Abstract
Background
Colistin is being administered as last-line therapy for patients that have failed to respond to other available antibiotics that are active against Escherichia coli. The underlying mechanisms of colistin resistance and heteroresistance remain largely uncharacterized. The present study investigated the mechanisms of resistance and heteroresistance to colistin in Escherichia coli isolates from Wenzhou, China.
Materials and Methods
Colistin resistance was detected by the broth microdilution method (BMD). Colistin heteroresistance was determined by population analysis profiles (PAPs). The polymerase chain reaction (PCR) was conducted to detect mcr-1, mcr-2, mcr-3, pmrA, pmrB, phoP, phoQ and mgrB, and quantitative real-time PCR (qRT-PCR) was used to determine the expression levels of mcr-1, pmrC, pmrA and pmrB. Lipid A characterization was conducted by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS).
Results
0.69% (2/291) of Escherichia coli strains were resistant to colistin, whereas the heteroresistance rate reached 1.37% (4/291). mcr-1, the mobile colistin-resistance gene, was present in the two resistant isolates. The substitutions in PmrB were detected in the two heteroresistant isolates. The transcripts levels of the pmrCAB operon were upregulated in two of the heteroresistant isolates. carbonylcyanide m-chlorophenylhydrazone (CCCP) was able to reverse colistin resistance of all isolates tested and exhibited a significantly higher effect on colistin-heteroresistant isolates. MALDI-TOF MS indicated that the additional phosphoethanolamine (PEtn) moieties in lipid A profiles were present both in colistin-resistant and heteroresistant isolates.
Conclusion
The present study was the first to investigate the differential mechanisms between colistin resistance and heteroresistance. The development of colistin heteroresistance should be addressed in future clinical surveillance.
Keywords: Escherichia coli, colistin, mcr-1, heteroresistance, lipid A
Introduction
Escherichia coli (E. coli) is one of the most commonly isolated microorganisms in clinical specimens, which is responsible for various nosocomial infections, such as meningitis, urinary tract infections (UTIs), wound infections and bloodstream infections.1–3 Treatment of multidrug-resistant (MDR) E. coli infections has become a serious clinical issue, notably with the emergence of resistance to last-resort antibiotics such as colistin.4
As a cationic antimicrobial polypeptide, colistin exerts antibacterial activity via interacting with the lipid A moiety of lipopolysaccharide (LPS) and subsequently disrupting the outer membrane of Gram-negative bacteria.5,6 Despite the potential of colistin as a high-efficient antimicrobial agent, its massive use in clinical therapeutics and as a feed additive has led to the development of colistin resistance in E. coli.7,8
To date, the most common reported mechanisms for colistin resistance in E. coli have involved the plasmid-borne mobile colistin resistance (mcr) gene, which was newly discovered in 2016 and denoted as mcr-1.9 Subsequently, several amino acid variants have been described, such as mcr-2 and mcr-3.10,11 This transferable mechanism of colistin plasmid-borne resistance could aggravate and contribute to the spread of resistance. In addition, chromosomally encoded systems, such as PhoPQ and PmrAB two-component regulatory systems (TCS) have been reported sporadically.7
Heteroresistance to colistin has been reported to be significantly increased in Gram-negative bacteria.12 This has been shown by a phenotypic characteristic demonstrating the presence of colistin-resistant subpopulations among a susceptible population.13 Several mechanisms of colistin heteroresistance have been proposed, including the activation of PmrAB and PhoPQ TCSs (Pseudomonas aeruginosa, Klebsiella pneumoniae and Enterobacter cloacae),14–17 overexpression of the efflux pumps (Acinetobacter baumannii, Enterobacter asburiae and Enterobacter cloacae),18,19 biofilm formation (Klebsiella pneumoniae)20 and putrescine/YceI communication (Burkholderia cenocepacia).21 However, only sporadic cases of colistin heteroresistance have been reported in E. coli,22 let alone the investigation of their mechanisms of heteroresistance against this compound.
The aim of our present study was to determine and compare the mechanisms that are responsible for resistance and heteroresistance to colistin in E coli strains isolated from a Chinese teaching hospital.
Materials and Methods
Bacterial Strains
A total of 291 non-duplicated E. coli clinical isolates were recovered from the First Affiliated Hospital of Wenzhou Medical University, China in 2018. Each isolate represented a single sample from one patient. The isolates were identified as E. coli by VITEK® MS automated system (BioMérieux, Hazelwood, MO, USA). The MICs of colistin were determined by the broth microdilution in cation-adjusted Mueller-Hinton Broth (CAMHB).23 The MIC breakpoint of colistin for E. coli was interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2019)24 (susceptible, ≤2 mg/L; resistant, >2 mg/L). The E. coli ATCC 25,922 served as the quality control for susceptibility testing.
Population Analysis Profiles (PAPs)
PAPs are used as the reference method to define antibiotic hetero-resistance.15 The analysis was performed among colistin-susceptible isolates. Bacterial cultures were grown overnight to the log-phase and subsequently plated using serial dilutions on Luria-Bertani (LB) agar with or without various concentrations of colistin (0, 0.25, 0.5, 1, 2, 4, 8 and 16 mg/L). The plates were incubated at 37°C and the Colony-Forming Units (CFU) were enumerated following 48h. The limit of detection was 20 CFU/mL. Colistin heteroresistance was defined as a colistin-susceptible isolate (MIC ≤2 mg/L) with subpopulations growing in the presence of ≥2 mg/L colistin.25 The percentage of colistin resistance was calculated as the number of bacterial colonies that grew on colistin plates divided by the number of bacteria that grew on LB broth without drug. For each isolate, a single colony was selected from the highest antibiotic concentration and the colistin MIC was reassessed following serial passaging on antibiotic-free medium to evaluate the stability of the hetero-resistant phenotype. Cultures with resistant or susceptible subpopulations were isolated from the highest colistin concentration or drug-free medium separately for further studies.
