Abstract
Circulating exosomal microRNAs (ExmiRNAs) provide an ideal non-invasive method for cancer diagnosis. In this study, we evaluated two circulating ExmiRNAs in NSCLC patients as a diagnostic tool for early-stage non-small lung cancer (NSCLC). The exosomes were characterized by qNano, transmission electron microscopy, and Western blot, and the ExmiRNA expression was measured by microarrays. The differentially expressed miRNAs were verified by RT-qPCR using peripheral blood specimens from NSCLC patients (n = 276, 0 and I stage: n = 104) and healthy donors (n = 282). The diagnostic values were measured by receiver operating characteristic (ROC) analysis. The results show that the expression of both ExmiR-20b-5p and ExmiR-3187-5p was drastically reduced in NSCLC patients. The area under the ROC curve (AUC) was determined to be 0.818 and 0.690 for ExmiR-20b-5p and ExmiR-3187-5p, respectively. When these two ExmiRNAs were combined, the AUC increased to 0.848. When the ExmiRNAs were administered with either carcinoembryonic antigen (CEA) or cytokeratin-19-fragment (CYFRA21-1), the AUC was further improved to 0.905 and 0.894, respectively. Additionally, both ExmiR-20b-5p and ExmiR-3187-5p could be used to distinguish early stages NSCLC (0 and I stage) from the healthy controls. The ROC curves showed that the AUCs were 0.810 and 0.673, respectively. Combination of ExmiR-20b-5p and ExmiR-3187-5p enhanced the AUC to 0.838. When CEA and CYFRA21-1 were administered with the ExmiRNAs, the AUCs were improved to 0.930 and 0.928, respectively. In summary, circulating serum exosomal miR-20b-5p and miR-3187-5p could be used as effective, non-invasive biomarkers for the diagnosis of early-stage NSCLC, and the effects were further improved when the ExmiRNAs were combined.
Impact statement
The high mortality of non-small cell lung cancer (NSCLC) is mainly because the cancer has progressed to a more advanced stage before diagnosis. If NSCLC can be diagnosed at early stages, especially stage 0 or I, the overall survival rate will be largely improved by definitive treatment such as lobectomy. We herein validated two novel circulating serum ExmiRs as diagnostic biomarkers for early-stage NSCLC to fulfill the unmet medical need. Considering the number of specimens in this study, circulating serum exosomal miR-20b-5p and miR-3187-5p are putative NSCLC biomarkers, which need to be further investigated in a larger randomized controlled clinical trial.
Keywords: Diagnostic biomarkers, non-small cell lung cancer, circulation, exosomes, miRNAs
Introduction
Lung cancer accounts for 14% of all newly diagnosed cancers worldwide in 2018.1 Despite the fact that the rapid development of current diagnostic methods and much improved treatment strategies, such as targeted molecular therapies, the survival rate has not been significantly improved (≤15%) for lung cancer patients.2,3 In current oncology studies, liquid biopsy has been extensively used in the diagnosis of cancers and the response measurements of cancer treatments.4–6 Liquid biopsy can be used to study circulating tumor cells (CTC), cell-free non-coding RNAs, cell-free DNA (cfDNA), and extracellular vesicles (EVs), including exosomes.7,8
Exosomes are produced by cells and secreted into many types of human body fluids, such as urine, hydrothorax, ascites, etc.9–11 Exosomes function as carriers for a variety of substances such as DNAs, proteins, and other genetic materials, which play essential roles in the regulations of their target genes and cells. In recent years, exosomes have been an emerging focus in scientific cancer research. Growing evidence has shown that exosomes regulate the progress and resistance of tumors by modulating a wide range of pathways.12–14 Additionally, one of the most principal and interesting respects of exosomes is its diagnostic and/or prognostic potential in the treatment of diseases, especially malignancies.
MiRNAs are short and non-coding RNAs; dysregulation of miRNAs is closely related with the derivation and progression of many diseases, such as cardiovascular diseases, nervous system diseases, and cancers.15–17 Increasing studies have reported that miRNAs are stably contained within exosomes rather than plasma or serum.18,19 In addition, exosomal miRNAs often respond differently to different pathological conditions, which directly reflect the physiological status of originally derived cells. Hence, exosomal miRNAs have been proposed as non-invasive diagnostic biomarkers for cancer.20–22 In colorectal cancer patients, poor prognosis is found to be related with the decreased expression of exosomal miR-150-5p in serum, suggesting it may be used in colorectal cancer diagnosis and prognosis.23 Ma et al.10 revealed that, in neuroblastoma patients, exosomal hsa-miR199a-3p shows an increased expression, which in turn promotes tumor proliferation and migration, indicating that it may be applied as a diagnostic biomarker for neuroblastoma. Compared with chronic pancreatitis (CP) patients, the relative expression of ex-miR21 and ex-miR-155 is dramatically elevated in pancreatic ductal adenocarcinoma (PDAC) patients, indicating these two Ex-miRNAs may be applied as diagnostic biomarkers for PDAC.24
The clinical significance of circulating serum exosomal miRNA-20b-5p and miRNA-3187-5p as biomarkers in NSCLC patients has yet to be investigated. In this study, we evaluated the expression signature and diagnostic power of these Ex-miRNAs in early-stage NSCLC patients.
Materials and methods
Patients, sample preparation, and study design
A total of 276 NSCLC patients (stage 0–I, n = 104 and stage II–IV, n = 172) and 282 healthy controls from Shandong Cancer Hospital Affiliated to Shandong First Medical University were enrolled between April 2019 to July 2019. All the enrolled patients had not received any treatment before sampling. The healthy control was selected from a healthy population who participated in the physical examination. Pathological and clinical characteristics of patients, such as gender and age, were obtained from the patients' records (Table S1). The participants who had a history of any tumor disease were excluded, as well as those with increased levels of serum tumor markers (Table S2).
