Abstract
Bacterial microcompartments (MCPs) are proteinaceous organelles consisting of a metabolic pathway encapsulated within a selectively permeable protein shell. Hundreds of species of bacteria produce MCPs of at least nine different types, and MCP metabolism is associated with enteric pathogenesis, cancer, and heart disease. This review focuses chiefly on the four types of catabolic MCPs (metabolosomes) found in Escherichia coli and Salmonella: the propanediol utilization (pdu), ethanolamine utilization (eut), choline utilization (cut), and glycyl radical propanediol (grp) MCPs. Although the great majority of work done on catabolic MCPs has been carried out with Salmonella and E. coli, research outside the group is mentioned where necessary for a comprehensive understanding. Salient characteristics found across MCPs are discussed, including enzymatic reactions and shell composition, with particular attention paid to key differences between classes of MCPs. We also highlight relevant research on the dynamic processes of MCP assembly, protein targeting, and the mechanisms that underlie selective permeability. Lastly, we discuss emerging biotechnology applications based on MCP principles and point out challenges, unanswered questions, and future directions.
GENERAL FEATURES OF BACTERIAL MICROCOMPARTMENTS (MCPs)
One often-cited mark of sophistication for eukaryotic cells is the ability to compartmentalize metabolic reactions. It is increasingly becoming appreciated, however, that prokaryotic cells also have the means to sequester cellular processes by the use of bacterial microcompartments (MCPs) (1–3). MCPs range in size from 100 to 400 nm in diameter and consist of a selectively permeable protein shell surrounding enzymes for a multistep metabolic pathway. The pathways encapsulated within MCPs have volatile or toxic intermediates, and the primary functions of MCPs are to prevent cellular toxicity, reduce carbon loss, concentrate reaction machinery, and increase process efficiency. The genes required for MCP formation are typically contained within large contiguous gene clusters (Fig. 1) that encode related elements and are frequently spread by horizontal gene transfer (3–8). All known MCPs include shells that are built from a conserved set of shell proteins (9), and many catabolic MCPs encapsulate pathways involving radical enzymes that produce aldehyde intermediates (1, 5–7, 10). MCPs are produced by hundreds of species of bacteria encompassing at least 23 phyla and have diverse physiological functions (4–8). The metabolism of MCPs provides a selective advantage to pathogens in host environments and impacts the ecology of the human gut (1, 11–13). Developments in characterizing the assembly and function of several classes of MCPs have revealed the complex nature of these particles and have motivated varied bioengineering applications (14–17). However, there are still a number of unanswered questions surrounding MCPs that promise the discovery of unusual biochemistry and will likely spur further biotechnology advances.
Figure 1.
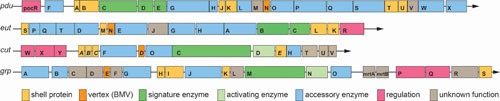
A comparison of selected MCP operons. Representative operons from S. enterica LT2 (pdu, eut), E. coli 536 (cut), and E. coli CFT073 (grp) are shown using one-letter abbreviations for genes and protein products. Gene sizes and spacing are roughly to scale. Note that in the cut operon, shell proteins are named with a different acronym, CmcA to -E to avoid redundancy with prior work.
Diverse Bacterial MCPs
Carboxysomes were the first bacterial MCP identified (18). Initially, these compartments were observed within cyanobacteria by electron microscopy (EM) and were thought to be viruses. However, later work showed that carboxysomes consisted of a protein shell filled with Rubisco and that their function was to enhance CO2 fixation as part of a CO2-concentrating mechanism (19–22). Over 20 years later, an operon required for 1,2-propanediol utilization (pdu) by Salmonella enterica serovar Typhimurium (referred to here as S. enterica) was found to contain multiple homologs of carboxysome shell genes (22). Further study showed that the pdu operon encoded a 100- to 150-nm polyhedral body (a new type of MCP) involved in 1,2-propanediol degradation (22). Shortly after this, searches of bacterial genomes for shell gene homologs began to identify diverse MCP operons which we now know are present in ∼20% of bacterial species (4–8). Further work found that catabolic MCPs (also referred to as metabolosomes) support bacterial growth on a variety of substrates, including 1,2-propanediol, ethanolamine, choline, glycerol, rhamnose, fucose, fucoidan, aminoacetone, and perhaps other metabolites (22–27).
This review focuses on metabolosomes found in Escherichia coli and Salmonella. These include the propanediol utilization (pdu), ethanolamine utilization (eut), choline utilization (cut), and glycyl radical propanediol (grp) MCPs. Each of these metabolosomes supports growth by breaking down substrates to alcohols and acids via acetaldehyde or propionaldehyde intermediates (Fig. 2). The Pdu and Eut MCPs are the best-studied metabolosomes, and much of this research has been conducted in Salmonella and E. coli (10). Both these MCPs use pathways that are dependent on coenzyme B12 (adenosylcobalamin), referred to here as Ado-B12. The Cut and Grp MCPs employ glycyl radical enzymes and are B12-independent (28–30). They are part of the widespread group of MCPs known as glycyl radical microcompartments (GRMs), which are absent from Salmonella but narrowly distributed among E. coli. These MCPs tend to be found in pathogenic strains where investigations aimed at dissecting their structure, function, mechanisms, and unique features have recently begun (29, 30).
Figure 2.
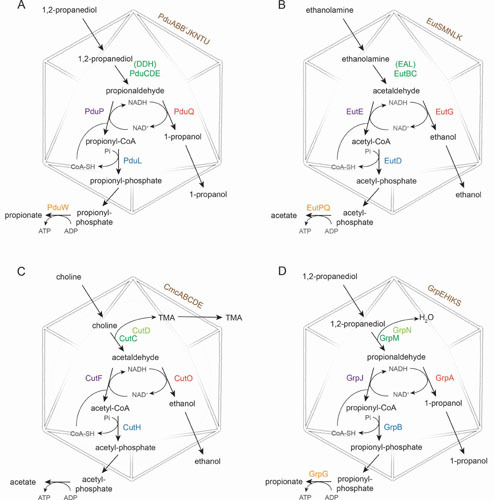
Schematic of MCP pathways discussed in the text. (A) Pdu MCP; (B) Eut MCP; (C) Cut MCP; (D) Grp MCP. Shell protein names are shown in brown, signature enzymes in green, accessory enzymes (where applicable) in light green, aldehyde dehydrogenase in purple, alcohol dehydrogenase in red, phosphotransacetylase in blue, and acetate kinase in gold.
Function of the MCP Shell
The defining feature of MCPs is the presence of a selectively permeable, polyhedral protein shell. The shell itself plays three important functional roles: (i) protecting the cell from toxic intermediates generated by MCP enzymes, (ii) sequestering volatile reaction intermediates, and (iii) concentrating substrates together with reaction machinery. Both genetic and bioinformatic analyses support the idea that metabolosome shells protect cells from toxic aldehydes during substrate degradation (31). Mutants unable to form the shell of the Pdu MCP accumulate propionaldehyde to toxic levels and incur DNA damage. Furthermore, polA (DNA repair polymerase) and gsh (glutathione biosynthesis) mutants are impaired for growth on 1,2-propanediol or ethanolamine, supporting aldehyde toxicity (32). In addition, under stringent conditions, S. enterica mutants unable to form the shell of the Eut MCP are incapable of growth due to acetaldehyde loss to the environment (33). Other experiments on ethanolamine degradation showed that overexpression of Eut catabolic enzymes in the absence of the Eut MCP supported wild-type growth rates, indicating that another function of the MCP may be to concentrate reaction-associated enzymes (34). Computational models of the Pdu MCP predict that the presence of the shell enhances pathway flux (35), similar to the role played by the carboxysome shell in increasing Rubisco activity (19). Although specific functions have not been extensively tested in glycyl radical microcompartments, a conserved function seems reasonable. Thus, studies support the model that bacterial MCPs enhance metabolic pathways by sequestering toxic or volatile intermediates and by concentrating enzymes together with their substrates.
Role of MCPs in Pathogenesis and Human Health
The metabolism of specific carbon substrates by MCPs confers a selective advantage to pathogenic bacteria in host environments and influences the ecology of the human gut in a manner that affects human health. Expression of the eut operon in Salmonella and E. coli is linked to virulence, including toxin production and increased transmission to new hosts (11, 36, 37), and is associated with urinary tract infections and Crohn’s disease (38, 39). Similarly, expression of the cobalamin (cob, B12) biosynthesis-propanediol utilization (cob-pdu) gene cluster in S. enterica is necessary for replication within macrophages (40) and required for colonization of the gut lumen of host organisms (41). Moreover, as a consequence of inflammation induced by S. enterica infection, chemical processes in the gut produce the unusual electron acceptor tetrathionate from thiosulfate, which supports anaerobic respiration of ethanolamine and 1,2-propanediol. This process provides a growth advantage to Salmonella, enabling it to outgrow other gut flora, thereby enhancing dissemination (42–44). Although less studied, the Grp MCP, which is found in some pathogenic E. coli, enables 1,2-propanediol metabolism, which has been associated with pathogenesis in Salmonella and Listeria (29, 45, 46).
Choline utilization MCPs are also closely associated with human health. The Cut MCP metabolizes choline, deficiencies of which have been linked to numerous diseases, including atherosclerosis and breast cancer (47, 48). The Cut MCP is a major source of trimethylamine (TMA) in the gut, which, upon exiting the MCP, travels through the bloodstream to the liver, where flavin-containing monooxygenase metabolizes it to trimethylamine-N-oxide (TMAO). TMAO is a marker for nonalcoholic fatty liver disease (49, 50), cardiovascular disease (12, 48, 50, 51), kidney disease (52, 53), and diabetes (54, 55), suggesting that MCP-associated metabolism in the gut plays an important role in physical health.
Distribution of the Pdu, Eut, Cut, and Grp MCPs
The Pdu MCP is thought to be particularly important for bacteria in anaerobic environments, such as soil, sediments, and the gut of higher organisms, where 1,2-propanediol is a major product of fucose and rhamnose fermentation (56). The pdu operon is unevenly distributed among many soil-dwelling and enteric bacteria, as well as Firmicutes, Actinobacteria, Deltaproteobacteria, Synergistetes, Fusobacteria, and Spirochaetes (4, 6). Among the Salmonella, growth on 1,2-propanediol in a B12-dependent fashion is nearly universal, whereas this ability is uncommon in E. coli (57). Studies suggest that Salmonella acquired this ability by horizontal gene transfer after its divergence from E. coli (57).
Ethanolamine, which is metabolized by the Eut MCP in an Ado-B12-dependent manner, is a component of cellular phospholipids and is present as a free compound within bodily fluids (58). The eut operon is found with high frequency throughout E. coli and Salmonella (39, 57) (Table 1). It is also located in a variety of other bacterial classes, including Firmicutes, Actinobacteria, Synergistetes, Fusobacteria, and Chloroflexi (4).
TABLE 1.
| Salmonella enterica | Escherichia coli | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NCBI taxon ID | Serovar | Strain | MCP Locus | NCBI Taxon ID | Serovar | Strain | MCP Locus | |||
| 41514 | Arizonae | 62:z4,z23:- | PDU | EUT | 216592 | 042 | EUT | |||
| 882884 | 62:z4,z23:-RSK2980 | PDU | EUT | 362663 | 536 | EUT | CUT | |||
| 454166 | Agona | SL483 | PDU | EUT | 655817 | ABU 83972 | EUT | |||
| 321314 | Choleraesuis | SC-B67 | PDU | EUT | 405955 | APEC O1 | EUT | GRP | ||
| 439851 | Dublin | CT_02021853 | PDU | EUT | 469008 | BL21(DE3) | EUT | |||
| 550537 | Enteritidis | P125109 | PDU | EUT | 199310 | CFT073 | EUT | GRP | ||
| 550538 | Gallinarum | 287/91 | PDU | EUT | 331111 | E24377A | PDU | EUT | ||
| 1081093 | Gallinarum/pullorum | PDU | EUT | 585397 | ED1a | EUT | GRP | |||
| 454169 | Heidelberg | SL476 | PDU | EUT | 585057 | IAI39 | EUT | CUT | ||
| 1267753 | Javiana | CFSAN001992 | PDU | EUT | 595495 | KO11FL | EUT | |||
| 423368 | Newport | SL254 | PDU | EUT | 591946 | LF82 | PDU | EUT | ||
| 295319 | Paratyphi A | ATCC9150 | PDU | EUT | 574521 | O127:H6 | E2348/69 | PDU | EUT | |
| 1016998 | Paratyphi B | SPB7 | PDU | EUT | 155864 | O157:H7 | EDL933 | EUT | ||
| 476213 | Paratyphi C | RKS4594 | PDU | EUT | 386585 | O157:H7 | Sakai | EUT | ||
| 439843 | Schwarzengrund | CVM19633 | PDU | EUT | 1072459 | O7:K1 | CE10 | EUT | CUT | |
| 220341 | Typhi | CT18 | PDU | EUT | 585035 | S88 | EUT | GRP | ||
| 1132507 | P-stx-12 | PDU | EUT | 409438 | SE11 | EUT | ||||
| 209261 | Ty2 | PDU | EUT | 439855 | SMS-3-5 | EUT | ||||
| 527001 | Ty21a | PDU | EUT | 511145 | K-12 | MG1655 | EUT | |||
| 588858 | Typhimurium | 14028S | PDU | EUT | 585056 | UMN026 | PDU | EUT | ||
| 568708 | D23580 | PDU | EUT | 696406 | UMNK88 | EUT | ||||
| 99278 | LT2 | PDU | EUT | 364106 | UTI89 | EUT | CUT | |||
| 216597 | SL1344 | PDU | EUT | |||||||
| 990282 | UK-1 | PDU | EUT | |||||||
| 936157 | Weltevreden | 2007-60-3289-1 | PDU | EUT | ||||||
See reference 4 methodology for details. Multiple strains of the same serovar are listed in the first instance.
