Abstract
Mismatch repair–deficient (dMMR) cancers generate a substantial number of immunogenic neoantigens, rendering them sensitive to immunotherapy. Yet, there is considerable variability in responses, and roughly one-half of dMMR cancers are refractory to immunotherapy. Here we study a patient with dMMR lung cancer refractory to immunotherapy. The tumor exhibited typical dMMR molecular features, including exceptionally high frameshift insertions and deletions (indels). Despite the treatment inducing abundant intratumoral T-cell infiltrates, it failed to elicit tumor regression, pointing to the T cells lacking cytotoxic activity. A post-treatment tumor demonstrated compound heterozygous frameshift deletions located upstream of the kinase domain in the gene encoding JAK1 protein, down-regulation of JAK1 and mediators of its signal transduction, and total loss of JAK1 phosphorylation. Importantly, one of the JAK1 mutations, despite not being detected in the pretreatment tumor, was found at low variant allele frequency in the pretreatment circulating tumor DNA, suggesting clonal selection of the mutation. To our knowledge, this report provides the most detailed look yet at defective JAK1 signaling in the context of dMMR and immunotherapy resistance. Together with observations of JAK1 frameshift indels being enriched in dMMR compared with MMR-proficient tumors, our findings demonstrate the critical function of JAK1 in immunological surveillance of dMMR cancer.
Keywords: neoplasm of the lung
INTRODUCTION
The mismatch DNA repair (MMR) system corrects errors in base insertion, deletion, or misincorporation that arise during DNA replication and recombination. Tumors with mismatch repair deficiency (dMMR)—due to germline or acquired deficiency of MMR proteins—represent ∼2%–4% of all cancers (Dudley et al. 2016). These tumors have exceptionally high numbers of somatic mutations (Cancer Genome Atlas Network 2012), especially frameshift insertion/deletions (indels) that generate a large proportion of mutant neoantigens, rendering them more responsive to programmed death-1 (PD-1)- or programmed death-ligand 1 (PD-L1)-directed therapy (Dudley et al. 2016). Immune checkpoint inhibitors yield durable tumor responses in 30%–40% of patients with dMMR cancers (Marabelle et al. 2020). Anti PD-1 antibody pembrolizumab is now approved by the United States Food and Drug Administration (FDA) for treatment of unresectable or metastatic dMMR tumors, irrespective of histology (Merck Sharp and Dohme 2018). Yet, one-half to two-thirds of dMMR cancers fail to respond or have disease progression within 6 mo of immunotherapy, suggesting primary resistance.
Here we report the identification of compound heterozygous Janus kinase 1 (JAK1) mutations in a dMMR cancer that result in functional JAK1 deficiency associated with resistance to immune checkpoint blockade. Furthermore, detailed analyses of a paired pre- and post-treatment tumor, circulating tumor DNA (ctDNA), plasma, and peripheral blood mononuclear cells (PBMCs) reveal dynamic changes in JAK1 allelic burden over time, which is integrally linked to systemic and tumor immune response dynamics and the patient's clinical trajectory. Our findings provide clinical evidence of how highly mutated tumors may grow unimpeded by immune recognition and have implications for primary resistance to immune checkpoint blockade in dMMR tumors.
RESULTS
Clinical Course and Mismatch Repair Deficiency
A 64-yr-old female ex-smoker presented to her primary care physician with left-sided chest and flank pain. Computed tomography (CT) scan revealed multiple pleural nodules, mediastinal lymphadenopathy, liver nodules, and a right adrenal mass. Liver biopsy showed small cells with scant cytoplasm, hyperchromatic nuclei, and inconspicuous nuclei, with extensive necrosis and mitotic activity. Immunohistochemistry (IHC) for the neuroendocrine markers chromogranin and synaptophysin was positive. The patient was diagnosed with extensive-stage small-cell lung cancer (SCLC) and treated with carboplatin and etoposide for six cycles (Fig. 1A). Although CT imaging during treatment showed a partial response, she developed progressive disease, with enlarging liver lesions and adrenal lesions soon after completion of chemotherapy. She then received a topotecan-based investigational drug combination with no response.
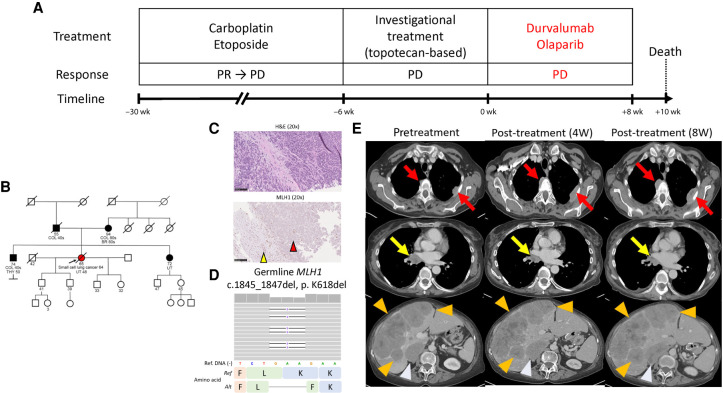
Figure 1.
