Summary
Many behaviors that are critical for survival and reproduction are expressed over extended time periods. The ability to inexpensively record and store large volumes of video data creates new opportunities to understand the biological basis of these behaviors and simultaneously creates a need for tools that can automatically quantify behaviors from large video datasets. Here, we demonstrate that 3D Residual Networks can be used to classify an array of complex behaviors in Lake Malawi cichlid fishes. We first apply pixel-based hidden Markov modeling combined with density-based spatiotemporal clustering to identify sand disturbance events. After this, a 3D ResNet, trained on 11,000 manually annotated video clips, accurately (>76%) classifies the sand disturbance events into 10 fish behavior categories, distinguishing between spitting, scooping, fin swipes, and spawning. Furthermore, animal intent can be determined from these clips, as spits and scoops performed during bower construction are classified independently from those during feeding.
Subject Areas: Piscine Behavior, Zoology, Computer Science, Artificial Intelligence
Graphical Abstract
Highlights
-
•
A dataset of more than 14,000 annotated animal behavior videos was created
-
•
3D residual networks can be used to classify animal behavior
-
•
Different intents of similar behavioral actions can be distinguished
-
•
A working solution to study long-term behaviors was established
Piscine Behavior; Zoology; Computer Science; Artificial Intelligence
Introduction
Animals respond to and interact with their environment using a rich repertoire of behavioral actions. A major challenge for neuroscientists is to understand how neural circuits coordinate these behaviors in response to sensory and internal stimuli. Automated identification and classification of behavioral actions will aid in this task, as most neural responses are stochastic, requiring a large number of replicates to accurately estimate relationships with complex behaviors (Egnor and Branson, 2016). In addition, many behaviors are executed over long timescales through the accumulated actions of thousands of individual decisions (e.g., foraging, construction, and social behaviors), making manual analysis of the full course of behavior entirely impractical (Russell et al., 2017; Feng et al., 2015; Mouritsen, 2018; Tucker, 1981).
One common approach is to manually observe snapshots of long-term behaviors over extended periods of time. This method is labor intensive and thus can severely limit the total number of animals that can be measured in a single study. Furthermore, it cannot provide a complete and detailed quantitative description of the full behavioral trajectory. An alternative approach is to design abbreviated assays that elicit the behavior of interest during a short period of time. One issue, however, is that many natural behaviors may be expressed differently over short timescales, and/or in unnatural/unfamiliar environments. Recently, deep learning approaches have revolutionized our ability to automatically analyze video and image data (Nath et al., 2019; Mathis et al., 2018; Weissbrod et al., 2013; Wild et al., 2018). Convolutional neural networks (CNNs) can be applied to images for the purpose of object detection, identifying all animals within an individual frame (Girshick, 2015; Ren et al., 2015; Redmon et al., 2016; Pereira et al., 2019). CNNs can also identify body parts, such as eyes, legs, or wings, allowing for the determination of an animal's posture at any specific time (Graving et al., 2019; Pereira et al., 2019; Kain et al., 2013; Petrou and Webb, 2012; Gunel et al., 2019; Andriluka et al., 2018; Nath et al., 2019). Although position and pose alone are not sufficient for defining animal behavior, postural time series can be used to define some behavioral actions. Such analysis has, for example, been used to quantitatively describe different types of stereotyped movements in Drosophila flies, such as different locomotor and grooming behaviors (Berman et al., 2014). Similar approaches have also been used successfully on other species (Stephens et al., 2008).
It remains challenging, however, to translate changes in position and posture into complex behaviors that are characterized by an animal's interaction with the environment. For example, many goal-directed behaviors involve significant manipulation of the physical environment in ways that are essential for survival or reproductive success, such as mice digging burrows, birds building nests, spiders weaving webs, and bowerbirds or cichlid fishes constructing courtship bowers (Hansell, 2000; Benjamin and Zschokke, 2000; Vollrath, 1992; Collias and Collias, 2014; Dawkins, 1982; McKaye et al., 2001). In such cases, information about changes to the physical environment itself is essential to fully describe the behavior.
One possible solution for analysis of these types of behaviors is to train a deep learning network that takes in videos as input and then outputs a prediction for the corresponding behavior type. For example, 3D Residual Networks (3D ResNets) have been successfully used to classify human behaviors, distinguishing between hundreds of different action classes (Hara et al., 2018; Qiu et al., 2017). These deep learning networks use 3D kernels with the ability to extract spatiotemporal features directly from videos. Videos are fed into these networks raw, without any individual body posture information beyond what can be learned from the training data. One major benefit of these 3D networks, when compared with networks that process each frame individually, is that they can integrate spatial and temporal information to recognize changes in the animal's environment that might indicate a particular behavior (e.g., digging, feeding, or construction behaviors). However, a significant challenge in applying action recognition to large videos is detecting actions of interest in a time and space frame, which is known as shot transition detection. This requires splitting large (e.g. 10-h-long) videos into small enough temporal units such that each unit contains only one action of interest and excludes as much irrelevant information as possible.
In this article, we describe an approach for analyzing construction, feeding, and mating behaviors from hundreds of hours of videos of Lake Malawi cichlids behaving freely in naturalistic and social home tank environments. Lake Malawi is the most species-rich freshwater lake on the Earth, and it contains 700–1,000 cichlid species that have rapidly evolved in the past 1–2 million years (Brawand et al., 2014). Approximately 200 of these species express long-term bower construction behaviors, in which males manipulate sand to create large courtship structures, or bowers, to attract female mates (York et al., 2015) (Figure 1A). Males construct pits and castles over the course of many days and make thousands of decisions about where to scoop up and then spit out mouthfuls of sand. Bower construction is therefore a useful model for understanding how goal-directed behaviors are executed over long time periods in environments that are physically and socially dynamic.
