Abstract
High-throughput DNA testing is becoming established as a standard diagnostic test in the renal clinic. Previously published studies on cohorts of patients with unexplained chronic kidney disease of a suspected genetic aetiology have suggested a diagnostic yield for genomic sequencing of up to 18%. Here we determine the yield of targeted gene panel in a clinically unscreened cohort of patients referred for percutaneous native renal biopsy. Patients who underwent renal biopsy for investigation of chronic kidney disease were sequenced using a genomic sequencing panel covering 227 genes in which variation is known to be associated with monogenic chronic kidney disease (CKD). Candidate disease-causing variants were assessed for pathogenicity using guidelines from the American College for Medical Genetics and Genomics. Fifty CKD patients were recruited and sequenced. A molecular diagnosis was obtained for two patients (4%). A molecular diagnosis is possible using genomic testing in ∼4% of clinically unscreened patients undergoing renal biopsy. Genetic screening may be useful for diagnosis in a subset of CKD patients but is most valuable when applied to patients with suspected heritable forms of kidney disease.
Keywords: acute tubulointerstitial nephritis, decreased glomerular filtration rate, elevated serum creatinine, glomerulonephritis, heavy proteinuria, hematuria, mild proteinuria, moderate proteinuria, stage 1 chronic kidney disease, stage 2 chronic kidney disease, stage 3 chronic kidney disease, stage 4 chronic kidney disease, stage 5 chronic kidney disease
INTRODUCTION
Genetic testing is becoming increasingly available as a viable first-line diagnostic test in chronic kidney disease (CKD) (Bullich et al. 2018; Harris 2018). Recent studies have shown that genomic sequencing may provide a molecular diagnosis in up to 10% of all patients with CKD and 18% of those who have CKD of unknown origin (Groopman et al. 2019). The diagnostic yield from genetic testing may be even higher in patients with a family history of renal disease (Connaughton et al. 2019). These recent studies sequenced patients referred by treating clinicians who suspected inherited kidney disease. Here, we assess the diagnostic yield of targeted genomic sequencing in a patient cohort early in their diagnostic journey, referred for percutaneous native renal biopsy, without clinical screening for suspected inherited kidney disease.
RESULTS
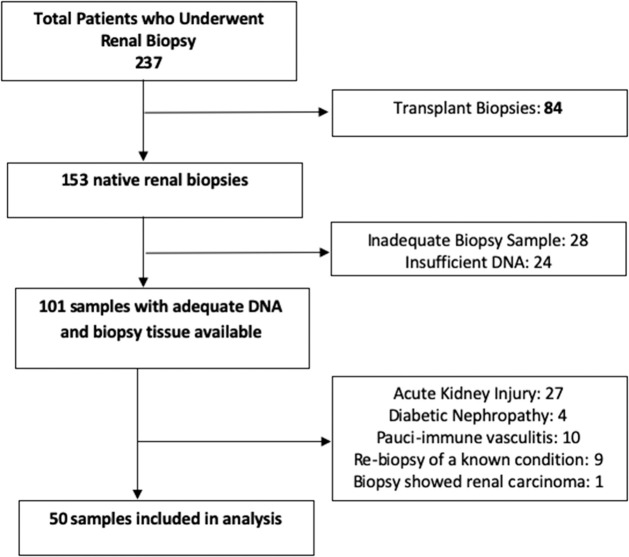
In total, 237 biopsy samples were screened, of which 84 were renal transplant biopsies. Of the remaining 153 native renal biopsies, a further 52 were excluded because DNA was unavailable or because sampling was inadequate for diagnosis at the time of biopsy, or biopsy was not ultimately performed. Following review, another 51 samples were excluded (see Fig. 1). Ultimately, 50 samples underwent sequencing.
Figure 1.
Schematic showing 237 biopsy samples were screened, of which 84 were renal transplant biopsies. Of the remaining 153 native renal biopsies, a further 52 were excluded because DNA was unavailable or because sampling was inadequate for diagnosis at the time of biopsy, or biopsy was not ultimately performed. Following review, another 51 samples were excluded. Ultimately, 50 samples underwent sequencing.