Patient clinical course and germline mismatch repair (MMR) deficiency. (A) Patient clinical course, treatment, response, and timelines. (B) Patient genogram. (C) Hematoxylin and eosin staining (top) and MLH1 IHC (bottom). The red arrowhead in the bottom panel indicates loss of MLH1 expression on tumor cells, whereas the yellow arrowhead indicates intact MLH1 expression in normal hepatic cells. Black bars, 100 µm. (D) Integrated Genomics Viewer panel of germline MLH1 mutation. (E) Contrast-enhanced computed tomography (CT) scans pretreatment (left), 4 wk (middle) after treatment, and 8 wk (right) after treatment. Red arrows, yellow arrows, orange arrowheads, and light blue arrowheads indicate pleural lesions, right mediastinal lymph nodes, liver lesions, and right adrenal mass, respectively. (dMMR) Mismatch repair deficiency, (W) week, (PR) partial response, (PD) progressive disease, (COL) colorectal cancer, (BR) breast cancer, (THY) thyroid cancer, (UT) endometrial cancer, (H&E) hematoxylin and eosin, (IHC) immunohistochemistry staining, (Ref) reference, (Alt) alteration.
At presentation to our clinic, she was noted to have a personal history of endometrial cancer at age 48, and a family history of early-onset colorectal cancers and endometrial cancer (Fig. 1B). Given the strong family history, which met the Amsterdam II clinical criteria for suspected Lynch syndrome (Vasen et al. 1999), we assessed for evidence of dMMR, revealing loss of MLH1 protein expression by IHC (Fig. 1C). Follow-up testing demonstrated an MLH1 deleterious germline mutation (c.1845_1847del, p. K618del; Fig. 1D). Given the dMMR tumor, she was enrolled on an investigational immunotherapy-combination of PD-L1 inhibitor and poly(ADP-ribose) polymerase (PARP) inhibitor (Thomas et al. 2019). The trial was designed based on preclinical observations of DNA damage caused by PARP inhibitors increasing cytosolic DNA, which activated the cyclic GMP-AMP synthase (cGAS)/stimulator of interferon genes (STING) innate immune pathway, ultimately yielding T-cell recruitment and PD-L1 expression (Sen et al. 2019). CT scan after 4 wk on treatment showed a mixed response, with increase in size of pleural, liver, and adrenal lesions and shrinkage of a right lung hilar mass (Fig. 1E). Given the possibility of immune-related pseudoprogression, and because the patient was asymptomatic, treatment was continued. She subsequently developed worsening right upper quadrant pain, nausea, and anorexia. A CT scan 8 wk after treatment showed continued worsening of pleural, hepatic, and right adrenal masses. The right hilar lymph node response was maintained (Fig. 1E). She was taken off treatment for disease progression and died 2 wk later. We sought to investigate the basis of primary resistance to immune checkpoint inhibitor combination in this patient, despite the tumor being dMMR (Fig. 2A).
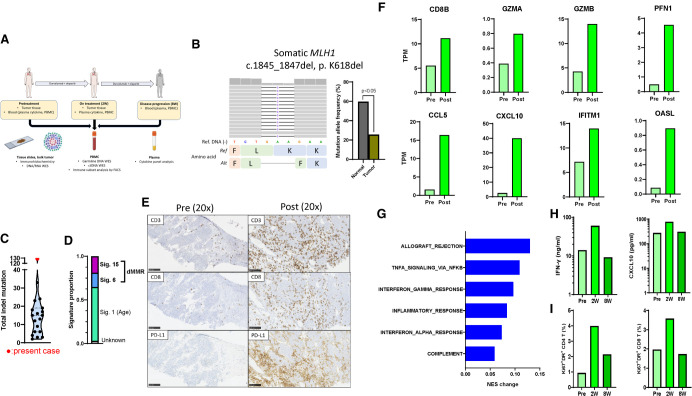
Figure 2.
