Abstract
Background
Microbial communities and their metabolic components in the gut are of vital importance for immune homeostasis and have an influence on the susceptibility of the host to a number of immune-mediated diseases like acute graft-versus-host disease (aGVHD) after allogeneic hematopoietic stem cell transplantation (allo-HSCT). However, little is known about the functional connections between microbiome and metabolome in aGVHD due to the complexity of the gastrointestinal environment.
Method
Initially, gut microbiota and fecal metabolic phenotype in aGVHD murine models were unleashed by performing 16S ribosomal DNA gene sequencing and ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS)-based metabolomics.
Findings
The group with aGVHD experienced a significant drop in Lachnospiraceae_unclassified but an increase in the relative abundance of Clostridium XI, Clostridium XIVa and Enterococcus. Meanwhile, a lower content of tyrosine was observed in the gut of aGVHD mice. The correlation analysis revealed that tyrosine-related metabolites were inversely correlated with Clostridium XIVa, besides, Blautia and Enterococcus also displayed the negative tendency in aGVHD condition. Apart from exploring the importance and function of tyrosine, different tyrosine diets were offered to mice during transplantation. Additional tyrosine supplements can improve overall survival, ameliorate symptoms at the early stage of aGVHD and change the structure and composition of gut microbiota and fecal metabolic phenotype. In addition, aGVHD mice deprived from tyrosine displayed worse manifestations than the vehicle diet group.
Interpretation
The results demonstrated the roles and mechanisms of gut microbiota, indispensable metabolites and tyrosine in the progression of aGVHD, which can be an underlying biomarker for aGVHD diagnosis and treatment.
Funding
This research was funded by the International Cooperation and Exchange Program (81520108002), the National Key R&D Program of China, Stem Cell and Translation Research (2018YFA0109300), National Natural Science Foundation of China (81670169, 81670148, 81870080 and 91949115) and Natural Science Foundation of Zhejiang Province (LQ19H080006).
Keywords: aGVHD, Tyrosine, Gut microbiome, Metabolomics, Correlation network
Research in context.
Evidence before this study
In recent years, gut microbiota has been observed to play a pivotal role in the pathobiology of various diseases like acute graft-versus-host disease (aGVHD) after allogeneic hematopoietic stem cell transplantation (allo-HSCT). Nevertheless, the functional connections between microbiome and metabolites in aGVHD remain largely elusive due to the complexity of the gastrointestinal environment.
Added value of this study
16S ribosomal DNA gene sequencing and ultra-high-performance liquid chromatography-mass spectrometry-based metabolomics were performed to systematically profile the mapping of both gut microbiota and fecal metabolites in murine aGVHD, demonstrating the functional connections between intestinal microbial flora and metabolites in the progression of aGVHD. Interestingly, it was found that tyrosine was dramatically declined in mice with aGVHD, which was closely associated with gut microbiota changes. Further, we supplied aGVHD mice with 2% tyrosine-supplemented diet, which markedly improved overall survival, ameliorated aGVHD symptoms and changed the structure and composition of gut microbiota and fecal metabolic phenotype at the early stage of aGVHD after allo-HSCT. Since tyrosine is of predominant importance as a constituent of protein nutrition status, findings pave the way for the use of tyrosine to improve the treatment of aGVHD and thus will have a broad impact on the field of general biomedicine.
Implications of all the available evidence
This research elucidated the roles and mechanisms of the interconnectivity between gut microbiota and metabolomes in the progression of aGVHD where tyrosine acted as the key regulators for reorganization of microbiome and the mitigation of this disease. Besides, these findings can provide a new reference for clinical application.
Alt-text: Unlabelled box
1. Introduction
Acute graft-versus-host disease (aGVHD) is one of the fatal and refractory complications after allogeneic hematopoietic stem cell transplantation (allo-HSCT). As the initially damaged organ, the gastrointestinal tract is particularly influenced by both activated donor T cells and pretransplant conditioning with chemotherapy and irradiation prior to allo-HSCT [1, 2]. While current treatments concentrate on how to inhibit the immunological attack of immune cells, gut microbiota and their metabolites are primarily and systemically exposed to epithelial cells, which are essential for the mediation of gastrointestinal immune responses and regarded as the potential future manipulation of controlling morbidity and mortality after allo-HSCT [3].
Decades ago, gut bacteria was recognized as an essential modulator of fine-tuning homeostasis in human health [4]. The dysbiosis of gut microbiota was demonstrated to be associated with several diseases, such as obesity, insulin resistance, chronic inflammation and even cancer [5, 6]. In addition, people intend to reveal the relationship between gut microbiota and aGVHD. The importance of microbiome diversity was first recognized in patients and murine aGVHD models after allo-HSCT [7], [8], [9], [10]. Some bacteria were defined as negative factors by a variety of experiments, such as Lactobacillus, Enterococcus, Streptococcus and Escherichia [11, 12]. Thereupon, researchers focused on studying the administration of beneficial microbes to modulate gut homeostasis via the direct supply of probiotics and fecal microbial transplantation (FMT) [13, 14].
In living organisms, circulating metabolites are generated by not only tissues but also gut microbiota [15], which has an impact on the bioavailability of metabolites in the gastrointestinal tract. Sufficient research was conducted on such a metabolite as short-chain fatty acids (SCFAs), like butyrate produced by certain bacteria and activating regulatory T cells [16]. Nowadays, an increasing number of people focus on amino acid metabolites involved in immune regulation and inflammation. For instance, Swimm et al. reported that GVHD is limited by tryptophan metabolite indole generated by the intestinal microbiome [17]. In addition, patients with aGVHD suffer from nutritional and metabolic disorders because of diarrhea, hypermetabolism and insufficient food intake, thereby turning nutritional support into a new therapeutic direction [18]. Researchers have initiated clinical trials where diets are modulated to maintain the sufficient provision of indispensable amino acids [19]. In the field of allo-HSCT, studies on mouse models and clinical implementation showed that supplementing glutamine promotes the healing of the intestinal tract and lowers the severity of GVHD and mucositis [20].
In our study, we firstly found that tyrosine markedly decreased in aGVHD mice by using 16S ribosomal DNA (rDNA) gene sequencing and ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS)-based metabolomics, which was closely correlated with gut microbiota changes. Tyrosine, a large neutral amino acid normally present in protein food, takes part in building essential proteins and providing energy [21] and constitutes the precursor of catecholamine helping people to effectively respond to acute stress and maintain homeostasis [22]. Therefore, a diet with a relatively high level of tyrosine was served, which can contribute to improving overall survival and ameliorating early aGVHD symptoms. Additionally, the addition of tyrosine shifted the structure of gut microbiota and led to the change of fecal metabolome, exhibiting a strong correlation between them. Therefore, it was speculated that gut microbiota interacting with metabolites is not only a potential therapeutic target but also a future biomarker for aGVHD diagnosis and therapeutic response.
2. Methods
2.1. Mice
Male BALB/c mice (H-2Kd) and C57BL/6 mice (H-2Kb) (SLAC Laboratory Supplies, Shanghai, China) at the age of 8–10 weeks were randomly grouped into TCD-BM or TCD-BM+T cell groups and the subsequently into different tyrosine supplement diets in accordance with the random numbers generated by Microsoft Excel by an investigator who was not involved in this study. All mice experiments were conducted under specific pathogen-free conditions in the Laboratory Animal Center of Zhejiang University (ZJU).