Choline, which is common in the diet of animals, is a necessary nutrient for humans and a growth substrate for various gut bacteria. Because the Cut MCP employs an oxygen-sensitive glycyl-radical enzyme, these MCPs occur exclusively in anaerobic and facultatively anaerobic bacteria. The cut operon appears narrowly distributed within the Gammaproteobacteria, but surprisingly, it is not found in Bacteroidetes, the largest class of gut-associated bacteria (59). Cut operons have also been identified in Firmicutes, Actinobacteria, and Deltaproteobacteria (4). Evaluation of the E. coli reference collection (ECOR), 72 isolates representing the diversity of the species (60), identified five strains (∼7% of the isolates) that were able to degrade choline and contained cut genes (30). In a related survey of gut metagenomic DNA, cutC genes were identified in 6.1% of E. coli tested (59), suggesting a narrow but important distribution of Cut MCPs. However, cut genes have not been found in Salmonella.
The Grp MCP, like the Pdu MCP, processes 1,2-propanediol but is less widely distributed. Grp MCPs are found only in Gammaproteobacteria and Firmicutes (4). Like the Cut MCP, it relies on an oxygen-sensitive glycyl radical enzyme and is only found within anaerobes and facultative anaerobes (61). The ECOR collection was assessed, and grp genes were identified in 10% of the isolates (29), suggesting that the grp operon is slightly more widespread than cut. Thus far, grp operons have not been found in Salmonella.
Lastly, we note that among bacteria that contain MCP gene clusters, about 40% contain two or more (5). Most Salmonella contain both pdu and eut operons (Table 1). In contrast, most E. coli contain an eut operon, and a small percentage contain pdu, cut, or grp operons, but these latter three have not been found to co-occur. Whether certain E. coli strains utilize B12-dependent or -independent 1,2-propanediol degradation is probably dependent on the environments encountered.
MCP OPERONS IN E. COLI AND SALMONELLA: AN OVERVIEW
Below, we provide brief descriptions of the four major MCPs characterized in E. coli and Salmonella, including general characteristics, operon layout, and regulation. Further details of structural and genetic studies are presented in the following sections on individual MCP components.
Pdu MCPs
The Pdu MCP of S. enterica serovar Typhimurium LT2 was the first metabolosome discovered, and it is the best understood. Pdu MCPs are used for Ado-B12-dependent growth of S. enterica on 1,2-propanediol under aerobic or fermentative conditions (62, 63). In addition, the Pdu MCP supports anaerobic respiration of 1,2-propanediol with tetrathionate as a terminal electron acceptor but not with other acceptors, including nitrate, dimethyl sulfoxide (DMSO), TMAO, and fumarate (63). S. enterica synthesizes the Ado-B12 needed for 1,2-propanediol metabolism de novo under anaerobic conditions but requires added vitamin B12 (or another complex precursor of Ado-B12) for aerobic 1,2-propanediol breakdown (10). A few strains of E. coli are able to degrade 1,2-propanediol in a B12-dependent manner, but B12-independent degradation is employed with higher frequency (4).
Figure 2 shows the accepted model of 1,2-propanediol catabolism by S. enterica. First, 1,2-propanediol diffuses across the MCP shell and is converted to the toxic intermediate propionaldehyde by an Ado-B12-dependent diol dehydratase (DDH, PduCDE) (22, 64, 65). The PduQ alcohol dehydrogenase converts propionaldehyde to 1-propanol, which exits the MCP and then leaves the cell (66). Propionaldehyde is also converted to propionyl-phosphate by aldehyde dehydrogenase (PduP) and phosphotransacylase (PduL) (67, 68). Propionyl-phosphate exits the MCP and is converted to propionate by propionate kinase (PduW), generating 1 ATP (69). Under respiratory conditions, propionyl-phosphate is thought to be converted back to propionyl-CoA (in the cytoplasm) and feed into the methylcitrate pathway (70). Within the Pdu MCP, NAD+ and coenzyme A (CoA) are recycled by the PduQ propanol dehydrogenase and the PduL phosphotransacylase, respectively. Ado-B12 (which is subject to breakdown during catalysis) is recycled by the PduGH reactivase, PduS cobalamin reductase, and the PduO ATP-cob(I)alamin adenosyltransferase (ACAT) (71–73). As mentioned above, the main function of the Pdu MCP is to prevent propionaldehyde toxicity during growth on 1,2-propanediol (31, 32), which presumably is achieved by balancing the rate of propionaldehyde production and consumption by MCP enzymes.
The genes for 1,2-propanediol utilization are found in the pdu locus (22). The main pdu operon contains genes for the MCP shell (pduA, pduB, pduB′, pduJ, pduK, pduN, pduT, pduU), the 1,2-propanediol degradative pathway (pduCDE, pduL, pduP, pduQ, pduW), Ado-B12 recycling and assimilation (pduGH, pduO, pduS), and two genes of uncertain function (pduM, pduV), which may be involved in MCP assembly (pduM) and MCP segregation at cell division (pduV) (22, 64, 67, 74–78). At the end of the pdu operon is a gene (pduX) which encodes an L-threonine kinase used for the assimilation of complex precursors of Ado-B12 (79, 80). The pdu locus also includes a regulatory protein (pocR) and a 1,2-propanediol diffusion facilitator (pduF) that are divergently transcribed from the main operon.
In S. enterica, the genes of the pdu locus are coregulated with the adjacent B12 (cobalamin, cob) biosynthetic genes. Both genes sets are induced by PocR in response to 1,2-propanediol (81, 82). This makes sense, as Ado-B12 is the required cofactor for the DDH that catalyzes the first step of 1,2-propanediol metabolism (10). pdu genes are also controlled by the global regulators CRP/cAMP, ArcA/B, CsrA, and Fnr (83–86). CRP/cAMP are needed for induction of the pdu genes under both aerobic and anaerobic conditions (83). ArcA/B, which monitor oxygen availability, further stimulate induction under anaerobic conditions (83, 84). Transcriptomic studies suggest CsrA is also needed for induction of the pdu operon during aerobic growth, and proteomics analysis suggests that Fnr represses the pdu operon in the absence of oxygen (85, 86).
Eut MCPs
The eut operon was first identified in S. enterica and encodes a bacterial MCP that mediates Ado-B12-dependent ethanolamine catabolism (10, 23, 87, 88). Under aerobic conditions, ethanolamine acts as the sole source of carbon, energy, and nitrogen (87). Under fermentative conditions, ethanolamine provides the sole nitrogen source and stimulates growth by providing an additional source of ATP (63). In the absence of oxygen, ethanolamine can also be respired with tetrathionate as a terminal electron acceptor, but better-known terminal electron accepters such as nitrate, DMSO, TMAO, and fumarate have not been shown to support ethanolamine respiration (63). S. enterica synthesizes the Ado-B12 needed for ethanolamine metabolism de novo under anaerobic conditions but requires a complex precursor under aerobic conditions (10, 87, 89). E. coli is unable to synthesize Ado-B12 de novo and requires a source of complex precursors such as cobinamide or vitamin B12 to grow on ethanolamine (57).
The most widely accepted model for the metabolism of ethanolamine begins with its diffusion into the lumen of the Eut MCP (23, 90). There, ethanolamine is converted to acetaldehyde and ammonia by Ado-B12-dependent ethanolamine ammonia-lyase (EAL, EutBC) (91). Subsequently, acetaldehyde is metabolized by a pathway that is analogous to that employed for propionaldehyde degradation by the Pdu MCP. The EutE aldehyde dehydrogenase, the EutG phosphotransacetylase, and the EutD alcohol dehydrogenase convert acetaldehyde to ethanol and acetyl-phosphate (23, 33, 34, 88, 92). Ethanol exits the MCP and then the cell. Acetyl-phosphate leaves the MCP and is converted to acetate, which is excreted into the environment. Under fermentative conditions, this pathway produces 1 ATP, provides a source of nitrogen, and stimulates growth but does not produce a source of cell carbon (63). Under respiratory conditions, acetyl-CoA generated by ethanolamine catabolism supports growth via the TCA cycle and glyoxylate shunt. NAD+ and HS-CoA are thought to be recycled internally within the Eut MCP by the EutG alcohol dehydrogenase and EutD phosphotransacetylase similarly to the process described above for the Pdu MCP (93). Ado-B12 is also thought to be recycled within the Eut MCP by the EutA reactivase and the EutT ACAT, which are unrelated to the B12 recycling enzymes of the Pdu MCP (94–97). Studies indicate that the function of the Eut MCP is to sequester acetaldehyde production to minimize toxicity and prevent carbon loss, as described in more detail above (32–34).
The 17-gene eut operon has been characterized mainly in S. enterica (23, 33, 34, 98). Ado-B12-dependent EAL is encoded by the eutBC genes (99, 100). Aldehyde dehydrogenase, alcohol dehydrogenase, and phosphotransacetylase enzymes are specified by eutE, eutG, and eutD, respectively (33, 34, 88, 92, 93, 101). The enzymes used for Ado-B12 recycling are encoded by eutA and eutT (94–97, 102). The eut operon contains genes for five shell proteins, eutS, eutM, eutN, eutK, and eutL, and a putative chaperone, eutJ, whose role in MCP metabolism is unknown (23, 103). Genes for additional proteins, including two novel acetate kinases (eutQ, eutP), a transcriptional regulator (eutR), and an ethanolamine permease (eutH) round out the operon. The EutQ acetate kinase is required for anaerobic respiration of ethanolamine with tetrathionate, but a specialized role for EutP is not currently known (102). The EutH permease is most important at lower pH or low external ethanolamine concentrations, where it mediates transport of protonated ethanolamine (104). EutR regulates eut transcription in the presence of both ethanolamine and Ado-B12 (the required cofactor for ethanolamine ammonia lyase) (87, 105). EutR also induces its own expression by a positive feedback loop, and this is thought to help it compete with EAL for available Ado-B12 (106). In Salmonella, the eut operon is globally regulated by CRP/cAMP, ArcA/B, and CsrA (107, 108). In other organisms, regulation of the Eut MCP does not involve EutR and instead employs an unusual noncoding RNA riboswitch (11).
As described above, a number of studies of eut regulation were conducted in S. enterica. However, S. enterica contains both eut and pdu operons, raising the question of how their regulation is coordinated since essential shell components are highly similar between these two MCPs. In vivo studies of S. enterica showed that 1,2-propanediol induces the pdu operon but acts as a repressor of the eut operon, thereby preventing detrimental mixing of the two systems (109). Therefore, while it is technically possible for MCPs to incorporate nonnative components (109–111), leading to some interesting protein design experiments, local regulation may be designed to prevent this in native hosts. We also point out that 40% of bacteria that produce MCPs make two or more; hence, coordinating their expression is broadly important to cell physiology.
Cut MCPs
Craciun and Balskus identified a glycyl radical choline TMA lyase in Desulfovibrio desulfuricans and other gut bacteria that converts choline to acetaldehyde and TMA via a novel C-N cleavage mechanism (112). Studies also found that the gene for choline-TMA lyase (cutC) clusters with MCP shell protein genes, suggesting that this enzyme is MCP associated (59, 112). Microcompartment formation linked to choline fermentation was later demonstrated in Proteus mirabilis (28) and subsequently in E. coli 536 (30).
In E. coli 536, choline stimulates anaerobic growth in medium supplemented with a usable carbon source such as yeast extract or fumarate (30). Although fumarate can function as a terminal electron acceptor for anaerobic respiration by E. coli, it is inconclusive whether it does so with choline. E. coli is not known to use choline as a nitrogen source, suggesting that TMA is simply excreted. The Cut MCP does not support aerobic growth, presumably because of the extreme oxygen sensitivity of the CutC choline TMA lyase. In P. mirabilus, choline supplementation is associated with colony expansion and biofilm formation, but E. coli has not been tested for these phenotypes (28). The Cut MCP has not been identified in Salmonella.
The first step of the proposed choline degradation pathway is the conversion of choline to acetaldehyde and TMA within the lumen of the Cut MCP (112). This reaction is catalyzed by a glycyl radical choline-TMA lyase (CutC) which must first be activated by CutD (30, 112). It is thought that TMA is excreted and acetaldehyde is converted to ethanol and acetyl-phosphate as in the Eut MCP (28, 30, 59). Ethanol is proposed to diffuse out of the MCP into the cytoplasm and then out of the cell. Acetyl-phosphate is thought to exit the MCP into the cytoplasm, where it is converted to acetate with the generation of 1 ATP. Although this pathway provides ATP, it does not provide a source of cell carbon. Hence, choline stimulates growth of E. coli only in medium supplemented with a usable carbon source (30). Based on analogy with the Pdu and Eut MCPs, the Cut MCP is proposed to mitigate acetaldehyde toxicity and carbon loss.
The choline-utilization MCPs are subdivided into two classes (type 1 and type 2) which have substantial differences in distribution and composition (7, 59). Gene clusters for type 1 Cut MCPs are found in obligate anaerobes and contain 20 genes, while type 2 loci are found in facultative anaerobes and contain 16 genes within two operons. These two classes also have lyases that differ in primary structure. Type 2 CutC enzymes contain an extended ∼350-residue N terminus relative to type 1 lyases which has been suggested to contribute to enzyme activity and/or play a role in targeting CutC to the MCP lumen (7, 113). E. coli strains that metabolize choline contain type 2 Cut MCPs.
In uropathogenic E. coli 536, the cut locus is located within a pathogenicity island, PAI-II536 (30, 114). Here, as is typical for type 2 clusters, there are two operons: the cutWXY operon and a “main” cut operon separated by an ∼519-bp intergenic region (Fig. 1). Within the main cut operon, cutC codes for the glycyl radical choline TMA lyase, and cutD codes for its partner activating enzyme. It also contains genes for downstream pathway enzymes, including an aldehyde dehydrogenase (cutF), alcohol dehydrogenase (cutO), and phosphotransacetylase (cutH), which were inferred by homology to enzymes found in the Pdu and Eut MCPs and likely convert the acetaldehyde to ethanol and acetate (Fig. 2). Similar to the Eut and Pdu MCPs, it is thought that NAD+ and HS-CoA are recycled internally within the Cut MCP, but conclusive experimental support is lacking (30). At the end of the cut operon are the genes cutU and cutV, which are homologs of efflux pumps that may be involved in product export or substrate uptake, but this has not been reported experimentally. The cut locus encodes five shell proteins: cmcA, cmcB, cmcC, cmcD, and cmcE; however, these do not include a trimeric BMC shell protein related to those found in the majority of MCP operons and hypothesized to transport larger metabolites and enzymatic cofactors through MCP shells (30). This unexpected finding raises the question of how larger compounds traverse the MCP shell. The presence of the CutU and CutV efflux pumps offers a possible solution.
