ABSTRACT
Hybrid seeds of several important crops with supreme qualities including yield, biotic and abiotic stress tolerance have been cultivated for decades. Thus far, a major challenge with hybrid seeds is that they do not have the ability to produce plants with the same qualities over subsequent generations. Apomixis, an asexual mode of reproduction by avoiding meiosis, exists naturally in flowering plants, and ultimately leads to seed production. Apomixis has the potential to preserve hybrid vigor for multiple generations in economically important plant genotypes. The evolution and genetics of asexual seed production are unclear, and much more effort will be required to determine the genetic architecture of this phenomenon. To fix hybrid vigor, synthetic apomixis has been suggested. The development of MiMe (mitosis instead of meiosis) genotypes has been utilized for clonal gamete production. However, the identification and parental origin of genes responsible for synthetic apomixis are little known and need further clarification. Genome modifications utilizing genome editing technologies (GETs), such as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (cas), a reverse genetics tool, have paved the way toward the utilization of emerging technologies in plant molecular biology. Over the last decade, several genes in important crops have been successfully edited. The vast availability of GETs has made functional genomics studies easy to conduct in crops important for food security. Disruption in the expression of genes specific to egg cell MATRILINEAL (MTL) through the CRISPR/Cas genome editing system promotes the induction of haploid seed, whereas triple knockout of the Baby Boom (BBM) genes BBM1, BBM2, and BBM3 cause embryo arrest and abortion, which can be fully rescued by male-transmitted BBM1. The establishment of synthetic apomixis by engineering the MiMe genotype by genome editing of BBM1 expression or disruption of MTL leads to clonal seed production and heritability for multiple generations. In the present review, we discuss current developments related to the use of CRISPR/Cas technology in plants and the possibility of promoting apomixis in crops to preserve hybrid vigor. In addition, genetics, evolution, epigenetic modifications, and strategies for MiMe genotype development are discussed in detail.
KEYWORDS: Hybrid vigor, flowering plants, apomixis, CRISPR/CAS system
1. Introduction
Scientific, logistical, and humanitarian approaches are simultaneously involved to ensure food security, starting with farmers and breeders and further extending to policymakers and governments. A universal solution for sustainable food security is difficult owing to differences in cultural values and geographical regions, environments, and technologies. Although these challenges are substantial, there is great potential to increase the efficiency and productivity of current agricultural products. The powerful breeding techniques that we have at our disposal are being used to exploit heterosis in commercially important crops. Hybrid seed production is a successful technology, and hybrid seeds produced either by three-line or two-line systems to achieve supreme qualities, i.e., premium yield and quality along with resistance against biotic and abiotic stresses, have been cultivated for a long time.1 Among the two-line and three-line hybrid development systems, the former is more advantageous than later because in this system, photoperiod-dependent/thermogenic male sterile lines have been successfully developed. Moreover, the application of CRISPR has further reduced the time required for hybrid seed development.2 Recently, a novel male fertility-related gene in wheat, Ms1, was identified, and the biallelic frameshift mutation resulted in complete male sterility that can be utilized for commercial hybrid seed production.3 Furthermore, the knockout of ZmTMS5 and OsTMS5 in maize and rice, respectively, resulted in thermosensitive lines. These lines contain fertile female parts and have been successfully utilized for hybrid seed production.4 Moreover, a transgene-free maize male sterile line was developed through the targeted mutagenesis of the MS8 gene and proved to be a valuable source for hybrid development5 (Fig. 1). However, a major challenge with hybrid crops thus far has been that unlike other crops, their progeny segregate, and the next generation is unable to maintain heterosis.6 These limitations of sexual reproduction make apomixis a feasible strategy for preserving hybrid seed qualities for multiple generations.7
Figure 1.
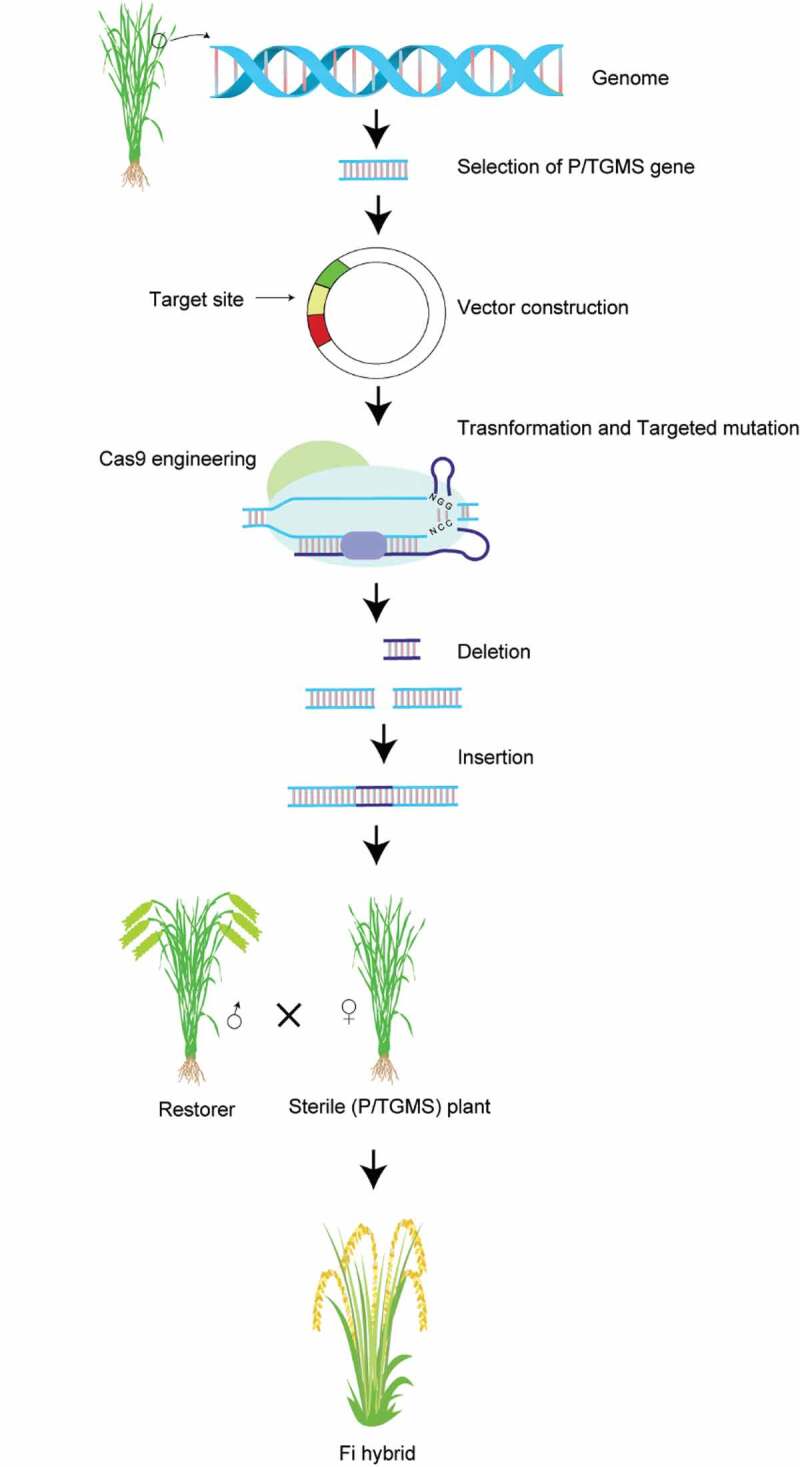
Schematic description of the hybrid development utilizing CRISPR/Cas genome editing system. The two line hybrid development through knockout of P/TGMS = photoperiod/thermo-sensitive genic male sterile genes from elite cultivar. The selection of target in any exon of gene under consideration and later targeted modification through CRISPR/Cas technology helps to induce mutations. The frame shift mutation (Deletion and Insertion) is mostly desirable. The screening of mutants to get male sterile plants, crossing with compatible restorer generate F1 hybrid.