MLH1 loss in the tumor and induction of cancer immunogenicity with treatment. (A) Schema of tissue sampling and time points. (B) Integrated Genomics Viewer panel of somatic MLH1 mutation and mutation allele frequency in normal (PBMC) and tumor samples. The difference of mutation allele frequency was statistically evaluated by a χ2 test. (C) Total number of indels in the present case and 18 other patients with relapsed small-cell lung cancer enrolled in the clinical trial of durvalumab and olaparib (Thomas et al. 2019). The present case is indicated by the red circle. (D) Proportion of mutational signatures (Alexandrov et al. 2020) in the present case. (E) CD3, CD8, and PD-L1 IHCs in pre- and post-treatment tumors. Scale bars, 100 µm. (F) Expression of cytotoxic T-cell genes (top panels) and interferon-related genes (bottom panels) in pre- and post-treatment tumors. (G) Changes of normalized enrichment scores by single sample gene set enrichment analysis of immune-related hallmark gene sets between pre- and post-treatment tumors. (H) Changes of plasma interferon-γ and CXCL10 levels at pretreatment, 2 wk and 8 wk after treatment. The y-axes are logarithm-transformed. (I) Percentage of activated (Ki67+, HLA-DR+) cells among either CD4+ or CD8+ T cells in PBMCs at pretreatment, 2 wk and 8 wk after treatment. (W) Week, (PBMC) peripheral blood mononuclear cells, (WES) whole-exome sequencing, (TMB) tumor mutational burden, (indel) insertion/deletion, (dMMR) mismatch repair deficiency, (IHC) immunohistochemistry staining, (TPM) transcripts per million, (PD-L1) programmed-death ligand-1, (NES) normalized enrichment score, (IFN-γ) interferon-gamma.
Tumor Mismatch Repair Deficiency and Immune Response Dynamics
Pretreatment tumor biopsy was obtained from one of liver lesions. First, we performed whole-exome sequencing (WES) and RNA sequencing (RNA-seq) of the pretreatment tumor. Consistent with the known loss of MLH1 protein, somatic MLH1 deleterious mutation was observed, with a significantly lower mutation allele frequency compared with the germline variant (Fig. 2B). The somatic copy-number landscape showed complete loss of minor allele of Chromosome 3 and displayed evidence of genomic instability (Supplemental Fig. S1; Cancer Genome Atlas Network 2012), together indicating MLH1 loss of heterozygosity. The pretreatment tumor harbored markedly high indel tumor mutational burden (TMB) (n = 129) compared with tumors of 18 other SCLC patients who enrolled on the same clinical trial (median [95% confidence interval]: 14 indels [9.0–17.4]; Fig. 2C) (Thomas et al. 2019). Indel TMB in this patient was also higher relative to papillary renal cell carcinoma, a cancer type with the highest indel TMB across 19 distinct cancers in a previous report (Turajlic et al. 2017). Total TMB was comparable to other SCLCs (Supplemental Fig. S2), although it remains unclear how total TMB associates with dMMR in SCLC. Further supporting the dMMR tumor phenotype, mutational signature analysis revealed a high proportion of dMMR-related signatures (35.3% signature 6 and 15; Fig. 2D; Alexandrov et al. 2020).
Next, we evaluated changes in tumor immune microenvironment in pre- and post-treatment tumors using exome, transcriptome, and IHC. The post-treatment tumor was obtained 2 wk after treatment from the same liver lesion that was biopsied pretreatment. Total and indel TMB were similar between pre- and post-treatment tumors (Supplemental Fig. S3). Despite the lack of clinical response, the post-treatment tumor was characterized by markedly high intratumoral and stromal CD8 T-cell infiltrates and high PD-L1 expression. In comparison, tumor infiltrating lymphocytes (TILs) were not observed and tumor cells focally expressed PD-L1 in the pretreatment tumor (Fig. 2E). Consistently, up-regulation of cytotoxic T cell–related genes (CD8B, GZMA, GZMB, and PFN) and interferon-stimulated genes (CCL5, CXCL10, IFITM1, and OASL) were observed in the post-treatment compared with the pretreatment tumor (Fig. 2F). Further, single sample gene set enrichment analysis also revealed up-regulation of multiple immune and inflammatory pathways after treatment (Fig. 2G; Supplemental Table S1).
To evaluate the corresponding changes in systemic immunity, we examined the patient's plasma and PBMCs pretreatment, 2 wk (corresponding to the post-treatment biopsy time point) and 8 wk (at disease progression) after treatment. Interferon-gamma (IFN-γ) and IFN-associated chemokine CXCL10 were transiently elevated at 2 wk but returned to pretreatment levels 8 wk after treatment (Fig. 2H). Similarly, activated (Ki67+ HLA-DR+) CD4+ and CD8+ T cells transiently increased at 2 wk but returned to pretreatment levels by 8 wk (Fig. 2I; Supplemental Fig. S4). Changes in other cytokines and immune subsets are shown in Supplemental Figures S5 and S6.
Discovery of Functional Loss of JAK1 Signaling by Compound Heterozygous Mutations
Despite induction of tumor immunity, the dMMR tumor in our patient proved refractory to immune checkpoint inhibitor combination. To investigate the resistance mechanisms, we examined WES and RNA-seq of pre- and post-treatment tumors. Given the discordant clinical response (progression of all lesions except the right hilar lymph node) and because small core biopsies may often not capture the tumor heterogeneity, we also examined ctDNA WES-sampled pretreatment.
