Abstract
The CXCL12 chemokine receptor CXCR4 belongs to the GPCR superfamily and is often overexpressed in cancer, being involved in tumor progression and metastasis. How CXCR4 signaling integrates with other relevant oncogenic transduction pathways and the role of GPCR regulatory mechanisms in such contexts are not well-understood. Recent data indicate concurrent upregulation in certain tumors of CXCR4, EGF receptor (EGFR), and G protein-coupled receptor kinase 2 (GRK2), a signaling node functionally linked to both receptor types. We have investigated in a model system the effect of the EGFR and GRK2 status on CXCL12/CXCR4-mediated activation of Gi, the earliest step downstream of receptor activation. We find that overexpressed and activated EGFR reduces CXCR4-mediated Gi1 activation and that GRK2 phosphorylation at tyrosine residues is required to exert its inhibitory actions on CXCR4–Gi stimulation, suggesting a shared path of modulation. Our data point to a role for GRK2 in the crosstalk of the CXCR4 and EGFR signal transduction pathways in pathological contexts characterized by concurrent overactivation of these proteins.
Keywords: CXCR4, EGF receptor, Gi proteins, GRK2, tyrosine phosphorylation, FRET
The CXCL12 chemokine receptor CXCR4 is overexpressed in different tumor types and has been suggested to play an important role in promoting proliferation, survival, invasion, and metastasis of tumor cells.1−4 CXCR4 belongs to the G protein-coupled receptor (GPCR) superfamily and preferentially couples to pertussis-toxin-sensitive heterotrimeric Gi proteins, thus eliciting the stimulation of effectors downstream Gαi and Gβγ subunits and leading to the subsequent modulation of calcium-, MAPK-, or PI3K/Akt-dependent cascades.1,5 Nonetheless, emerging evidence indicates that the molecular mechanisms linking the CXCL12/CXCR4 axis to relevant cancer cell hallmarks are very complex and include intricate crosstalk mechanisms with other transduction networks acting within the tumor microenvironment. The crosstalk between CXCR4 and members of the epidermal growth factor receptor (EGFR) family is of particular interest given the frequent concurrent upregulation of these receptors in a variety of tumors and their shared relevance for tumor growth and metastasis occurrence (see refs (2), (6), and (7) and references therein).
EGFR family members have been suggested to impact CXCR4 signaling by several potential mechanisms, including hijacking of the canonical GPCR signaling machinery (as reviewed in refs (8) and (9)), upregulation of CXCR4 expression levels,10,11 and promotion of CXCR4 phosphorylation at serine and/or tyrosine residues, leading to CXCL12-independent receptor activation of downstream cascades.2,6,12,13 Conversely, the CXCL12/CXCR4 axis may enhance EGFR functionality by activation of membrane-bound proteases, leading to the release of different EGFR ligands,14,15 or by triggering direct tyrosine phosphorylation and EGFR transactivation via Gi/Src-mediated pathways.16,17
Interestingly, recent evidence suggests that CXCR4 and EGFR share G protein-coupled receptor kinase 2 (GRK2) as a common component of their transduction cascades. GRK2 is emerging as a key oncomodulator node.18,19 GRK2 levels are enhanced in a subset of breast cancer patients and in breast cancer cell models, and GRK2 upregulation is able to foster EGF-dependent proliferation and survival cascades and favor tumor growth in vivo in both xenograft and orthotopic mice models.18,20 EGFR stimulation has also been reported to recruit GRK2 and promote stimulatory GRK2 phosphorylation on tyrosine residues (see refs (21) and (22) and references therein). On the other hand, ligand-activated CXCR4 can be phosphorylated by GRK2 at specific serine/threonine intracellular residues,2,23 thus causing recruitment of β-arrestins, which in turn leads to β-arrestin-dependent signaling, receptor uncoupling from G proteins, internalization, and recycling.24
In order to better understand the potential crosstalk mechanisms among these signaling components, we have investigated the effect of EGFR and GRK2 on CXCL12/CXCR4-mediated activation of Gi proteins, the earliest step downstream of receptor activation, by the use of fluorescence resonance energy transfer (FRET)-based Gi protein sensors in a suitable model system.
Results and Discussion
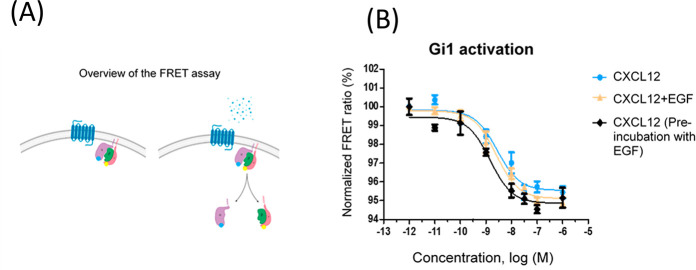
We sought to investigate potential interactions between the EGFR and CXCR4 signaling pathways at a level close to GPCR activation, thus avoiding the influence of potential additional players that might play a role if more downstream readouts (e.g., MAPK stimulation, cell proliferation, or migration) are analyzed. In order to specifically study the CXCR4-mediated G protein activation step, FRET-based Gi protein sensors for Gi1, Gi2, and Gi3 (developed as explained in ref (25)) were employed. These sensors consist of the Gαi subunit fused to mTurquoise (the donor fluorophore), the Gγ subunit fused to cpVenus (the acceptor fluorophore), and the unlabeled Gβ subunit, all in a single plasmid. Under basal conditions, the proximity between the subunits is highest, whereas upon CXCL12 stimulation, activation of CXCR4 mediates dissociation of the labeled subunits, leading to a decreased FRET ratio (Figure 1A scheme). In our experimental setting, HEK293 cells were transfected with the receptor of interest and the desired Gi protein sensor and then stimulated with increasing concentrations of the ligand in a 96-well plate format. Using this model, we have reported that CXCL12 activates Gi1, Gi2, or Gi3 heterotrimeric G proteins in a concentration-dependent manner (Perpiñá-Viciano et al., submitted).
