Graphical abstract
Keywords: COVID-19, Kinins, Kallikrein-kinin system, Renin-angiotensin system, Coagulation system
Highlights
-
•
SARS-CoV-2 urged at end of 2019 as the causative of (COVID-19) causing a worldwide dissemination of the disease.
-
•
Its pathophysiology is complex with the involvement of a diversity of systems and can evolve to death.
-
•
Among the various systems involved in the disease, the renin-angiotensin (RAS) and kallikrein-kinin (KKS) systems stand out.
-
•
Here we described RAS and KKS in COVID-19 suggesting their importance in the evolution of the disease.
-
•
Some possibilities for drug repositioning in treating COVID-19 is also discussed.
Abstract
In November 2019 the first cases of a novel acute respiratory syndrome has been reported in Wuhan province, China. Soon after, in January 2020 the World Health Organization declared a pandemic state due to the dissemination of a virus named SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), the cause of coronavirus disease 2019 (COVID-19). Being an unknown disease, it is essential to assess not only its main characteristic features and overall clinical symptomatology but also its patient infection mode and propagation to design appropriate clinical interventions and treatments. In this review the pathophysiology of SARS-CoV-2 infection and how the virus enters the cells and activates the immune system are described. The role of three systems involved in the SARS- CoV-2 infection (renin-angiotensin, kinin and coagulation systems) is discussed with the objectives to identify and try to explain several of the events observed during the evolution of the disease and to suggest possible targets for therapeutic interventions.
1. Introduction
In late 2019, numerous cases of pneumonia of unidentified cause emerged in Wuhan, Hubei province, China [1,2]. With the increase of cases the epidemiological and aetiological studies showed a correlation between number of patients with pneumonia and their exposure to a wholesale seafood market in Wuhan, where, in addition to seafood, wild animals such as snakes, pangolins, rats, bats were traded [3]. The infection was characterized as COVID-19 (Coronavirus Disease 19) caused by a new Coronavirus named SARS-Cov-2 [1,4]. The high contagion rate of SARS-CoV-2 occurs through direct transmission (coughing, sneezing, and inhalation of viral particles) or through contact (oral, nasal or ocular mucous membranes) that occur between humans, thus favoring their rapid spread around the world ([5,6]; After more than 35,000 confirmed cases and 700 deaths in China and other countries, in late February 2020 the World Health Organization declared a pandemic state [6,7]. By September 28, more than 33,500,000 cases of COVID-19 were identified, with more than 1,000,000 deaths worldwide [8].
About 80 % of patients infected with SARS-CoV-2 have mild symptoms of the disease (with no or only a mild picture of pneumonia), 14 % develop a severe picture of the disease with dyspnea, hypoxia or lung injury greater than 50 % in imaging tests, and 5% develop a critical condition characterized by respiratory failure, systemic shock or multiple organ failure [9]. It is important to note that patients may present other symptoms within the mentioned clinical conditions including fever (98 %), dry cough (76 %), tiredness (44 %) and less common symptoms such as headache (8%) and diarrhea (3%) [1]. Pan and collaborators [10] have identified the maximum viral load which occurs between the 5th and 7th days post-symptoms appearance and that progressively decreases after 9 days. In moderate and severe cases the presence of lymphopenia has been detected and associated with a drastic reduction in CD4+ and CD8+ T cells, high levels of C-reactive protein, ferritin, polymorphonuclear and lymphocyte cells, inflammatory cytokines and chemokines [9,11,12].
