Abstract
Coronavirus outbreak in December 2019 (COVID-19) is an emerging viral disease that poses major menace to Humans and it’s a crucial need to find the possible treatment strategies. Spike protein (S2), a envelop glycoprotein aids viral entry into the host cells that corresponds to immunogenic ACE2 receptor binding and represents a potential antiviral drug target. Several drugs such as antimalarial, antibiotic, anti-inflammatory and HIV-protease inhibitors are currently undergoing treatment as clinical studies to test the efficacy and safety of COVID-19. Some promising results have been observed with the patients and also with high mortality rate. Hence, there is a need to screen the best CoV inhibitors using insilico analysis. The Molecular methodologies applied in the present study are, Molecular docking, virtual screening, drug-like and ADMET prediction helps to target CoV inhibitors. The results were screened based on docking score, H-bonds, and amino acid interactions. The results shows HIV-protease inhibitors such as cobicistat (-8.3kcal/mol), Darunavir (-7.4kcal/mol), Lopinavir (-9.1kcal/mol) and Ritonavir (-8.0 kcal/mol), anti-inflammatory drugs such as Baricitinib (-5.8kcal/mol), Ruxolitinib (-6.5kcal/mol), Thalidomide (-6.5kcal/mol), antibiotic drugs such as Erythromycin(-9.0kcal/mol) and Spiramycin (-8.5kcal/mol) molecules have good affinity towards spike protein compared to antimalarial drugs Chloroquine (-6.2kcal/mol), Hydroxychloroquine (-5.2kcal/mol) and Artemisinin (-6.8kcal/mol) have poor affinity to spike protein. The insilico pharmacological evaluation shows that these molecules exhibit good affinity of drug-like and ADMET properties. Hence, we propose that HIVprotease, anti-inflammatory and antibiotic inhibitors are the potential lead drug molecules for spike protein and preclinical studies needed to confirm the promising therapeutic ability against COVID-19.
Keywords: COVID-19, Anti-inflammatory drugs, Coronavirus, Molecular docking, Homology modeling, Antiviral drugs
Graphical abstract
The Target spike protein docking with antimalarial, antimicrobial, anti-inflammatory and HIV-Protease inhibitors. The docking processed using AutoDock Vina and the binding affinity score is noted based on kcal/mol and estimated inhibitory constant (KI).
Highlights
-
•
Corona virus infection is a pandemic disease caused from Coronaviridae.
-
•
The Spike protein is the major target to attached to the host-receptor ACE2.
-
•
Computational Approaches to build the 3D structure of protein.
-
•
Drug selection for preclinical trials to study the efficacy and disease treatment.
-
•
Molecular docking approaches to screen the compounds based on binding energy.
1. Introduction
COVID19 (Corona virus disease 2019) outbreak is a pandemic respiratory disease caused by Coronavirus (CoV) it belongs to the family of Coronaviridae [1,2]. Initially most of the infected people reported in China, Wuhan city in 2019 with a large extent of seafood and animal meat market [3,4]. Many researchers have reported that the disease is distributed among humans, mammals and birds with the large implications on respiratory, gastrointestinal and neurological infections [5]. In the beginning of January 2020, the disease is increasing in China and distributed throughout world with quantifiable speed [6]. Based on the patient observation, it is reported that the disease similar to the signs and symptoms of Severe Acute Respiratory Syndrome Corona Virus (SARS-CoV) emerged in 2002–03 from southern China and Middle East Respiratory Syndrome Corona virus (MERS-CoV) emerged in 2012–13 from Saudi Arabia and spread around 26 countries throughout the world [[7], [8], [9]]. In April 12, 2020, the disease infected patients 1,896,156; deaths 117,671 and recovered 438,205 cases were reported globally [10]. The most common symptoms observed with the patients are fever, runny nose, sore throat, diarrhea, tiredness, dry cough and some patients have observed aches and pains in the body [11]. Some people become infected but have not develop any symptoms; about 80% of population was recovered from the disease without needing special treatment [12]. Till date, there are no approved drugs and therapeutic protocols recommended to prevent the disease. It is the greatest challenge of both pharmaceutical companies and research organizations to develop novel anti-corona viral drug.
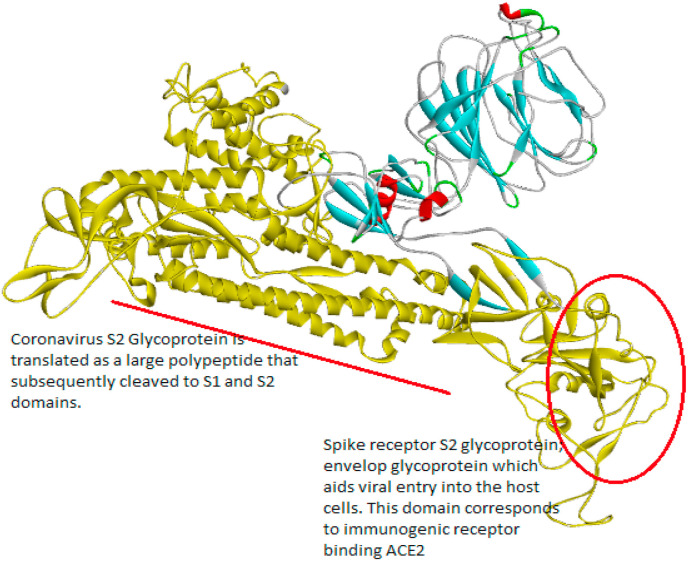
Understanding the molecular structure of the virus is important to researchers to develop targeted therapies to the disease. Based on the taxonomy of corona virus, SARS-CoV2 is causing disease in 2020 [13]. SARS-CoV2 belongs to the group of single stranded RNA (++ssRNA) virus associated with nucleoprotein within the capsid comprised of matrix protein. The virus has spherical or pleomorphic enveloped proteins surrounded by a fatty outer layer covered with a thin layer of crown like structure (spike protein) made of glycoprotein projections. It also associated with membrane glycoprotein, hemagglutinin-acetylesterase glycoprotein, and small envelope glycoprotein [[14], [15], [16]]. The target spike glycoprotein has two importance functions (1) receptor binding domain (RBD), (2) cleavage site. The RBD grapping hook that grip onto host receptor such as zinc peptidase angiotensin-converting enzyme 2 (ACE2) [17,18], aminopeptidase N (APN) [19,20], and dipeptidyl peptidase 4 (DPP4) [21,22] and the cleave site that can opens host receptor that allows the virus to enter host cells. The envelop (E) is small, integral membrane structural protein involved in virus’ life cycle and pathogenesis, but still the complete structural and functional information remains unknown [23]. The membrane (M) protein is most abundant structural protein involved in viral envelop and integrate in pathogenesis. The integration of S with M proteins in necessary for viral envelope and cause pathogenesis [24,25]. The nucleocapsid protein relating to viral genome involved in CoV replication cycle and the host cellular response to viral infection [26]. Although, the interaction between viral and human proteins is suggested as potential targets for identification of therapeutic protocols.
