Abstract
We report a case of a slow-growing, diffuse, infiltrating glioma in the right brainstem of a 9-yr-old boy. The tumor was negative by immunohistochemical staining for histone H3 K27M, BRAF V600E, and IDH1 R132H mutations. Fluorescence in situ hybridization did not reveal a BRAF duplication. Genomic profiling of the tumor, by DNA methylation array and cancer whole-exome and transcriptome sequencing, was performed. This analysis showed copy-number alterations, including gains of several chromosomes. In addition, a novel fusion involving the first 17 exons of FGFR2 fused to exon 2 of VPS35 was identified. This novel fusion is predicted to result in activation of fibroblast growth factor receptor (FGFR) signaling and is potentially targetable using FGFR inhibitors. This tumor expands the spectrum of pediatric diffuse gliomas.
Keywords: neoplasm of the central nervous system
CASE PRESENTATION
The patient is a 9-yr-old boy who was found to have abnormal eye movements at 1 yr of age. At that time, magnetic resonance imaging (MRI) revealed a right-sided brainstem lesion (not shown). The patient was monitored by annual MRIs that showed slow progression, and he remained clinically stable. At age 9, the episodes of abnormal eye movements, including his eyes “turning in” and horizontal nystagmus, increased in frequency with no other change in mental status or behavior. No other symptoms such as diplopia, blurry vision, headache, dizziness, or nausea were reported. MRI showed a heterogeneous, nonenhancing lesion in the right medulla with an exophytic component in the fourth ventricle (Fig. 1A,B). The cystic components within the lesion had increased compared to an MRI from the year before (not shown). Partial resection of the lesion was performed. Postoperatively, the patient is clinically stable and imaging studies show no progression (not shown).
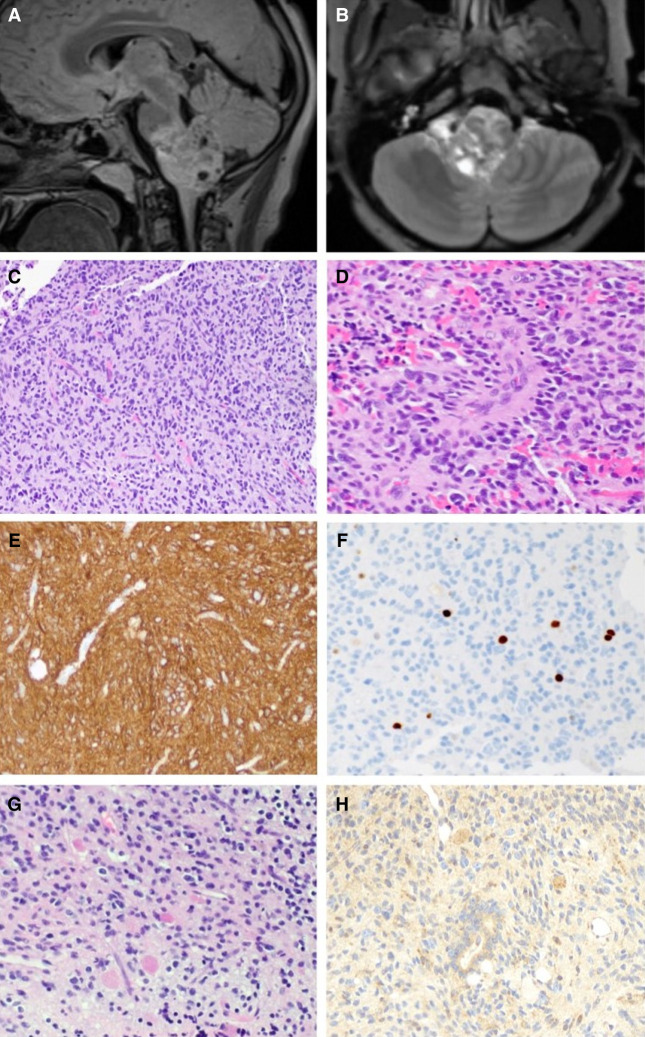
Figure 1.
Radiographic and histologic features of the tumor. Preoperative sagittal T2 FLAIR (A) and axial postcontrast (B) views of preoperative MRI showing a heterogeneous mass centered in the medulla and projecting into the fourth ventricle. (C) A representative hematoxylin and eosin–stained section reveals a diffusely infiltrating glial neoplasm with round-to-oval nuclei and variable perinuclear clearing amid thin-walled capillaries. (D) Some neoplastic cells are arranged around blood vessels. Several tumor cells have vesicular nuclei and mild-moderate pleomorphism. (E) The neoplastic cells are strongly and diffusely positive for glial fibrillary acidic protein (GFAP). (F) The Ki-67 antibody labels a subset of cells, with 2.4% of cells staining positive in this field. (G) Abundant eosinophilic granular bodies are scattered throughout the neoplasm. (H) A VPS35 monoclonal antibody strongly highlights the eosinophilic granular bodies, and variable immunoreactivity is seen in the tumor cells. Magnifications, 20× (C,E–H) and 40× (D).