Figure 1.
Gi1 activation by CXCR4 in response to increasing concentrations of CXCL12 is not affected by EGF at endogenous levels of EGFR expression. (A) Schematic overview of the FRET-based sensors for monitoring Gi activation. The agonist-activated receptors mediate the activation and dissociation of G protein subunits, leading to a loss of FRET between Gα-mTurquoise and Gγ-Venus in the sensor. (B) Normalized FRET ratio/dose–response curves (0.001 nM to 1 μM CXCL12) were obtained as detailed in Methods in HEK293 cells expressing CXCR4 and the Gi sensor in the presence of the chemokine alone, in combination with EGF (0.001 nM to 1 μM, same concentration of each ligand, EGF+CXCL12), or upon prior preincubation of cells with 1 μM EGF for 60 min. Data are reported as mean ± SEM of quadruplicate determinations in paired FRET assays representative of n = 12 independent experiments. In this particular experiment, the EC50 values for CXCL12 were 2.8 nM (endogenous EGFR, cells challenged with CXCL12), 2.5 nM (cells challenged simultaneously with CXCL12+EGF), and 1.5 nM (cells subjected to EGF preincubation prior to CXCL12 challenge).
The Presence of Overexpressed and Activated EGFR Decreases CXCR4-Mediated Gi1 Activation
Consistent with previous observations, concentration-dependent activation of Gi1 by CXCR4 in response to CXCL12 was detected (Figure 1B). Simultaneous stimulation with EGF of EGFR, reported to be endogenously expressed at low levels in HEK293 cells,26,27 did not affect this pattern. Under control conditions, stimulation of CXCR4 with CXCL12 resulted in a decrease in the FRET signal with an average amplitude of 3.27 ± 0.13 and EC50 values of 15.5 [9.3–25.9] nM (mean [asymmetric 95% CI]), similar to data obtained in cells challenged simultaneously with CXCL12+EGF (average amplitude 3.46 ± 0.18 and EC50 = 7.43 [0.15–61.3] nM). A similar trend was observed upon preincubation of cells with EGF prior to CXCL12 addition (Figure 1B). As experimental controls, we verified that CXCL12, EGF, or a combination of the two does not activate Gi1 in the absence of overexpressed receptors (Figure S1) and also that EGF does not affect CXCR4-mediated Gi2 or Gi3 stimulation under such endogenous EGFR experimental conditions (Figure S2).
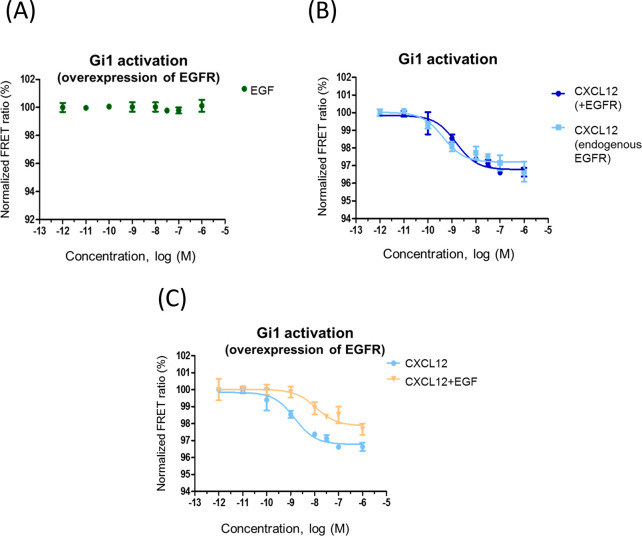
As indicated in the Introduction, different cancer cells are characterized by concurrent overexpression of EGFR and CXCR4.6,7 In order to mimic such an enhanced receptor expression context, we overexpressed EGFR and CXCR4 in HEK293 cells together with the Gi1 sensor (Figure S3). Of note, although it has been suggested that receptor tyrosine kinases (RTKs) may signal in part through the GPCR machinery in certain contexts,8 EGF alone did not activate Gi1 in the presence of overexpressed EGFR (Figure 2A), therefore ruling out the possibility that this receptor hijacks Gi signaling in our experimental setting. EGFR overexpression per se did not modify CXCL12/CXCR4-mediated Gi activation (Figure 2B). With endogenous EGFR, an average amplitude of 3.36 ± 0.25 and EC50 = 4.8 [0.2–32] nM was observed upon CXCL12 stimulation, whereas an amplitude of 3.07 ± 0.33 and EC50 = 5.3 [0.2–32.5] nM were determined under conditions of EGFR overexpression. However, concomitant EGFR activation by EGF markedly attenuated Gi1 activation by CXCL12 (Figure 2C), as indicated by a decreased observed amplitude of 2.25 ± 0.16 (p = 0.07 vs CXCL12 stimulation alone) and a significantly higher EC50 of 32.4 [2.4–197] nM (p < 0.05 vs CXCL12 stimulation alone). These results suggested that overexpressed and active EGFR (mirroring an oncogenic context) would affect CXCR4 signaling via Gi1. It might be of interest to compare the effect of EGFR overexpression and activation on CXCR4 signaling to Gi1, Gi2, and Gi3, particularly if changes in the expression patterns of these Gi proteins are identified in specific cancer contexts.
