ABSTRACT
Outbreaks of infection by novel avian influenza virus strains in humans cause public health issues worldwide, and the development of vaccines against such novel strains is the most effective method for the prevention of these virus outbreaks. All types of vaccines must be tested for potency before use; thus, quantitative potency assays are needed for influenza vaccines. The single radial immunodiffusion (SRID) assay is considered the gold standard for quantification of influenza virus antigens, and the SRID reference reagents are essential for the determination of vaccine potency. However, it remains debatable whether reference reagents derived from egg-based vaccine platforms can be used to precisely quantify non-egg-derived vaccines; thus, influenza vaccine production using cell-based platforms has attracted increasing attention. To evaluate the utility of reference reagents derived from a cell-based influenza vaccine platform, we prepared cell-based reference reagents from MDCK cell-grown viruses and compared them with egg-derived reference reagents. A primary liquid standard (PLS) was purified from cell-derived candidate influenza vaccine viruses, and hemagglutinin (HA) antigen content was determined by a densitometric method. The produced PLS could be stored at 4°C for more than 10 months. We also established a simple HA protein purification method for goat antiserum preparation, and the performance of the resulting antiserum was compared to that of standard reagents obtained using different production platforms. The results of this study indicate that these reference reagents can be used for both cell-based and egg-based production platforms and that the differences between these two types of platforms are negligible.
KEYWORDS: Standard reagents, hemagglutinin, H7N9 influenza virus, quantification, validation
Introduction
Influenza viruses cause millions of infections every year. According to their serotypes, influenza viruses can be divided into A, B, C and D types, and influenza A viruses can be further classified into different subtypes based on the types of hemagglutinin (HA, H1~ H18) and neuraminidase (NA, N1~ N11) they express.1 Changes in the antigenicity of influenza viruses are typically caused by antigenic shift and antigenic drift. Antigenic shift occurs when two or more viruses exchange genetic material (a process called reassortment), such as when avian or swine influenza viruses introduce new antigens, usually HA and NA, into human influenza viruses. Reassorted human influenza viruses bearing foreign antigens have high potential to cause pandemics, such as the H1N1 pandemic in 2009.2 In contrast, antigenic drift refers to a mutation caused by viral replication that leads to a change in antigenicity, which could cause influenza vaccines to lose efficacy over time.3 Given the occurrence of such mutation, the WHO renews seasonal influenza vaccines every few years.
Vaccination is the most effective method for the prevention of viral infection. Therefore, a standard method for quantifying the active antigen(s) of a vaccine is essential. The single radial immunodiffusion (SRID) assay is the gold standard for HA protein quantification in influenza vaccines. In this technique, which was developed by John Wood in 1972, the formation of a precipitation ring by influenza HA antigens and corresponding strain-specific anti-HA polyclonal antibodies is quantified in an agarose gel.4 The SRID assay is a relatively simple method for the quantification of biologically active HA proteins. This technique is used for vaccines obtained using the product-release method recommended by the WHO and national regulatory agencies.5 The reference reagents used in the SRID assay are produced primarily by egg-derived candidate vaccine viruses (CVVs).
Several cell-based production systems have been developed to rapidly respond to the possibility of a pandemic influenza outbreak.6-8 The influenza virus vaccine FLUCELVAX, produced by Seqirus using an MDCK cell culture system, was recently approved by the US FDA.9 Because an increasing number of influenza vaccines are produced using mammalian cells, it is important to determine whether egg-derived reference reagents can be used to precisely quantify vaccine products obtained from cell-based platforms.10,11 In this study, we prepared an influenza HA antigen for H7N9 influenza vaccines using a cell-based platform and compared it with an antigen produced using an egg-based platform. HA was quantified by SDS-PAGE. We also prepared a corresponding strain-specific anti-HA polyclonal antiserum by vaccinating goats. We compared the performances of the HAs produced using the cell-based and egg-based platforms by SRID assay. The results of this study illustrate the suitability of cell-derived antigen could be used as a reference reagent to quantify egg-derived influenza antigen, vice versa.
Materials and methods
Preparation of an influenza virus (H7N9) vaccine in bulk
The H7N9 candidate vaccine strains virus (CCVs) (NIBRG-268) was obtained from the NIBSC. This virus contains six internal genes from the egg-adapted high-growth A/PR8 virus and two surface protein genes (HA and NA) from A/Anhui/1/2013 (H7N9). This strain was further adapted to MDCK cells (ATCC CCL-34), which were purchased from the Food Industry Research and Development Institute, Hsinchu, Taiwan. The influenza strain H7N9 were originally derived from the egg process. The egg-derived CCVs do not grow well in MDCK cell culture. The re-adapted process was done by choosing better replication colonies in the plaque assay. Generally, the process needs to be repeated 3 to 10 times before obtaining the high growth CVVs in the MDCK cell culture. For preparation of the vaccine standard antigen, OptiPro serum-free medium (Invitrogen) was used for MDCK cell growth, and OptiPro supplemented with 2 μg/mL TPCK-trypsin (Sigma) was used for viral replication.12 The harvested virus was inactivated with formaldehyde and then purified by clarification and chromatography.13 This final bulk-produced H7N9 virus was considered the cell-derived PLS.
