Abstract
Pathogenic sequence variants (PSV) in BRCA1 or BRCA2 (BRCA1/2) are associated with increased risk and severity of prostate cancer (PCa). We evaluated whether PSVs in BRCA1/2 were associated with risk of overall PCa or high grade (Gleason 8+) PCa using an international sample of 65 BRCA1 and 171 BRCA2 male PSV carriers with PCa, and 3,388 BRCA1 and 2,880 BRCA2 male PSV carriers without PCa. PSVs in the 3’ region of BRCA2 (c.7914+) were significantly associated with elevated risk of PCa compared with reference bin c.1001-c.7913 (HR=1.78, 95%CI: 1.25–2.52, p=0.001), as well as elevated risk of Gleason 8+ PCa (HR=3.11, 95%CI: 1.63–5.95, p=0.001). c.756-c.1000 was also associated with elevated PCa risk (HR=2.83, 95%CI: 1.71–4.68, p=0.00004) and elevated risk of Gleason 8+ PCa (HR=4.95, 95%CI: 2.12–11.54, p=0.0002). No genotype-phenotype associations were detected for PSVs in BRCA1. These results demonstrate that specific BRCA2 PSVs may be associated with elevated risk of developing aggressive PCa.
Keywords: BRCA1, BRCA2, Prostate Cancer, Pathogenic sequence variant location, Risk estimation
Introduction
Inherited pathogenic sequence variants (PSVs) in DNA repair pathway genes including BRCA1 and BRCA2 (BRCA1/2) are associated with prostate cancer (PCa) risk and severity(1–15). Carriers of BRCA2 PSVs have been reported to have increased levels of serum prostate-specific antigen (PSA) at diagnosis, increased proportion of high Gleason (7+) tumors, less favorable tumor stage, increased rates of nodal and distant metastases, and increased rate of recurrence after treatment(2,11–18). BRCA2 PSVs confer lower overall survival and PCa specific survival(13–15). Ashkenazi Jewish carriers of BRCA1 PSVs have been reported to have elevated rates of Gleason 7+ tumors, higher rates of recurrence, and a five-fold increase in PCa death(5,19), although the association of BRCA1 and PCa has not been replicated in all studies(20). Distinct tumor PSV, methylation, and expression patterns have been identified in BRCA2 compared with non-BRCA2 mutant prostate tumors. These data suggest that BRCA2 mutant tumors have features that are more similar to metastatic castrate resistant disease than localized PCa(21–23).
Specific genotype-phenotype correlations have been reported(24), including BRCA1/2-associated breast and ovarian cancers(25–27), APC PSVs and severity of familial adenomatous polyposis (FAP)(28,29), and RET PSVs in multiple endocrine neoplasia type 2 (MEN2) and Familial Medullary Thyroid Carcinoma(30). There have been suggestions in the literature that similar patterns exist for BRCA1 or BRCA2 and PCa. Liede et al.(31) reported that early-onset PCa (<age 65 years) was more frequent in men with BRCA2 PSVs outside of the ovarian cancer cluster region. More recently, Roed Nielsen et al.(32), using a sample of 37 PCa cases, 19 of whom had BRCA2 PSVs, identified a region in BRCA2 at c.6373-c.6492 in which PSVs were associated with an increased risk of PCa.
We analyzed a large international cohort of 3,453 BRCA1 and 3,051 BRCA2 PSV male carriers to evaluate the distribution of germline PSVs in men diagnosed with PCa and men without prior PCa diagnosis. We hypothesized that specific PSVs in BRCA1 or BRCA2 might influence development of PCa and be associated with PCa severity.
Materials and Methods
Study Sample
The Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) is an international collaboration of centers on six continents that has collected information about carriers of BRCA1/2 PSVs(33). All carriers participated in clinical assessment and/or research studies at a participating institution after providing informed consent under protocols approved by local institutional review boards. Participants ascertainment date was defined as the time of study interview (e.g., enrollment in a research study). Forty-eight centers and multicenter consortia (Supplementary Table 1) in 31 countries submitted de-identified data that met the CIMBA inclusion criteria as previously described. No races/ethnicities were excluded from this study. Self-reported race/ethnicity data were collected across the various centers using either fixed categories or open-ended questions.
We analyzed only male carriers with clearly pathogenic BRCA1/2 PSVs that occurred 3’ of nucleotide position 1 (A of the ATC translation initiation codon in either BRCA1 and BRCA2. This excluded 101 males who had a PSV occurring 5’ translation start site. Definitions of these PSVs are shown in Supplementary Table 2. PSVs were defined using CIMBA criteria as follows: (1) PSVs generating a premature termination codon, except those in exon 27 at or after codon 3310 of BRCA2; (2) large in-frame deletions that spanned ≥1 exons; and (3) deletions of transcription regulatory regions (promoter and/or first exon) expected to cause lack of mutant allele expression(33–35). We also included missense variants considered pathogenic as determined by using multifactorial likelihood approaches(35,36). PSVs are described using the Human Genome Variation Society (HGVS) nomenclature (Supplementary Table 2).
Authors have obtained written informed consent.
Pathogenic Sequence Variant Binning
To identify segments across the intronic and exonic regions of BRCA1 and BRCA2 associated with different PCa risks, we created PSV bins by base pair location within each gene. These genomic sequence bins contained all PSVs regardless of category or function, except for large genomic rearrangements, which were excluded from this analysis since they may span multiple bins. Bins were constructed in two ways. First, we used an algorithm in which each bin contained approximately equal numbers of participants (including all cases and controls) with bin length defined by distance in base pairs. Thus, bin length for common PSVs (e.g., the Icelandic founder PSV c.771_775del) were small compared to bins with a wider range of PSVs. We divided the number of PSVs across the span of BRCA1 or BRCA2 into deciles of PSVs observed in cases and non-cases (i.e., “decile” bins). Second, we identified putative functional domains in BRCA1 or BRCA2 and created bins that captured these domains, as well as bins that contained no functional domain. These domains were determined by boundaries reported in the pfam database(37). The resulting bin boundaries are presented in Supplementary Table 3 and shown graphically in Figure 1 for BRCA1 and Figure 2 for BRCA2. We chose to use these two binning methods based on our earlier published research(24) that indicated the inferences about mutation risk association differences were similar regardless of the binning approach used. After the initial evaluation across all bins (Supplementary Table 3), we further collapsed bins that were inferred to have homogeneous PCa, either elevated above or not different from the reference bin.
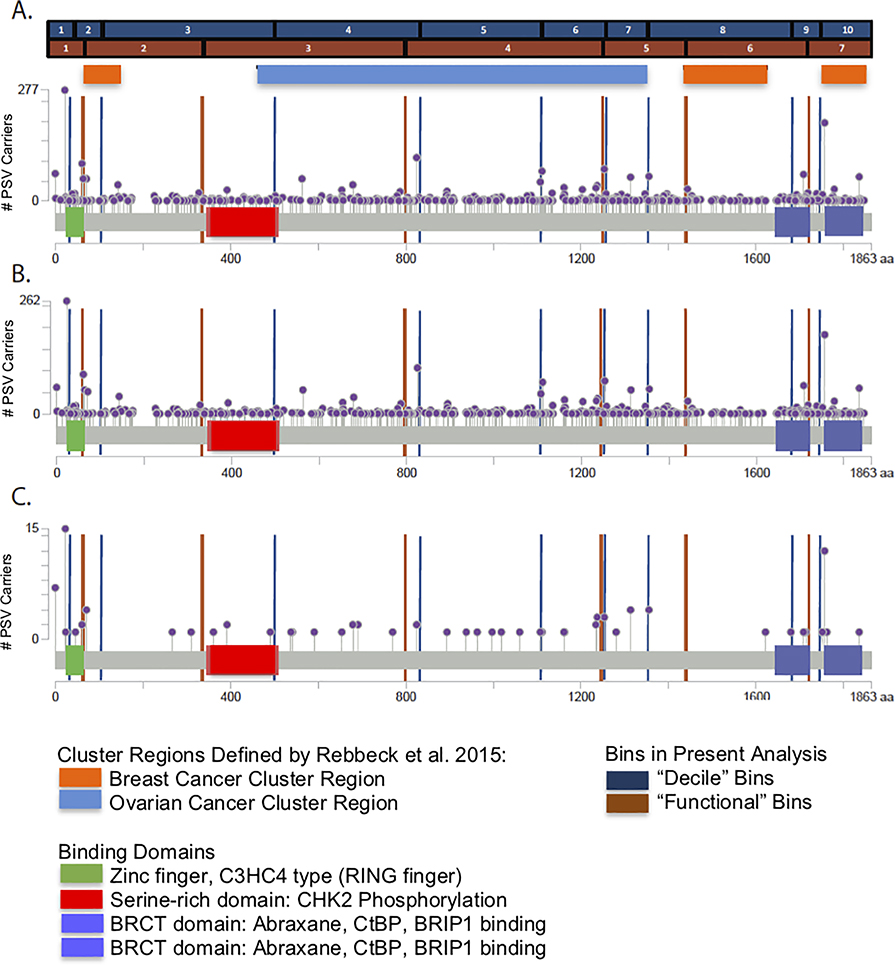
Figure 1: BRCA1 Pathogenic Sequence Variant Distribution.