The Cut microcompartment contains a potentially unique local regulation system that utilizes three proposed regulatory genes (cutWXY) located in an operon adjacent to the main cut cluster (Fig. 1) (30). Deletions of individual genes showed that cutX, which contains sequence similarity to transcriptional regulators, and cutY, which does not, are necessary for induction of the cut operon by choline. On the other hand, deletion of the cutW gene, which encodes a protein with a DNA-binding domain, did not affect cut operon induction, and its function is currently unknown.
Grp MCPs
The Grp MCP processes the same substrate as the Pdu MCP: 1,2-propanediol. In contrast to the Pdu MCP, the Grp MCP employs a B12-independent glycyl radical diol dehydratase (GR-DDH, GrpM) in the first catabolic step (6, 29, 61). GR-DDH is extremely oxygen sensitive, and the metabolism of 1,2-propanediol by the Grp MCP only occurs under strict anaerobic conditions. In E. coli CFT073, the anaerobic metabolism of 1,2-propanediol by the Grp MCP stimulates growth if a source of cell carbon such as yeast extract or fumarate is also present, but 1,2-propanediol does not serve as a sole carbon and energy source (29). Similar to the cut operon, 1,2-propanediol was not respired when DMSO, TMAO, or nitrate were provided as terminal electron acceptors (29).
The proposed model for 1,2-propanediol metabolism by the Grp MCP is similar to the model for the Pdu MCP but with GR-DDH (which requires activation by GrpN) replacing B12-dependent DDH in the first catabolic step, the conversion of 1,2-propanediol to propionaldehyde (29, 115). From there, the downstream steps of the pathway that convert propionaldehyde to 1-propanol and propionic acid are thought to be directly analogous to the Pdu MCP. Cofactors are thought to be recycled within the Grp MCP as described for the Pdu MCP, but more supporting work is needed (29). While the function of the Grp MCP has not been studied, it is presumed to sequester propionaldehyde to mitigate its toxicity.
The grp operon has been genetically characterized in uropathogenic E. coli CFT073 (29). The grp locus contains 21 genes that encode six pathway enzymes (grpA, grpB, grpG, grpJ, grpM, grpN), five shell proteins (grpE, grpH, grpI, grpK, grpS), two regulators (grpP, grpQ), a permease (grpO), and several proteins of unknown function (grpC, grpD, grpF, grpL, grpR, mrtA, mrtB). Sequence homology to other MCP components suggests that grpC encodes a protein that resembles EutJ, a chaperone of unknown function, while grpD, grpF, grpL, and grpR encode proteins with homology to a putative flavoprotein, heme-binding protein, protease, and methionine adenosyltransferase, respectively. The grp cluster contains three large intergenic regions (IGRs) (284 to 549 bp) which is unusual for bacterial gene clusters. Two of these IGRs may be remnants of the horizontal gene transfer, because they flank two reverse transcriptase maturase genes (mrtAB) which are not involved in 1,2-propanediol metabolism. However, these IGRs might also be involved in gene regulation.
The Grp MCP has been classified as a type 3 glycyl radical microcompartment (GRM 3) (7). It is distinguished from other classes of GRM by the substrate processed, gene content of its encoding operon, and protein phylogeny. Additionally, in type 3 GRMs, GR-DDH contains a 50- to 60-residue region between its two structural domains which may be involved in enzyme localization to the MCP lumen.
Regulatory studies found that the grp operon in E. coli CFT073 is induced by 1,2-propanediol and is regulated by a two-component regulatory system, grpP (histidine kinase) and grpQ (response regulator), located near the end of the operon (29). Related studies in Rhodobacter capsulatus also identified a similar regulatory pair, pointing out partial sequence conservation with PocR, which regulates the pdu operon in response to 1,2-propanediol (115). The operon organization of R. capsulatus differs from that of E. coli CFT073 and other identified GRMs. In most GRMs, the gene for the activating enzyme is adjacent to the GRE, possibly indicating coregulation. However, these two components are located near opposite ends of the operon in R. capsulatus (115). Whether this difference affects enzyme activation and function is unknown.
MAIN COMPONENTS OF MCPs
Targeting Sequences
The question of how cargo is directed to the MCP interior during assembly is essential to a complete understanding of these organelles. An early clue regarding MCP assembly came from bioinformatic studies that identified terminal extensions of roughly 20 amino acids present on a subset of MCP proteins but absent from homologs that are not MCP associated (116). Subsequent genetic and biochemical work showed that a number of such extensions (referred to throughout the literature as “signal sequences,” “targeting sequences,” or “encapsulation peptides”) were necessary and sufficient for directing enzymes to the lumen of bacterial MCPs. Functional N-terminal targeting sequences were demonstrated for DDH (PduCDE) (117) and EAL (EutC) (118), as well as two MCP-associated aldehyde dehydrogenases (PduP and EutE) (119, 120). In addition, bioinformatic studies indicated that targeting sequences are widely used for the encapsulation of proteins into diverse MCPs and share a similar pattern of alternating polar and hydrophobic residues, although their sequences are poorly conserved (110, 121, 122). An NMR structure of the 18-residue N-terminal targeting sequence of PduP revealed that this region forms an amphipathic helix (123), and it was suggested that other targeting sequences adopt a similar fold. Due to their widespread and generic character, targeting sequences do not appear to be overly specific for particular MCPs. In fact, targeting sequences from varied MCPs as well as nonnative sequences are able to direct enzymes to the Pdu MCP simply through a common hydrophobic motif (110). However, many MCP-associated proteins lack recognizable targeting sequences, indicating that other mechanisms are also used to facilitate assembly and encapsulation (116, 121).
How targeting sequencing mediates encapsulation of MCP core enzymes and participates in shell assembly is not fully understood. Presumably, targeting sequences bind MCP shell proteins to mediate encapsulation during assembly. Supporting this idea, studies indicate that targeting sequences found on Eut and Pdu enzymes bind to the PduA, PduJ, PduK, and EutS shell proteins (103, 116, 123). Furthermore, in the case of the PduP aldehyde dehydrogenase, evidence indicates that its N-terminal targeting sequence binds to the C-terminal helix of PduA and PduJ. Recent work suggests that PduP is still trafficked to the MCP shell in the absence of PduA, indicating that multiple partner proteins may be involved in binding cargo to the protein shell (124). The interplay between shell proteins and shell contents remains an important yet underexplored area of MCP research.
Signature Enzymes
Enzymes that determine MCP substrates are sometimes referred to as signature enzymes. With regard to the MCPs found in E. coli and Salmonella, the signature enzymes are the PduCDE Ado-B12-dependent diol dehydratase (DDH) of the Pdu MCP, the EutBC Ado-B12-dependent ethanolamine ammonia lyase (EAL) of the Eut MCP, the CutC glycyl radical choline TMA lyase of the Cut MCP, and the GrpM glycyl radical diol dehydratase (GR-DDH) of the Grp MCP. In each of these cases, the signature enzyme catalyzes the first step of substrate breakdown.
B12-Dependent Enzymes
Ado-B12-dependent ethanolamine ammonia lyase (EAL) in the eut system and diol dehydratase (DDH) in the pdu system catalyze the first steps of ethanolamine and 1,2-propanediol degradation, respectively (23, 64, 88). While there are distinguishing features between these two enzymes, they both utilize the same basic mechanism (125). These enzymes use Ado-B12 in the “base-on” form, meaning that the dimethylbenzimidizole (DMB) ring is coordinated to the central cobalt atom (126). Binding coenzyme B12 by the apoenzyme allows for activation through homolytic cleavage of the Co-C bond creating cob(II)alamin and the 5′-deoxyadenosyl radical. Binding substrate shifts the equilibrium toward activation from 1% to 60% and 90% for DDH and EAL, respectively (127). The generalized reaction mechanism is as follows: the adenosyl radical abstracts a hydrogen from the substrate, and the substrate radical rearranges to form the product radical, which abstracts a hydrogen from 5′-deoxyadenosine, allowing cob(II)alamin and the adenosyl radical to recombine and regenerate the starting state of the enzyme.
B12-Dependent Enzyme Reactivases
Both EAL and DDH are subject to substrate-dependent and substrate-independent inactivation by the formation of catalytically inactive forms of cobalamin (125, 128). In the case of substrate-independent inactivation, O2 reacts with activated enzyme forming HO-B12, which remains tightly bound in the active site (129). In substrate and substrate analog inactivation, 5′-deoxyadenosine and a catalytically inactive cobalamin are formed and degraded to HO-B12 (130–132). The reactivation of enzyme and recycling of the HO-B12 to Ado-B12 is understood to be handled inside the MCP by MCP-associated proteins. The reactivase mechanism has been heavily studied in both Pdu and Eut and is the subject of detailed reviews (125, 133); here, we briefly highlight the key steps. Removal of HO-B12 is mediated by PduGH or EutA reactivases in the relevant MCPs, both of which require ATP and Mg2+ (96, 134). The PduGH structure in Klebsiella oxytoca was solved by X-ray crystallography, and homology analysis identified similarity to the ATPase domain of the E. coli HSP70 DnaK, suggesting that it may function as a chaperone that partly unfolds DDH, releasing HO-B12 (135). After release from the inactive enzyme, HO-B12 is converted back to Ado-B12. This begins with reduction of HO-B12 to cob(I)alamin by the PduS cobalamin reductase in the case of the Pdu MCP or by unknown means in the case of the Eut MCP (71, 136). Cob(I)alamin is adenosylated to form Ado-B12 by the PduO (72, 128) or by the EutT ACAT (94, 95) in a reaction where ATP is the adenosyl-group donor. Lastly, Ado-B12 reassociates with DDH or EAL apoenzymes to form active holoenzyme, completing the reactivation cycle. In S. enterica, deletion of PduS cobalamin reductase partly impairs 1,2-propanediol degradation (71). This may be due to functional redundancy with NAD(P)H:flavin oxidoreductase (Fre) or NADP+:ferredoxin(flavodoxin) reductase (Fpr) (137, 138), although genetic analyses have not yet fully worked this out. Deletion of the PduO or EutT ACAT partially impairs 1,2-propanediol or ethanolamine degradation, respectively (72, 95). Both enzymes are thought to be partially redundant with the CobA housekeeping ACAT since cobA eutT or cobA pduO double mutants are unable to grow on ethanolamine or 1,2-propanediol, respectively (72, 94, 95). Presumably, the PduO and EutT ACATs are more efficient at recycling Ado-B12 internally within their respective MCPs, but experimental evidence for this has not yet been reported.
Ethanolamine ammonia lyase (EAL, EutBC)
While the overall mechanism is the same for both EAL and DDH, the substrate specificity and structure of these two enzymes are not. EAL (PDB: 3ABO, crystal structure from E. coli K-12 [91]) is composed of two subunits (EutB and EutC) that come together as a dodecameric ring (α2β2)3. Using modeling and validated by biochemical methods, Bovell and Warncke put forward a model of assembly where EutB dimerizes and then trimerizes to form an EutB hexameric ring, allowing EutC to dimerize with EutB to form the final [(EutB-EutC)2]3 complex (139). The Ado-B12 is sandwiched between the EutB and EutC subunits, oriented with the DMB liganded face toward EutC (91). Genetic knockout studies have shown that mutants lacking eutBC are unable to grow on ethanolamine (33, 88). Studies indicate that the EutC subunit of EAL has a functional targeting sequence that presumably directs its encapsulation into the Eut MCP (118).
Diol dehydratase (DDH, PduCDE)
In Salmonella, three genes encode the diol dehydratase encapsulated in the Pdu MCP (PduCDE) (64). Unlike EAL, which makes a trimer of dimers, PduCDE from K. oxytoca (96% amino acid similarity to Salmonella PduCDE) forms a dimer of trimers (αβγ)2, where PduC makes up the full dimer interface, and PduD and PduE are on opposite sides of PduC from the dimer interface (140). Both PduD and PduE have N-terminal extensions compared to homologs that are not MCP associated; however, only the N-terminal extension of PduD has been shown to function as a targeting sequence (117). A number of in vivo and in vitro studies established that PduCDE localizes to the lumen of the Pdu MCP (22, 78, 117, 141), and knockouts of PduCDE abolish 1,2-propanediol metabolism by Salmonella (73).
Glycyl radical enzymes (GREs)
A widespread, but less well-studied, class of MCPs (which includes the Cut and Grp MCPs of E. coli) uses glycyl radical enzymes to initiate substrate metabolism. These enzymes employ a glycyl radical instead of a 5′-deoxyadenosyl radical generated from Ado-B12 for catalysis (142, 143). GREs, unlike B12-dependent enzymes, need to be initially activated by a radical S-adenosylmethionine (SAM) enzyme (142). Each GRE has a specific partner activating enzyme (AE) (144). The AE generates a 5′-deoxyadenosyl radical (from SAM) which abstracts a hydrogen from the active site glycine in the GRE. In the presence of substrate, the glycyl radical abstracts a hydrogen from a nearby cysteine, generating a thiyl radical. The thiyl radical abstracts a hydrogen from the substrate to form the substrate radical, allowing for radical rearrangement to produce the product radical. At this point, the thiyl radical is regenerated, producing the final product, and lastly, the glycyl radical is regenerated (143). While the AE is necessary, it is only needed for initial activation, after which the catalytic cycle regenerates the glycyl radical. The glycyl radical is extremely oxygen sensitive and can react with molecular oxygen, cleaving the peptide backbone at the site of the radical. In E. coli, GRE “spare parts” proteins have been identified, which replace the portion of enzyme missing after O2 inactivation (145), but these are not encoded within the MCP operons.