Approximately 4 mL peripheral blood from each patient and healthy donor was collected and stored in separating gel coagulating-promoting tubes. Within 1 h, the serums were centrifuged at 1000 × g at 4°C for 10 min and stored at −80°C.
A four-phase study was conducted to obtain serum exosomal miRNAs for the diagnosis of early-stage NSCLC. In the screening phase, differentially expressed ExmiRNAs was identified between one pool of healthy control (HC) and two pools of NSCLC samples (five peripheral serum samples from the healthy control, five non-metastatic NSCLC patients, and five metastatic NSCLC patients) using microarray. Five samples were combined together as one pool sample. A total of 28 miRNAs (19 down-regulated and nine up-regulated, Table 1) were selected using the criteria of >2.0-fold difference. In the training stage, these 28 dysregulated ExmiRNAs were determined by qRT-PCR using 48 HCs and 48 NSCLC samples. In the testing stage, the identified ExmiRNAs were validated in serum exosome samples of 72 HCs and 72 NSCLC patients. In the last stage, 318 samples (162 controls and 156 NSCLC) were collected to further evaluate the ExmiRNAs in NSCLC.
Table 1.
miRNAs expression profiling (9 up-regulated and 19 down-regulated).
| miRNA | Fold change | Description | miRNA | Fold change | Description |
|---|---|---|---|---|---|
| hsa-miR-764 | 3.9459 | Up | hsa-miR-26b-5p | 0.3113 | Down |
| hsa-miR-520c-3p | 3.8483 | Up | hsa-miR-5197-5p | 0.3084 | Down |
| hsa-miR-936 | 3.1372 | Up | hsa-miR-3136-3p | 0.3048 | Down |
| hsa-miR-593-3p | 2.8005 | Up | hsa-miR-107 | 0.3008 | Down |
| hsa-miR-522-3p | 2.7530 | Up | hsa-miR-146a-5p | 0.2972 | Down |
| hsa-miR-204-3p | 2.3536 | Up | hsa-miR-185-5p | 0.2721 | Down |
| hsa-miR-588 | 2.1621 | Up | hsa-miR-103a-3p | 0.2712 | Down |
| hsa-miR-96-3p | 2.1256 | Up | hsa-miR-375 | 0.2622 | Down |
| hsa-miR-642a-5p | 2.0915 | Up | hsa-miR-186-5p | 0.2554 | Down |
| hsa-miR-335-3p | 0.4292 | Down | hsa-miR-491-3p | 0.2531 | Down |
| hsa-miR-425-5p | 0.3811 | Down | hsa-miR-4764-3p | 0.2413 | Down |
| hsa-miR-320a | 0.3643 | Down | hsa-miR-20b-5p | 0.2387 | Down |
| hsa-miR-30c-5p | 0.3313 | Down | hsa-miR-502-3p | 0.2335 | Down |
| hsa-miR-126-5p | 0.3236 | Down | hsa-miR-3187-5p | 0.1324 | Down |
Isolation of exosomes
The exosomes were collected by ultracentrifugation according to the procedure described previously.25 In summary, the serum was thawed on ice, and subject to centrifugation at 10,000 × g at 4°C for 30 min. Then, the exosomes were isolated by ultracentrifuge at 100,000 × g at 4°C for 2 h.
Characterization of exosomes
The size and distribution of the nanoparticle were detected on a qNano (New Zealand) according to the operation manual, which was then analyzed using the Izon Control Suite. Then, exosomes samples (15 µL) were added on a formvar-coated copper grid. The remaining liquid was removed using a filter paper, and the exosomes were stained in 15 µL 2% uranyl acetate staining solution at 20°C for 1 min. After wiping off the excessive liquid, the copper grid was baked for 10 min. Then, the exosomes were observed on a FEI Tecnai T20 transmission electron microscope (FEI Company, USA).
Western immunoblotting was performed using rabbit primary antibodies against GM130, TSG101 and CD9 at 4°C, respectively. The PVDF membrane was then incubated with HRP-conjugated secondary antibodies for 1 h at 20°C. The ECL blotting detection reagent (Bio-Rad, USA) was used for the visualization of the immunoreactive bands.
RNA isolation
All experiments were conducted in an RNase-free area. The total RNAs were isolated using the TRIzol reagent (CA, USA). The RNAs was quantified using a NanoDrop spectrophotometer (MA, USA). Then, complementary DNA (cDNA) was obtained by reverse-transcription using the Mix-X miRNA first strand synthesis kit (Kusatsu, Japan).
MiRNA profiling and data analyses
Serum exosomal RNAs were extracted from 5 additional healthy controls and 10 NSCLC patients. The samples were hybridized onto a miRCURYTM LNA array (v.19.0), after the exosomal RNAs were labeled by the miRCURY™ Hy3™/Hy5™ power labeling kit (Vedbaek, Denmark), The expression data were normalized using the miRNAs with intensities ≥30. The significantly differentially expressed miRNAs were obtained using the criteria of fold change ≥2.0. MiRNA expression levels between the samples were distinguished by hierarchical clustering.