2. SARS-C0V-2
2.1. Isolation, classification and genome
SARS-CoV-2 was first isolated from bronchoalveolar lavage of five patients with severe pneumonia hospitalized in Wuhan [13]. Metagenomic analyses in RNA identified the virus as belonging to Coronaviridae family, Nidovirales order [14]. Coronaviruses (CoVs) are enveloped, single- and positive-stranded RNA viruses with a genome of approximately 26−32 kb [15]. Genome and gene expression organizations are similar for all CoVs, most of them have 8–10 open reading frames (ORFs). ORF1a/b located at the 5′ end encodes 16 non-structural proteins (NSPs) called NSP1 to NSP16. In addition, other ORFs at the 3′ end encode accessory and structural proteins [[16], [17], [18]], including spike (S) protein responsible for anchoring the virus to the receptor and subsequent fusion of the membrane, thereby mediating the entry of the virus into the cell; the envelope protein (E) plays a pivotal role in the assembly and release of the virus in addition to exercising the ion channel function necessary for the pathogenesis of SARS-CoV-2. The membrane protein (M) assists in maintenance of the membrane curvature and binding to the nucleocapsid whereas the nucleocapsid protein (N) is involved in the packaging of the viral genome encapsulated in the viral particles and its interaction with the M and NSP3 proteins [19].
Currently there are four genera of CoVs: α-CoV, β-CoV, γ-CoV and δ-CoV, with only the genera α and β with strains considered pathogenic for humans. These are α-CoV: HCoV-229E, HCoV-NL63 and β-CoV: HCoV−OC43, HCoV-HKU1, SARS-CoV, MERS-CoV and the recently discovered SARS-CoV-2 [20] with approximately 79 % homology with SARS-CoV and 50 % homology with MERS-CoV [12].
α-CoV and β-CoV mainly infect respiratory and gastrointestinal tracts and the central nervous system of mammals whereas the γ-CoV and δ-CoV mainly infect birds [6].
2.2. Transmission
Bats have been identified as natural hosts for several emerging viruses that can cause serious human diseases. These viruses are RNA viruses such as the Marburg, Hendra, Sosuga and Nipah virus. Evidence suggests that other emerging viruses such as Ebola, the coronavirus that causes severe acute respiratory syndrome (SARS-CoV), the coronavirus that causes middle east respiratory syndrome (MERS-CoV) also originated from bats [21].
Similar to SARS-CoV and MERS-CoV, SARS-CoV-2 showed 96 % genomic compatibility with the bat coronavirus RaTG13 (Bat CoV RaTG13) of the species Rhinolophus affinis found in Yunnan province, thus indicating the origin of SARS- CoV-2 [22,23].
The first corona virus epidemic was caused by SARS-CoV in 2002–2003 with 916 deaths in 37 countries. MERS-CoV epidemia which occurred in 2012 has infected 1791 people causing 640 deaths in 27 countries [24].
CoVs that infect bats cannot directly infect humans, unless they mutate or recombine in animals such as civets and camels, which are intermediate hosts for SARS-CoV and MERS-CoV respectively [83].
Through metagenomic sequencing it was observed that the CoV present in pangolins exhibited strong genomic similarities with SARS-CoV-2 as well as its relation to the receptor binding domain region [25]. In view of this fact, Zhang et al. [23] demonstrated that the CoV that infects the pangolin had a 91.02 % and 90.55 % ratio of genetic similarity to SARS-CoV-2 and Bat CoV RaTG13 respectively, which suggested that pangolins are the intermediate hosts of SARS-CoV-2.
2.3. Mechanisms of entry into the host cells
The entry of CoVs into host cells is mediated by the transmembrane S glycoprotein, which forms homotrimers projected from the viral surface. This protein is composed of two functional subunits, the S1 subunit, which has a specific region called the receptor binding domain (RBD) responsible for binding to the host cell receptor and the S2 subunit responsible for the fusion of the viral and cellular membranes, that occurs after protein S activation [[26], [27], [28]].
Previous studies with SARS-CoV and MERS-CoV have shown that protein S activation is a complex process that involves several cleavage events that occur in different locations and with the involvement of several host proteases [29]. CoV protein S can be cleaved by one or more proteases, including furin, trypsin, cathepsin, type 2 transmembrane serine protease (TMPRSS-2), TMPRSS-4 or human airway trypsin-like protease (HAT) [30].