The solidarity trails of World Health Organization (WHO) and Indian Council of Medical research (ICMR) proposed the FDA approved drugs, trails with the combination of antimalarial drugs chloroquine and Hydroxychloroquine can helps to prevent the disease. Chloroquine prevents the viral attachment itself to the ACE2 receptors but it causes several side effects in this regard the current trail made with the less toxic derivative Hydroxychloroquine [[27], [28], [29]]. The effect of these two drugs is still being studied to understand the inhibition of SARS-CoV2. Another set of trails include antiHIV drug combination such as Lipinovir-Ritonavir regulates inflammation in the body that can split HIV proteins [30], these combinations also recommended to inhibit SARS-CoV2. The another trailed drug remdesivir is a nucleotide analog originally created to inhibit Ebola virus, but this drug also recommended to inhibit the novel Coronavirus by targeting the action of a key enzyme that facilitates its replication [31]. Some studies are looking at the investigation of viral protein structure and its behavior as a potential target for future drugs.
In recent study suggested that the recommended drugs might help patients with mild cased of COVID19, but that study has limitations. Researchers also evaluating the results of the disease in severe condition go into overdrive with inflammation that can damage the lungs and other organs, but so far there is no proof that it has that effect. Based on these limitations, the current research is designed to evaluate the drugs with target proteins. The major objective is to design the target proteins of both structural and non-structural proteins. Second objective is to study the modeling and evaluating the target proteins to predict active site amino acids. Third objective is focused o screening of recommended drugs based on pharmacophore and pharmacokinetic analysis. Fourth objective is to predict the protein-ligand interaction and virtual screening to understand how strong the drug can interact with target proteins to predict the potential effect against SARS-CoV2.
2. Materials and methods
2.1. Computational features
The insilico analyses were performed using HP Z440 workstation with Next-generation HP Intel Xeon E5-1630v4 3.70 Hz processor. The antimalarial, antibiotic, anti-inflammatory and HIV-protease inhibitor drugs were retrieved from Drug Bank database [32] and the conformational structures were observed in Chemsketch v12.0 [33]. Pharmacophore properties are analyzed using Molinspiration [34]. Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties of chemical structure were analyzed using admetSAR tool [35]. Further, the docking studies were carried out using AutoDock 4.2 [36] and Virtual screening using AutoDock Vina [37]. The intermolecular interactions were analyzed using Pymol [38] and BIOVIA Discovery Studio (DS) 2017 R2 [39].
2.2. Retrieving chemical structures
Based on data mining approaches and clinical practicing antimalarial drugs such as Chloroquine (DB00608), Hydroxychloquine (DB01611), Pyrimethamine (DB00205), Artemisinin (DB13132), and Mefloquine (DB00358). HIV-protease inhibitors such as Lopinavir (DB01601), Ritonavir (DB00503), Darunavir (DB01264), Cobicistat (DB09065). Anti-inflammatory drugs such as Baricitinib (DB11817), ruxolitinib (DB08877), and Thalidomide (DB01041), antimicrobial drugs such as Azithromycin (DB00207), Clarithromycin (DB01211), Erythromycin (DB00199), Spiramycin (DB06145), Camostat (DB13729), Fingolimod (DB08868), and Umifenovir (DB13609) chemical structures retrieved from Drug Bank database by PDB format [32].
2.3. Prediction of target protein
The experimental spike protein sequence was retrieved from GenBank database (YP_009724390). The protein sequence has the length of 1273 amino acids contains two functional domains. (1) Spike receptor binding domain (330–583) which corresponds to immunogenic ACE2 receptor binding domain. (2) Coronavirus S2 glycoprotein (662–1270) is translated as a large polypeptide that is subsequently cleaved to S1 and S2 domains [40,41]. Using C-ITASSER pipeline to create three dimensional protein models based on deep convolutional neural-network guide to the I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) fragment assembly simulations [42]. The 3D protein structure is evaluated using Structure analysis and verification server (SAVES v5.0)36 (https://servicesn.mbi.ucla.edu/SAVES/) and RAMAPAGE37. SAVES is used to understand the complexity of protein structure based on atom-to-atom interaction of Ψ versus Φ conformational angels of 3D macromolecule measures the torsion angels of Cα (ideal) -N-Cβ (obs) and the results were represented in Ramachandran Plot. Active site amino acids were predicted using the CastP calculation server based on the delineating measures of surface regions such as surface area and surface volume of 3D protein structure.