MicroRNA detection by real-time PCR
RT-PCR was performed on the Roche LightCycler480 System (Basel, Switzerland). The relative expression of miRNAs was calculated using the following equation, ΔCT = CtmiRNA-CtU6, as described previously.26 Each experiment was performed in triplicate with U6 as an internal control.27
Statistical analyses
Statistical analyses were performed by GraphPad Prism 6.0 (CA, USA) and SPSS 22.0 (Ehningen, Germany). The Mann–Whitney unpaired test and paired t-test were used to validate different miRNAs among groups. The paired-samples t test and χ2 test were employed to evaluate the relationship between miRNAs and clinical characteristics. The diagnostic power of the candidate predictors was calculated by receiver operating characteristic (ROC) and area under the curve (AUC). A P-value of < 0.05 was considered to be statistically significant.
Results
Identification of exosomes
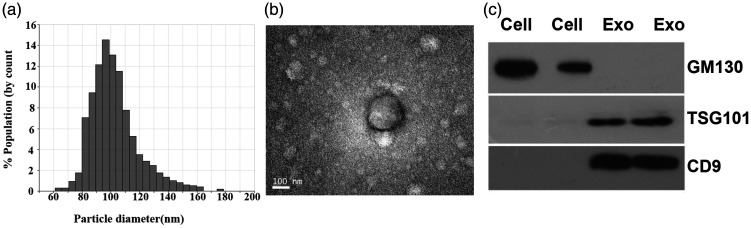
The size of exosome nanoparticles was distributed between 50 nm and 150 nm in diameter as shown by the qNano analysis (Figure 1(a)). Similarly, the exosomes exhibited a spherical morphology with a diameter of 50–150 nm (Figure 1(b)). Two well-known markers, TSG101 and CD9,27 were detected in the exosomes but absent in cells as revealed by Western blot analysis (Figure 1(c)). However, the negative control, GM130, was only found in the cells. These results indicated that the exosomes were successfully obtained by ultracentrifugation.
Figure 1.
Characteristics of the exosomes. (a) Size distribution of the exosomes (50–150 nm in diameter). (b) Representative exosomes with a diameter of 50–150 nm from NSCLC patients by transmission electron microscopy (high voltage (HV) = 20 kV; scale bar: 100 nm). (c) Western blot of TSG101 and CD9, and GM130.
Exosomal miRNA profiling
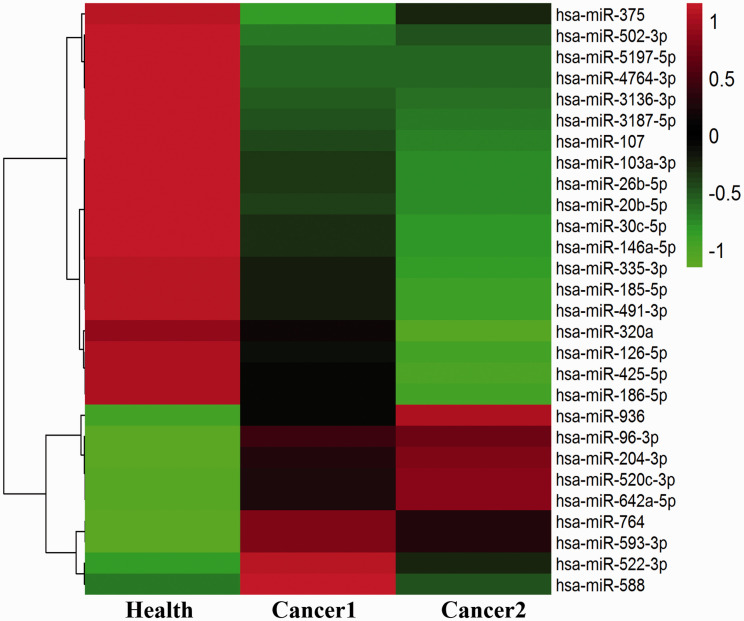
According to the raw exosomal miRNA expression profile, a total of 1937 differentially expressed ExmiRNAs were screened, among which, 28 (nine up-regulated and 19 down-regulated, Table 1) were selected using the criteria of >2.0-fold difference between the healthy donors (H) and NSCLC patients (C) (Figure 2).
Figure 2.
Exosomal miRNAs from NSCLC patients. The down-regulated and up-regulated miRNAs are shown in green and red, respectively. (A color version of this figure is available in the online journal.)
Correlations between exosomal miRNAs and clinical pathological and clinical characteristics
The relationships between the clinical pathological and clinical characteristics of NSCLC patients and the expression of ExmiRNAs are summarized in Table 2. Neither miR-20b-5p nor miR-3187-5p had correlation with gender, age, smoking, drinking, lymph node metastasis, histological type, and distant metastasis.
Table 2.
Characteristics of NSCLC patients for differentially expressed ExmiR-20b-5p and miR-3187-5p.