Virological assays
HA titrations were conducted in conical-bottom 96-well microplates using turkey red blood cells (RBCs) following a standard technique.14 Subsequently, 0.5% turkey RBCs were added to each well, and the plates were examined 40 min later for agglutination. The total protein volume was detected using a Lowry protein assay kit (Thermo).13 For the HI assay, serum was treated with cholera filtrate (receptor-destroying enzyme, RDE; Sigma) overnight at 37°C and inactivated by RDE at 56°C for 30 min using an initial dilution of 1:40 in PBS and two-fold serial dilutions in PBS. Mixed antigen samples (8 HA units/50 μL) were then incubated at room temperature (RT) for 15 min, and 0.5% turkey RBCs were added to each well. Agglutination was observed after 40 min. The reciprocal values of the highest serum dilutions that completely inhibited HA represented the HI titers.15
Calibration of the PLS
The method used for calibration of the PLS has been described in a WHO technical report.16 The PLS was analyzed by deglycosylation treatment using a modified method based on previous reports.17-19 Approximately 5 μg of viral protein was deglycosylated using PNGase F (New England Biolabs). The deglycosylated samples were separated by 10% SDS-PAGE and stained using a Colloidal Blue Staining Kit (Invitrogen). The densitometry quantification was carried out using the scanner.
The HA content of PLS was calculated based on the ratio between HA (HA1 and HA2) was quantified using TotalLab software (10-0100-70 v11.4) and the total protein content which was determined by Lowry assay. This analyzed mothed followed by Ruth Harveya et al., 2008.20
Trypsin digestion and protein identification by LC-MS/MS
Reduction and alkylation of the PLS were performed with DTT and iodoacetamide (IAM), respectively. The denatured PLS was further mixed with trypsin at a weight ratio of 1:20 for digestion at 37°C overnight. The resulting peptide fragments were analyzed using an ESI-Q-TOF mass spectrometer (Waters SynaptTM G1 HDMS) equipped with an ultra-performance liquid chromatography (UPLC) system (nanoACQUITY, Waters). A sample volume of 5 μL was injected, concentrated with a C18 trap column (180 mm id x 20 mm, 5 μm, Waters) and then separated with a C18 column (75 μm id × 10 mm, 1.7 μm, Waters). The survey scan was performed from m/z 400 to 1600, and the MS/MS scan was performed from m/z 50 to 1990. The threshold to switch from MS to MS/MS was 40 counts, and the run was then switched back to MS until the signal reached less than 10 counts or until 2.4 s had passed. For protein identification, the raw data from the MS/MS spectra were transferred to the peak list (PKL) using MassLynx 4.0 Global ProteinLynx. The Mascot server (Matrix Science, version 2.4.1) was used for the database search. The mass tolerance for both precursor and fragment ions was set to 0.2 Da. Carbamidomethyl and oxidization were set as the fixed and variable modifications, respectively. One missed trypsin cleavage was allowed in the MS/MS ion search.
Preparation of XHA protein from H7N9 influenza virus
The preparation method was modified based on a previous report.21 Cell-derived H7N9 influenza virus was mixed with Triton X-100 (Sigma) at a concentration of 0.5% containing β-mercaptoethanol (1:1000, v/v) and allowed to react with gentle shaking for 3 h at RT. The treated virus was centrifuged at 50,000 rpm and 4°C for 90 min using a benchtop ultracentrifuge (Beckman), and the supernatant was subjected to continuous sucrose gradients of 10% to 50% for 18 h at 4°C using an ultracentrifuge. The X-100-treated HA protein, XHA, was collected and dialyzed, and the HA content was measured using an SRID assay.
Preparation of BHA protein from H7N9 influenza virus
The preparation method was modified based on a previous report.18 Cell-derived H7N9 influenza virus was mixed with bromelain enzyme at a concentration of 0.1 U/mL (Sigma-Aldrich) containing β-mercaptoethanol (1:1000, v/v) and reacted for 3 h at RT with gentle shaking. The subsequent steps used for BHA preparation were identical to the steps used for XHA protein purification and SRID confirmation.
Preparation of H7 antiserum
All immunization and blood collection steps were performed based on an animal protocol approved by the NHRI. Two three-month-old goats that were confirmed to be seronegative for H7N9 influenza virus were used in this study. For the primary immunization, the goats were inoculated intramuscularly with 20 μg of XHA mixed with Freund’s complete adjuvant (FCA). A further dose of 10 μg of XHA mixed with Freund’s incomplete adjuvant (FIA) was administered 2 weeks later. Serum was collected 4 weeks after inoculation, and the HI titer was then measured. Final, the serum materials were stored at −20°C until later experiments.
