Abstract
In a short time, humanity has experienced two pandemics: the influenza A virus pandemic (pH1N1) in 2009 and the coronavirus disease 2019 (COVID-19) pandemic in 2020. Therefore, it is likely that the general population will erroneously seek to compare the two pandemics and adopt similar attitudes in facing them. However, the two pandemics have their intrinsic characteristics that distinguish them considerably; for example, the virulence of the infectious agents and the availability of treatment and vaccine. Consequently, given this knowledge gap between the pH1N1 and COVID-19 pandemics, we conducted this review to clarify and summarize, above all, the epidemiological historical aspects of these two viruses of great importance to global public health.
Keywords: COVID-19 pandemic, 2009 H1N1 pandemic, Emerging viruses, SARS-CoV-2, Global epidemics
Introduction
At the beginning of the 21st century, human public health has been marked by the emergence of new viruses and pandemics [1]. Approximately a decade ago, the World Health Organization (WHO) declared the first of two global epidemics: on June 11, 2009, an influenza A H1N1 pandemic (pH1N1) was declared; the second and current pandemic of coronavirus disease 2019 (COVID-19) was declared on March 11, 2020 [2,3]. Besides, other major developments have raised extreme concern for human public health authorities, such as the emergence of nonrespiratory virus epidemics, including Zika, Chikungunya and Ebola [[4], [5], [6]]. Therefore, because of this evidence, it is clear that viruses will continue to play an increasing role in the field of infectious diseases throughout this century.
Acute viral respiratory diseases are extremely common among outpatients. Consequently, infections of the respiratory system, such as influenza-like diseases, colds, bronchitis and pneumonia, are the main causes of morbidity and mortality worldwide [7]. Influenza viruses, which often cause epidemics and pandemics, are also included in this scenario [8,9]. Thus, it is not surprising that the first pandemic of the 21st century (pH1N1) had, as its etiological agent, a new strain of the influenza A virus (A/H1N1pdm09), which initially began circulating in North America [[10], [11], [12], [13]]. This virus represented a new strain that originated through the regrouping of genes from human, avian and swine type influenza A viruses. Soon after the completion of molecular studies, the term 'swine flu' was coined for pH1N1 because the viral strain was probably transmitted from pigs to humans, although pigs were not involved in the worldwide spread of the virus during the pandemic [[14], [15], [16]].
Unexpectedly, the year 2020 has been deeply affected by the COVID-19 pandemic, which emerged from the city of Wuhan, China, in late December 2019 [17,18]. The current COVID-19 pandemic is caused by a novel coronavirus (SARS-CoV-2) that usually causes severe acute respiratory syndrome (SARS), which is why it is part of the SARS taxonomy [19,20]. Surprisingly, there have been warnings that coronaviruses are potential causes of a pandemic, as had been previously demonstrated by the occurrence of the SARS-CoV epidemic in 2002–2003, and the Middle East Respiratory Syndrome (MERS) epidemic arising in 2012 [21,22].
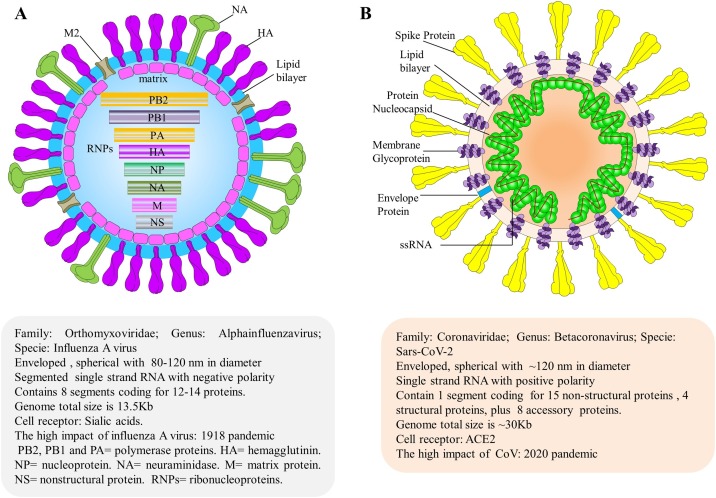
In summary, the two pandemics of the 21st century originated from different viruses, but they have some points in common, such as the fact that they were both caused by enveloped RNA viruses, usually of spherical morphology (see Fig. 1 ) [23,24]. Other points that call attention are related to the frequent mutations and diversity of the hosts that can be infected [19,25,26]. Thus, due to the importance of epidemiological knowledge of the two recent pandemics, this study was carried out to review and synthesize the epidemiological aspects while showing the particularities that differentiate between the pH1N1 and COVID-19 pandemics regarding clinical findings, laboratory diagnosis, and prevention/treatment.
Fig. 1.
Structure and genome organization of human influenza A and SARS-CoV-2. (A) Virion structure of influenza A particles. The organization of influenza A particle is shown schematically to indicate the protein locations and genome viral organization, along with additional information. The virus uses sialic acid cell receptors to adhere to target cells. The highest impact of this virus was in 1918, when the flu pandemic, sometimes referred to as the “Spanish flu,” killed an estimated 50 million people worldwide. (B) Virion structure of SARS-CoV-2. The relative size and organization of the SARS-CoV-2 genome are shown. ACE-2 is the host cell receptor responsible for mediating infection by SARS-CoV-2, the novel coronavirus responsible for the COVID-19 pandemic [23,24].