The x-axis displays the amino acid sequence of the BRCA1 gene. The violet markers indicate the position of PSVs found in the BRCA1 PSV carriers. The vertical position of the markers on the y-axis indicates the frequency of the PSV found in the cohort. Additionally, the blue and tan bars with corresponding axis markers delineate the bins of the BRCA1 PSVs that were created using the “decile” binning strategy and the “functional” binning strategy. Orange and light blue bars indicate the position of breast and ovarian cancer cluster regions, respectively, as identified in the CIMBA breast cancer cohort (Rebbeck et al. JAMA 2015). Lastly, known functional domains within the BRCA1 gene are highlighted.
A. Distribution of total BRCA1 PSVs in carriers, B. Distribution of BRCA1 PSVs in carriers who did not develop prostate cancer, C. Distribution of BRCA1 PSVs in carriers who developed prostate cancer.
Figure 2. BRCA2 Pathogenic Sequence Variant Distribution.
The x-axis displays the amino acid sequence of the BRCA2 gene. The violet markers indicate the position of PSVs found in the BRCA2 PSV carriers. The vertical position of the markers on the y-axis indicates the frequency of the PSV found in the cohort. Additionally, the blue and tan bars with corresponding axis markers delineate the bins of the BRCA2 PSVs that were created using the “decile” binning strategy and the “functional” binning strategy. Orange and light blue bars indicate the position of breast and ovarian cancer cluster regions, respectively, as identified in the CIMBA breast cancer cohort (Rebbeck et al. JAMA 2015). Lastly, known functional domains within the BRCA2 gene are highlighted.
A. Distribution of total BRCA2 PSVs in carriers, B. Distribution of BRCA2 PSVs in carriers who did not develop prostate cancer, C. Distribution of BRCA2 PSVs in carriers who developed prostate cancer.
Pathogenic Sequence Variant Type and Function
In addition to the binning analyses described above, we also considered whether the predicted type and function of heritable BRCA1/2 PSVs in the CIMBA database were associated with PCa. The definition of these PSV types and their functions are presented in Supplementary Table 2. PSVs were grouped by type and function as frameshift (FS), nonsense (NS), missense (MS), and splice site (SP) (Supplementary Table 2). PSVs expected to generate stable or unstable, or no proteins were designated into previously reported classes 1, 2, or 3(38–40). Missense PSVs in BRCA1 were combined into one group that contained PSVs in the RING(41,42) and BRCT domains(43–46). We compared PSVs predicted to produce nonsense-mediated decay (NMD) vs. those that were not. PSVs predicted not to cause NMD were defined as those creating a stop codon within 50 nucleotides before or within the last exon(47). Premature termination codons comprised all PSVs leading to a truncated open reading frame.
Statistical Methods
For the first set of analyses assessing all bins across the genes, a different reference group was defined for each combination of gene (BRCA1 or BRCA2) and binning scheme (decile or functional). Reference bins were chosen based on analysis of each bin’s association with PCa compared with all other bins as a group and found to have the lowest hazard of PCa for each gene. The reference bins used in each analysis are shown in Table 1. An exploration of other reference bins did not change the inferences of this analysis.
Table 1:
Association Analyses of Prostate Cancer by bin for BRCA1 and BRCA2 PSVs
| BRCA1 – All Prostate Cancer | ||||||||
| Grouping | Bin | Nucleotide Range | PC+ | PC− | HR | 95% CI | p-value | |
| BRCA1 Decile | 1 | ≤c.81 | 11 | 339 | 1.06 | 0.36–3.13 | 0.917 | |
| 2 | c.82-c.302 | 5 | 325 | REF | ||||
| 3 | c.303 – c.1504 | 4 | 331 | 0.82 | 0.23–2.92 | 0.761 | ||
| 4 | c.1505 – c.2475 | 8 | 431 | 1.09 | 0.34–3.43 | 0.888 | ||
| 5 | c.2476 – c.3319 | 3 | 274 | 0.33 | 0.15–2.60 | 0.526 | ||
| 6 | c.3320 – c.3710 | 5 | 308 | 1.32 | 0.36–4.89 | 0.677 | ||
| 7 | c.3711 – c.4065 | 9 | 318 | 1.96 | 0.65–5.86 | 0.230 | ||
| 8 | c.4066 – c.5030 | 1 | 333 | 0.16 | 0.02–1.27 | 0.084 | ||
| 9 | c.5031 – c.5266 | 13 | 425 | 1.68 | 0.58–4.84 | 0.339 | ||
| 10 | c.5267+ | 2 | 231 | 0.49 | 0.10–2.49 | 0.389 | ||
| BRCA1 Functional | 1 | ≤c.181 | 13 | 515 | 0.72 | 0.34–1.53 | 0.396 | |
| 2 | c.182-c.1287 | 6 | 433 | 0.83 | 0.32–2.20 | 0.713 | ||
| 3 | c.1288-c.2475 | 9 | 478 | 0.93 | 0.37–2.36 | 0.887 | ||
| 4 | c.2476-c.3607 | 5 | 487 | 0.58 | 0.19–1.75 | 0.333 | ||
| 5 | c.3608-c.4183 | 12 | 462 | 1.19 | 0.54–2.64 | 0.671 | ||
| 6 | c.4184-c.5194 | 5 | 485 | 0.38 | 0.12–1.25 | 0.112 | ||
| 7 | c.5195+ | 11 | 455 | REF | ||||
| BRCA2 – All Prostate Cancer | ||||||||
| Grouping | Bin | Nucleotide Range | PC+ | PC− | HR | 95% CI | p-value | |
| Decile | 1 | ≤c.755 | 12 | 296 | 1.71 | 0.66–4.46 | 0.268 | |
| 2 | c.756-c.1813 | 25 | 277 | 3.38 | 1.24–9.19 | 0.017 | ||
| 3 | c.1814-c.3530 | 6 | 293 | REF | ||||
| 4 | c.3531-c.4965 | 13 | 296 | 2.00 | 0.69–5.76 | 0.202 | ||
| 5 | c.4966-c.5909 | 13 | 307 | 2.14 | 0.66–7.00 | 0.207 | ||
| 6 | c.5910-c.6275 | 30 | 334 | 2.83 | 1.21–6.58 | 0.016 | ||
| 7 | c.6276-c.7007 | 12 | 214 | 2.69 | 0.89–8.13 | 0.079 | ||
| 8 | c.7008-c.7913 | 10 | 285 | 2.12 | 0.60–7.42 | 0.240 | ||
| 9 | c.7914-c.8953 | 26 | 281 | 3.32 | 1.28–8.65 | 0.014 | ||
| 10 | c.8954+ | 23 | 274 | 4.26 | 1.60–11.37 | 0.004 | ||
| Functional | 1 | ≤c.1000 | 27 | 398 | 1.39 | 0.74–2.64 | 0.307 | |
| 2 | c.1001-c.3005 | 14 | 397 | 0.80 | 0.39–1.63 | 0.535 | ||