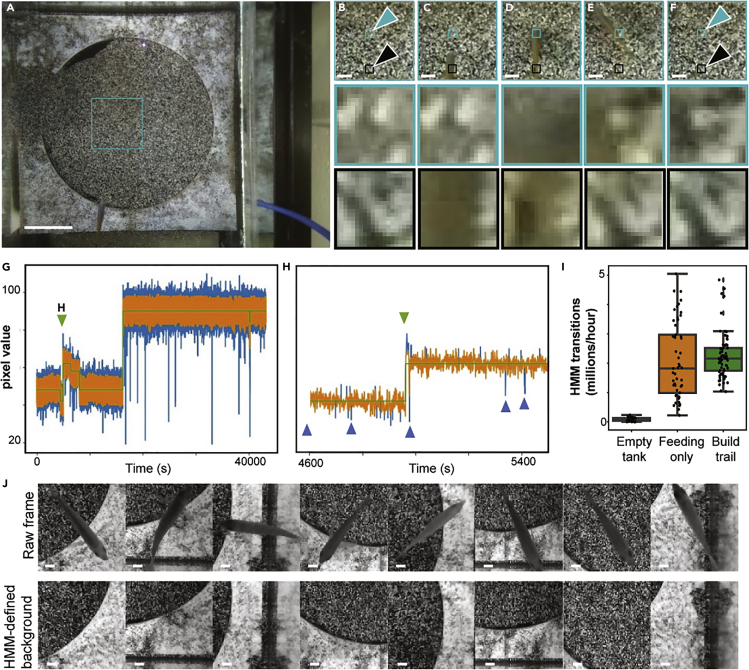
Figure 1.
Measurement of Lake Malawi Cichlid Bower Behaviors in Laboratory Aquariums
(A–C) Approximately 200 species of Lake Malawi cichlids exhibit bower behaviors. In these species, sociosexual cues trigger reproductive adult males to construct large courtship structures by manipulating sand with their mouths. The geometric structure of the bower is species-specific. (A) Example of a castle structure built in Lake Malawi. (B) Example of a castle structure built in a standard aquatics facility aquarium. (C) Top down view of acrylic tray used to constrain bower building to a third of the tank. Video recordings using this view were used to characterize bower building behaviors throughout this article. Scale bar, 10 cm. Photo credit to Dr. Isabel Magalhaes (A).
See also Figure S1.
To measure bower construction, we first develop an action detection algorithm that uses hidden Markov models (HMMs) and density-based spatial clustering to identify regions of the video where the fish has manipulated sand using its mouth, fins, or other parts of its body. Then, after generating small video clips that encompass these events, we use a 3D ResNet to classify each sand disturbance event into one of ten action categories. We demonstrate that this approach can be used to quickly, accurately, and automatically identify hundreds of thousands of behaviors across hundreds of hours of video. Through this approach, we measure the times and locations of construction, mating, and feeding behaviors expressed over the course of many days in three different species and one hybrid cross.
Results
Collection of Video Recordings of Male Cichlid Fish Constructing Bowers
If provided access to an appropriate type of sand and gravid females, Lake Malawi male cichlids will typically construct species-typical bower structures in standard aquarium tanks within 1–2 weeks (York et al., 2015) (Figure 1B). After testing sand from different suppliers, we found that males construct most vigorously with a sand mixture composed of black and white grains. Only half of each home tank was accessible for top-down video recording, due to physical constraints such as support beams and water supply/drain lines. To restrict sand manipulation behaviors to the video-accessible region of each tank, we placed a custom-built acrylic tray containing sand directly within the video camera field of view. We then introduced a single male individual to four female individuals (Figure 1C). For each trial, we collected 10 h of video daily for approximately 10 consecutive days, resulting in the collection of ∼100 h of video per trial. We analyzed two types of trials: construction trials, in which males constructed bowers in the presence of four females, and control trials, in which tanks were empty or tanks contained four females but no male. We analyzed eight construction trials, encompassing three different species and one F1 hybrid cross: pit-digger Copadichromis virginalis, CV, n = 1; castle-builder Mchenga conophoros, MC, n = 3; pit-digger Tramitichromis intermedius, TI, n = 2; and pit-castle hybrid MCxCV F1, n = 2. Visual inspection confirmed that each male built a species-typical bower. We also analyzed three empty tank control trials, and five female-only control trials. Table 1 summarizes all trials that were analyzed in this article.
Table 1.
Summary of Video Recordings Used in This Report
| Species | Type | n | Bower Shape | Training Set | Description |
|---|---|---|---|---|---|
| – | Empty | 3 | – | No | No fish in tank |
| CV | Feeding only | 2 | – | No | Four female fish |
| MC | Feeding only | 2 | – | No | Four female fish |
| TI | Feeding only | 1 | – | No | Four female fish |
| CV | Building | 1 | Pit | Yes | One male and four female fish |
| MC | Building | 3 | Castle | Yesa | One male and four female fish |
| TI | Building | 2 | Pit | Yes | One male and four female fish |
| MC/CV F1 | Building | 2 | Pit/castleb | Yes | One male and four female fish |
CV, Copachromis virganialis; MC, Mchenga conophoros; TI, Tramitichromis intermedius
Only two of the three MC trials was manually labeled for training.
F1 hybrids display codominant phenotype. Males initially build a pit structure and then transition to build a castle structure nearby the original pit.
To initially analyze these videos, we first focused on a single MC trial, identifying 3 days when castle building occurred. A trained observer manually annotated all spit and scoop events for a single hour on each day. In total, 1,104 spit events and 1,575 scoop events were observed (there is not a 1:1 correspondence between spit and scoop events because males sometimes scoop sand from multiple locations before performing a single spit) (Figure S1). Analysis of these 3 h of videos required 36 h of human time, or a 12:1 ratio of human time to video time. Full analysis of a single trial would require 1,200 h of human time, or approximately 30 weeks of full-time work dedicated to manual annotation. Automated analysis of these videos was thus necessary for the full characterization of bower construction even in this limited set of trials, let alone any larger-scale investigation of bower behavior.
Automatic Identification of Sand Disturbance Events
In the process of manually annotating these videos, we noticed that spit and scoop events resulted in an enduring and visually apparent spatial rearrangement of the black and white grains of sand. Scoop and spit events were associated with a permanent transition between pixel values. To test whether events could be identified despite the frequent occurrences of fish swimming over the sand, we inspected local regions in which a large number of pixels underwent permanent transitions. We observed temporary changes in pixel values when fish or shadows occluded the sand and permanent changes in pixel values at the locations of sand disturbance events (Figures 2A–2F). Consistent with this, we plotted the grayscale value of every pixel over entire 10-h videos and found that individual pixel values were, in general, fixed around a specific mean value for extended periods of time, but showed small oscillations and large but temporary deviations about this value across video frames (Figures 2G and 2H). In addition, we identified permanent transitions in pixel value, in which the mean grayscale value of a single pixel would change to, and retain, a new value.