Epidemiologic description of the pH1N1 and COVID-19 pandemics
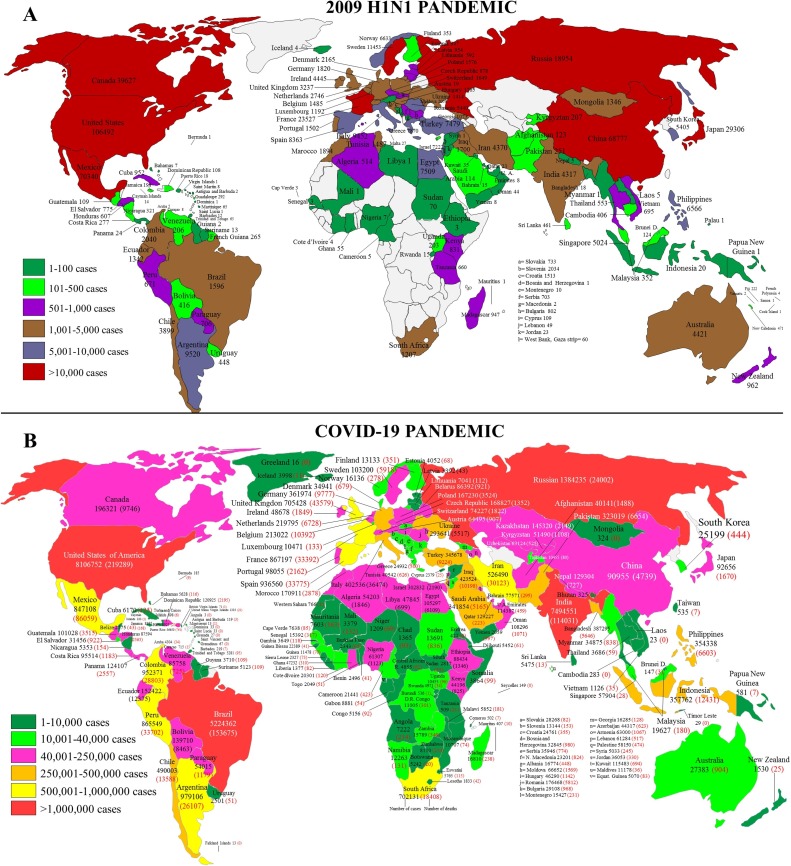
The pH1N1 began at the end of March 2009 and continued until August 2010. The first cases were recorded in Mexico, with subsequent spread to the USA and then the whole world. Thus, after three months, the number of confirmed cases by the WHO was 94,512 in 110 countries, with at least 429 deaths registered [27]. Later, with nine months of influenza A (H1N1) virus circulation, the number of detected cases increased significantly, as can be seen in Fig. 2 A [28]. At the end of pH1N1, 18,500 deaths were confirmed by WHO [29]. However, the study by Dawood and colleagues suggests that these numbers were underestimated and suggested the total number of deaths was actually between 151,700 and 575,400 [30]. Currently, the influenza A(H1N1)pdm09 virus has continued to circulating in human seasonally since the end of flu pandemic [31,32].
Fig. 2.
Geographic distribution of the H1N1 (A) and COVID-19 (B) pandemics. Data were collected about nine months from the initial cases [27,28,38]. Data are from the World Health Organization (https://www.who.int/csr/don/2009_07_06/en/, https://www.who.int/influenza/resources/charts/en/), European Centre for Disease Prevention and Control. COVID-19 (https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases), and were collected through October 18, 2020.
During the pH1N1, it was observed that the most affected age group comprised of children and young adults. Dawood and colleagues found that out of hundreds of confirmed cases of influenza, a total of 40% of patients were aged 10–18 years, while only 5% of cases were adults over 51 years of age [33]. One explanation for this age distribution suggests partial immunity to the virus in the older population [34].
The comorbidity was a risk factor for swine flu. Although some studies only show that about half of deaths from the disease have other cofactors, it is important to note that the incidence of comorbidity increases with age, but the incidence of influenza tended to be higher in young groups. The most common comorbidities identified in patients who died of A H1N1 influenza were hypertension (23%), diabetes (5.7%), obesity (4.2%), neurological diseases (4%), renal failure (3.8%), and pneumopathy (3.7%) [35,36].
An epidemiological study in the USA, it was observed that the swine flu hospitalization rate decreases according to age of patients increases. Thus, the hospitalization rate per 100,000 population was 117, 84, 70, and 90 for peoples with aged 0–17 years, 18–64 years, over 65 years, and all ages, respectively [37].