| Characteristic | No. cases |
miR-20b-5p |
miR-3187-5p |
|||
|---|---|---|---|---|---|---|
| Median with interquartile range | P-value | Median with interquartile range | P-value | |||
| Age (year) | ≤62 | 145 | 3.9400 (3.2650–4.7125) | 0.6691 | 3.2250 (2.4750–3.9000) | 0.2207 |
| >62 | 131 | 4.1200 (3.4250–4.7600) | 3.0100 (2.4225–3.6800) | |||
| Gender | Male | 162 | 4.4125 (3.4500–4.7200) | 0.2399 | 3.2275 (2.5550–3.8550) | 0.0801 |
| Female | 114 | 3.9025 (3.2100–4.7550) | 3.0725 (2.4150–3.5350) | |||
| Histological | SCC | 63 | 3.9500 (3.5050–4.6900) | 0.6935 | 3.2250 (2.6125–3.8000) | 0.4299 |
| type | AD | 197 | 4.0050 (3.2925–4.7200) | 3.1300 (2.4150–3.7525) | ||
| Others | 16 | |||||
| Smoking | Smoker | 142 | 4.1125 (3.4000–4.7200) | 0.4552 | 3.2850 (2.5800–3.8250) | 0.0859 |
| history | Non-smoker | 134 | 3.9375 (3.2650–4.7550) | 3.0125 (2.4150–3.5600) | ||
| Drinking | Drinker | 90 | 4.1000 (3.3250–4.9400) | 0.3899 | 3.2000 (2.3900–3.7350) | 0.4626 |
| history | Non-drinker | 186 | 3.9825 (3.3600–4.6650) | 3.1200 (2.4900–3.8100) | ||
| Lymph node metastasis | Yes | 130 | 3.9425 (3.2000–4.7050) | 0.3056 | 3.0775 (2.4100–3.7300) | 0.4324 |
| No | 139 | 4.1150 (3.4200–4.7200) | 3.2050 (2.5650–3.7750) | |||
| Unknown | 7 | |||||
| Distant metastasis | Yes | 190 | 3.9825 (3.4150–4.7150) | 0.7788 | 3.0825 (2.4150–3.6800) | 0.1992 |
| No | 80 | 4.0900 (3.2075–4.7200) | 3.3050 (2.5675–3.9100) | |||
| Unknown | 6 | |||||
SCC: squamous cell carcinoma; AC: adenocarcinoma.
Exosomal miR-20b-5p and miR-3187-5p for diagnosis of NSCLC
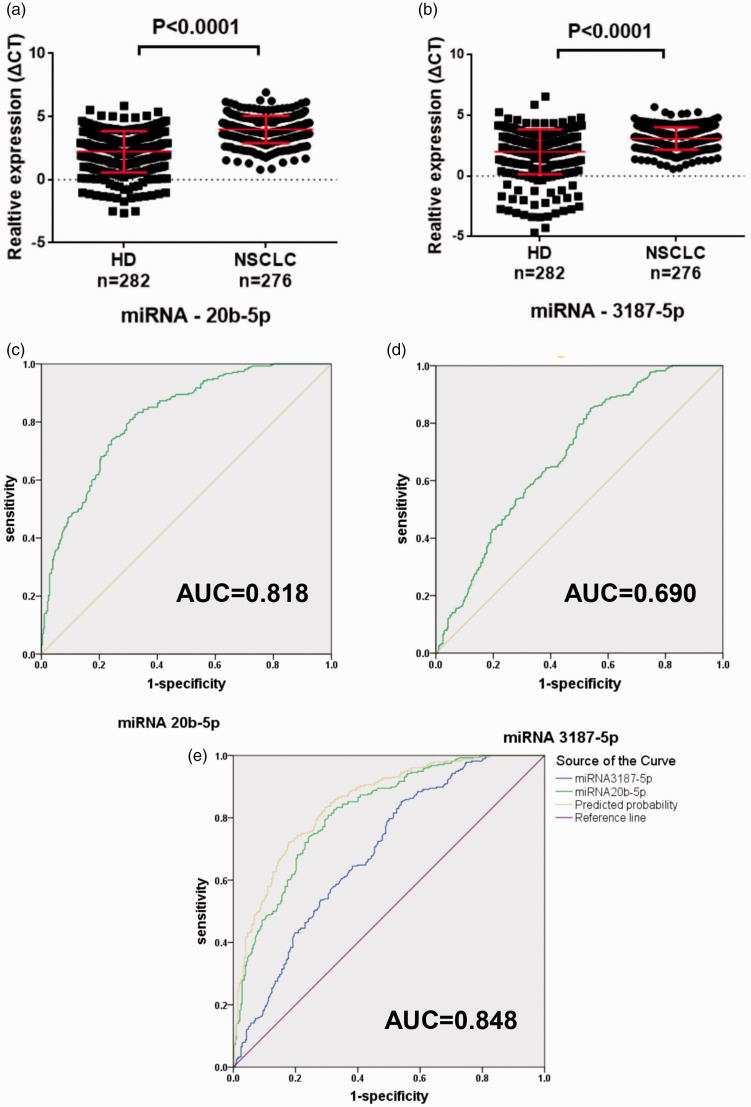
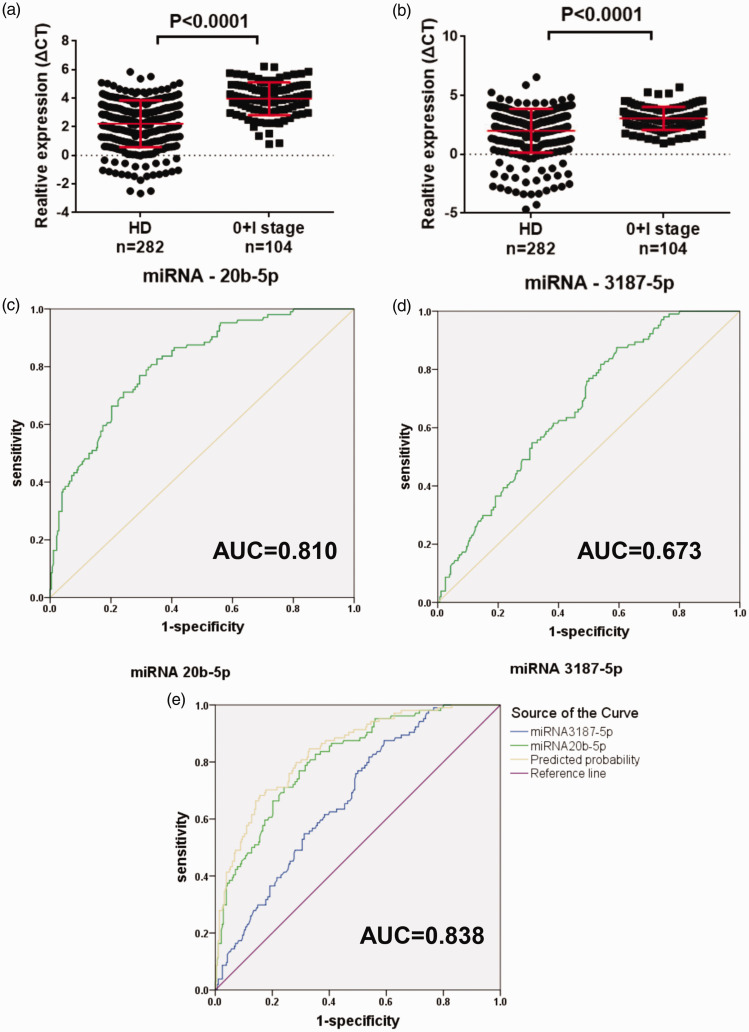
The expression of the exosomal miRNAs was verified in both serum exosomes and cells by qPCR. Out of 28 selected miRNAs, 16 were not further studied because of low specificity of the primers. The other 12 miRNAs were further verified using a larger set of samples (Table S3). Among these miRNAs, miR-20b-5p and miR-3187-5p were greatly down-regulated (P < 0.0001), both of which showed differential expressions between healthy controls and NSCLC patients (Figure 3(a) and (b)). The diagnostic power of these two ExmiRNAs was evaluated by the ROC curve, which showed that the AUC was 0.818 (95% CI: 0.784–0.853) and 0.690 (95% CI: 0.646–0.733) for miR-20b-5p and