Figure 2.
Automated Detection of Sand Change from Video Data
(A−H) The sand in this behavioral paradigm is composed of black and white grains (e.g., as seen in A), and therefore sand manipulation events during bower construction cause permanent rearrangement of the black and white grains at specific locations. We aimed to detect these events by processing whole video frames (A, with turquoise box indicating an example region of interest. Scale bar, 10 cm) sampled once per second and tracking the values of individual pixels throughout whole trials. Fish swimming over sand cause transient changes in pixel values (e.g., B−F, black arrows indicate an example location of a fish swimming over the sand; the bottom row depicts a zoomed in 20 × 20-pixel view of a location that the fish swims over, sampled from representative frames across 4 s). In contrast, sand manipulation behaviors cause enduring changes in pixel values (e.g., B−F, turquoise arrows indicate an example location of a fish scooping sand. Scale bar, 2cm; the middle row contains a zoomed in 20 × 20-pixel view of a location where the fish scoops sand). We used a custom hidden Markov model to identify all enduring state changes for each pixel throughout entire videos (G, green line indicates HMM-predicted state, orange line indicates raw grayscale pixel value, and blue lines indicate transient fluctuations beyond the pixel's typical range of values likely caused by fish swimming or shadows). Because fish swim over the sand frequently, a large number of transient changes are ignored (e.g., pixel value fluctuations indicated by blue arrows in H), while enduring changes are identified (e.g., pixel value change indicated by green arrow in (H).
(I) Number of HMM transitions identified per hour based on trial type. “Feeding only” are trials containing four females. “Build trial” contains four females and one male that builds a bower. The boxplot shows quartiles of the dataset while the whiskers show the rest of the distribution unless the point is an outlier.
(J) The HMM could be used to calculate a background image at a given time point, resulting in removal of the fish and the associated shadow from the image. Scale bar, 1cm.
To automatically identify permanent transitions in pixel color, we used an HMM to calculate a hidden state for each pixel that represented its current color. In general, the HMM partitioned each pixel into a small set of values (∼10–50) over the course of each 10-h video (Figures 2G and 2H). In testing the speed and accuracy of the HMM on raw data, we observed that large temporary deviations greatly impacted the accuracy of the HMM calls, potentially due to their violation of the assumption of a Gaussian distribution. Therefore, these large deviations were excluded using a rolling mean filter. We also found a speed/resolution trade-off to the frame rate. As we wanted to perform this analysis for the entire 1,296 × 972-pixel video, we needed to analyze over 1 million pixels through time. We found that 1 frame per second, sampled from the 30 frames per second in the raw video, was a reasonable trade-off—an entire video could be analyzed in approximately 2 h on a 24-core machine. After setting appropriate parameters, manual inspection of the HMM fits showed reasonable agreement with our expectations: enduring transitions in pixel values were associated with sand disturbance by fish, and three orders of magnitude more of these transitions were detected in tanks with fish versus empty tanks (Figure 2I). In addition to identifying when and where a sand disturbance occurs, this approach also allowed us to create background images that excluded fish and shadows from each frame (Figure 2J).
Clustering of HMM Changes to Detect Sand Disturbance Events
When a fish scoops up or spits out sand, HMM transitions occur in hundreds to thousands of nearby pixels. To group individual pixel transitions together, we applied density-based spatial clustering to the x, y, and time coordinate of each individual HMM transition (Ester et al., 1996) (Figure 3A). To find the best parameters, we used exploratory data analysis and parameter grid search (Figure S2). This approach allowed us to determine the spatial and temporal location of thousands of individual fish-mediated sand disturbances on each day of each bower trial. We also calculated distributions for the width, height, temporal span, and number of HMM transitions for every cluster (Figure S3). These data further demonstrated that clusters were associated with the presence of fish, as empty tanks showed a minimal number of clusters (Figure 3B). Inspection of a subset of these clusters confirmed that the clusters were associated with regions of sand that were undergoing lasting change (Figure 3C).
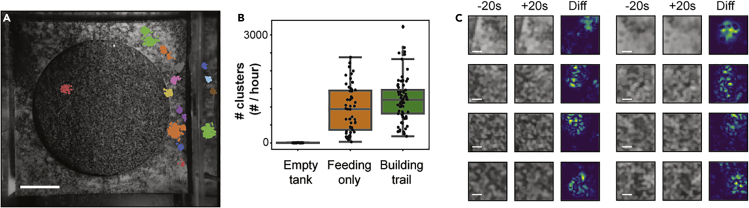
Figure 3.
Clustering HMM Events Identifies Sand Disturbance Events
(A) Example of clusters identified in a 60 s period. HMM transitions are color coded based on their cluster membership. Scale bar, 10 cm.
(B) Number of clusters identified per hour based on trial type. “Feeding only” are trials containing four females. “Build trial” contain four females and one male that builds a bower. The boxplot shows quartiles of the dataset while the whiskers show the rest of the distribution unless the point is an outlier.
(C) Before and after images (20 s) for eight example clusters. Left and middle panels show raw grayscale images. Right panel shows heatmap displaying pixel value differences in the left and middle frames. Yellow indicates large changes. Blue indicates no change. Scale bar, 1 cm.
See also Figures S2−S4.
To further validate these clusters, we generated 200 × 200-pixel, 4-s video clips for a randomly sampled subset of clusters centered over their mean spatial and temporal position (∼2,000 per trial for seven of the eight bower construction trials). Manual review of these video clips revealed that the majority (>90%, 13,288/14,234 analyzed events) of clusters were true sand change events caused by fish behaviors, with the remaining portion including reflections of events in the glass, shadows caused by stationary or slow-moving fish, or small bits of food, feces, or other debris settling on the sand surface.