The current COVID-19 pandemic began in the middle of December 2019 after several cases of pneumonia of unknown etiology had been reported in Wuhan, China. It is possible that the outbreak started in early December or November, with the number of cases increasing rapidly in the coming months, eventually spreading to more than 203 countries and territories (Fig. 2B). As of October 18, 2020, a total of 39,774,852 cases of COVID-19 were confirmed worldwide, with 1,110,902 deaths [17,18,38].
The causative agent of the outbreak was quickly identified, and its genomic sequence was made available on January 12, 2020. Because the genomic sequence was closely related to the 2003 SARS-CoV, the novel CoV was identified as SARS-CoV-2 [19]. Besides, studies have found that this virus probably originated from bats but may have been amplified in an intermediate host before transmission to humans [39]. Thus, pangolins have been proposed as a potential intermediate host; however, the zoonosis of pangolins-SARS-CoV-2 needs to be clarified [40]. In addition, although much has been learned about COVID-19 in the nine months since this outbreak began, but there remain significant knowledge gaps in understanding COVID-19 at current stage [41].
The age group most affected by COVID-19 has been adults over 40 years old (∼73%), while only 10% of cases are under 30 years old. Thus, COVID-19 hospitalization rate increases according to age of the patients. This positive association shows that individuals over the age of 85 years have the highest rate of hospitalization (833 positive cases in each group of 100,000). Soon after, the hospitalization rates are 460/100,000, 116/100,000, and 17/100,000 for patients over the age of 65 years, 18–49 years, and 0–4 years, respectively. The overall cumulative hospitalization rate was 170 per 100,000 population in the USA [[42], [43], [44]].
The prevalence of comorbidity is high in people hospitalized with COVID-19. It is noteworthy that a large proportion of patients aged 60 years and older presenting multiple chronic diseases. The most common conditions contributing to deaths involving COVID-19 were influenza and pneumonia (42%), respiratory failure (34%), hypertension (21.7%), cardiac arrest (12.3%), diabetes (16%), ischemic heart (11%), and vascular and unspecified dementia (11%) [45].
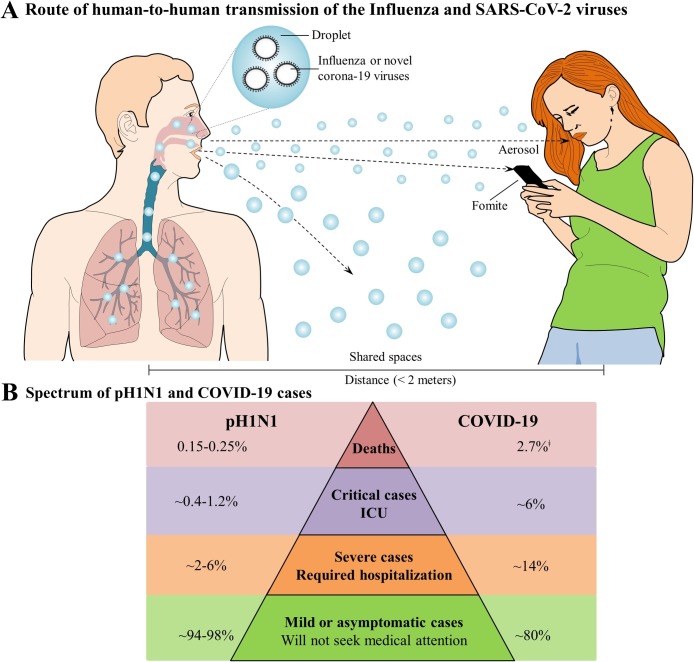
One of the key factors in the epidemiology of infectious diseases refers to the transmission efficiency of the pathogen. In this regard, the influenza viruses and SARS-CoV-2 are both efficient in causing respiratory disease because they easily spread among humans through oral and nasal droplets. Additionally, they can also be transmitted via indirect contact (see Fig. 3 A) [[46], [47], [48]].
Fig. 3.
Transmission and spectrum of human influenza A and SARS-CoV-2 viruses. (A) Route of human-to-human transmission of influenza and SARS-CoV-2 viruses. The SARS-CoV-2 primary transmission mode is person-to-person contact through respiratory droplets generated by breathing, sneezing, coughing, etc. In addition, the viruses may be transmitted among humans by direct contact with an infected subject or indirect contact, trough hand-mediated transfer of the virus from contaminated fomites to the mouth, nose, or eyes [[46], [47], [48]]. (B) The spectrum of pH1N1 and COVID-19 cases. Although the transmission route for both viruses is the same, the spectrum of cases for both differ greatly: mild or asymptomatic infections are most common for both viruses; however, a higher percentage of mild or asymptomatic cases were observed for pH1N1 (approximately 94–98%) than for SARS-CoV-2 (80%) [11,38,89]. The estimate of the COVID-19 case fatality rates has varied widely, with the data in Fig. 3B representing the current overall number of reported deaths divided by the number of reported cases at a specific time point. ǂRemains to be determined.
The course of an epidemic is defined by several key factors, some of which are represented by the effective reproduction number R0 (also called the basic reproduction number), defined as the average number of secondary cases produced by a typical case in a particular population [49]. Based on this epidemiological concept, the COVID-19 transmission patterns have been better identified. Consequently, it is known that the R0 of COVID-19 has been approximately 3 (95% CI 2.65–3.39), which is superior to that of influenza A (R0 = 1.3–1.7) [[50], [51], [52], [53]].