2.2. Bone marrow transplantation model
Bone marrow transplantation (BMT) recipients receiving a tail vein injection of 5 × 106 T-cell-depleted bone marrow (TCD-BM) cells (Mouse CD3 Positive Selection Kit, Biolegend) with or without 5 × 105 splenic T cells (Pan T Cell Isolation Kit, Miltenyi Biotech) on day 0 after 8-Gy lethal irradiation (split doses of 2 × 4.0 Gy apart to 4 h) formed C57BL/6→BALB/c at ZJU, China. A group of investigators who were blinded to the treatments estimated the clinical score according to the criteria described by Geoffrey et al. [23]. Slides of different organs specimens were collected on 14 days after BMT and stained with hematoxylin and eosin by professional staff who were blinded to the groups. Histopathology scoring for aGVHD was determined in terms of previously published systems by another group of investigators who were blinded to the treatment [23, 24]. All animal experiments were carried out in accordance with approved animal protocols, the guideline and ethical approval of the Institutional Animal Care and Use Committee of ZJU.
2.3. Dietary tyrosine dosing of mouse models
A pilot study showed that an additional 2% tyrosine within the standard safety margin was established as the highest and safe concentration compared with the normal standard (0.7% tyrosine) [25]. Tyrosine intervention groups commenced on 0% or 2% tyrosine diet (Hangzhou LiLeng Biotechnology Co., Ltd) a week before BMT and were given fresh foods every two to three days until the end of survival time observation or sacrifice.
2.4. PCR amplification and 16S rDNA gene sequence analysis
DNA was extracted from around 50-100mg of fecal samples utilizing E.Z.N.A.® Soil DNA Kit (Omega), frozen by liquid nitrogen and then stored at -80°C. The V3-V4 region of the prokaryotic (bacterial and archaeal) small-subunit (16S) rRNA gene was amplified by slightly modified versions of primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′- GGACTACHVGGGTWTCTAAT-3′) whose 5′ ends were tagged with specific barcodes per sample and universal sequencing primer. Polymerase chain reaction (PCR) products were confirmed with 2% agarose gel electrophoresis, purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by Qubit (Invitrogen, USA). PhiX Control library (v3) (Illumina) and amplicon library (expected to be 30%) were combined. Either of them was sequenced on 300PE MiSeq runs, while the other was sequenced with two protocols through the use of standard Illumina sequencing primers, which eliminated the need for the reading of a third (or fourth) index.
As suggested by the manufacturer, an Illumina MiSeq platform was used for sequencing samples obtained from LC-Bio. Paired-end reads were assigned to samples on the basis of their unique barcodes, truncated through the cutoff of barcodes and primer sequences and merged by use of FLASH. Clean tags were obtained through the quality filtering of raw tags under specific conditions of filtering in accordance with the fqtrim (V 0.94). In addition, the software Vsearch (v2.3.4) was applied to filter chimeric sequences and assign sequences with ≥ 97% similarity to the same operational taxonomic units (OTUs) [26]. Every OTU was given representative sequences, each of which was then assigned with taxonomic data using the Ribosomal Database Project (RDP) classifier. The software MAFFT (V 7.310) was used to conduct a multiple sequence alignment of the differences of dominant species in all groups to explore the phylogenetic relationship of various OTUs. The abundance information of OTUs was normalized with a standard of sequence number which corresponded to the sample with the fewest sequences. The complex species diversity of each sample was revealed by alpha-diversity through Chao1, Observed_species, Goods_coverage, Shannon and Simpson, which was analyzed by QIIME (Version 1.8.0). Beta-diversity was analyzed to evaluate the differences of samples in the complexity of species by the software QIIME (Version 1.8.0). Distance-based statistical tests (weighted and unweighted UniFrac distances clustered) had been applied to test the association of the cluster composition with environmental and biological covariates [27].
2.5. LC-MS analysis
All samples were acquired by the LC-MS system and all chromatographic separations were performed using a UPLC system (SCIEX, UK). Maintaining at 35°C, an ACQUITY UPLC T3 column (100mm*2.1mm, 1.8µm, Waters, UK) was used for a reversed-phase separation. Gradient elution parameters and conditions were set as recommended [28]. Metabolites were detected by a high-resolution tandem mass spectrometer TripleTOF5600plus (SCIEX, UK) operating in 5 kV positive and -4.5 kV negative modes respectively. The mass spectrometry data were acquired in Interactive Disassembler Professional (IDA) mode and the time-of-flight (TOF) mass ranged from 60~1200 Da. Survey scans were acquired in 150 ms and as many as 12 product ion scans were collected if exceeding a threshold of 100 counts per second with a 1+ charge state. The total cycle time was fixed to 0.56 s. Four time bins were summed for each scan at a pulser frequency value of 11 kHz through the monitoring of the 40 GHz multichannel TDC detector with four-anode/channel detection. Dynamic exclusion was set as 4 s [29]. The mass accuracy of every 20 samples was calibrated and the quality control (QC) sample was acquired from every 10 samples.
After being transformed into mzXML format, LC−MS raw data files were processed by XCMS, CAMERA as well as metaX toolbox realized by the R software. Retention time (RT) was combined with m/z data to identify each ion. The intensity of every peak was recorded to generate a three-dimensional matrix which contained randomly assigned peak indices (RT-m/z pairs), sample names (observations) as well as the intensity information of ions (variables). Human Metabolome Database (HMDB) and online Kyoto Encyclopedia of Genes and Genomes (KEGG) were adopted to annotate metabolites through the matching between the precise molecular mass data (m/z) of samples and those from database. A difference of mass between observed and database values below 10 ppm would contribute to annotating metabolites, further identifying and validating their molecular formula through the measurement of isotopic distribution. Besides, an in-house fragment spectrum library of metabolites was used for validating metabolite identification. MetaX was applied to further preprocess the intensity of peak data. Those features detected in below 50% of QC samples or 80% of biological ones were excluded. In order to further improve the quality of data, the k-nearest neighbor algorithm was used to impute the rest of peaks with missing values. Principal component analysis (PCA) was performed to detect outliers and evaluate batch effects by means of the pre-processed dataset. Robust LOESS signal correction based on QC was fitted to QC data on the order of injection so as to minimize the drift of signal intensity over time. Additionally, the relative standard deviations (SDs) of metabolic features among all QC samples were calculated, among which those > 30% were excluded.
2.6. Correlation network analysis and pathway analysis
Spearman's correlation analysis was conducted to identify the relationship between fecal microbiome abundance and expression of metabolites. Consideration was only given to the correlation value (rho)>0.6 and p<0.05. Cytoscape was used to construct the correlation network [30]. The KEGG analysis of metabolites was performed by uploading selected metabolites to the database MetaboAnalyst.