To ensure this procedure recovered most sand disturbance events, we leveraged our manually annotated spit/scoop dataset (Figure S1) and performed HMM and density-based spatial clustering. For each event, the spatial and temporal center of the event was compared. The differences in time between human annotations and machine annotations follow a Gaussian distribution (Figure S4A). 95.6% (725/758) human annotations have at least one machine annotation in the (-1s, +1s) interval. Of these 725 events, 93.7% (679/725) also had a machine annotation event within 70 pixels (approximately 3.5 cm), which is approximately one-third of the oval fish length (Figure S4B). By these criteria, 89.6% (679/758) of human-annotated events can be retrieved by the automated machine HMM/clustering process.
Automatic Classification of Cichlid Behaviors with 3D Residual Networks
On average, ∼1,000 clusters were identified in each hour of video (Figure 3B). We aimed to automatically identify the subset of these events corresponding to bower construction behaviors in each video. However, scooping and spitting sand during bower construction represents only a subset of behaviors that cause sand change in our paradigm. For example, feeding behaviors performed by both males and females also involve scooping and spitting sand, and are expressed frequently throughout trials. Quivering and spawning behavior, in which a male rapidly circles and displays his fins for a gravid female, are less frequent, and also cause significant sand change. Identifying bower construction behaviors therefore requires identifying other behaviors that cause sand change. To achieve this, we turned to a deep learning approach and assessed whether 3D Residual Networks (3D ResNets), which have been recently shown to accurately classify human actions from video data (Qiu et al., 2017), could accurately distinguish fish behaviors that cause sand change in our paradigm.
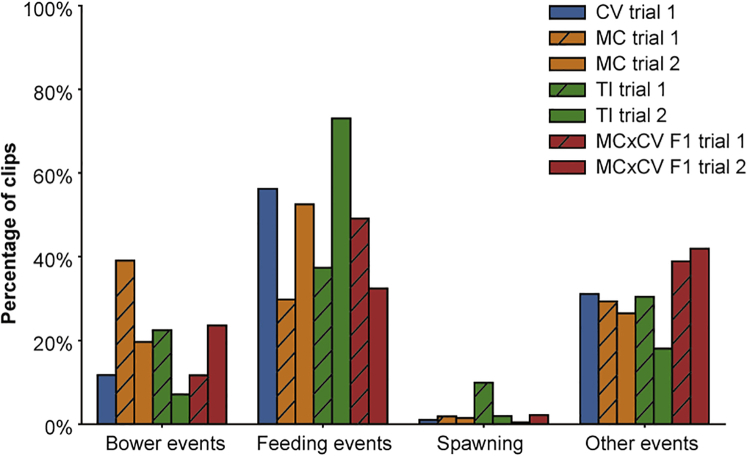
To create a training set for the 3D ResNet, we first generated a video clip for each cluster based on its location in space and time. This process narrowed down each event to a 4-s, 200 × 200-pixel video clip from the original 1,296 × 972-pixel video. To create a training set for the 3D ResNet, a trained observer manually classified 14,172 of these video clips (∼2,000 per trial) into one of ten categories (bower scoop, bower spit, bower multiple, feed scoop, feed spit, feed multiple, drop sand, quivering, other, and shadow/reflection) (Videos S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10). Feeding was the most frequently observed behavior, accounting for nearly half of all clips (47.0%, 6,659/14,172 annotated clips; feeding scoops, 15.2%; feeding spits, 11.6%; multiple feeding events, 20.2%). Bower construction behaviors were the next most common (19.5%; bower scoops, 9.4%; bower spits, 8.1%; multiple bower construction events, 1.9%). Quivering and spawning events were the least frequently observed, accounting for just 2.6% of all clips. The remainder of sand change events were annotated as sand dropping behavior (5.6%), “other” behaviors (e.g., brushing the sand surface with the fins or the body; 18.7%), or shadows/reflections (6.6%) (Figure 4).
Figure 4.
Distribution of Different Sand Perturbation Events
A human observer manually classified 14,234 video clips into one of ten categories. Bower events are scoops, spits, or multiple scoop/spit events associated with bower construction. Feeding events are scoops, spits, or multiple scoop/spit events associated with feeding behaviors. Spawning events involve male fish quivering to attract females. Other events include sand perturbations caused by fins or the body, reflections of events in the aquarium glass, or sand dropped from above.
A 3D ResNet was then trained on 80% (∼11,200 clips) of the data, and the remaining 20% of the data was reserved for validation (2,752 clips) (Table 2 and Figures 5A and 5B). To place the ResNet predictions in the context of human performance, we also measured the accuracy of a previously naive human observer who underwent 12 h of training and then manually annotated a test set of 3,039 clips from three trials and all 10 behavior categories (Table S1). The 3D ResNet achieved ∼76% accuracy on the validation set, which was better than the accuracy of the newly trained human observer (∼73.9% accuracy, 2,246/3,039 clips). Confidence for 3D ResNet predictions on the validation set ranged from 22.1% to 100%, and confidence tended to be greater for correct predictions (mean confidence 92.93% ± 0.279%) than for incorrect predictions (mean confidence 78.28% ± 0.074%) (Figure S5A, C). We found an imbalance in the distribution of incorrect predictions across categories (Table 2). For some categories, such as “build multiple,” “feed multiple,” and “fish other,” video clips could contain behaviors that also fit into other categories. For example, a “feed multiple” clip by definition contains multiple feeding scoop and/or feeding spit events, a “bower multiple” clip contains multiple bower scoops and/or bower spits, and a “fish other” clip may contain a bower scoop and a fin swipe (or some other combination of behaviors). We found that erroneous “within building” category predictions for build multiple, “within feeding” predictions for feed multiple, and “fish other” predictions accounted for ∼82% of all incorrect predictions. We further found that the area under the precision recall curve was 0.91 (Figure S5B). By setting a confidence threshold of 90%, most (∼62%) incorrect predictions were excluded, whereas most (70%) correct predictions were included. At the same time, ∼86% of correct bower scoop predictions and ∼88% of correct bower spit predictions were included. Of all predictions, 69% were above the 90% confidence threshold, and overall accuracy for these high-confidence predictions was ∼87% (Figure S5C).
Table 2.
