for hepatocellular carcinoma: 2017 Update. Oncology. 2017;93:1135–46. doi: 10.1159/000481244. [DOI] [PubMed] [Google Scholar]
- 6.Kataoka M, Wang DZ. Non-coding RNAs including miRNAs and lncRNAs in cardiovascular biology and disease. Cells. 2014;3(3):883–98. doi: 10.3390/cells3030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin MT, Song HJ, Ding XY. Long non-coding RNAs involved in metastasis of gastric cancer. World J Gastroenterol. 2018;24(33):3724–37. doi: 10.3748/wjg.v24.i33.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi B, Zhang X, Chao L, et al. Comprehensive analysis of key genes, microRNAs and long non-coding RNAs in hepatocellular carcinoma. FEBS Open Bio. 2018;8(9):1424–36. doi: 10.1002/2211-5463.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X, Chen Y. Identification of potentially functional circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70–78. doi: 10.12659/MSMBR.909737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Guo ZY, Zhang R, et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34(8):1773–81. doi: 10.1093/carcin/bgt139. [DOI] [PubMed] [Google Scholar]
- 11.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA. language? Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan M, Li X, Jiang W, et al. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5(4):1143–46. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–21. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20(19):R858-61. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–8. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 18.Ala U, Karreth FA, Bosia C, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A. 2013;110(18):7154–9. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figliuzzi M, Marinari E, De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys J. 2013;104(5):1203–13. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarnow P, Jopling CL, Norman KL, et al. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4(9):651–9. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- 21.Slezak-Prochazka I, Kluiver J, de Jong D, et al. Cellular localization and processing of primary transcripts of exonic microRNAs. PLoS One. 2013;8(9):e76647. doi: 10.1371/journal.pone.0076647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56(3):347–59. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 25.He N, Zheng H, Li P, et al. miR-485-5p binding site SNP rs8752 in HPGD gene is associated with breast cancer risk. PLoS One. 2014;9(7):e102093. doi: 10.1371/journal.pone.0102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 27.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 28.Bish R, Vogel C. RNA binding protein-mediated post-transcriptional gene regulation in medulloblastoma. Mol Cells. 2014;37(5):357–64. doi: 10.14348/molcells.2014.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epis MR, Barker A, Giles KM, et al. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem. 2011;286(48):41442–54. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HH, Kuwano Y, Srikantan S, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23(15):1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T, Wang J, Liu C, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34(Database issue):D150-2. doi: 10.1093/nar/gkj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41(Database issue):D239-45. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231-8. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20(11):1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong J, Liu C, Liu W, et al. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017;45(D1):D79-d84. doi: 10.1093/nar/gkw835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu SM, Liu H, Huang PJ, et al. circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience. 2018;7(1):1–10. doi: 10.1093/gigascience/gix118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S, Feng J, Chen K, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925-d29–D925-d29. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furio-Tari P, Tarazona S, Gabaldon T, et al. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44(W1):W176-80. doi: 10.1093/nar/gkw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44(D1):D1005-10. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das S, Ghosal S, Sen R, et al. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9(6):e98965. doi: 10.1371/journal.pone.0098965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–3. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–51. doi: 10.1042/BCJ20170079. [DOI] [PubMed] [Google Scholar]
- 51.He T, Zhang L, Kong Y, et al. Long non-coding RNA CASC15 is upregulated in hepatocellular carcinoma and facilitates hepatocarcinogenesis. Int J Oncol. 2017;51(6):1722–30. doi: 10.3892/ijo.2017.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–40. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 53.Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–46. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Chen F, Zhao M, et al. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292(37):15395–407. doi: 10.1074/jbc.M117.783738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng D, Deng J, Zhang B, et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;361:59–70. doi: 10.1016/j.ebiom.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang T, He X, Chen A, et al. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–22. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 57.Pan Y, Tong S, Cui R, et al. Long Non-Coding MALAT1 Functions as a Competing Endogenous RNA to Regulate Vimentin Expression by Sponging miR-30a-5p in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;50(1):108–20. doi: 10.1159/000493962. [DOI] [PubMed] [Google Scholar]