miR-3187-5p, respectively (Figure 3(c) and (d)). Combining the results of miR-20b-5p and miR-3187-5p could improve the AUC to 0.848 (95% CI: 0.817–0.880) (Figure 3(e)). Furthermore, both ExmiR-20b-5p and miR-3187-5p had a significant correlation with early stage NSCLC (0 and I stage, n = 104) compared to the controls (P < 0.0001) (Figure 4(a) and (b)). AUCs were 0.810 (95% CI: 0.764–0.865) and 0.673 (95% CI: 0.617–0.728) for miR-20b-5p and miR-3187-5p, respectively (Figure 4(c) and (d)). Using the combination of miR-20b-5p and miR-3187-5p, the AUC was improved to 0.838 (95% CI: 0.794–0.881) (Figure 4(e)).
Figure 3.
Characterization of miR-20b-5p and miR-3187-5p in NSCLC. (a, b) Expression of miR-20b-5p and miR-3187-5p. (c, d) AUC for miR-20b-5p and miR-3187-5p. (e) AUC for the combination of miR-20b-5p and miR-3187-5p. (A color version of this figure is available in the online journal.)
Figure 4.
Characterization of miR-20b-5p and miR-3187-5p in the early-stage NSCLC (0+I stage). (a, b) Expression of miR-20b-5p and miR-3187-5p. (c, d) AUC for miR-20b-5p and miR-3187-5p. (e) AUC for the combination of miR-20b-5p and miR-3187-5p. (A color version of this figure is available in the online journal.)
Diagnostic value of the combination panel of exosomal miRNAs with conventional biomarkers
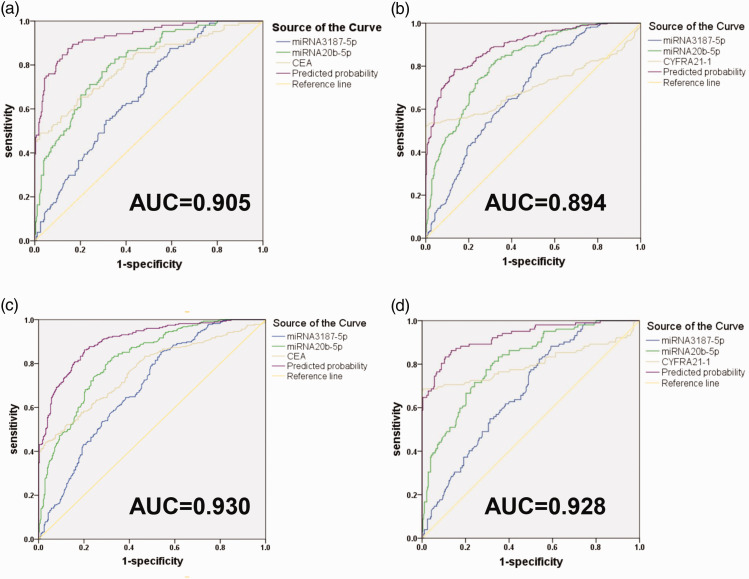
We further assessed the effects of CYFRA21-1 and CEA with ExmiR-20b-5p and miR-3187-5p on the diagnostic power for NSCLC. As shown in Figure 5(a) and (b), diagnosis of NSCLC was greatly improved by using CEA in combination with ExmiR-20b-5p and ExmiR-3187-5p, and the AUC was increased to 0.905 (95% CI: 0.880–0.929) in 282 healthy donors and 276 NSCLC patients. Similarly, combination of ExmiR-20b-5p, ExmiR-3187-5p, and CYFRA21-1 also improved the diagnosing power of NSCLC, and the AUC was increased to 0.894 (95% CI: 0.868–0.920). In Figure 5(c) and (d), combination of exosomal miR-20b-5p, miR-3187-5p, and CEA for diagnosis of early-stage NSCLC was evaluated, which showed that the AUC was increased to 0.930 (95% CI: 0.900–0.959) in 104 early-stage NSCLC patients (0+I stage) and 282 healthy donors. Similarly, the combination of exosomal miR-20b-5p, miR-3187-5p, and CYFRA21-1 was evaluated, and the results show that the AUC was increased to 0.928 (95% CI: 0.896–0.959).