Confusion Matrixa for Sand Disturbance Events Classified with a 3D Residual Network
| Predicted Label | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human label | Category | Build scoop | Feed scoop | Build spit | Feed spit | Build mult | Feed mult | Spawn | Shadow Reflect |
Other | Sand drop | Total | Percent | Acc |
| Build scoop | 187 | 25 | 1 | 0 | 2 | 1 | 0 | 0 | 26 | 2 | 244 | 9% | 77% | |
| Feed scoop | 23 | 303 | 0 | 3 | 0 | 48 | 0 | 1 | 28 | 0 | 406 | 15% | 75% | |
| Build spit | 1 | 0 | 194 | 5 | 6 | 1 | 1 | 0 | 10 | 2 | 220 | 8% | 88% | |
| Feed spit | 1 | 6 | 15 | 225 | 0 | 38 | 2 | 1 | 27 | 26 | 341 | 12% | 66% | |
| Build mult | 8 | 0 | 9 | 0 | 29 | 1 | 1 | 0 | 7 | 0 | 55 | 2% | 53% | |
| Feed mult | 16 | 73 | 4 | 22 | 2 | 402 | 0 | 0 | 17 | 1 | 537 | 20 | 75% | |
| Spawn | 0 | 0 | 0 | 0 | 1 | 0 | 56 | 0 | 15 | 1 | 73 | 3% | 77% | |
| Shadow/reflect | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 169 | 11 | 1 | 184 | 7% | 92% | |
| Other | 26 | 26 | 4 | 10 | 10 | 11 | 8 | 9 | 388 | 20 | 512 | 19% | 76% | |
| Sand drop | 1 | 0 | 1 | 12 | 0 | 1 | 0 | 2 | 14 | 149 | 180 | 7% | 83% | |
| Total | 263 | 433 | 229 | 278 | 50 | 504 | 68 | 182 | 543 | 202 | 2,752 | 100% | 76% | |
Each row of the matrix represents the instances in an actual class while each column represents the instances in a predicted class.
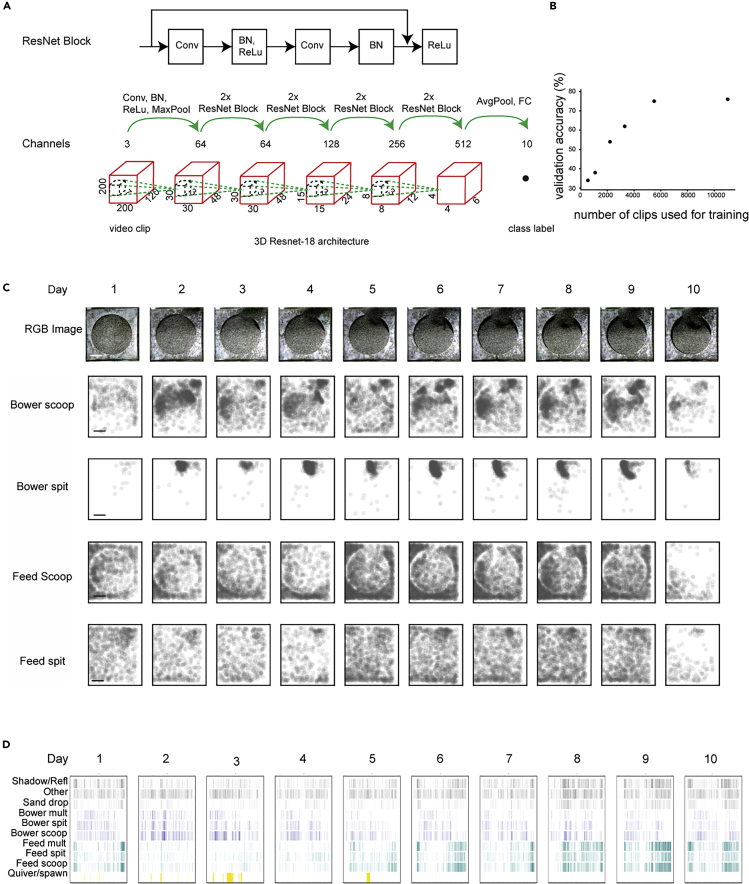
Figure 5.
Automated Classification of Sand Disturbance Events Using a 3D Residual Network
(A) Schematic of 3D residual network.
(B) Validation accuracy of machine-learning labels as a function of number of videos used to train the model.
(C) Spatial position of four categories of sand manipulation events over 10 days of building by a Mchenga conophoros. A castle structure was built in the top middle of the field of view. Scale bar, 10 cm.
(D) Raster plot of time of each sand disturbance event by classification. (C and D) Building events occur at different spatial and temporal positions than feeding events.
See also Figure S5.
We tested how many annotated clips are required to achieve a similar accuracy by inputting 5%–50% of the original labeled clips into the training dataset. A similar accuracy was achieved using just 50% of the clips, but a significant decrease in accuracy (75%–62%) was observed when the number of training clips was decreased from 50% to 30% of the original set (Figure 5B).
As there was an uneven distribution of the different categories in the training data, we were curious if our model might be biased toward the more frequent categories and have lower overall accuracy than a model trained on data with a balanced distribution of the different categories. To test this hypothesis, we built a model from an equal sampling of all 10 categories. First, we created an “equal sampling” set where each category in the training dataset has the same number of clips. Because the category “build multiple” has the least number of clips (277 clips out of 14,172 annotated video clips), for each category, we used 220 clips for training and 50 clips for validation. As a “random sampling” control, we randomly sampled an equal number of clips for training and validation as the uniform set. Specifically, there are 2,200 randomly sampled clips for training and 500 for testing. Overall, the accuracy was comparable, although we did note some large differences in accuracy based upon category (Figure S5F). For example, build multiple, a very rare category, was much less accurately predicted in the “random sampling” model comipared to the “equal sampling” model (18% versus 75%). When we compared the overall accuracy of each model, we noted that the higher-performing model depended on the validation set. When an equal sampling of validation clips was used, the equal sampling model performed better (64.0% accuracy compared to 60.5% accuracy). When a random sampling of validation clips was used, the random sampling method was more accurate (62.2% accuracy compared to 57.7% accuracy).