CutC (a type 2 lyase) is the only microcompartment GRE characterized in detail (7, 112, 113, 146). Crystal structures of CutC from Klebsiella pneumoniae were determined in both the presence and absence of choline substrate, revealing an unexpected topological change upon choline binding (113, 147), in contrast to other non-MCP-associated GREs. As expected, deletion of either the choline TMA lyase gene (cutC) or the gene for its activating enzyme (cutD) eliminates choline degradation by the Cut MCP (30). Similarly, deletion of GR-DDH (grpM) or its activating enzyme (grpN) prevent 1,2-propanediol metabolism by the Grp MCP (29).
MCP Pathway Enzymes
In the Pdu, Eut, Cut, and Grp MCPs discussed here, the signature enzyme generates an aldehyde (acetaldehyde or propionaldehyde) product which is metabolized to the corresponding alcohol and organic acid by a conserved pathway using the enzymes described below (Fig. 2).
Aldehyde dehydrogenase
Aldehyde dehydrogenases (Ald) are used for substrate metabolism by the Pdu, Eut, Cut, and Grp MCPs. These enzymes convert an aldehyde, HS-CoA, and NAD+ to the corresponding acyl-CoA and NADH internally within MCPs. Aldehyde dehydrogenases are essential to functional MCPs; when the pduP, eutE, cutF, or grpJ aldehyde dehydrogenase genes are individually deleted, growth on the respective substrates is eliminated (29, 30, 67, 88). In vitro assays of purified enzyme verify that PduP and EutE have CoA-acylating aldehyde dehydrogenase activity (92). The PduP and EutE aldehyde dehydrogenases were shown to have N-terminal targeting sequences that mediate their encapsulation into the Pdu and Eut MCPs (93, 116). In addition, PduP was shown to copurify with Pdu MCPs (67, 119), and a series of genetics studies localized EutE to the interior of the Eut MCP (93). CutF and GrpJ also have aldehyde dehydrogenase activities as shown by in vivo assays, and a structure of GrpJ from Rhodopseudomonas palustris has been solved (30, 61). Like their EutE and PduP counterparts, the CutF and GrpJ aldehyde dehydrogenases have N-terminal extensions predicted to mediate MCP encapsulation, but experimental support for this function has not been reported.
Phosphotransacetylase
Nearly all metabolosome operons encode phosphotransacylase (PATC) enzymes or homologs thereof. Within the metabolosome, PATC enzymes accept the acyl-CoA products of aldehyde dehydrogenases and convert them to acyl-phosphates and HS-CoA (68, 75). This not only produces a high-energy phosphate compound, but it regenerates HS-CoA for the aldehyde dehydrogenases. Genetic deletion of the PATC genes found in the pdu, eut, cut, and grp operons results in partial growth impairment under standard growth conditions and, in the case of eut, no growth under stringent conditions (29, 30, 75, 93). The explanation for these phenotypes is that MCP-associated PATC enzymes have activities that are redundant with housekeeping enzymes but are still needed for regenerating HS-CoA internally within the MCP. Fairly extensive genetic studies of both the PduL and EutD PATC enzymes support a role in internal cofactor recycling, the key finding being that breaking the MCP shell corrects the phenotypes of PATC deletions (75, 93). Both PduL and EutD have been recombinantly expressed and purified, and biochemical assays verify that they possess PATC activity (75, 148). The PduL class of PATC, which is the most common version encoded by MCP operons, is unrelated in sequence to the housekeeping enzymes (Pta), which are widespread in biology. The crystal structure of PduL from R. palustris has been solved, and it forms a dimer in the crystals, as well as by size exclusion chromatography (SEC). However, Phe116, which is important for dimerization, is poorly conserved and is not present in S. enterica PduL. This brings into question the oligomeric state of PduL generally (149). The EutD enzyme of S. enterica is related in sequence to the C-terminal PTA_PTB domain of housekeeping Pta enzymes (101) and is less common in MCP operons compared to PduL-type PATC enzymes. The PTAC enzymes in the cut and grp operons (CutH, GrpB) which are PduL-type have not been assessed in any detail. However, deletion of their encoding genes results in partial impairment of growth consistent with a role in internal cofactor recycling (29, 30).
Acetate/propionate kinase
Acetate/propionate kinases are often encoded by MCP operons but are thought to localize to the cytoplasm of the cell rather than the MCP interior. These enzymes catalyze the conversion of an acyl-phosphate, ADP, and Pi to the corresponding organic acid and ATP. The eut operon encodes two acetate kinases (EutP and EutQ). Both show activity in vitro. EutQ is necessary for growth of S. enterica on ethanolamine under anoxic conditions, while EutP deletions have no known phenotype (97). The analogous Pdu protein PduW shows propionate kinase activity and catalyzes the conversion of propionyl-phosphate to propionate with the production of ATP from ADP and Pi (69). However, PduW is not necessary for growth on 1,2-propanediol under any tested condition. This suggests that PduW is functionally redundant with the housekeeping acetate kinase (Ack) (69). Presumably, PduW contributes to the degradation of 1,2-propanediol under as yet unidentified conditions. The type 2 cut operon of E. coli 536 is not known to contain an acetate kinase and likely relies on the housekeeping Ack enzyme for this function (7). For the Grp MCP, grpG is believed to be a propionate kinase by homology, and knocking out grpG partially impairs growth on 1,2-propanediol (29). This suggests that GrpG might have an MCP-specific role, but the details are unclear.
Alcohol dehydrogenase
Most MCP operons encode alcohol dehydrogenases (or homologs) that convert aldehydes and NADH to alcohols and NAD+. The primary function of these enzymes in MCP metabolism is to regenerate NAD+ from NADH (formed by the aldehyde dehydrogenase reaction) internally within the MCP (66, 75, 93). Deletion of MCP-associated alcohol dehydrogenases typically only partially impairs growth due to functional redundancy with housekeeping enzymes under fermentative conditions or the electron transport chain under respiratory conditions. Internal NAD+ recycling by MCP alcohol dehydrogenases is well supported by genetic studies on both the Eut (93) and Pdu (66) MCPs. The alcohol dehydrogenases encoded by the cut and grp operons have not been as extensively studied. However, genetic deletion of the cutO or grpA alcohol dehydrogenase partially impaired substrate metabolism, suggesting a role in NAD+ recycling as well as partial redundancy with housekeeping Adh enzymes (29, 30). Above, we mentioned genetic analyses that indicated functional redundancy between MCP-associated alcohol dehydrogenases and housekeeping Adh enzymes or the electron transport chain. However, regenerating NAD+ outside the MCP for use within the MCP requires that NADH and NAD+ move across the MCP shell by a means that has not been defined but which may involve gated channels in trimeric shell proteins as described further below.
Microcompartment Shell Proteins
The outer boundary of bacterial MCPs is a polyhedral protein shell 100 to 400 nm in diameter, which is similar in morphology across the MCPs considered here (29, 30, 103, 141) (Fig. 3). MCP shells form a selectively permeable barrier which allows the passage of substrates and products while sequestering toxic and/or volatile pathway intermediates. Remarkably, the shells of diverse MCPs are built primarily from a single family of small, functionally diversified proteins known as bacterial microcompartment (BMC) domain proteins (Pfam00936) (150). There are two main groups of BMC domain proteins: hexameric (BMC-H) and trimeric (BMC-T). As the name suggests, BMC-H proteins assemble into hexamers in solution, with each monomer contributing a BMC domain. Each BMC-T monomer, by contrast, comprises two fused BMC domains and three monomers assemble into pseudohexameric trimers. Both BMC-H and BMC-T shell proteins share a similar flat hexagonal quaternary structure. MCP shells are typically built from two to ten different types of BMC domain proteins (BMC-H and BMC-T combined) that tile edge to edge into mixed sheets, forming the facets of the shell. Variations in BMC domain proteins are thought to allow the metabolism of diverse substrates in a range of host organisms (151–153). The diversification of BMC domain proteins includes circular permutations, [Fe-S] clusters, and fusions to a number of different domains of unknown function (154–157). In addition to BMC domain proteins, MCPs also contain conserved pentameric proteins that cap the vertices and impart shell curvature. These are known as bacterial MCP vertex proteins (BMV) or BMC-P proteins (Pfam0916) (151–153, 158).
Figure 3.
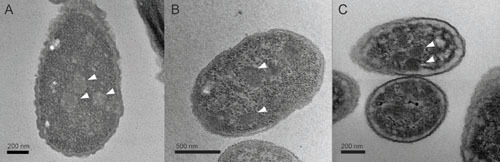
Electron micrographs of bacteria expressing MCPs, indicated by arrows. (A) Cut MCPs from E. coli 536; (B) Grp MCPs from E. Coli CFT073; (C) Pdu MCPs from S. enterica. Scale bars are located on each image. Images were obtained as described (76).
Numerous BMC shell protein structures have been solved by crystallography (2, 9, 151, 153, 155, 159–164). Together with genetic and biochemical studies, these findings have substantially advanced our understanding of shell assembly and function (Fig. 4). In addition, several structures of empty recombinant shells have been reported recently, providing further insights into MCP architecture (147, 165–167). Below, we provide more detailed information on MCP shell proteins focusing on those from the Pdu, Eut, Cut, and Grp MCPs of E. coli and Salmonella.
Figure 4.
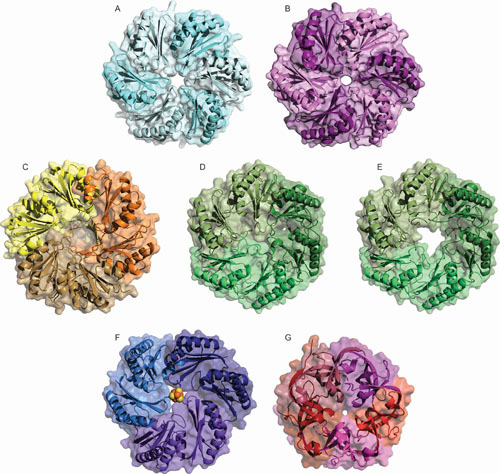
Representative crystal structures of select MCP shell proteins. (A) PduA BMC-H protein from S. enterica typhimurium (PDB ID: 3NGK) (155); (B) EutM, the orthologous BMC-H protein from E. coli K-12 (3I6P) (159); (C) PduU permuted BMC-H protein from S. enterica serovar Typhimurium (3CGI) (154); (D) EutL BMC-T closed form from E. coli K12 (3GFH) (160); (E) EutL BMC-T open form from E. coli K12 (3I87) (159); (F) PduT [Fe-S]-containing BMC-T protein from C. freundii (3PAC) [156]; (G) GrpN BMV pentamer from R. rubrum (4I7A) (158).
Hexameric shell proteins (BMC-H)
Hexameric BMC shell proteins are the most prevalent class, and the pdu, eut, cut, and grp loci of Salmonella and E. coli all encode multiple BMC-H proteins. The structure of BMC-H proteins is highly conserved among diverse MCPs (Fig. 4A and B). In crystals, BMC-H proteins are usually flat, hexagonally shaped cyclic hexamers (159, 168–170) which interact edge to edge to form sheets. Two positively charged residues at the hexamer edge (K26 and R79 in the canonical BMC-H protein PduA) are crucial for hexamer-hexamer tiling and are widely conserved across BMC-H (and BMC-T) proteins, suggesting a common manner of assembly (152, 171–173).
The primary opening in the protein sheets formed by BMC-H proteins is a 5- to 10-Å diameter pore at the center of each hexamer. Studies indicate that these pores allow the selective diffusion of small metabolites into and out of MCPs. In vivo, in vitro, and in silico studies showed that the PduA hexamer preferentially allowed the diffusion of 1,2-propanediol (compared to propionaldehyde) across the shell of the Pdu MCP (163, 174). Although the topology of BMC-H hexamers is highly conserved, the residues lining the pores may vary in their chemical and physical properties, suggesting that they mediate selective transport of different compounds, presumably MCP substrates and products. This might explain in part why MCP operons often encode multiple BMC-H proteins. For example, altering the chemical properties of the PduA pore residues by site-directed mutagenesis was sufficient to change the permeability of the Pdu MCP (111, 163). Additionally, both in vivo and in vitro studies have highlighted the importance of relative gene position in shell function and formation, which could also account for multiple BMC-H genes within a single MCP operon (78, 162).
BMC-H proteins are understood to make up a large portion of the microcompartment shell and, as a result, play a pivotal role in MCP formation (124, 141, 165). For the Pdu MCP of S. enterica, the PduA and PduJ shell hexamers are estimated to comprise ∼40 to 60% of the protein of the shell (124, 141), and deletion of the pduA or pduJ genes individually results in the formation of aberrant microcompartments and propionaldehyde toxicity (76). Similarly, a knockout of the EutM BMC-H component of the Eut MCP causes the buildup of acetaldehyde and cellular toxicity, most likely as a result of a defective shell (33). The effects of BMC-H gene knockouts on the Cut MCP are currently uncertain, as relatively limited work has been reported, and characteristic phenotypes were not observed (30). Knockouts of BMC-H protein GrpH (as well as other shell proteins of the Grp MCP) revealed the surprising phenotype of inhibition of growth stimulation by substrate, suggesting that the Grp MCP shell is essential for enzymatic activity in vivo, in contrast to other MCPs studied (29).
The pdu, eut, cut, and grp operons also contain variations of canonical BMC-H proteins, including circular permutations (found in EutS, PduU, and GrpK) where the quaternary structures of permuted hexamers align well with canonical hexamers but their secondary structural elements are in a different order. A unique feature of some permuted hexamers is an N-terminal extension from each subunit which forms a six-stranded beta barrel “capping” the pore, as first observed in the crystal structure of PduU from S. enterica (Fig. 4C) (155). If and how occlusion of the central pore is controlled by pathway flux or interactions with specific metabolites remains unknown.