| 3 | c.3006-c.5172 | 16 | 408 | REF | ||||
| 4 | c.5173-c.6255 | 32 | 498 | 1.01 | 0.53–1.93 | 0.967 | ||
| 5 | c.6256-c.7436 | 24 | 400 | 1.44 | 0.74–2.82 | 0.286 | ||
| 6 | c.7437-c.8616 | 28 | 390 | 1.68 | 0.91–3.13 | 0.100 | ||
| 7 | c.8617+ | 29 | 366 | 1.64 | 0.90–3.01 | 0.106 | ||
| Elevated vs. No Elevated PCa Risk | 1 | ≤c.755 | 12 | 296 | 0.73 | 0.40–1.31 | 0.288 | |
| 2* | c.756-c.1000 | 15 | 102 | 2.83 | 1.71–4.68 | 4×10−5 | ||
| 3 | c.1001-c.7913 | 94 | 1904 | REF | ||||
| PCCR | c.7914+ | 49 | 555 | 1.78 | 1.25–2.52 | 0.001 | ||
| Elevated vs. No Elevated PCa Risk | No Elevated PCa Risk | ≤c.755, c.1001-c.7913 | 106 | 2,200 | REF | |||
| Elevated PCa Risk | c.756-c.1000, c.7914+ | 65 | 657 | 2.02 | 1.48–2.77 | 9×10−6 | ||
| BRCA2 –Prostate Cancer by Gleason Grade | ||||||||
| Gleason 8+ | Bin | Nucleotide Range | PC+ | PC− | HR | 95% CI | p-value | |
| Bins with Elevated Risk | 1 | ≤c.755 | 2 | 299 | 0.53 | 0.12–2.32 | 0.399 | |
| 2 | c.756-c.1000 | 6 | 108 | 4.95 | 2.12–11.54 | 2×10−4 | ||
| 3 | c.1001-c.7913 | 19 | 1940 | REF | ||||
| PCCR | c.7914+ | 18 | 572 | 3.11 | 1.63–5.95 | 0.001 | ||
| Bins with Elevated Risk | No Elevated PCa Risk | ≤c.755, c.1001-c.7913 | 21 | 2239 | REF | |||
| Elevated PCa Risk | c.756-c.1000, c.7914+ | 24 | 680 | 3.80 | 2.10–6.89 | 1×10−5 | ||
| Gleason ≤ 7 | ||||||||
| Bins with Elevated Risk | 1 | ≤c.755 | 3 | 298 | 0.47 | 0.14–1.57 | 0.221 | |
| 2* | c.756-c.1000 | 6 | 108 | 3.29 | 1.38–7.83 | 0.007 | ||
| 3 | c.1001-c.7913 | 36 | 1923 | REF | ||||
| PCCR | c.7914+ | 17 | 573 | 1.56 | 0.88–2.78 | 0.130 | ||
| Bins with Elevated Risk | No Elevated PCa Risk | ≤c.755, c.1001-c.7913 | 39 | 2221 | REF | |||
| Elevated PCa Risk | c.756-c.1000, c.7914+ | 23 | 681 | 1.89 | 1.14–3.14 | 0.014 | ||
Bin containing Icelandic Founder PSV c.771_775del
To estimate the relative hazards associated with each bin compared with the reference bin, we fitted Cox proportional hazards regression models separately in BRCA1 and BRCA2 PSV carriers. The primary outcomes of interest were diagnosis of PCa (vs. no PCa) or Gleason 8+ PCa (vs. no PCa) and Gleason ≤7 (vs. No PCa). Time to event was computed from birth to age at PCa diagnosis or age at ascertainment (which ever occurred first). No time or events were considered after time of ascertainment. All analyses were adjusted for confounding by race (African American vs. any other ethnicity) and birth cohort, defined as those born before or after median birth date of the total sample. We also adjusted all analyses by country of ascertainment. We computed the PCa hazard ratio for each defined bin relative to the common reference bin. To account for intra-cluster dependence due to multiple individuals from the same family, a robust sandwich variance estimate was specified in Cox proportional hazards models(48).
Hypothesis tests were judged to be statistically significant based on 2-sided tests with P-value < 0.05. All P-values were corrected for multiple hypothesis testing within each table of results by controlling the false discovery rate (FDR) using the method of Benjamini and Hochberg(49). Analyses were conducted in STATA v14, SPSS, or R version 2.7.2 (R Foundation for Statistical Computing).
Results
A total of 3,453 male BRCA1 and 3,051 male BRCA2 PSV carriers were eligible for analysis. (see Table 2). The median PCa diagnosis ages were 64 years in both BRCA1 and BRCA2 PSV carriers. Among BRCA1/2 PSV carriers, 74% and 81%, respectively, self-reported their race as white.
Table 2:
Characteristics of Study Participants
| Carriers of PSV in BRCA1 | Carriers of PSV in BRCA2 | ||||||
|---|---|---|---|---|---|---|---|
| Total | N 3,453 |
% | N 3,051 |
% | |||
| Region | Asia | 76 | 2.2 | 90 | 2.9 | ||
| Australia | 386 | 11.2 | 292 | 9.6 | |||
| Europe | 2,287 | 66.2 | 2,165 | 71.0 | |||
| North America | 662 | 19.2 | 497 | 16.3 | |||
| South America | 42 | 1.2 | 7 | 0.2 | |||
| Self-Identified Race/Ethnicity | Caucasian | 2,557 | 74.1 | 2,455 | 80.5 | ||
| African American | 20 | 0.6 | 14 | 0.5 | |||
| Asian | 76 | 2.2 | 101 | 3.3 | |||
| Hispanic | 54 | 1.6 | 16 | 0.5 | |||
| Jewish | 124 | 3.6 | 94 | 3.1 | |||
| Other | 45 | 1.3 | 10 | 0.3 | |||
| Unknown | 575 | 16.7 | 358 | 11.7 | |||
| Ascertainment | Clinic-based | 3,352 | 97.1 | 2,969 | 97.3 | ||
| Population-based | 101 | 2.9 | 82 | 2.7 | |||
| PCa | Yes | 65 | 1.9 | 171 | 5.5 | ||
| No | 3,388 | 98.1 | 2,880 | 94.4 | |||
| Gleason Score | ≤6 | 16 | 24.6 | 32 | 18.7 | ||
| 7 | 9 | 13.8 | 30 | 17.5 | |||
| 8 | 3 | 4.6 | 16 | 3.1 | |||
| 9 | 7 | 10.8 | 26 | 15.2 | |||
| 10 | 0 | 0.0 | 5 | 2.9 | |||
| Missing | 30 | 46.2 | 62 | 36.2 | |||
| M Stage | M0 | 18 | 27.7 | 33 | 19.3 | ||
| M1 | 2 | 3.1 | 14 | 8.2 | |||
| MX | 8 | 12.3 | 28 | 16.4 | |||
| Missing | 37 | 56.9 | 96 | 56.1 | |||
| Other Cancer Diagnosis | Yes | 332 | 9.6 | 657 | 21.5 | ||
| No | 3,121 | 90.4 | 2,389 | 78.5 | |||
| N | Median | Range | N | Median | Range | ||
| Age at Ascertainment (yrs) | 3,453 | 50 | 18–91 | 3,051 | 51 | 18–101 | |
| Time to PCa or Censoring (yrs) | 3,453 | 50 | 18–91 | 3,051 | 54 | 18–101 | |
| Age at PCa Diagnosis (yrs) | 65 | 64 | 30–85 | 171 | 64 | 29–87 | |
| Age at Other Cancer Diagnosis (yrs) | 332 | 59 | 19–88 | 657 | 60 | 21–88 | |
BRCA1
As shown in Table 1a, there were no statistically significant associations between PSVs in any BRCA1 bin and elevated PCa risk. There was also no association of BRCA1 PSVs with Gleason 8+ disease with region.