Antimicrobial Susceptibility Test
The MICs of clinical routine antimicrobial agents, including aztreonam (ATM), ceftazidime (CAZ), fosfomycin (FOS), gentamicin (GEN), levofloxacin (LEV), meropenem (MEM), amikacin (AMK), ciprofloxacin (CIP), imipenem (IMP) and cefotaxime (CTX) were determined using the agar dilution method (BMD) in accordance with the guidelines of CLSI 2019.26 To examine the effects of efflux pump on colistin resistance and heteroresistance, MICs were measured via the broth microdilution method (BMD) by adding 10 mg/L of carbonylcyanide m-chlorophenylhydrazone (CCCP) into CAMHB.27
Polymerase Chain Reaction (PCR) and Sequencing
Whole-cell DNA samples of colistin-resistant and -heteroresistant isolates were extracted using the Biospin Bacterial Genomic DNA Extraction kit (Bioflux, Tokyo, Japan) according to the manufacturer’s instructions. The genes of mcr-1, mcr-2, mcr-3, pmrA, pmrB, phoP, phoQ and mgrB among the E. coli strains were investigated by PCR and the primers are described in Table 1.28–31 The amplicons of mcr-1, mcr-2, mcr-3, pmrA, pmrB, phoP and phoQ and mgrB were sequenced by Shanghai BGI Technology Co., Ltd. and subsequently analyzed by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of resistant isolates were compared with those of ATCC 25,922, while the comparison was carried out between susceptible and resistant subpopulations in heteroresistant isolates.
Table 1.
PCR Primers Used in This Study
| Gene | Sequence (5ʹ→3ʹ) | Amplicon Size (bp) |
|---|---|---|
| Primers for sequencing | ||
| mcr-1 | F: GCTCGGTCAGTCCGTTTG | 162628 |
| R: GAATGCGGTGCGGTCTTT | ||
| mcr-2 | F: TGTTGCTTGTGCCGATTGGA | 161729 |
| R: AGATGGTATTGTTGGTTGCTG | ||
| mcr-3 | F: AAATAAAAATTGTTCCGCTTATG | 92930 |
| R: AATGGAGATCCCCGTTTTT | ||
| pmrA | F: AGTTTTCCTCATTCGCGACCA | 71431 |
| R: TACCAGGCTGCGGATGATATTCT | ||
| pmrB | F: GGATGGCCTGATGTGACGCTGTC | 131231 |
| R: GCGCGGCTTTGGCTATA | ||
| phoP | F: GCCAGTACCGCCAGCTTAA | 179831 |
| R: CTCGCCACGTAACAGCCGAA | ||
| phoQ | F: GGCACAATATCCCCAAGAAGT | 159531 |
| R: ATCCACAGGCTGGTATCTGCA | ||
| mgrB | F: CACGAATATCGACATAGTTAG | 27531 |
| R: TATTCTACCACTGCTGGAGAG | ||
| Primers for qRT-PCR | ||
| gapA | F: CGTTAAAGGCGCTAACTTCG | 1167 |
| R: ACGGTGGTCATCAGACCTTC | ||
| mcr-1 | F: GCAGCATACTTCTGTGTGGTAC | 14533 |
| R: ACAAAGCCGAGATTGTCCGCG | ||
| pmrC | F: CTGATGACGCCACGAATG | 1347 |
| R: TTGTAGAACAGTGCGGCAAC | ||
| pmrA | F: CCTTTTGCGCTGGAAGAG | 1567 |
| R: TGGGCGTCAGAATCAACTC | ||
| pmrB | F: TCCCCTCGTATGACGAACTC | 1127 |
| R: TCATAATGTTGCTGCCTTGC | ||
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA samples from heteroresistant isolates (including susceptible and resistant subpopulations) were extracted from the log-phase bacterial inoculum using the Bacterial RNA Miniprep Kit (Biomiga, Shanghai, China). An aliquot of RNA from each isolate was subjected to cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher, Shanghai, China). The expression levels of pmrC, pmrA and pmrB were assessed by the qRT-PCR assay.7 The gapA housekeeping gene was used as a calibrator. The expression levels of the three genes in the heteroresistant isolates were performed by comparing their susceptible and resistant subpopulations. The ∆∆Ct method was used to quantify gene expression. The experiments for each gene were conducted in triplicate.
In order to evaluate the impact of CCCP on colistin resistance and heteroresistance, mcr-1 gene transcription and pmrC gene expression were quantified in colistin-resistant and colistin-heteroresistant isolates, respectively. RNA was extracted from LB broth cultures containing 2 mg/L colistin +10 mg/L CCCP, as described previously.32 The RNA was used to quantify gene expression as previously mentioned. The primers used in the present study are listed in Table 1.7,33
Lipid A Isolation from Whole Cells
Lipid A was isolated by an optimized large-scale mild acid hydrolysis-based protocol.34 Overnight cultures (200 mL at 37°C) in LB broth were harvested via 3220×g for 30min. Bacterial pellets were washed by single-phase Bligh–Dyer mixture (chloroform/methanol/water; 1:2:0.8, v/v) and centrifuged at 3220×g for 15min. The LPS pellets were suspended with sodium acetate buffer (50 mM, PH 4.5) and incubated at 100°C for 30–45min. The reactions were converted into a two-phase Bligh–Dyer mixture (chloroform/methanol/water; 1:1:0.9, v/v) and centrifuged at 3220×g for 15min. The lower phases were removed to clean tubes and dried using rotary evaporation. The dried samples contained whole-cell extracts of lipid A.