The median age at biopsy was 48 yr; 60% were male. The most common reason for renal biopsy was a deterioration in renal function (22%) measured as a rise in serum creatinine concentration or a fall in glomerular filtration rate (GFR), and the development of nephritic syndrome (30%), followed by nephrotic syndrome (12%), hematuria and proteinuria (12%), and isolated proteinuria (8%) or hematuria (8%). On biopsy, the most common histological diagnosis was IgA nephropathy, accounting for 20 patients (40%), eight (16%) with other forms of glomerulonephritis, six (12%) with arteriosclerosis, eight (16%) with chronic thrombotic microangiopathy (TMA), five (10%) with thin basement membrane nephropathy (TBMN), and one with Alport syndrome (2%). Two patients (4%) had mixed pathological findings. Diagnostic variants were identified in two patients (2/50, 4%) (see Table 1).
Table 1.
Variants considered for pathogenicity (n = 50 total patients)
| Manuscript ID | Gene | GT | Gene inheritance pattern | ACMG classification | ACMG evidence | MAF (gnomAD) | Type | RefSeq | HGVS | AA position | ClinVar ID | Total coverage | Average coverage | % Above 20× | Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 213 | EHHADH | Het | AD | VUS | PVS1 | 7.37E-05 | Frameshift deletion | NM_001966.4 | c.594_595del | p.L198fs | SCV001328294 | 77686 | 194.7 | 100 | IgA nephropathy |
| 8 | FN1 | Comp het (dom) | AD | VUS | PP2, PP3, BS1 | 0.0047 | Missense | NM_212482.4 | c.4486C > T | p.R1496W | SCV001328286 | 75174 | 310.64 | 100 | Nonspecific |
| 8 | FN1 | Comp het (dom) | AD | VUS | PP2, PP3, BS1 | 0.0024 | Missense | NM_212482.4 | c.1070G > A | p.G357E | SCV001328267 | 94718 | 306.53 | 100 | Nonspecific |
| 249 | COL4A1 | Het | AD | VUS | PP2, PP3, BP6 | 0.0029 | Missense | NM_001845.6 | c.161C > T | p.P54L | SCV001328270 | 32220 | 216.24 | 100 | IgA nephropathy |
| 256 | COL4A3 | Het | AR/AD | VUS | PP3 | 4.47E-05 | Missense | NM_000091.5 | c.4700T > G | p.I1567S | SCV001328289 | 35707 | 212.54 | 100 | Arteriosclerosis |
| 261 | CFH | Het | AD | Likely pathogenic | PVS1, PM2 | 0 | Stop-gain | NM_000186.4 | c.2517C > A | p.C839Ter | SCV001305469 | 51994 | 245.25 | 100 | Arteriosclerosis |
| 261 | COL4A4 | Het | AR/AD | Pathogenic | PVS1, PM2, PP3 | 0 | Frameshift deletion | NM_000092.5 | c.4603_4604del | p.Q1535fs | SCV001305364 | 79477 | 228.38 | 100 | Arteriosclerosis |
| 264 | SLC9A3R1 | Het | AD | VUS | PP3, PP5 | 0.0019 | Missense | NM_004252.5 | c.458G > A | p.R153Q | SCV001328287 | 53389 | 232.13 | 100 | TMA |
| 286 | COL4A4 | Het | AR/AD | Pathogenic | PVS1, PS1 | 6.90E-05 | Stop-gain | NM_000092.5 | c.2906C > G | p.S969Ter | SCV001305529 | 35958 | 210.28 | 100 | TBMN |
| 14 | APOA1 | Het | AD | VUS | PM4, BS1 | 0.0002 | Nonframeshift deletion | NM_001318018.2 | c.391_393del | p.131_131del | SCV001328284 | 146178 | 220.15 | 100 | Nonspecific |
| 203 | CFHR5 | Het | AD | VUS | PVS1, BS1 | 0.0021 | Frameshift insertion | NM_030787.4 | c.480dupA | p.P160fs | SCV001328290 | 46564 | 232.82 | 100 | IgA nephropathy |