SRID assay for HA protein determination
The commercial standard reagents, including antigen H7N9 (NIBSC 14/250) and H7 antiserum (NIBSC 13/180), were purchased from the NIBSC. The test samples and HA standard antigens were first treated with 1% Zwittergent 3–14 (Lonza) for 30 min. The treated antigens were then serially diluted and introduced into a slab of 1% agarose gel containing the antiserum, and the reaction was allowed to proceed overnight. The agarose gel was dried and stained with Coomassie blue, and the precipitation ring size was measured. The HA content was further calculated as described previously.22
Results
Analysis of the cell-derived primary liquid standard (PLS) by SDS-PAGE
The cell-derived PLS RG268-01 was purified in bulk from an MDCK cell-adapted RG268 virus (derived from A/Anhui/1/2013 from the National Health Research Institutes [NHRI], Taiwan). Before the SDS-PAGE analysis, RG268-01 was treated with PNGase F to remove surface glycan proteins.17 The ratio of each viral protein was determined by densitometry, and the following six viral proteins were separated on the SDS-PAGE gel (Figure 1 and Table 1): PB1 (79 kDa), NP (59 kDa), NA (54 kDa), HA1 (35 kDa), M1 (27 kDa), and HA2 (23 kDa). The protein size of the PNGase F enzyme was 32 kDa, similar to that of the HA1 subunit protein.19 The viral proteins accounted for 82.5% of the total proteins (PB1: 6.2%, NP: 29.5%, NA: 2.4%, HA1: 22.1%, M1: 12.2%, and HA2: 10.1%), and the HA proteins (both the HA1 and HA2 subunits) accounted for 32.2% of the total proteins. The total viral proteins of RG268 (egg-derived antigen, National Institute of Biological Standard and Control [NIBSC] code: 14/250] accounted for 92.1% of the total proteins (NP: 25.3%, NA: 3.7%, HA1: 21.0%, M1: 32.8%, and HA2: 9.3%), and the HA protein of RG268 accounted for 30.3% of the total proteins. Thus, the percentages of total viral and HA proteins of RG268-01 were similar to those of the H7N9 reference antigen, which showed that the cell-derived PLS RG268-01 had a protein content similar to that of the egg-derived H7N9 reference antigen. Additionally, we identified each band on the SDS-PAGE gel by mass spectrometry (Table 1). The protein bands were searched for unique peptide signals, and their Mascot scores were evaluated, which showed significant reliability (a confidence level of p < .05). Thus, the identification of these proteins was confirmed with the NCBI database (Table 1).
Figure 1.
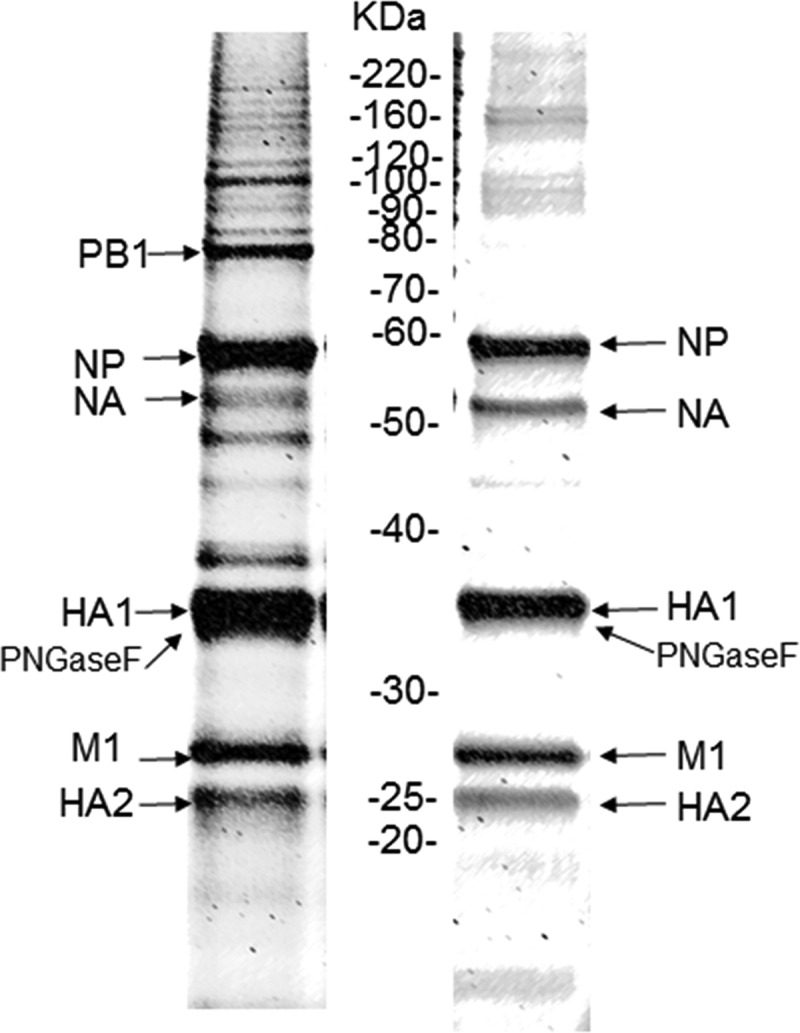
SDS-PAGE analysis of H7N9 influenza virus vaccine antigens.