Figure 2.
Overexpressed and activated EGFR decreases CXCR4-mediated Gi1 activation. Normalized FRET ratio/dose–response curves were obtained as detailed in Methods in HEK293 cells expressing EGFR and the Gi sensor (panel A) or the indicated combinations of CXCR4 and EGFR (panels B and C). Cells were challenged with 0.001 nM to 1 μM EGF (A), CXCL12 (B and C), or EGF+CXCL12 (0.001 nM to 1 μM, same concentration of each ligand) (C). Data are reported as mean ± SEM of quadruplicate determinations in paired FRET assays representative of n = 6 independent experiments run in parallel for the different conditions. In this particular experiment, the EC50 values for CXCL12 were 1.5 nM (overexpressed EGFR), 0.43 nM (endogenous EGFR), and 11 nM (overexpressed EGFR and cells challenged simultaneously with CXCL12+EGF).
GRK2 Levels, Kinase Activity, and Tyrosine Phosphorylation Status Affect CXCR4-Mediated Gi1 Activation
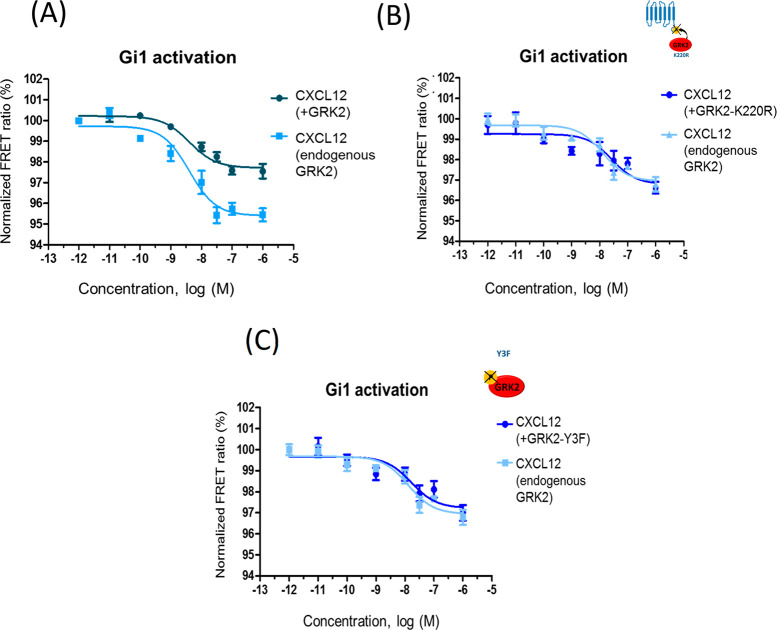
GRK2 is a canonical negative modulator of GPCR signaling that is able to phosphorylate agonist-activated CXCR4 receptors, leading to β-arrestin recruitment and uncoupling from G proteins.23 GRK2 expression is enhanced in some tumor contexts along with EGFR and CXCR4, such as in breast cancer cells.20 We observed that GRK2 overexpression notably decreases Gi1 activation by CXCL12 (Figure 3A). With endogenous GRK2, an average amplitude of 3.22 ± 0.15 and EC50 = 7 [0.1–55] nM was detected upon CXCL12 stimulation, whereas GRK2 overexpression markedly reduced the amplitude to 1.78 ± 0.16 (p < 0.01 vs endogenous GRK2) and led to increased EC50 values (49.3 [1–417] nM, p < 0.001 vs endogenous GRK2). Such GRK2-dependent reduction in the FRET ratio is abolished when a catalytically inactive mutant of GRK2 (GRK2-K220R) is overexpressed at similar levels as wild-type protein (Figures 3B and S3), indicating that its effect on CXCR4 signaling to G proteins is phosphorylation-dependent. In this setting, no significant differences in amplitudes or EC50 values were apparent (3.09 ± 0.17 and EC50 = 5.1 [0.1–48] nM for endogenous conditions and 2.84 ± 0.19 and EC50 = 17 [9–24] nM for K220R overexpression, respectively).
Figure 3.
GRK2 activity and tyrosine phosphorylation status is key for inhibiting CXCR4-mediated Gi1 activation. Normalized FRET ratio/dose–response curves were obtained as detailed in Methods in HEK293 cells expressing CXCR4, Gi1 sensor, and either wild-type GRK2 (panel A), inactive GRK2 (GRK2-K220R) (panel B), or a mutant GRK2 unable to be phosphorylated at the specific tyrosine residues 13, 86, and 92 (GRK2-Y3F) (panel C). Cells were challenged with 0.001 nM to 1 μM CXCL12 as in previous figures. Data are reported as mean ± SEM of quadruplicate determinations in paired FRET assays representative of n = 11 (A) or 4 (B, C) independent experiments. In these particular experiments, the EC50 values for CXCL12 were 2.8 nM (endogenous GRK2) and 9 nM (overexpression of GRK2) in panel A and 13 nM (endogenous GRK2), 27 nM (overexpression of GRK2-K220R), and 17 nM (overexpression of GRK2 Y3F) in panels B and C.