Figure 5.
Diagnostic value of the combination of miR-20b-5p, miR-3187-5p, serum CEA, and CYFRA21-1 for NSCLC. (a) Combination of miR-20b-5p, miR-3187-5p, and CEA for NSCLC. (b) Combination of miR-20b-5p, miR-3187-5p, and CYFRA21-1 for NSCLC. (c) Combination of Exosomal miR-20b-5p, miR-3187-5p, and CEA for diagnosing early-stage NSCLC. (d) Combination of miR-20b-5p, miR-3187-5p, and CYFRA21-1 for diagnosing early-stage NSCLC. (A color version of this figure is available in the online journal.)
Discussion
The development of lung cancer is not only driven by genetic changes of neoplasm, but also regulated by other mechanisms, one of which is through cell interactions in the tumor microenvironment.10,28 Exosomes, specially the enclosed miRNA contents, have significant effects on cell-to-cell communication.29 Emerging studies have demonstrated that exosomes function in lung cancer initiation, progression, and metastasis. Thus, ExmiRNAs could be used as potential no-invasive biomarkers for NSCLC.
It has been found that the expression levels of plasma miRNA-23a and miRNA-21 are significantly different between patients with different histological grades of lung adenocarcinoma, suggesting they may be used as prognostic biomarkers for lung adenocarcinoma.30 We conducted correlation analysis between serum ExmiRNAs and different clinical and pathological characteristics, such as age, drinking, histological type, etc. However, the expression level of ExmiR-20b-5p and ExmiR-3187-5p showed no significant difference among NSCLC patients with different pathological and clinical characteristics (P > 0.05).
In our study, we investigated the vital role of serum exosomal miRNAs. We observed the expression of ExmiR-20b-5p and ExmiR-3187-5p was greatly decreased in NSCLC patients compared with that of the controls, reflecting their role in tumor suppression during the development of NSCLC.
MiR-20b-5p has been reported for its roles in inflammation, immunity and, in particular, human malignancies.6,31 A four-miRNA panel, including miR-20b-5p, has been found as a diagnostic biomarker for patients with breast cancer.32 Reduced expression of miR-20b-5p attenuates apoptosis by up-regulating Mcl-1, thereby promoting the survival of M. tuberculosis in RAW 264.7 macrophages, which may be a potential treatment for tuberculosis.33 A previous study has found that five serum miRNAs show elevated expressions in esophageal squamous cell carcinoma (ESCC), which may also be employed as diagnostic markers for ESCC.34 Taken together, these studies reveal the oncogenic features of miR-20b-5p. In our study, the expression of ExmiR-20b-5p was greatly down-regulated, which could be used to distinguish healthy controls from NSCLC patients, especially the patients with 0 and I stage NSCLC.
miR-3187-3p has been linked to carcinogenesis by a number of studies. However, no report has shown that miR-3187-5p plays a similar function. In previous studies, exosomal miR-3187-3p can reduce the responses of T-cell through down-regulating TCR signaling and lowering the secretion of cytokines and granzyme B, suggesting that melanoma-derived exosomal miRNAs facilitate immune escape and can be employed as a therapeutic target.35 In addition, down-regulation of miR-3187-3p is found to be related with shorter recurrence-free survival (RFS) of nonmuscle-invasive bladder cancer.36 In our study, we demonstrated that the expression of ExmiR-3187-5p was significantly reduced in NSCLC patients, which could be used to distinguish from the healthy controls.
Many reports confirmed that circulating miRNAs (CmiRNAs) are more stable in vesicles such as exosomes. Moreover, miRNAs are highly increased in exosomes in plasma and serum.37 A study conducted by Tanaka et al.18 demonstrated that CmiR-21 stems from exosomes. However, after exosomes were extracted, the expression of miR-21 was evidently higher in exosomes. These findings indicate ExmiRs are more suitable as diagnostic markers than CmiRs and provide a theoretical basis for precise assessment of the clinical diagnostic power of serum ExmiRNAs in NSCLC.
Elevated levels of serum ExmiR-20b-5p and ExmiR-3187-5p (with an AUC of 0.818 and 0.690, respectively) were observed in NSCLC patients, indicating these ExmiRNAs might be used as non-invasive circulatory biomarkers for the early detection of NSCLC patients, especially in stage 0 and I. Therefore, if patients with NSCLC are identified and diagnosed early, there will be more treatment options, which will ensure better overall survival of NSCLC.
Serum biomarkers for NSCLC, such as CEA, CA125, NSE, CYFRA21–1, and CA153, have been widely studied and applied in clinical practices. Among them, CEA and CYFRA21 have been commonly investigated for lung cancer.38 The combination of CEA with CYFRA 21 greatly improves the diagnosis power of lung cancer.39 However, no effective biomarker has been discovered for the diagnosis of early-stage NSCLC probably because the biomarkers such as CYFRA21-1 and CEA have only been revealed as meaningful prognostic markers to measure the therapeutic effects or metastasis and recurrence. Our study showed that CEA and CYFRA21-1 could be combined for the diagnosis of early-stage NSCLC patients. These two ExmiRNAs (miR-20b-5p and miR-3187-5p) in combination with CEA and CYFRA21-1 could improve the diagnostic power for early-stage NSCLC to 0.930 and 0.928, respectively. In summary, our study provided a new method for effective diagnosis of early-stage NSCLC.