| 203 | CFHR5 | Het | AD | VUS | PP3, PP5, BS1 | 0.0014 | Missense | NM_030787.4 | c.622T > C | p.C208R | SCV001328296 | 74760 | 318.13 | 100 | IgA nephropathy |
| 16 | DSTYK | Het | AD | VUS | PP3, BS1 | 0.0009 | Missense | NM_015375.3 | c.2776G > T | p.D926Y | SCV001328277 | 63745 | 252.96 | 100 | TMA |
| 16 | WT1 | Het | AD | VUS | PM2, PP3 | 0 | Missense | NM_024426.6 | c.576G > T | p.Q192H | SCV001328293 | 148187 | 212.61 | 100 | TMA |
| 43 | ZNF423 | Het | AR/AD | VUS | PP2, PP3 | 6.92E-05 | Missense | NM_015069.4 | c.2251C > T | p.R751C | SCV001328275 | 944847 | 290.19 | 100 | MPGN |
| 20 | APOA1 | Het | AD | VUS | PM1, PP3 | 0 | Missense | NM_001318018.2 | c.625G > A | p.G209S | SCV001328297 | 944847 | 290.19 | 100 | TBMN |
| 139 | ACTN4 | Het | AD | VUS | PP2, PP3 | 4.51E-05 | Missense | NM_004924.6 | c.928C > T | p.R310W | SCV001328306 | 176954 | 266.5 | 100 | Minimal change disease |
| 142 | DSTYK | Het | AD | VUS | PP3, BS1 | 0.0009 | Missense | NM_015375.3 | c.2776G > T | p.D926Y | SCV001328277 | 75958 | 257.48 | 100 | AIN |
| 152 | EHHADH | Het | AD | VUS | PP3, BS1 | 0.0097 | Missense | NM_001966.4 | c.2108C > T | p.S703F | SCV001328274 | 27071 | 172.43 | 100 | Membranous |
| 158 | UMOD | Het | AD | VUS | PP2, PP3 | 2.03E-05 | Missense | NM_003361.3 | c.1243C > T | p.R415C | SCV001328268 | 31225 | 151.58 | 100 | IgA nephropathy |
| 159 | FREM1 | Het | AD | VUS | PP3, BS1 | 0.0001 | Missense | NM_144966.7 | c.1640C > G | p.A547G | SCV001328271 | 74568 | 191.2 | 100 | IgA nephropathy |
| 159 | SOX17 | Het | AD | VUS | PM4, BS1 | 0.0072 | Nonframeshift insertion | NM_022454.4 | c.948_949insCACCAG | p.Q316delinsQHQ | SCV001328307 | 105825 | 108.32 | 100 | IgA nephropathy |
| 1 | CFHR1 | Hom | AR/AD | VUS | PVS1, PP3 | 0.0018 | Splicing | NM_002113.3 | c.790 + 1G > A | SCV001328302 | 42400 | 170.97 | 100 | IgA nephropathy | |
| 1 | COL4A3 | Het | AR/AD | VUS | PP3 | 0.0003 | Missense | NM_000091.5 | c.1886C > T | p.T629M | SCV001328273 | 64232 | 302.98 | 100 | IgA nephropathy |
| 1 | FN1 | Het | AD | VUS | PP2, PP3, BS1 | 0.0002 | Missense | NM_212482.4 | c.5954C > A | p.P1985H | SCV001328295 | 86861 | 277.51 | 100 | IgA nephropathy |
| 1 | FN1 | Het | AD | VUS | PP2, PP3 | 5.28E-05 | Missense | NM_212482.4 | c.3130G > A | p.V1044M | SCV001328281 | 36521 | 262.74 | 100 | IgA nephropathy |
| 175 | PKD1 | Het | AD | VUS | PP3, PP5 | 0.0009 | Missense | NM_001009944.3 | c.12460C > T | p.R4154C | SCV001328269 | 141278 | 263.58 | 100 | TBMN |
| 186 | SLC7A9 | Het | AR/AD | VUS | PVS1, PM2 | 0 | Stop-gain | NM_001985.3 | c.292C > T | p.R98C | SCV001328279 | 83790 | 301.4 | 100 | IgA nephropathy |
(GT) Genotype, (ACMG) American College of Medical Genetics and Genomics, (MAF) minor allele frequency, (HGVS) Human Genome Variation Society, (AA) amino acid, (AD) autosomal dominant, (VUS) variant of uncertain significance, (AR) autosomal recessive, (Het) heterogeneous, (TMA) thrombotic microangiopathy, (MPGN) membranoproliferative glomerulonephritis, (TBMN) thin basement membrane nephropathy, (Hom) homogeneous, (AIN) acute interstitial nephritis.