GRK2 phosphorylation in key tyrosine residues has been shown to take place downstream of EGFR or GPCR/β-arrestin/Src cascades and to foster its kinase activity toward both GPCR and non-GPCR substrates.22,28,29 Since such GRK2 tyrosine phosphorylation status is likely to be enhanced in oncogenic contexts as a consequence of increased RTKs and/or GPCR expression and activity, we tested the effects on CXCL12/CXCR4-stimulated Gi1 activation of overexpressing a GRK2 construct mutated in previously identified key target tyrosine residues (Y13F, Y86F, and Y92F, hereafter termed GRK2-Y3F).30 Strikingly, the effect observed upon wild-type GRK2 expression is notably attenuated when similar levels of GRK2-Y3F are present (Figures 3C and S3). No significant differences in amplitudes or EC50 values were apparent in the presence of this mutant construct (3.13 ± 0.29 and EC50 = 3.8 [0.4–36] nM for endogenous conditions and 2.58 ± 0.29 and EC50 = 3.8 [0.7–42] nM for GRK2-Y3F overexpression, respectively). These data suggest that a certain cellular pool of GRK2 phosphorylated at these tyrosine residues is required to exert its modulatory actions on CXCR4-triggered Gi activation. Of note, coexpression of wild-type GRK2 or these mutants does not significantly alter the total (Figure S1) or surface levels (as assessed by flow cytometry, data not shown) of transfected CXCR4 in our model system.
EGFR and GRK2 Appear to Modulate CXCR4-Mediated Gi1 Activation via a Shared Path
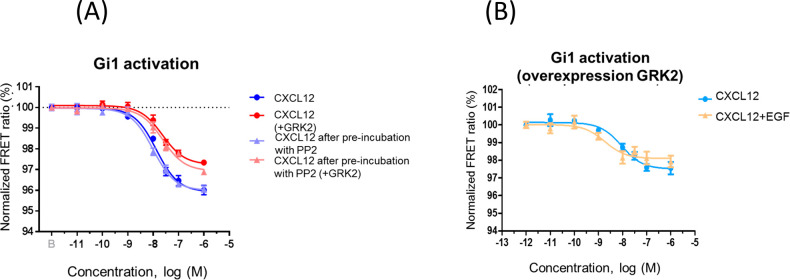
Of note, preincubation with PP2, a c-Src inhibitor, did not affect CXCL12/CXCR4-mediated Gi stimulation or the ability of overexpressed GRK2 to inhibit this pathway (Figure 4A). This indicates that the GPCR/Src pathway reported to lead to GRK2 tyrosine phosphorylation30 does not play a relevant role in our conditions and suggests that basal cell culture conditions in the presence of serum growth factors acting via RTKs may be sufficient to attain the required threshold of tyrosine-phosphorylated GRK2 upon kinase overexpression. Consistent with this notion, the presence of EGF acting on endogenous receptors does not further inhibit Gi1 activation by CXCL12 when GRK2 is overexpressed (Figure 4B). Stimulation of CXCR4 with CXCL12 alone in cells overexpressing GRK2 resulted in a decrease in the signal amplitude (1.86 ± 0.24 and EC50 = 10 [1.6–41] nM) and thus was not significantly different from data obtained when these cells were challenged simultaneously with CXCL12+EGF (average amplitude 1.70 ± 0.12 and EC50 = 20 [0.2–57] nM). When we overexpressed both GRK2 and EGFR together with CXCR4, the ability of CXCL12 to promote Gi1 activation was compromised, and EGF did not further enhance this effect (only ca. 1% displacement of the FRET ratio at 100 nM CXCL12 under both conditions). Overall, these data suggest that GRK2 and EGFR do not display an additive effect upon CXCR4/Gi coupling and that they may act via the same regulatory pathway (see the scheme in the abstract graphic).
Figure 4.
Effect of tyrosine kinase modulators on the ability of GRK2 to regulate CXCR4/Gi coupling. Normalized FRET ratio/dose–response curves were obtained as detailed in Methods in HEK293 cells expressing CXCR4 and the Gi1 sensor as well as wild-type GRK2 where indicated. Cells were challenged with 0.001 nM to 1 μM CXCL12 under different conditions. (A) Preincubation for 1 h with 5 μM PP2 (a Src kinase inhibitor) does not alter the response to CXCL12 alone or prevents the decreased Gi1 activation upon GRK2 overexpression. Data are reported as mean ± SEM of quadruplicate determinations in paired FRET assays representative of n = 2 independent experiments. In this particular experiment, the EC50 values for CXCL12 stimulation were 13 nM (control), 9.9 nM (after PP2 preincubation), 23 nM (overexpressed GRK2), and 24 nM (after PP2 preincubation with overexpressed GRK2). (B) EGF does not further affect CXCL12-mediated Gi1 activation under conditions of GRK2 overexpression. Data are reported as mean ± SEM of quadruplicate determinations in paired FRET assays representative of n = 6 independent experiments. In this particular experiment, the EC50 values were 9 nM (CXCL12 challenge under conditions of overexpression of GRK2 alone) and 2 nM (CXCL12 plus EGF challenge).