BRCA2
In BRCA2, we identified a “prostate cancer cluster region” (PCCR) in which PSVs were associated with elevated PCa risk. The risk estimates were obtained by considering all PSVs within the region of interest defined by the overlap of bins generated using the “decile” and functional binning methods described above. The PCCR included all PSVs 3’ of c.7914 and associated with HR=1.78 (95%CI: 1.25–2.52, p=0.001) when compared with PSVs in the reference bin c.1001-c.7913 (Table 1b). In addition, we identified a region bounded by c.756 and c.1000 (Supplementary Table 3 and Figure 2) that was associated with elevated PCa risk with HR=2.83 (95% CI: 1.71–4.68, p=4×10−5) compared with PSVs in the reference bin c.1001-c.7913. This region contains the c.771_775del Icelandic founder PSV, which is the dominant PSV in this bin (n=92 of 117 total PSVs in this bin). Comparison of the risk in carriers of c.771_775del to the risk in carriers of PSVs in c.1001-c.7913 gave HR=3.34 (95%CI: 2.01–5.55, p=3×10−6). Due to the small number of carriers of other PSVs in this bin (N=25), it was not possible to estimate risk of PCa for carriers of the other (non-c.771_775del) PSVs in this bin. Risk of PCa among those without a PCCR PSV was not elevated except for carriers of PSVs in bin 6 (c.5910-c.6275) (HR=2.83, 95%CI: 1.21–6.58, p=0.016) (Table 2b). Both the PCCR and region c.756-c.1000 were contained almost entirely within the previously identified breast cancer cluster regions (BCCRs)(24). Collectively, regions in which PSVs were associated with a significantly increased risk of PCa development contained the BRCA2 helical plasma domain, the oligonucleotide/oligosaccharide-binding domain 1 (OB1), the Tower domain (OB2), and the N-terminal PALB2 binding site (Figure 2). Highest risk was associated with PSVs affecting OB1 and OB2 (Figure 2).
Risk of high-grade PCa (Gleason 8+) was even more strongly associated with PSVs in the PCCR (HR=3.11, 95% CI: 1.63–5.95, p=0.001; Table 1c). A similar association was also observed for PSVs in the region containing the Icelandic founder PSV, c.771_775_del (HR=4.95, 95% CI: 2.12–11.54, p=2×10−4), and the c.771_775del PSV itself (HR=5.66, 95% CI: 2.43–13.22, p=6×10−5) Together, these regions were associated with increased Gleason 8+ PCa risk (HR=3.80, 95% CI: 2.10–6.89, p=1×10−5). Risk of Gleason ≤7 PCa was elevated for carriers of c.771_775del (HR=3.29, 95%CI: 1.38–7.83, p=0.007), but not elevated for those with PSVs in the PCCR (HR=1.56, 95%CI: 0.88–2.78, p=0.130; Table 1c).
To ensure that the inferred effects were not due to the common Jewish founder PSV c.5946del that was included in the reference bin, we repeated calculations after excluding carriers these PSVs from the reference bin. After excluding these PSV carriers from the reference bin, the association with PSVs in the bin containing the c.771_775del and in the PCCR remained statistically significant (HR=3.03, 95%CI: 1.83–5.04, p=2×10−5 and HR=1.89, 95%CI: 1.34–2.66, p=3×10−4, respectively). Similarly, we repeated the analysis including only self-identified Caucasians. In part because of the small number of non-Caucasians in the study, the point estimates did not change to the second decimal place compared with the total sample that included non-Caucasians. Finally, we corrected for correlation due to the presence of multiple individuals in a family. With and without this correction, no change in the inferences were observed.
Pathogenic Sequence Variant Type and Function
In addition to seeking for regional variation in PCa risk associated with PSVs across BRCA1/2, we also evaluated potential genotype-phenotype correlations by PSV type or function (Table 3). No PSV groups defined by type or function were significantly associated with prostate cancer for either BRCA1 or BRCA2.
Table 3:
Association of Pathogenic Sequence Variant (PSV) Type or Function with risk of prostate cancer. Hazard Ratios (HRs) represent the comparison of PSVs with a certain type or function designation vs. all other PSVs. HRs are adjusted for year of birth cohort, race, and country of ascertainment.
| BRCA1 mutation carriers | BRCA2 mutation carriers | |||||||
|---|---|---|---|---|---|---|---|---|
| PSV type | N | PCa | HR (95% CI) | p-value | N | PCa | HR (95% CI) | p-value |
| Premature Truncating Codon | 2,720 | 54 (2.0%) | 1.04 (0.47–2.28) | 0.931 | 2,699 | 151 (5.6%) | 0.90 (0.40–2.04) | 0.805 |
| Nonsense-Mediated Decay | 1,996 | 31 (1.6%) | 0.65 (0.38–1.11) | 0.117 | 2,692 | 150 (5.6%) | 0.86 (0.41–1.82) | 0.698 |
| Class 1 | 2,489 | 48 (1.9%) | 0.80 (0.44–1.47) | 0.474 | 2,712 | 151 (5.6%) | 0.81 (0.37–1.78) | 0.596 |
| Deletion | 279 | 5 (1.8%) | 0.79 (0.32–1.95) | 0.606 | 57 | 5 (8.8) | 1.25 (0.51–3.08) | 0.469 |
| Frameshift | 1,845 | 43 (2.3%) | 1.66 (0.99–2.77) | 0.055 | 2,040 | 115 (5.6%) | 1.01 (0.74–1.41) | 0.910 |
| Insertion | 61 | 0 | * | * | 21 | 0 | * | * |
| Missense | 283 | 3 (1.1%) | 0.66 (0.21–2.11) | 0.488 | 60 | 4 (7%) | 1.08 (0.37–3.17) | 0.886 |
| Nonsense | 679 | 9 (1.3%) | 0.68 (0.34–1.36) | 0.271 | 591 | 32 (5.4%) | 0.94 (0.64–1.39) | 0.740 |
| Splicing | 306 | 5 (1.6%) | 0.94 (0.39–2.30) | 0.896 | 282 | 15 (5.3%) | 1.00 (0.57–1.76) | 0.994 |
Could not be estimated.
Discussion
Using a multinational data resource of ~6,500 men carrying a BRCA1/2 PSV, we identified 2 regions in BRCA2 (c.756-c 1000 and c.7914+) that were associated with increased risk of PCa diagnosis and of Gleason 8+ PCa. These data suggest that PSV-specific PCA-risks exist for BRCA2 PSV carriers. This observation is consistent with earlier studies reporting a PSV-specific increase in PCa risk among BRCA1/2 PSV carriers(31,32). However, most studies that have made these observations have estimated the prevalence of BRCA1/2 mutations in PCa cases. Few studies have evaluated PCa incidence in mutation BRCA1/2 carriers. Roed Nielsen et al. (32) reported an elevated PCa relative risk in BRCA2 mutation carriers whose mutations fell in c.6373-c.6492 with a relative risk of 3.7 for mutations within this region compared with mutations outside this region. This elevated relative risk was not observed in the larger current analysis, which included the carriers reported by Roed Nielsen. We also demonstrated a remarkable similarity between PSVs conferring increased PCa risk and those associated with increased breast cancer risk in female BRCA2 PSV carriers(24).