2.7. Statistical analysis
For 16S rDNA gene sequencing, top 20 microbiomes at the genus level were chosen to compare their differences in relative abundance. Statistical analysis was performed by conducting an unpaired (two-sided) Student's t-test and Mann-Whitney U (MWU) test was conducted if the data failed to meet normality criteria by Graphpad software. Metabolites were analyzed by conducting t-test to detect the differences between two phenotypes in metabolite concentration. A false discovery rate (FDR) (Benjamini–Hochberg) was used to adjust the p value for all multiple tests. Supervised partial least squares discriminant analysis (PLS-DA) was performed through metaX to discriminate different inter-group variables. The calculation was conducted for the value of variable important for the projection (VIP). Important features were selected using a VIP cut-off value of 1.0. Only the ratio of metabolites ≥2 or ≤1/2, VIP (obtained by performing multivariate statistical analysis with PLS-DA)≥1 and q value ≤0.05 and annotated at MS2 level were taken into account. Graphs and significance test were prepared with GraphPad Software. To explore the correlation between metabolites and the microbiota, we performed Spearman's correlation analysis to test the abundance of microbiota against the altered expression of metabolites in R software.
The size of samples in murine GVHD experiments was not predetermined with statistical methods, which however resembled those generally used in this field [31, 32]. Log-rank test was conducted to analyze the differences in animal survival (Kaplan-Meier survival curves, GraphPad Software). To acquire unbiased data, the investigator who administered the BMT was the only person aware of the treatment group allocation. Another two groups of investigators evaluated the clinical and histopathological scores of aGVHD severity respectively, who were blind to the types of treatments. If necessary, data were expressed as mean ± standard error of the mean (SEM) or standard deviation (SD). It was considered that differences were significant when P<0.05. No other randomization methods were adopted. 16S rDNA amplicon sequencing data were deposited in the Sequence Read Archive (SRA) database: PRJNA637751.
3. Result
3.1. Gut microbiome structure shifted in aGVHD murine models
To determine whether aGVHD affects the parameters of the gut environment, a well-established C57BL/6 (H-2Kb) was initially put into a BALB/c (H-2Kd) major histocompability complex (MHC) mismatch model (Fig. 1a) [31]. The TCD-BM+T cells group showed severe aGVHD symptoms including shortened survival time, rapid weight loss, severe diarrhea and injury in multiple organs such as liver, intestine and skin (Fig. 1b-e and Supplementary Fig.S1). The fecal microbiome was detected on days 14 and 28 after allo-HSCT to see whether the changes of bacterial flora diversity were associated with the occurrence and development of aGVHD.
Fig. 1.
Relative abundance of gut microbiota at phylum and genus levels.
(a) BALB/c host mice receiving C57BL/6 TCD-BM (n=12) or TCD-BM+T cells (n=12) to establish aGVHD murine models. (b-e) Significant changes of (b) survival time (log-rank test, p<0.05), (c) weight changes (two-tailed Student's t-test, p<0.01), (d) clinical scores (two-tailed Student's t-test, p<0.01) and (e) pathology scores of each group on day 14. (f-g) 16S rDNA gene sequencing of fecal microbiota from TCD-BM (n=6) and TCD-BM+T cells groups (n=6) and the relative abundance of gut microbiota with the histogram of fold change (TCD-BM+T/TCD-BM) at the phylum level on days 14 (f) and 28 (g). (h-i) Relative abundance of top 20 genera with the histogram of fold change (TCD-BM+T/TCD-BM) on days 14 (h) and 28 (i). The number of mice in each group was indicated (n) within the bracket. The scores displayed mean ± SEM from three to four independent experiments. *p < 0.05, **p < 0.01.
Firstly, we evaluated the bacterial alpha-diversity for the richness and evenness. No significant differences were found in either Chao1 (MWU test, day14: p>0.05, day 28: p>0.05) or Shannon (MWU test, day14: p>0.05, day28: p>0.05) (Table 1, Supplementary Fig.S2a), but TCD-BM group presented higher alpha-diversity than TCD-BM+T cells group especially on day 28. This comparison revealed that the TCD-BM+T cells group had lower abundance and uniformity (Table 1). To guide our sampling depth, we further plotted rarefaction curve of each sample that reaching a plateau and saturation stage, which verified that most of the species were observed in all the groups (Supplementary Fig.S2b). In beta-diversity analysis, the unweighted UniFrac indexes strengthened the difference between two groups were widening (Supplementary Fig.S2c).
Table 1.
Statistics of the alpha diversity index between TCD-BM and TCD-BM+T groups.
| Gut microbiota Diversity Index | Day 14 |
Day 28 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TCD-BM | TCD-BM+T |
p-value | TCD-BM |
TCD-BM+T |
p-value | |||||
| Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | |||
| Observed species | 82.33±5.95 | 82.00 (61.00-100.00) | 109.67±7.92 | 113.00 (76.00-135.00) | 0.0260 | 58.67±7.71 | 61.00 (31.00-79.00) | 91.00±21.70 | 78.00 (25.00-161.00) | 0.2597 |
| Chao1 | 130.83±12.61 | 124.00 (90.25-172.30) | 104.71±24.28 | 93.00 (39.25-208.38) | 0.3095 | 170.00±14.49 | 175.00 (116.62-213.24) | 138.39±27.03 | 155.00 (43.33-203.25) | 0.5880 |
| Shannon | 1.83±0.23 | 1.57 (1.35-2.83) | 1.91±0.26 | 2.11 (1.01-2.59) | 0.9990 | 2.63±0.21 | 2.84 (1.78-3.11) | 2.46±0.41 | 2.53 (0.70-3.80) | 0.6990 |
| Simpson | 0.50±0.18 | 0.44 (0.33-0.77) | 0.57±0.20 | 0.64 (0.32-0.77) | 0.8240 | 0.70±0.14 | 0.74 (0.45-0.81) | 0.64±0.24 | 0.69 (0.18-0.89) | 0.6991 |
| Good coverage | 1.00±0.00 | 1.00 (1.00-1.00) | 1.00±0.00 | 1.00 (1.00-1.00) | >0.9999 | 0.998±0.00 | 0.995 (0.99-1.00) | 0.99±0.00 | 1.00 (0.99-1.00) | >0.9999 |
Data are mean ± standard deviation (SD) or median and interquartile range (IQR) . Mann-Whitney U (MWU) test was employed for values comparisons.
Despite the difficulty in obtaining conspicuous differences at the phylum level, some changes between TCD-BM and TCD-BM+T cells groups were observed from the data of fecal microbial 16S rDNA gene sequencing. Firmicutes, Bacteroidetes, Verrucomicrobia and Proteobacteria are top four dominant components of fecal microbes (Fig. 1f and g). Firmicutes, accounting for the largest proportion in the microbiota compositions, was decreased in the TCD-BM+T cells group, while the other three phylum levels increased. These variation trends remained constant throughout the progression of aGVHD. It was notable that some significant changes were observed at the genus level. Altered floras predominantly belonged to the phylum Firmicutes. One of the most abundant genus, Lachnospiraceae_unclassified, maintained a high proportion in the TCD-BM group, but decreased in the TCD-BM+T cells group (the mean of TCD-BM and TCD-BM+T cells groups, day 14: 54. 96% and 9.45%, MWU test, p<0.05; day 28: 38. 28% and 11.90%, MWU test, p<0.05). In addition, the tendency was even more convincing due to consistent and significant differences in fold change on days 14 (two-tailed Student's t-test, p<0.05) and 28 (two-tailed Student's t-test, p<0.05) (Fig. 1 h and i, Supplementary Table S1). Other microbiota belonging to Firmicutes were up-regulated in the TCD-BM+T cells group, such as Blautia, Clostridium XI, Clostridium XIVa on day 14 and Enterococcus on day 28. Besides, the TCD-BM+T cells group saw an increase in other altered genera like Proteus belonging to Proteobacteria and high-occupied Bacteroides belonging to Bacteroidetes. Alistipes, another component of Bacteroidetes was down-regulated in the TCD-BM+T cells group (MWU test, p<0.05 on day 28). All the tendencies were consistent suggesting that the composition of gut microbiome exhibited remarkable changes with the occurrence and development of aGVHD.