Our results are consistent with an effect of GRK2 or EGFR at the step of CXCR4-mediated Gi1 activation at the plasma membrane. However, the possibility that changes in CXCR4 cell surface expression under these experimental conditions may cooperate with the observed changes in receptor response cannot be completely ruled out. It would be also of interest to compare the effects of varying expression levels of EGFR on CXCR4/Gi1 coupling in order to determine the occurrence of a threshold level of EGFR overexpression for observation of a modulatory effect, above which a limiting factor might operate. According to our hypothesis, GRK2 might be one such limiting factor mediating the EGFR effects, but proving this would require future research using varying doses of EGFR and GRK2.
A Role for GRK2 Tyrosine Phosphorylation in the Crosstalk of the CXCR4 and EGFR Signal Transduction Pathways in Pathological Contexts
In view of the reported concomitant overexpression of EGFR, CXCR4, and GRK2 in certain cancer cell types, namely, breast cancer (see refs (7), (11), and (20) and references therein), it is tempting to hypothesize that in such pathological contexts activated EGFR would favor the recruitment and subsequent tyrosine phosphorylation of a significant pool of GRK2, resulting in more efficient uncoupling of CXCR4 from Gi proteins, which may foster “biased” signaling downstream of the GRK2/β-arrestin axis in such contexts.31 In parallel, GRK2 upregulation would potentiate EGF-dependent proliferation and survival cascades, as reported in breast cancer cell models, thus favoring tumor growth.18
In order to support the notion that EGFR and GRK2 modulate CXCR4-mediated Gi1 activation via a shared path, it would be of interest to investigate whether GRK2 silencing, its pharmacological inhibition, or the expression of the tyrosine-phosphorylation-deficient mutant Y3F attenuate the observed effects of activated EGFR on CXCR4-mediated Gi1 activation. Conversely, it would be interesting to investigate the effect of inhibiting endogenous EGFR on the effect of overexpressed GRK2 on CXCR4/Gi1 coupling.
In this regard, it has been reported that EGFR-mediated GRK2 phosphorylation on these specific tyrosine residues promotes membrane recruitment and enhanced GRK2-mediated desensitization of opioid receptors22 or dopamine D3 receptors32 in an EGF-dependent manner. In other cell types, TCR-activated c-Src leads to tyrosine phosphorylation of GRK2 and stimulation of GRK2-dependent CXCR4-Ser339 phosphorylation and TCR–CXCR4 complex formation, leading to subsequent recruitment of PREX1, which is required for fostering of cytokine secretion upon T cell activation.33 Whether EGFR activation leads to GRK2 phosphorylation on tyrosine residues in our experimental setting remains to be confirmed.
Both c-Src and EGFR cascades phosphorylate GRK2 on tyrosine residues located within the αN-helix (Tyr13) or the RH region (Tyr86 and Tyr92).28,29 The mechanisms by which such modifications result in enhanced catalytic activity toward both soluble and membrane-bound substrates likely involve allosteric effects that are not fully understood. It has been suggested that the side chain of Tyr13 is packed against the active-site tether (AST) of the kinase, thus contributing to both receptor docking and activation. In fact, the Y13A mutant shows reduced substrate phosphorylation, consistent with defective stabilization of the active state of GRK2.34 The gain of a negative charge in phosphorylated Y13, adjacent to acidic residues (D10, E476) involved in the interaction with the AST loop might thus modulate receptor docking. In addition, residues Y86 and Y92 are localized close to a hydrophobic interface shaped between the RH domain and the kinase large lobe, suggesting that their phosphorylation might modulate the closure of the kinase domain (as reviewed in ref (28)).
Overall, our data suggest a new example of the relevance of the GRK2 tyrosine phosphorylation status in its GPCR modulatory function. Such GRK2 post-translational modification may also constitute an important integrative node for the crosstalk between EGFR and CXCR4 receptors in pathological contexts of concomitant overexpression and/or overstimulation of these proteins. Investigation of GRK2 phosphorylation barcodes and the functional interactions between the EGFR and CXCR4 signaling networks in such situations is an interesting venue for future studies.
Methods
Plasmid DNA Constructs
The G protein sensors for Gi1, Gi2, and Gi3 have been previously described.25 Human CXCR4 and human CXCR4 with three hemagglutinin (HA) tags fused to the N-terminus (3HA-CXCR4) are in pcDEF3 (Vrije Universiteit, Amsterdam, The Netherlands). Flag EGFR is in pFLAG-CMV3 (Addgene). GRK2, GRK2-K220R, and GRK2-Y3F are in pcDNA3 (generated in our lab).
Cellular Treatments and Antibodies
Recombinant human CXCL12 was purchased from Peprotech (cat. no. 100-03) and human EGF from Miltenyi Biotec (cat. no. 130-097-749). PP2 was purchased from Sigma (cat. no. P0042). Affinity-purified rabbit polyclonal antibodies raised against GRK2 (C-15, cat. no. SC-562) and the HA tag (F7, cat. no. SC-7392) were purchased from Santa Cruz Biotechnology. Flag M1 monoclonal antibody was obtained from Sigma (cat. no. F3040). Secondary antibodies were obtained from Nordic Immunology (goat anti-rabbit IgG- HRP, cat. no. GAR/IgG(H+L)/PO, and goat anti-mouse IgG-HRP, cat. no. GAM/IgG(H+L)/PO).