- 58.Hou Z, Xu X, Zhou L, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017;39(7):1010428317718135. doi: 10.1177/1010428317718135. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Yao H, Wang K, et al. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118(12):4836–43. doi: 10.1002/jcb.26158. [DOI] [PubMed] [Google Scholar]
- 60.Huang X, Gao Y, Qin J, et al. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2018;314(5):G559-g65. doi: 10.1152/ajpgi.00242.2017. [DOI] [PubMed] [Google Scholar]
- 61.Tang J, Zhuo H, Zhang X, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng L, Yang SB, Xu FF, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 64.Guo D, Li Y, Chen Y, et al. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Prolif. 2019;52(4):e12628. doi: 10.1111/cpr.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Pu J, Zhang Y, et al. DANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12. J Cell Physiol. 2019;234(6):9408–16. doi: 10.1002/jcp.27625. [DOI] [PubMed] [Google Scholar]
- 66.Tsang FH, Au SL, Wei L, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35(5):1597–606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 67.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao JN, Yan TH, Yu RM, et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143(6):981–90. doi: 10.1007/s00432-017-2370-1. [DOI] [PubMed] [Google Scholar]
- 70.Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–29. doi: 10.1016/j.lfs.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Yan X, Zhang D, Wu W, et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77(23):6704–16. doi: 10.1158/0008-5472.CAN-17-1915. [DOI] [PubMed] [Google Scholar]
- 72.Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong J, Teng F, Guo W, et al. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(5):2262–74. doi: 10.1159/000495871. [DOI] [PubMed] [Google Scholar]
- 74.Huang Y, Xiang B, Liu Y, et al. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi: 10.1016/j.canlet.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 75.Zhang K, Zhao Z, Yu J, et al. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119(7):6045–56. doi: 10.1002/jcb.26802. [DOI] [PubMed] [Google Scholar]
- 76.Li T, Xie J, Shen C, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75(15):3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 77.Zhang XN, Zhou J, Lu XJ. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol. 2018;233(9):6733–41. doi: 10.1002/jcp.26371. [DOI] [PubMed] [Google Scholar]
- 78.He C, Liu Z, Jin L, et al. lncRNA TUG1-Mediated Mir-142-3p Downregulation Contributes to Metastasis and the Epithelial-to-Mesenchymal Transition of Hepatocellular Carcinoma by Targeting ZEB1. Cell Physiol Biochem. 2018;48(5):1928–41. doi: 10.1159/000492517. [DOI] [PubMed] [Google Scholar]
- 79.Lv J, Kong Y, Gao Z, et al. LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma. Int J Biochem Cell Biol. 2018;101:19–28. doi: 10.1016/j.biocel.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 80.Ma J, Li T, Han X, et al. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144(2):205–14. doi: 10.1007/s00432-017-2543-y. [DOI] [PubMed] [Google Scholar]
- 81.Fan H, Lv P, Mu T, et al. LncRNA n335586/miR-924/CKMT1A axis contributes to cell migration and invasion in hepatocellular carcinoma cells. Cancer Lett. 2018;429:89–99. doi: 10.1016/j.canlet.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Xu J, Zhang S, et al. HOXA-AS2 Promotes Proliferation and Induces Epithelial-Mesenchymal Transition via the miR-520c-3p/GPC3 Axis in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;50(6):2124–38. doi: 10.1159/000495056. [DOI] [PubMed] [Google Scholar]
- 83.Ren Y, Shang J, Li J, et al. The long noncoding RNA PCAT-1 links the microRNA miR-215 to oncogene CRKL-mediated signaling in hepatocellular carcinoma. J Biol Chem. 2017;292(43):17939–49. doi: 10.1074/jbc.M116.773978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao C, Zhang T, Zhang D, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36(8):1112–22. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 86.Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Sun L, Wang L, et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/beta-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018;9(9):851. doi: 10.1038/s41419-018-0937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tu J, Zhao Z, Xu M, et al. LINC00707 contributes to hepatocellular carcinoma progression via sponging miR-206 to increase CDK14. J Cell Physiol. 2019;234(7):10615–24. doi: 10.1002/jcp.27737. [DOI] [PubMed] [Google Scholar]