Apomixis exists naturally and produces offspring that are genetically identical to the mother plant. Naturally, the phenomena of apomixis are widespread but exceptional. In 400 families of flowering plants, approximately 10% undergo the phenomenon of apomixis, which constitutes only 1% of the 40,000 species in those families.8 Under natural conditions, multiple developmental pathways controlled by various molecular mechanisms play an integral role in achieving apomixis.9 Apomictic pathways have been categorized as examples of adventitious embryony, a sporophytic type of apomixis, while megaspore mother cells (diplospory) and nearby nucellar cells (apospory) exhibit two gametophytic forms of apomixis. In gametophytic apomixis, a chromosomally unreduced embryo sac develops from diplospory or from apospory in a process termed apomeiosis.10 Parthenogenesis, the development of the unreduced, unfertilized egg into an embryo, constitutes the second step of the apomictic process.11
Synthetic apomixis has been proposed by researchers to overcome the genetic segregation of F1 hybrids of different crops. Genetic analysis of the inheritance of apomixis in different plant species, cereals, grasses, and related genera, i.e., Tripsacum, Pennisetum, Panicum, Bracchiaria, and Paspalum, detected a single chromosome segment responsible for inducing apomixis.12 Nevertheless, efforts to transfer chromosomal segments responsible for the promotion of apomixis remain elusive owing to genetic evolution load.11 In addition, induced mutations cause complete male or female sterility and seldom produce unreduced gametes.13 However, the molecular mechanisms controlling apomixis for clonal seed formation are largely unknown, and novel plant breeding strategies are desirable to unearth the genetic mechanism controlling such interesting phenomena. If synthetic apomixis is introduced in any crop vital for food security, it can help fix and propagate genotypes regardless of the genetic complexity in controlling a particular phenotype, ultimately enhancing the application of heterosis fixation in agriculture.14 Because of these enormous prospects, ingress of apomixis in commercially important crops is considered a hotspot to be studied and explored by plant biologists and the seed industry 15,16 introduced synthetic apomixis in a few fruit crops, i.e., citrus and apple; however, epigenetic modifications involve severe seed abortion, leading to the failure to transfer apomixis in major crops.10 Recent studies have discussed genetic and biotechnological approaches,17 integration of knowledge from various aspects of plant breeding,18 and methods for exploiting genome editing, especially the CRISPR/Cas genome editing system, for improving yield and preservation of heterosis in hybrids.19,20 The scientific community, equipped with genome editing expertise, has played a significant role by transferring relevant and reliable information to beginners in genome editing, in contrast to the proprietary nature of methods utilizing zinc-finger nucleases (ZFNs). In addition, several online platforms are now freely accessible to assist researchers with their efforts related to genome editing, especially using CRISPR.21 Here, in the present review, we discussed details of apomixis along with the utilization of the CRISPR/Cas genome editing system to develop synthetic apomixis as an alternative method to preserve hybrid seed vigor in economically important crops.
2. Evolution of Apomixis
Apomixis is an interesting plant attribute that allows maternal clones via seed production. Apomixis is an intangible but revolutionary characteristic used in plant breeding and for preserving hybrid seed qualities. Recent findings lead to the conclusion that apomicts are useful, with the potential to develop more research interest in the evolutionary process of asexual seed production in flowering plants. It is believed among researchers that apomixis alone has the ability to revolutionize twenty-first century agriculture in both developed and developing countries. Apomixis has been reported in approximately 35 angiosperm families and in more than 300 plant species.22 Unfortunately, except for a few forage grasses and fruit trees, apomixis phenomena are not obvious in crop species. It is commonly believed that apomixis phenomena mostly occur in polyploid genotypes23; however, the discovery of apomixis in diploid species24 abolished the notion that polyploidy was necessary for apomixis.25 Four perennial Cenchrus spp. were found to exhibit natural apomixis, i.e., C. myosuroides is an obligate sexual, diploid apomictic, and C. setigerus, C. echinatus, and C. myosuroides are polyploid and sexual in nature, indicating apomixis without polyploidy. Thus, there is still a quest for in-depth research to challenge the strong but less understood correlation between polyploidy and apomixis. Moreover, the reliable transmission of maternal genotypes to the next generations also needs sufficient scientific investigation due to genetic variability among apomictic populations. There are at least three proposed mechanisms that may cause genetic variability in apomictic plants or species.26 First, new mutations accumulate22; second, irregular sexual reproduction causes recombinant genotypes in populations, a phenomenon often called leaky apomixis27; and third, facultative reproduction occurs.28 Many researchers around the globe have analyzed and discussed these mechanisms in detail29 (Fig. 2).
Figure 2.
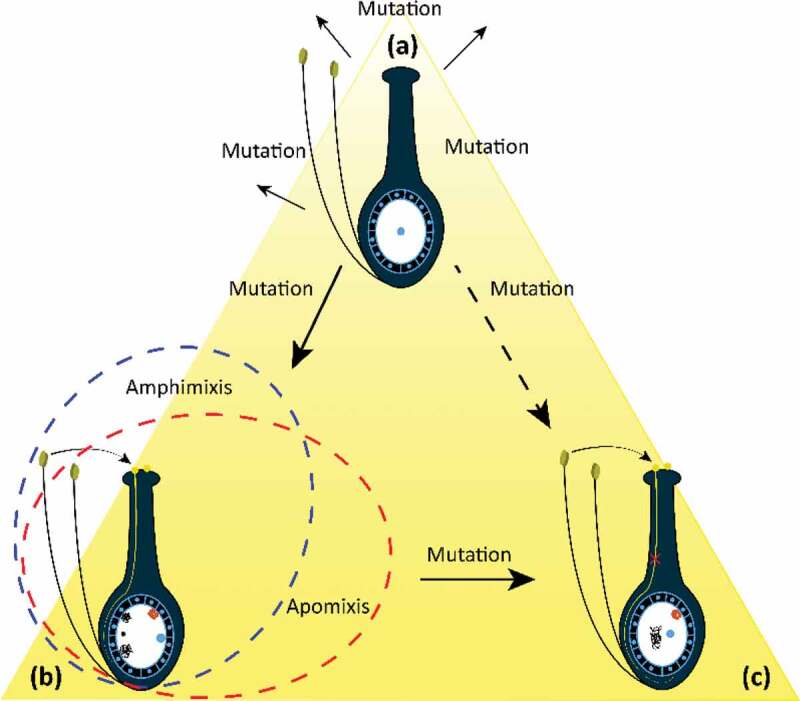
Illustration of the emergence of apomictic types in plants modified from Fei et al. (2019).30 (a) Amphimixis, (b) facultative apomixis, and (c) obligate apomixis.