2.4. Molecular docking
Molecular docking studies used to find the binding affinity of the ligand molecule with target protein. The homology modeled protein structure docked with selected chemical structures using AutoDock 4.2 and virtual screening by AutoDock Vina. The protein structure is selected to add Gasteiger chargers and hydrogen atoms to the polar group of amino acids the macromolecule, whereas ligand structure by adding torsion counts of amide bonds rotatable and all active bonds non-rotatable and generated the PDBQT file for both protein and ligand structures. Grid space was set in Autogrid by selecting important residues with the grid box size x = 54 Å, y = 54 Å and z = 54 Å and grid spacing of 0.886 Å that provides search space, the grid centre was selected at dimensions x = −22.885, y = −10.008, z = 514.693 used to calculate grid parameters that help to understand the grid energy with equilibrated energy distribution. AutoDock was used to dock protein and ligand structures by adding Lamarckian genetic algorithm (LGA) with default parameters. The best docking conformation of protein-ligand interactions is predicted with the energy value in kcal/mol and followed by the analysis of hydrogen bonding interaction and the hydrophobic interaction. The best docking complex of protein-ligand was screened based on clustering analysis and visualized using Pymol and BIOVIA Discovery Studio (2017V).
2.5. Drug-like properties and ADMET properties
Pharmacophore analysis is performed to understand the drug-like character of the chemicals based on Lipinski and Veber’s Rule with selected parameters such as logP, TPSA, molecular weight, hydrogen bond donor, hydrogen bond acceptor, volume, number of rotatable bonds and total number of atoms, bioactive properties such as GPCR, ion channel, kinase inhibitor, nuclear receptor, protease inhibitor and enzyme inhibitor properties are predicted using molinspiration. ADMET analysis is performed to the selected compounds using admetSAR tool to screen the compounds based on absorption, distribution, metabolism, excretion and toxicity prediction. The most important parameters such as blood-brain barrier, acute toxicity, carcinogenicity, LD50, maximum recommended daily dose and Mutagenicity are predicted by lazar toxicity predictions server.
3. Results
3.1. Protein structure prediction
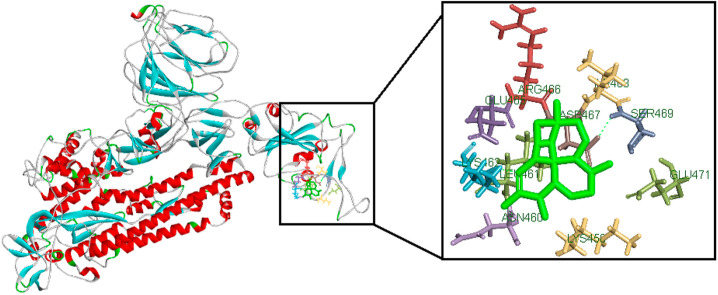
Prediction of protein structure is important in molecular docking, using I-TASSER to build three dimensional protein structure and used for homology modeling using SwissPDBViewer (SPDBV) software. Ramachandran plot were predicted to understand the complexity of amino acids within allowed region, energy is minimized to the protein structure with e-value = −39051.781. Using SAVES to predict the ERRAT value of 89.2% and Verify3D of 81.6%. CastP is used to predict ligand binding sites with Fmoc-amino acids Lys353, Arg355, Arg403, Lys417, Ile418, Asp424, Pro426, Asp427, Phe429, Tyr453, Leu455, Asn460, Leu461, Lys462, Pro463, Phe464, Ser469, Gln493, Tyr495, Gly496, Phe515, Leu517 amino acids (Fig. 1 ).
Fig. 1.
Spike protein structure predicted using I-TASSER, the red label represents (1) Spike receptor binding domain (330–583) which corresponds to immunogenic ACE2 receptor binding domain. (2) Coronavirus S2 glycoprotein (662–1270) is translated as a large polypeptide that is subsequently cleaved to S1 and S2 domains from. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Molecular docking plays vital role in computer-aided drug discovery to predict best active molecules to the target protein. Using AutoDock 4.2 and AutoDock Vina to predict the best drug binding sites towards the affinity of active site amino acids are screened based on binding energy and number of hydrogen bonds formed to the target amino acids.
3.2. Molecular docking of antimalarial inhibitors
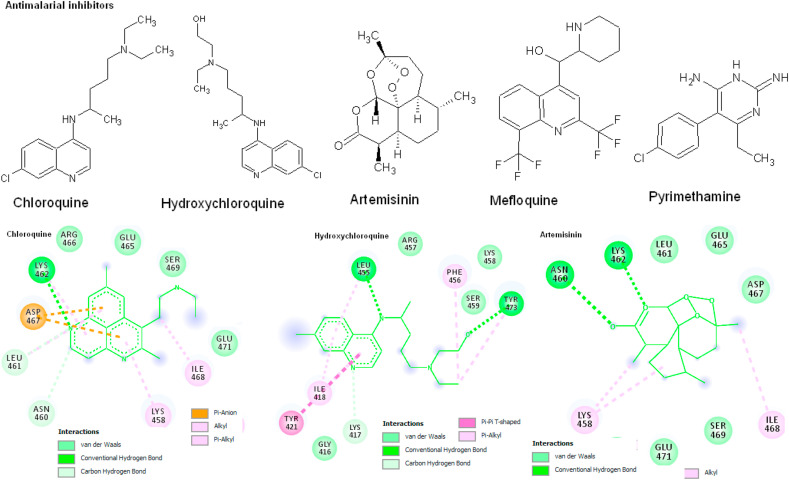
The antimalarial drugs such as Chloroquine, Hydroxychloroquine, Artemisinin, Mefloquine and Pyrimethamine chemical structures were docking with target spike protein and the results shows the best interaction towards the target amino acids. The results shows chloroquine, Hydroxychloroquine and Artemisinin formed 2 hydrogen bonds with highest binding energy of −6.2, −5.2 and −6.8 kcal/mol, estimated inhibitory constant (KI) of 7.46 mM, 7.53 mM and 15.37 μM respectively towards the binding amino acids Tyr453, Leu455, Asn460, Lys462, Ser469 respectively. The conformational energy of the chloroquine is minimized by Pi-cation interaction with Leu455, Asn460, Lys462 and Tyr473 amino acids. Hydroxychloroquine has one Pi-anion interaction with Asp467. There are 12 Van der Waals interactions with Gly416, Leu455, Arg457, Lys458, Ser459, Leu461, Glu465, Arg466, Asp467, Ser469, Glu471 and Tyr473 amino acids. The Mefloquine and Pyrimethamine compounds formed one hydrogen bond with binding affinity of −6.7 and −5.8 kcal/mol respectively within the active site amino acid Lys462 (Fig. 2 ; Table 1 ).