Lipid A Characterization by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS)
Dried lipid A samples were resuspended in 100 μL chloroform/methanol mixture (1:1, v/v). A total of 3 μL DHB matrix (20 mg/mL in TA30) were mixed with 3 μL lipid A. Aliquots of mixture were spotted directly onto the well of the MALDI-TOF plate (Ground Steel). The mass spectra were recorded for optimal ion signals in negative mode using a Brukerautoflex MALDI-TOF mass spectrometer (BrukerDaltonics Inc., Billerica, MA, USA). The data were acquired and processed by flexControl and flexAnalysis 3.4 (BrukerDaltonics Inc.).
Statistical Analysis
The differences in the expression levels of each gene of interest were tested using the Student’s t-test. For all analyses, a P value lower than 0.05 (P<0.05) was used for significant differences. All statistical calculations were conducted with the SPSS v.22.0 software (SPSS Inc., Chicago, IL, USA).
Results
Antibiotic Susceptibility Profiles of Colistin-Resistant and Heteroresistant Isolates
Of the 291 E. coli strains, two isolates (DC 8277 and DC 8313) were resistant to colistin with Minimum inhibitory concentration (MIC) of 4 mg/L and 8 mg/L, respectively. PAPs were used as a standard method for determining colistin heteroresistance. Four isolates (DC 8243, DC 8253, DC8446 and DC8471) were detected out of colistin-susceptible E. coli strains. The results indicated that the growth of subpopulations with 16- to 64-fold dilutions exhibited higher colistin MIC than their parental populations. The frequency of heteroresistant subpopulations ranged from 4.0×10−7 to 4.0×10−6. The MICs of colistin against the resistant subpopulations did not change following 1 week of subculturing in antibiotic-free medium. All colistin-resistant isolates and two of four colistin-heteroresistant isolates exhibited multidrug-resistance (Table 2).
Table 2.
Susceptibility Characteristics of the Six Escherichia coli Isolates Studied
| Isolate | Broth MIC (mg/L) | Highest Concentration of Growth in PAPs (mg/L) |
Proportion of Resistant Subpopulations | Resistant Colonies MIC After 1 Week Daily Passages onto Colistin-Free Medium (mg/L) |
Population Analysis Profiling | The MICs of Clinical Routine Antimicrobial Agents (mg/L) a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | FOS | GEN | LEV | MEM | AMK | CIP | IPM | CTX | COL +CCCP |
||||||
| DC8277 | 4 | 8 | NA | NA | resistant | 2 | 1 | 16 | ≥8 | 0.06 | 1 | ≥8 | 0.5 | 32 | 0.5 |
| DC8313 | 8 | 8 | NA | NA | resistant | 32 | 1 | 1 | ≥8 | 0.015 | 2 | ≥8 | 0.25 | 8 | 1 |
| DC8243 | 0.25 | 16 | 2.2×10−6 | 16 | Hetero-resistant | 8 | 2 | 1 | 0.25 | 0.03 | 2 | 0.125 | 0.25 | 16 | 0.06 |
| DC8253 | 0.25 | 16 | 4.0×10−7 | 8 | Hetero-resistant | 1 | 2 | 1 | 0.015 | 0.06 | 2 | 0.125 | 0.25 | 0.06 | 0.06 |
| DC8446 | 0.5 | 8 | 4.0×10−7 | 8 | Hetero-resistant | 16 | 2 | 64 | 4 | 0.06 | 8 | ≥8 | 0.125 | ≥128 | 0.125 |
| DC8471 | 0.5 | 8 | 4.0×10−6 | 8 | Hetero-resistant | 16 | 1 | 64 | 0.5 | 0.06 | 2 | 1 | 0.25 | ≥128 | 0.25 |
Notes: aFor heteroresistant isolates, the objects of MICs detection are resistant subpopulations selected from PAPs.
Abbreviations: CAZ, ceftazidime; FOS, fosfomycin; Gen, gentamicin; LEV, levofloxacin; MEM, meropenem; AMK, amikacin; CIP, ciprofloxacin; IPM imipenem; CTX, cefotaxime; COL, colistin; CCCP, carbonylcyanide m-chlorophenylhydrazone.
Analysis of Mobile Colistin-Resistance (mcr) Mechanism
The mcr genes in the colistin-resistant and heteroresistant isolates were analyzed by PCR. Both of the two resistant isolates carried the mcr-1 gene, which was not present in any of the heteroresistant isolates, while mcr-2 and mcr-3 were not detected in all studied isolates (Table 3).
Table 3.