3. Genetic Basis and Molecular Control of Apomixis
Apomixis is qualitative in nature and involves components of genetic analysis. Molecular studies to unearth the genetic basis for apomixis are unable to explain mysterious phenomena because most apomicts are not important crops.31 There is a well-established association between polyploidy and gametophytic apomixis.32 The discoveries of apomictic phenomena in diploid plant populations disassociated the absolute necessity for polyploidy to occur along with apomixis.25 However, some researchers have suggested that the genetic control of apomixis is not an independent trait but is triggered by epigenetic temporal and spatial modifications of the sexual system.33 Earlier, the genetic control of apomixis was thought to be controlled by recessive genes, while their balance may change after each successful crossing. Recently, a new concept for the inheritance process predicted the involvement of a single major regulatory gene or a set of key dominant genes, allowing megaspore mother cells or somatic nuclear cells to form an embryo sac and an embryo from unreduced egg cells without fertilization.,32,34 However, the spatial and temporal distribution of these genes during developmental processes is still not well understood. The genetic mechanism of asexual reproduction is complicated in nature; therefore, interspecific hybrids or intraspecific hybrids within agamic complexes are widely studied in the context of the segregation (apomeiosis, parthenogenesis, and functional endosperm development) of apomixis. Moreover, the phenotypic analysis of plant generation reproduction either asexually or sexually involves time-consuming cytology or progeny testing. A single gene with regulatory function was initially proposed to promote asexual reproduction. Recently 31 reported that three key components (apomeiotic megaspores, parthenogenic-unreduced egg cells, and modified endosperms) of gametophytic apomixis depend on three independent Mendelian loci of major influence. The relationship between rich retrotransposon regions of heterochromatin and apomictic mechanisms has elevated the possibility for the role of DNA and/or RNA interference in regulating the expression of apomictic genes.35 Some researchers have reported the influence of specific genes encoding novel proteins for new functions that are absent from plants with sexual reproduction and ultimately the identification of candidate genes/loci. Some genes originally fine mapped and characterized in or from species that reproduce sexually may play an important role in promoting apomixis through loss of function or change in function of genes, differentiating between apomictic and sexual pathways. However, the genetic factors controlling apomixis are also associated with some lethal genes influencing the normal functioning of both male and female gametes, which needs further investigation.
Apomictic species exhibit suppressed recombination; however, the genes controlling the various components of apomixis have been identified, and sequencing of these loci has revealed a number of genes with the potential to have critical roles in apomixis. Random amplified polymorphic DNA-based analysis helped in the identification of the apospory-specific genomic region (ASGR) of Pennisetum in both sexual and apomictic plants.36 ASGR sequences revealed that ASGR contains both gene-rich and gene-poor segments, several genes that may play a role in apomictic development, and many classes of transposable elements, and ASGR does not exhibit large-scale synteny with either rice or sorghum genomes but does contain multiple regions of microsynteny with these species.37 The bacterial artificial chromosome (BAC) clone sequencing of ASGR from Pennisetum and Cenchrus revealed 40 putative protein-coding regions, two of which showed sequence similarity to the BABY BOOM (BBM) gene. The BBM gene was identified in Brassica napus as an AP2-domain transcription factor, and overexpression analysis in Arabidopsis showed its involvement in embryo development from vegetative tissues.38 The ASGR-BBM possesses potential and is a candidate gene for the induction and/or maintenance of apomictic events.37 Amplified fragment length polymorphism (AFLP) marker screening in apomictic and sexual plants of Hypericum led to the identification of the locus HYPERICUM APOSPORY. The perfect co-segregation of the AFLP was further utilized to screen the BAC library of Hypericum. The screening helped in the identification of a single clone containing aubiquitin-mediated E3 ligase.39 This ARIADNE 7-like E3 ligase (HpARI) has been proposed as a candidate for the HAPPY locus, as one of the four HpARI alleles within the tetraploid apomict is truncated compared to that in sexual plants. HpARI acts as a negative regulator and interacts with the remaining three alleles. The E3 ligase is responsible for ubiquitin-mediated protein degradation involved in embryo sac development. With roles in mitotic spindle function and nuclear fate in the developing maize embryo sac, the MATH-BTB protein MAB interacts with an E3 ubiquitin ligase component (Cullin3a).40 Any modification in the expression or function of the HpARI E3 ligase may impact embryo sac development and apomixis in Hypericum. Spatial and temporal alterations in the expression of sexual pathway-controlling genes may cause apomixis; therefore, comparative gene expression analysis may provide an opportunity to identify such genes. The low depth express sequence tag (EST) libraries consisting of the apomictic and sexually propagated aposporous grass Brachiaria brizantha revealed sequence similarity among genes involved in female embryo sac development.41 Characterization of identified genes mainly expressed in aposporous initial cells was performed. The helicase BbrizHelic, BbrizAGL6 transcription factor, and BbrizSti1 stress-inducing protein were found to play integral roles in aposporous initial cells.42 Apomixis-associated APOLLO locus is suggested to be one of the important apomixis-related genes in Boechera. APOLLO transcripts are down-regulated in sexual ovules when they enter meiosis and are up-regulated in apomeiotic ovules at the same stage of development.43 APOLLO has both ‘apoalleles’ and ‘sexalleles,’ implying apomixis-associated polymorphisms. In Poa pratensis, cDNA-AFLP helped in the identification and characterization of two candidate genes: SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (PpSERK) and APOSTART.44 Both the PpSERK and APOSTART genes were isolated in two copies, and there were several alleles present within the P. pratensis genome. The PpSERK gene is a tyrosine kinase that switches aposporous initial cells to form and develop embryo sacs in somatic cells.44 APOSTART contains a lipid-binding START domain and is believed to have a role in meiosis. APOSTART may also be related to programmed cell death and the degeneration of nonfunctional megaspores, with one of the two isolated copies (APOSTART1) overexpressed in sexual lines relative to those in apomicts. The Arabidopsis APOSTART1 ortholog is expressed in mature female embryo sacs and developing embryos, and the phenotype of APOSTART1/APOSTART2 double mutants suggests that this gene has a role in embryo and seed development.11 Several other genes identified by several researchers play important roles in controlling asexual reproduction and can be utilized in the future to understand their regulatory functions (Table 1).
Table 1.
Apomixis-related candidate genes and related information.