H7N9 RG268-01 was produced using an MDCK cell platform (cell-based platform, left) at the NHRI, Taiwan. The H7N9 NIBRG-268 reference antigen (NIBSC code: 140/250) was produced using an egg-based platform (right).
Table 1.
Purified H7N9 influenza whole-virus standard antigens identified by mass spectrometry.
| Protein name | LC-MS/MS |
||||
|---|---|---|---|---|---|
| Accession number | Protein hit | No. of observed peptidesb | Mascot scorec | ||
| PB1 | gi | 525338806 | Influenza A virus (A/duck/Anhui/SC702/2013 [H7N9]) | 13 | 42 | 23 | |
| NP | gi | 256259586 | Influenza A virus (Puerto Rico/8/1934) | 2 | 430 | 1 | |
| NA | gi | 459252891 | Influenza A virus (A/duck/Anhui/SC702/2013 [H7N9]) | 12 | 334 | 1 | |
| HA subunit 1 | gi | 475662454 | Influenza A virus (A/duck/Anhui/SC702/2013 [H7N9]) | 7 | 476 | 45 | |
| PNGase F | gi | 157833480 | Glycosylasparaginase from Flavobacterium meningosepticum | 15 | 602 | 57 | |
| Matrix protein 1 | gi | 8486123 | Influenza A virus (A/Puerto Rico/8/34 [H1N1]) | 4 | 150 | 1 | |
| HA subunit 2 | gi | 475662454 | Influenza A virus (A/duck/Anhui/SC702/2013 [H7N9]) | 5 | 412 | 59 | |
aThe masses of proteins in SDS-PAGE gels (Figure 1) were calculated using TotalLab quantification software.
bThe observed peptides include all peptides that showed differences in sequence, modification or charge.
cThe protein hit is the protein with the highest protein score.
dThe Mascot score was used to identify the protein with the highest score, and the number after “|” indicates the significant protein score (p < 0.05).
Quantification and stability analysis of the cell-derived PLS
To evaluate the stability of the cell-derived PLS, the deviation from the protein quantity obtained by densitometry was first established. The HA levels of the reference antigens RG268, RG6 (NIBSC code: 07/290) and RG14 (NIBSC code: 05/204) were quantified by SDS-PAGE and compared with the levels given by the NIBSC, and the results showed that the HA levels for RG268, RG6, and RG14 deviated by 15%, 1.6%, and 4.3%, respectively, from those specified by the NIBSC (Table 2). These findings suggested that the deviations in protein levels obtained using this quantification method are less than 15%. The HA levels of the PLS and RG268-01 measured by SDS-PAGE and the SRID assay using the RG268 reference reagent were 111.9 and 109.3 μg/mL, respectively, representing a deviation of 8.3% (Table 3). In addition, the stability of the PLS was tested, and the proportion and concentration of HA protein remained consistent over time. After 10 months of storage at 4°C, the HA level of RG268-01 was determined to be 103.1 μg/mL by SDS-PAGE and 108.9 μg/mL by SRID. The deviation was 4.6% (Table 3). This result indicated that the cell-derived PLS RG268-01 was stable during long-term storage.
Table 2.
Calibration of the primary liquid standard of the influenza antigen.
| Component | Method | Strain |
H7N9 NIBRG-268a |
H5N1 IBCDC-RG6b |
H5N1 NIBRG-14c |
|---|---|---|---|---|---|
| ID | NIBSC code: 14/250 | NIBSC code: 07/290 | NIBSC code: 05/204 | ||
| Primary liquid standard (PLS) | Lowry total protein assay (μg/mL) | Mean | 155 | 292.0 | 532.3 |
| Hemagglutination assay (% total protein)b |
Mean ± SD | 20.7 ± 0.5 | 33.8 ± 0.6 | 25.9 ± 1.0 | |
| Hemagglutination assay (μg/mL) | Mean ± SD | 32.1 ± 0.8 | 97.4 ± 1.6 | 57.5 ± 1.7 | |
| Reference antigen | Hemagglutinin (μg/mL)d | 37 | 99 | 60 | |
| PLS deviation (%) | 15 | 1.6 | 4.3 |
aThe H7N9 influenza reference antigen (A/Anhui/01/2013 NIBRG-268) was obtained from the NIBSC.
bThe H5N1 influenza Cadel II reference antigen (A/Anhui/1/05 IBCDC-RG6) was obtained from the NIBSC.
cThe H5N1 influenza Cadel I reference antigen (A/Vietnam/1194/04 NIBRG-14) was obtained from the NIBSC.
dThe concentration of HA was determined using the NIBSC datasheet.
Table 3.