3.2. Gut metabolome revealed a dramatic reduction of tyrosine and related metabolites in aGVHD mice
As microbiota alteration in aGVHD has been extensively explored, some studies stated that the structure of microbiome has a strong influence on the metabolite profiles of multiple diseases [33, 34]. Then, the differences in fecal metabolome after BMT were observed. To confirm the implication of these metabolites at the onset of aGVHD, PCA was used to verify whether aGVHD could be discriminated in multivariate analysis (Supplementary Fig. S2c and d). In PLS-DA, PLS-DA score plots indicated the reliable discrepancies between TCD-BM and TCD-BM+T cells groups and suggested rejection reactions leading to significant biochemical changes (Supplementary Fig.S2d and e). Based on the pairwise comparisons of different metabolites between two groups and different development stages of aGVHD, the fecal metabolic profiles of the two groups displayed depression in the early stage on day 14. However, the infertile situation improved on day 28, which corresponded to BM reconstruction after irradiation (Supplementary Fig.S2f).
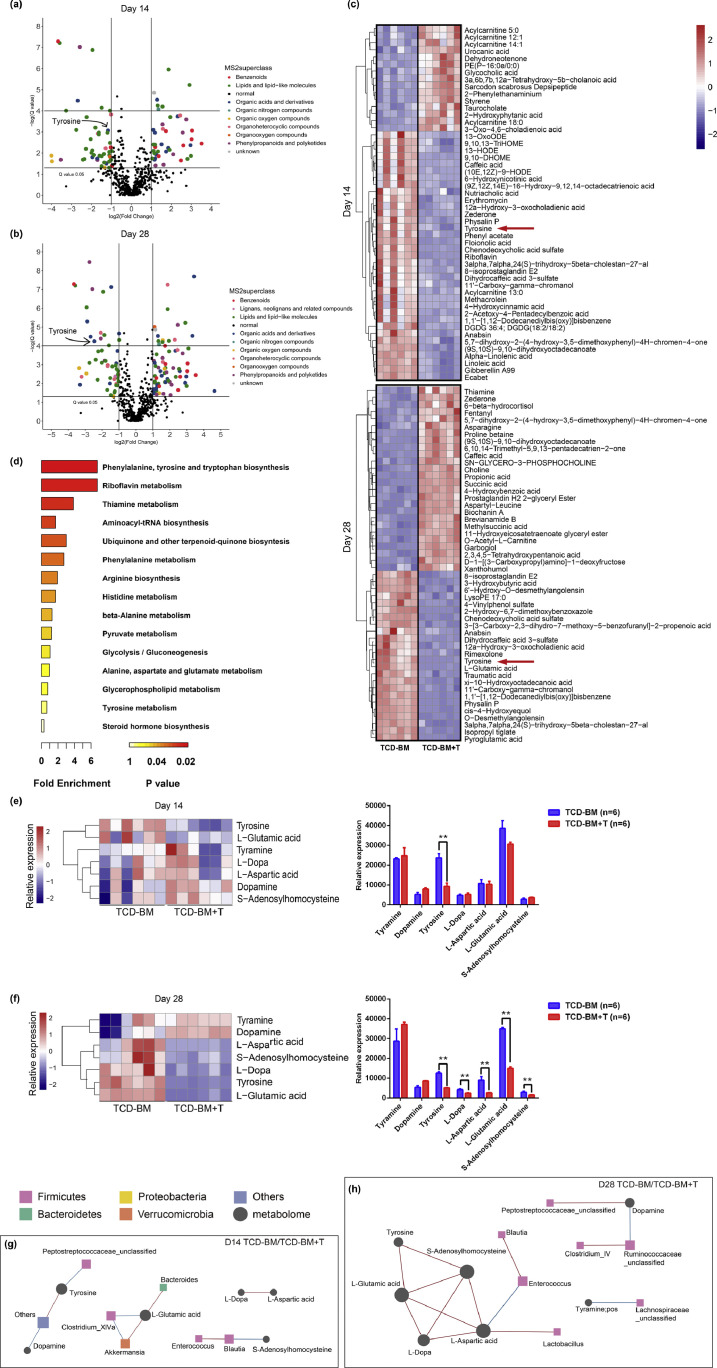
Metabolites with putative biological relevance were identified by comparing the amount of each metabolite between TCD-BM and TCD-BM+T cells groups on days 14 (Fig. 2a) and 28 (Fig. 2b). Compared with the TCD-BM group only featuring transfusion, the TCD-BM+T cells group was mainly characterized by a significant decrease in the number of lipids and lipid-like molecules, especially physalin P, acylcarnitine and 8-isoprostaglandin E2. Additionally, organic acids and derivatives, especially dihydrocaffeic acid 3-sulfate and tyrosine, showed a downward trend in the TCD-BM+T cells group (Fig. 2c, Supplementary Table S2). Later, KEGG analysis was performed to analyze the pathways with all significantly different metabolites, whose results showed that pathways related to several amino acids attracted attention. Pathways of phenylalanine, tyrosine and tryptophan biosynthesis ranked on the top (Fig. 2d). Strikingly, the following several pathways including ubiquinone and other terpenoid-quinone biosynthesis and aminoacyl-tRNA biosynthesis, shared the same metabolite, namely tyrosine. Thus, it was found that the TCD-BM+T cells group presented an overall downtrend in not only tyrosine but also metabolites interacting with tyrosine. While tyrosine remarkably diminished on days 14 (two-tailed Student's t-test, p<0.01) and 28 (two-tailed Student's t-test, p<0.01) (Fig. 2e; Supplementary Table S3), L-Aspartic acid, L-Glutamic acid and S-Adenosylhomocysteine significantly increased on day 28, as shown in the heatmap (Fig. 2f; Supplementary Table S3).
Fig. 2.
Changes in the metabolomics of TCD-BM and TCD-BM+T cells groups.
(a, b) A comparison was made between detected metabolites with paired Student's t-test and then Bonferroni correction. The volcano plot shows the variation in the number of metabolites between TCD-BM (n=6) and TCD-BM+T cells (n=6) groups according to the –log (q value) and log2 (fold change) on days 14 (a) and 28 (b). (c) The most important 50 metabolites after Student's t-test and the hierarchical clustering of samples shown in the heatmap. (d) KEGG-annotated metabolic pathways between TCD-BM and TCD-BM+T cells groups. (e) On day 14, the heatmap summarizes the altered fecal metabolites related to tyrosine between TCD-BM and TCD-BM+T cells groups and only displays the decrease of tyrosine in the TCD-BM+T cells group (two-tailed Student's t-test, p<0.01). (f) On day 28, the TCD-BM+T cells group showed a significant decrease in tyrosine, L-dopa, L-aspartic acid, L-glutamic acid and S-adenosylhomocysteine (two-tailed Student's t-test, p<0.01). (g, h) Correlation network between altered metabolites (circular) and differentially abundant microbiota (square). A connection means that a microbe is correlated with a metabolite; red and blue lines indicate positive and negative correlations respectively; volume size refers to the strength of the correlation. Networks between TCD-BM and TCD-BM+T cells groups were shown on days 14 (g) and 28 (h). The number of mice in each group was indicated.