| Genes | Function/Process involved | Description | Species | References |
|---|---|---|---|---|
| ASGR–BBML | Apospory | Multiple copies of the PsASGRBABY BOOM-like (PsASGRBBML) gene reside within the apospory-specific genomic region | Pennisetum squamulatum | 9 |
| LOSS OF APOMEIOSIS (LOA) |
Apospory | Responsible for apospory | Hieracium Praealtum | 45 |
| Deficiens | Apospory | Deficiens is a member of the B class of floral homeotic regulators that have been shown to specify petal and stamen identity according to the ABC model of floral development | Hieracium piloselloides | 46 |
| PnTgs1-like | Apospory | PnTgs1-like (formerly N69) encodes a trimethylguanosine synthase-like protein whose function in mammals and yeast is critical for development, including reproduction | Paspalum notatum | 47 |
| GID1 | Apospory | Gibberellin-insensitive DWARF1 | Brachiaria brizantha | 48 |
| MSP1 | Adventitious embryony | Multiple sporocyte controls early sporogenic development | Oryza sativa | 49 |
| RWP | Adventitious embryony | RWP is the key gene controlling polyembryony | Citrus reticulate | 14 |
| SERK | Adventitious embryony | Somatic embryogenesis receptor-like kinase | Poa pratensis/Paspalum notatum | 50 |
| ORC | Adventitious embryony | ORC (ORIGIN RECOGNITION COMPLEX) is a multiprotein complex which controls DNA replication and cell differentiation in eukaryotes | Paspalum simplex | 51 |
| FIE | Autonomous endosperm | Fertilization independent endosperm | Malus hupehensis/Solanum lycopersicum | 52 |
| DYAD/SWITCH1 | Diplospory | Result in defects in female meiosis/affect meiosis in megasporocytes | Arabidopsis, dyad mutant | 53 |
| Apostart | Diplospory | Speculated to participate in the formation of 2 n eggs in apomixis | Medicago sativa | 44 |
| DMC1 | Diplospory | Participation in meiosis | Arabidopsis | 54 |
| Argonaute 9 (AGO 9) | Diplospory | AGO9 controls female gamete formation by limiting the specification of gametophyte precursors in a non-cell autonomous manner | Arabidopsis, AGO9 mutant | 55 |
| AGO 104 | Diplospory | AGO 104 and AGO9 are homologous genes, and their defects can produce a phenotype similar to apomixis, giving rise to up to 70% of functional unreduced female gametes | Tripsacum | 56 |
| AGAMOUS-LIKE 62 | Inhibit endosperm cellularization | Agamous-like 62 (AGL62) is a member of the MADS-box gene, an increase in its expression level can inhibit the development of the germ and inhibit endosperm cellularization | Arabidopsis, fis2 mutant | 57 |
| lorelei-like/n20gap-1 | Might play a role in the final stages of the apomixis developmental cascade. | Encodes a GPI-anchored protein previously associated with apomixis. | Paspalum notatum | 58 |
| dmt | May be involved in differentiation between apomictic and sexual reproduction | The expression of the dmt (DNA methyl transferases) gene was significantly different between sexual and apomictic reproduction. The absence of dmt102 and dmt103 can produce a phenotype similar to apomixis | maize-Tripsacum hybrid | 59 |
| APOLLO | Apomixis-associated polymorphisms | Has ‘apoalleles’ and ‘sexalleles’, apomictic Boechera spp. are heterozygous for the APOLLO gene, sexual genotypes are homozygous for sex alleles | Boechera retrofracta | 43 |
| DEMETER (DME) | DNA demethylase | Component of PRC2, in mutants autonomous endosperm forms. Responsible for endosperm maternalallele-specific hypomethylation at the MEDEA (MEA), regulation of gene expression by genetic imprinting |
Arabidopsis | 60 |
4. Epigenetic Control of Apomixis
The variable developmental processes and cells involved in apomictic mechanisms indicate the influence of epigenetic regulatory mechanisms in controlling apomictic processes. To investigate the hypothesis of cytosine methylation histone modification and comparative analysis of small-RNA have been undertaken both in apomictic and sexually propagated plants. In higher plants, the activation or movement of transposable elements (TEs) covers a significant proportion of the genome, influencing the evolution of the genome, alternation in gene expression, and frame shift mutations.61 To maintain the genomic integrity of the host genome, TE movement is important for regulation. The centromeric or telemetric regions are generally saturated with TEs with a high rate of methylation and packed as heterochromatic regions. In maize, it has been proven that in heterochromatin regions, TEs are specific for methylation, indicating their integral role in the alternation of structural and functional properties in plants. The role of retrotransposons in apomictic development has also been proposed; however, further proof of concept from both apomictic and sexually propagated plants is needed. From the Cenchrus ciliaris public database of the EST/BAC sequence, several classes of retrotransposons were identified, and their possible role in apomixis was elucidated. Based on expression analysis from 19 identified retrotransposons, six were found to be associated with apomictic activity. Among the six retrotransposons, C-105 was further utilized to understand epigenetic regulation based on its differential activity in both apomictic and sexually propagated plants of C. ciliaris. Bisulfite sequencing of retrotransposons (~0.3 kb fragment) showed 95% methylation of cytosine in sexually propagated plants compared to apomictic plants. The reversible ability of epigenetic modification in sexual pathways has also been hypothesized. The analysis of Arabidopsis mutant ovules epigenetically controlled by the sRNA-mediated silencing pathway involving the ARGONAUTE 9 (AGO9) protein showed a transition from apomictic to sexual reproduction.49 The loss of function of the mutant protein ago104 homologue to maize AGO9 showed apomictic-like characteristics, with ~70% functional female gametes. The comparative analysis of sexually reproductive maize with its apomictic wild relative Tripsacum further supported the epigenetic regulation of apomixis. A small number of chromatin enzymes, e.g., CMT3 and DRM2, showed differences in expression among apomictic and sexual plants. Moreover, the involvement of mutant alleles of genes responsible for DNA methylation and siRNA pathways in apospory or diplospory of Arabidopsis and maize has been recently revealed. Moreover, epigenetic modifications in the apomictic hybrids and polyploids lead to a collapse of the timing of developmental pathways, where developmental steps that are normally expressed successively occur simultaneously causing apomeiotic development of the embryo sac and full expression of apomixis.62 MEDEA (MEA) gene is involved in the control of fertilization-independent development of endosperm. This gene is associated with the epigenetic regulatory polycomb repressive complex 2 (PRC2). The function of PRC2 is silencing of gene expression via trimethylation of histone H3 at lysine 27 (H3K27me3). Loss of function of the MEA in Arabidopsis mea mutants resulted in the development of seeds independent of fertilization in Arabidopsis,63 the trait that is observed in some apomicts. However, there is still a quest to understand the underlying mechanism linking epigenetic modification with the establishment of apomixis in plants. Thus, the underlying mechanism/theory controlling epigenetic regulation of apomixis is worth studying, especially with regard to its ability to facilitate reversion to sexual propagation from the apomictic mode.22 The genetics, genomics, and epigenetic modifications that promote apomixis in flowering plants are well studied, whereas studies on induced apomixis through genome editing techniques, i.e., the CRISPR/Cas system, are at the foundation stage, as only a few reports have been published. The reports indicated the potential role of engineered apomixis in preserving hybrid seed vigor, which will ultimately lower the cost of hybrid seeds that farmers must buy every year.