Fig. 2.
Antimalarial inhibitors docking with spike protein using AutoDock Vina. The 2D structures of protein-ligand interactions are visualized using DS visualize and the interactions are predicted based on binding energy (kcal/mol) and hydrogen bonds.
Table 1.
Molecular docking of Antimalarial inhibitors with spike protein using AutoDock Vina. The interactions are predicted based on binding energy (ΔG = Kcal/Mol).
| Ligands | H-bonds | Binding Energy (ΔG = Kcal/Mol) | KI | Amino acids |
|---|---|---|---|---|
| Chloroquine | 2 | −6.2 | 7.46 mM | Lys462, Ser469 |
| Hydroxychloroquine | 2 | −5.2 | 7.53 mM | Tyr453, Leu455 |
| Artemisinin | 2 | −6.8 | 15.37 μM | Asn460, Lys462 |
| Mefloquine | 1 | −6.7 | 835.56 μM | Lys462 |
| Pyrimethamine | 1 | −5.8 | 93.59 μM | Lys462 |
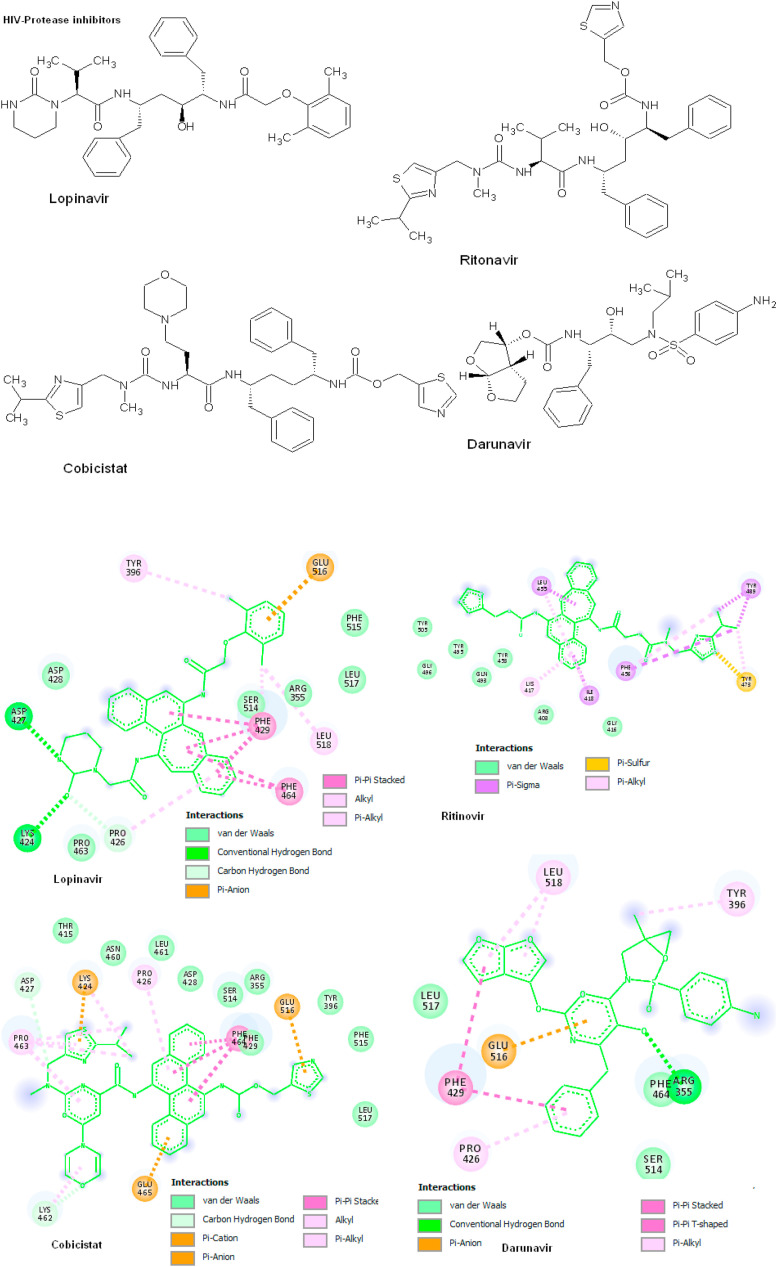
3.3. Molecular docking of HIV-protease inhibitors
The HIV protease inhibitors are interacting with target spike protein by forming 4–6 hydrogen bonds with the binding affinity of −7.4 to −9.1 kcal/mol. The conformational binding of Cobicistat has Pi-Anion with Glu465 and Lys424 along with the Pi-cation of Glu516 amino acids. The other side chain interactions of Pi-Pi stacked with Phe464 amino acids and non-bonding Van der Waals interaction of Asp427, Thr415, Asn460, Leu461, Asp428, Ser514, Arg355, Tyr369, Phe515 and Leu513 amino acids. The binding interaction of cobicistat has −8.3 kcal/mol with KI of 17.40 μM is strongly binds with target protein. The compound Darunavir has one hydrogen bond with Arg355 amino acid with binding energy of −8.3 kcal/mol. The compound has Pi-Anion interaction with Glu516 amino acids and Pi-Pi stacked to Phe429 amino acid, Pi-Pi T-shaped to Leu518, Tyr396 and Pro426 amino acids, Van der Waals interaction of Leu517 and Ser514 amino acids, based on the ΔG and KI the Darunavir is strongly recommended to interact with spike protein. Lopinavir forming strong hydrogen bonding with Lys424 and Asp427 amino acids with the binding interaction of −9.1 kcal/mol and KI = 39.22 mM. The compounds has one Pi-anion interaction with Glu516, Pi-Pi stacked with Phe429 and Phe464, Alkyl interaction with Tyr396, Van der Waals interaction with Asp428, Ser514, Arg355, Leu517, Phe515 and Pro463 amino acids. Another compound such as Ritonavir forming 4 hydrogen bonds within active site amino acid Gly496 with the binding energy of −8.0 kcal/mol, KI = 30.44 μM. The compound has Pi-sigma interaction of Ile418, Leu455, Phe456, Tyr489 amino acids, and Van der Waals interaction of Tyr505, Gly496, Tyr495, gln493, Tyr453, Arg403 and Gly416 amino acids (Fig. 3 ; Table 2 ).