Regarding the pre-symptomatic transmission of pH1N1, few studies have documented about infectiousness during the pre-symptomatic phase. Consequently, only three studies had this purpose, with two of them finding evidence of pre-symptomatic transmission of the virus among close contacts [[54], [55], [56]]. On the other hand, it appears that pre-symptomatic transmissions of SARS-CoV-2 infections could be somewhat common and would have a significant contribution to the overall SARS-CoV-2 transmission rate. In this context, highlight the study by He et al., which inferred that 44% of secondary cases were infected during pre-symptomatic stages of disease [57].
Referring to preliminary data on the overall case-fatality ratio, calculated from total numbers of reported deaths by the number de reported cases, in the first three months of each pandemic, the case-fatality ratio were 0.45% and 4.7% for pH1N1 and COVID-19, respectively [27,38]. Subsequently in the pH1N1, the values were 0.15–0.25%, while during the COVID-19 pandemic; the overall case-fatality ratio was estimated to be 2.7%, which was calculated by using different populations (Fig. 3B). For the two pandemics, the case fatality rates vary widely, and over time, possibly because of adequate/inadequate healthcare, coinfections, comorbidities, and patient demographics (i.e., older patients), which suggests considerable uncertainty over the exact COVID-19, and pH1N1 case fatality rates [11,27,58].
A brief comparison between clinical aspects and laboratory diagnoses: pH1N1 vs COVID-19
Among the pH1N1 and COVID-19 cases, there are similarities in the clinical spectra. However, COVID-19 cases tend to be more severe as they often evolve to SARS, with these patients needing hospital care for several days [59], mainly because they need mechanical ventilation support.
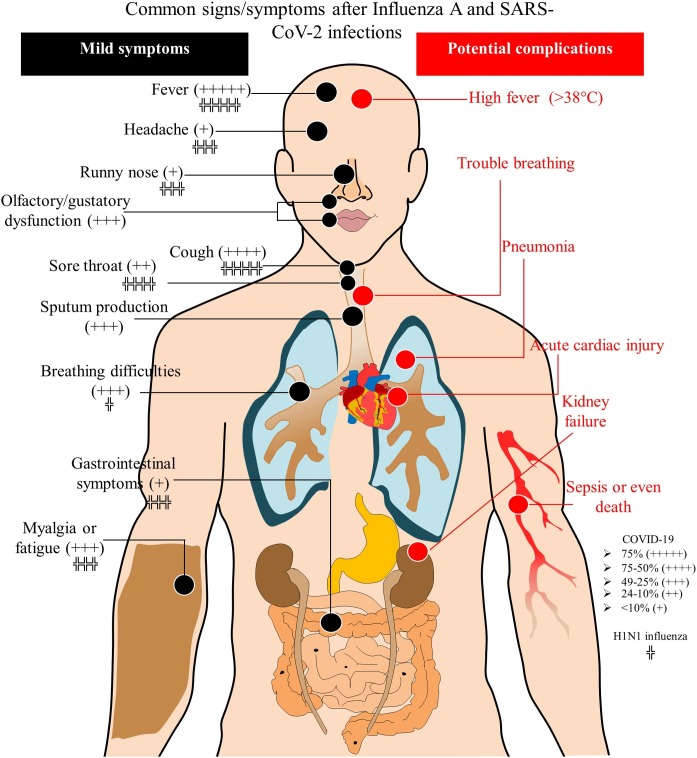
For pH1N1, the incubation period was 1.5–3 days, but sometimes it extended to 7 days [60]. For COVID-19, the incubation period is usually longer (2–14 days), with an average of 5.2 days. The clinical picture usually tends to start with fever and cough that is sometimes accompanied by a sore throat and myalgia (see Fig. 4 ) [[60], [61], [62], [63], [64]].
Fig. 4.
Schematic image of the clinical picture of influenza A and COVID-19 [18,60,64,[90], [91], [92], [93], [94]].
For the laboratory diagnosis of influenza A and SARS-CoV-2, clinical samples including a swab of the throat or nasopharynx, a saliva sample or aspirate of the lower respiratory system are indicated, with the collections being representative of the acute phase of the disease [63]. Additionally, samples of whole blood or serum/plasma can be collected in the acute and convalescent phase to be paired with other samples and provide evidence of serological conversion. Rapid techniques for detecting the influenza virus include immunofluorescence and immunoenzymatic assays [[65], [66], [67]]. For both viruses, real-time RT-PCR represents the gold standard technique [68]. Rapid immunochromatographic tests will be extremely important for mass screening of individuals in the COVID-19 pandemic [69,70]. Another option for detecting SARS-CoV-2 includes diagnostic imaging, which is obtained by computed tomography of the chest, but this technique is limited by its low specificity [71].