Analysis of Mobile Colistin Resistance (mcr) Genes and TCS Regulatory Pathways in (Hetero)resistant E. coli Isolates
| Isolates | Mobile Colistin Genes (mcr) a |
TCS Regulatory Pathways b | ||||||
|---|---|---|---|---|---|---|---|---|
| mcr-1 | mcr-2 | mcr-3 | pmrA | pmrB | phoP | phoQ | mgrB | |
| Colistin resistant | ||||||||
| DC 8277 | + | – | – | / | / | / | / | / |
| DC 8313 | + | – | – | / | / | / | / | / |
| Colistin heteroresistant | ||||||||
| DC 8243 | – | – | – | / | L14R | / | / | / |
| DC 8253 | – | – | – | / | / | / | / | / |
| DC 8446 | – | – | – | / | / | / | / | / |
| DC 8471 | – | – | – | / | P95L | / | / | |
Notes: In colistin-resistant group, the sequences were compared with the colistin-susceptible Escherichia coli ATCC 25,922. In colistin-heteroresistant group, the comparison was carried on between susceptible and resistant subpopulations. a +, mcr-positive. -, mcr-negative. b/, mutation was not detected. L, Leu, leucine; R, Arg, arginine; P, Pro, proline. L14R, corresponding to the 14th amino acid substitution of pmrB from Leu to Arg; P95L, corresponding to the 95th amino acid substitution of pmrB from Pro to Leu.
Analysis of TCS Mechanism-Involvement in Colistin (Hetero)Resistance
In the present study, pmrA, pmrB, phoP, phoQ and mgrB were sequenced in DNA samples extracted from two resistant isolates and four heteroresistant isolates. No mutation was detected in the two resistant isolates. In the heteroresistant group, we observed a mutation of PmrB (L14R) in DC 8243 and PmrB (P95L) in DC 8471 (Table 3).
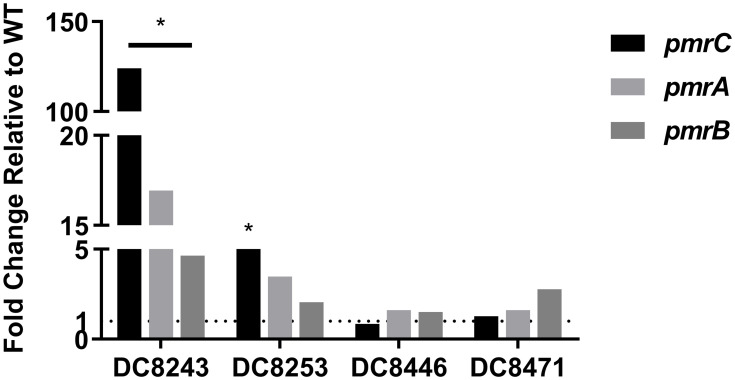
To connect the molecular genetic mutations with the transcriptional expression level, the pmrC, pmrA and pmrB expression levels of four colistin-heteroresistant isolates were analyzed. The results are presented in Figure 1. Compared to its native isolate, DC 8243 indicated significantly higher relative expression levels in pmrC, pmrA and pmrB, which were upregulated by 123.99-, 16.93- and 4.64-fold, respectively (P<0.05). In DC 8253, the expression levels of pmrC, pmrA and pmrB were slightly increased by 5.29- (P<0.05), 3.48- and 2.05-fold. In the isolates DC 8446 and DC 8471, the increment of the expression levels was not found.
Figure 1.
Expression levels of regulatory genes in colistin-heteroresistant isolates. *P<0.05; WT, native isolates.
The Effect of CCCP on Colistin (Hetero)Resistance
We tested the effects of the efflux pump inhibitor CCCP on both colistin-resistant and heteroresistant E. coli isolates. All strains were found to rescue colistin susceptibility (MIC ranged from 0.06 to 1 mg/L) following exposure to CCCP at a concentration of 10 mg/L (Table 2).
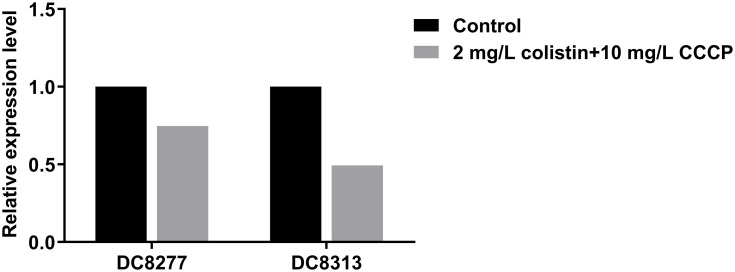
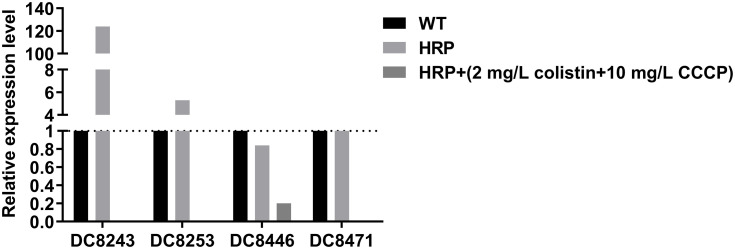
Subsequently, the effects of CCCP on colistin-related gene expression were analyzed on two resistant isolates and four heteroresistant isolates. In the colistin-resistant group, the addition of 2mg/L colistin + 10 mg/L CCCP led to a 0.75-fold and 0.49-fold decrease in the expression levels of the mcr-1 gene with regard to the isolate DC 8277 and the isolate DC 8313, respectively (Figure 2). However, the results were not statistically significant (P>0.05). In the colistin-heteroresistant group, the expression levels of pmrC were considered an indicator and all the isolates exhibited significantly decreased expression levels, ranging from 5- to 1000-fold (P<0.05) (Figure 3).
Figure 2.
Mcr-1 gene expression in colistin-resistant E. coli in the absence of antibiotics and following 2 mg/L colistin +10 mg/L CCCP.