Fig. 3.
HIV-Protease inhibitors docking with spike protein using AutoDock Vina. The 2D structures of protein-ligand interactions are visualized using DS visualize and the interactions are predicted based on binding energy (kcal/mol) and hydrogen bonds.
Table 2.
Molecular docking of HIV-protease inhibitors with spike protein using AutoDock Vina. The interactions are predicted based on binding energy (ΔG = Kcal/Mol).
| Ligands | H-bonds | Binding Energy (Kcal/Mol) | KI | Amino acids |
|---|---|---|---|---|
| Cobicistat | 6 | −8.3 | 17.40 μM | Phe464, Pro426, Pro463, Leu461 |
| Darunavir | 5 | −7.4 | 603.1 μM | Phe464, Arg355, Leu517 |
| Lopinavir | 5 | −9.1 | 39.22 mM | Phe515, Phe426, Asp427, Lys424, Phe429 |
| Ritonavir | 4 | −8.0 | 30.44 μM | Gly496, Tyr453, Ile418, Leu455 |
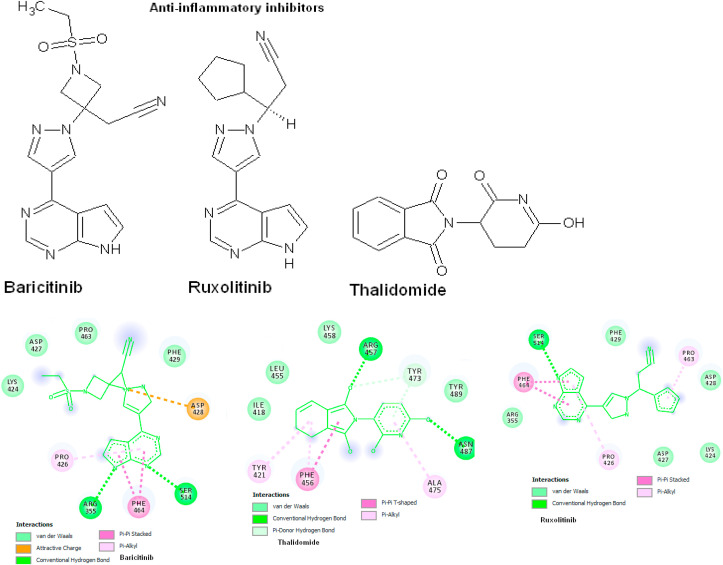
3.4. Molecular docking of anti-inflammatory inhibitors
The anti-inflammatory drugs are strong interaction with spike protein by forming 3–6 hydrogen bonds with the binding energy of −5.8 to −6.5 kcal/mol. The Baricitinib has two hydrogen bonds with Arg355 and Ser514 amino acids, ΔG = −5.8 kcal/mol, KI = 17.40 μM. The compound has attractive charge with Asp428, Pi-Pi stacked to Phe464, Pi-Alkyl interaction to Pro426 and Van der Waals interaction of Lys424, Asp427, Pro463 and Phe429 amino acids. The Ruxolitinib has one hydrogen bond to Ser514 and ΔG = −6.5 kcal/mol, KI = 1.21 mM, the Pi-Pi stacked with Phe464 amino acid, Pi-Alkyl interaction to Pro426 and Pro463 amino acids, Van der Waals interaction to the amino acids Arg355, Asp427, Lys424, Asp428 and Phe429. Thalidomide has 2 strong hydrogen bonds with Arg457 and Asn487 amino acids, Pi-donor-H-bond of Tyr473, Pi-Pi T-stacked to Phe456, Pi-Alkyl to Tyr421, Ala475 and Van der Waals interaction to Ile418, Leu455, Lys458, Tyr489 amino acids (Fig. 4 , Table 3 ).
Fig. 4.
Anti-inflammatory inhibitors docking with spike protein using AutoDock Vina. The 2D structures of protein-ligand interactions are visualized using DS visualize and the interactions are predicted based on binding energy (kcal/mol) and hydrogen bonds.
Table 3.
Molecular docking of anti-inflammatory inhibitors with spike protein using AutoDock Vina. The interactions are predicted based on binding energy (ΔG = Kcal/Mol).