Cell Line and Cell Culture
The human embryonic kidney 293 (HEK293) cell line (ATCC; CRL-1573) was cultured using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 4.5 g/L glucose (Gibco), 10% v/v fetal bovine serum (Biochrom), 1% penicillin/streptomycin (Gibco), and 1% l-glutamine (PanBiotech). Cells were maintained in a humidified 5% CO2 atmosphere at 37 °C. Cells were split every 3 days by washing with Dulbecco’s phosphate-buffered saline (DPBS) (Gibco) and using trypsin-EDTA (PanBiotech) to detach.
Transfection
HEK293 cells were plated in 100 mm plates. When 60–65% confluency was reached, the cell medium was replaced with fresh medium, and the cells were transfected with 1.5 μg of receptor (CXCR4, 3HA-CXCR4, or Flag-EGFR), 1.2 μg of GRK2 WT or mutants, and 3 μg of FRET-based G protein sensor (Gi1, Gi2, Gi3) plasmids using Effectene (Qiagen), following the manufacturer’s instructions.
G Protein Activation in 96-Well Plates
Cells were transferred to a black flat-bottom 96-well plate coated with poly-d-lysine (1 mg/mL, 30 min) at a density of 30 000 cells/well 24 h after transfection. After 24 h, the medium was replaced by 90 μL/well of imaging buffer (140 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, pH 7.3, 0.1% BSA), and the cells were incubated at 37 °C for 30 min. To generate concentration–response curves in a microplate reader, the fluorescence was read every 39 s for 5 min to determine the basal (buffer-treated-cells) signal. Afterward, 10 μL of buffer or ligands (concentrations indicated in the figure) were added to the wells for a total assay volume of 100 μL. Fluorescence was read again every 39 s for a total of 20 min to determine the response signal. Experiments were performed using a Synergy Neo2 multimode microplate reader (Biotek) with Gen5 data analysis software. During the measurements, cells were excited at 420/50 nm (Biotek CFP-YFP filter, 1035013), and emission was monitored at 485/20 nm and 540/25 nm (Biotek CFP-YFP filter, 1035043). Ligands were prepared in imaging buffer containing 0.1% BSA. The FRET change produced by each concentration of ligand tested was normalized by dividing the response signal (average of time points read after ligand addition) by the basal signal (nonligand; buffer-treated cells). Experiments were performed in quadruplicate. To determine the EC50 values for G protein activation, the data were fitted to a three-parameter sigmoidal model using GraphPad. The EC50 values are reported in the figure legends.
Western Blot Analysis
Cellular lysates were prepared by washing twice in cold PBS followed by solubilization in RIPA buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1% Triton-X100, 0.1% SDS, 0.5% sodium deoxycholate, and 1 mM NaF, supplemented with 1 mM sodium orthovanadate plus a mixture of protease inhibitors). Proteins were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with specific antibodies (Flag-M1, 1:1000 dilution; HA, 1:500 dilution; GRK2, 1:500 dilution). Secondary antibodies (rabbit and mouse) were used at a 1:50000 dilution. Blots were developed using the chemiluminescence method (ECL, Amersham Pharmacia Biotech).
Quantification and Statistical Analysis
The means of the EC50 values and the asymmetric 95% CIs reported in the text were calculated on the basis of logarithmic values (logEC50). To determine the amplitudes of the signals, maximum and minimum activation values were considered, and the data are reported as mean ± SEM. Statistical significance was assessed using the t test on the logEC50 values or Emax/amplitude data. The minimum criterion for statistical significance was p < 0.05.
Acknowledgments
Our laboratories are supported by the European Union (H2020-MSCA Program, Grant Agreement 860229-ONCORNET 2.0 to F.M. and C.H.), Agencia Estatal de Investigación of Spain (Grant SAF2017-84125-R to F.M.), CIBERCV-Instituto de Salud Carlos III, Spain (Grant CB16/11/00278 to F.M., cofunded with a European FEDER contribution), Fundación Ramón Areces (to F.M.), Instituto de Salud Carlos III (Grant PI17-00576 to P.P., cofunded with a European FEDER contribution), and Programa de Actividades en Biomedicina de la Comunidad de Madrid-B2017/BMD-3671-INFLAMUNE (to F.M.). F.M. and C.H. are members of the ERNST CA18133 Cost Action.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00021.
CXCL12, EGF, or combination of the two ligands does not activate Gi1 in the absence of overexpressed receptors; EGF at endogenous levels of EGFR expression does not affect activation of Gi2 or Gi3 by CXCR4 in response to increasing concentrations of CXCL12; expression levels of 3HA-CXCR4 and GRK2 constructs and Flag-EGFR in HEK293 cells (PDF)
Author Contributions
M.N. and C.P.-V. designed and performed the experiments, prepared the figures, analyzed the data, and revised the manuscript; P.P. helped with the study concept and revised the manuscript; C.H. and F.M. provided the study concept and design, interpreted the experiments, supervised the study, wrote and revised the manuscript, and obtained funding.
The authors declare no competing financial interest.