Spearman's correlation analysis of tyrosine-related metabolites and altered microbiota was performed to explore the internal connections of microbiota and metabolome in the host system. In general, tyrosine located in the center of the correlation network in both comparisons, and bacteria belonging to Firmicutes were the closest to selected metabolites (Fig. 2g and h). The components of the network in TCD-BM+T cells and TCD-BM groups were smaller and more scattered on day 14 than day 28. Still, tyrosine-related metabolites were correlated inversely with Clostridium XIVa (Spearman's correlation analysis, day14: rho -0.8, p<0.01), besides, Blautia (Spearman's correlation analysis, day28: rho -0.29, p>0.05) and Enterococcus (Spearman's correlation analysis, day28: rho -0.41, p>0.05) displayed the negative tendency. Interestingly, it was found that all the genera in the network belonged to Firmicutes on day 28, implying that this phylum might play a prominent role in the development of aGVHD. Collectively, these results indicated that the low level of tyrosine in the gut was likely to be correlated with the occurrence and development of aGVHD.
3.3. Tyrosine supplement partially ameliorated aGVHD phenotype
The results demonstrated that tyrosine declined in the gut with the alteration of microbiota in aGVHD. Subsequently, an evaluation was made on whether tyrosine replenishment can have a positive effect on the alleviation of aGVHD and further affect microbiome structure and metabolite profiles. To test this, TCD-BM and TCD-BM+T cells groups were supplied with 2% tyrosine diet higher than normal but within safety content and 0% tyrosine diet as the deprivation group respectively (Fig. 3a). The administration of tyrosine supplement contributed to lots of improvements, such as the extension of total survival time, a significant reduction in weight loss and a decrease in aGVHD clinical scores in the early period of aGVHD (Fig. 3b-e), especially slighter diarrhea and better physical status (Supplementary Fig.S3g). It is worth noting that the TCD-BM+T cells + 2% tyrosine diet group displayed lower scores of intestinal and skin pathology with improved intestinal epithelial structure and kept intact microvilli and tight junctions and more complete skin structure on day 14 after BMT (Fig. 3f, Supplementary Fig.S3h). However, these advantages became weaker at the later stage of aGVHD as it was shown that the weight variances and clinical scores of the TCD-BM+T cells + 2% tyrosine diet group approached to those of the TCD-BM+T cells + vehicle diet group near the end of day 40. When deprived from tyrosine by being given 0% tyrosine diet, mice with aGVHD displayed even worse survival rate and clinical scores (day 30, two-tailed Student's t-test, p<0.01) than the TCD-BM+T cells + vehicle diet group.
Fig. 3.
Amelioration of aGVHD by tyrosine supplement.
(a) 0% or 2% tyrosine diets were served to TCD-BM (n=18) and TCD-BM+T cells (n=18) groups from day -7 of irradiation until the end of experiments. Compared with TCD-BM+T cells + vehicle, 2% tyrosine diet can result in (b) the prolonging of survival time, (c) the loss of less weight (day 6 to 24, two-tailed Student's t-test, p<0.01; day 27 to 33, two-tailed Student's t-test, p<0.05) and (d) the decrease of clinical scores (day 12 to 30, two-tailed Student's t-test, p<0.01; day 9 to 33, two-tailed Student's t-test, p<0.05) in early aGVHD. The administration of 2% tyrosine can decrease the (e) pathological scores of the large intestine and skin on day 14. (f) 2% tyrosine diet protected intestines and skins from the attack of allogenic T-cells, as shown in HE images. The pathology scores reflected mean ± SEM from three to four independent experiments, with n = 6 in each group.
Mice receiving TCD-BM only presented no signs of aGVHD, whose survival rate thus was not affected by deprived or extra tyrosine diets until the end (Fig. 3b). In comparison, the TCD-BM + 0% tyrosine diet group exhibited delayed weight regain and slight aGVHD symptoms at the initial stage of aGVHD after BMT. Moreover, the histopathologic analysis displayed mild lesions on intestine and skin on day 14 (Fig. 3e and f, Supplementary Fig.S3h). Above all, these results implied that additional tyrosine diets only played a role in aGVHD status and had an influence at the early stage of aGVHD after allo-HSCT.
3.4. Tyrosine supplement amended gut microbiome and metabolome
Tyrosine supplement was observed to play a role in ameliorating aGVHD, whose effects on the gut environment thus were further verified. The analysis of the fecal microbiome showed that changes after the intervention of tyrosine significantly increased according to alpha-diversity on day 28, as demonstrated by indexes Chao1 (MWU test, day28 p<0.05) and Shannon (MWU test, day28 p<0.05) (Table 2, Supplementary Fig. S3a, b). In addition, both weighted and unweighted UniFrac exhibited greater differences (MWU test, p<0.01) on day 28 (Supplementary Fig.S3c). The above results indicated that the group intervened by tyrosine exhibited dysbiosis and remained potential uncertainty.
Table 2.
Statistics of the alpha diversity index between TCD-BM+T+vehicle and TCD-BM+T+2% Tyrosine diet.
| Gut microbiota Diversity Index | Day 14 |
Day 28 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TCD-BM+T +vehicle |
TCD-BM+T +2%Tyrosine diet |
p-value | TCD-BM+T +vehicle |
TCD-BM+T +2%Tyrosine diet |
p-value | |||||
| Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | |||
| Observed species | 1695.17±88.90 | 1718.50 (1399.00-1964.00) | 1697.83±111.67 | 1704.50 (1402.00-2074.00) | 0.8182 | 554.00±39.28 | 549.00 (431.00-680.00) | 935.16±126.03 | 868.00 (642.00-1405.00) | 0.0087 |
| Chao1 | 2048.91±209.97 | 2085.00 (1712.46-2259.63) | 2066.21±270.11 | 2076.97 (1779.83-2365.80) | 0.8182 | 760.54±197.04 | 740.71 (528.63-1061.80) | 1315.56±389.49 | 1246.45 (872.24-1868.01) | 0.0152 |
| Shannon | 6.64±0.73 | 6.83 (5.71-7.45) | 6.63±0.58 | 6.60 (5.74-7.44) | 0.9372 | 4.11±0.46 | 4.26 (3.25-4.47) | 4.71±0.45 | 4.75 (3.92-5.23) | 0.0411 |
| Simpson | 0.93±0.05 | 0.96 (0.86-0.97) | 0.96±0.02 | 0.96 (0.92-0.97) | 0.6320 | 0.83±0.08 | 0.86 (0.67-0.90) | 0.88±0.03 | 0.88 (0.83-0.92) | 0.1190 |
| Good coverage | 0.99±0.00 | 0.99 (0.99-0.99) | 0.99±0.00 | 0.99 (0.99-0.99) | >0.9999 | 0.99±0.00 | 1.00 (0.99-1.00) | 0.99±0.00 | 1.00 (0.99-1.00) | >0.9999 |
Data are mean ± standard deviation (SD) or median and interquartile range (IQR) . Mann-Whitney U (MWU) test was employed for values comparisons.