Prevention and treatment strategies for pH1N1 vs COVID-19
For infectious diseases of epidemic potential, several mitigation measures are indicated to reduce transmission and delay the spread of the infectious agent. These measures are extremely important when the transmissibility of the disease is high and when a high proportion of cases require hospitalization [72]. These measures to contain the rapid viral spread are even more important when there is the emergence of a new highly contagious disease for which there are no drugs or vaccines available. There are ten measures to reduce the transmission of infectious diseases: (1) frequent hand hygiene with soap and water/or disinfection with 70% alcohol gel; (2) periodic disinfection of surfaces with 70% alcohol or 1% sodium hypochlorite targeted at different segments of society and public places; (3) social distancing to keep a distance of approximately 2 m between people and instruction to avoid touching the eyes, nose and mouth; (4) isolation of sick people; (5) contact tracking; (6) quarantine of exposed people; (7) school closing; (8) changes in the workplace (home office, delivery works); (9) prohibition of all types of gatherings; and (10) restricted travel [73].
In 2009, pH1N1 was combated with the dissemination of daily reports by the media. In these reports, proper handwashing procedures and avoiding close contact among people were highlighted. However, some regions of the countries most affected by pH1N1 adopted other measures to contain the virus, including the social isolation of some segments of society. In this scenario, the United States, Mexico, Singapore, Canada and China (Hong Kong) adopted measures of social distancing in the workplace and in schools [74,75]. Additionally, school closures were one of the main pillars of the response to prevention of pH1N1 because the age group most affected was children and young people [30].
In the pH1N1, Mexico was one of the few countries that adopted the most stringent measures of social isolation from April 23 to May 11, 2009. Initially, these measures were implemented in the metropolitan region of Mexico City, with social isolation persisting for approximately two weeks, including forced school closures and the closure of all economic activities considered nonessential for approximately ten days. Subsequently, part of these measures was adopted across the country [76]. However, for the COVID-19 pandemic, a lockdown was adopted with the closing of companies, schools and public places, and social isolation and travel restrictions were intensified in various regions of the world to reduce the spread of SARS-CoV-2 [77,78]. Consequently, due to these factors adopted in several countries, the current pandemic will plunge the world into a serious economic crisis.
The vaccine against H1N1 infection was the most effective prevention and control measure, as it prevented the virus from spreading and mitigated the severity and impact of the disease. Thus, the experience with vaccines against seasonal influenza allowed for small adjustments and the addition of the representative pH1N1 strain to the vaccination. The result was immediate, and approximately five months after the first cases started, China was the first country to start mass vaccination [79].
Another factor that mitigated the impact of pH1N1 was the existence of guiding principles for the treatment of the disease. Thus, due to the knowledge generated from combating the circulation of different strains of seasonal influenza, antiviral treatment was already well established, such as the use of oseltamivir (Tamiflu) and zanamivir (Relenza). Therefore, in the first cases of pH1N1, safe and effective antiviral drugs were already available for use in the treatment and prophylaxis of H1N1. This was extremely important, as it contributed to reducing the length of hospital stay, reducing the risk of disease progression to a severe clinical condition and reducing the rate of contagion [80].
In contrast to pH1N1, the COVID-19 pandemic does not have an antiviral drug or vaccine available. Thus, given the high number of hospitalized and severe cases of the disease, added to the high contagiousness of the virus and the entire population being exposed to infection, it is extremely urgent to establish treatments and vaccines against the current pandemic. Consequently, a rapid search for potential antiviral drugs against SARS-CoV-2 was initiated a few months ago [81,82]. The most practical way to obtain antivirals is to test other known antivirals or other established drugs. This contributes to a time gain, since some safety parameters of the medication are already known, for prompt availability of the medication on the market. Initially, remdesivir, (hydroxy)chloroquine, lopinavir/ritonavir, ribavirin, nitazoxanide and nelfinavir were proposed as antiviral therapies against SARS-CoV-2 [83,84]. Many of them have been tested in compassionate therapies, including the use of convalescent plasma, in patients with severe COVID-19 [85,86]. However, for a better understanding of their effectiveness, randomized, placebo-controlled clinical trials are required.
It is believed that the humanitarian and economic impact of the COVID-19 pandemic will contribute to new platforms for the next generation of vaccines aimed at accelerating the development of vaccines. Following this path of producing something with unprecedented speed, the first candidate for the COVID-19 vaccine entered human clinical trials on March 16, 2020. Since then, 78 vaccine candidates have been confirmed as active. Some of these are already in the clinical development phase (Phase III) and include (1) an encapsulated mRNA vaccine encoding S protein; (2) an adenovirus type 5 vector that expresses S protein; (3) a DNA plasmid-encoding S protein delivered by electroporation; (4) dendritic cells modified with a lentiviral vector expressing synthetic minigene based on domains of selected viral proteins of selected proteins; and (5) artificial antigen-presenting cells modified with a lentiviral vector expressing synthetic minigene of selected viral proteins. Given the emergence of the pandemic, it is estimated that a vaccine will be available in early 2021 [87,88].
Funding
VGC was supported by scholarships from Fundação de Apoio à Pesquisa do Distrito Federal (FAPDF) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
None declared.
Ethical approval
Not required.