Figure 3.
pmrC gene expression in colistin-heteroresistant E. coli in the absence of antibiotics and following 2 mg/L colistin +10 mg/L CCCP; WT, native isolates; HRP, heteroresistant subpopulations.
Lipid A Modifications in Colistin-Resistant and Heteroresistant Isolates
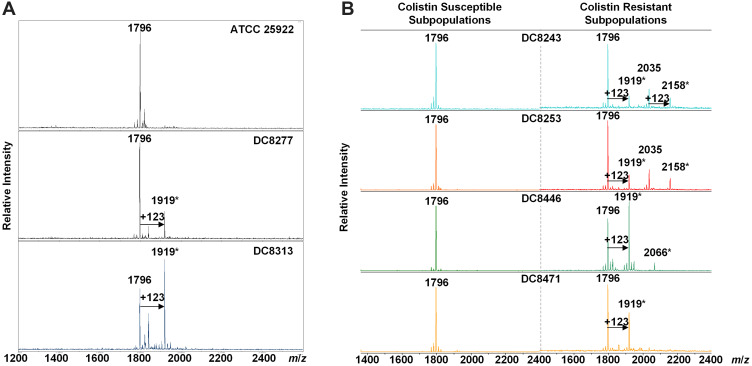
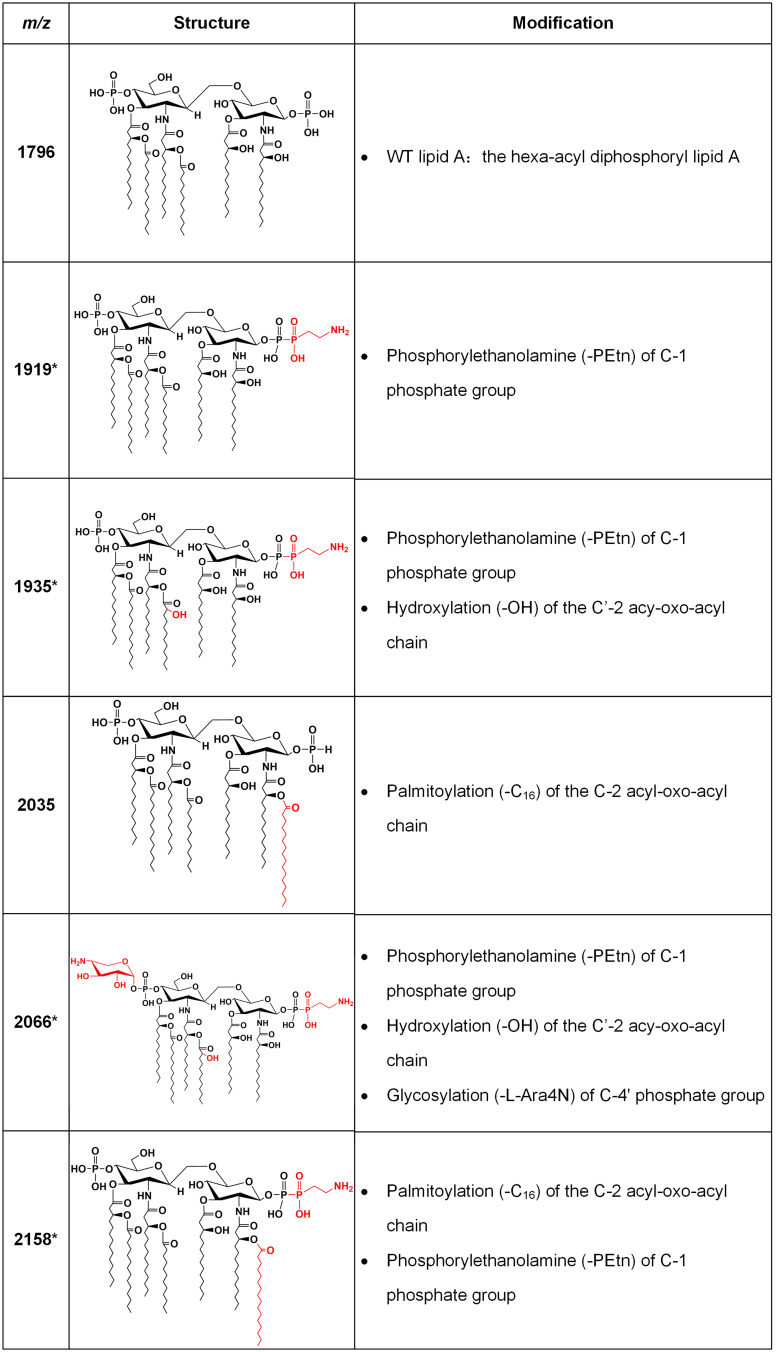
Lipid A was extracted from (hetero)resistant strains and profiled using MALDI-TOF MS to analyze modifications associated with observed genetic alterations. The mass spectrum for lipid A from the colistin-susceptible and reference strain ATCC 25,922 indicated major peaks at the mass/charge ratios (m/z) of 1796, which have been known to correspond to the hexa-acyl diphosphoryl lipid A.