Another variation on the BMC-H theme is canonical hexamers with C-terminal extensions. At least one copy of this protein type is found in the four operons discussed here (PduK, CmcE, EutK, GrpS). The exact role of these extensions is unknown. In the case of PduK, its C-terminus includes an [Fe-S] cluster binding motif, suggesting a role in electron transfer (7, 155, 157). Biochemical and structural analyses of EutK show that it does not purify as a hexamer and contains a putative DNA binding domain whose role in shell structure and/or MCP assembly is not known (159). With regard to CmcE and GrpS, so far, there have been no reports on the function of their C-terminal domains. In addition to examples mentioned here, extended BMC-H proteins are widespread among diverse MCPs and warrant further study.
Trimeric shell proteins (BMC-T)
BMC-T proteins have a similar topology to BMC-H proteins and are thought to have arisen by gene duplication and divergence of BMC-H proteins. Each BMC-T monomer contains two circularly permuted or two canonical Pfam00936 domains and assembles into trimeric pseudohexamers that have six-fold symmetry (150). Like BMC-H oligomers, BMC-T trimers contain a central pore region; however, this region may be occluded by an [Fe-S] cluster or may be dynamic, opening and closing in response to environmental conditions. The pdu, eut, and grp loci of Salmonella and E. coli all encode BMC-T proteins, but the cut locus is unusual in that it does not.
PduB and PduB′ are the major BMC-T proteins of the Pdu MCP of S. enterica, together comprising about 25 to 50% of the total shell protein content (124, 141). PduB and PduB′ are permuted trimers transcribed from overlapping genes and are identical in amino acid sequence except that PduB′ is shorter by 37 amino acids on the N-terminus. These 37 residues play a key role in assembly of the Pdu MCP. When removed, large MCPs lacking lumen enzymes result, indicating that the N-terminus of PduB is needed for the encapsulation of enzymatic cargo (175). A crystal structure of PduB from Lactobacillus reuteri (62% sequence similarity to S. enterica) displays the major structural hallmarks of BMC-T oligomers and is discussed in more detail below (161). S. enterica strains with pduBB′ deletions fail to make MCPs, instead producing polar bodies which are aggregates of other MCP proteins (76). Such mutations also result in propionaldehyde accumulation, cellular toxicity, and DNA damage during growth of S. enterica on 1,2-propanediol.
The Eut operons of E. coli and Salmonella encode a single permuted trimeric BMC domain protein, EutL. Interestingly, EutL crystallized in both “open” and “closed” conformations where a 10-Å central pore present in the open form is absent in the closed form (Fig. 4D and E) (153). This suggests that EutL may allow selective transport of metabolites to and from the MCP interior through a gating mechanism (159, 160). In addition to the gated central pore, the crystal structures of EutL revealed allosteric binding pockets between the tandem repeat domains, which could serve to “unlock” the central gate under specific conditions (161, 164). In a crystal structure of the closed form of EutL from Clostridium perfringens (73% sequence similarity to S. enterica), ethanolamine was shown to bind in these pockets, presumably as a means of negative allosteric regulation (164). Glycerol was shown to bind in a similar location in the closed crystal structure of PduB from L. reuteri, likely fulfilling the same function in the Pdu system (161). BMC-T proteins from β-carboxysomes have also been crystallized in open and closed forms. In addition, these trimeric rings have been shown to stack face-to-face, forming a central cavity that might function by an airlock mechanism in metabolite transport (176–178). A working model suggests that larger metabolites such as ATP or cobalamin could traverse the shell using this method, while aldehyde intermediates remain trapped within (164). However, active gating has not yet been demonstrated in vivo or in vitro.
Another type of BMC-T protein, exemplified by PduT from the Pdu MCP, is composed of canonical BMC domains but surprisingly contains a [4Fe-4S] cluster blocking the central pore (155, 156) (Fig. 4F). This arrangement, coordinated by loops containing conserved Cys residues contributed by each subunit, may function in facilitating redox chemistry such as regenerating encapsulated cofactors via electron transfer across the microcompartment shell (155). In a recent study, site-directed mutagenesis of the conserved Cys residues to remove the [Fe-S] clusters (leaving behind pores) resulted in greater diffusion of substrate, intermediate, and NAD+/NADH cofactors, when small charged or uncharged residues were substituted (179). These findings indicate that alteration of the pore chemistry is a reasonable method for fine-tuning transport through BMC-T oligomers.
The grp locus of E. coli CFT073 encodes a single BMC-T protein, GrpI. It is in the permuted class, suggesting a role in dynamic solute transport. Structural characterization of GrpI has not been reported; however, a genetic knockout of grpI in E. coli CFT073 eliminated growth on 1,2-propanediol (29). This phenotype was unexpected since shell gene knockouts in other MCPs result in aldehyde toxicity and DNA damage but typically do not prevent growth on MCP substrates.
As mentioned above, the cut operon of E. coli 536 lacks a BMC-T gene, but this organism is still able to metabolize choline, extrude product, and form MCPs which are similar in size to Pdu MCPs (30). Other proteins may compensate for the role of BMC-T proteins in this system. Genetic knockouts of cutU and cutV, putative efflux pumps which might fulfill the function of BMC-T proteins in the Cut MCP, abolish choline metabolism, suggesting a different method of large metabolite transport in this class of B12-independent MCPs. Alternatively, it is possible that other yet uncharacterized elements of the Cut MCP system may be involved in large metabolite transport.
Bacterial MCP vertex (BMV) proteins
Most (or all) MCPs are thought to use proteins with conserved Pfam03319 domains as vertices (151, 158). BMV proteins generally occur in low abundance in MCPs, since fewer copies relative to other proteins are needed for shell assembly. Because of this, BMV proteins were not initially identified in preparations of purified MCPs (141, 180). Subsequently, the PduN vertex protein was found to be a component of both purified Pdu MCPs and recombinant empty shells by Western blotting (76) and green fluorescent protein (GFP) tagging (78). More recently, the role of various BMV proteins as vertices was demonstrated when the structures of several empty MCP shells were assigned (147, 165, 167).
Unlike other shell proteins, BMV proteins are pentameric in arrangement. The first BMV proteins to be identified, CcmL and CsoS4A from two types of carboxysomes, were found to be pentamers whose size and shape were compatible with vertex formation (151). Later, GrpN from Rhodospirillum rubrum was also shown to form pentamers (158) (Fig. 4G). Surprisingly, the BMV protein from the eut operon (EutN) crystallized as a hexamer with pseudo-pentameric symmetry (151). However, additional biochemical studies showed that EutN forms a pentamer in solution, in agreement with other BMV structures (158). In some instances outside of E. coli and Salmonella, multiple copies of BMV genes may exist in a single MCP operon (181). Although the rationale for this diversity has not been determined, it suggests additional roles for BMV proteins beyond capping MCP vertices.
Genetics studies have shown that BMV proteins are essential for proper formation of the Pdu, Eut, Cut, and Grp MCPs. In S. enterica, deletion of pduN resulted in Pdu MCPs with unusual shapes by EM as well as propionaldehyde toxicity during growth on 1,2-propanediol (76). Similarly, increased acetaldehyde accumulation was observed when eutN was genetically deleted from the S. enterica eut operon, again indicating a role in proper MCP formation (33). Recent work on the cut operon in K. pneumoniae revealed that recombinant empty MCP shells were not formed without the cmcD pentamer (147). And finally, in the grp operon of E. coli CFT073, deletion of the grpE gene prevented growth on 1,2-propanediol, indicating a role in MCP function (29).
Specifically if and how vertex proteins are able to impart curved structure to MCPs has been explored computationally but not experimentally (182, 183). It is possible that vertex capping occurs as a later step in metabolosome formation, similarly to β-carboxysome assembly (184, 185). In keeping with this hypothesis, vertex proteins have been utilized in a novel strategy for the purification of synthetic MCPs with a complete shell. By attaching a strep affinity tag to recombinantly produced vertex proteins, MCPs expressed in the absence of BMV proteins may be “sealed” and purified in a single step on an affinity column (186). Whether this technique can be employed across diverse metabolosomes has not been investigated.
Shell-associated proteins
Some shell-associated proteins are also predicted to play a role in MCP architecture and assembly. Deletion of the pduM gene, which lacks a BMC domain, leads to the formation of highly aberrant MCPs and cells that undergo propionaldehyde toxicity during growth on 1,2-propanediol (74). Similarly, the EutQ acetate kinase (also lacking a BMC domain) is necessary for shell integrity and acetaldehyde degradation under anoxic conditions but also is able to bind to the N-terminus of the BMC-H protein EutM and increases the cellular copy number of Eut shells by an unknown mechanism (97, 103). How and when these various interactions occur during MCP assembly is unknown.
Although not well studied, some MCP-associated proteins are predicted to interact with the external face of the shell, providing clues regarding native MCP intercellular localization. PduV, a protein of unknown function with sequence similarity to RAS-type GTPases, was shown by GFP tagging to localize to the external face of empty Pdu MCPs (78). Additionally, based on its dynamics over time, PduV was speculated to be involved with anchoring MCPs to the cytoskeleton in growing bacterial cells. Further evidence from computational docking studies and bacterial two-hybrid experiments predicted an interaction between the N terminus of PduV and the C-terminal extended β-barrel of PduU, placing PduV on the external face of the MCP (187); these findings were supported by recent GFP localization studies (124). Whether other MCP operons also include external shell-associated proteins remains to be determined, although a similar role has been suggested for shell protein PduK (76), and cytoskeletal movement of MCPs has been detected but not localized in Eut (118). Details of the global spatial organization of MCP-containing cells beyond cytoskeletal attachment has remained elusive due to challenges with EM visualization (188) and the findings that knockouts of individual MCP genes may produce profound effects on MCP abundance, structure, and assembly (76, 103, 175). Understanding these complex interactions could provide additional details on MCP function and their specific role in cellular metabolism.
Cryo-EM and crystal structures of empty MCP shells
Recently, recombinant empty MCP shells amenable to crystallography and cryo-EM have been produced in E. coli and characterized structurally (147, 165, 167). The recombinant shells that have been analyzed thus far were composed of a subset of shell proteins and lacked core enzymes, resulting in structures that were much smaller but more homogeneous than wild-type MCPs. It seems reasonable that these structures assembled in a physiologically relevant manner, but studies with native MCPs will be needed to rigorously establish the overall structure with all components present.
A subset of MCP shell proteins was utilized to obtain the first high-resolution structure of an intact empty metabolosome shell from Haliangium ochraceum (165, 166). Mixed sheets of BMC-H and BMC-T proteins formed the facets of the shell, which was icosahedral in shape. Interestingly, the permuted BMC-T proteins were arranged into a two-BMC-T stack, presumed to act as an “airlock,” potentially allowing tight control of metabolite flux (165, 166). Additionally, BMV proteins formed the vertices, as anticipated (151, 159). The global arrangement of proteins forming this shell resembles the cryo-EM structure of an empty β-carboxysome (167), suggesting structural similarities in the overall construction of carboxysomes and metabolosomes. Recently, an empty Cut MCP shell was characterized in K. pneumoniae using a subset of shell proteins: CmcABC′D (CmcC′ contained an extended C-terminus) (147). Like other empty shells, the structure was icosahedral and highly regular, although the authors note that including all five native shell proteins from the cut operon resulted in larger and less homogeneous shell preparations. To date, a structure of a native MCP (containing cargo) has not been determined; how the structural organization differs from the empty shells would shed light on a number of important questions.
APPLICATIONS OF BACTERIAL MICROCOMPARTMENTS
As more information has been revealed about the structure and function of MCPs, attempts to use this knowledge for biotechnology applications have also advanced. Diverse MCPs, including carboxysomes and metabolosomes, have been heterologously expressed in E. coli and other organisms, allowing the transfer of MCP functions to new hosts. In addition, these applications have allowed a better understanding of component parts of MCPs and revealed key rules in particle assembly and stability (186, 189–192).
Along these lines, a number of groups are working on engineering carboxysomes into C3 plants, with the goal of improving CO2 fixation and reducing photorespiration (2, 191–193). Additionally, a great deal of effort has also gone into repurposing metabolosomes as enzyme scaffolds, nanobioreactors, containers for toxic proteins, and hybrid enzyme-inorganic catalysts (14–17, 194). Here, we briefly outline the main research directions in MCP bioengineering, including the design and production of empty protein shells from MCP components, the use of targeting sequences to drive novel cargo encapsulation, efforts to control shell permeability, and repurposing MCPs toward novel functions. For more information, we refer readers to a number of excellent reviews on this subject (14, 16, 195–198).
“Empty” MCPs and Production in Industrial Hosts
Parsons and colleagues were the first to demonstrate the assembly of a cargo-free MCP shell by developing a fusion plasmid containing all essential shell genes in the pdu operon from C. freundii and expressing these recombinantly in E. coli (78). Later, empty protein shells from carboxysomes (167), a Hochraceum MCP of unknown function (165), the Eut MCP (103, 118), and the Cut MCP (147) were also produced in E. coli. Protein shells have also been assembled in vitro from single shell proteins at an oil-water interface (199) and from multiple shell proteins modified with cleavable SUMO tags to block premature assembly of individual shell proteins into protein sheets (186).
Another approach to the production of protein shells has been the redesign of native MCP shell proteins. A circularly permuted variant of PduA was found to form a 60-subunit nanocage (200). Interestingly, this novel compartment was sensitive to changes in pH and salt concentration, environmental factors which could provide biological tuning for further applications. Also along these lines, empty shells were formed by a synthetic tandem duplication of a BMC-H protein (201) and a BMC-T shell protein with inverted sidedness (202), resulting in relatively homogeneous protein shells.