Our study provides the first survey of serum ExmiR-20b-5p and ExmiR-3187-5p as biomarkers for NSCLC. Considering the size of the specimens in our study, the results we obtained were convincing. However, the present study has several limitations. On the one hand, due to the lack of long-term clinical follow-up data, we could not evaluate the prognostic power of ExmiR-20b-5p and ExmiR-3187-5p. On the other hand, this study focused on healthy donors and NSCLC patients, but did not include cases with benign lung diseases, such as pneumonia, pulmonary fibrosis, etc. A further analysis that includes patients with those diseases should be conducted to demonstrate if these biomarkers could be used to distinguish NSCLC patients from healthy individuals and patients benign lung diseases.
In conclusion, down-regulation of circulating ExmiR-20b-5p and ExmiR-3187-5p was critical for the diagnosis of NSCLC, especially the early-stage NSCLC (0 and I stage). Combination of these two ExmiRs could improve the diagnostic power. These serum exosomal miRNAs might be used as a new strategy for the diagnosis of early-stage NSCLC.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220945987 for Circulating serum exosomal miR-20b-5p and miR-3187-5p as efficient diagnostic biomarkers for early-stage non-small cell lung cancer by Zhi-Jun Zhang, Xing-Guo Song, Li Xie, Kang-Yu Wang, You-Yong Tang, Miao- Yu, Xiao-Dong Feng and Xian-Rang Song in Experimental Biology and Medicine
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; ZJZ and XRS designed the experiments; KYW, YYT, MY, and XDF performed the experiments; LX did the statistical analysis; ZJZ and XGS wrote the paper. The final manuscript has the approval of all authors.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval
The studies were reviewed and approved in accordance with the Helsinki Declaration of 1975 and the guideline by the ethical committee of Shandong Cancer Hospital Affiliated to Shandong First Medical University and Shandong Academy of Medical Sciences and. Written informed consent was provided by the participants’ legal guardian/next of kin.
FUNDING
This work was supported by Shandong Provincial Natural Science Foundation (ZR2019MH004 and ZR2019LZL016), the Shandong Provincial Key Research and Development Program (2016GSF201146, 2016GSF201151, 2017GSF18183 and 2017CXGC1207), and the National Natural Science Foundation of China (81773237, 81672104, 81972014).
ORCID iDs
Li Xie https://orcid.org/0000-0003-4715-136X
Xiao-Dong Feng https://orcid.org/0000-0003-2700-8304
Xian-Rang Song https://orcid.org/0000-0003-1722-8207
SUPPLEMENTAL MATERIAL
Supplemental material for this article is available online.
References
- 1.Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers 2019; 11:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oellerich M, Christenson RH, Beck J, Walson PD. Plasma EGFR mutation testing in non-small cell lung cancer: a value proposition. Clin Chim Acta 2019; 495:481–6 [DOI] [PubMed] [Google Scholar]
- 3.Alipoor SD, Mortaz E, Varahram M, Movassaghi M, Kraneveld AD, Garssen J, Adcock IM. The potential biomarkers and immunological effects of tumor-derived exosomes in lung cancer. Front Immunol 2018; 9:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanciu J, Tariman JD. Liquid biopsy: a tool for the diagnostic and prognostic evaluation of cancers. Clin J Oncol Nurs 2020; 24:19–21 [DOI] [PubMed] [Google Scholar]
- 5.Najminejad H, Kalantar SM, Abdollahpour-Alitappeh M, Karimi MH, Seifalian AM, Gholipourmalekabadi M, Sheikhha MH. Emerging roles of exosomal miRNAs in breast cancer drug resistance. IUBMB Life 2019; 71:1672–84 [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Zou X, Wu L, Zhang S, Wang T, Liu P, Zhu W, Zhu J. Identification of a 7-microRNA signature in plasma as promising biomarker for nasopharyngeal carcinoma detection. Cancer Med 2020; 9:1230–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem 2017; 41:755–68 [DOI] [PubMed] [Google Scholar]
- 8.Lai J, Du B, Wang Y, Wu R, Yu Z. Next-generation sequencing of circulating tumor DNA for detection of gene mutations in lung cancer: implications for precision treatment. Onco Targets Ther 2018; 11:9111–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghoreishy A, Khosravi A, Ghaemmaghami A. Exosomal microRNA and stroke: a review. J Cell Biochem 2019; 120:16352–61 [DOI] [PubMed] [Google Scholar]
- 10.Ma J, Xu M, Yin M, Hong J, Chen H, Gao Y, Xie C, Shen N, Gu S, Mo X. Exosomal hsa-miR199a-3p promotes proliferation and migration in neuroblastoma. Front Oncol 2019; 9:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, Xu T, Xu M, Wang S. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomarkers Prev 2018; 27:746–54 [DOI] [PubMed] [Google Scholar]
- 12.Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer 2017; 141:220–30 [DOI] [PubMed] [Google Scholar]
- 13.Yuwen D, Ma Y, Wang D, Gao J, Li X, Xue W, Fan M, Xu Q, Shen Y, Shu Y. Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to Platinum-Based chemotherapy. Cancer Epidemiol Biomarkers Prev 2019; 28:163–73 [DOI] [PubMed] [Google Scholar]
- 14.Alharbi M, Zuniga F, Elfeky O, Guanzon D, Lai A, Rice GE, Perrin L, Hooper J, Salomon C. The potential role of miRNAs and exosomes in chemotherapy in ovarian cancer. Endocr Relat Cancer 2018; 25:R663–R85 [DOI] [PubMed] [Google Scholar]