Tumor cellularity was high in both pre- and post-treatment tumors (Supplemental Fig. S7). We identified 530 and 539 variants in pre- and post-treatment tumors, respectively. Consistent with dMMR, frameshift indels were prevalent both before and after treatment (65 [12.3%] and 64 [11.9%], respectively). Most variants (n = 425) were shared between the two time points (Supplemental Fig. S8; Supplemental Table S2). We discovered a JAK1 frameshift deletion (c.1289delC p. Pro430Argfs*2) in both pre- and post-treatment tumors (variant allele frequency [VAF] = 43.8% and 39.7%, respectively) (Table 1; Fig. 3A). Notably, another JAK1 frameshift deletion (c.2580delA, p. Lys860Asnfs*16) was found in the post-treatment tumor (VAF = 39.7%) but was not detected in the pretreatment tumor (Table 1; Fig. 3A,B). Notably, both JAK1 variants were found in the pretreatment ctDNA, but with low VAF (Pro430Argfs*2 variant: 0.5%; Lys860Asnfs*16 variant: 8.6%, respectively). Both JAK1 variants were located upstream of the kinase domains and likely truncate the protein or cause nonsense-mediated decay (Fig. 3A). JAK1 Pro430Argfs*2 and Lys860Asnfs*16 have previously been reported as recurrent mutations (Fig. 3A), especially in microsatellite unstable tumors (Albacker et al. 2017), and reported as nonfunctional mutations (Ren et al. 2013). We also assessed changes in VAF of tumor-derived RB1 mutation in pre- and post-treatment tumors. The RB1 p.Pro374fs variant decreased in the post-treatment tumor compared with the pretreatment tumor (77.6% and 63.2% pre- and post-treatment, respectively), suggesting that this major clone did not expand, in contrast to the subclone with JAK1 composite mutations.
Table 1.
Variant table
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | COSMIC genomic mutation ID | Genotype | Samples |
|---|---|---|---|---|---|---|---|---|
| JAK1 | Chr 1 |
NC_000001.10:g.65325833delG c.1289del |
p.P430Rfs | Frameshift | Deletion | COSV61087940 | Heterozygous | Pretreatment tumor, post-treatment tumor, pretreatment ctDNA |
| JAK1 | Chr 1 |
NC_000001.10:
65306997delT c.2580del |
p.P860Nfs | Frameshift | Deletion | COSV61086989 | Heterozygous | Post-treatment tumor, pretreatment ctDNA |
(JAK1) Janus kinase 1, (COSMIC) Catalogue of Somatic Mutations in Cancer, (ctDNA) circulating tumor DNA.
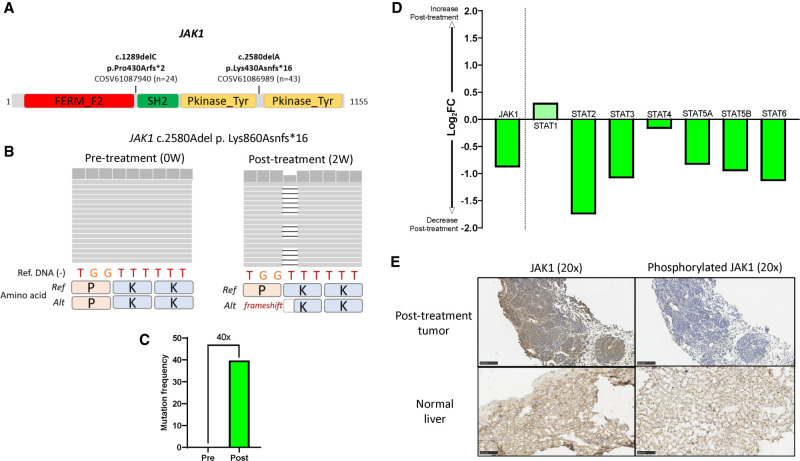
Figure 3.
Functional loss of signaling by compound heterozygous JAK1 mutation. (A) Schematic representation of JAK1 gene and the two variants observed in pre- and post-treatment tumors. The numbers next to the COSMIC identifiers indicate the numbers of mutations found in COSMIC (Tate et al. 2019) (https://cancer.sanger.ac.uk/cosmic/). (B) Integrated Genomics Viewer panels of the JAK1 Lys860Asn*16 variant in pre- and post-treatment tumors. (C) Mutation frequency comparison of JAK1 Lys860Asn*16 variant between pre- and post-treatment tumors. (D) Changes in gene expressions of JAK1 and STATs between pre- and post-treatment tumors. The y-axis shows logarithm 2 transformed fold changes of gene expression between pre- and post-treatment tumors. (E) Immunohistochemistry stainings (IHCs) of JAK1 (left) and phosphorylated JAK1 (right) proteins in the post-treatment tumors in the present case (top) and hepatic cells in a healthy donor (bottom). Scale bars, 100 µm. (JAK1) Janus kinase 1, (COSMIC) Catalogue of Somatic Mutations in Cancer, (W) weeks, (Ref) reference, (Alt) alteration, (STAT) signal transducers and activators of transcription, (FC) fold change.