Supplementary Material
References
- Guo F.; Wang Y.; Liu J.; Mok S. C.; Xue F.; Zhang W. (2016) CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene 35, 816–826. 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- Fumagalli A.; Zarca A.; Neves M.; Caspar B.; Hill S. J.; Mayor F. Jr.; Smit M. J.; Marin P. (2019) CXCR4/ACKR3 Phosphorylation and Recruitment of Interacting Proteins: Key Mechanisms Regulating Their Functional Status. Mol. Pharmacol. 96, 794–808. 10.1124/mol.118.115360. [DOI] [PubMed] [Google Scholar]
- Heuninck J.; Perpina Viciano C.; Isbilir A.; Caspar B.; Capoferri D.; Briddon S. J.; Durroux T.; Hill S. J.; Lohse M. J.; Milligan G.; Pin J. P.; Hoffmann C. (2019) Context-Dependent Signaling of CXC Chemokine Receptor 4 and Atypical Chemokine Receptor 3. Mol. Pharmacol. 96, 778–793. 10.1124/mol.118.115477. [DOI] [PubMed] [Google Scholar]
- Neves M.; Fumagalli A.; van den Bor J.; Marin P.; Smit M. J.; Mayor F. (2019) The Role of ACKR3 in Breast, Lung, and Brain Cancer. Mol. Pharmacol. 96, 819–825. 10.1124/mol.118.115279. [DOI] [PubMed] [Google Scholar]
- Scala S. (2015) Molecular Pathways: Targeting the CXCR4-CXCL12 Axis--Untapped Potential in the Tumor Microenvironment. Clin. Cancer Res. 21, 4278–4285. 10.1158/1078-0432.CCR-14-0914. [DOI] [PubMed] [Google Scholar]
- Sosa M. S.; Lopez-Haber C.; Yang C.; Wang H.; Lemmon M. A.; Busillo J. M.; Luo J.; Benovic J. L.; Klein-Szanto A.; Yagi H.; Gutkind J. S.; Parsons R. E.; Kazanietz M. G. (2010) Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol. Cell 40, 877–892. 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa P. C.; Humphries B. A.; Lewin Mejia D.; Buschhaus J. M.; Linderman J. J.; Luker G. D.; Luker K. E. (2019) Short-term cellular memory tunes the signaling responses of the chemokine receptor CXCR4. Sci. Signaling 12, eaaw4204. 10.1126/scisignal.aaw4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt N.; Bockaert J.; Marin P. (2007) GPCR-jacking: from a new route in RTK signalling to a new concept in GPCR activation. Trends Pharmacol. Sci. 28, 602–607. 10.1016/j.tips.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Crudden C.; Ilic M.; Suleymanova N.; Worrall C.; Girnita A.; Girnita L. (2015) The dichotomy of the Insulin-like growth factor 1 receptor: RTK and GPCR: friend or foe for cancer treatment?. Growth Horm. IGF Res. 25, 2–12. 10.1016/j.ghir.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Tsai M. F.; Chang T. H.; Wu S. G.; Yang H. Y.; Hsu Y. C.; Yang P. C.; Shih J. Y. (2015) EGFR-L858R mutant enhances lung adenocarcinoma cell invasive ability and promotes malignant pleural effusion formation through activation of the CXCL12-CXCR4 pathway. Sci. Rep. 5, 13574. 10.1038/srep13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. M.; Pan Y.; Wei Y.; Cheng X.; Zhou B. P.; Tan M.; Zhou X.; Xia W.; Hortobagyi G. N.; Yu D.; Hung M. C. (2004) Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell 6, 459–469. 10.1016/j.ccr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Woerner B. M.; Warrington N. M.; Kung A. L.; Perry A.; Rubin J. B. (2005) Widespread CXCR4 activation in astrocytomas revealed by phospho-CXCR4-specific antibodies. Cancer Res. 65, 11392–11399. 10.1158/0008-5472.CAN-05-0847. [DOI] [PubMed] [Google Scholar]
- Mueller W.; Schutz D.; Nagel F.; Schulz S.; Stumm R. (2013) Hierarchical organization of multi-site phosphorylation at the CXCR4 C terminus. PLoS One 8, e64975 10.1371/journal.pone.0064975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi R.; Dougherty U.; Mustafi D.; Ayaloglu-Butun F.; Fletcher M.; Adhikari S.; Sadiq F.; Meckel K.; Haider H. I.; Khalil A.; Pekow J.; Konda V.; Joseph L.; Hart J.; Fichera A.; Li Y. C.; Bissonnette M. (2017) ADAM17 is a Tumor Promoter and Therapeutic Target in Western Diet-associated Colon Cancer. Clin. Cancer Res. 23, 549–561. 10.1158/1078-0432.CCR-15-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. (2016) Transactivation of Epidermal Growth Factor Receptor by G Protein-Coupled Receptors: Recent Progress, Challenges and Future Research. Int. J. Mol. Sci. 17, 95. 10.3390/ijms17010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattarozzi A.; Gatti M.; Barbieri F.; Wurth R.; Porcile C.; Lunardi G.; Ratto A.; Favoni R.; Bajetto A.; Ferrari A.; Florio T. (2008) 17beta-estradiol promotes breast cancer cell proliferation-inducing stromal cell-derived factor-1-mediated epidermal growth factor receptor transactivation: reversal by gefitinib pretreatment. Mol. Pharmacol. 73, 191–202. 10.1124/mol.107.039974. [DOI] [PubMed] [Google Scholar]
- Cabioglu N.; Summy J.; Miller C.; Parikh N. U.; Sahin A. A.; Tuzlali S.; Pumiglia K.; Gallick G. E.; Price J. E. (2005) CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu in breast cancer cells by a novel pathway involving Src kinase activation. Cancer Res. 65, 6493–6497. 10.1158/0008-5472.CAN-04-1303. [DOI] [PubMed] [Google Scholar]