Stability of A/Anhui/01/2013 (H7N9) RG268-01 from the MDCK cell-derived influenza reference standard antigen.
| Component | Method | Initial | 1 month | 2 months | 10 months | |
|---|---|---|---|---|---|---|
| Primary liquid standard | Lowry total protein assay (μg/mL) | Mean SD | 316.8 ± 5.2 | 285.1 ± 4.5 | 290.7 ± 0.1 | 297.2 ± 6.1 |
| Hemagglutination assay (% total protein) | Mean SD | 35.3 ± 0.7 | 36.6 ± 0.2 | 33.9 ± 1.1 | 34.7 ± 2.4 | |
| Hemagglutination assay (μg/mL) | Mean SD | 111.9 ± 2.1 | 104.3 ± 0.7 | 98.5 ± 3.1 | 103.1 ± 14.7 | |
| H7N9 antigen | SRID assay (μg/mL) | Mean SD CV (%) | 109.3 ± 9.1 8.3 | 110.8 ± 7.2 13.1 | 109.2 ± 17.7 16.1 | 108.9 ± 6.0 4.6 |
Preparation of standard antiserum using cell-derived H7N9 viruses
For standard serum preparation, HA proteins were further purified from cell-derived H7N9 viruses. According to the NIBSC protocol, bromelain can be used for the removal of HA proteins from whole virus particles. In addition, the nonionic detergent Triton X-100 has also considered an alternative material for the splitting of whole viruses. After treatment with either bromelain or Triton X-100, the HA protein can be purified by sucrose gradient centrifugation. In this study, we compared these two HA protein purification methods. The percent recovery of HA protein obtained through purification with bromelain treatment (BHA) was 16.1%, while that of HA protein obtained with Triton X-100 treatment (XHA) was 46% (Table 4). These data showed that the recovery rate of the XHA protein was higher than that of BHA protein.
Table 4.
Comparison of purified H7N9 standard antigens obtained after various cleavage treatments with cell-based antigens.
| Treatment | HA content (μg/mL)a |
HA recovery (%) | |
|---|---|---|---|
| Before | After | ||
| 0.5% X-Triton 100 | 279.1 | 128.7 | 46.0 |
| Bromelain (0.1 U/mL) | 279.1 | 44.9 | 16.1 |
aThe HA content was determined by SRID assay.
For antiserum preparation, goats were immunized with 20 μg of XHA and boosted with 10 μg of XHA every 2 weeks until the HA inhibition (HI) titer peaked, at which time Y-291 antiserum was collected from the immunized goats. An HI assay for evaluation of immunogenicity revealed that the titer of the antiserum against the PLS was approximately 640 units/50μl. The antigenicity of Y-291 antiserum were compared to those of the antiserum NIBRG-268 (NIBSC code 13/180), and the data showed that there was no difference in HI titer between the two antisera (Table 5).
Table 5.
HI titers of goat antisera generated by immunization with purified H7 antigens.
| Immunogen | Antiserum |
|
|---|---|---|
| NIBSC code: 13/180a | NHRI: Y-291b | |
| H7N9 (RG268-01) cell-derived antigen | 640 | 640 |
| H7N9 (NIBRG-268) egg-derived antigen c | 640 | 640 |
aNIBSC code: 13/180: An antiserum reagent prepared in sheep SH596 cells yielded the purified cell-derived HA of NIBRG-270 (A/Anhui/1/2013 x A/PR/8/34).
bY-291: The purified cell-derived HA was prepared by immunization of goats with the antiserum reagent (A/Anhui/01/2013 (H7N9), RG268-01).
cThe egg-derived antigen of influenza A/Anhui/1/2013 (H7N9) (NIBRG-268) was purchased from the NIBSC (code: 14/250).
Comparison of SRID reagents obtained using cell-based and egg-based platforms
To evaluate the SRID reagents obtained using cell-based and egg-based platforms, cell-derived reference reagents (C-antigen and C-serum) and egg-derived reference reagents (E-antigen and E-serum) were assessed by SRID assay in various combinations (Table 6): C-antigen + C-serum, C-antigen + E-serum, E-antigen+ C-serum, and E-antigen + E-serum. The R-squared values of the four combinations were approximately 0.99, the HA levels in the tested samples ranged from 108.9 to 120.9 μg/mL, and the deviation for the four combinations was less than 10%. These data indicated that both cell-derived and egg-derived reagents could be used for the SRID assay and yield similar results. Therefore, the production platform (egg-based or cell-based) used to generate the reference reagents did not affect HA quantification.
Table 6.
Analysis of the cross-relation between cell-derived and egg-derived SRID reagents.
| Standard antiserum | Standard antigen |
Standard curve R-squared value |
H7N9 antigen (μg/mL) |
CV (%) |
|---|---|---|---|---|
| Cell-derivedb | Cell-derived | 0.9965 | 120.9 ± 8.9 | 7.3 |
| Cell-derived | Egg-derivedd | 0.9971 | 112.4 ± 11.7 | 10.1 |
| Egg-derived | Egg-derived | 0.9960 | 108.9 ± 6.0 | 5.5 |
| Egg-deriveda | Cell-derivedc | 0.9911 | 111.9 ± 19.0 | 16.9 |
aThe egg-derived standard antiserum reagent (NIBSC code: 13/180) was purchased from the NIBSC.
bThe cell-derived standard antiserum reagent was Y-291, which was obtained using purified cell-derived HA prepared for goat immunization with the antiserum reagent.
cThe cell-derived standard antigen (NHRI RG268-01) was prepared from MDCK cells. The HA protein content was found to be 120 μg/mL by SDS-PAGE analysis.
dThe egg-derived antigen of influenza A/Anhui/1/2013 (H7N9) (NIBRG-268) was purchased from the NIBSC (code: 14/250).