When phylum showed no difference between groups on day 14, Firmicutes persisted at a low level in the TCD-BM+T cells + 2% tyrosine diet group (MWU test, p<0.05) on day 28, while Bacteroidetes (MWU test, p<0.05) and Verrucomicrobia showed higher levels (Fig. 4a and b). Through the analysis of genus level, however, it was supposed that tyrosine treatment could not only ameliorate aGVHD symptoms, but also restore gut microflora to aGVHD-free condition. 2% tyrosine supplement led to an increase in the abundance of Lachnospiraceae_unclassified (MWU test, p<0.05, on day 28) but a decrease in the fold changes of Bacteroides (two-tailed Student's t-test, p<0.05, on day 14), Clostridium XI (two-tailed Student's t-test, p<0.01, on day 28), Clostridium XIVa (two-tailed Student's t-test, p<0.05, on day 14) and Enterococcus (Fig. 4c and d; Supplementary Table S4). Notably, some microbiota that was insignificant in the comparison between TCD-BM and TCD-BM+T cells groups became striking after tyrosine intervention. For instance, the TCD-BM+T cells + 2% tyrosine diet group experienced a significant increase in both the proportion (MWU test, p<0.05) and fold change (two-tailed Student's t-test, p<0.01) of Barnesiella on day 14. In spite of only occupying a small proportion in all aGVHD groups, Escherichia decreased after 2% tyrosine intervention on day 14 (two-tailed Student's t-test, p<0.01), whereas the opposite trend was observed on day 28 (two-tailed Student's t-test, p<0.05). Besides, Lactobacillus saw a sudden increase on day 28 (TCD-BM+T cells + vehicle diet group and TCD-BM+T cells + 2% tyrosine diet group: 37.48% and 15.34%, MWU test, p<0.01), which was significantly down-regulated in the 2% tyrosine diet group.
Fig. 4.
Tyrosine supplement altered microbiome and metabolome.
(a-d) The relative abundance of gut microbiota between TCD-BM+T cells + vehicle (n=6) and 2% tyrosine diet (n=6) groups and the histogram of fold change (TCD-BM+T cells + 2% tyrosine diet group/TCD-BM+T cells + vehicle group) at the phylum level on days 14 (a) and day 28 (b). Relative abundance of top 20 genera shown in the histogram of fold change (TCD-BM+T cells + 2% tyrosine diet group/TCD-BM+T cells + vehicle group) on days 14 (c) and 28 (d). (e-f) The heatmap summarizes the different fecal metabolites related to tyrosine between TCD-BM+T cells + vehicle (n=6) and 2% tyrosine diet (n=6) groups. The 2% tyrosine-diet group showed an increase in tyrosine saw (two-tailed Student's t-test, p<0.01) on day 14 (e), but no significant difference in tyrosine and other metabolites on day 28 (f). (g-h) The same correlation analysis was performed to build a network according to Fig. 2 g-h where nodes represent microbiota or metabolites between TCD-BM+T cells + vehicle and TCD-BM+T cells + 2% tyrosine diet groups on days 14 (g) and 28 (h).
Both PCA and PLS-DA revealed grander different metabolites between TCD-BM+T cells groups with vehicle and 2% tyrosine diets (Supplementary Fig.S3d and e; Supplementary Table S5). The comparison statistics found that the number of different metabolites on day 14 was larger than that on day 28 (Supplementary Fig.S3f). However, most of the different metabolites belong to the superclass of lipids and lipid-like molecules followed by organheterocyclic compounds, organic acids and derivatives (Supplementary Fig.S4a-c; Supplementary Table S5). Fecal metabolomic profiles showed that the TCD-BM+T cells + 2% tyrosine diet group maintained a high level of tyrosine on day 14 (two-tailed Student's t-test, p<0.01). Moreover, its related metabolites including L-Glutamic, oxalacetic acid, S-Adenosylhomocysteine and homogentisic acid presented an upward trend (Fig. 4e; Supplementary Table S6). Nevertheless, these differences disappeared on day 28 in accordance with unsatisfied clinical and microbiota changes (Fig. 4f; Supplementary Table S6). Taken together, these data exemplified that the replenishment of tyrosine can ameliorate aGVHD in the early stage and change the structure of gut microbiota and fecal metabolic phenotype.
Larger network components were displayed in the comparison between TCD-BM+T cells groups with vehicle and 2% tyrosine diets. On day 14, L-Aspartic acid was positively correlated with Escherichia and Akkermansia. Tyramine, another attractive network node, a monoamine compound and trace amine derived from tyrosine, had a positive relationship with Bacteroides, Escherichia and Clostridium_sensu_stricto (Fig. 4g). Meanwhile, Bacteroides was negatively associated with tyrosine (Spearman's correlation analysis, rho -0.79, p<0.01) and S-Adenosylhomocysteine (Spearman's correlation analysis, rho -0.86, p<0.01). Moreover, oxalacetic acid displayed a positive correlation with Mucispirillum (Spearman's correlation analysis, rho 0.75, p<0.01), which however only occupied a small proportion. The correlation network showed that tyrosine faded out of the center at the later stage of aGVHD just as the treatment of tyrosine failed to meet the expectation on day 28. As only a negative correlation was found between tyrosine and Firmicutes microbiota like Peptostreptococcaceae_unclassified (Spearman's correlation analysis, rho -0.70, p<0.05) and Blautia (Spearman's correlation analysis, rho -0.72, p<0.01). S-Adenosylhomocysteine became the central node of five types of microbiota, exhibiting a positive relationship with Lactobacillus (Spearman's correlation analysis, rho 0.66, p<0.05) and Clostridium_XI (Spearman's correlation analysis, rho 0.66, p<0.05) and a negative relationship with Blautia (Spearman's correlation analysis, rho -0.77, p<0.05) (Fig. 4h). Overall, it was still believed that gut microbiota was closely correlated with tyrosine in aGVHD models.
4. Discussion
In this study, an approach integrating 16S rDNA gene sequencing and LC-MS-based metabolomics was taken to explore the impact of tyrosine supplement in murine aGVHD after BMT. The results indicated for the first time that tyrosine-replenished diets affected the composition of gut microbiota in stool samples and the dynamic changes of gut metabolomics in aGVHD.