Patient 261
This 40-yr-old female patient had an American College of Medical Genetics and Genomics (ACMG)-classified pathogenic (PVS1, PM2, PP3) heterozygous frameshift deletion in Collagen Type IV Alpha 4 Chain (COL4A4) (NM_000092.4:exon47:c.4603_4604del: p.Q1535fs), suggestive of autosomal dominant Alport-type disease (Phenotype MIM number 203780) or autosomal dominant TBMN. This variant was absent from the gnomAD database (Karczewski et al. 2019). The patient presented with microscopic hematuria and proteinuria during pregnancy, along with pregnancy-induced hypertension. Her hypertension resolved postpregnancy, but she had persistent proteinuria and hematuria. Her renal function was preserved. The patient also carried an ACMG- Pathogenic (PVS1 (null variant), PS1 (previously established pathogenic variant), PM2 (absent from controls)) stop-gain variant in Complement Factor H (CFH) (NM_000186.3:exon16:c.C2517A:p.C839Ter). Patient 261 presented with hematuria and proteinuria and notably had a low C3 level (consistent with the presence of the pathogenic CFH variant). Her proteinuria improved with renin-angiotensin-aldosterone-system (RAAS) blockade but did not entirely resolve. Biopsy showed TBMN as well as mild arteriosclerosis and atherosclerosis, prominent double contour formation, and TMA, with 2/10 sclerosed glomeruli. The patient had no history of hearing loss and no family history of kidney disease or hearing loss. Sixty months of follow-up did not reveal significant loss of renal function over time. Parents were unavailable for further testing. We conclude that this patient has a thin basement membrane causing hematuria as a result of a pathogenic variant in COL4A4 as well as low C3 as a result of a pathogenic variant in CFH.
Patient 286
A heterozygous stop-gain variant in COL4A4 (NM_000092.4:exon32:c.2906C > G: p.S969Ter) was identified in a 39-yr-old female patient that was classified using ACMG guidelines as Pathogenic (PVS1 [null variant], PS1 [previously established pathogenic variant]). The patient had normal renal function, with a history of loin pain and microscopic hematuria. She had been treated multiple times for urinary tract infection on the basis of dipstick hematuria but did not self-report any other symptoms of urinary tract infections. The patient reported an extensive family history of loin pain and at least one relative with advanced CKD. She did not have any self-reported hearing or visual disturbance. Biopsy results indicated TBMN. We conclude that this patient has thin basement membrane nephropathy as a result of the presence of a heterozygous COL4A4 variant.
Variants of Unknown Significance
Additionally, 25 ACMG-classified variants of uncertain significance (VUSs) were identified in 18 patients, including four loss-of-function variants and two truncating variants (see Table 1). Five patients without a molecular diagnosis carried more than one VUS. An SLC7A9 (solute carrier family 7, member 9) stop-gain variant (NM_001985:exon3: c.C292T:p.R98C) in patient 186, was classified as Likely Pathogenic using ACMG guidelines, but following clinical review was determined to be a poor phenotypic match and reclassified as a VUS.