CRediT authorship contribution statement
Vivaldo Gomes da Costa: Conceptualization, Data curation, Formal analysis, Methodology, Writing - original draft, Writing - review & editing. Marielena Vogel Saivish: Data curation, Formal analysis, Methodology, Writing - original draft, Writing - review & editing. Dhullya Eduarda Resende Santos: Formal analysis, Writing - review & editing. Rebeca Francielle de Lima Silva: Formal analysis, Writing - review & editing. Marcos Lázaro Moreli: Writing - review & editing.
References
- 1.Bedford J., Farrar J., Ihekweazu C., Kang G., Koopmans M., Nkengasong J. A new twenty-first century science for effective epidemic response. Nature. 2019;575(7):130–136. doi: 10.1038/s41586-019-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) 2009. H1N1 pandemic timeline.https://www.cdc.gov/flu/pandemic-resources/2009-pandemic-timeline.html [Google Scholar]
- 3.World Health Organization (WHO). WHO timeline-COVID-19. https://www.who.int/news-room/detail/08-04-2020-who-timeline---covid-19. Accessed Apr 2020.
- 4.Gubler D.J., Vasilakis N., Musso D. History and emergence of zika virus. J Infect Dis. 2017;216(S10):S860–S867. doi: 10.1093/infdis/jix451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahid B., Ali A., Rafique S., Iddres M. Global expansion of chikungunya virus: mapping the 64-year history. Int J Infect Dis. 2017;58:69–76. doi: 10.1016/j.ijid.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Malvy D., McElroy A., Clerck H., Günther S., Griensven J.V. Ebola virus disease. Lancet. 2019;393:936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 7.Ruopp M., Chiswell K., Thaden J.T., Merchant K., Tsalik E.L. Respiratory tract infection clinical trials from 2007 to 2012: a systematic review of ClinicalTrials.gov. Ann Am Thorac Soc. 2015;12:1852–1863. doi: 10.1513/AnnalsATS.201505-291OC. [DOI] [PubMed] [Google Scholar]
- 8.Zimmer S.M., Burke D.S. Historical perspective - emergence of influenza a (H1N1) viruses. N Engl J Med. 2009;361:279–285. doi: 10.1056/NEJMra0904322. [DOI] [PubMed] [Google Scholar]
- 9.Paules C., Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow D.L., Finelli L., Bridges C.B. H1N1 influenza pandemic: field and epidemiologic investigations in the United States at the start of the first pandemic of the 21st century. Clin Infect Dis. 2009;2009(52):S1–S3. doi: 10.1093/cid/ciq005. [DOI] [PubMed] [Google Scholar]
- 11.Girard M.P., Tam J.S., Assossou O.M., Kieny M.P. The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine. 2010;28:4895–4902. doi: 10.1016/j.vaccine.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Scalera N.M., Mossad S.B. The first pandemic of the 21st century: review of the 2009 pandemic variant influenza a (H1N1) virus. Postgrad Med. 2015;121(5):43–47. doi: 10.3810/pgm.2009.09.2051. [DOI] [PubMed] [Google Scholar]
- 13.Smith G.J.D., Vijaykrishna D., Bahl J., Lycett S.J., Worobey M., Pybus O.G. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1126. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 14.Peiris J.S.M., Poon L.L.M., Guan Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol. 2009;45(3):169–173. doi: 10.1016/j.jcv.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallat B. Flu: no sign so far that the human pandemic is spread by pigs. Nature. 2009;460:683. doi: 10.1038/460683b. [DOI] [PubMed] [Google Scholar]
- 16.Rambaut A., Pybus O.G., Nelson M.I., Viboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya A.E., Baker S.C., Baric R.S., Groot R.J., Drosten C., Gulyaeva A.A. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arabi Y.M., Balkhy H.H., Hayden F.G., Bouchama A., Luke T., Baillie J.K. Middle east respiratory syndrome. N Engl J Med. 2017;376:6. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouchier R.A.M., Kuiken T., Schutten M., Amerongen G.v., Doornum G.J.J., v., Hoogen B.G.v.d. Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw M.L., Palese P. Orthomyxoviridae. In: Knipe D.M., Howley P.M., editors. Fields virology. 6th ed. Lippincott Williams and Wilkins; Philadelphia, PA: 2013. pp. 1151–1184. [Google Scholar]
- 24.Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon S.W., Webby R.J., Webster R.G. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol. 2014;385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 26.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. PNAS. 2020 doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO. Pandemic (H1N1) 2009 - update 58. https://www.who.int/csr/don/2009_07_06/en/. [Accessed April 2020].
- 28.WHO Influenza surveillance report. Combined surveillance graphs by country, date range: 01/04/2009-20/11/2009. https://www.who.int/influenza/resources/charts/en/. [Accessed July 2020].
- 29.WHO. Pandemic (H1N1) 2009 - update 112. https://www.who.int/csr/don/2010_08_06/en/. [Accessed April 2020].