5. Synthetic Apomixis through the MiMe Strategy
There are three most common approaches in plant breeding to convert crop plants from sexual to apomictic modes of reproduction: i) wide crosses with apomictic wild relatives, ii) mutation breeding, and iii) genetic transformation techniques. Sexual hybridization to transfer apomixis through interspecific hybridization relies heavily on the availability of wild relatives. In maize, apomixis was transferred from its wild relative Tripsacum through hybridization, whereas the hybrid produced after a series of backcrosses was sterile in nature, and facultative apomicts were not produced to recover the maize genome.64 Thus far, it is generally believed that the transfer of apomixis through wide crosses is not a successful approach. Moreover, the application of artificial mutation studies has provided significant evidence regarding the genetic architecture of apomixis. Meiosis and mitosis are distinguished based on three integral characteristics. First, DNA double standard breaks (DSBs) are induced with recombination and pairing between homologous chromosomes. Second, the first cell division leads to monopolar orientation of kinetochores of sister chromatids. Third, after genome duplication, the division of cells takes place two times.65 The genetic factors controlling these three developmental processes have been identified in plants. The genes SPO11-1, SPO11-2, HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1), PUTATIVE RECOMBINATION INITIATION DEFECT1 (PRD1), PUTATIVE RECOMBINATION INITIATION DEFECT2 (PRD2), MEIOTIC TOPOISOMERASE VIB-LIKE (MTOPVIB), DSB FORMATION (DFO), CENTRAL REGION COMPONENT 1 (CRC1), and P31comet are indispensable for the induction of DSBs in plants. The induced mutations in the abovementioned genes disrupt the normal pairing and recombination of homologous chromosomes.66 The monopolar orientation of kinetochores during the first meiotic division is mainly regulated by the meiotic cohesion complex controlled by a major component REC8.67 The genes OMISSION OF SECOND DIVISION (OSD1) and TARDY ASYNCHRONOUS MEIOSIS (TAM) control entry into the second division of meiosis. Mutations in any of these genes impact the second division, leading to the production of diploid gametes in both males and females.68 The spo11-1 and rec8 double mutant produced a mitotic-like first meiotic division.69 The osd1, rec8, and spo11-1 triple mutant and tam, rec8 and spo11-1 triple mutant exhibited apomeiosis phenotypes in which meiosis switched into mitosis-like division, and this genotype was called MiMe.65 The MiMe strategy helped in the development of viable diploid gametes and their successful transfer into rice by combining mutations in the genes (PAIR1, REC8, and OSD1) involved in the same process, suggesting that this strategy could be widely applied in flowering plants.8 Synthetic apomixis can be achieved by eliminating one set of chromosomes from either parent that has to be eliminated. Centromeres are the loci where the microtubule of the spindle is located and are essential for chromosome segregation during cell division. Centromeres are specified by a centromere-specific histone 3 (CENH3) protein in plants. The engineered genome elimination line (GEM) can be achieved through the application of CENH3 and further crossed with wild-type plants. Clonal seeds were generated by crossing MiMe plants with the GEM line and ultimately opening a plethora of options for clonal reproduction through seeds with mechanisms similar to apomixis.70,71Nonomura et al. (2007) reported the ortholog AGO5 in rice that proved to play an integral role in premeiotic mitosis and meiosis; similarly, in maize, the lack of the ortholog AGO9 leads to the production of viable gametes without meiosis.56 In Arabidopsis, the screening of parallel mutants for apomixis helped in the identification of genes responsible for the initiation of fertilization-independent seed (FIS) development. These FIS genes encode proteins that resemble members of the polycomb-related complex.72 Mutants of FIS genes are known to trigger endosperm development through an asexual mode of reproduction to varying extents. However, embryo initiation for most fis mutants is either low or does not occur. However, the epistatic effect of genes conferring the natural process of seed development is believed to be a limitation to obtaining apomictic plants. To date, several genes and their mutants have been developed to promote apomixis in various plant species and are well reviewed by Hand and Koltunow (2014); Barcaccia and Albertini (2013).10,11 The genetic transformation approach contains much potential, but limited genetic information is the only limiting factor. Map-based cloning for genes controlling apomixis in crop plants is relatively slow owing to the suppressed recombinants and excess repeated DNA sequences in apomixis-associated genomic regions. However, in a few plant species, apomictic genes were detected utilizing deletion studies.45 Recently, another approach called “conditional apomixis” has gained scientific ground, **switching73 the default mode of the plant reproduction system to an apomictic or sexual mode of reproduction.74 Conditional apomixis in plants can be achieved utilizing special promoters whose expression can be altered by certain chemicals or through epigenetic factors.35 However, the classical methods in plant breeding are time-consuming, labor-intensive, costly, unreliable, and not flexible. Due to these limitations in classical breeding approaches, modern genome editing technologies have gained scientific ground and have been widely utilized by plant breeders to meet their objectives with more ease and precision.
6. Synthetic Apomixis through Genome Editing to Preserve Hybrid Vigor
Currently, CRISPR technology is widely utilized with tremendous success in agriculture, especially for crops essential for food security, i.e., rice, wheat, maize, Arabidopsis, cotton, tomato, and potato. The user friendliness and low cost of genome editing have spurred research on crop trait development not only in academia but also with private companies dealing with agricultural products.75 Moreover, genome editing technology has not only accelerated breeding efforts to improve yield, grain quality, resistance against abiotic and biotic stresses and domestication in plants but also to develop plants that can withstand diverse environments with high-value traits, such as hybrid vigor or heterosis, which has proven to be a major challenge for researchers and farmers. These are significantly desirable goals that could change the agricultural production system. De novo targeted modification can alter the sexual mode of reproduction to one of apomixis and has been successfully employed in Arabidopsis.70 The BBM gene belongs to the APETALA 2/ETHYLENE RESPONSE FACTOR (AP2/ERF) DNA-binding domain family and is expressed in sperm cells. The AP2/ERF gene family is divided into the ERF-like category, which has a single AP2 domain, and the AP2-like category, which contains two AP2 domains (repeats 1 and 2) that are similar to each other and separated by a linker region. In Pennisetum squamulatum, apomixis is transmitted by a physically large, hemizygous, non-recombining chromosomal region, the ASGR.12 Several copies of PsASGR-BBML genes are considered candidate genes promoting parthenogenesis due to their strong linkage between apomictic Pennisetum/Cenchrus species. 37,76 Conner et al. (2013) reported a mutant of the CcASGR-BBML gene in Cenchrus ciliaris that ultimately lost its ability to experience parthenogenesis; moreover, ASGR-BBML has also been found in Arabidopsis and Brassica., 38,9successfully reduced the expression of PsASGR-BBML in apomictic F1 RNAi transgenic plants and endorsed the key role of PsASGR-BBML in promoting parthenogenesis in Pennisetum squamulatum, which can further lead toward haploid induction to vigorously obtain homozygous transgenic lines for breeding.7 Khanday et al. (2019) reported that triple knockout of the genes BBM1, BBM2, and BBM3 caused embryo arrest and abortion, phenotypes which were fully rescued by the male-transmitted BBM1. These findings suggest that the requirement for fertilization in embryogenesis is mediated by male genome transmission of pluripotency factors. When genome editing to substitute the mitosis for meiosis (MiMe) phenotype7,48 is combined with the expression of BBM1 in the egg cell, clonal progeny can be obtained that retain genome-wide parental heterozygosity. The synthetic asexual-propagation trait is heritable through multiple generations of clones, also known as the clonal fix strategy. Moreover, Wang (2019)14 reported that multiplex editing of three (REC8, PAIR1, and OSD1) key meiotic genes in hybrid rice led to the production of clonal diploid gametes and tetraploid seeds. Next, editing of the MTL 77,4gene involved in fertilization resulted in the induction of haploid seeds in hybrid rice. By simultaneous editing of these four endogenous genes in hybrid rice using the CRISPR/Cas9 system, plants that are able to propagate clonally through seeds were obtained. The quadruple aop (Apomictic Offspring Producer) mutants obtained through the knockout of OsSPO11‐1, OsREC8, OsOSD1, and OsMATL produced the MiMe phenotype. These mutants have the ability to produce apomictic plants.47 However, thus far, the number of viable clone seeds with their original hybrid genetics intact has been much lower than expected, and this area needs further research.
The only drawback to hybrid technology includes the loss of hybrid vigor along with other desired traits due to genetic segregations in subsequent generations. Moreover, the cost of new seeds for every sowing is a major challenge faced by subsistence farmers.78 The preservation of hybrid seed qualities has significant implications for ensuring food security, environmental preservation, and employment. Ensuring that crops pass on hybrid qualities to seeds has been a major challenge, but researchers equipped with modern plant breeding technologies are striving to overcome this challenge. The findings in the abovementioned studies revealed a straightforward strategy to promote apomixis to preserve hybrid vigor in rice. Moreover, this approach can also work for other cereal crops that have equivalent genes, such as wheat, corn, barley, and millet, by increasing the percentage of seeds with the same hybrid vigor, and ultimately, these seeds can reach farmers’ fields. Moreover, identifying genes controlling the molecular mechanism of apomictic reproduction is critical to understanding its genetic architecture, which is still little understood (Fig. 3).