Hematoxylin and eosin–stained sections of the resection specimen reveal a diffusely infiltrating glial neoplasm consisting of pleomorphic tumor cells, with irregular, round-to-ovoid, hyperchromatic nuclei and variable perinuclear clearing, in a background with thin-walled capillaries (Fig. 1C) and calcifications. Rare mitoses are present (not shown); however, glomeruloid vascular proliferation and necrosis are not identified. Scattered perivascular pseudorosettes are seen (Fig. 1D). Abundant eosinophilic granular bodies (Fig. 1G) and occasional Rosenthal fibers (not shown) are present. A glial fibrillary acidic protein (GFAP) immunostain confirms the identity of the tumor as a glioma (Fig. 1E). Immunohistochemical staining with a p53 antibody is positive in a small subset of cells (not shown). ATRX is preserved in tumor nuclei (not shown). Antibodies against mutant BRAF (V600E), IDH1 (R132H), and histone H3 (K27M) do not show any staining (not shown). The Ki67 proliferation index is focally up to 2.4% (Fig. 1F). A diagnosis of diffuse glioma was rendered, and molecular studies were then performed to determine the genomic alterations present in this low-grade glioma.
TECHNICAL ANALYSIS
Whole-exome and transcriptome sequencing were performed on DNA and RNA from normal (buccal swab) and tumor (formalin-fixed paraffin-embedded) samples, as previously described (Oberg et al. 2016). In brief, exonic sequences were captured using Agilent SureSelect v5 + UTR reagents and RNA sequencing was performed using TruSeq Stranded Total RNA LT reagents. DNA and RNA libraries were sequenced on Illumina HiSeq2500 at 100 bp x2. All clinical sequencing quality metrics were met. One hundred and eighty-seven million reads were obtained on the normal and tumor samples for whole-exome sequencing, resulting in an average coverage of 188× and 186×, respectively. One hundred and ninety-two million reads for tumor transcriptome were obtained, of which 104 million were uniquely mapping reads. Alignment and variant calling in 674 genes found in COSMIC (https://www.pathology.columbia.edu/file/2522/download?token=jAQS2HOe) was performed using NextGENe software, with 10% cutoff for somatic missense mutations and indels, including actionable somatic mutations, somatic mutations in targeted pathways, and somatic variants of uncertain significance. Copy-number calling was performed with Excavator software and fusion calling with FusionMap software. Confirmation of the fusion was performed using anchored multiplex polymerase chain reaction (PCR), as previously described (Pagan et al. 2019). DNA methylation profiling was performed using the Illumina EPIC Array 850 Bead-Chip (850K) array as previously described (Serrano and Snuderl 2018).
VARIANT INTERPRETATION
Whole-exome sequencing of the tumor did not identify any somatic missense mutations or indels. Consistent with the immunohistochemical analysis, no variants in BRAF, IDH1, H3F3A, or HIST1H3B were found. Gains of multiple chromosomes (5, 7, 12, 14, 19, and X) were seen in the tumor. DNA methylation profiling was performed and was not able to classify this tumor definitively. The closest match was “low grade glioma, ganglioglioma” (calibrated score 0.33) (Supplemental Fig. 1). Copy-number analysis of the DNA methylation data showed the same chromosomal gains as seen by whole-exome analysis (Supplemental Fig. 2). Transcriptome analysis showed the presence of a novel FGFR2–VPS35 fusion, involving the first 17 exons of FGFR2 fused to exon 2 of VPS35 (Table 1; Supplemental Fig. 3). To gain further insight on this fusion, pathway and process enrichment analysis on differentially expressed genes was performed using Metascape (Zhou et al. 2019). The “transmembrane receptor protein tyrosine kinase signaling pathway” was found among the top enriched pathways (P-value 3.9 × 10−9), supporting the hypothesis that this fusion results in activation of signaling. The presence of this novel fusion and breakpoints was confirmed by anchored multiplex PCR. In addition, an anti-VPS35 antibody showed up-regulated expression and was particularly strong in tumor cells and eosinophilic granular bodies (Fig. 1H). Taken together, these results underscore the importance of a multifaceted approach for the analysis of genomic architecture. In this case, the analysis of transcriptome data was used to identify a novel fusion in a situation in which interrogation of DNA was unrevealing.
Table 1.
Genomic findings
| Gene | Genomic coordinates | Fusion description | Variant type | Predicted effect | Target coverage |
|---|---|---|---|---|---|
| FGFR2–VPS35 | 10: 123243212; 16: 46717518 | NM_022970:r.1_2304_NM_018206:r.4_2388 | Fusion | Oncogenic | 65× |
SUMMARY
FGFR2 is a member of the fibroblast growth receptor family. Ligand-dependent dimerization leads to tyrosine phosphorylation and activation of MAPK and PI-3K/AKT pathways. The case reported here represents a novel member of the “diffuse glioma, other MAPK pathway alteration” category that was recently established for pediatric diffuse gliomas (Ellison et al. 2019). FGFR2 fusions have been previously reported in diffuse gliomas (Huse et al. 2017). The FGFR2–VPS35 rearrangement identified in this case is novel, to the best of our knowledge. The FGFR2 breakpoint at exon 17 is a previously reported breakpoint and results in a fusion protein that retains the FGFR2 kinase domain and nearly the entire coding sequence of the partner gene, VPS35. This FGFR fusion is targetable by FGFR tyrosine kinase inhibitors because of its intact FGFR tyrosine kinase domain. Improvement of some patients with high-grade gliomas containing FGFR alterations after treatment with ponatinib (Linzey et al. 2017) or infigratinib (Lassman et al. 2019) suggests that treatment with FGFR tyrosine kinase inhibitors may be warranted in this patient if the tumor undergoes transformation.