BRCA2 is among the few known clinically relevant loci, in which many deleterious variants cause a highly penetrant PCa predisposition(50). Our work addressed the hypothesis that germline PSVs in BRCA1/2 that influence development of overall PCa and PCa severity demonstrate nonrandom distribution by location and/or function of the gene. Since PCa patients with Gleason 8+ disease are far more likely than men with Gleason <8 PCa to have unfavorable clinical outcome(2,11–18), the observation that PCCR PSVs are associated with elevated Gleason score suggests that PCCR PSVs may be associated with poorer prognosis than other BRCA2 PSVs. However, this needs to be investigated in future studies. We observe an elevated risk of both Gleason 8+ and Gleason ≤7 cancers, although the magnitude of association for Gleason 8+ is higher than that for Gleason ≤7. Thus, it is possible that the PCCR reported here is associated with PCa in general, and not only with high grade PCa. This observation requires additional research to confirm. Additionally, knowledge of the importance of DNA damage repair suggests that the mechanism of prostate carcinogenesis is broadly modified by BRCA2-related pathways(23). The IMPACT trial reported that PSA screening may be more informative in detecting PCa in BRCA2 PSV carriers compared with non-carriers(51). Additional research is needed to evaluate whether the PCCR PSVs reported here also influence the results of different management strategies.
In addition to its co-location with a previously-identified breast cancer cluster region(24), PSVs in the PCCR (3’ of c.7914) are focused within two of the principal DNA binding domains of the OB1 (i.e., oligonucleotide/oligosaccharide-binding domain 1; amino acids 2670 – 2796) and OB2 (i.e., Tower ssDNA and dsDNA binding domain 2; amino acids 2831 – 2872). However, the present data set does not allow us to understand the mechanism that might explain why BRCA2 PCCR PSVs are associated with elevated PCa risks. Additional mechanistic research will be required to elucidate the biological basis for risk heterogeneity implied by the present results.
The most common PSV in the c.756-c.1000 region was the Icelandic and Finnish founder PSV, c.771_775del, which has long been known as a PCa predisposition PSV(52–54) and is associated with a rapid progression to fatal PCa(10). Thus, our results regarding the association of this founder PSV with PCa severity are consistent with this prior report. We were not able to infer if c.756-c.1000 is a second PCCR region, or if the observed effect is due solely to c.771_775del. We returned to the original data from participants with this PSV to identify any potential bias in ascertainment that may have influenced this result. Based on original records from the Icelandic clinics from which these men were ascertained, no individual was ascertained based on genetic testing of prostate cancer. The carriers of this PSV were identified through family studies of breast cancer, mainly by screening unselected breast cancer patients and then, if mutation positive, by screening their close relatives. There was no ascertainment preference for prostate cancer in Icelandic male carriers, and there was no instance of a BRCA2 carrier identified by testing prostate cancer cases (Aðalgeir Arason, Personal communication).
Our present results complement the growing body of knowledge that cancer susceptibility PSVs demonstrate clinically relevant genotype-phenotype relationships. PSV location within APC is associated with polyposis severity and prevalence of extracolonic features, such as desmoid fibromas(55). Similarly, genotype-phenotype relationships have been reported for (missense) PSVs in RET in multiple endocrine neoplasia type 2 (MEN2) and Familial Medullary Thyroid Carcinoma(30). These findings have shaped the Neuroendocrine Tumor Society consensus guidelines, which now suggest thyroidectomy before age five years for individuals with PSVs within these high-risk regions, providing insight into the structure and function of cancer susceptibility PSVs in these genes and guiding clinical risk assessment and management. Despite evidence of genotype-phenotype relationships at multiple loci, the characteristics and mechanistic influences on cancer risk are likely quite different for PVSs in APC, RET, BRCA1/2, and others.
In contrast to prior work that evaluated prevalence of PSVs in BRCA1/2 in various PCa case series, we have leveraged a large, international multicenter consortium study of BRCA1/2 PSV carriers, irrespective of PCa status. However, our analysis has some limitations. The CIMBA study uses a non-standardized recruitment strategy from multiple referral centers. Thus, our data may not represent either the full spectrum of PCa patients or BRCA1/2 PSV carriers in the general population. Similarly, we were not able to assess issues of survival bias in our data that may be related to cancer screening or treatment.
While the present study identifies potentially interesting PSV-specific PCa associations, there are limitations in the data and analysis that require future validation. We used two binning approaches to identify relevant regions of BRCA1/2 that could have different risk or penetrance effects on PCa based on our earlier research that undertook a similar analysis for breast and ovarian cancer(24). In that analysis, we determined that the combination of these two approaches were complementary and identified similar regions of interest. While this approach points toward genomic regions that may confer different PCa risks, a full understanding of the causes of the effects we report will require experimental and mechanistic studies to further define the boundaries of the relevant domains and to understand the underlying mechanisms that lead to the observations reported here. In addition, the choice of the reference bin in our analysis will affect estimates of the hazard ratios reported here. Thus, the present report focuses on the identification of genomic regions that may confer elevated PCa risks in BRCA2 mutation carriers, and the hazard ratio estimates presented here should be interpreted with caution and not used for clinical risk estimation purposes.
Studies in female PSV carriers using a study design similar to that used here applied analytical corrections to account for the possibility that affected individuals (particularly those affected at younger ages) are more likely to be sampled than unaffected individuals. Unlike prior breast and ovarian cancer studies in BRCA1/2 mutation carriers, the present sample did not ascertain specific PCa cases (e.g., those diagnosed at an early age). Our median age at diagnosis is 64 years, which is similar to that reported in other non-BRCA1/2 populations. Our case sample is substantially older than PCa cases ascertained for BRCA1/2 screening studies, which tend to have a large proportion of cases diagnosed before age 55 (56). Thus, while there is limited evidence that ascertainment of cases conferred a major bias to the present results, future research is required to determine the extent of bias in our relative risk estimates arising from these issues.
Finally, pathology review of prostate tumors was neither centralized or available for all cases. A relatively large proportion of Gleason score and tumor stage data were also missing from the present sample, since many cases were based on self-report only. Cases with missing tumor stage and grade were excluded from those analyses, so any differential reporting of tumor traits could have caused bias in those results.
The present study indicates that personalized PCa risk assessment may be a future option, as well as individualized clinical management based on the specific BRCA2 PSV status. Additional research is required to fully understand the implication of carrying specific BRCA2 PSVs. Further characterization of the relationship between these PSVs and various cancer outcomes might help direct the future use of DNA repair-directed treatments and radiation therapy in men carrying these PSVs.
Supplementary Material
Statement of Significance.