Figure 3.
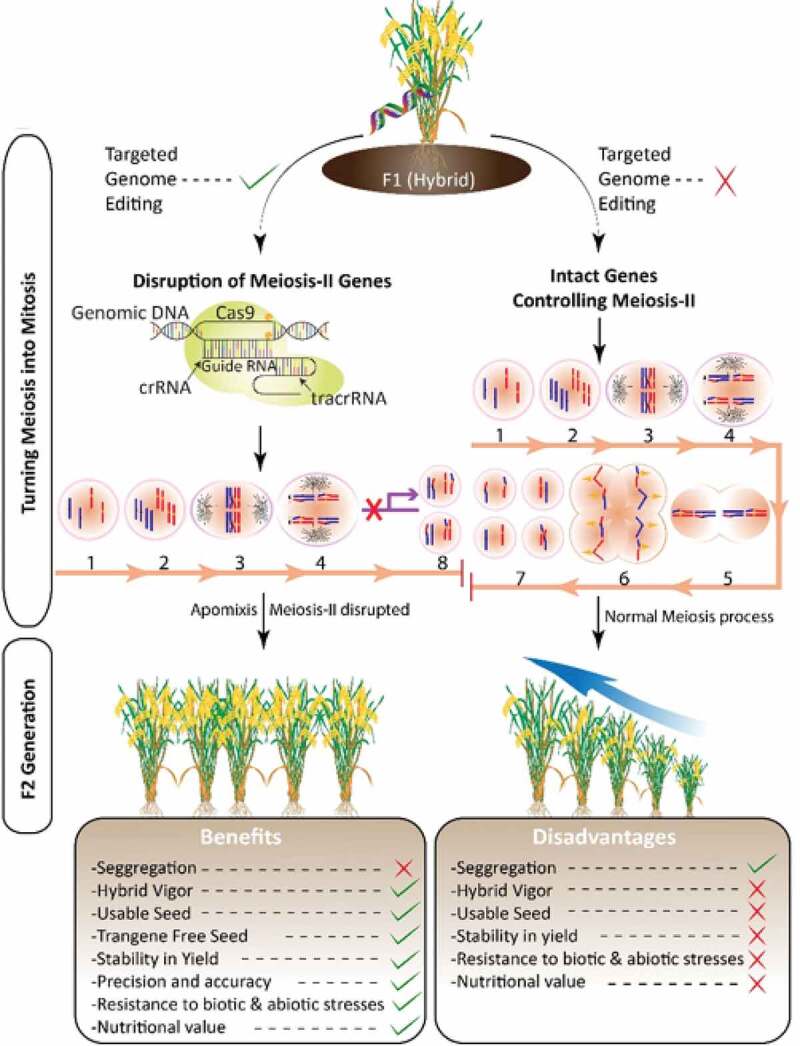
An illustration of promoting apomixis for hybrid vigor preservation through the CRISPR/Cas9 system. To fix hybrid vigor in F2 scientist are promoting apomixis in hybrid seed to keep intact the characteristics of F1 hybrid seed. The MiMe phenotypes is achieved through disruption of gene-controlling meiosis II which leads toward unreduced embryo sac essential for apomeiosis. However, in other case the hybrid segregate in F2 plant populations and all the important characteristics are lost. 1, Interphase; 2, Prophase-I; 3, Metaphase-I; 4, Anaphase-I; 5, Metaphase-II; 6, Anaphase-II; 7, n gametes; 8, 2 n gametes; NBTs, (new breeding techniques); P/TGMS, Cas9; CRISPR- associated protein 9, tracrRNA (trans-activating RNA).
7. Limitations to Synthetic Apomixis
The utilization of the CRISP/Cas system for synthetic apomixis has been successfully employed in rice; however, seed production with intact hybrid vigor was significantly reduced.,13reported that MiMe does not significantly reduce seed production, whereas a mutation in the MTL gene leads to haploid induction at the expense of seed production in rice. Similarly, in maize, large-scale chromosome fragmentation caused a reduction in fertility owing to post meiosis in the mtl gametophyte.4 Fragmentation leads to an imbalance between the genomes of both the maternal and paternal sides. Several genes involved in embryo and endosperm development have been investigated in Arabidopsis and apomictic plants. A novel strategy, i.e., combining the Fix (Fixation of hybrids) strategy with genes promoting autoendosperm development, may resolve the issue of low fertility caused by the Fix strategy. In the Fix strategy, clonal diploid gametes are produced, similar to the MiMe strategy. After self-fertilization, in addition to tetraploid hybrids, clonal diploid hybrids are produced in the offspring. Using the Fix strategy, we successfully introduced synthetic apomixis into hybrid rice. However, seed production, which is one of the most important traits for rice cultivation, was significantly reduced in both Fix and clonal Fix plants. Therefore, the Fix strategy can be successfully employed in crops where seed production is a secondary objective, e.g., vegetables and forage crops, but this also needs further investigation.,9reported that the engineered Fix strategy was not as effective as naturally occurring apomixis because of penetration ability. Therefore, further studies, such as the identification of new parthenogenesis-inducing genes, are required to determine how to increase the frequency of clonal seeds. In contrast, techniques to distinguish clonal seeds or plants among offspring could be developed to further expand the application of the Fix strategy in agriculture. The mechanism of heterosis is less known, and it is generally believed that the degree of genetic distance contributes to heterosis. 79 However, recent studies have shown that epigenetic involvement also contributes to heterosis, 80,14 re-sequenced the whole genome of parent plants and clonal plants and reported the reliable transmission of genetic information, which demonstrated that epigenetic information was also transferred normally across generations. Although epigenetic modification cannot be ruled out, the modifications were too minor to induce detectable phenotypic changes in rice under field conditions. Further studies are thus required in several other crops to investigate phenotypic stability over multiple generations.
8. Conclusion and Future Perspectives
Understanding the phenomenon of apomixis remains challenging for plant geneticists; however, its potential benefits remain a focus of enormous interest for plant breeders. Both naturally existing genes and induced mutations can divert the natural sexual pathway toward apomixis, which can be further investigated. A combined strategy that can target apomeiosis, parthenogenesis, and seed formation might be of the utmost importance to exploit the full potential of apomixis to eliminate breeding barriers. It has been historically proven that the development of hybrids for different crops has helped farmers obtain benefits, such as maximum crop yields and grain quality as well as increased resistance to biotic and abiotic stresses, but hybrid seed lacks the ability to produce plants with the same qualities in subsequent generations. Hence, farmers have had no option other than to buy expensive hybrid seeds every year. The identification of QTLs/genes through new mapping technologies, that is, comparative mapping, linkage disequilibrium mapping, deletion mapping, and new high-throughput sequencing methods, will help to reveal core apomixis chromosomal regions. Similarly, high-throughput technologies can expose or remove the evolutionary genetic load of genes and epigenetic modifications related to apomixis and can be further utilized in agriculture as a tool to fix elite genotypes that are important for food security. In summary, the successful application of genome editing technology for targeted mutagenesis has opened further avenues of research for understanding the molecular mechanisms controlling apomixis. The efficiency of clonal propagation in crops, particularly in rice, has been limited by the frequency of parthenogenesis. However, this could be improved in the future by studying different promoters of genes or incorporating specific alleles that exhibit full or partial dominance.