- 89.Liu Z, Wang Y, Wang L, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):194. doi: 10.1186/s13046-019-1188-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Zhao L, Hu K, Cao J, et al. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging. 2019;11(17):7098–122. doi: 10.18632/aging.102240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji D, Hu G, Zhang X, et al. Long non-coding RNA DSCAM-AS1 accelerates the progression of hepatocellular carcinoma via sponging miR-338-3p. Am J Transl Res. 2019;11(7):4290–302. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Kong Q, Zhang S, Liang C, et al. LncRNA XIST functions as a molecular sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular carcinoma cell. J Cell Biochem. 2018;119(6):4458–68. doi: 10.1002/jcb.26540. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Y, Zhu Z, Huang S, et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 2019;19:198. doi: 10.1186/s12935-019-0909-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhuang LK, Yang YT, Ma X, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu B, Cai H, Zheng R, et al. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol. 2017;92:34–42. doi: 10.1016/j.biocel.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 96.Liu Z, Chen JY, Zhong Y, et al. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res. 2019;52(10):e8631. doi: 10.1590/1414-431x20198631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu F, Zha G, Wu Y, et al. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855–63. doi: 10.2147/OTT.S182005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Liu Z, Yao B, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16(1):123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan S, Tang Z, Chen K, et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37(1):214. doi: 10.1186/s13046-018-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu F, Yuan JH, Huang JF, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35(41):5422–34. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 101.Suzuki H, Zuo Y, Wang J, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–42. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu ZQ, Zhou SL, Li J, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2019 doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 104.Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11(19):8182–203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu L, Gong X, Sun L, et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Bai N, Peng E, Xia F, et al. CircABCC2 regulates hepatocellular cancer progression by decoying MiR-665. J Cancer. 2019;10(17):3893–98. doi: 10.7150/jca.31362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond) 2019;133(13):1487–503. doi: 10.1042/CS20190394. [DOI] [PubMed] [Google Scholar]
- 108.Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091. doi: 10.1038/s41419-018-1132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172. doi: 10.1186/s13046-018-0838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang XY, Huang ZL, Zhang PB, et al. CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Front Oncol. 2019;9:392. doi: 10.3389/fonc.2019.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–32. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 112.Xie B, Zhao Z, Liu Q, et al. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–61. doi: 10.1016/j.gene.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 113.Wei Y, Chen X, Liang C, et al. A noncoding eegulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2019 doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 114.Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–59. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17(19–20):2349–59. doi: 10.1080/15384101.2018.1526599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu H, Xue L, Song C, et al. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem Biophys Res Commun. 2018;503(4):2659–65. doi: 10.1016/j.bbrc.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 117.Liu L, Yang X, Li NF, et al. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18(21):2939–53. doi: 10.1080/15384101.2019.1664223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang B, Chen H, Zhang C, et al. Effects of hsa_circRBM23 on hepatocellular carcinoma cell viability and migration as produced by regulating miR-138 expression. Cancer Biother Radiopharm. 2018;33(5):194–202. doi: 10.1089/cbr.2017.2424. [DOI] [PubMed] [Google Scholar]
- 119.Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500–08. doi: 10.1002/jcp.26612. [DOI] [PubMed] [Google Scholar]
- 120.Jiang W, Wen D, Gong L, et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun. 2018;500(2):211–16. doi: 10.1016/j.bbrc.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 121.Li MF, Li YH, He YH, et al. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed Pharmacother. 2018;99:848–58. doi: 10.1016/j.biopha.2018.01.150. [DOI] [PubMed] [Google Scholar]
- 122.Yang X, Song H, Zi Z, et al. Circ_0005075 promotes hepatocellular carcinoma progression by suppression of microRNA-335. J Cell Physiol. 2019;234(12):21937–46. doi: 10.1002/jcp.28757. [DOI] [PubMed] [Google Scholar]
- 123.Yang G, Wang X, Liu B, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle. 2019;18(9):976–89. doi: 10.1080/15384101.2019.1601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo J, Duan H, Li Y, et al. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513(4):812–19. doi: 10.1016/j.bbrc.2019.03.214. [DOI] [PubMed] [Google Scholar]
- 125.Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. doi: 10.1016/j.abb.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 126.Lin T, Dai Y, Guo X, et al. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag Res. 2019;119:133–43. doi: 10.2147/CMAR.S222716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He S, Guo Z, Kang Q, et al. Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 2020;20:360. doi: 10.1186/s12935-020-01447-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Jia C, Yao Z, Lin Z, et al. circNFATC3 sponges miR-548I acts as a ceRNA to protect NFATC3 itself and suppressed hepatocellular carcinoma progression. J Cell Physiol. 2020 doi: 10.1002/jcp.29931. [DOI] [PubMed] [Google Scholar]