- 15.Perez-Hernandez J, Olivares D, Forner MJ, Ortega A, Solaz E, Martinez F, Chaves FJ, Redon J, Cortes R. Urinary exosome miR-146a is a potential marker of albuminuria in essential hypertension. J Transl Med 2018; 16:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JJ, Yang G, Yan QQ, Zhao J, Li S. Exosome-encapsulated microRNAs as promising biomarkers for Alzheimer's disease. Rev Neurosci 2019; 31:77–87 [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Ni M, Su Y, Wang H, Zhu S, Zhao A, Li G. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur Urol Focus 2018; 4:412–9 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013; 119:1159–67 [DOI] [PubMed] [Google Scholar]
- 19.Sanz-Rubio D, Martin-Burriel I, Gil A, Cubero P, Forner M, Khalyfa A, Marin JM. Stability of circulating exosomal miRNAs in healthy subjects. Sci Rep 2018; 8:10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurahashi R, Kadomatsu T, Baba M, Hara C, Itoh H, Miyata K, Endo M, Morinaga J, Terada K, Araki K, Eto M, Schmidt LS, Kamba T, Linehan WM, Oike Y. MicroRNA-204-5p: a novel candidate urinary biomarker of Xp11.2 translocation renal cell carcinoma. Cancer Sci 2019; 110:1897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, Dahiya R, Tanaka Y, Saini S. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res 2018; 78:1833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Li PW, Yang WQ, Mi H, Pan JL, Huang YC, Hou ZK, Hou QK, Luo Q, Liu FB. Identification of non-invasive biomarkers for chronic atrophic gastritis from serum exosomal microRNAs. BMC Cancer 2019; 19:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou SL, Chen YL, Ge ZZ, Qu YY, Cao Y, Kang ZX. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark 2019; 26:69–77 [DOI] [PubMed] [Google Scholar]
- 24.Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, Saeki K, Mori Y, Nakata K, Miyasaka Y, Onishi H, Oda Y, Goggins M, Nakamura M. Pancreatic juice exosomal MicroRNAs as biomarkers for detection of pancreatic ductal adenocarcinoma. Ann Surg Oncol 2019; 26:2104–11 [DOI] [PubMed] [Google Scholar]
- 25.Niu L, Song X, Wang N, Xue L, Song X, Xie L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci 2019; 110:433–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen X, Xue Y, Cong H, Wang X, Ju S. Dysregulation of serum microRNA-574-3p and its clinical significance in hepatocellular carcinoma. Ann Clin Biochem 2018; 55:478–84 [DOI] [PubMed] [Google Scholar]
- 27.Zhao YJ, Song X, Niu L, Tang Y, Song X, Xie L. Circulating exosomal miR-150-5p and miR-99b-5p as diagnostic biomarkers for colorectal cancer. Front Oncol 2019; 9:1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singal G, Miller PG, Agarwala V, Li G, Kaushik G, Backenroth D, Gossai A, Frampton GM, Torres AZ, Lehnert EM, Bourque D, O'Connell C, Bowser B, Caron T, Baydur E, Seidl-Rathkopf K, Ivanov I, Alpha-Cobb G, Guria A, He J, Frank S, Nunnally AC, Bailey M, Jaskiw A, Feuchtbaum D, Nussbaum N, Abernethy AP, Miller VA. Association of patient characteristics and tumor genomics with clinical outcomes among patients with Non-Small cell lung cancer using a clinicogenomic database. JAMA 2019; 321:1391–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kia V, Paryan M, Mortazavi Y, Biglari A, Mohammadi-Yeganeh S. Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. J Cell Biochem 2019; 120:5666–76 [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Mei H, Xu C, Tang H, Wei W. Circulating microRNA-339-5p and -21 in plasma as an early detection predictors of lung adenocarcinoma. Pathol Res Pract 2018; 214:119–25 [DOI] [PubMed] [Google Scholar]
- 31.Kawai S, Fujii T, Kukimoto I, Yamada H, Yamamoto N, Kuroda M, Otani S, Ichikawa R, Nishio E, Torii Y, Iwata A. Identification of miRNAs in cervical mucus as a novel diagnostic marker for cervical neoplasia. Sci Rep 2018; 8:7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Zhou Y, Xia T, Zhou X, Huang Z, Zhang H, Zhu W, Ding Q, Wang S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat 2018; 170:257–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Yi Z, Fu Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by mcl-1 upregulation. J Cell Biochem 2019; 120:5889–96 [DOI] [PubMed] [Google Scholar]
- 34.Huang Z, Zhang L, Zhu D, Shan X, Zhou X, Qi LW, Wu L, Zhu J, Cheng W, Zhang H, Chen Y, Zhu W, Wang T, Liu P. A novel serum microRNA signature to screen esophageal squamous cell carcinoma. Cancer Med 2017; 6:109–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vignard V, Labbe M, Marec N, Andre-Gregoire G, Jouand N, Fonteneau JF, Labarriere N, Fradin D. MicroRNAs in tumor exosomes drive immune escape in melanoma. Cancer Immunol Res 2020; 8:255–67 [DOI] [PubMed] [Google Scholar]
- 36.Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, Wang C. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer 2015; 136:854–62 [DOI] [PubMed] [Google Scholar]
- 37.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012; 7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang R, Zhu Y, Khadka VS, Zhang F, Jiang B, Deng Y. The evaluation of serum biomarkers for non-small cell lung cancer (NSCLC) diagnosis. Front Physiol 2018; 9:1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamura K, Takayama K, Izumi M, Harada T, Furuyama K, Nakanishi Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer 2013; 80:45–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220945987 for Circulating serum exosomal miR-20b-5p and miR-3187-5p as efficient diagnostic biomarkers for early-stage non-small cell lung cancer by Zhi-Jun Zhang, Xing-Guo Song, Li Xie, Kang-Yu Wang, You-Yong Tang, Miao- Yu, Xiao-Dong Feng and Xian-Rang Song in Experimental Biology and Medicine