Acknowledgments
The authors are thankful to Mr. Shakeel Ahmad from State Key Laboratory of Rice Biology, China National Rice research Institute, Hangzhou, P.R. China for providing illustration (Fig. 3) and insights to improve the manuscript.
Funding Statement
The publication of the present work is supported by the National Key Research and Development Program of China (grant no. 2017YFC0504704) and the National Natural Science Foundation of China (51669034, 41761068, 51809224).
Disclosure Of Potential Conflicts Of Interest
No potential conflicts of interest were disclosed.
Author Contributions
SF and XW conceived the idea and drafted the manuscript. SF and AY collected the literature review. XW, AY, and AR provided with technical assistance. AR provided the illustration BA and HA helped in proofreading and final editing of the manuscript. All authors listed have made substantial, direct, intellectual contributions to the work and have approved it for publication.
References
- 1.Sheng Z, Wei X, Shao G, Chen M, Song J, Tang S, Luo J, Hu Y, Hu P, Chen L.. Genetic analysis and fine mapping of tms9, a novel thermosensitive genic male‐sterile gene in rice (Oryza sativa L.). Plant Breed. 2013;132:159–64. [Google Scholar]
- 2.Barman HN, Sheng Z, Fiaz S, Zhong M, Wu Y, Cai Y, Wang W, Jiao G, Tang S, Wei X. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019;19:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada A, Arndell T, Borisjuk N, Sharma N, Watson HNS, E J T, Baumann U, Langridge P, Whitford R. CRISPR/Cas9‐mediated knockout of Ms1 enables the rapid generation of male‐sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J. 2019;17:1905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Meng D, Chen S, Luo H, Zhang Q, Jin W, Yan J. Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat Commun. 2017;8:991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen R, Xu Q, Liu Y, Zhang J, Ren D, Wang G, Liu Y. Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 2018;9:1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X, Yang S, Gong J, Zhao Q, Feng Q, Zhan Q, Zhao Y, Li W, Cheng B, Xia J. Genomic architecture of heterosis for yield traits in rice. Nature. 2016;537:629–33. [DOI] [PubMed] [Google Scholar]
- 7.Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature. 2019;565:91–95. [DOI] [PubMed] [Google Scholar]
- 8.Mieulet D, Jolivet S, Rivard M, Cromer L, Vernet A, Mayonove P, Pereira L, Droc G, Courtois B, Guiderdoni E. Turning rice meiosis into mitosis. Cell Res. 2016;26:1242–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conner JA, Mookkan M, Huo H, Chae K, Ozias-Akins P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci. 2015;112:11205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hand ML, Koltunow AM. The genetic control of apomixis: asexual seed formation. Genet. 2014;197:441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barcaccia G, Albertini E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod. 2013;26:159–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozias-Akins P, van Dijk PJ. Mendelian genetics of apomixis in plants. Annu Rev Genet. 2007;41:509–37. [DOI] [PubMed] [Google Scholar]
- 13.Ravi M, Marimuthu MP, Siddiqi I. Gamete formation without meiosis in Arabidopsis. Nature. 2008;451:1121. [DOI] [PubMed] [Google Scholar]
- 14.Wang K. Fixation of hybrid vigor in rice: synthetic apomixis generated by genome editing. A Biotech. 2020;1:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sailer C, Schmid B, Grossniklaus U. Apomixis allows the transgenerational fixation of phenotypes in hybrid plants. Curr Bio. 2016;26:331–37. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Yuantao X, Zhang S, Cao L, Hunag Y, Cheng J, Wu G, Tian S, Chen C, Liu Y, et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet. 2017;49:765. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Zhu G, Li R, Yan S, Fu D, Zhu B, Tian H, Luo Y, Zhu H. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 2017;175:1690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Jiao G, Liu Z, Zhang X, Li J, Guo X, Du W, Du J, Francis F, Zhao Y, et al. Generation of high amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci. 2017;8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 2016;7:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie E, Li Y, Tang D, Yanli LV, Shen Y, Cheng Z. A strategy for generating rice apomixis by gene editing. J Integr Plant Biol. 2019;61:911–16. [DOI] [PubMed] [Google Scholar]
- 21.Xu R, Yang Y, Qi R, Li H, Qiu C, Li L, Wei P, Yang J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genetics and Genomics. 2016;43:529–32. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S. Epigenetic control of apomixis: a new perspective of an old enigma. Adv Plants Agric Res. 2017;7:15406. [Google Scholar]
- 23.Carman JG. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc. 1997;61:51–94. [Google Scholar]
- 24.Voigt-Zielinski ML, Piwczyński M, Sharbel TF. Differential effects of polyploidy and diploidy on fitness of apomictic Boechera. Sex Plant Reprod. 2012;25:97–109. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Leal D, Vielle-Calzada JP. Regulation of apomixis: learning from sexual experience. Curr Opin Plant Biol. 2012;15:549–55. [DOI] [PubMed] [Google Scholar]
- 26.Adolfsson S, Bengtsson BO. The spread of apomixis and its effect on resident genetic variation. J Evol Biol. 2007;20(5):1933–40. doi: 10.1111/j.1420-9101.2007.01371.x. [DOI] [PubMed] [Google Scholar]
- 27.Balloux F, Lehmann L, de Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengtsson BO. Genetic variation in organisms with sexual and asexual reproduction. J Evol Biol. 2003;16:189–99. [DOI] [PubMed] [Google Scholar]
- 29.Yadav CB, Suresh YQ, Kumar MG, Bhat GV. Genetic linkage maps of the chromosomal regions associated with apomictic and sexual modes of reproduction in Cenchrus ciliaris. Mol Breed. 2012;30:239–50. [Google Scholar]
- 30.Fei X, Shi J, Liu Y, Niu J, Wei A. The steps from sexual reproduction to apomixis. Planta. 2019;249:1715–30. [DOI] [PubMed] [Google Scholar]
- 31.Albertini E, Barcaccia G, Mazzucato A, Sharbel TF, Falcinelli M. Apomixis in the era of biotechnology. In: Plant developmental biology, Biotechnological perspectives. Springer-Verlag Berlin Heidelberg; 2010. p. 405–36. [Google Scholar]
- 32.Savidan Y. Apomixis: genetics and breeding. Plant Breed Rev. 2000;18:13–86. [Google Scholar]
- 33.Grimanelli D. Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr Opi Plant Biol. 2012;15:57–62. [DOI] [PubMed] [Google Scholar]
- 34.Grossniklaus U, Spillane C, Page DR, Kohler C. Genomic imprinting and seed development: endosperm formation with and without sex. Curr Opi Plant Biol. 2001;4:21–27. [DOI] [PubMed] [Google Scholar]
- 35.Pupilli F, Barcaccia G. Cloning plants by seeds: inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J Biotech. 2012;159:291–311. [DOI] [PubMed] [Google Scholar]
- 36.Ozias-Akins P, Roche D, Hanna WW. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci. 1998;95:5127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conner JA, Goel S, Gunawan G, Cordonnier-Pratt MM, Johnson VE, Chun L, Wang H, Pratt LH, Mullet JE, DeBarry J, et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008;147:1396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schallau A, Arzenton F, Johnston AJ, Hähnel U, Koszegi D, Blattner FR, Altschmied L, Haberer G, Barcaccia G, Bäumlein H. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010;62:773–84. [DOI] [PubMed] [Google Scholar]