VPS35 is a component of the retromer cargo-recognition complex, which functions in the retrograde transport of cargo proteins from endosomes to the trans-Golgi network or to the plasma membrane. In a very recent study, VPS35 mutations and copy-number alterations were identified in a subset of hepatocellular carcinomas. It was found that VPS35 activated the PI-3K/AKT signaling pathway at least in part via recycling of FGFR3 (Zhang et al. 2020). The possibility of increased FGFR2 at the plasma membrane in the glioma described in this case report remains to be determined. In potential support of a role for VPS35 in protein turnover, VPS35 localized to EGBs in the tumor (Fig. 1H). EGBs are immunoreactive for α-1-antichymotrypsin and α-1-antitrypsin and thought to be related to lysosomes (Hitotsumatsu et al. 1994). Finally, VPS35 has also been implicated in regulation of the subcellular localization and signaling of NRAS (Zhou and Phillips 2019). This raises the question of whether the FGFR2–VPS35 fusion can potentially be involved in RAS signaling as well.
ADDITIONAL INFORMATION
Data Deposition and Access
The variant has been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with the accession number SCV001366116. Patient permission was not granted to deposit WES and transcriptome data.
Ethics Statement
Written patient consent was obtained for this clinical testing. Publication was approved by the Columbia University Medical Center Institutional Review Board (IRB# AAAR8988).
Funding
This work was supported by a grant from the Sohn Conference Foundation and by start-up departmental funds (K.R.).
Competing Interest Statement
The authors have declared no competing interest.
Referees
Stephen Yip
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Ellison DW, Hawkins C, Jones DTW, Onar-Thomas A, Pfister SM, Reifenberger G, Louis DN. 2019. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol 137: 683–687. 10.1007/s00401-019-01987-0 [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu T, Iwaki T, Fukui M, Tateishi J. 1994. Cytoplasmic inclusions of astrocytic elements of glial tumors: special reference to round granulated body and eosinophilic hyaline droplets. Acta Neuropathol 88: 501–510. 10.1007/BF00296486 [DOI] [PubMed] [Google Scholar]
- Huse JT, Snuderl M, Jones DT, Brathwaite CD, Altman N, Lavi E, Saffery R, Sexton-Oates A, Blumcke I, Capper D, et al. 2017. Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): an epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 133: 417–429. 10.1007/s00401-016-1639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassman AB, Sepúlveda-Sánchez JM, Cloughesy T, Gil-Gil JM, Puduvalli VK, Raizer J, De Vos FY, Wen PY, Butowski N, Clement P, et al. 2019. OS10.6 Infigratinib (BGJ398) in patients with recurrent gliomas with fibroblast growth factor receptor (FGFR) alterations: a multicenter phase II study. NeuroOncology 21: iii21–iii22. 10.1093/neuonc/noz126.072 [DOI] [Google Scholar]
- Linzey JR, Marini B, McFadden K, Lorenzana A, Mody R, Robertson PL, Koschmann C. 2017. Identification and targeting of an FGFR fusion in a pediatric thalamic “central oligodendroglioma”. NPJ Precis Oncol 1: 29 10.1038/s41698-017-0036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg JA, Glade Bender JL, Sulis ML, Pendrick D, Sireci AN, Hsiao SJ, Turk AT, Dela Cruz FS, Hibshoosh H, Remotti H, et al. 2016. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med 8: 133 10.1186/s13073-016-0389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan C, Barua S, Hsiao SJ, Mansukhani M, Saqi A, Murty V, Fernandes H. 2019. Targeting SLMAP-ALK—a novel gene fusion in lung adenocarcinoma. Cold Spring Harb Mol Case Stud 5: a003939 10.1101/mcs.a003939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano J, Snuderl M. 2018. Whole genome DNA methylation analysis of human glioblastoma using Illumina BeadArrays. Methods Mol Biol 1741: 31–51. 10.1007/978-1-4939-7659-1_2 [DOI] [PubMed] [Google Scholar]
- Zhang G, Tang X, Liang L, Zhang W, Li D, Li X, Zhao D, Zheng Y, Chen Y, Hao B, et al. 2020. DNA and RNA sequencing identified a novel oncogene VPS35 in liver hepatocellular carcinoma. Oncogene 10.1038/s41388-020-1215-6 [DOI] [PubMed] [Google Scholar]
- Zhou M, Philips MR. 2019. Where no Ras has gone before: VPS35 steers N-Ras through the cytosol. Small GTPases 10: 20–25. 10.1080/21541248.2016.1263380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant has been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with the accession number SCV001366116. Patient permission was not granted to deposit WES and transcriptome data.