- 129.Li X, Shen M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci. 2019;23(2):555–66. doi: 10.26355/eurrev_201902_16868. [DOI] [PubMed] [Google Scholar]
- 130.Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499(4):1044–49. doi: 10.1016/j.bbrc.2018.03.221. [DOI] [PubMed] [Google Scholar]
- 131.Wang YG, Wang T, Ding M, et al. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–38. doi: 10.1016/j.canlet.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 132.Su Y, Xu C, Liu Y, et al. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging. 2019;11(10):3362–75. doi: 10.18632/aging.101988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–64. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 134.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 135.Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. doi: 10.1186/s13046-019-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang PF, Wei CY, Huang XY, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1):105. doi: 10.1186/s12943-019-1031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–55. doi: 10.1002/1878-0261.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song C, Li D, Liu H, et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J Cell Physiol. 2019;234(3):2460–70. doi: 10.1002/jcp.26764. [DOI] [PubMed] [Google Scholar]
- 139.Zhang X, Luo P, Jing W, et al. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 2018;11:2853–63. doi: 10.2147/OTT.S158008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen Z, Zuo X, Pu L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(2):568–81. doi: 10.1111/cas.13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8(27):43878–88. doi: 10.18632/oncotarget.16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–9. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 143.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–37. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71(1):130–47. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 145.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim MS, Pinto SM, Getnet D, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang MY, Chen DP, Qi B, et al. Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis. 2019;10(6):426. doi: 10.1038/s41419-019-1666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peng H, Ishida M, Li L, et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget. 2015;6(8):5666–77. doi: 10.18632/oncotarget.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pan GJ, Chang ZY, Scholer HR, et al. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12(5–6):321–9. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 150.Xie DY, Ren ZG, Zhou J, et al. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr. 2020;9(4):452–63. doi: 10.21037/hbsn-20-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Trevisani F, D’Intino PE, Morselli-Labate AM, et al. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34(4):570–5. doi: 10.1016/S0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 152.EASL Clinical Practice Guidelines Management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 153.Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–80. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 154.Luo P, Wu S, Yu Y, et al. Current Status and Perspective Biomarkers in AFP Negative HCC: Towards Screening for and Diagnosing Hepatocellular Carcinoma at an Earlier Stage. Pathol Oncol Res. 2020;26(2):599–603. doi: 10.1007/s12253-019-00585-5. [DOI] [PubMed] [Google Scholar]
- 155.Xie F, Feng S, Sun L, et al. The first-line treatment for unresectable hepatocellular carcinoma patients: lenvatinib versus sorafenib, or beyond? Hepatobiliary Surg Nutr. 2018;7(3):221–24. doi: 10.21037/hbsn.2018.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abd El Gwad A, Matboli M, El-Tawdi A, et al. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem. 2018;119(10):8600–10. doi: 10.1002/jcb.27109. [DOI] [PubMed] [Google Scholar]
- 157.Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37(1):206. doi: 10.1186/s13046-018-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu YH, Deng JL, Wang G, et al. Long non-coding RNAs in prostate cancer: Functional roles and clinical implications. Cancer Lett. 2019;464:37–55. doi: 10.1016/j.canlet.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 159.Ming XL, Feng YL, He DD, et al. Role of BCYRN1 in hepatocellular carcinoma pathogenesis by lncRNA-miRNA-mRNA network analysis and its diagnostic and prognostic value. Epigenomics. 2019;11(10):1209–31. doi: 10.2217/epi-2018-0218. [DOI] [PubMed] [Google Scholar]
- 160.Ding S, Jin Y, Hao Q, et al. LncRNA BCYRN1/miR-490-3p/POU3F2, served as a ceRNA network, is connected with worse survival rate of hepatocellular carcinoma patients and promotes tumor cell growth and metastasis. Cancer Cell Int. 2020;20:6. doi: 10.1186/s12935-019-1081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–48. doi: 10.1002/jcp.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Long J, Bai Y, Yang X, et al. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi: 10.1186/s12935-019-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liao X, Wang X, Huang K, et al. Integrated analysis of competing endogenous RNA network revealing potential prognostic biomarkers of hepatocellular carcinoma. J Cancer. 2019;10(14):3267–83. doi: 10.7150/jca.29986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Seo H, Kim W, Lee J, et al. Network-based approaches for anticancer therapy (Review) Int J Oncol. 2013;43(6):1737–44. doi: 10.3892/ijo.2013.2114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.