Novel Enzyme Design and Targeting
Concomitant with the development of empty protein shells derived from MCP proteins, methods for targeting novel cargo to those shells were developed. A number of groups were able to target fluorescent proteins to empty MCP shells using native targeting sequences (such as the 18 N-terminal residues of PduP) or synthetically designed targeting sequences with specific motifs (78, 117, 118, 120, 122, 203–206). In addition, native targeting sequences were used to direct multiple heterologous proteins to the Pdu shell, thereby constructing a proof-of-concept bioreactor for the production of ethanol (123). Another attractive direction for the design of bespoke MCP shells is the sequestration of toxic products. Yung and colleagues were able to increase expression of lysis protein E from bacteriophage Φx174 by fusing it to an MCP targeting sequence and recombinantly coexpressing it with Pdu shell proteins (207). In another example, an engineered MCP for the removal of phosphate contaminants from wastewater was developed by targeting polyphosphate kinase (PPK1) to an empty Pdu MCP through fusion with a PduP targeting sequence (203). This allowed an 8-fold increase in cellular phosphate accumulation, apparently by blocking competing reactions that break down polyphosphate (199). More recently, approaches to improve the targeting efficiency of cargo directed to MCP-based protein shells were pioneered by fusing shell and cargo proteins to nonnative binding partners, including scaffolding domains (SH3, PDZ, and GBD), a split coiled-coil domain, or the spy-tag spy-catcher system (186, 204, 208). Both proteins and small molecules have also been encapsulated within minimal shells derived from MCPs by in vitro loading during assembly on an oil-water interface (199). Lastly, we mention that empty MCP shells and protein targeting systems have been engineered into industrially important organisms, including the thermophilic bacilli and corynebacteria (204, 206). Thus, substantial progress has been made on targeting desired cargo to protein shells built from MCP proteins.
Current models for MCPs propose that optimal function requires a selectively permeable protein shell. Accordingly, work aimed at controlling shell permeability has begun. So far, this work has focused on modifying the pores present in shell proteins by site-directed mutagenesis and by constructing hybrid shells using BMC proteins derived from different MCPs (111, 163, 179, 181). As mentioned above, there are many aspects of shell permeability that are poorly understood. Hence, further work will be needed before shells with desired permeability can be engineered.
Another notable biotechnology application is the use of MCPs to construct hybrid enzyme-inorganic catalysts (209). In a recent study, gold nanoparticles were fabricated onto the surface of purified Pdu MCPs. The gold nanoparticle-MCPs retained the native diol dehydratase activity in the MCP interior and possessed inorganic activity from the gold nanoparticles on the MCP exterior. This is a first step toward developing hybrid catalysts based on MCPs.
CURRENT CHALLENGES AND OPEN QUESTIONS
Although many aspects of MCP structure, function, and physiology have been elucidated, open questions remain. Genetic characterization has revealed many MCP components and their general roles in product production, assembly, and bacterial growth. However, in many cases their particular mechanisms of action, as well as specific locations within (or in some cases, potentially outside) the microcompartments remain to be determined. Additionally, while crystal structures of individual MCP proteins have advanced the understanding of MCP composition, function, and assembly, how MCP parts work together, both structurally and enzymatically, to build and maintain a complete bioreactor has yet to be determined.
The pathways by which metabolosomes assemble remain only partly understood. Current findings suggest different assembly mechanisms between classes of carboxysomes (185, 210, 211); therefore, it is possible that each metabolosome class possesses its own assembly framework. Relatedly, while many protein-protein interactions that govern MCP assembly have been determined, many more, particularly those that mediate assembly of the core and its interaction with the shell, are only beginning to be understood. Moreover, once an MCP is assembled, it is unclear how MCP structure enhances substrate processing. Despite an understanding of the mechanistic pathway taking place within the MCP, there are many unanswered questions about how MCP interiors are organized, and in turn, how these compartments interact with host cells.
A particularly intriguing question about MCPs is how their shells mediate the selective movement of small molecules. The size and chemical properties of the central pores of BMC-H proteins are thought to allow the selective diffusion of MCP substrates. However, the manner in which MCP products and enzymatic cofactors move across the shell has not been worked out. Dynamic gates in BMC-T proteins have been proposed but not demonstrated experimentally. Furthermore, many MCPs include only a single gated trimer, and how a single gate would mediate the movement of several different cofactors is unclear. In addition, electron transfer through shell proteins with [Fe-S] clusters has been suggested but not experimentally demonstrated.
It is also of note that as large, complex organelles, MCPs are energetically costly to the cell. The stability and longevity of these compartments remain unclear, and these aspects, in particular, may have implications for inhibiting pathogenic and commensal bacteria to optimize health. Further, an understanding of MCP stability would allow improved engineering for designing compartments, allowing them to survive in harsh environments to sequester toxic species or dissociate predictably at the site of drug delivery.
The diversity of bacterial MCPs is another area where further study is likely to yield rewards. Many diverse MCPs found throughout the domains of bacteria have been studied very little or not at all. In some cases, MCP substrates are unknown, and almost no work has been done on how MCPs fit into the broader metabolic schemes of their host organisms. Future work in these areas promises to uncover interesting biochemistry, advance bioengineering and biomedical applications, and broaden our understanding of bacterial physiology and pathogenesis. Furthermore, we expect that ongoing studies of these sophisticated organelles in the context of the Salmonella and E. coli hosts will drive advances in many of the areas mentioned above.
Contributor Information
Katie L. Stewart, The Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, USA 50011
Andrew M. Stewart, The Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, USA 50011
Thomas A. Bobik, The Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, USA 50011
James M. Slauch, The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL
Gregory Phillips, College of Veterinary Medicine, Iowa State University, Ames, IA.
REFERENCES
- 1.Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. 2014. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78:438–468 10.1128/MMBR.00009-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M. 2018. Bacterial microcompartments. Nat Rev Microbiol 16:277–290 10.1038/nrmicro.2018.10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77:357–379 10.1128/MMBR.00061-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput Biol 10:e1003898 10.1371/journal.pcbi.1003898. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdul-Rahman F, Petit E, Blanchard JL. 2013. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J Phylo Evo Bio 01:1–7. [Google Scholar]
- 6.Jorda J, Lopez D, Wheatley NM, Yeates TO. 2013. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci 22:179–195 10.1002/pro.2196. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarzycki J, Erbilgin O, Kerfeld CA. 2015. Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl Environ Microbiol 81:8315–8329 10.1128/AEM.02587-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobik TA. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol 70:517–525 10.1007/s00253-005-0295-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Yeates TO, Jorda J, Bobik TA. 2013. The shells of BMC-type microcompartment organelles in bacteria. J Mol Microbiol Biotechnol 23:290–299 10.1159/000351347. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50:137–181 10.1146/annurev.micro.50.1.137. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8:290–295 10.1038/nrmicro2334. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63 10.1038/nature09922. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae S, Ulrich CM, Neuhouser ML, Malysheva O, Bailey LB, Xiao L, Brown EC, Cushing-Haugen KL, Zheng Y, Cheng TYD, Miller JW, Green R, Lane DS, Beresford SAA, Caudill MA. 2014. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer Res 74:7442–7452 10.1158/0008-5472.CAN-14-1835. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank S, Lawrence AD, Prentice MB, Warren MJ. 2013. Bacterial microcompartments moving into a synthetic biological world. J Biotechnol 163:273–279 10.1016/j.jbiotec.2012.09.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Kim EY, Tullman-Ercek D. 2013. Engineering nanoscale protein compartments for synthetic organelles. Curr Opin Biotechnol 24:627–632 10.1016/j.copbio.2012.11.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Plegaria JS, Kerfeld CA. 2018. Engineering nanoreactors using bacterial microcompartment architectures. Curr Opin Biotechnol 51:1–7 10.1016/j.copbio.2017.09.005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai SJ, Yeates TO. 2011. Bacterial microcompartments: insights into the structure, mechanism, and engineering applications. Prog Mol Biol Transl Sci 103:1–20 10.1016/B978-0-12-415906-8.00008-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Drews G, Niklowitz W. 1956. Beiträge zur cytologie der blaualgen. II. Mittelung zentroplasma und granuläre einschlüsse von phormidium uncinatum. Arch Mikrobiol 24:147–162 10.1007/BF00408629. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Badger MR, Price GD. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54:609–622 10.1093/jxb/erg076. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Cannon GC, Heinhorst S, Bradburne CE, Shively JM. 2002. Carboxysome genomics: a status report. Funct Plant Biol 29:175–182 10.1071/PP01200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586 10.1126/science.182.4112.584. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. J Bacteriol 181:5967–5975 10.1128/JB.181.19.5967-5975.1999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5329 10.1128/JB.181.17.5317-5329.1999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erbilgin O, McDonald KL, Kerfeld CA. 2014. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80:2193–2205 10.1128/AEM.03887-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, Warren MJ, Prentice MB. 2008. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol 190:4559–4567 10.1128/JB.01535-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ. 1990. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD+ oxidoreductase. Appl Environ Microbiol 56:943–948 10.1128/AEM.56.4.943-948.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. 2013. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337 10.1371/journal.pone.0054337. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jameson E, Fu T, Brown IR, Paszkiewicz K, Purdy KJ, Frank S, Chen Y. 2016. Anaerobic choline metabolism in microcompartments promotes growth and swarming of Proteus mirabilis. Environ Microbiol 18:2886–2898 10.1111/1462-2920.13059. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundin AP, Stewart KL, Stewart AM, Herring TI, Chowdhury C, Bobik TA. 2020. Genetic characterization of a glycyl radical microcompartment used for 1,2-propanediol fermentation by uropathogenic Escherichia coli CFT073. J Bacteriol 202:202 10.1128/JB.00017-20. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herring TI, Harris TN, Chowdhury C, Mohanty SK, Bobik TA. 2018. A bacterial microcompartment is used for choline fermentation by Escherichia coli 536. J Bacteriol 200:1–13 10.1128/JB.00764-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971 10.1128/JB.01925-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rondon MR, Horswill AR, Escalante-Semerena JC. 1995. DNA polymerase I function is required for the utilization of ethanolamine, 1,2-propanediol, and propionate by Salmonella typhimurium LT2. J Bacteriol 177:7119–7124 10.1128/JB.177.24.7119-7124.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penrod JT, Roth JR. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 188:2865–2874 10.1128/JB.188.8.2865-2874.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinsmade SR, Paldon T, Escalante-Semerena JC. 2005. Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol 187:8039–8046 10.1128/JB.187.23.8039-8046.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobson CM, Tullman-Ercek D, Slininger MF, Mangan NM. 2017. A systems-level model reveals that 1,2-propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLOS Comput Biol 13:e1005525 10.1371/journal.pcbi.1005525. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio 3:1–10 10.1128/mBio.00050-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maadani A, Fox KA, Mylonakis E, Garsin DA. 2007. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect Immun 75:2634–2637 10.1128/IAI.01372-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ormsby MJ, Logan M, Johnson SA, McIntosh A, Fallata G, Papadopoulou R, Papachristou E, Hold GL, Hansen R, Ijaz UZ, Russell RK, Gerasimidis K, Wall DM. 2019. Inflammation associated ethanolamine facilitates infection by Crohn’s disease-linked adherent-invasive Escherichia coli. EBioMedicine 43:325–332 10.1016/j.ebiom.2019.03.071. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dadswell K, Creagh S, McCullagh E, Liang M, Brown IR, Warren MJ, McNally A, MacSharry J, Prentice MB. 2019. Bacterial microcompartment-mediated ethanolamine metabolism in Escherichia coli urinary tract infection. Infect Immun 87:e00211–e00219 10.1128/IAI.00211-19. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiol (Read) 153:1207–1220 10.1099/mic.0.2006/004747-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A Jr, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121 10.1128/IAI.01390-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108:17480–17485 10.1073/pnas.1107857108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 10.1038/nature09415. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakobson CM, Tullman-Ercek D. 2016. Dumpster diving in the gut: bacterial microcompartments as part of a host-associated lifestyle. PLoS Pathog 12:e1005558 10.1371/journal.ppat.1005558. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ. 2017. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:e1006129. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romano KA, Martinez-Del Campo A, Kasahara K, Chittim CL, Vivas EI, Amador-Noguez D, Balskus EP, Rey FE. 2017. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22:279–290.e7 10.1016/j.chom.2017.07.021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang WHW, Kitai T, Hazen SL. 2017. Gut microbiota in cardiovascular health and disease. Circ Res 120:1183–1196 10.1161/CIRCRESAHA.117.309715. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YM, Liu Y, Zhou RF, Chen XL, Wang C, Tan XY, Wang LJ, Zheng RD, Zhang HW, Ling WH, Zhu HL. 2016. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep 6:19076 10.1038/srep19076. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845 10.1038/s41467-017-00900-1. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trøseid M, Ueland T, Hov JR, Svardal A, Gregersen I, Dahl CP, Aakhus S, Gude E, Bjørndal B, Halvorsen B, Karlsen TH, Aukrust P, Gullestad L, Berge RK, Yndestad A. 2015. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med 277:717–726 10.1111/joim.12328. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Moraes C, Fouque D, Amaral ACF, Mafra D.2015. Trimethylamine N-oxide from gut microbiota in chronic kidney disease patients: focus on diet. J Ren Nutr 25:459–465. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Xu KY, Xia GH, Lu JQ, Chen MX, Zhen X, Wang S, You C, Nie J, Zhou HW, Yin J. 2017. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci Rep 7:1445 10.1038/s41598-017-01387-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang WHW, Wang Z, Li XS, Fan Y, Li DS, Wu Y, Hazen SL. 2017. Increased trimethylamine N-oxide portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin Chem 63:297–306 10.1373/clinchem.2016.263640. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dambrova M, Latkovskis G, Kuka J, Strele I, Konrade I, Grinberga S, Hartmane D, Pugovics O, Erglis A, Liepinsh E. 2016. Diabetes is associated with higher trimethylamine N-oxide plasma levels. Exp Clin Endocrinol Diabetes 124:251–256 10.1055/s-0035-1569330. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Obradors N, Badía J, Baldomà L, Aguilar J. 1988. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella Typhimurium. J Bacteriol 170:2159–2162 10.1128/JB.170.5.2159-2162.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawrence JG, Roth JR. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaval KG, Gebbie M, Goodson JR, Cruz MR, Winkler WC, Garsin DA. 2019. Ethanolamine utilization and bacterial microcompartment formation are subject to carbon catabolite repression. J Bacteriol 201:1–13 10.1128/JB.00703-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martínez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ, Balskus EP. 2015. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio 6:1–12 10.1128/mBio.00042-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157:690–693 10.1128/JB.157.2.690-693.1984. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zarzycki J, Sutter M, Cortina NS, Erb TJ, Kerfeld CA. 2017. In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme-associated bacterial microcompartment. Sci Rep 7:42757 10.1038/srep42757. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeter RM. 1990. Cobalamin-dependent 1,2-propanediol utilization by Salmonella Typhimurium. J Gen Microbiol 136:887–896 10.1099/00221287-136-5-887. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol 183:2463–2475 10.1128/JB.183.8.2463-2475.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. 1997. Propanediol utilization genes (pdu) of Salmonella Typhimurium: three genes for the propanediol dehydratase. J Bacteriol 179:6633–6639 10.1128/JB.179.21.6633-6639.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abeles RH, Lee HA Jr. 1961. An intramolecular oxidation-reduction requiring a cobamide coenzyme. J Biol Chem 236:2347–2350. [PubMed] [Google Scholar]