16S rDNA gene sequencing was used to reveal the robust linkage between dietary intake and gut microbial community structure in murine aGVHD models. Researchers emphasized that Firmicutes was of importance in patients after allo-HSCT, which thus was believed to hold the balance on gut microbiota because of its largest proportion in each group. Both TCD-BM and TCD-BM+T cells groups contained a larger percentage of Firmicutes, but displayed opposite prognoses, which indicated the difficulty in judging it properly or obtaining the overall trend at the phylum level because they harbored dense and diverse microbial communities with advantageous and disadvantageous components. What's more, the individual difference and limitation of case number were potential contributing factors that might explain differences in response. Hence, lower levels like family and genus are worth discussing. Many results of this study were consistent with previous publications. Many researchers described a relative shift towards Enterococcus in gastrointestinal aGVHD after allo-HSCT, and the degree of dysbiosis dominated by this Enterococcus was correlated with the severity of aGVHD [35, 36]. Our research results showed that groups with severe aGVHD accumulated a large proportion of Enterococcus. On the contrary, Jenq RR et al. considered that a high proportion of Blautia was associated with the reduced death rate of aGVHD patients, while the death rate of aGVHD groups was found to increase [37]. Reaching a broad consensus on standards may be difficult due to temporal and geographical variations, differences in diet and antibiotic administration. Moreover, the different dietary structures of human and animal models play a decisive role [38]. Recently, it has been hypothesized that the increased abundance of Clostridiales was associated with reduced lethal GVHD and improved overall survival following allo-HSCT [39]. Likewise, some original findings were obtained in this research. Groups with a higher proportion of Lachnospiraceae_unclassified, one of major microbiota in data, showed less aGVHD. Besides, Porphyromonadaceae_unclassified occupied a large proportion in groups after tyrosine intervention on day 14. It was remarkable that both dominant microbiota mentioned above were not classified at the genus level, which made difficulties for follow-up work.
After allo-HSCT, extra nutrients were required to not only repair tissues damaged from chemotherapy or radiation in the initial period of transplantation, but also reconstruct BM and face the challenges from the body such as infection, wound healing and hypermetabolism after transplantation [40]. Nutritional supplementation appeared to be a more suitable treatment as the loss of beneficial microbiota was avoided mainly through antibiotic selection or probiotics administration. Restoring butyrate in the intestinal tract or enhancing the sensor of SCFAs can improve the junctional integrity of intestinal epithelial cells and up-regulate regulatory T cells to mitigate murine aGVHD [16, 41]. Similar ideal results can be obtained by the replenishment of Clostridia, a butyrate-producing bacterium, highlighting the mutual influence of gut microbiota and metabolism [16, 42]. As recent discoveries have underscored those changes in the microbiota, modulating the host immune system by modulating tryptophan metabolism [43], whether another metabolite bore resemblance to it in the same pathway was suspected in this research. Some researchers illustrated that the presence of gut microbiota increased the levels of tyrosine, glutamate, alanine and aspartate in the small intestine and creatine, glutamine and aspartate in the colon [44]. This study was the first to find the reduction of tyrosine in the gut with aGVHD. It was possible that the lack of tyrosine resulted from the decrease of absorption capacity attributed to the damage of the intestinal epithelial cellular membrane or dysbacteriosis caused by myeloablative management. Further, it was found that a comparatively high content of tyrosine could attenuate the clinical manifestations of aGVHD and improve the metabolites belonging to tyrosine biosynthesis pathway. Despite the difficulty in differentiating whether tyrosine was from self-synthesis or external uptake so far, it was certain that a comparatively high level of tyrosine played an active role in preventing damages during aGVHD. It was possible that the less weight loss during the early stage of aGVHD might be due to other mechanisms such as decreased caloric intake besides tyrosine intervention. However, since we changed the food every three days, the consumption of food was very low and the weight variations of feed were hard to measure. In addition, the non-aGVHD control groups, TCD-BM+ 0%/ 2% tyrosine diet, showed unobvious weight changes overall, which indicating that the content changes of tyrosine could not affect the total calories of the feed. Even though the content of tyrosine was the unique intervention, the alterations of certain metabolites can influence the whole metabolic network through their interactions with other elements leading to the change of the metabolic homeostasis of the host organism [45].
Correlation network analysis is an emerging field capturing the relations between different parts of a complex biological system, such as molecules, processes, organs and even microbes and metabolites [46]. The relations between changes in microbiota members and metabolites were charted, and the specific groups of correlations relevant to the development of aGVHD were identified. Specifically, many changes were observed to take place in microbiota belonging to Firmicutes and Bacteroidetes correlated to tyrosine and its biosynthesis in the host. Zhang et al. suggested that the levels of tyrosine and SCFAs were negatively associated with Enterococcus, [47] which was in line with the results of this study. Moreover, it was found tyrosine-related metabolites like L-Glutamic and L-Aspartic acids were inversely correlated with Clostridium XIVa and Enterococcus. Furthermore, Bacteroides displayed opposite correlations with different selected metabolites during aGVHD, which can explain the possible importance of tyrosine metabolism in gut microbiota and underlying metabolic interactions in host immune homeostasis.
However, we do acknowledge that there were some limitations in this study. Firstly, the tyrosine-supplemented diets displayed a striking protective role in the early stage of aGVHD but the sustained effectiveness were not observed, which possibly because the dose of tyrosine was incapable of remedying aGVHD at a later stage or the compensatory mechanism of the organism under the stimulation of pathological stress. Therefore, further studies are warranted to verify whether tyrosine and curative property have a dose-dependent relationship, or the combination therapy of complex amino acids is needed to control the progression of aGVHD. Secondly, it was supposed to find a certain gut microbiota that were sensitive to tyrosine and consider it to be a regulator for the treatment of aGVHD, and meanwhile provide molecular evidence on the relationship between tyrosine gene expression and the microbiota function. Nevertheless, decisive and dominant microbiota in this study was mostly unclassified at the genus level, such as Porphyromonadaceae_unclassified, Lachnospiraceae_unclassified and Peptostreptococcaceae_unclassified, which formed the obstacles of the further genetic and molecular experiments. Deep sequencing technology has made it possible to characterize the composition of mixed bacterial samples and distinguish thousands of bacteria, but complexity leads to many limitations in the identification and functional verification of all species [48, 49]. Consequently, further studies remain to be conducted to explore unknown but exceptional species mainly through the separation, purification and research of their genomes.
In all, these results provide a broader view of the effect of tyrosine on gut microbiome and metabolome. Further studies are essential to uncover underlying mechanisms and potential biological significances.
Data sharing statement
The datasets generated during the current study have been deposited in Sequence Read Archive (SRA). The SRA accession is PRJNA637751. The Submission ID is SUB7556797. The links is https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB7556797/overview.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
We thank for the support and help from members in core facilities of Zhejiang University School of medicine and laboratory animal center of Zhejiang University. We also thank LC-Bio Technology co.ltd. (Hangzhou, China) for technical support.
This work was supported by grants from International Cooperation and Exchange Program (81520108002), the National Key R&D Program of China, Stem Cell and Translation Research (2018YFA0109300), National Natural Science Foundation of China (81670169, 81670148, 81870080, 91949115) and Natural Science Foundation of Zhejiang Province (LQ19H080006). The Funders did not have any role in study design, data collection, data analyses, interpretation, or writing of this manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103048.
Contributor Information
Pengxu Qian, Email: axu@zju.edu.cn.
He Huang, Email: huanghe@zju.edu.cn.