| Ligands | H-bonds | Binding Energy (Kcal/Mol) | KI | Amino acids |
|---|---|---|---|---|
| Baricitinib | 6 | −5.8 | 17.40 μM | Pro426, Ser514, Arg355, Phe464, Asp428 |
| Ruxolitinib | 4 | −6.5 | 1.21 mM | Asp428, Ser514, Arg355 |
| Thalidomide | 3 | −6.5 | 99.85 μM | Asn487, Tyr473, Arg457 |
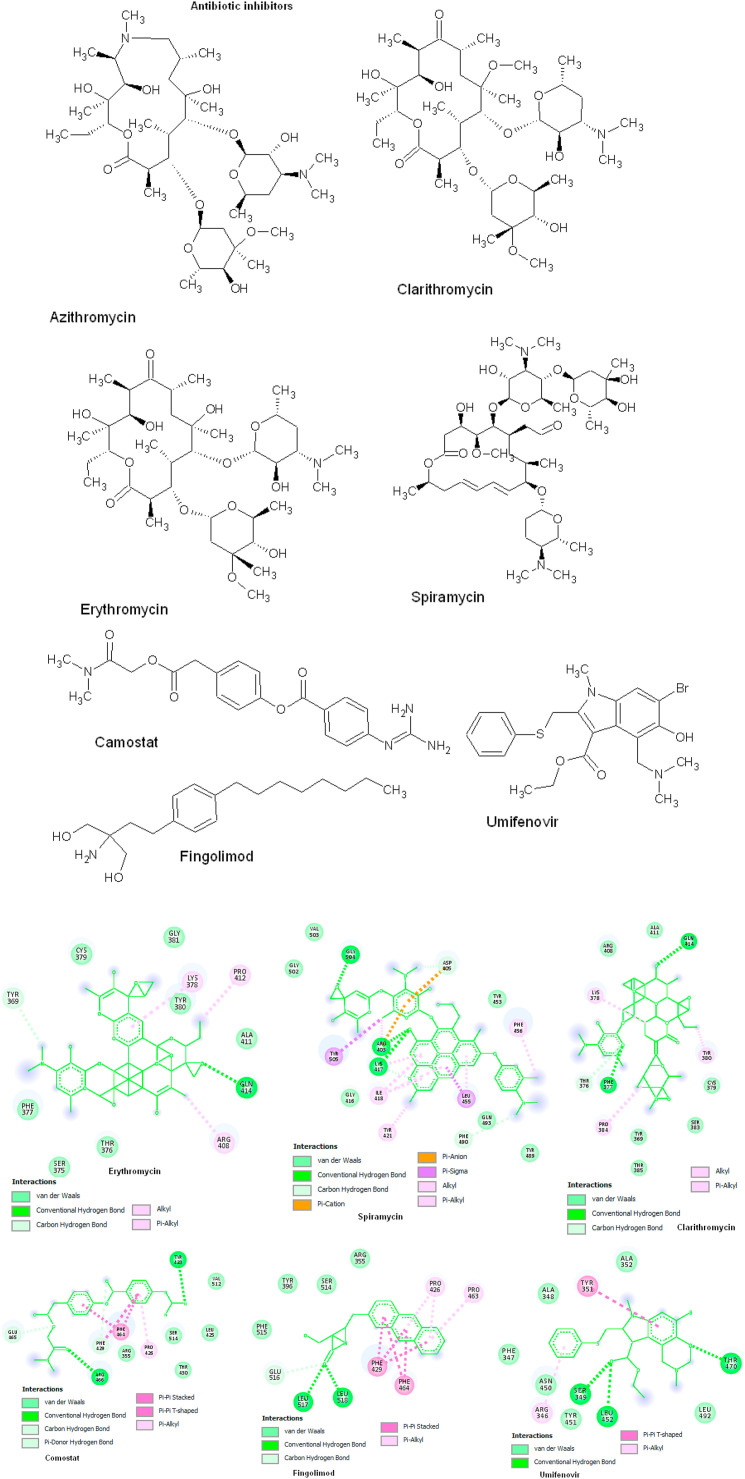
3.5. Molecular docking of antimicrobial inhibitors
The antibiotic drugs is also docked with target receptors by forming 3–6 hydrogen bonds and the interaction energy ranges from −6.0 to −9.0 kcal/mol. The Azithromycin is strong interaction with target receptor by forming 2 hydrogen bonds within active site amino acids Tyr380 and Gln414, ΔG = −8.7 kcal/mol, KI = 63.94 μM The Van der Waals interaction to Arg408, Thr376, Lys378 and Ala411 amino acids. Clarithromycin has two strong hydrogen bonds to Phe377 and Gln414, ΔG = −8.2 kcal/mol, KI = 5.46 μM, Pi-Alkyl interaction with Lys378 and Tyr380. Van der Waals interact to Arg408, Ala411, Thr376, Tyr369, Thr385, ser383, Cys379 amino acids. Erythromycin has 4 strong hydrogen bonds with binding energy of −9.0 kcal/mol, KI = 16.47 μM, within active site amino acid Gln414. The Alkyl interaction of Pro412, Lys378 and Arg408 amino acids, Van der Waals interaction of Tyr369, Cys379, Gly381, Phe377, Ser375, Thr376 and Ala411 amino acids. Spiramycin has 3 strong hydrogen bonds to Arg403, Lys417 and Gly504 amino acids with ΔG = −8.5 kcal/mol, KI = 2.95 μM, one Pi-cation to Asp405 and one Pi-anion to Arg403 amino acids, Pi-sigma of Tyr505 and Leu455, Alkyl interaction Ile418, Tyr421 and Van der Waals interactions to Gly502, Val503, Tyr453, Gly416, Gln493 and Tyr489 amino acids. Comostat has 2 strong hydrogen bonds to Tyr423 and Arg466 amino acids with ΔG = −6.5 kcal/mol, KI = 17.40 μM, Pi-Pi stacked interaction to Phe464, Pi-Alkyl interaction to Pro426 and Van der Waals interaction to Glu465, Arg355, Ser514, Thr430, Leu425 and Val512 amino acids. Fingolimod has 2 hydrogen bonds to Leu517 and Leu518 amino acids and ΔG = −7.9 kcal/mol, KI = 306.05 μM, Pi-Pi stacked to Phe429, Phe464, Pi-alkyl interaction to Pro426, Pro463 and Van der Waals interaction to Tyr396, Arg355, Ser514, Phe515 and Glu516 amino acids. Umifenovir has 3 hydrogen bonds to Ser349, Leu452 and Thr470 amino acids and binding energy of ΔG = −6.0 kcal/mol, KI = 2.95 μM, Pi-Pi stacked to Tyr351 amino acid, Pi-alkyl interaction to Arg346, Van der Waals interaction to Ala352, Ala348, Phe347, Asn450, Tyr451 and Leu492 amino acids (Fig. 5 ; Table 4 ).
Fig. 5.