The spatiotemporal dimension of each video clip is a very important hyperparameter to tune to achieve the best classification accuracy. The spatial scale is optimal when the whole behavior is captured, and when irrelevant pixels are excluded from the frame as much as possible. The same is true for temporal scale. Therefore, we decided to test the amount of spatial and temporal data necessary to accurately classify each clip. We found that the classification accuracy initially increased with frame size and peaked at 120 × 120 pixels per frame (Figure S5D). The accuracy decreased from 76% to 74% when the entire 200 × 200-pixel frame was used as input. This decrease in accuracy further affirmed our choice to crop the clips before feeding them to the ResNet, as it eliminates information unrelated to action in the frame. Next, we tested the relationship between the number of frames per clip and the classification accuracy. We tuned two parameters, including the total number of frames sampled (n = 16, 32, 48) and the interval between each sampled frame (i = 0, 1, 2, 3, 4). Classification accuracy steadily increased from 67% (n = 16) to 72% (n = 48) when more frames were used. When the total number of frames was fixed, accuracy was greater when the sampling interval (i) was larger (Figure S5E).
Distinguishing between Feeding and Bower Construction Events
The differences between construction behaviors and feeding behaviors are subtle and are often indistinguishable to inexperienced observers (Table S1). Feeding and construction both involve scooping sand into the mouth, swimming, and then spitting sand from the mouth. Feeding behaviors are also typically performed more frequently relative to bower construction behaviors (Figure 4). We were therefore concerned that the 3D ResNet would be unable to accurately distinguish between feeding versus construction behaviors, which would prevent accurate measurement of spatial patterns associated with bower construction.
Much to our surprise, the ResNet reliably distinguished between feeding and construction events; for construction and feeding scoops and spits, the model achieved F1 scores of 0.74, 0.86, 0.72, and 0.73 (for build scoops, build spits, feeding scoop, and feeding spits, respectively), with balanced precision and recall scores. These F1 scores were comparable to the overall averaged F1 score (or accuracy) of the model, 0.76. Remarkably, the model outperformed a newly trained human observer in distinguishing between construction and feeding: the average F1 score for the model across these categories was 0.76, whereas the newly trained observer's average F1 score was 0.71. The difference in performance was further evident when we quantified the proportion of build-feed false-positives (build scoops mis-classified as feed scoops, build spits mis-classified as feed spits, and vice versa); these erroneous predictions were 2.5× more frequent in the newly trained observer's annotations compared with the model's predictions (201/1,469 predictions in these categories, or 13.7% for the human; 68/1,211 predictions in these categories, or 5.6% for the model). Differences in the spatial position and relative timing were readily observed among bower construction, feeding, and spawning events (Figures 5C and 5D).
Accuracy of Model on New Trials
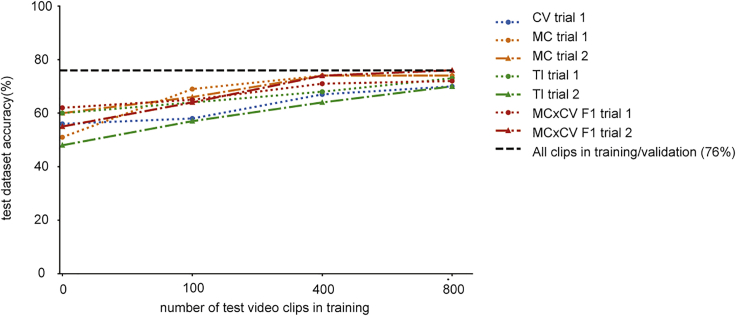
The trained model showed high validation accuracy on the seven trials. However, our ultimate goal is to measure bower construction in hundreds of independent trials. To that end, we tested how generalizable this model is when applied to previously unobserved individuals. To test this, we retrained models using six of the seven trials and tested their ability to classify video clips from the remaining trial. In all seven cases, we saw a significant drop in accuracy, from 76% to between 48% and 62% depending on the trial (Figure 6). We could, however, recapture much of this accuracy loss by including a small subset of videos from the excluded trial (∼100–400), suggesting that including a limited number of labeled videos from new trials could dramatically increase the accuracy. Interestingly, trials that had the largest imbalance in the frequency of different categories compared with the mean also showed the largest decrease in accuracy (Figure S6).
Figure 6.
Application of the Model on New Trials
Accuracy of model on different trials using 0, 100, 400, or 800 training videos. We restricted training data for one of the seven trials and then tested the accuracy of the model on video clips specific to that trial. Models that used zero video clips for training showed a decrease in accuracy.
See also Figure S6.
Discussion
Automated classification of behavior is an important goal for many areas of basic, applied, and translational research. Advances in hardware have made collection of large amounts of video data cost effective, using small, battery-operated cameras (e.g., Go Pro), mobile phones, or small microcomputers (e.g., Raspberry Pi). The small size and inexpensive nature of these hardware systems makes it possible to collect large volumes of data from many individuals. For example, here we collected hundreds of hours of video from seven home tanks in standard aquatics housing facilities. Inexpensive cloud data storage systems additionally allowed for the transfer and archiving of this data. We used Dropbox to store all video data for these experiments, and our long-term goal is to collect data from hundreds of trials in many species.
In this paradigm, our recording system collects >300 gigabytes of video data for each behavioral trial. A major challenge is thus designing a pipeline to identify behaviors of interest through large amounts of background noise. Here we demonstrate one possible solution that involves first identifying environmental disturbances and then classifying the behavioral causes of those disturbances. This approach is rooted in two main advances: first, an action detection algorithm for identifying times and locations of sand disturbances, allowing small video clips containing behaviors of interest to be generated on a large scale; and second, custom-trained 3D Residual Networks to classify each clip into one of ten possible categories.
For action detection, we tailored an HMM to recognize permanent changes in pixel color that occur whenever fish alter the sand. The core approach involves identification of lasting changes in the sand, which manifest as permanent changes in pixel color within specific subfields of view. This approach may be useful for studying many other animal behaviors that are defined by manipulation of the environment, such as nest construction, burrow digging, or web weaving. Many animals also disturb the environment as they forage and feed; for example, they may disturb or dig through ground substrate or ingest physical components of the environment such as leaves, fruits, berries, or algae. Thus, this approach may facilitate measurement of many behaviors in more complex and naturalistic environments. One limitation to this approach is that individual pixels must be focused on a specific region of the background. In our paradigm, we accomplished this by fixing the camera, but slightly moving cameras could also be used if an accurate means of registration is available to align frames at different time points.