- 30.Dawood F.S., Iuliano A.D., Reed C., Meltzer M.I., Shay D.K., Cheng P.Y. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 31.Rabaan A.A., Alshaikh S.A., Bazzi A.M. Influenza a(H1N1)pdm09 epidemiology in the Eastern Province of Saudi Arabia. J Infect Public Heal. 2018;11(5):636–639. doi: 10.1016/j.jiph.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention. Influenza: Types of Influenza Viruses. https://www.cdc.gov/flu/about/viruses/types.htm. [Accessed July 2020].
- 33.Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J. Emergence of a novel swine-origin influenza a (H1N1) virus in humans. N Engl J Med. 2009;360(25):2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention (CDC) 2009 Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. [PubMed] [Google Scholar]
- 35.Jain S., Kamimoto L., Bramley A.M., Schmitz A.M., Benoit S.R., Louie J. Hospitalized patients with 2009 H1N1 influenza in the United States, April–June 2009. N Engl J Med. 2009;361:1935–1944. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Flores E., Izquierdo-Puente J., Castilho-Pérez J.J., Ramírez-Rosales G., Grijalva-Otero I., Lopez-Macias C. Quantifying the mortality caused by the H1N1 influenza virus during the 2009 pandemic in Mexico. J Infect Dev Ctries. 2014;8(6):742–748. doi: 10.3855/jidc.3622. [DOI] [PubMed] [Google Scholar]
- 37.Shrestha S.S., Swerdlow D.L., Borse R.H., Prabhu V.S., Finelli L., Atkins C.Y. Estimating the Burden of 2009 Pandemic Influenza A (H1N1) in the United States (April 2009-April 2010) Clin Infect Dis. 2011;52:75–82. doi: 10.1093/cid/ciq012. [DOI] [PubMed] [Google Scholar]
- 38.European Centre for Disease Prevention and Control. COVID-19. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases. [Accessed 18 October 2020].
- 39.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 41.Damme W.V., Dahake R., Delamou A., Ingelbeen B., Wouters E., Vanham G. The COVID-19 pandemic: diverse contexts; different epidemics—how and why? BMJ Glob Health. 2020;5(7) doi: 10.1136/bmjgh-2020-003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z., Bing X., Zhi X.Z. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)-China, 2020. CCDC Weekly. 2020;10(41):145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [Google Scholar]
- 43.Zhou F., Yu T., Du R., Fan G., Liu Z., Xiang J. Clinical course and risk factors for mortality of adult inpatients with Covid-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonnet A., Chetboun M., Poissy J., Raverdy V., Noulette J., Duhamel A. High prevalence of obesity in severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) requiring invasive mechanical ventilation. Obesity. 2020;28:1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CDC. Coronavirus Disease 2019 (covid-19). https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html. [Accessed September 2020].
- 46.CDC Weekly Updates by Select Demographic and Geographic Characteristics. https://www.cdc.gov/nchs/nvss/vsrr/covid_weekly/index.htm#Comorbidities. [Accessed September 2020].
- 47.Lakdawala S.S., Subbarao K. The ongoing battle against influenza: the challenge of flu transmission. Nature Med. 2012;18(10):1468–1470. doi: 10.1038/nm.2953. [DOI] [PubMed] [Google Scholar]
- 48.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:1–3. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boëlle P.Y., Ansart S., Cori A., Valleron A.J. Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza Other Respir Viruses. 2011;5:306–316. doi: 10.1111/j.1750-2659.2011.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y., Sugimoto J.D., Halloran M.E., Basta N.E., Chao D.L., Matrajt L. The transmissibility and control of pandemic influenza a (H1N1) virus. Science. 2009;326(5953):729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majumder M.S., Mandl K.D. Early in the epidemic: impact of preprints on global discourse about COVID-19 transmissibility. Lancet Glob Health. 2020;8(5):e627–630. doi: 10.1016/S2214-109X(20)30113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2):1–4. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freitas F.T.M., Cabral A.P.S., Barros E.N.C., Burigo M.J.O., Prochnow R.D., Silva L.A. Pre-symptomatic transmission of pandemic influenza H1N1 2009: investigation of a family cluster, Brazil. Epidemiol Infect. 2012;141(4):763–766. doi: 10.1017/S0950268812001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu Y., Komiya N., Kamiya H., Yasui Y., Taniguchi K., Okabe N. Pandemic (H1N1) 2009 transmission during presymptomatic phase, Japan. Emerg Infect Dis. 2011;17(9):1737–1739. doi: 10.3201/eid1709.101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hermes J., Bernard H., Buchholz U., Spackova M., Löw J., Loytved G. Lack of evidence for pre‐symptomatic transmission of pandemic influenza virus A(H1N1) 2009 in an outbreak among teenagers; Germany, 2009. Influenza Other Respir Viruses. 2011;5(6):e499–e503. doi: 10.1111/j.1750-2659.2011.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 59.WHO. Coronavirus disease 2019 (COVID-19). Situation Report 70. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200330-sitrep-70-covid-19.pdf?sfvrsn=7e0fe3f8_4. [Accessed April 2020].
- 60.Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf. [Accessed April 2020].