Homozygous JAK1 mutations are notable given reports of immunotherapy resistance linked to emergence of mutations in the IFN-γ signaling pathway (Zaretsky et al. 2016). However, JAK1 heterozygous mutations would not carry the same functional significance as homozygous mutations, as signaling would still occur upon IFN exposure through the wild-type JAK1 protein from the nonmutated allele. To examine whether the JAK1 mutations we found were present in different alleles, resulting in a compound heterozygote with likely loss of function versus two variants on the same allele, with less definitive impact, we examined the two JAK1 frameshift variants in RNA-seq. Post-treatment tumor RNA-seq showed only the JAK1 frameshift Pro430Argfs*2 variant, whereas the other JAK1 Lys860Asnfs*16 variant was not detected in transcripts (Supplemental Fig. S9), suggesting that the two JAK1 variants are located in different alleles.
To assess the impact of the mutations on JAK1 function, we examined mRNA expression of JAK1 and downstream signal transducers and activators of transcription (STAT). Expression of JAK1 itself and most STAT family genes except STAT1 were down-regulated in the post-treatment compared with the pretreatment tumor (Fig. 3D). Upon activation, JAKs trans-phosphorylate each other at tyrosines within the kinase domain and phosphorylate the cytoplasmic tail of the receptor (O'Shea et al. 2015). To further understand the effect of the mutations on JAK1, we assessed JAK1 and phosphorylated JAK1 protein expression in the post-treatment tumor. Whereas JAK1 was diffusely expressed on the tumor cells, phosphorylation of JAK1 was totally absent compared with hepatic cells in a healthy donor (Fig. 3E). Considering that the composite JAK1 mutations are located upstream of the kinase domain (Fig. 3A), it is likely that they affected the function and downstream signaling of JAK1, rather than JAK1 protein expression.
DISCUSSION
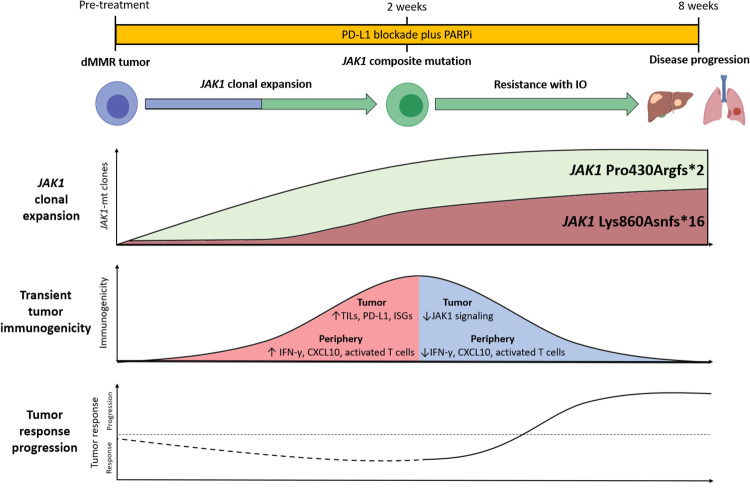
Patients with dMMR tumors experience highly variable responses, and roughly one-half to two-thirds are refractory to PD-1/PD-L1-targeted therapies. Understanding the molecular mechanisms of primary resistance may help identify dMMR tumors that are unlikely to respond to immunotherapy. Here we study a patient with dMMR SCLC and JAK1 compound heterozygous mutations that were associated with primary resistance to immunotherapy combination treatment. Loss of JAK1 function has previously been implicated in primary and acquired immunotherapy resistance in melanoma (Zaretsky et al. 2016), colorectal cancer (Grasso et al. 2018), and endometrioid cancer (Gulhan et al. 2020). To our knowledge, this report provides the most detailed look yet of impaired JAK1 signaling in the context of immunotherapy-resistant dMMR tumors (Fig. 4). Our findings support the notion that tumors growing out under immune selective pressure likely start out as a heterogeneous mixture, in which sensitive tumor cells are eliminated and resistant tumor cells grow progressively (Fig. 4).
Figure 4.
Schema of JAK1 function impacting mutations leading to immunotherapy resistance. (JAK) Janus kinase, (PD-L1) programmed death-ligand 1, (PARPi) Poly (ADP-ribose) polymerase inhibition, (dMMR) mismatch repair deficiency, (IO) immune checkpoint blockade, (mt) mutant, (TILs) tumor infiltrating lymphocytes, (ISGs) interferon stimulated genes.
Mechanisms underlying the substantial diversity among responses of dMMR tumors to immunotherapy are poorly understood. Mandal et al. (2019) reported that indel mutational load rather than the burden of missense mutations were associated with the extent of immunotherapy response among dMMR tumors. dMMR tumors also display frequent mutations in immune genes, such as those related to antigen presentation machinery, including biallelic losses of B2M and HLA genes (Grasso et al. 2018), suggesting that these tumors frequently undergo immunoediting driven by selection for genetic events allowing immune escape. Consistently, recent reports have identified nonfunctional genomic alterations of B2M and JAK1 in dMMR tumors resistant to immune checkpoint blockade (Gurjao et al. 2019; Gulhan et al. 2020). Indeed, JAK1 frameshift indels including the variants identified in the present study are found more frequently in dMMR tumors than in MMR-proficient ones (Albacker et al. 2017; Gulhan et al. 2020). Although several other mechanisms regulating immune response have been proposed in dMMR tumors, none of them have been shown to impact clinical outcomes to immunotherapy (Supplemental Table S3).