Following identification of sand disturbance events, we used action recognition to classify events into ethologically meaningful behavioral categories. Previous machine learning strategies have relied on positional tracking and/or pose estimation data to classify animal behaviors (Hong et al., 2015; Anderson and Perona, 2014; Robie et al., 2017). In contrast, our paradigm was poorly suited to these methods, due to a lack of conspicuous stereotypical joint movement from top-down video and an abundance of stereotypical interactions between subjects and their environment, which were critical for defining the behaviors of interest. We found that a 3D ResNet classified video clips of animal behavior into 10 categories more accurately than a newly trained human observer, demonstrating that these networks can effectively quantify many different behaviors of interest from large volumes of raw video data. This result suggests that 3D ResNets may be a powerful tool for measuring animal behavior in naturalistic settings, and may drastically increase the scale of experimental designs in systems that, historically, have been constrained by the amount of human observation time required to measure behaviors of interest.
Feeding and construction behaviors have different underlying goals and are critical for survival and reproduction in the wild, but their physical execution is often indistinguishable to inexperienced observers. Separating these behaviors is critically important in our paradigm, where there is a high risk that decisions to scoop and spit during bower construction will be undetectable through a high volume of feeding actions. Remarkably, the 3D ResNet was able to distinguish between feeding and construction behaviors more accurately than a newly trained human observer. The ability to accurately identify subtle differences in behavior may have important implications for better understanding the progression of neurological diseases characterized by subtle changes in locomotor function over extended periods of time, or monitoring efficacy of different treatment strategies for improving locomotor function.
Striking differences in the relative spatial positions and timing of predicted feeding versus construction behaviors were readily apparent across whole trials. For example, in castle-building MC males, bower spits were more spatially concentrated than bower scoops, feeding spits, and feeding scoops, consistent with the idea that castle construction is driven by scooping sand from dispersed locations and spitting into a concentrated region. Similarly, feeding behaviors and bower construction behaviors were expressed daily but at different times, whereas spawning events occurred infrequently in punctuated bursts. These data show that our system can be used to map tens of thousands of behavioral events in time and space, allowing future studies to unravel how these different complex behaviors are expressed in dynamic environments over extended time periods. It will be important to link these sand manipulation events with information about the actual structure being built by the fish. Development of an approach to characterize the shape of the bower at any given time will be necessary for connecting building behaviors with the structure.
The 3D ResNet accurately classified behaviors across three different species and one interspecies hybrid cross that all differ in morphology and color patterning. For example, Copadichromis virginalis males exhibit black body coloration, yellow heads and dorsal fins, and narrower jaws relative to Tramitichromis intermedius males, which exhibit yellow and red body coloration, blue heads, and a relatively wide jaw apparatus. Bower construction in particular has evolved in hundreds of species, and the evolution of feeding morphology and behavior is thought to be central to the explosive radiation of cichlids into thousands of species. Our results suggest that our system will likely be effective for measuring natural variation in these behaviors among hundreds of species. Lake Malawi cichlids can also be hybridized across species boundaries, enabling powerful genetic mapping approaches (e.g., quantitative trait loci mapping) to be applied in subsequent hybrid generations to identify genetic variants that influence complex traits. Our data show that our system is effective for phenotyping hybrid individuals, allowing future studies to identify specific regions of the genome that are responsible for pit versus castle building. Last, high prediction accuracy for quivering, a conserved and stereotyped sexual behavior expressed by many fish, supports that action recognition may be useful for analyzing mating behaviors in many fish species. More broadly, our results suggest that 3D ResNets may be effective tools for measuring complex behaviors in other systems even when individuals vary substantially in physical traits.
Limitations of the Study
It is important to note that although this report demonstrates that 3D Resnets should be useful for behavioral classification, it does not immediately generalize to other systems without additional work. For example, mouse behavior scientists could not immediately apply this to their behavior of choice.
Resource Availability
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact Patrick T. McGrath (patrick.mcgrath@biology.gatech.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Original data for 14,000 annotations and video clips used to train and validate the 3D ResNet is available at Mendeley Data: https://doi.org/10.17632/3hspb73m79.1
All codes, including a ReadMe file explaining how to use them, for running action detection is available on GitHub at https://github.com/ptmcgrat/CichlidActionDetection. All codes for training the 3D ResNet for action classification is available on GitHub at https://github.com/ptmcgrat/CichlidActionRecognition.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Tucker Balch for suggestions on tank setup. We thank Andrew Gordus for helpful comments on writing the manuscript. This work was supported in part by NIH R01GM101095 to J.T.S., NIH R01GM114170 to P.T.M., NIH F32GM128346 to Z.V.J., and Georgia Tech Graduate Research Fellowships to J.L., V.A., and M.A.
Author Contributions
Conceptualization, L.L., Z.V.J., and P.T.M.; Methodology, L.L., Z.V.J., and P.T.M.; Investigation, L.L., Z.V.J., J.L., T.L., V.A., and M.A.; Writing – Original Draft, L.L., Z.V.J., and P.T.M.; Writing – Review & Editing, L.L., Z.V.J., and P.T.M.; Funding Acquisition, J.T.S., P.T.M., and Z.V.J.
Declaration of Interests
The authors declare no competing interests.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101591.
Contributor Information
Jeffrey T. Streelman, Email: todd.streelman@biology.gatech.edu.
Patrick T. McGrath, Email: patrick.mcgrath@biology.gatech.edu.