- Nogues L.; Reglero C.; Rivas V.; Salcedo A.; Lafarga V.; Neves M.; Ramos P.; Mendiola M.; Berjon A.; Stamatakis K.; Zhou X. Z.; Lu K. P.; Hardisson D.; Mayor F. Jr.; Penela P. (2016) G Protein-coupled Receptor Kinase 2 (GRK2) Promotes Breast Tumorigenesis Through a HDAC6-Pin1 Axis. EBioMedicine 13, 132–145. 10.1016/j.ebiom.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogues L.; Reglero C.; Rivas V.; Neves M.; Penela P.; Mayor F. Jr. (2017) G-Protein-Coupled Receptor Kinase 2 as a Potential Modulator of the Hallmarks of Cancer. Mol. Pharmacol. 91, 220–228. 10.1124/mol.116.107185. [DOI] [PubMed] [Google Scholar]
- Nogues L.; Palacios-Garcia J.; Reglero C.; Rivas V.; Neves M.; Ribas C.; Penela P.; Mayor F. Jr. (2018) G protein-coupled receptor kinases (GRKs) in tumorigenesis and cancer progression: GPCR regulators and signaling hubs. Semin. Cancer Biol. 48, 78–90. 10.1016/j.semcancer.2017.04.013. [DOI] [PubMed] [Google Scholar]
- Penela P.; Murga C.; Ribas C.; Lafarga V.; Mayor F. Jr. (2010) The complex G protein-coupled receptor kinase 2 (GRK2) interactome unveils new physiopathological targets. Br. J. Pharmacol. 160, 821–832. 10.1111/j.1476-5381.2010.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Long H.; Wu Z.; Jiang X.; Ma L. (2008) EGF transregulates opioid receptors through EGFR-mediated GRK2 phosphorylation and activation. Mol. Biol. Cell 19, 2973–2983. 10.1091/mbc.e07-10-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busillo J. M.; Armando S.; Sengupta R.; Meucci O.; Bouvier M.; Benovic J. L. (2010) Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J. Biol. Chem. 285, 7805–7817. 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A. (2014) Endocytic trafficking of chemokine receptors. Curr. Opin. Cell Biol. 27, 72–77. 10.1016/j.ceb.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Unen J.; Stumpf A. D.; Schmid B.; Reinhard N. R.; Hordijk P. L.; Hoffmann C.; Gadella T. W. Jr.; Goedhart J. (2016) A New Generation of FRET Sensors for Robust Measurement of Galphai1, Galphai2 and Galphai3 Activation Kinetics in Single Cells. PLoS One 11, e0146789 10.1371/journal.pone.0146789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavas S.; Machan R.; Wohland T. (2016) The Epidermal Growth Factor Receptor Forms Location-Dependent Complexes in Resting Cells. Biophys. J. 111, 2241–2254. 10.1016/j.bpj.2016.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H. H.; Furnari F. B.; Cavenee W. K.; Bogler O. (2003) Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc. Natl. Acad. Sci. U. S. A. 100, 6505–6510. 10.1073/pnas.1031790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penela P.; Ribas C.; Sánchez-Madrid F.; Mayor F. (2019) G protein-coupled receptor kinase 2 (GRK2) as a multifunctional signaling hub. Cell. Mol. Life Sci. 76, 4423–4446. 10.1007/s00018-019-03274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnago S.; Elorza A.; Mayor F. Jr. (1999) Agonist-dependent phosphorylation of the G protein-coupled receptor kinase 2 (GRK2) by Src tyrosine kinase. J. Biol. Chem. 274, 34411–34416. 10.1074/jbc.274.48.34411. [DOI] [PubMed] [Google Scholar]
- Penela P.; Elorza A.; Sarnago S.; Mayor F. Jr. (2001) Beta-arrestin- and c-Src-dependent degradation of G-protein-coupled receptor kinase 2. EMBO J. 20, 5129–5138. 10.1093/emboj/20.18.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. J.; Ziarek J. J.; Gravel S.; Veldkamp C. T.; Takekoshi T.; Hwang S. T.; Heveker N.; Volkman B. F.; Dwinell M. B. (2011) Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 108, 17655–17660. 10.1073/pnas.1101133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N.; Zhang X.; Zhang X.; Kim K. M. (2017) The EGF receptor inhibits the signaling of dopamine D3 receptor through the phosphorylation of GRK2 on tyrosine residues. Biochem. Biophys. Res. Commun. 489, 515–522. 10.1016/j.bbrc.2017.05.183. [DOI] [PubMed] [Google Scholar]
- Dinkel B. A.; Kremer K. N.; Rollins M. R.; Medlyn M. J.; Hedin K. E. (2018) GRK2 mediates TCR-induced transactivation of CXCR4 and TCR-CXCR4 complex formation that drives PI3Kgamma/PREX1 signaling and T cell cytokine secretion. J. Biol. Chem. 293, 14022–14039. 10.1074/jbc.RA118.003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beautrait A.; Michalski K. R.; Lopez T. S.; Mannix K. M.; McDonald D. J.; Cutter A. R.; Medina C. B.; Hebert A. M.; Francis C. J.; Bouvier M.; Tesmer J. J.; Sterne-Marr R. (2014) Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. J. Biol. Chem. 289, 25262–25275. 10.1074/jbc.M114.593178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.