Compared with the mass assigned to native lipid A, the additional peak was observed at the m/z of 1919 on MALDI-TOF spectra obtained from all two resistant isolates and four heteroresistant isolates, which was associated with an addition of phosphoethanolamine (PEtn) moiety (m/z123 mass units) at the C-1 of the native lipid A (Figure 4A). The colistin-heteroresistant isolates, DC 8243 and DC 8253 demonstrated the presence of ions at m/z 2035 and 2158, which resulted from a palmitoylation (m/z239 mass units) at the C-2 position of the structures identified with m/z of 1796 and 1919. An additional peak at m/z 2066 was observed for the isolate DC 8446. This peak was assigned to the addition of a 4-amino-4-deoxy-L-arabinose (L-Ara4N) moiety (m/z131 mass units) onto the phosphate group at position C’-4 of native lipid A with concomitant addition of the hydroxylation group (m/z16 mass units) at C’-2 (Figure 4B). These findings were consistent with previous studies.35 The lipid A structures and the corresponding m/z values of these signature ions are shown in Figure 5.
Figure 4.
MALDI-TOF of E. coli with differential colistin susceptibility. (A) The MS profiles of colistin-resistant isolates. *, Lipid A modification; (B) The MS profiles of colistin-heteroresistant isolates. *Lipid A modification.
Figure 5.
Lipid A structures with corresponding m/z values found in clinical isolates. Lipid A m/z values and molecular structures found in mass spectra of the E. coli clinical isolates are shown, with descriptions of the modifications responsible for the observed mass shifts. The asterisks denote ions associated with colistin resistance. The modified moieties are highlighted in red.
Discussion
Colistin has been proposed as a last-line antibiotic for the treatment of MDR Gram-negative pathogens infections. Despite its superior bacterial killing, colistin resistance and heteroresistance have gained global attention and pose a new threat to public health.36,37 In the present study, we presented the first report of the mechanisms of resistance and heteroresistance to colistin in E. coli clinical isolates.
Two colistin-resistant isolates and four colistin-heteroresistant isolates were acquired from E. coli clinical isolates in 2018 by antimicrobial sensitivity testing and PAP, respectively. The resistance rate of E. coli to colistin was 0.69% (2/291), which was slightly higher than that of 2002–2016 noted in our previous study (0.35%, 12/3434).38 Furthermore, the rate of heteroresistance was 1.37% (4/291), which was considerably lower than that noted in other reports.22 The reasons for the differences may be attributed to the sample size, single location and the detection methods used in different regions of the study.39 The four heteroresistant isolates exhibited MICs for colistin at 8–16 mg/L, two of which indicated MDR phenotypes (Table 2).
The mechanisms of colistin resistance were mainly due to chromosomal and plasmid-mediated resistance. In the E. coli, the presence of the plasmid-mediated mcr gene is the most common mechanism of resistance, notably for the case of mcr-1.40–44 We found that both of the resistant isolates possessed mcr-1 (Table 3), which was consistent with previous findings.40–44
By contrast, colistin-heteroresistant strains were more common than resistant strains.45 El- Halfawy et al demonstrated that heteroresistance was an emerging field that required further investigation.46 Despite certain reports that have been published regarding heteroresistance in the past years, the perception of colistin heteroresistance in E. coli is relatively limited and remains undiscovered. Therefore, we examined the possible mechanisms by analyzing the role of PmrAB and PhoPQ TCS. The results indicated that the strains DC 8243 and DC 8471 exhibited mutations in pmrB and led to amino acid substitutions (L14R and P95L in PmrB, respectively)(Table 3). Mutations in the pmrB gene can activate PmrAB TCS, which causes the upregulation of the pmrCAB operon.47 Additional studies indicated that the expression levels of pmrC, pmrA and pmrB in strain DC 8243 were significantly upregulated (P<0.05). In comparison, strain DC 8471 did not show the corresponding changes, indicating that the substitution in the PmrB P95L was a synonymous mutation, or included unknown compensation mutants. It is interesting to note that pmrCAB levels were upregulated in strain DC 8253 and contained no mutations of the aforementioned genes, suggesting that other genes seem to be involved in colistin heteroresistance (Figure 1).
Sophie et al identified that the efflux pump inhibitor CCCP could reduce colistin susceptibility in Gram-negative bacteria with diverse molecular mechanisms.33 The results demonstrated that the reversal effects with a colistin MIC were changed ≥8 fold in all the studied isolates (Table 2). The effect of CCCP on heteroresistant isolates was much greater than that noted on resistant isolates, speculating that the inhibitory role of CCCP was more critical for colistin heteroresistance. Furthermore, transcriptomic analysis of regulatory genes was conducted. In the colistin-resistance group, the association of colistin and CCCP inhibited the transcription of the mcr-1 gene (Figure 2), as previously reported.26 However, in the colistin heteroresistance group, the higher inhibitory activity with regard to the expression of pmrC ranged from 5- to 1000-fold (Figure 3). Interestingly, a certain correlation was noted between the decreased MIC and the altered transcription of the genes. However, it remains unknown how CCCP contributed to the reversal of the susceptibility of colistin. This may have been caused either by inhibition of efflux pumps19,48–50 or by an unknown action of CCCP,51 as previously described. Thorough studies are still warranted on the underlying inhibition of efflux pumps in order to fully elucidate the complex mechanisms of colistin resistance and heteroresistance.
It has been reported that the modification of lipid A, such as the addition of L-Ara4N, PEtn and GalN, is associated with colistin resistance.52,53 In E. coli, the addition of PEtn and/or L-Ara4N was associated with chromosome-encoded resistance mechanisms, such as mutations in the PmrAB or PhoPQ TCS.35 In addition, PEtn can also occur in strains with plasmid-borne mcr genes.54 However, until now, a limited number of studies have reported that the lipid A profile is associated with colistin heteroresistance, notably in E. coli. In the present study, all strains were subjected to MALDI-TOF analysis of extracted lipid A to characterize modifications (Figure 4). The results indicated that two colistin-resistant isolates displayed the PEtn peaks (m/z 1919). The heteroresistant isolate DC 8471 has also demonstrated the PEtn peaks at the m/z of 1919. The isolates DC 8243 and DC 8253, shared the same MS profile with the PEtn peaks at m/z of 1919 and 2158, respectively. It is interesting to note that the isolate DC 8446 exhibited PEtn (m/z 1919) and L-Ara4N (m/z 2066) peaks, whereas it did not contain non-synonymous mutations of the aforementioned genes. Additional studies regarding the mechanisms of colistin heteroresistance should be performed.