DISCUSSION
Genetic testing using DNA from peripheral blood in an undifferentiated population of patients undergoing renal biopsy led to a diagnostic rate of 4% in a cohort of 50 patients. Testing in a larger group of patients is merited. Whole-exome and -genome sequencing may increase the rate of diagnosis; however, panel-based sequencing is already in widespread use as a first-line test for screening those with suspected inherited kidney disease and reflects current clinical practice (Mallett et al. 2017; Bullich et al. 2018; Lata et al. 2018; Heyne et al. 2019). We expect the diagnostic yield to rise as databases mature and further evidence emerges in the literature.
Both patients carried heterozygous, diagnostic variants in COL4A4. Both had histology consistent with TBMN. Although this gene has typically been associated with recessive Alport syndrome, multiple reports (Longo et al. 2002; Marcocci et al. 2009; Hines et al. 2018) have demonstrated that patients with heterozygous COL4A4 or COL4A3 variants can develop significant renal disease and can benefit from early initiation of RAAS blockade (Stock et al. 2017).
The diagnostic yield obtained (4%) is lower than that reported in previous studies of CKD patients (10%–18%) (Groopman et al. 2019), but this yield is notable given that these patients are not clinically screened for suspected heritable forms of kidney disease. These results suggest that genetic screening may be useful for diagnosis in a subset of CKD patients. However, without careful selection, clinical acumen, and examination of the pedigree by an experienced clinician or clinical geneticist, the yield of testing in an unscreened cohort may be low. An effort to define the health economic as well as clinical utility of genomic testing in unscreened cohorts of CKD patients may be an area for future research.
METHODS
Samples were obtained from the North Dublin Renal Biobank (NDRBB), which was established in 2010 to obtain tissue samples from patients with renal disease. Blood, urine, and renal tissue samples were collected prospectively from those undergoing percutaneous renal biopsy. We recruited sequential individuals who underwent renal biopsy for investigation of chronic kidney disease in Beaumont Hospital between 2010 and 2018, if they were over the age of 18 and capable of giving informed consent. Patients were excluded from analysis if:
They underwent transplant renal biopsy.
They underwent rebiopsy to reassess a known condition.
They were thought to have an acute kidney injury (AKI) secondary to a defined insult. This was considered to be the case if they had an acute rise in creatinine and a diagnosis on biopsy of acute tubular necrosis (ATN) or acute interstitial nephritis (AIN).
They had a positive anti-neutrophilic cytoplasmic autoantibody (ANCA) test and a biopsy showing pauci-immune vasculitis.
They were a known diabetic and had a diagnosis consistent with diabetic nephropathy.
DNA was extracted from blood lymphocytes. Genomic sequencing was performed in-house, with library preparation using a previously described targeted renal disease gene panel (Supplemental Table 1) (Cormican et al. 2019). This sequencing method is unable to detect small insertions and deletions in the variable number tandem repeat region of MUC1. Exonic and splicing variants were prioritized for multidisciplinary team discussion if they had a minor allele frequency (MAF) of <1%. Synonymous variants were excluded from analysis. Variant pathogenicity was classified according to the ACMG guidelines (Richards et al. 2015). A variant was classified as diagnostic if it was categorized by the ACMG guidelines as “Likely Pathogenic” or “Pathogenic” and was a good phenotypic match. Sequencing and variant interpretation was conducted in a research (nonaccredited) capacity; diagnostic variants were confirmed using an accredited test from a service provider and reported back to the patients.
ADDITIONAL INFORMATION
Data Deposition and Access
All variants discussed in this manuscript have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (see Table 1) and requests for access to raw sequence data can be made via direct contact with the corresponding author.
Ethics Statement
Written, informed consent was obtained from all participants of this study. Ethical approval was provided from the Ethics committee of Beaumont Hospital, Dublin, Ireland (REC 12/75).
Acknowledgments
We also acknowledge the participation of the patients and their families in this study.
