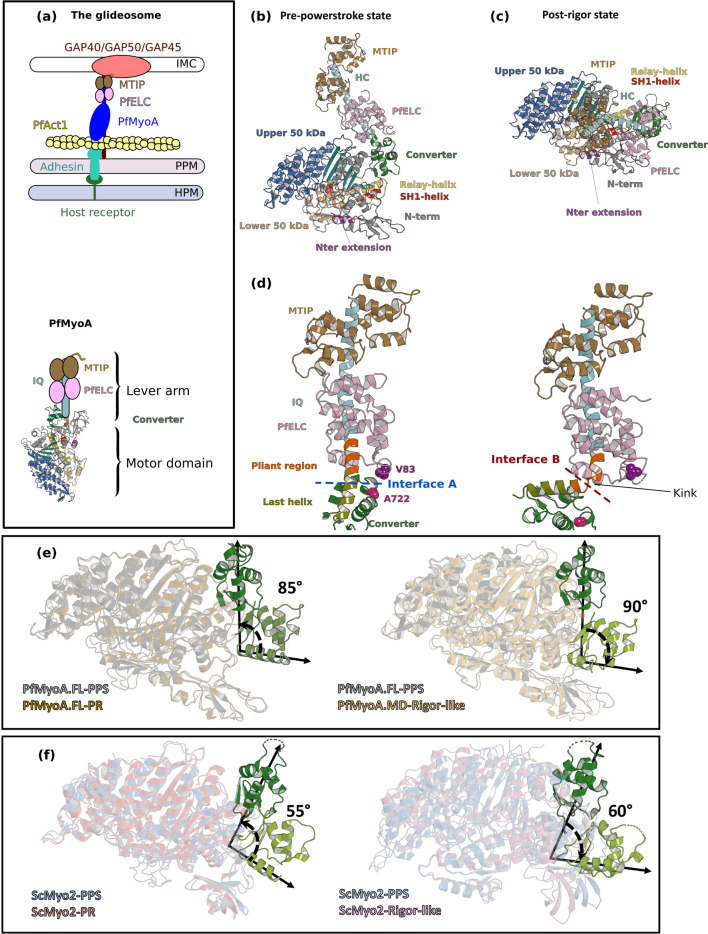
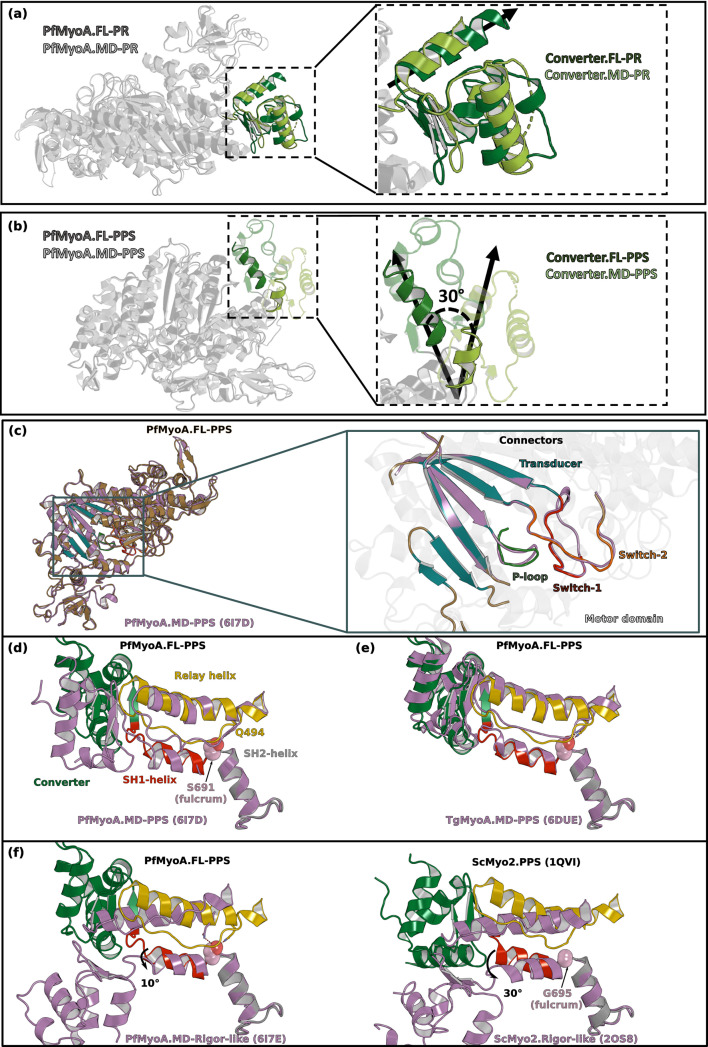
Figure 1. Crystal structures of PfMyoA in the Post-rigor (PR) and the Pre-Powerstroke (PPS) states.
(a) (Left) The PfMyoA motor is located in the intermembrane space of the parasite. PfMyoA (blue) binds two light chains, PfELC and myosin tail interacting protein (MTIP). MTIP connects the motor to the glideosome-associated proteins (GAP) complex, which is anchored in the inner membrane complex (IMC). PfMyoA cyclically interacts with PfAct1 filaments, which are bound to adhesins from the parasite plasma membrane (PPM); these adhesins also bind receptors from the host cell plasma membrane (HPM). The displacement of PfAct1 filaments by PfMyoA drives parasite gliding motility. (Right) The crystal structure of the motor domain of PfMyoA has been solved (Robert-Paganin et al., 2019), but the lever arm structure was not known. (b,c) Overall structures of the full-length PfMyoA motor in the PPS and PR states, displayed so that their N-terminal subdomains adopt a similar orientation. As expected, the orientation of the converter and lever arm differs in these two states. (d) The lever arm has been built in these two states of the motor, revealing the structure of the two bound light chains, PfELC and MTIP, displayed here in a similar orientation. The kink in the lever arm helix at the end of the converter (pliant region in orange; last helix of the converter in deep olive green) induces different converter/PfELC interfaces in the PPS (interface A, left) compared to the PR state (interface B, right). To illustrate that the two interfaces are different, two reporter residues are displayed as spheres, A722 from the converter and V83 from PfELC. These residues are the part of interface A but not the part of interface B. (e) and (f) represents the recovery stroke (left) and the powerstroke (right) for PfMyoA and scallop myosin 2 (ScMyo2), respectively. Structures of ScMyo2 used: PR (PDB code 1S5G); PPS (PDB code 1QVI).