- 66.Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7:e47144 10.1371/journal.pone.0047144. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leal NA, Havemann GD, Bobik TA. 2003. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol 180:353–361 10.1007/s00203-003-0601-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Leal NA, Sampson EM, Johnson CLV, Havemann GD, Bobik TA. 2007. PduL is an evolutionarily distinct phosphotransacylase involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. J Bacteriol 189:1589–1596 10.1128/JB.01151-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palacios S, Starai VJ, Escalante-Semerena JC. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J Bacteriol 185:2802–2810 10.1128/JB.185.9.2802-2810.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horswill AR, Escalante-Semerena JC. 1999. Salmonella Typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol 181:5615–5623 10.1128/JB.181.18.5615-5623.1999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng S, Bobik TA. 2010. Characterization of the PduS cobalamin reductase of Salmonella enterica and its role in the Pdu microcompartment. J Bacteriol 192:5071–5080 10.1128/JB.00575-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson CLV, Pechonick E, Park SD, Havemann GD, Leal NA, Bobik TA. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol 183:1577–1584 10.1128/JB.183.5.1577-1584.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walter D, Ailion M, Roth J. 1997. Genetic characterization of the pdu operon: use of 1,2-propanediol in Salmonella Typhimurium. J Bacteriol 179:1013–1022 10.1128/JB.179.4.1013-1022.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinha S, Cheng S, Fan C, Bobik TA. 2012. The PduM protein is a structural component of the microcompartments involved in coenzyme B(12)-dependent 1,2-propanediol degradation by Salmonella enterica. J Bacteriol 194:1912–1918 10.1128/JB.06529-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y, Jorda J, Yeates TO, Bobik TA. 2015. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J Bacteriol 197:2392–2399 10.1128/JB.00056-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. 2011. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J Bacteriol 193:1385–1392 10.1128/JB.01473-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Havemann GD, Sampson EM, Bobik TA. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 184:1253–1261 10.1128/JB.184.5.1253-1261.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. 2010. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 38:305–315 10.1016/j.molcel.2010.04.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Fan C, Fromm HJ, Bobik TA. 2009. Kinetic and functional analysis of l-threonine kinase, the PduX enzyme of Salmonella enterica. J Biol Chem 284:20240–20248 10.1074/jbc.M109.027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan C, Bobik TA. 2008. The PduX enzyme of Salmonella enterica is an l-threonine kinase used for coenzyme B12 synthesis. J Biol Chem 283:11322–11329 10.1074/jbc.M800287200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Rondon MR, Escalante-Semerena JC. 1996. In vitro analysis of the interactions between the PocR regulatory protein and the promoter region of the cobalamin biosynthetic (cob) operon of Salmonella Typhimurium LT2. J Bacteriol 178:2196–2203 10.1128/JB.178.8.2196-2203.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bobik TA, Ailion M, Roth JR. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol 174:2253–2266 10.1128/JB.174.7.2253-2266.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ailion M, Bobik TA, Roth JR. 1993. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol 175:7200–7208 10.1128/JB.175.22.7200-7208.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersson DI. 1992. Involvement of the Arc system in redox regulation of the Cob operon in Salmonella typhimurium. Mol Microbiol 6:1491–1494 10.1111/j.1365-2958.1992.tb00869.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. 2003. Global regulation by CsrA in Salmonella Typhimurium. Mol Microbiol 48:1633–1645 10.1046/j.1365-2958.2003.03535.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, Sun J, Tian M, Xu Z, Liu Y, Fu J, Yan A, Liu X. 2019. Proteomic analysis of FNR-regulated anaerobiosis in Salmonella Typhimurium. J Am Soc Mass Spectrom 30:1001–1012 10.1007/s13361-019-02145-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Roof DM, Roth JR. 1988. Ethanolamine utilization in Salmonella Typhimurium. J Bacteriol 170:3855–3863 10.1128/JB.170.9.3855-3863.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roof DM, Roth JR. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella Typhimurium. J Bacteriol 171:3316–3323 10.1128/JB.171.6.3316-3323.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jeter RM, Olivera BM, Roth JR. 1984. Salmonella Typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol 159:206–213 10.1128/JB.159.1.206-213.1984. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bobik TA, Lehman BP, Yeates TO. 2015. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98:193–207 10.1111/mmi.13117. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shibata N, Tamagaki H, Hieda N, Akita K, Komori H, Shomura Y, Terawaki S, Mori K, Yasuoka N, Higuchi Y, Toraya T. 2010. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J Biol Chem 285:26484–26493 10.1074/jbc.M110.125112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu H, Gonzalez R, Bobik TA. 2011. Coproduction of acetaldehyde and hydrogen during glucose fermentation by Escherichia coli. Appl Environ Microbiol 77:6441–6450 10.1128/AEM.05358-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huseby DL, Roth JR. 2013. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol 195:2864–2879 10.1128/JB.02179-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buan NR, Suh SJ, Escalante-Semerena JC. 2004. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J Bacteriol 186:5708–5714 10.1128/JB.186.17.5708-5714.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheppard DE, Penrod JT, Bobik T, Kofoid E, Roth JR. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J Bacteriol 186:7635–7644 10.1128/JB.186.22.7635-7644.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mori K, Bando R, Hieda N, Toraya T. 2004. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J Bacteriol 186:6845–6854 10.1128/JB.186.20.6845-6854.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moore TC, Escalante-Semerena JC. 2016. The EutQ and EutP proteins are novel acetate kinases involved in ethanolamine catabolism: physiological implications for the function of the ethanolamine metabolosome in Salmonella enterica. Mol Microbiol 99:497–511 10.1111/mmi.13243. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stojiljkovic I, Bäumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella Typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA. cchB. eutE. eutJ. eutG. eutH gene cluster. J Bacteriol 177:1357–1366 10.1128/JB.177.5.1357-1366.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bandarian V, Poyner RR, Reed GH. 1999. Hydrogen atom exchange between 5′-deoxyadenosine and hydroxyethylhydrazine during the single turnover inactivation of ethanolamine ammonia-lyase. Biochemistry 38:12403–12407 10.1021/bi9906219. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Bandarian V, Reed GH. 1999. Hydrazine cation radical in the active site of ethanolamine ammonia-lyase: mechanism-based inactivation by hydroxyethylhydrazine. Biochemistry 38:12394–12402 10.1021/bi990620g. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Bologna FP, Campos-Bermudez VA, Saavedra DD, Andreo CS, Drincovich MF. 2010. Characterization of Escherichia coli EutD: a phosphotransacetylase of the ethanolamine operon. J Microbiol 48:629–636 10.1007/s12275-010-0091-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Buan NR, Escalante-Semerena JC. 2006. Purification and initial biochemical characterization of ATP:Cob(I)alamin adenosyltransferase (EutT) enzyme of Salmonella enterica. J Biol Chem 281:16971–16977 10.1074/jbc.M603069200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Held M, Kolb A, Perdue S, Hsu SY, Bloch SE, Quin MB, Schmidt-Dannert C. 2016. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Sci Rep 6:24359 10.1038/srep24359. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Penrod JT, Mace CC, Roth JR. 2004. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J Bacteriol 186:6885–6890 10.1128/JB.186.20.6885-6890.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roof DM, Roth JR. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella Typhimurium. J Bacteriol 174:6634–6643 10.1128/JB.174.20.6634-6643.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sheppard DE, Roth JR. 1994. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella Typhimurium. J Bacteriol 176:1287–1296 10.1128/JB.176.5.1287-1296.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shimada T, Fujita N, Yamamoto K, Ishihama A. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081 10.1371/journal.pone.0020081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213 10.1006/jmbi.1999.3161. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Sturms R, Streauslin NA, Cheng S, Bobik TA. 2015. In Salmonella enterica, ethanolamine utilization is repressed by 1,2-propanediol to prevent detrimental mixing of components of two different bacterial microcompartments. J Bacteriol 197:2412–2421 10.1128/JB.00215-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jakobson CM, Kim EY, Slininger MF, Chien A, Tullman-Ercek D. 2015. Localization of proteins to the 1,2-propanediol utilization microcompartment by non-native signal sequences is mediated by a common hydrophobic motif. J Biol Chem 290:24519–24533 10.1074/jbc.M115.651919. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Slininger Lee MF, Jakobson CM, Tullman-Ercek D. 2017. Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synth Biol 6:1880–1891 10.1021/acssynbio.7b00042. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 109:21307–21312 10.1073/pnas.1215689109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kalnins G, Kuka J, Grinberga S, Makrecka-Kuka M, Liepinsh E, Dambrova M, Tars K. 2015. Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J Biol Chem 290:21732–21740 10.1074/jbc.M115.670471. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun 70:6365–6372 10.1128/IAI.70.11.6365-6372.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schindel HS, Karty JA, McKinlay JB, Bauer CE. 2019. Characterization of a glycyl radical enzyme bacterial microcompartment pathway in Rhodobacter capsulatus. J Bacteriol 201:e00343-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. 2010. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA 107:7509–7514 10.1073/pnas.0913199107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan C, Bobik TA. 2011. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193:5623–5628 10.1128/JB.05661-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. 2012. Engineered protein nano-compartments for targeted enzyme localization. PLoS One 7:e33342 10.1371/journal.pone.0033342. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fan C, Cheng S, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci USA 109:14995–15000 10.1073/pnas.1207516109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quin MB, Perdue SA, Hsu SY, Schmidt-Dannert C. 2016. Encapsulation of multiple cargo proteins within recombinant Eut nanocompartments. Appl Microbiol Biotechnol 100:9187–9200 10.1007/s00253-016-7737-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 287:17729–17736 10.1074/jbc.M112.355305. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim EY, Tullman-Ercek D. 2014. A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments. Biotechnol J 9:348–354 10.1002/biot.201300391. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ. 2014. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3:454–465 10.1021/sb4001118. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang M, Simpson DM, Wenner N, Brownridge P, Harman VM, Hinton JCD, Beynon RJ, Liu L-N. 2020. Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nat Commun 11:1976 10.1038/s41467-020-15888-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Toraya T. 2014. Cobalamin-dependent dehydratases and a deaminase: radical catalysis and reactivating chaperones. Arch Biochem Biophys 544:40–57 10.1016/j.abb.2013.11.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Akita K, Hieda N, Baba N, Kawaguchi S, Sakamoto H, Nakanishi Y, Yamanishi M, Mori K, Toraya T. 2010. Purification and some properties of wild-type and N-terminal-truncated ethanolamine ammonia-lyase of Escherichia coli. J Biochem 147:83–93 10.1093/jb/mvp145. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Toraya T. 2003. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem Rev 103:2095–2127 10.1021/cr020428b. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Johnson CLV, Buszko ML, Bobik TA. 2004. Purification and initial characterization of the Salmonella enterica PduO ATP:Cob(I)alamin adenosyltransferase. J Bacteriol 186:7881–7887 10.1128/JB.186.23.7881-7887.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wagner OW, Lee HA Jr, Frey PA, Abeles RH. 1966. Studies on the mechanism of action of cobamide coenzymes. Chemical properties of the enzyme-coenzyme complex. J Biol Chem 241:1751–1762. [PubMed] [Google Scholar]