Discussion
Vaccination is an important technology for the prevention of influenza outbreaks. In the past, egg-based platforms have been used to produce influenza vaccines because of manufacturing strategies;23 however, recent studies have shown that egg-based platforms may not be suitable for the production of vaccines for particular influenza strains, particularly H3N2 virus strains.24 Cell-based platforms are associated with more flexible production timelines and higher capacities and are thus considered alternatives to egg-based platforms.6,7,25 Some vaccines produced through alternative strategies, such as FLUCELVAX9 and Flublok,26 have already been approved by the FDA.
The HA proteins on the influenza virus surface have been recognized as the active ingredients in influenza vaccines. Thus, the establishment of a precise method for HA protein quantification is a critical issue for the vaccine industry. The current standard method for HA quantification, the SRID assay, has been accepted by national regulatory agencies.27 The HA standard antigen and antiserum used for SRID assays are provided by Essential Regulatory Laboratories (ERLs). In this study, an SRID reagent was prepared with a cell-based platform and used to clarify whether reference reagents derived using egg-based platforms can precisely quantify cell-derived products.11 In our previous study, we successfully established a production and purification process using MDCK cells.12,13 In this study, we prepared cell-derived PLS as a purified bulk material from MDCK cell-adapted RG268 virus and quantified the HA protein concentration by SDS-PAGE. In accordance with the WHO16 guidelines, the HA protein ratios were calculated as the ratios of measured HA1 and HA2 subunit protein levels to total protein levels. Deglycosylation of the PLS significantly improved the separation of the HA1 and HA2 subunit protein bands from the other protein bands.17 However, the protein band of HA1 (35 kDa) was very close to that of PNGase F (deglycosylase, 33 kDa) in the SDS-PAGE gel (Figure 1), and, each band was confirmed by MS, and this method was found to be very reproducible. Fortunately, the statistical analysis software could effectively eliminate the noise from the PNGase F signal in the SDS-PAGE gel. The HA1 and PNGase F mixed band signal deduction the only PNGase F group band signal (control group) which the results for quantified the HA1 from mixed protein band in these calculations on the SDS-PAGE. The results showed similar to reported that has very reproducible and consistent.20
In this study, the PLS satisfied the WHO criteria and the protein content, with variability less than ± 15% CV, accounts for between 20 and 50% of the total protein content.16,28 The longevity of a PLS is also important for influenza vaccine preparations; the reference PLSs used in this study are very stable for at least 28 months when stored at 4°C, as demonstrated in our previous study.29 Additionally, the virus strain was re-adapted may cause the change of the antigenicity due to the host-cell dependency. Hence, the antigenicity always is confirmed by the HI assay. The tested virus strains always compared with the standard reference antigen and antibody from the NIBSC, UK. (Table 5). The HA gene could mutate in certain virus strains during a mammalian-cell passage, but no changes in antigenicity. This phenomenon also was referred to by a previous study.30 The H7N9 antigen used as a reagent for the SRID assay was identified by SDS-PAGE and LC-MS/MS. Based on the above studies, the PLS exhibited a purity similar to that of the reference antigen from the NIBSC, showed high quality, and was predicted to be stable for at least 2 years.
Suitable antisera are needed as reagents for SRID assays. The preparation process of these reagents, from virus production to animal immunization, is time-consuming (usually requiring 3 ~ 6 months) and difficult.5 The suitability of SRID assays for quantification of cell-based vaccines remains debatable;11 indeed, it is possible that such assays are inaccurate when used for vaccines obtained using cell-based production platforms. In this study, an HA reference antiserum was prepared and calibrated according to NIBSC protocols. For preparation of the antiserum, the PLS was first subjected to truncation and purification the HA protein; the purified HA protein preparations were subsequently injected into goats for antiserum generation. A previous study showed that some influenza strains or production platforms might not be conducive to purification of HA protein from whole influenza viruses, particularly pandemic viruses, and that immunized animals might not always generate specific antisera at high titers.24,31 We established a simple approach for obtaining HA immunogens using a cell-derived platform with 0.1% Triton X-100. Notably, we successfully obtained the H7N9 antiserum Y-291 from a goat at a high titer; thus, we obtained specific immunization antisera from both cell- and egg-derived platforms, both platforms yielded the same titer. These data showed that the reference antiserum was suitable for the determination of the potency of H7N9 vaccines using SRID assays and that the protocol met the criteria established by the WHO guidelines.28 Similar studies have previously been reported by other scientists.5,32,33
Additionally, the cell-derived antiserum was compared with the egg-derived antiserum through SRID assays. Cross-relation analysis by SRID assay revealed good consistency among the vaccine antigens from different sources. This study illustrated that SRID reagents from different sources exhibited similar effectiveness and suitability, and the cross-relation between cell-derived and egg-derived SRID reagents was confirmed based on this phenomenon. Besides, these cell-based SRID reagents also illustrate the questionable raised by Manceur et al. 2015 regarding the suitability of different platforms.11
In this study, the results showed that cell-derived or egg-derived reference reagents were enabled to quantify both cell- and egg-derived influenza antigens. However, some reported studies showed that these H3N2 vaccine antigens from cell-derived and egg-derived could be protectively different in humans.24,25,34 Thus, the effect of influenza H3N2 quantification on the reference reagent platform was still unclear. In the future, we plan to analyze influenza virus vaccine strains produced through different platforms; we will collect sera from these vaccine antigens and analyze their potencies using the SRID assays. Interested investigators can contact the authors for the Y-291 antiserum reagents for H7N9 influenza and RG268 for use in exploratory research.