Aggressive prostate cancer risk in BRCA2 mutation carriers may vary according to the specific BRCA2 mutation inherited by the at-risk individual
Acknowledgements
Funding Support
ELB was supported by grants from the National Cancer Institute (5T32CA009001, P60-CA105641). The CIMBA data management and data analysis were supported by Cancer Research – UK grants C12292/A20861, C12292/A11174. ACA is a Cancer Research -UK Senior Cancer Research Fellow. GCT and ABS are NHMRC Research Fellows. iCOGS: the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer (CRN-87521), and the Ministry of Economic Development, Innovation and Export Trade (PSR-SIIRI-701), Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The PERSPECTIVE project was supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministry of Economy, Science and Innovation through Genome Québec, and The Quebec Breast Cancer Foundation. BCFR: UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. BFBOCC: Lithuania (BFBOCC-LT): Research Council of Lithuania grant SEN-18/2015. BIDMC: Breast Cancer Research Foundation. BMBSA: Cancer Association of South Africa (PI Elizabeth J. van Rensburg). CNIO: Spanish Ministry of Health PI16/00440 supported by FEDER funds, the Spanish Ministry of Economy and Competitiveness (MINECO) SAF2014-57680-R and the Spanish Research Network on Rare diseases (CIBERER). COH-CCGCRN: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number R25CA112486, and RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CONSIT: Associazione Italiana Ricerca sul Cancro (AIRC; IG2014 no.15547) to P. Radice. Italian Association for Cancer Research (AIRC; grant no.16933) to L. Ottini. Associazione Italiana Ricerca sul Cancro (AIRC; IG2015 no.16732) to P. Peterlongo. Associazione Italiana Ricerca sul Cancro (AIRC grant IG17734) to G. Giannini. DEMOKRITOS: European Union (European Social Fund - ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program of the General Secretariat for Research & Technology: SYN11_10_19 NBCA. Investing in knowledge society through the European Social Fund. DFKZ: German Cancer Research Center. EMBRACE: Cancer Research UK Grants C1287/A10118 and C1287/A11990. Fiona Lalloo is supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Elizabeth Bancroft id supported by Cancer Research UK Grant C5047/A8385FCCC: The University of Kansas Cancer Center (P30 CA168524) and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. was funded by R0 1CA140323, R01 CA214545, and by the Chancellors Distinguished Chair in Biomedical Sciences Professorship. FPGMX: FISPI05/2275 and Mutua Madrileña Foundation (FMMA). GC-HBOC: German Cancer Aid (grant no 110837, Rita K. Schmutzler) and the European Regional Development Fund and Free State of Saxony, Germany (LIFE - Leipzig Research Centre for Civilization Diseases, project numbers 713-241202, 713-241202, 14505/2470, 14575/2470). GEMO: Ligue Nationale Contre le Cancer; the Association “Le cancer du sein, parlons-en!” Award, the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program and the French National Institute of Cancer (INCa). GEORGETOWN: the Non-Therapeutic Subject Registry Shared Resource at Georgetown University (NIH/NCI grant P30-CA051008), the Fisher Center for Hereditary Cancer and Clinical Genomics Research, and Swing Fore the Cure. G-FAST: Bruce Poppe is a senior clinical investigator of FWO. Mattias Van Heetvelde obtained funding from IWT. HCSC: Spanish Ministry of Health PI15/00059, PI16/01292, and CB-161200301 CIBERONC from ISCIII (Spain), partially supported by European Regional Development FEDER funds. HEBCS: Helsinki University Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society and the Sigrid Juselius Foundation. HEBON: the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research grant NWO 91109024, the Pink Ribbon grants 110005 and 2014-187.WO76, the BBMRI grant NWO 184.021.007/CP46 and the Transcan grant JTC 2012 Cancer 12-054. HEBON thanks the registration teams of Dutch Cancer Registry (IKNL; S. Siesling, J. Verloop) and the Dutch Pathology database (PALGA; L. Overbeek) for part of the data collection. HRBCP: Hong Kong Sanatorium and Hospital, Dr Ellen Li Charitable Foundation, The Kerry Group Kuok Foundation, National Institute of Health1R 03CA130065, and North California Cancer Center. HUNBOCS: Hungarian Research Grants KTIA-OTKA CK-80745 and OTKA K-112228. ICO: The authors would like to particularly acknowledge the support of the Asociación Española Contra el Cáncer (AECC), the Instituto de Salud Carlos III (organismo adscrito al Ministerio de Economía y Competitividad) and “Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa” (PI10/01422, PI13/00285, PIE13/00022, PI15/00854, PI16/00563 and CIBERONC) and the Institut Català de la Salut and Autonomous Government of Catalonia (2009SGR290, 2014SGR338 and PERIS Project MedPerCan). IHCC: PBZ_KBN_122/P05/2004. ILUH: Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. INHERIT: Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program - grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade - grant # PSR-SIIRI-701. IOVHBOCS: Ministero della Salute and “5×1000” Istituto Oncologico Veneto grant. IPOBCS: Liga Portuguesa Contra o Cancro. kConFab: The National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. MAYO: NIH grants CA116167, CA192393 and CA176785, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201),and a grant from the Breast Cancer Research Foundation. MCGILL: Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. MSKCC: the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Cancer Genetics Initiative, the Andrew Sabin Research Fund and a Cancer Center Support Grant/Core Grant (P30 CA008748). NAROD: 1R01 CA149429-01. NCI: the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts NO2-CP-11019-50, N02-CP-21013-63 and N02-CP-65504 with Westat, Inc, Rockville, MD. NICCC: Clalit Health Services in Israel, the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), NY. NNPIO: the Russian Federation for Basic Research (grants 15-04-01744, 16-54-00055 and 17-54-12007). NRG Oncology: U10 CA180868, NRG SDMC grant U10 CA180822, NRG Administrative Office and the NRG Tissue Bank (CA 27469), the NRG Statistical and Data Center (CA 37517) and the Intramural Research Program, NCI. OSUCCG: Ohio State University Comprehensive Cancer Center. PBCS: Italian Association of Cancer Research (AIRC) [IG 2013 N.14477] and Tuscany Institute for Tumors (ITT) grant 2014-2015-2016. SEABASS: Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation. SMC: the Israeli Cancer Association. SWE-BRCA: the Swedish Cancer Society. UCHICAGO: NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women’s Cancer Research Alliance and the Breast Cancer research Foundation. OIO is an ACS Clinical Research Professor. UCLA: Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. UCSF: UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. UKFOCR: Cancer Researc h UK. UPENN: Breast Cancer Research Foundation; Susan G. Komen Foundation for the cure, Basser Center for BRCA. UPITT/MWH: Hackers for Hope Pittsburgh. VFCTG: Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. WCP: Dr Karlan is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124. S. Gutiérrez-Enríquez is supported by the Miguel Servet Progam (CP10/00617).