Conclusion
In conclusion, the present study provided the first report for colistin-resistance and heteroresistance in E. coli. Colistin exhibited a strong bacteriostatic activity against the tested isolates. The presence of the mcr-1 gene was the main mechanism of colistin resistance, while the PmrAB TCS mainly contributed to heteroresistance against colistin. The present study further suggested that the action of CCCP was particularly important for colistin heteroresistance. Nevertheless, it remains unclear how relevant this phenotype is with reduced treatment efficacy and a poor clinical outcome. Additional studies are required in the future to fully understand the extent of colistin heteroresistance.
Funding Statement
We thank the Planned Science and Technology Project of Wenzhou (no. Y20170204) for providing financial funding.
Ethical Statement
No samples were collected specifically for this research; only anonymized clinical residual samples were collected during routine hospital procedures and subsequently used for this study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed on the journal to which the article will be submitted; gave final approval of the manuscript version to be published; and agreed to be accountable for all aspects of the work. Jianming Cao and Tieli Zhou are joint corresponding authors.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Tian X, Zheng X, Sun Y, et al. Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002–2017. Infect Drug Resist. 2020;13:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajdacs M, Abrok M, Lazar A, Burian K. Comparative epidemiology and resistance trends of common urinary pathogens in a tertiary-care hospital: a 10-year surveillance study. Medicina (Kaunas). 2019;55(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behzadi P, Urban E, Gajdacs M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): an in vitro study. Diseases. 2020;8(2):17. doi: 10.3390/diseases8020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flament-Simon SC, de Toro M, Mora A, et al. Whole genome sequencing and characteristics of mcr-1-harboring plasmids of porcine Escherichia coli isolates belonging to the high-risk clone O25b: H4-ST131Clade B. Front Microbiol. 2020;11:387. doi: 10.3389/fmicb.2020.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caniaux I, van Belkum A, Zambardi G, Poirel L, Gros MF. MCR: modern colistin resistance. Eur J Clin Microbiol Infect Dis. 2017;36(3):415–420. doi: 10.1007/s10096-016-2846-y [DOI] [PubMed] [Google Scholar]
- 6.Cai Y, Lee W, Kwa AL. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther. 2015;13(12):1481–1497. doi: 10.1586/14787210.2015.1093933 [DOI] [PubMed] [Google Scholar]
- 7.Phan MD, Nhu NTK, Achard MES, et al. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother. 2017;72(10):2729–2736. doi: 10.1093/jac/dkx204 [DOI] [PubMed] [Google Scholar]
- 8.Shen C, Feng S, Chen H, et al. Transmission of mcr-1-producing multidrug-resistant enterobacteriaceae in public transportation in Guangzhou, China. Clin Infect Dis. 2018;67(suppl_2):S217–S224. doi: 10.1093/cid/ciy661 [DOI] [PubMed] [Google Scholar]
- 9.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 10.Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surveill. 2016;21(27). doi: 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- 11.Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajdacs M. The concept of an ideal antibiotic: implications for drug design. Molecules. 2019;24(5):892. doi: 10.3390/molecules24050892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Rayner CR, Nation RL, et al. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2006;50(9):2946–2950. doi: 10.1128/AAC.00103-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Xu C, Fang R, et al. Resistance and heteroresistance to colistin in Pseudomonas aeruginosa Isolates from Wenzhou, China. Antimicrob Agents Chemother. 2019;63(10). doi: 10.1128/AAC.00556-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charretier Y, Diene SM, Baud D, et al. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018;62(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio. 2018;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong YK, Ko KS. PmrAB and PhoPQ variants in colistin-resistant enterobacter spp. isolates in Korea. Curr Microbiol. 2019;76(5):644–649. doi: 10.1007/s00284-019-01672-1 [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Lin J, Lu H, et al. Deciphering colistin heteroresistance in Acinetobacter baumannii clinical isolates from Wenzhou, China. J Antibiot (Tokyo). 2020. [DOI] [PubMed] [Google Scholar]
- 19.Telke AA, Olaitan AO, Morand S, Rolain JM. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother. 2017;72(10):2715–2721. doi: 10.1093/jac/dkx215 [DOI] [PubMed] [Google Scholar]
- 20.Silva A, Sousa AM, Alves D, Lourenco A, Pereira MO. Heteroresistance to colistin in Klebsiella pneumoniae is triggered by small colony variants sub-populations within biofilms. Pathog Dis. 2016;74(5). doi: 10.1093/femspd/ftw036 [DOI] [PubMed] [Google Scholar]
- 21.El-Halfawy OM, Valvano MA. Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One. 2013;8(7):e68874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juhasz E, Ivan M, Pinter E, Pongracz J, Kristof K. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J Glob Antimicrob Resist. 2017;11:167–170. doi: 10.1016/j.jgar.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 23.Gajdács M, Ábrók M, Lázár A, Burián K. Differential epidemiology and antibiotic resistance of lactose-fermenting and non-fermenting Escherichia coli: is it just a matter of taste? Biol Futur. 2020;71(1–2):175–182. doi: 10.1007/s42977-020-00016-6 [DOI] [PubMed] [Google Scholar]
- 24.The European committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of mics and zone diameters. version 9.0; 2019. Available from: http://www.eucast.org. Accessed September23, 2020.