- 40.Juranić M, Srilunchang K, Krohn NG, Levanić DL, Sprunck S, Dresselhaus T. Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell. 2012;24:4974–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silveira ED, Guimarães LA, de Alencar Dusi DM, da Silva FR, Martins NF, Do Carmo Costa MM, Alves-Ferreira M, de Campos Carneiro VT. Expressed sequence-tag analysis of ovaries of Brachiaria brizantha reveals genes associated with the early steps of embryo sac differentiation of apomictic plants. Plant Cell Rep. 2012;31:403–16. [DOI] [PubMed] [Google Scholar]
- 42.Guimarães LA, Dusi DMA, Masiero S, Resentini F, Gomes ACMM. BbrizAGL6 is differentially expressed during embryo sac formation of apomictic and sexual Brachiaria brizantha plants. Plant Mol Biol Rep. 2013;31:1397–406. [Google Scholar]
- 43.Corral JM, Vogel H, Aliyu OM, Hensel G, Thiel T, Kumlehn J, Sharbel TF. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013;163:1660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albertini E, Marconi G, Reale L, Barcaccia G, Porceddu A, Ferranti F, Falcinelli M. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiol. 2005;138:2185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotani Y, Henderson ST, Suzuki G, Johnson SD, Okada T, Siddons H, Mukai Y, Koltunow AM. The loss of apomeiosis (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. 2014;201:973–81. [DOI] [PubMed] [Google Scholar]
- 46.Guerin J, Rossel JB, Robert S, Tsuchiya T, and Koltunow A. A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta. 2000;76:914–20. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira LG, de Alencar Dusi DM, Irsigler AST, Gomes A, Mendes MA, Colombo L, de Campos Carneiro VT. GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Rep. 2018;37:293–306. [DOI] [PubMed] [Google Scholar]
- 48.Siena LA, Ortiz JPA, Leblanc O, Pessino AS. PnTgs1-like expression during reproductive development supports a role for RNA methyltransferases in the aposporous pathway. BMC Plant Biol. 2014;14:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nonomura KI. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell Online. 2003;15:1728–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podio M, Caceres ME, Samoluk SS, Seijo JG, Pessino SC, Ortiz JP, Pupilli F. A methylation status analysis of the apomixisspecific region in Paspalum spp. Suggests an Epigenetic Control of Parthenogenesis. J Exp Bot. 2014;65:6411–24. [DOI] [PubMed] [Google Scholar]
- 51.Siena LA, Ortiz JP, Calderini O, Paolocci F, Caceres ME, Kaushal P, Grisan S, S C P, Pupilli F. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J Exp Bot. 2016;67:1965–78. [DOI] [PubMed] [Google Scholar]
- 52.Liu DD, Dong QL, Sun C, Wang QL, You CX, Yao YX, Hao YJ. Functional characterization of an apple apomixisrelated MhFIE gene in reproduction development. Plant Sci. 2012;185–186. [DOI] [PubMed] [Google Scholar]
- 53.Boateng KA, Yang X, Dong F, Owen HA, Makaroff CA. SWI1 is required for meiotic chromosome remodeling events. Mol Plant. 2008;1:620–33. [DOI] [PubMed] [Google Scholar]
- 54.Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y. Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 2014;80:615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olmedo-Monfil V, Figueroa ND, Vázquez MA, Arévalo MD, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Calzada JPV. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E D H, Kradolfer D, Köhler C. Endosperm cellularization defines an important developmental transition for embryo development. Development. 2012;139:2031–39. [DOI] [PubMed] [Google Scholar]
- 58.Felitti SA, Seijo JG, González AM, Podio M, Laspina NV, Siena L, Ortiz JPA, Pessino SC. Expression of lorelei-like genes in aposporous and sexual Paspalum notatum plants. Plant Mol Biol. 2011;77:337. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell. 2010;22:3249–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA. 2011;108:8042–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan H, Kikuchi S, Neumann P, Zhang W, Wu Y, Chen F, Jiang J. Genome‐wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 2010;63:353–65. [DOI] [PubMed] [Google Scholar]
- 62.Carman JG. Do duplicate genes cause apomixis? In: Horandl E, Grossniklaus U, van Dijk P, Sharbel T, editors. Apomixis: evolution, mechanisms and perspectives. Lichtenstein: ARG Gantner Verlag KG; 2007. p. 169–94. [Google Scholar]
- 63.Schmidt A, Wo¨hrmann HJ, Raissig MT, Arand J, Gheyselinck J, Gagliardini V, Heichinger C, Walter J, Grossniklaus U. The polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J. 2013;73:776–87. [DOI] [PubMed] [Google Scholar]
- 64.Kandemir N, Saygili I. Apomixis: new horizons in plant breeding. Turk J Agri For. 2015;39:549–56. [Google Scholar]
- 65.d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. Turning meiosis into mitosis. PLoS Biol. 2009;7:1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Y, Yin Z, Zeng Y, Zhang Q, Chen L, He Y, Lu P, De Y, Zhang X. MTOPVIB interacts with AtPRD1 and plays important roles in formation of meiotic DNA double-strand breaks in Arabidopsis. Sci Rep. 2017;7:10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 2011;156:1386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.d'Erfurth I, Cromer L, Jolivet S, Girard C, Horlow C, Sun Y, Jennifer PC, Berchowitz TLE, Copenhaver GP, Mercier R. The CYCLIN-A CYCA 1;2/TAM Is Required for the Meiosis I to Meiosis II Transition and Cooperates with OSD1 for the Prophase to First Meiotic Division Transition. PLoS Genet. 2010;6:e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–32. [DOI] [PubMed] [Google Scholar]
- 70.Marimuthu MP, Jolivet S, Ravi M, Pereira L, Davda JN, Cromer L, Wang L, Nogué F, Chan SWL, Siddiq I. Synthetic clonal reproduction through seeds. Sci. 2011;331:876–876. [DOI] [PubMed] [Google Scholar]
- 71.Nonomura KI, Morohoshi K, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell–specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koltunow AM, Grossniklaus U. Apomixis: a developmental perspective. Annu Rev Plant Biol. 2003;54:547–74. [DOI] [PubMed] [Google Scholar]
- 73.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Gene. 2010;11:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spillane C, Curtis MD, Grossniklaus U. Apomixis technology development virgin births in farmers’ fields? Nat Biotech. 2004;22:687. [DOI] [PubMed] [Google Scholar]
- 75.Fiaz S, Ahmad S, Noor MA, Wang X, Younas A, Riaz A, Ali F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Inter J Mol Sci. 2019;20:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conner JA, Gunawan G, Ozias-Akins P. Recombination within the apospory specific genomic region leads to the uncoupling of apomixis components in Cenchrus ciliaris. Planta. 2013;238:51–63. [DOI] [PubMed] [Google Scholar]
- 77.Kelliher T, Starr D, Richbourg L, Chintamanani S, Delzer B, Nuccio ML, Green J, Chen Z, McCuiston J, Wang W. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature. 2017;542:105. [DOI] [PubMed] [Google Scholar]
- 78.Janaiah A, Hossain M, Husain M. Hybrid rice for tomorrow’s food security: can the chinese miracle be replicated in other countries? Outlook Agri. 2002;31:23–33. [Google Scholar]
- 79.Birchler JA. Genetic rules of heterosis in plants. In: Polyploid and hybrid genomics. John Wiley & Sons, Inc; 2013. p. 313. [Google Scholar]
- 80.Dapp M, Reinders J, Bédiée A, Balsera C, Bucher E, Theiler G, Granier C, Jerzy P. Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants. 2015;1:15092. [DOI] [PubMed] [Google Scholar]