- 61.Bautista E., Chotpitayasunondh T., Gao Z., Harper S.A., Shaw M., Uyeki T.M. Clinical aspects of pandemic 2009 influenza a (H1N1) virus infection. N Engl J Med. 2010;362(18):1708–1719. doi: 10.1056/NEJMx100025. [DOI] [PubMed] [Google Scholar]
- 62.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–Infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain R., Goldman R.D. Novel Influenza A (H1N1) clinical presentation, diagnosis, and management. Pediatr Emerg Care. 2009;25:791–796. doi: 10.1097/PEC.0b013e3181c3c8f8. [DOI] [PubMed] [Google Scholar]
- 65.Agyeman A.A., Chin K.L., Landersdorfer C.B., Liew D., Ofori-Asenso R. Smell and taste dysfunction in patients with COVID-19: a systematic review and meta-analysis. Mayo Clin Proc. 2020;95(8):1621–1631. doi: 10.1016/j.mayocp.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jernigan D.B., Lindstrom S.L., Johnson J.R., Miller J.D., Hoelscher M., Humes R. Detecting 2009 Pandemic Influenza A (H1N1) Virus Infection: Availability of Diagnostic Testing Led to Rapid Pandemic Response. Clin Infect Dis. 2011;52(S1):S36–S43. doi: 10.1093/cid/ciq020. [DOI] [PubMed] [Google Scholar]
- 67.Centers for Disease Control and Prevention. Information for Clinicians on Rapid Diagnostic Testing for Influenza. https://www.cdc.gov/flu/professionals/diagnosis/rapidclin.htm. [Accessed April 2020].
- 68.Lu C.Y., Chang L.Y., Chen P.J., Xia N.S., Shao P.L., Huang L.M. A highly specific ELISA for diagnosis of 2009 influenza A (H1N1) virus infections. J Formos Med Assoc. 2012;111:693–697. doi: 10.1016/j.jfma.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 69.WHO. Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance. [Accessed April 2020].
- 70.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M. Diagnostic testing for severe acute respiratory syndrome–related Coronavirus-2: a narrative review. Ann Intern Med. 2020;4:1–10. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81(1):e28–32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 case. Radiology. 2020;296(2):e32–40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahtani K.R., Heneghan C., Aronson J.K. What is the evidence for social distancing during global pandemics? A rapid summary of current knowledge. www.cebm.net/oxford-covid-19/. [Accessed April 2020].
- 74.Guidance on social distancing for everyone in the UK. https://www.gov.uk/government/publications/covid-19-guidance-on-social-distancing-and-for-vulnerable-people/guidance-on-social-distancing-for-everyone-in-the-uk-and-protecting-older-people-and-vulnerable-adults. [Accessed April 2020].
- 75.Ahmed F., Zviedrite N., Uzicanin A. Effectiveness of workplace social distancing measures in reducing influenza transmission: a systematic review. BMC Public Health. 2018;18:518. doi: 10.1186/s12889-018-5446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uscher-Pines L., Schwartz H.L., Ahmed F., Zheteyeva Y., Meza E., Baker G. School practices to promote social distancing in K-12 schools: review of influenza pandemic policies and practices. BMC Public Health. 2018;18:406. doi: 10.1186/s12889-018-5302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Velasco-Hernandez J.X., Leite M.C.A. A model for the A(H1N1) epidemic in Mexico, including social isolation. Salud Públ Méx. 2009;53(1):1–8. doi: 10.1590/s0036-36342011000100007. [DOI] [PubMed] [Google Scholar]
- 78.Sjodin H., Wilder-Smith A., Osman S., Farooq Z., Rocklov J. Only strict quarantine measures can curb the coronavirus disease (COVID-19) outbreak in Italy, 2020. Euro Surveill. 2020;25(13) doi: 10.2807/1560-7917.ES.2020.25.13.2000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicola M., O’Neill N., Sohrabi C., Khan M., Agha M., Agha R. Evidence based management guideline for the COVID-19 pandemic-review article. Int J Surg. 2020;77:206–216. doi: 10.1016/j.ijsu.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stone R. Swine flu outbreak. China first to vaccinate against novel H1N1 virus. Science. 2009;325:1482–1483. doi: 10.1126/science.325_1482. [DOI] [PubMed] [Google Scholar]
- 81.Rewar S., Mirdha D., Rewar P. Treatment and prevention of pandemic H1N1 influenza. Ann Glob Health. 2015;81(5):645–653. doi: 10.1016/j.aogh.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 82.Barlow A., Landolf K.M., Barlow B., Yeung S.Y.A., Heavner J.J., Claassen C.W. Review of emerging pharmacotherapy for the treatment of coronavirus disease 2019. Pharmacotherapy. 2020;40(5):416–437. doi: 10.1002/phar.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baden L.R., Rubin E.J. Covid-19-the search for effective therapy. N Engl J Med. 2020;382(19):1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir–Ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. PNAS. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 89.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen N., Zhau M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang W., Cao Q., Qin L., Wang X., Cheng Z., Pan A. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):a multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80:388–393. doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian S., Hu N., Lou J., Chen K., Kang X., Xiang Z. Characteristics of COVID-19 infection in Beijing. J Infect. 2020;80:401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]