Antimicrobial inhibitors docking with spike protein using AutoDock Vina. The 2D structures of protein-ligand interactions are visualized using DS visualize and the interactions are predicted based on binding energy (kcal/mol) and hydrogen bonds.
Table 4.
Molecular docking of antimicrobial inhibitors with spike protein using AutoDock Vina. The interactions are predicted based on binding energy (ΔG = Kcal/Mol).
| Ligands | H-bonds | Binding Energy (Kcal/Mol) | KI | Amino acids |
|---|---|---|---|---|
| Azithromycin | 4 | −8.7 | 63.94 μM | Lys378, Cys379, Tyr369, Pro384 |
| Clarithromycin | 5 | −8.2 | 5.46 μM | Phe377, Lys378, Tyr380, Cys379, Gln414 |
| Erythromycin | 4 | −9.0 | 16.47 μM | Cys378, Cys379, Tyr380, Gln414, Phe377, Arg408 |
| Spiramycin | 6 | −8.5 | 2.95 μM | Gly504, Arg403, Lys417, Asp405, Gly416, Ile418 |
| Comostat | 3 | −6.5 | 17.40 μM | Tyr423, Leu425, Ser514, Arg466 |
| Fingolimod | 3 | −7.9 | 306.05 μM | Leu517, Leu518, Ser514, |
| Umifenovir | 4 | −6.0 | 2.95 μM | Ala352, Thr470, Leu452, Ser349, Tyr351, |
3.6. Drug-like prediction and ADMET investigation
Drug-like properties and ADMET investigation also important in screening of compounds based on the protein-ligand interactions. The molinspiration results shows the compound Ritonavir and Cobicistat has miLogP >5 and these compounds has less dissolution with water-octonol solution, Lopinavir, Ritonavir, Darunavir and Cobicistat has high molecular weight (MW) > 500kda and these compounds can select only with small concentration with the cells to best activity. The bioavailability properties also predicted based on GPCR ligand, Ion channel modulator, kinase inhibitor, nuclear receptor ligand, protease inhibitor and enzyme inhibitor with the probability of acceptance >0.5 accepts as strong inhibitors. ADMET properties of drug compounds also screened based on human intestinal absorption (HIA) > 0.5 and all selected chemicals have very good HIA absorption. Blood brain barrier (BBB) > 0.5 shows strong BBA absorption, Human oral bioavailability of HIV-protease inhibitors, antimicrobial inhibitors such as Remdesivir, comostat, Fingolimod and Umifenovir has poor absorption towards oral. All chemicals strings have no carcinogenicity and these compounds are strongly recommended as best preclinical molecules for Corona virus infection (COVID-19) (Supplementary Table: 1).
4. Discussion
The outbreak of research is to finding multiple treatment plans for Coronavirus infection (COVID-19). Computational drug discovery and virtual screening approaches helps to predict possible active molecules against Coronavirus targets. Based on WHO and ICMR trails antimalarial drugs, HIV-Protease inhibitors, antiviral and anti-inflammatory drugs have strongly recommended treating COVID-19. Based on the observation, the mortality rate is still increasing, in this regard there is a need to understand the compounds and screen the best molecules to treat COVID-19. Form the present study, computational analysis was formed in step wise tasks to understand the host-pathogen interaction to drug screening. From the initial study, spike surface glycoprotein is the major target receptor binds with ACE2 receptor from the host cell and is represent as potential target molecule. The protein structure were predicted using I-TASSER and the structure are used for homology modeling shows 96% of amino acids is accepted in the complex structure and 4% of amino acids are glycine residues is not allowed in complex structure. The active site of amino acids are identified using CastP calculation server and results shows Lys353, Arg355, Arg403, Lys417, Ile418, Asp424, Pro426, Asp427, Phe429, Tyr453, Leu455, Asn460, Leu461, Lys462, Pro463, Phe464, Ser469, Gln493, Tyr495, Gly496, Phe515, Leu517 amino acids are used as drug binding sites.
The antimalarial, antiviral, anti-inflammatory and HIV-protease inhibitor chemical structures were retrieved from Drug Bank database. Using pharmacophore and pharmacokinetic analysis to predict the drug-like properties and also helps to understand bioactive properties of the chemical structures. Using molecular docking studies of each compound against the target receptor and the results were screened based on number of hydrogen bonds and binding energy within active site amino acids. The antimalarial drugs such as chloroquine, Hydroxychloroquine and Artemisinin is forming 2 hydrogen bonds within active site amino acids and these compounds are not strongly recommended to spike protein inhibitor. We also docked with HIV protease inhibitors such as Cobicistat, Darunavir, Lopinavir and Ritonavir are forming 4–6 hydrogen bonds within active sites amino acids of spike protein. Based on the literature and present computational docking interactions these compounds are strongly recommended to spike protein interactions. We also screened anti-inflammatory drugs such as Baricitinib, Ruxolitinib and Thalidomide compounds also strong binding with spike protein by forming 3–6 hydrogen bonds and these compounds also strongly recommended to spike protein interactions. The antiviral drugs such as Azithromycin, Clarithromycin, Erythromycin, Spiramycin, Comostat, Fingolimod and Umifenovir are strongly binds to target protein by forming 3–6 hydrogen bonds and these compounds also strongly recommends targeting protein interactions.
In our docking study, antimalarial drugs such as chloroquine, Hydroxychloroquine and Artemisinin shows weak interaction to the target receptor. The antiviral drugs such as Erythromycin and Spiramycin, anti-inflammatory drugs such as Baricitinib, Ruxolitinib, Thalidomide and HIV-protease inhibitors such as Cobicistat, Darunavir, Lopinavir and Ritonavir compounds shows strong interaction to the target receptor and these compounds are strongly recommended to the spike protein inhibitors in COVID-19. We hope the comprehensive protein structure; drug-like property and drug binding modes include number of binding hydrogen bonds, Pi-anion, Pi-cation, Pi-Pi stacking, Pi-alkyl and Van der Waals interactions provide valuable insights to screen potential drug compounds for COVID-19. Hence, our present study suggests the use of HIV-protease, anti-inflammatory and antibiotic inhibitors are the potential lead drug molecules for spike protein and further it should be validated with the preclinical studies needed to confirm the promising therapeutic ability against COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr.C.N.Prashantha, Assistant Professor, Department of Biotechnology, School of Applied Sciences, REVA University for providing overall guidance towards the docking studies.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2020.107769.