- 25.Bergen PJ, Forrest A, Bulitta JB, et al. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother. 2011;55(11):5134–5142. doi: 10.1128/AAC.05028-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100 Wayne, PA: Clinical and Laboratory Standards Institute; 2019 [Google Scholar]
- 27.Spengler G, Kincses A, Gajdacs M, Amaral L. New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules. 2017;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao L, Li X, Xu Y, Shen J. Prevalence and molecular characteristics of mcr-1 colistin resistance in Escherichia coli: isolates of clinical infection from a Chinese University hospital. Infect Drug Resist. 2018;11:1597–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surveill. 2016;21. [DOI] [PubMed] [Google Scholar]
- 30.Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Q, Yu W, Zhou K, et al. Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front Microbiol. 2017;8:2262. doi: 10.3389/fmicb.2017.02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron SA, Rolain JM. Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and gram-negative bacteria. J Antimicrob Chemother. 2018;73(7):1862–1871. doi: 10.1093/jac/dky134 [DOI] [PubMed] [Google Scholar]
- 33.Chabou S, Leangapichart T, Okdah L, Le Page S, Hadjadj L, Rolain JM. Real-time quantitative PCR assay with Taqman((R)) probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016;13:71–74. doi: 10.1016/j.nmni.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hankins JV, Madsen JA, Needham BD, Brodbelt JS, Trent MS. The outer membrane of gram-negative bacteria: lipid A isolation and characterization. Methods Mol Biol. 2013;966:239–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dortet L, Bonnin RA, Pennisi I, et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J Antimicrob Chemother. 2018;73(12):3359–3367. [DOI] [PubMed] [Google Scholar]
- 36.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17(8):479–496. doi: 10.1038/s41579-019-0218-1 [DOI] [PubMed] [Google Scholar]
- 38.Lu H, Wang C, Dong G, et al. Prevalence and molecular characterization of Escherichia coli clinical isolates carrying mcr-1 in a Chinese teaching hospital from 2002–2016. Antimicrob Agents Chemother. 2018;62(9). doi: 10.1128/AAC.02623-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeannot K, Bolard A, Plesiat P. Resistance to polymyxins in gram-negative organisms. Int J Antimicrob Agents. 2017;49(5):526–535. doi: 10.1016/j.ijantimicag.2016.11.029 [DOI] [PubMed] [Google Scholar]
- 40.Shen Y, Wu Z, Wang Y, et al. Heterogeneous and flexible transmission of mcr-1 in hospital-associated Escherichia coli. mBio. 2018;9(4). doi: 10.1128/mBio.00943-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daly SM, Sturge CR, Felder-Scott CF, Geller BL, Greenberg DE, Bonomo RA. MCR-1 inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in Escherichia coli. mBio. 2017;8(6). doi: 10.1128/mBio.01315-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulman ZP, Chen L, Walsh TJ, et al. Polymyxin combinations combat Escherichia coli harboring mcr-1 and blaNDM-5: preparation for a postantibiotic era. mBio. 2017;8(4). doi: 10.1128/mBio.00540-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuge X, Ji Y, Tang F, et al. Population structure and antimicrobial resistance traits of avian-origin mcr-1-positive Escherichia coli in Eastern China, 2015 to 2017. Transbound Emerg Dis. 2019;66(5):1920–1929. doi: 10.1111/tbed.13222 [DOI] [PubMed] [Google Scholar]
- 44.Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 2016;7(4). doi: 10.1128/mBio.01191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawley JS, Murray CK, Jorgensen JH. Colistin heteroresistance in acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother. 2008;52(1):351–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28(1):191–207. doi: 10.1128/CMR.00058-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quesada A, Porrero MC, Tellez S, Palomo G, Garcia M, Dominguez L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J Antimicrob Chemother. 2015;70(1):71–74. doi: 10.1093/jac/dku320 [DOI] [PubMed] [Google Scholar]
- 48.Poole K, Lau CH, Gilmour C, Hao Y, Lam JS. Polymyxin susceptibility in Pseudomonas aeruginosa linked to the MexXY-OprM multidrug efflux system. Antimicrob Agents Chemother. 2015;59(12):7276–7289. doi: 10.1128/AAC.01785-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez A, Poza M, Fernandez A, et al. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother. 2012;56(4):2084–2090. doi: 10.1128/AAC.05509-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machado D, Antunes J, Simoes A, et al. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J Med Microbiol. 2018;67(6):740–749. doi: 10.1099/jmm.0.000741 [DOI] [PubMed] [Google Scholar]
- 51.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biochim Biophys Acta. 2011;1807(12):1507–1538. doi: 10.1016/j.bbabio.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 52.Leung LM, Fondrie WE, Doi Y, et al. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci Rep. 2017;7(1):6403. doi: 10.1038/s41598-017-04793-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powers MJ, Herrera CM, Tucker AT, Davies BW, Trent MS. Isolation of lipid cell envelope components from Acinetobacter baumannii. Methods Mol Biol. 2019;1946:233–252. [DOI] [PubMed] [Google Scholar]
- 54.Sun J, Xu Y, Gao R, et al. Deciphering MCR-2 colistin resistance. mBio. 2017;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]