All the families and clinicians who contribute to the studies; Sue Healey, in particular taking on the task of PSV classification with the late Olga Sinilnikova; Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis; members and participants in the New York site of the Breast Cancer Family Registry; members and participants in the Ontario Familial Breast Cancer Registry; Vilius Rudaitis and Laimonas Griškevičius; Drs Janis Eglitis, Anna Krilova and Aivars Stengrevics; Rosario Alonso and Guillermo Pita; Milena Mariani, Daniela Zaffaroni, Monica Barile, Irene Feroce, Riccardo Dolcetti, Laura Papi, Gabriele Lorenzo Capone, Viviana Gismondi, Daniela Furlan, Antonella Savarese, Aline Martayan, Brunella Pilato; the personnel of the Cogentech Cancer Genetic Test Laboratory, Milan, Italy. Ms. JoEllen Weaver and Dr. Betsy Bove; Marta Santamariña, Ana Blanco, Miguel Aguado, Uxía Esperón and Belinda Rodríguez; IFE - Leipzig Research Centre for Civilization Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter, Ronny Baber); We thank all participants, clinicians, family doctors, researchers, and technicians for their contributions and commitment to the DKFZ study and the collaborating groups in Lahore, Pakistan (Muhammad U. Rashid, Noor Muhammad, Sidra Gull, Seerat Bajwa, Faiz Ali Khan, Humaira Naeemi, Saima Faisal, Asif Loya, Mohammed Aasim Yusuf) and Bogota, Colombia (Ignacio Briceno, Fabian Gil). Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) study is a study from the National Cancer Genetics Network UNICANCER Genetic Group, France. We wish to pay a tribute to Olga M. Sinilnikova, who initiated and coordinated GEMO until she sadly passed away on the 30th June 2014. The team in Lyon (Olga Sinilnikova, Mélanie Léoné, Laure Barjhoux, Carole Verny-Pierre, Sylvie Mazoyer, Francesca Damiola, Valérie Sornin) managed the GEMO samples until the biological resource centre was transferred to Paris in December 2015 (. We want to thank all the GEMO collaborating groups for their contribution to this study: Coordinating Centre, Service de Génétique, Institut Curie, Paris, France: Ophélie Bertrand, Anne-Marie Birot, Sandrine Caputo, Anaïs Dupré, Emmanuelle Fourme, Lisa Golmard, Claude Houdayer, Marine Le Mentec, Virginie Moncoutier, Antoine de Pauw, Dominique yen yand Inserm U900, Institut Curie, Paris, France: Fabienne Lesueur, Noura Mebirouk.Contributing Centres : Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon - Centre Léon Bérard, Lyon, France: Nadia Boutry-Kryza, Alain Calender, Sophie Giraud, Mélanie Léone. Institut Gustave Roussy, Villejuif, France: Brigitte Bressac-de-Paillerets, Olivier Caron, Marine Guillaud-Bataille. Centre Jean Perrin, Clermont-Ferrand, France: Yves-Jean Bignon, Nancy Uhrhammer. Centre François Baclesse, Caen, France: Pascaline Berthet, Laurent Castera, Dominique Vaur. Institut Paoli Calmettes, Marseille, France: Violaine Bourdon, Catherine Noguès, Tetsuro Noguchi, Cornel Popovici, Audrey Remenieras, Hagay Sobol. CHU Arnaud-de-Villeneuve, Montpellier, France: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille, France: Claude Adenis, Aurélie Dumont, Françoise Révillion. Centre Paul Strauss, Strasbourg, France: Danièle Muller. Institut Bergonié, Bordeaux, France: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Michel Longy, Nicolas Sevenet, Institut Claudius Regaud, Toulouse, France: Laurence Gladieff, Rosine Guimbaud, Viviane Feillel, Christine Toulas. CHU Grenoble, France: Hélène Dreyfus, Christine Dominique Leroux, Magalie Peysselon, Rebischung. CHU Dijon, France: Amandine Baurand, Geoffrey Bertolone, Fanny Coron, Caroline Jacquot, Sarab Lizard. CHU St-Etienne, France: Caroline Kientz, Marine Lebrun, Fabienne Prieur. Hôtel Dieu Centre Hospitalier, Chambéry, France: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice, France: Véronique Mari. CHU Limoges, France: Laurence Vénat-Bouvet. CHU Nantes, France: Stéphane Bézieau, Capucine Delnatte. CHU Bretonneau, Tours and Centre Hospitalier de Bourges France: Isabelle Mortemousque. Groupe Hospitalier Pitié-Salpétrière, Paris, France: Chrystelle Colas, Florence Coulet, Florent Soubrier, Mathilde Warcoin. CHU Vandoeuvre-les-Nancy, France: Myriam Bronner, Johanna Sokolowska. CHU Besançon, France: Marie-Agnès Collonge-Rame, Alexandre Damette. CHU Poitiers, Centre Hospitalier d’Angoulême and Centre Hospitalier de Niort, France: Hakima Lallaoui. CHU Nîmes Carémeau, France : Jean Chiesa. CHI Poissy, France: Denise Molina-Gomes. CHU Angers, France : Olivier Ingster; Ilse Coene en Brecht Crombez; Ilse Coene and Brecht Crombez; Alicia Tosar and Paula Diaque; Sofia Khan, Taru A. Muranen, Carl Blomqvist, Irja Erkkilä and Virpi Palola; The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) consists of the following Collaborating Centers: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: F.E. van Leeuwen, S. Verhoef, M.K. Schmidt, N.S. Russell, D.J. Jenner; Erasmus Medical Center, Rotterdam, NL: J.M. Collée, A.M.W. van den Ouweland, M.J. Hooning, C.H.M. van Deurzen, I.M. Obdeijn; Leiden University Medical Center, NL: J.T. Wijnen, R.A.E.M. Tollenaar, P. Devilee, T.C.T.E.F. van Cronenburg; Radboud University Nijmegen Medical Center, NL: C.M. Kets; University Medical Center Utrecht, NL: M.G.E.M. Ausems, C.C. van der Pol; Amsterdam Medical Center, NL: C.M. Aalfs, T.A.M. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz,;; University Medical Center Groningen, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits, G.H. de Bock; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen; The Netherlands Comprehensive Cancer Organization (IKNL): S. Siesling, J.Verloop; The Dutch Pathology Registry (PALGA): L.I.H. Overbeek; Hong Kong Sanatorium and Hospital; the Hungarian Breast and Ovarian Cancer Study Group members (Janos Papp, Aniko Bozsik, Timea Pocza, Zoltan Matrai, Miklos Kasler, Judit Franko, Maria Balogh, Gabriella Domokos, Judit Ferenczi, Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary) and the clinicians and patients for their contributions to this study; the Oncogenetics Group (VHIO) and the High Risk and Cancer Prevention Unit of the University Hospital Vall d’Hebron, and the Cellex Foundation for providing research facilities and equipment; the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella; the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella; Dr Martine Dumont for sample management and skillful assistance; Pedro Pinto; members of the Center of Molecular Diagnosis, Oncogenetics Department and Molecular Oncology Research Center of Barretos Cancer Hospital; Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab; the KOBRA Study Group; Csilla Szabo (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA Eva Machackova (Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute and MF MU, Brno, Czech Republic); and Michal Zikan, Petr Pohlreich and Zdenek Kleibl (Oncogynecologic Center and Department of Biochemistry and Experimental Oncology, First Faculty of Medicine, Charles University, Prague, Czech Republic); Anne Lincoln, Lauren Jacobs; the NICCC National Familial Cancer Consultation Service team led by Sara Dishon, the lab team led by Dr. Flavio Lejbkowicz, and the research field operations team led by Dr. Mila Pinchev; the investigators of the Australia New Zealand NRG Oncology group; members and participants in the Ontario Cancer Genetics Network; Kevin Sweet, Caroline Craven, Julia Cooper, and Michelle O’Conor; Yip Cheng Har, Nur Aishah Mohd Taib, Phuah Sze Yee, Norhashimah Hassan and all the research nurses, research assistants and doctors involved in the MyBrCa Study for assistance in patient recruitment, data collection and sample preparation, Philip Iau, Sng Jen-Hwei and Sharifah Nor Akmal for contributing samples from the Singapore Breast Cancer Study and the HUKM-HKL Study respectively; the Meirav Comprehensive breast cancer center team at the Sheba Medical Center; Christina Selkirk; Håkan Olsson, Helena Jernström, Karin Henriksson, Katja Harbst, Maria Soller, Ulf Kristoffersson; from Gothenburg Sahlgrenska University Hospital: Anna Öfverholm, Margareta Nordling, Per Karlsson, Zakaria Einbeigi; from Stockholm and Karolinska University Hospital: Gisela Barbany Bustinza; from Umeå University Hospital: Beatrice Melin, Christina Edwinsdotter Ardnor, Monica Emanuelsson; from Uppsala University: Maritta Hellström Pigg, Richard Rosenquist; from Linköping University Hospital: Marie Stenmark-Askmalm, Sigrun Liedgren; Cecilia Zvocec, Qun Niu; Joyce Seldon and Lorna Kwan; Dr. Robert Nussbaum, Beth Crawford, Kate Loranger, Julie Mak, Nicola Stewart, Robin Lee, Amie Blanco and Peggy Conrad and Salina Chan; Simon Gayther, Susan Ramus, Paul Pharoah, Carole Pye, Patricia Harrington and Eva Wozniak; Geoffrey Lindeman, Marion Harris, Martin Delatycki, Sarah Sawyer, Rebecca Driessen, and Ella Thompson for performing all DNA amplification.
Footnotes
Financial support and conflict of interest statements are included at the end of the manuscript.