Despite the lack of clinical response, we noted a striking increase in tumor and systemic immunogenicity 2 wk after treatment, as demonstrated by the marked increases in TILs and IFN-related gene expression and accompanied by increases in tumor PD-L1 expression, plasma IFN-γ, and CXCL10, and activated T cells in the peripheral blood. Although a tumor sample from beyond the 2-wk time point was not available, the blood-based markers point to the immune response being transient; IFN-γ, CXCL10, and activated T cells returned to pretreatment levels 8 wk after treatment. Clonal expansion of JAK1-mutated resistant tumor cells may explain the brief and nonsustained nature of the immune activation. Transient immunogenicity may also have resulted from PARP inhibition, given activation of the cGAS and STING pathways and enhanced IFN-stimulated gene expression observed in preclinical models following PARP inhibition (Pantelidou et al. 2019; Sen et al. 2019). However, clinical studies to date suggest that the STING pathway activation may not be a dominant effect of PARP inhibition in patients (Lampert et al. 2020).
The present study had several limitations. First, we did not observe genomic alterations in genes with known roles in immune evasion (e.g., STK11 and LKB1) or antigen presentation (e.g., B2M, TAP1, TAP2, and HLA) in tumors or ctDNA. However, other underlying resistance mechanisms cannot be excluded (Supplemental Table S3). Although the changes in allelic burden of the compound heterozygous JAK1 mutation suggest tumor immunoediting driven by selection of the JAK1 variant as the basis of immunotherapy resistance, the studies here do not causally implicate the alteration for the clinical phenotype. Second, WES of pre- and post-treatment core needle tumor biopsies from the liver lesion may not capture the biological characteristics of widespread metastatic cancer involving mediastinal lymph nodes, pleura, and adrenals. This limitation may partly be offset by profiling of ctDNA at multiple time points. We observed a moderate level of concordance between ctDNA and tumor, with most of the tumor variants detected in ctDNA (368 variants were shared including 69.4% of tumor variants and 41.4% of ctDNA variants; Supplemental Fig. S10). Further, consistent with the notion of ctDNA providing a global view representing genomic changes from diverse metastatic sites (Moreno et al. 2019), ctDNA also captured variants that were not detected in the tumor (58.6% of ctDNA variants were unique to ctDNA; n = 521). Third, the tumor transcriptome data generally supported reduced post-treatment JAK1 signaling, with the notable exception of STAT1 expression, which increased post-treatment (Fig. 3D). Several cell-intrinsic pathways activate STAT1 independently of interferons (Ramsauer et al. 2002; Dempoya et al. 2012; Luu et al. 2014; Collins-McMillen et al. 2017), including the cGAS-STING pathway (Kreienkamp et al. 2018) up-regulated by PARP inhibition (Sen et al. 2019), and may have contributed to the isolated STAT1 up-regulation. Finally, the impact of dMMR on pathogenesis, prognosis, and patterns of response and resistance to anticancer therapies in SCLC is not defined, in contrast to colorectal or endometrial carcinomas, which are often considered as “proof of principle” of the dMMR paradigm. Thus, the putative resistance mechanism identified here in a dMMR SCLC patient needs validation in the setting of these more typical dMMR tumors.
In conclusion, we describe a patient with dMMR tumor resistant to immune checkpoint blockade despite exhibiting molecular features of dMMR tumors. Functional characterization and allelic burden tracking of the compound heterozygous JAK1 mutation suggests tumor immunoediting driven by selection of the JAK1 variant may underlie immune escape. Our findings have implications for primary resistance to immune checkpoint blockade in dMMR tumors.
METHODS
Patient and Treatment
The patient was enrolled in a single-arm phase II study (NCT02484404) and was treated with PD-L1 inhibitor durvalumab (1500 mg administered every 4 wk) and PARP inhibitor olaparib (300 mg administered twice daily) (Thomas et al. 2019). Written informed consent was obtained from the patient. Tumor response was assessed using Response Evaluation Criteria in Solid Tumors (RECIST v 1.1). The study was approved by the Office for Human Research Protections at the Department of Health and Human Services.
Tumor, Germline, and ctDNA Sequencing
DNA and RNA were sequenced from formalin-fixed, paraffin-embedded (FFPE) tumor tissue samples using WES and RNA-seq, and whole-exome sequenced from ctDNA, and PBMCs for germline control. Detailed methods of sequencing and analyses are provided in the Supplemental Methods.