Supplemental Information
References
- Anderson D.J., Perona P. Toward a science of computational ethology. Neuron. 2014;84:18–31. doi: 10.1016/j.neuron.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Andriluka M., Iqbal U., Insafutdinov E., Pishchulin L., Milan A., Gall J., Schiele B. Posetrack: a benchmark for human pose estimation and tracking. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2018:5167–5176. [Google Scholar]
- Benjamin S.P., Zschokke S. A computerised method to observe spider web building behaviour in a semi-natural light environment. Eur. Arachnol. 2000:117–122. [Google Scholar]
- Berman G.J., Choi D.M., Bialek W., Shaevitz J.W. Mapping the stereotyped behaviour of freely moving fruit flies. J. R. Soc. Interf. 2014;11:20140672. doi: 10.1098/rsif.2014.0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D., Wagner C.E., Li Y.I., Malinsky M., Keller I., Fan S., Simakov O., Ng A.Y., Lim Z.W., Bezault E. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collias N.E., Collias E.C. Princeton University Press; 2014. Nest Building and Bird Behavior. [Google Scholar]
- Dawkins R. Oxford University Press; 1982. The Extended Phenotype. [Google Scholar]
- Egnor S.E., Branson K. Computational analysis of behavior. Annu. Rev. Neurosci. 2016;39:217–236. doi: 10.1146/annurev-neuro-070815-013845. [DOI] [PubMed] [Google Scholar]
- Ester M., Kriegel H.-P., Sander J., Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Kdd. 1996:226–231. [Google Scholar]
- Feng N.Y., Fergus D.J., Bass A.H. Neural transcriptome reveals molecular mechanisms for temporal control of vocalization across multiple timescales. BMC Genomics. 2015;16:408. doi: 10.1186/s12864-015-1577-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girshick R. Fast r-cnn. Proc. IEEE Int. Conf. Comput. Vis. 2015:1440–1448. [Google Scholar]
- Graving J.M., Chae D., Naik H., Li L., Koger B., Costelloe B.R., Couzin I.D. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. Elife. 2019;8:e47994. doi: 10.7554/eLife.47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunel S., Rhodin H., Morales D., Campagnolo J., Ramdya P., Fua P. DeepFly3D, a deep learning-based approach for 3D limb and appendage tracking in tethered, adult Drosophila. Elife. 2019;8:e48571. doi: 10.7554/eLife.48571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell M. Cambridge University Press; 2000. Bird Nests and Construction Behaviour. [Google Scholar]
- Hara K., Kataoka H., Satoh Y. Can spatiotemporal 3d cnns retrace the history of 2d cnns and imagenet? Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2018:6546–6555. [Google Scholar]
- Hong W., Kennedy A., Burgos-Artizzu X.P., Zelikowsky M., Navonne S.G., Perona P., Anderson D.J. Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning. Proc. Natl. Acad. Sci. U S A. 2015;112:E5351–E5360. doi: 10.1073/pnas.1515982112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain J., Stokes C., Gaudry Q., Song X., Foley J., Wilson R., De Bivort B. Leg-tracking and automated behavioural classification in Drosophila. Nat. Commun. 2013;4:1910. doi: 10.1038/ncomms2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A., Mamidanna P., Cury K.M., Abe T., Murthy V.N., Mathis M.W., Bethge M. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018;21:1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- McKaye K.R., Stauffer J., Turner G., Konings A., Sato T. Fishes, as well as birds, build bowers. J. Aquaricult. Aquat. Sci. 2001;9:121–128. [Google Scholar]
- Mouritsen H. Long-distance navigation and magnetoreception in migratory animals. Nature. 2018;558:50–59. doi: 10.1038/s41586-018-0176-1. [DOI] [PubMed] [Google Scholar]
- Nath T., Mathis A., Chen A.C., Patel A., Bethge M., Mathis M.W. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 2019;14:2152–2176. doi: 10.1038/s41596-019-0176-0. [DOI] [PubMed] [Google Scholar]
- Pereira T.D., Aldarondo D.E., Willmore L., Kislin M., Wang S.S., Murthy M., Shaevitz J.W. Fast animal pose estimation using deep neural networks. Nat. Methods. 2019;16:117–125. doi: 10.1038/s41592-018-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrou G., Webb B. Detailed tracking of body and leg movements of a freely walking female cricket during phonotaxis. J. Neurosci. Methods. 2012;203:56–68. doi: 10.1016/j.jneumeth.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Qiu Z., Yao T., Mei T. Learning spatio-temporal representation with pseudo-3d residual networks. Proc. IEEE Int. Conf. Comput. Vis. 2017:5533–5541. [Google Scholar]
- Redmon J., Divvala S., Girshick R., Farhadi A. You only look once: Unified, real-time object detection. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016:779–788. [Google Scholar]
- Ren S., He K., Girshick R., Sun J. Faster r-cnn: towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015:91–99. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- Robie A.A., Seagraves K.M., Egnor S.R., Branson K. Machine vision methods for analyzing social interactions. J. Exp. Biol. 2017;220:25–34. doi: 10.1242/jeb.142281. [DOI] [PubMed] [Google Scholar]
- Russell A.L., Morrison S.J., Moschonas E.H., papaj D.R. Patterns of pollen and nectar foraging specialization by bumblebees over multiple timescales using RFID. Sci. Rep. 2017;7:42448. doi: 10.1038/srep42448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G.J., Johnson-Kerner B., Bialek W., Ryu W.S. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput. Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R. The digging behavior and skin differentiations in Heterocephalus glaber. J. Morphol. 1981;168:51–71. doi: 10.1002/jmor.1051680107. [DOI] [PubMed] [Google Scholar]
- Vollrath F. Analysis and interpretation of orb spider exploration and web-building behavior. Adv. Study Behav. 1992;21:147–199. [Google Scholar]
- Weissbrod A., Shapiro A., Vasserman G., Edry L., Dayan M., Yitzhaky A., Hertzberg L., Feinerman O., Kimchi T. Automated long-term tracking and social behavioural phenotyping of animal colonies within a semi-natural environment. Nat. Commun. 2013;4:2018. doi: 10.1038/ncomms3018. [DOI] [PubMed] [Google Scholar]
- Wild B., Sixt L., Landgraf T. Automatic localization and decoding of honeybee markers using deep convolutional neural networks. arXiv. 2018 arXiv:1802.04557. [Google Scholar]
- York R.A., Patil C., Hulsey C.D., Anoruo O., Streelman J.T., Fernald R.D. Evolution of bower building in Lake Malawi cichlid fish: phylogeny, morphology, and behavior. Front. Ecol. Evol. 2015;3:18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data for 14,000 annotations and video clips used to train and validate the 3D ResNet is available at Mendeley Data: https://doi.org/10.17632/3hspb73m79.1
All codes, including a ReadMe file explaining how to use them, for running action detection is available on GitHub at https://github.com/ptmcgrat/CichlidActionDetection. All codes for training the 3D ResNet for action classification is available on GitHub at https://github.com/ptmcgrat/CichlidActionRecognition.