Appendix. Supplementary materials
References
- 1.Thoo L, Noti M, Krebs P. Keep calm: the intestinal barrier at the interface of peace and war. Cell Death Dis. 2019;10(11):849. doi: 10.1038/s41419-019-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shono Y, Docampo MD, Peled JU, Perobelli SM, Jenq RR. Intestinal microbiota-related effects on graft-versus-host disease. Int J Hematol. 2015;101(5):428–437. doi: 10.1007/s12185-015-1781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18(1):2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi M, Yu S, Qin S, Liu Q, Xu H, Zhao W. Gut microbiome modulates efficacy of immune checkpoint inhibitors. J Hematol Oncol. 2018;11(1):47. doi: 10.1186/s13045-018-0592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 6.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Frank DN, Horch M, Chau S, Ir D, Horch EA. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant. 2017;52(12):1643–1650. doi: 10.1038/bmt.2017.200. [DOI] [PubMed] [Google Scholar]
- 8.Laterza L, Rizzatti G, Gaetani E, Chiusolo P, Gasbarrini A. The gut microbiota and immune system relationship in human graft-versus-host disease. Mediterr J Hematol Infect Dis. 2016;8(1) doi: 10.4084/MJHID.2016.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy S, Nguyen VH. Role of gut microbiota in graft-versus-host disease. Leuk Lymphoma. 2011;52(10):1844–1856. doi: 10.3109/10428194.2011.580476. [DOI] [PubMed] [Google Scholar]
- 10.Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K. Stool microbiota at neutrophil recovery is predictive for severe acute graft vs host disease after hematopoietic cell transplantation. Clin Infect Dis. 2017;65(12):1984–1991. doi: 10.1093/cid/cix699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut. 2010;59(8):1079–1087. doi: 10.1136/gut.2009.197434. [DOI] [PubMed] [Google Scholar]
- 12.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakihana K. Fecal microbiota transplantation for acute graft-versus-host disease of the gut. Rinsho Ketsueki. 2017;58(5):499–505. doi: 10.11406/rinketsu.58.499. [DOI] [PubMed] [Google Scholar]
- 14.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4) doi: 10.1053/j.gastro.2012.06.031. 913-6 e7. [DOI] [PubMed] [Google Scholar]
- 15.Michonneau D, Latis E, Dubouchet L, Peffault De Latour R, Robin M, Sicre De Fontbrune F. Metabolomics profiling after allogeneic hematopoietic stem cell transplantation unravels a specific signature in human acute GVHD. Blood. 2018;132(Supplement 1):69-. [Google Scholar]
- 16.Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17(5):505–513. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swimm A, Giver CR, DeFilipp Z, Rangaraju S, Sharma A, Ulezko Antonova A. Indoles derived from intestinal microbiota act via type I interferon signaling to limit graft-versus-host disease. Blood. 2018;132(23):2506–2519. doi: 10.1182/blood-2018-03-838193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Meij BS, de Graaf P, Wierdsma NJ, Langius JA, Janssen JJ, van Leeuwen PA. Nutritional support in patients with GVHD of the digestive tract: state of the art. Bone Marrow Transplant. 2013;48(4):474–482. doi: 10.1038/bmt.2012.124. [DOI] [PubMed] [Google Scholar]
- 19.Morris CR, Hamilton-Reeves J, Martindale RG, Sarav M, Ochoa Gautier JB. Acquired amino acid deficiencies: a focus on arginine and glutamine. Nutr Clin Pract. 2017;32(1_suppl):30S–47S. doi: 10.1177/0884533617691250. [DOI] [PubMed] [Google Scholar]
- 20.Noe JE. L-glutamine use in the treatment and prevention of mucositis and cachexia: a naturopathic perspective. Integr Cancer Ther. 2009;8(4):409–415. doi: 10.1177/1534735409348865. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn S, Duzel S, Colzato L, Norman K, Gallinat J, Brandmaier AM. Food for thought: association between dietary tyrosine and cognitive performance in younger and older adults. Psychol Res. 2019;83(6):1097–1106. doi: 10.1007/s00426-017-0957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman HR, Georgelis JH, Maher TJ, Yeghiayan SK. Tyrosine prevents effects of hyperthermia on behavior and increases norepinephrine. Physiol Behav. 2005;84(1):33–38. doi: 10.1016/j.physbeh.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JL. Total body irradiation and acute graft-versus-host disease: the role of gastrointestinal damage and inflammatory cytokines. Blood. 1997;90(8):3204–3213. [PubMed] [Google Scholar]
- 24.Kaplan DH, Anderson BE, McNiff JM, Jain D, Shlomchik MJ, Shlomchik WD. Target antigens determine graft-versus-host disease phenotype. J Immunol. 2004;173(9):5467–5475. doi: 10.4049/jimmunol.173.9.5467. [DOI] [PubMed] [Google Scholar]
- 25.Messineo AM, Gineste C, Sztal TE, McNamara EL, Vilmen C, Ogier AC. L-tyrosine supplementation does not ameliorate skeletal muscle dysfunction in zebrafish and mouse models of dominant skeletal muscle alpha-actin nemaline myopathy. Sci Rep. 2018;8(1):11490. doi: 10.1038/s41598-018-29437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uebanso T, Kano S, Yoshimoto A, Naito C, Shimohata T, Mawatari K. Effects of consuming xylitol on gut microbiota and lipid metabolism in mice. Nutrients. 2017;9(7) doi: 10.3390/nu9070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao X, Guo F, Zhou Q, Hu F, Xiang H, Xiao GG. Bacterial community mapping of the intestinal tract in acute pancreatitis rats based on 16S rDNA gene sequence analysis. RSC Adv. 2019;9(9):5025–5036. doi: 10.1039/c8ra09547g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5(6):1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 29.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion–switching, targeted mass spectrometry–based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spoerl S, Mathew NR, Bscheider M, Schmitt-Graeff A, Chen S, Mueller T. Activity of therapeutic JAK 1/2 blockade in graft-versus-host disease. Blood. 2014;123(24):3832–3842. doi: 10.1182/blood-2013-12-543736. [DOI] [PubMed] [Google Scholar]
- 32.Tugues S, Amorim A, Spath S, Martin-Blondel G, Schreiner B, De Feo D. Graft-versus-host disease, but not graft-versus-leukemia immunity, is mediated by GM-CSF-licensed myeloid cells. Sci Transl Med. 2018;10(469) doi: 10.1126/scitranslmed.aat8410. [DOI] [PubMed] [Google Scholar]
- 33.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4(7):e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker A, Pfitzner B, Neschen S, Kahle M, Harir M, Lucio M. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J. 2014;8(12):2380–2396. doi: 10.1038/ismej.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 2019;366(6469):1143–1149. doi: 10.1126/science.aax3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transpl. 2014;20(5):640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21(8):1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8(339) doi: 10.1126/scitranslmed.aaf2311. 339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Brink M, Jenq R, Pamer EG, Taur Y, Shono Y. Intestinal microbiota and gvhd. Google Patents. 2017 [Google Scholar]
- 40.Roberts S, Thompson J. Graft-vs-host disease: nutrition therapy in a challenging condition. Nutrition in Clinical Practice. 2017;20(4):440–450. doi: 10.1177/0115426505020004440. [DOI] [PubMed] [Google Scholar]
- 41.Fujiwara H, Docampo MD, Riwes M, Peltier D, Toubai T, Henig I. Microbial metabolite sensor GPR43 controls severity of experimental GVHD. Nat Commun. 2018;9(1):3674. doi: 10.1038/s41467-018-06048-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 43.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barabasi A-L, Oltvai ZN. Network biology: understanding the cell's functional organization. Nature reviews genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9(4):e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sentausa E, Fournier PE. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19(9):790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.