Acknowledgments
The authors would like to thank the Ministry of Science and Technology for funding support. Chia-Chun Lai conducted his thesis research under the auspices of the Graduate Program of Biotechnology in Medicine at National Tsing Hua University and the National Health Research Institutes.
Funding Statement
This work was supported by the Ministry of Science and Technology, Taiwan (R.O.C. 102-2622-B-400-001-CC2; 103-2622-B-400-001-CC2).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Freidl GS, Binger T, Muller MA, de Bruin E, van Beek J, Corman VM, Rasche A, Drexler JF, Sylverken A, Oppong SK, et al. Serological evidence of influenza A viruses in frugivorous bats from Africa. PLoS One. 2015;10:e0127035. doi: 10.1371/journal.pone.0127035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrat F, Flahault A.. Influenza vaccine: the challenge of antigenic drift. Vaccine. 2007;25:6852–62. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Saunders-Hastings PR, Krewski D.. Reviewing the history of pandemic influenza: understanding patterns of emergence and transmission. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood JMSG, Newman RW, Seagroatt V. An improved single-radial-immunodiffusion technique for the assay of influenza haemagglutinin antigen: application for potency determinations of inactivated whole virus and subunit vaccines. J Biol Stand. 1977;5:237–47. doi: 10.1016/S0092-1157(77)80008-5. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Xu K, Hashem A, Shao M, Liu S, Zou Y, Gao Q, Zhang Y, Yuan L, Xu M, et al. Collaborative studies on the development of national reference standards for potency determination of H7N9 influenza vaccine. Hum Vaccin Immunother. 2015;11:1351–56. doi: 10.1080/21645515.2015.1032490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MS, Hu AY. A cell-based backup to speed up pandemic influenza vaccine production. Trends Microbiol. 2012;20:103–05. doi: 10.1016/j.tim.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Milian E, Kamen AA. Current and emerging cell culture manufacturing technologies for influenza vaccines. Biomed Res Int. 2015;2015:504831. doi: 10.1155/2015/504831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey S, Vesikari T, Szymczakiewicz-Multanowska A, Lattanzi M, Izu A, Groth N, Holmes S. Clinical efficacy of cell culture-derived and egg-derived inactivated subunit influenza vaccines in healthy adults. Clin Infect Dis. 2010;51:997–1004. doi: 10.1086/656578. [DOI] [PubMed] [Google Scholar]
- 9.Manini I, Domnich A, Amicizia D, Rossi S, Pozzi T, Gasparini R, Panatto D, Montomoli E. Flucelvax (Optaflu) for seasonal influenza. Expert Rev Vaccines. 2015;14:789–804. doi: 10.1586/14760584.2015.1039520. [DOI] [PubMed] [Google Scholar]
- 10.Johannsen RMH, Hinz J, Friesen HJ, Gruschkau H. Quantification ofhaemagglutinin of influenza Tween–ether split vaccines by immunodiffusion. Vaccine. 1985;235–40. doi: 10.1016/0264-410X(85)90114-8. [DOI] [PubMed] [Google Scholar]
- 11.Manceur AP, Kamen AA. Critical review of current and emerging quantification methods for the development of influenza vaccine candidates. Vaccine. 2015;33:5913–19. doi: 10.1016/j.vaccine.2015.07.104. [DOI] [PubMed] [Google Scholar]
- 12.Hu AY, Tseng YF, Weng TC, Liao CC, Wu J, Chou AH, Chao H-J, Gu A, Chen J, Lin S-C, et al. Production of inactivated influenza H5N1 vaccines from MDCK cells in serum-free medium. PLoS One. 2011;6:e14578. doi: 10.1371/journal.pone.0014578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng YF, Weng TC, Lai CC, Chen PL, Lee MS, Hu AY. A fast and efficient purification platform for cell-based influenza viruses by flow-through chromatography. Vaccine. 2018;36:3146–52. doi: 10.1016/j.vaccine.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 14.The WHO Manual on Animal Influenza Diagnosis and Surveillance. [accessed 2002 May 1]. https://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.