Author Contributions
K.A.B., S.L.M., G.L.C., C.G., and P.J.C. conceived the idea and planned the experiments. S.L.M., P.J.C., and D.S. facilitated recruitment of patients to this study. A.M.D., B.D., and S.L.M. assessed the renal biopsy samples. S.L.M. and K.A.B. drafted the manuscript with support from P.J.C. and G.L.C. K.A.B. conducted the sequencing and bioinformatic analysis of sequencing data. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Funding
This research was funded in part by the Irish Research Council and Punchestown Kidney Research Fund (EPSPD/2019/213) and the Royal College of Surgeons in Ireland STAR Hermitage MD programme.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Andrew Mallett
Hugh J. McCarthy
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Bullich G, Domingo-Gallego A, Vargas I, Ruiz P, Lorente-Grandoso L, Furlano M, Fraga G, Madrid Á, Ariceta G, Borregán M, et al. 2018. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 94: 363–371. 10.1016/j.kint.2018.02.027 [DOI] [PubMed] [Google Scholar]
- Connaughton DM, Kennedy C, Shril S, Mann N, Murray SL, Williams PA, Conlon E, Nakayama M, van der Ven AT, Ityel H, et al. 2019. Monogenic causes of chronic kidney disease in adults. Kidney Int 95: 914–928. 10.1016/j.kint.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormican S, Connaughton DM, Kennedy C, Murray S, Živná M, Kmoch S, Fennelly NK, O'Kelly P, Benson KA, Conlon ET, et al. 2019. Autosomal dominant tubulointerstitial kidney disease (ADTKD) in Ireland. Ren Fail 41: 832–841. 10.1080/0886022X.2019.1655452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, Li Y, Zhang J, Nestor J, Krithivasan P, et al. 2019. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 380: 142–151. 10.1056/NEJMoa1806891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PC. 2018. The time for next-generation molecular genetic diagnostics in nephrology is now! Kidney Int 94: 237–239. 10.1016/j.kint.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyne HO, Artomov M, Battke F, Bianchini C, Smith DR, Liebmann N, Tadigotla V, Stanley CM, Lal D, Rehm H, et al. 2019. Targeted gene sequencing in 6994 individuals with neurodevelopmental disorder with epilepsy. Genet Med 21: 2496–2503. 10.1038/s41436-019-0531-0 [DOI] [PubMed] [Google Scholar]
- Hines SL, Agarwal A, Ghandour M, Aslam N, Mohammad AN, Atwal PS. 2018. Novel variants in COL4A4 and COL4A5 are rare causes of FSGS in two unrelated families. Hum Genome Var 5: 15 10.1038/s41439-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 [DOI] [Google Scholar]
- Lata S, Marasa M, Li Y, Fasel DA, Groopman E, Jobanputra V, Rasouly H, Mitrotti A, Westland R, Verbitsky M, et al. 2018. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med 168: 100–109. 10.7326/M17-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo I, Porcedda P, Mari F, Giachino D, Meloni I, Deplano C, Brusco A, Bosio M, Massella L, Lavoratti G, et al. 2002. COL4A3/COL4A4 mutations: from familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int 61: 1947–1956. 10.1046/j.1523-1755.2002.00379.x [DOI] [PubMed] [Google Scholar]
- Mallett AJ, McCarthy HJ, Ho G, Holman K, Farnsworth E, Patel C, Fletcher JT, Mallawaarachchi A, Quinlan C, Bennetts B, et al. 2017. Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int 92: 1493–1506. 10.1016/j.kint.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Marcocci E, Uliana V, Bruttini M, Artuso R, Silengo MC, Zerial M, Bergesio F, Amoroso A, Savoldi S, Pennesi M, et al. 2009. Autosomal dominant Alport syndrome: molecular analysis of the COL4A4 gene and clinical outcome. Nephrol Dial Transplant 24: 1464–1471. 10.1093/ndt/gfn681 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J, Kuenanz J, Glonke N, Sonntag J, Frese J, Tonshoff B, Hocker B, Hoppe B, Feldkotter M, Pape L, et al. 2017. Prospective study on the potential of RAAS blockade to halt renal disease in Alport syndrome patients with heterozygous mutations. Pediatr Nephrol 32: 131–137. 10.1007/s00467-016-3452-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants discussed in this manuscript have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (see Table 1) and requests for access to raw sequence data can be made via direct contact with the corresponding author.