Author contributions
Prashantha C·N designed the research, Gouthami K, Lavanya L, Sivaramireddy Bhavanam, Ajay Jakhar, Shakthiraju, Suraj V, Sahana K·V, Sujana H·S performed the in silico study and analyzed the results, Prashantha C·N, Guruprasad N.M and Ramachandra R prepared the manuscript. All the authors reviewed the manuscript.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cherian Sarah S., Agrawal Megha, Basu Atanu, Abraham Priya. Perspectives for repurposing drugs for the coronavirus disease 2019. Indian Journal of Medical Research. 2020;5:1–12. doi: 10.4103/ijmr.IJMR_585_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burrell Christopher J., Howard Colin R., Murphy Frederick A. fifth ed. Academic Press; 2017. Chapter 31 -Coronaviruses, Fenner and White’s Medical Virology; pp. 437–446. [Google Scholar]
- 3.Zhu N. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382 doi: 10.1056/NEJMoa2001017. 2020 Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO) WHO; China. Beijing: 2020. WHO Statement Regarding Cluster of Pneumonia Cases in Wuhan. [Google Scholar]
- 5.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munster A novel coronavirus emerging in China - key questions for impact assessment. N. Engl. J. Med. 2020;382 doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 7.Huang et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. Published Online January 24, 2020. [DOI] [PMC free article] [PubMed]
- 8.Graham Carlos W., Dela Cruz Charles S., Cao Bin, Pasnick Susan, Jamil Shazia. novel wuhan (2019-nCoV) coronavirus. Am. J. Respir. Crit. Care Med. 2020;201(4):P7–P8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 9.Ryu S., Chun B.C. Korean Society of Epidemiology 2019-nCoV Task Force Team. An interim review of the epidemiological characteristics of 2019 novel coronavirus. Epidemiol Health. 2020;42 doi: 10.4178/epih.e2020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25(5):2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai N., Dorigatti I., Cori A., Donnelly C., Riley S., Ferguson N.M. 2020. Report 2: Estimating the Potential Total Number of Novel Coronavirus Cases in Wuhan City, China. London. [Google Scholar]
- 13.Li Fang. Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes Kathryn V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 15.Duncan Debbie, Lyall Gillian. Understanding the coronavirus. Br. J. Midwifery. 2020;28(3):146–148. [Google Scholar]
- 16.Olivia Li Ji-Peng, Chiu Lam Dennis Shun, Chen Youxin, Wei Ting Daniel Shu. Novel Coronavirus disease 2019 (COVID-19): the importance of recognising possible early ocular manifestation and using protective eyewear. Br. J. Ophthalmol. 2020;104(3):297–298. doi: 10.1136/bjophthalmol-2020-315994. [DOI] [PubMed] [Google Scholar]
- 17.Li W.H., Moore M.J., Vasilieva N., Sui J.H., Wong S.K. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome Coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmas B., Gelfi J., Lharidon R., Vogel L.K., Sjostrom H. Aminopeptidase-N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delmas B., Gelfi J., Sjostrom H., Noren O., Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 1993;342:293–298. doi: 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- 21.Raj V.S., Mou H.H., Smits S.L., Dekkers D.H.W., Muller M.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y., Du L., Liu C., Wang L., Ma C. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh P.-K., Chang S.C., Huang C.-C., Lee T.-T., Hsiao C.-W., Kou Y.-H. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79(22):13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr. 2020;14(3):241–246. doi: 10.1016/j.dsx.2020.03.011. [published online ahead of print, 2020 Mar 26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Tian Z., Breakthrough Yang X. Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1) doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 30.Bin Cao M.D., Yeming Wang M.D., Wen Danning. A trial of lopinavir–ritonavir in adults hospitalized with severe covid-19. The New England journal of medicine, March. 2020:1–13. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J Gordon Calvin, Tchesnokov Egor P., Woolner Emma, Perry Jason K., Feng Joy Y., Porter Danielle P., Gotte Matthias. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20) doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart David S., Craig Knox. An chi guo, savita shrivastava, murtaza hassanali, Paul stothard, zhan chang, jennifer woolsey, DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Issue suppl_1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter Allen D. ACD/ChemSketch 1.0 (freeware); ACD/ChemSketch 2.0 and its tautomers, dictionary, and 3D plug-ins; ACD/HNMR 2.0; ACD/CNMR 2.0. J. Chem. Educ. 1997;74(8):905. [Google Scholar]
- 34.Di L., Kerns E.H. Application of pharmaceutical profiling assays for optimization of drug-like properties. Curr. Opin. Drug Discov. Dev. 2005;8:495–504. [PubMed] [Google Scholar]
- 35.van de Waterbeemd H., Gifford E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 36.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Computational Chemistry. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrödinger L.L.C. The PyMOL- molecular graphics system. Version∼1.8. 2015 [Google Scholar]
- 39.Dassault Systèmes BIOVIA . Dassault Systèmes, 2016; San Diego: 2017. Discovery Studio Modeling Environment, Release. [Google Scholar]
- 40.Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Jr., Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3(1):e5. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binns Matthew M., Boursnell Michael E.G., Cavanagh David, Darryl J., Pappin C., Brown T. David K. Cloning and sequencing of the gene encoding the spike protein of the coronavirus IBV. J. Gen. Virol. 1985;66(Pt 4):719–726. doi: 10.1099/0022-1317-66-4-719. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Chengxin, Zheng Wei, Huang Xiaoqiang, Bell Eric W., Zhou Xiaogen, Zhang Yang. Protein structure and sequence re-analysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.