- 15.Wang W, Suguitan AL Jr., Zengel J, Chen Z, Jin H. Generation of recombinant pandemic H1N1 influenza virus with the HA cleavable by bromelain and identification of the residues influencing HA bromelain cleavage. Vaccine. 2012;30:872–78. doi: 10.1016/j.vaccine.2011.11.101. [DOI] [PubMed] [Google Scholar]
- 16.Organization WH . Generic protocol for the calibration of seasonal and pandemic influenza antigen working reagents by WHO essential regulatory laboratories. WHO Technical Report Series. 2011.
- 17.Li C, Shao M, Cui X, Song Y, Li J, Yuan L, Fang H, Liang Z, Cyr TD, Li F, et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals. 2010;38:284–89. doi: 10.1016/j.biologicals.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Jing X, Soto J, Gao Y, Phy K, Ye Z. Assessment of viral replication in eggs and HA protein yield of pre-pandemic H5N1 candidate vaccine viruses. Vaccine. 2013;31:4091–97. doi: 10.1016/j.vaccine.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Harvey R, Hamill M, Robertson JS, Minor PD, Vodeiko GM, Weir JP, Takahashi H, Harada Y, Itamura S, Bamford P, et al. Application of deglycosylation to SDS PAGE analysis improves calibration of influenza antigen standards. Biologicals. 2012;40:96–99. doi: 10.1016/j.biologicals.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Harvey R, Wheeler JX, Wallis CL, Robertson JS, Engelhardt OG. Quantitation of haemagglutinin in H5N1 influenza viruses reveals low haemagglutinin content of vaccine virus NIBRG-14 (H5N1). Vaccine. 2008;26:6550–54. doi: 10.1016/j.vaccine.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 21.Herrler GNA, Meier-Ewert H, Bhown AS, Compans RW. Isolation and structural analysis of influenza C virion glycoproteins. Virology. 1981;439–51. doi: 10.1016/0042-6822(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 22.Kluge S, Benndorf D, Genzel Y, Scharfenberg K, Rapp E, Reichl U. Monitoring changes in proteome during stepwise adaptation of a MDCK cell line from adherence to growth in suspension. Vaccine. 2015;33:4269–80. doi: 10.1016/j.vaccine.2015.02.077. [DOI] [PubMed] [Google Scholar]
- 23.Wong SS, Webby RJ. Traditional and new influenza vaccines. Clin Microbiol Rev. 2013;26:476–92. doi: 10.1128/CMR.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, Fonseca K, Winter A-L, Gubbay JB, Krajden M, et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One. 2014;9:e92153. doi: 10.1371/journal.pone.0092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin D, Park KJ, Lee H, Cho EY, Kim MS, Hwang MH, Kim SI, Ahn DH. Comparison of immunogenicity of cell-and egg-passaged viruses for manufacturing MDCK cell culture-based influenza vaccines. Virus Res. 2015;204:40–46. doi: 10.1016/j.virusres.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Manon MJ, Cox RI, Post P, Dunkle L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv Vaccines. 2015;3:97–108. doi: 10.1177/2051013615595595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi Y, Lee S, Kwon SY, Lee Y, Park YK, Ban SJ. Analysis of the proficiency of single radial immunodiffusion assays for quality control of influenza vaccines in Korea. Biologicals. 2017;50:137–40. doi: 10.1016/j.biologicals.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 28.WHO . Generic protocol for the calibration of seasonal and pandemic influenza antigen working reagents by WHO essential regulatory laboratories. Sixty‐second report of the Expert Committee on Biological Standardization, Annex 5 WHO Technical Report Series 979. 2013.
- 29.Tzeng TL, Lai CC, Weng TC, Cyue MH, Tsai SY, Tseng YF, Sung WC, Lee MS, Hu AYC. The stability and immunogenicity of inactivated MDCK cell-derived influenza H7N9 viruses. Vaccine. 2019. doi: 10.1016/j.vaccine.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 30.Lai CC, Weng TC, Tseng YF, Chiang JR, Lee MS, Hu AY. Evaluation of novel disposable bioreactors on pandemic influenza virus production. PLoS One. 2019;14:e0220803. doi: 10.1371/journal.pone.0220803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, Martin W. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother. 2013;9:950–56. doi: 10.4161/hv.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vodeiko GM, Weir JP. Determination of H5N1 vaccine potency using reference antisera from heterologous strains of influenza. Influenza Other Respir Viruses. 2012;6:176–87. doi: 10.1111/j.1750-2659.2011.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmeisser F, Jing X, Joshi M, Vasudevan A, Soto J, Li X, Choudhary A, Baichoo N, Resnick J, Ye Z, et al. A novel approach for preparation of the antisera reagent for potency determination of inactivated H7N9 influenza vaccines. Influenza Other Respir Viruses. 2016;10:134–40. doi: 10.1111/irv.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barr IG, Donis RO, Katz JM, McCauley JW, Odagiri T, Trusheim H, Tsai TF, Wentworth DE. Cell culture-derived influenza vaccines in the severe 2017–2018 epidemic season: a step towards improved influenza vaccine effectiveness. NPJ Vaccines. 2018;3:44. doi: 10.1038/s41541-018-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