Immunohistochemistry
Five-micron sections of FFPE tissue were assessed for PD-L1 expression, TILs, JAK1, and phosphorylated JAK1 protein expression using the following antibodies and detection methods following the manufacturer's protocol: PD-L1 (1:3 dilution, clone SP142 [Springer Biosciences], Leica Bond [Leica Biosystems]), CD3 (predilute, clone 2GV6, Ventana and Ventana BenchMark Ultra [Ventana Medical Systems]), and CD8 (1:25, clone CD8/144B [Dako], Ventana BenchMark Ultra [Ventana Medical Systems]), JAK1 (1:500), and phosphorylated JAK1Tyr1034/1035 (1:100) (Cell Signaling Technologies Inc.).
Plasma Cytokine Analyses
Plasma cytokine levels were measured pretreatment, 2 and 8 wk after treatment using the Meso-Scale Discovery (MSD) Human Biomarker Group 1 U-Plex and Human TGF-beta Combo kits. Initially 74 cytokines were measured and 18 cytokines (IFN-γ, IL-8, IL-9, IL-10, IL-13, CXCL10, I-TAC, IL-21, IL-29/IFN-λ1, MCP-2, MCP-3, MCP-4, MIP-1α, MIP-1β, FLT3L, IL-1RA, MIF, M-CSF) were finally analyzed after quality control.
PBMC Immune Subset Analysis
Blood samples were collected in an 8-mL sodium citrate CPT tube (BD) pretreatment, 2 wk, and 8 wk after treatment. PBMCs were isolated and analyzed as previously described (Tomita et al. 2016). Flow cytometric analyses were performed using a MACSQuant Analyzer (Miltenyi Biotec). Data were analyzed using FlowJo software (FlowJo, LLC) (Ziegler-Heitbrock et al. 2010; Zawada et al. 2011). Please refer to Supplemental Methods for more details of methods.
Additional Information
Data Deposition and Access
Raw WES and RNA-seq data are deposited in the database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap/) under accession number phs002089.v1.p1. The JAK1 variants were submitted to COSMIC (https://cancer.sanger.ac.uk/cosmic) and can be found using COSMIC identifier COSP48518.
Ethics Statement
Written informed consent was obtained from the patient. The study was approved by the Office for Human Research Protections at the Department of Health and Human Services.
Acknowledgments
We thank Dr. Johnathan M. Hernandez of Surgical Oncology Branch, National Cancer Institute (NCI), for providing normal liver tissue. This study was supported by the Center for Cancer Research, the Intramural Program of the NCI (ZIA BC 011793).
Author's Contributions
N.T. and A.T. conceptualized the study. N.T., V.N.R., L.P., C.T., and A.T. curated the data. N.T., V.N.R., and L.P. performed the formal analysis. A.T. supervised. N.T., V.N.R., L.P., C.T., and A.T. validated the data. N.T., V.N.R., L.P., C.T., and A.T. investigated. N.T., V.N.R., L.P., S.K., R.E.C., H.A.Y., J.B.T., and A.T. visualized the study. N.T. and A.T. wrote the original draft. N.T. and A.T. administered the project. All authors came up with the methodology and performed writing review and editing.
Funding
A.T. reports grants from AstraZeneca and EMD Serono to the National Cancer Institute/National Institutes of Health. The other authors declare no conflict of interest.
Competing Interest Statement
A.T. reports grants from AstraZeneca and EMD Serono to the National Cancer Institute/National Institutes of Health. The other authors declare no conflict of interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Albacker LA, Wu J, Smith P, Warmuth M, Stephens PJ, Zhu P, Yu L, Chmielecki J. 2017. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS One 12: e0176181 10.1371/journal.pone.0176181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-McMillen D, Stevenson EV, Kim JH, Lee BJ, Cieply SJ, Nogalski MT, Chan GC, Frost RW III, Spohn CR, Yurochko AD. 2017. Human cytomegalovirus utilizes a nontraditional signal transducer and activator of transcription 1 activation cascade via signaling through epidermal growth factor receptor and integrins to efficiently promote the motility, differentiation, and polarization of infected monocytes. J Virol 91: e00622-17 10.1128/JVI.00622-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempoya J, Matsumiya T, Imaizumi T, Hayakari R, Xing F, Yoshida H, Okumura K, Satoh K. 2012. Double-stranded RNA induces biphasic STAT1 phosphorylation by both type I interferon (IFN)-dependent and type I IFN-independent pathways. J Virol 86: 12760–12769. 10.1128/JVI.01881-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JC, Lin MT, Le DT, Eshleman JR. 2016. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 22: 813–820. 10.1158/1078-0432.CCR-15-1678 [DOI] [PubMed] [Google Scholar]
- Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, Nowak JA, Nishihara R, Qian ZR, Inamura K, et al. 2018. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov 8: 730–749. 10.1158/2159-8290.CD-17-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulhan DC, Garcia E, Lee EK, Lindemann NI, Liu JF, Matulonis UA, Park PJ, Konstantinopoulos PA. 2020. Genomic determinants of de novo resistance to immune checkpoint blockade in mismatch repair-deficient endometrial cancer. JCO Precis Oncol 4: 492–497. 10.1200/PO.20.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurjao C, Liu D, Hofree M, AlDubayan SH, Wakiro I, Su MJ, Felt K, Gjini E, Brais LK, Rotem A, et al. 2019. Intrinsic resistance to immune checkpoint blockade in a mismatch repair-deficient colorectal cancer. Cancer Immunol Res 7: 1230–1236. 10.1158/2326-6066.CIR-18-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp R, Graziano S, Coll-Bonfill N, Bedia-Diaz G, Cybulla E, Vindigni A, Dorsett D, Kubben N, Batista LFZ, Gonzalo S. 2018. A cell-intrinsic interferon-like response links replication stress to cellular aging caused by progerin. Cell Rep 22: 2006–2015. 10.1016/j.celrep.2018.01.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert EJ, Zimmer AS, Padget MR, Cimino-Mathews A, Nair JR, Liu Y, Swisher EM, Hodge JW, Nixon AB, Nichols E, et al. 2020. Combination of PARP inhibitor olaparib, and PD-L1 inhibitor durvalumab, in recurrent ovarian cancer: a proof-of-concept phase 2 study. Clin Cancer Res 26: 4268–4279. 10.1158/1078-0432.CCR-20-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu K, Greenhill CJ, Majoros A, Decker T, Jenkins BJ, Mansell A. 2014. STAT1 plays a role in TLR signal transduction and inflammatory responses. Immunol Cell Biol 92: 761–769. 10.1038/icb.2014.51 [DOI] [PubMed] [Google Scholar]
- Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, Sabio EY, Makarov V, Kuo F, Blecua P, et al. 2019. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364: 485–491. 10.1126/science.aau0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, Geva R, Gottfried M, Penel N, Hansen AR, et al. 2020. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 38: 1–10. 10.1200/JCO.19.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merck Sharp and Dohme. 2018. KEYTRUDA (pembrolizumab) full prescribing information. Merck Sharp & Dohme Corp, Whitehouse Station, NJ. [Google Scholar]
- Moreno F, Gayarre J, López-Tarruella S, del Monte-Millán M, Picornell AC, Álvarez E, García-Saenz JÁ, Jerez Y, Márquez-Rodas I, Echavarría I, et al. 2019. Concordance of genomic variants in matched primary breast cancer, metastatic tumor, and circulating tumor DNA: the MIRROR study. JCO Preci Oncol 1–16. 10.1200/PO.18.00263 [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. 2015. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med 66: 311–328. 10.1146/annurev-med-051113-024537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelidou C, Sonzogni O, De Oliveria Taveira M, Mehta AK, Kothari A, Wang D, Visal T, Li MK, Pinto J, Castrillon JA, et al. 2019. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov 9: 722–737. 10.1158/2159-8290.CD-18-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsauer K, Sadzak I, Porras A, Pilz A, Nebreda AR, Decker T, Kovarik P. 2002. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci 99: 12859–12864. 10.1073/pnas.192264999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, Wu J. 2013. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep 3: 3042 10.1038/srep03042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, Cristea S, Nguyen T, Diao L, Li L, et al. 2019. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 9: 646–661. 10.1158/2159-8290.CD-18-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Vilimas R, Trindade C, Erwin-Cohen R, Roper N, Xi L, Krishnasamy V, Levy E, Mammen A, Nichols S, et al. 2019. Durvalumab in combination with olaparib in patients with relapsed SCLC: results from a phase II study. J Thorac Oncol 14: 1447–1457. 10.1016/j.jtho.2019.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Lee MJ, Lee S, Tomita S, Chumsri S, Cruickshank S, Ordentlich P, Trepel JB. 2016. The interplay of epigenetic therapy and immunity in locally recurrent or metastatic estrogen receptor-positive breast cancer: correlative analysis of ENCORE 301, a randomized, placebo-controlled phase II trial of exemestane with or without entinostat. Oncoimmunology 5: e1219008 10.1080/2162402X.2016.1219008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M, et al. 2017. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 18: 1009–1021. 10.1016/S1470-2045(17)30516-8 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT. 1999. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116: 1453–1456. 10.1016/S0016-5085(99)70510-X [DOI] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. 2016. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375: 819–829. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawada AM, Rogacev KS, Rotter B, Winter P, Marell RR, Fliser D, Heine GH. 2011. SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood 118: e50–e61. 10.1182/blood-2011-01-326827 [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ, et al. 2010. Nomenclature of monocytes and dendritic cells in blood. Blood 116: e74–e80. 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw WES and RNA-seq data are deposited in the database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap/) under accession number phs002089.v1.p1. The JAK1 variants were submitted to COSMIC (https://cancer.sanger.ac.uk/cosmic) and can be found using COSMIC identifier COSP48518.