References
- 1.Giusti RM, Rutter JL, Duray PH, Freedman LS, Konichezky M, Fisher-Fischbein J, et al. A twofold increase in BRCA mutation related prostate cancer among Ashkenazi Israelis is not associated with distinctive histopathology. J Med Genet 2003;40:787–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallagher DJ, Gaudet MM, Pal P, Kirchhoff T, Balistreri L, Vora K, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 2010;16:2115–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchhoff T, Kauff ND, Mitra N, Nafa K, Huang H, Palmer C, et al. BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin Cancer Res 2004;10:2918–21 [DOI] [PubMed] [Google Scholar]
- 4.Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control 2010;21:289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agalliu I, Gern R, Leanza S, Burk RD. Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder mutations. Clin Cancer Res 2009;15:1112–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agalliu I, Karlins E, Kwon EM, Iwasaki LM, Diamond A, Ostrander EA, et al. Rare germline mutations in the BRCA2 gene are associated with early-onset prostate cancer. Br J Cancer 2007;97:826–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agalliu I, Kwon EM, Zadory D, McIntosh L, Thompson J, Stanford JL, et al. Germline mutations in the BRCA2 gene and susceptibility to hereditary prostate cancer. Clin Cancer Res 2007;13:839–43 [DOI] [PubMed] [Google Scholar]
- 8.Eerola H, Heikkila P, Tamminen A, Aittomaki K, Blomqvist C, Nevanlinna H. Histopathological features of breast tumours in BRCA1, BRCA2 and mutation-negative breast cancer families. Breast Cancer Res 2005;7:R93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cybulski C, Wokołorczyk D, Kluźniak W, Kashyap A, Gołąb A, Słojewski M, et al. A personalised approach to prostate cancer screening based on genotyping of risk founder alleles. Br J Cancer 2013;108:2601–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tryggvadottir L, Vidarsdottir L, Thorgeirsson T, Jonasson JG, Olafsdottir EJ, Olafsdottir GH, et al. Prostate cancer progression and survival in BRCA2 mutation carriers. J Natl Cancer Inst 2007;99:929–35 [DOI] [PubMed] [Google Scholar]
- 11.Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer 2011;105:1230–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, et al. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer 2012;106:1697–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA Mutations Are Associated With Higher Risk of Nodal Involvement, Distant Metastasis, and Poor Survival Outcomes in Prostate Cancer. J Clin Oncol 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorne H, Willems AJ, Niedermayr E, Hoh IM, Li J, Clouston D, et al. Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev Res (Phila) 2011;4:1002–10 [DOI] [PubMed] [Google Scholar]
- 15.Gleicher S, Kauffman EC, Kotula L, Bratslavsky G, Vourganti S. Implications of High Rates of Metastatic Prostate Cancer in BRCA2 Mutation Carriers. Prostate 2016;76:1135–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson D, Easton D. Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet 2001;68:410–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker R, Louis A, Berlin A, Horsburgh S, Bristow RG, Trachtenberg J. Prostate cancer screening characteristics in men with BRCA1/2 mutations attending a high-risk prevention clinic. Can Urol Assoc J 2014;8:E783–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier C, Herkommer K, Luedeke M, Rinckleb A, Schrader M, Vogel W. Subgroups of familial and aggressive prostate cancer with considerable frequencies of BRCA2 mutations. Prostate 2014;74:1444–51 [DOI] [PubMed] [Google Scholar]
- 19.Laitman Y, Keinan Boker L, Liphsitz I, Weissglas-Volkov D, Litz-Philipsborn S, Schayek H, et al. Cancer risks in Jewish male BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat 2015;150:631–5 [DOI] [PubMed] [Google Scholar]
- 20.Streff H, Profato J, Ye Y, Nebgen D, Peterson SK, Singletary C, et al. Cancer Incidence in First- and Second-Degree Relatives of BRCA1 and BRCA2 Mutation Carriers. Oncologist 2016;21:869–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor RA, Fraser M, Livingstone J, Espiritu SM, Thorne H, Huang V, et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun 2017;8:13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015;313:1347–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, et al. Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nature Genetics 1995;11:428–33 [DOI] [PubMed] [Google Scholar]
- 26.Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, et al. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet 1997;15:103–5 [DOI] [PubMed] [Google Scholar]
- 27.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017;317:2402–16 [DOI] [PubMed] [Google Scholar]
- 28.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, et al. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A 2000;97:3352–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci U S A 2007;104:4036–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krampitz GW, Norton JA. RET gene mutations (genotype and phenotype) of multiple endocrine neoplasia type 2 and familial medullary thyroid carcinoma. Cancer 2014;120:1920–31 [DOI] [PubMed] [Google Scholar]
- 31.Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J Clin Oncol 2004;22:735–42 [DOI] [PubMed] [Google Scholar]
- 32.Roed Nielsen H, Petersen J, Therkildsen C, Skytte AB, Nilbert M. Increased risk of male cancer and identification of a potential prostate cancer cluster region in BRCA2. Acta Oncol 2016;55:38–44 [DOI] [PubMed] [Google Scholar]
- 33.Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 2007;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claes K, Vandesompele J, Poppe B, Dahan K, Coene I, De Paepe A, et al. Pathological splice mutations outside the invariant AG/GT splice sites of BRCA1 exon 5 increase alternative transcript levels in the 5’ end of the BRCA1 gene. Oncogene 2002;21:4171–5 [DOI] [PubMed] [Google Scholar]
- 35.Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet 2004;75:535–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein JL, Teraoka S, Southey MC, Jenkins MA, Andrulis IL, Knight JA, et al. Population-based estimates of breast cancer risks associated with ATM gene variants c.7271T>G and c.1066–6T>G (IVS10–6T>G) from the Breast Cancer Family Registry. Hum Mutat 2006;27:1122–8 [DOI] [PubMed] [Google Scholar]
- 37.PFam v.31.0. 2017.
- 38.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Human Molecular Genetics 2002;11:2805–14 [DOI] [PubMed] [Google Scholar]
- 39.Mikaelsdottir EK, Valgeirsdottir S, Eyfjord JE, Rafnar T. The Icelandic founder mutation BRCA2 999del5: analysis of expression. Breast Cancer Res 2004;6:R284–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buisson M, Anczukow O, Zetoune AB, Ware MD, Mazoyer S. The 185delAG mutation (c.68_69delAG) in the BRCA1 gene triggers translation reinitiation at a downstream AUG codon. Hum Mutat 2006;27:1024–9 [DOI] [PubMed] [Google Scholar]
- 41.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 2001;8:833–7 [DOI] [PubMed] [Google Scholar]
- 42.Bienstock RJ, Darden T, Wiseman R, Pedersen L, Barrett JC. Molecular modeling of the amino-terminal zinc ring domain of BRCA1. Cancer Res 1996;56:2539–45 [PubMed] [Google Scholar]
- 43.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 1996;14:430–40 [DOI] [PubMed] [Google Scholar]
- 44.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 1997;11:68–76 [PubMed] [Google Scholar]
- 45.Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol 2001;8:838–42 [DOI] [PubMed] [Google Scholar]
- 46.Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res 2000;460:319–32 [DOI] [PubMed] [Google Scholar]
- 47.Palacios IM. Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genomics 2013;12:25–36 [DOI] [PubMed] [Google Scholar]
- 48.Lin DY, Wei LJ. The robust inference for the Cox proportional hazards model. JASA 1989;84:1074–8 [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 50.Giri VN, Knudsen KE, Kelly WK, Abida W, Andriole GL, Bangma CH, et al. Role of Genetic Testing for Inherited Prostate Cancer Risk: Philadelphia Prostate Cancer Consensus Conference 2017. J Clin Oncol 2017:JCO2017741173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bancroft EK, Page EC, Castro E, Lilja H, Vickers A, Sjoberg D, et al. Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol 2014;66:489–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigurdsson S, Thorlacius S, Tomasson J, Tryggvadottir L, Benediktsdottir K, Eyfjord JE, et al. BRCA2 mutation in Icelandic prostate cancer patients. Journal of Molecular Medicine 1997;75:758–61 [DOI] [PubMed] [Google Scholar]
- 53.Gudmundsson J, Johannesdottir G, Bergthorsson JT, Arason A, Ingvarsson S, Egilsson V, et al. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res 1995;55:4830–2 [PubMed] [Google Scholar]
- 54.Barkardottir RB, Sarantaus L, Arason A, Vehmanen P, Bendahl PO, Kainu T, et al. Haplotype analysis in Icelandic and Finnish BRCA2 999del5 breast cancer families. Eur J Hum Genet 2001;9:773–9 [DOI] [PubMed] [Google Scholar]
- 55.Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol 2007;61:153–61 [DOI] [PubMed] [Google Scholar]
- 56.Cavanagh H, Rogers KM. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hered Cancer Clin Pract 2015;13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.