- 130.Bachovchin WW, Eagar RG Jr, Moore KW, Richards JH. 1977. Mechanism of action of adenosylcobalamin: glycerol and other substrate analogues as substrates and inactivators for propanediol dehydratase: kinetics, stereospecificity, and mechanism. Biochemistry 16:1082–1092 10.1021/bi00625a009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Yamanishi M, Kinoshita K, Fukuoka M, Saito T, Tanokuchi A, Ikeda Y, Obayashi H, Mori K, Shibata N, Tobimatsu T, Toraya T. 2012. Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols. FEBS J 279:793–804 10.1111/j.1742-4658.2012.08470.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Daniel R, Bobik TA, Gottschalk G. 1998. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol Rev 22:553–566 10.1111/j.1574-6976.1998.tb00387.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Toraya T. 2000. Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell Mol Life Sci 57:106–127 10.1007/s000180050502. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mori K, Hosokawa Y, Yoshinaga T, Toraya T. 2010. Diol dehydratase-reactivating factor is a reactivase: evidence for multiple turnovers and subunit swapping with diol dehydratase. FEBS J 277:4931–4943 10.1111/j.1742-4658.2010.07898.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Mori K, Obayashi K, Hosokawa Y, Yamamoto A, Yano M, Yoshinaga T, Toraya T. 2013. Essential roles of nucleotide-switch and metal-coordinating residues for chaperone function of diol dehydratase-reactivase. Biochemistry 52:8677–8686 10.1021/bi401290j. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Sampson EM, Johnson CLV, Bobik TA. 2005. Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiol Read 151:1169–1177 10.1099/mic.0.27755-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Fonseca MV, Escalante-Semerena JC. 2001. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J Biol Chem 276:32101–32108 10.1074/jbc.M102510200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Fonseca MV, Escalante-Semerena JC. 2000. Reduction of Cob(III)alamin to Cob(II)alamin in Salmonella enterica serovar typhimurium LT2. J Bacteriol 182:4304–4309 10.1128/JB.182.15.4304-4309.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bovell AM, Warncke K. 2013. The structural model of Salmonella Typhimurium ethanolamine ammonia-lyase directs a rational approach to the assembly of the functional [(EutB-EutC)2]3 oligomer from isolated subunits. Biochemistry 52:1419–1428 10.1021/bi301651n. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shibata N, Masuda J, Tobimatsu T, Toraya T, Suto K, Morimoto Y, Yasuoka N. 1999. A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: x-ray structure of diol dehydratase. Structure 7:997–1008 10.1016/S0969-2126(99)80126-9. [DOI] [PubMed] [Google Scholar]
- 141.Havemann GD, Bobik TA. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 185:5086–5095 10.1128/JB.185.17.5086-5095.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Frey PA. 2001. Radical mechanisms of enzymatic catalysis. Annu Rev Biochem 70:121–148 10.1146/annurev.biochem.70.1.121. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Backman LRF, Funk MA, Dawson CD, Drennan CL. 2017. New tricks for the glycyl radical enzyme family. Crit Rev Biochem Mol Biol 52:674–695 10.1080/10409238.2017.1373741. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shisler KA, Broderick JB. 2014. Glycyl radical activating enzymes: structure, mechanism, and substrate interactions. Arch Biochem Biophys 546:64–71 10.1016/j.abb.2014.01.020. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bowman SEJ, Backman LRF, Bjork RE, Andorfer MC, Yori S, Caruso A, Stultz CM, Drennan CL. 2019. Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme. J Biol Inorg Chem 24:817–829 10.1007/s00775-019-01681-2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bodea S, Balskus EP. 2018. Purification and characterization of the choline trimethylamine-lyase (CutC)-activating protein CutD. Methods Enzymol 606:73–94 10.1016/bs.mie.2018.04.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Kalnins G, Cesle E-E, Jansons J, Liepins J, Filimonenko A, Tars K. 2020. Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nat Commun 11:388 10.1038/s41467-019-14205-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Starai VJ, Garrity J, Escalante-Semerena JC. 2005. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiol (Reading) 151:3793–3801 10.1099/mic.0.28156-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Erbilgin O, Sutter M, Kerfeld CA. 2016. The structural basis of coenzyme A recycling in a bacterial organelle. PLoS Biol 14:e1002399 10.1371/journal.pbio.1002399. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yeates TO, Thompson MC, Bobik TA. 2011. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol 21:223–231 10.1016/j.sbi.2011.01.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083–1086 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 152.Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA. 2014. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426:2328–2345 10.1016/j.jmb.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Takenoya M, Nikolakakis K, Sagermann M. 2010. Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J Bacteriol 192:6056–6063 10.1128/JB.00652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Crowley CS, Sawaya MR, Bobik TA, Yeates TO. 2008. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 16:1324–1332 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. 2010. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem 285:37838–37846 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pang A, Warren MJ, Pickersgill RW. 2011. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr 67:91–96 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- 157.Thompson MC, Wheatley NM, Jorda J, Sawaya MR, Gidaniyan SD, Ahmed H, Yang Z, McCarty KN, Whitelegge JP, Yeates TO. 2014. Identification of a unique Fe-S cluster binding site in a glycyl-radical type microcompartment shell protein. J Mol Biol 426:3287–3304 10.1016/j.jmb.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. 2013. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 22:660–665 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–84 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 160.Sagermann M, Ohtaki A, Nikolakakis K. 2009. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci USA 106:8883–8887 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pang A, Liang M, Prentice MB, Pickersgill RW. 2012. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 68:1642–1652 10.1107/S0907444912039315. [DOI] [PubMed] [Google Scholar]
- 162.Chowdhury C, Chun S, Sawaya MR, Yeates TO, Bobik TA. 2016. The function of the PduJ microcompartment shell protein is determined by the genomic position of its encoding gene. Mol Microbiol 101:770–783 10.1111/mmi.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. 2015. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci USA 112:2990–2995 10.1073/pnas.1423672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thompson MC, Cascio D, Leibly DJ, Yeates TO. 2015. An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Sci 24:956–975 10.1002/pro.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sutter M, Greber B, Aussignargues C, Kerfeld CA. 2017. Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356:1293–1297 10.1126/science.aan3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Greber BJ, Sutter M, Kerfeld CA. 2019. The plasticity of molecular interactions governs bacterial microcompartment shell assembly. Structure 27:749–763.e4 10.1016/j.str.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sutter M, Laughlin TG, Sloan NB, Serwas D, Davies KM, Kerfeld CA. 2019. Structure of a synthetic β-carboxysome shell. Plant Physiol 181:1050–1058 10.1104/pp.19.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Garcia-Alles LF, Lesniewska E, Root K, Aubry N, Pocholle N, Mendoza CI, Bourillot E, Barylyuk K, Pompon D, Zenobi R, Reguera D, Truan G. 2017. Spontaneous non-canonical assembly of CcmK hexameric components from β-carboxysome shells of cyanobacteria. PLoS One 12:e0185109 10.1371/journal.pone.0185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yeates TO, Crowley CS, Tanaka S. 2010. Bacterial microcompartment organelles: protein shell structure and evolution. Annu Rev Biophys 39:185–205 10.1146/annurev.biophys.093008.131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 171.Pang A, Frank S, Brown I, Warren MJ, Pickersgill RW. 2014. Structural insights into higher order assembly and function of the bacterial microcompartment protein PduA. J Biol Chem 289:22377–22384 10.1074/jbc.M114.569285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, Liu LN. 2016. Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett 16:1590–1595 10.1021/acs.nanolett.5b04259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Faulkner M, Zhao LS, Barrett S, Liu LN. 2019. Self-assembly, stability, and variability of bacterial microcompartment shell proteins in response to environmental change. Nano Res Lett 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Park J, Chun S, Bobik TA, Houk KN, Yeates TO. 2017. Molecular dynamics simulations of selective metabolite transport across the propanediol bacterial microcompartment shell. J Phys Chem B 121:8149–8154 10.1021/acs.jpcb.7b07232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lehman BP, Chowdhury C, Bobik TA. 2017. The N-terminus of the PduB protein binds the protein shell of the Pdu microcompartment to its enzymatic core. J Bacteriol 199:1–13 10.1128/JB.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. 2013. The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem 288:16055–16063 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Larsson AM, Hasse D, Valegård K, Andersson I. 2017. Crystal structures of β-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. J Exp Bot 68:3857–3867 10.1093/jxb/erx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392:319–333 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 179.Chowdhury C, Bobik TA. 2019. Engineering the PduT shell protein to modify the permeability of the 1,2-propanediol microcompartment of Salmonella. Microbiol Read 165:1355–1364 10.1099/mic.0.000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cannon GC, Shively JM. 1983. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol 134:52–59 10.1007/BF00429407. [DOI] [Google Scholar]
- 181.Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA. 2014. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J Mol Biol 426:2217–2228 10.1016/j.jmb.2014.02.025. [DOI] [PubMed] [Google Scholar]
- 182.Mohajerani F, Hagan MF. 2018. The role of the encapsulated cargo in microcompartment assembly. PLOS Comput Biol 14:e1006351 10.1371/journal.pcbi.1006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rotskoff GM, Geissler PL. 2018. Robust nonequilibrium pathways to microcompartment assembly. Proc Natl Acad Sci USA 115:6341–6346 10.1073/pnas.1802499115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kerfeld CA, Melnicki MR. 2016. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31:66–75 10.1016/j.pbi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 185.Dai W, Chen M, Myers C, Ludtke SJ, Pettitt BM, King JA, Schmid MF, Chiu W. 2018. Visualizing individual RuBisCO and its assembly into carboxysomes in marine cyanobacteria by cryo-electron tomography. J Mol Biol 430:4156–4167 10.1016/j.jmb.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hagen A, Sutter M, Sloan N, Kerfeld CA. 2018. Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nat Commun 9:2881 10.1038/s41467-018-05162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jorda J, Liu Y, Bobik TA, Yeates TO. 2015. Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment. PLOS Comput Biol 11:e1004067 10.1371/journal.pcbi.1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kennedy NW, Hershewe JM, Nichols TM, Roth EW, Wilke CD, Mills CE, Jewett MC, Tullman-Ercek D. 2020. Apparent size and morphology of bacterial microcompartments varies with technique. PLoS One 15:e0226395 10.1371/journal.pone.0226395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lünsdorf H, Rambach A, Gass MH, Bleloch A, McClean KJ, Munro AW, Rigby SEJ, Warren MJ, Prentice MB. 2008. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 283:14366–14375 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 190.Graf L, Wu K, Wilson JW. 2018. Transfer and analysis of Salmonella pdu genes in a range of Gram-negative bacteria demonstrate exogenous microcompartment expression across a variety of species. Microb Biotechnol 11:199–210 10.1111/1751-7915.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fang Y, Huang F, Faulkner M, Jiang Q, Dykes GF, Yang M, Liu LN. 2018. Engineering and modulating functional cyanobacterial CO2-fixing organelles. Front Plant Sci 9:739 10.3389/fpls.2018.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. 2012. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA 109:478–483 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, Lim YL, Nguyen ND, Massey B, Bala S, von Caemmerer S, Badger MR, Price GD. 2018. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nature Coms 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee MJ, Palmer DJ, Warren MJ. 2019. Biotechnological advances in bacterial microcompartment technology. Trends Biotechnol 37:325–336 10.1016/j.tibtech.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 195.Young EJ, Burton R, Mahalik JP, Sumpter BG, Fuentes-Cabrera M, Kerfeld CA, Ducat DC. 2017. Engineering the bacterial microcompartment domain for molecular scaffolding applications. Front Microbiol 8:1441 10.3389/fmicb.2017.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chessher A, Breitling R, Takano E. 2015. Bacterial microcompartments: biomaterials for synthetic biology-based compartmentalization strategies. ACS Biomater Sci Eng 1:345–351 10.1021/acsbiomaterials.5b00059. [DOI] [PubMed] [Google Scholar]
- 197.Corchero JL, Cedano J. 2011. Self-assembling, protein-based intracellular bacterial organelles: emerging vehicles for encapsulating, targeting and delivering therapeutical cargoes. Microb Cell Fact 10:92 10.1186/1475-2859-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Planamente S, Frank S. 2019. Bio-engineering of bacterial microcompartments: a mini review. Biochem Soc Trans 47:765–777 10.1042/BST20170564. [DOI] [PubMed] [Google Scholar]
- 199.Bari NK, Kumar G, Hazra JP, Kaur S, Sinha S. 2020. Functional protein shells fabricated from the self-assembling protein sheets of prokaryotic organelles. J Mater Chem B Mater Biol Med 8:523–533 10.1039/C9TB02224D. [DOI] [PubMed] [Google Scholar]
- 200.Jorda J, Leibly DJ, Thompson MC, Yeates TO. 2016. Structure of a novel 13 nm dodecahedral nanocage assembled from a redesigned bacterial microcompartment shell protein. Chem Commun (Camb) 52:5041–5044 10.1039/C6CC00851H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sutter M, McGuire S, Ferlez B, Kerfeld CA. 2019. Structural characterization of a synthetic tandem-domain bacterial microcompartment shell protein capable of forming icosahedral shell assemblies. ACS Synth Biol 8:668–674 10.1021/acssynbio.9b00011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ferlez B, Sutter M, Kerfeld CA. 2019. A designed bacterial microcompartment shell with tunable composition and precision cargo loading. Metab Eng 54:286–291 10.1016/j.ymben.2019.04.011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liang M, Frank S, Lünsdorf H, Warren MJ, Prentice MB. 2017. Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol J 12:12 10.1002/biot.201600415. [PubMed] [DOI] [PubMed] [Google Scholar]
- 204.Huber I, Palmer DJ, Ludwig KN, Brown IR, Warren MJ, Frunzke J. 2017. Construction of recombinant Pdu metabolosome shells for small molecule production in Corynebacterium glutamicum. ACS Synth Biol 6:2145–2156 10.1021/acssynbio.7b00167. [PubMed] [DOI] [PubMed] [Google Scholar]
- 205.Nichols TM, Kennedy NW, Tullman-Ercek D. 2019. Cargo encapsulation in bacterial microcompartments: methods and analysis. Methods Enzymol 617:155–186 10.1016/bs.mie.2018.12.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wade Y, Daniel RA, Leak DJ. 2019. Heterologous microcompartment assembly in Bacillaceae: establishing the components necessary for scaffold formation. ACS Synth Biol 8:1642–1654 10.1021/acssynbio.9b00155. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Yung MC, Bourguet FA, Carpenter TS, Coleman MA. 2017. Re-directing bacterial microcompartment systems to enhance recombinant expression of lysis protein E from bacteriophage ϕX174 in Escherichia coli. Microb Cell Fact 16:71 10.1186/s12934-017-0685-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lee MJ, Mantell J, Brown IR, Fletcher JM, Verkade P, Pickersgill RW, Woolfson DN, Frank S, Warren MJ. 2018. De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments. Nat Commun 9:3413 10.1038/s41467-018-05922-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bari NK, Kumar G, Bhatt A, Hazra JP, Garg A, Ali ME, Sinha S. 2018. Nanoparticle fabrication on bacterial microcompartment surface for the development of hybrid enzyme-inorganic catalyst. ACS Catal 8:7742–7748 10.1021/acscatal.8b02322. [DOI] [Google Scholar]
- 210.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155:1131–1140 10.1016/j.cell.2013.10.044. [PubMed] [DOI] [PubMed] [Google Scholar]
- 211.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. 2010. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol 396:105–117 10.1016/j.jmb.2009.11.019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