Hoffmann and collaborators [31] were the first to report that the spread of the SARS-CoV-2 virus was dependent on the action of the serine protease TMPRSS2, responsible for cleaving the S protein between the S1/S2 subunit. In addition, studies with specific protease inhibitors such as E64d, a lysosomal cathepsin inhibitor, have also significantly reduced the entry of the virus into HeLa cells. Overall, these results demonstrate that cell surface proteases and lysosomal proteases can cleave protein S, thus favoring the entry of the SARS-CoV-2 virus [32].
SARS-CoV and SARS-CoV-2 S proteins are known to have approximately 76 % aminoacid similarity [33,34]. However, SARS-CoV-2 has a distinct sequence of 4 aminoacids (SPRRAR) located between the S1/S2 subunit. This insertion, also known as a polybasic or multibasic site, is a potent cleavage site for the furin protein. Polybasic sites are a highly pathogenic characteristic feature expressed in protein S of MERS-CoV and influenza virus [28,29,31,35].
To determine whether the insertion of amino acids in the S1/S2 subunits of SARS-CoV and SARS-CoV-2 would have different patterns of infectivity and pathogenicity between strains, it was observed that furin cleaves the S1/S2 subunit of SARS-CoV-2 but not of SARS-CoV; however other proteases such as PC1, matriptase, trypsin and cathepsin B can efficiently recognize and cleave the S1/S2 cleavage site of the SARS-CoV-2 protein [29]. Furthermore, furin pre-activation allows SARS-CoV-2 to be less dependent on target cells, improving its entry particularly into cells with relatively low expressions of TMPRSS2 and/or lysosomal cathepsins corroborating the high degree of pathogenicity observed for the SARS-CoV-2 virus [32].
After binding the RBD to the host cell receptor and subsequent cleavage of the S1/S2 protein by proteases, a structural change occurs in the S2 subunit, causing the heptad repeat 1 (HR1) and 2 (HR2) domains to interact between itself, forming a packet of six helices (6-HB), thus approaching peptide with the cell membranes, resulting in the subsequent fusion and infection of the host cell [36,37].
2.4. Receptors
Just like the SARS-CoV virus, SARS-CoV-2 S protein binds directly to angiotensin converting enzyme 2 (ACE2) facilitating virus entry into cells demonstrating that the ACE2 enzyme acts as a functional receptor for both virus [28,38]. It is also known that the RBD from SARS-CoV-2 has a 10 to 20-fold higher affinity to human ACE2 than the SARS-CoV RBD [30,32,38].
Immune cells can also be infected with SARS-CoV-2, as well as with MERS-CoV and SARS-CoV. Thus, it is possible that there are other receptors that allow the virus to enter different cell types [39]. In fact, as seen with SARS-CoV, another receptor CD147 also called basigin (encoded by BSG) has shown affinity for the SARS-CoV-2 protein S [40]. CD147 is a transmembrane glycoprotein belonging to the immunoglobulin superfamily and is related to the development of tumors and viral infection [37]. Radzikowska et al. [41] showed the expression of CD147 in epithelial tissues and immune cells such as macrophages, monocytes, innate lymphoid cells, natural killer, T and B cells, which can be infected in the lungs or can carry the SARS-CoV-2 virus from infected epithelial cells via CD147 and participate in the local and systemic spread of the virus. It is important to note that the expression of CD147 is regulated by high concentrations of glucose; this may reflect its correlation with obesity and potentially with diabetes, two comorbidities related to the development of more severe COVID-19 [42].
2.5. Evasion of SARS-CoV-2 and the widespread inflammation
In a similar process to the SARS-CoV infection, after SARS-CoV-2 entry into the infected cell, the presence of viral replication is detected through pattern recognition receptors (PRRs) which include the family of Toll-like receptors (TLRs). In particular, for RNA viruses such as CoVs, the molecular patterns associated with viral pathogens (PAMPs) such as viral genomic RNA, dsRNA (intermediate formed during viral replication) and mRNA, are recognized by endosomal RNA receptors, TLR3 and TLR7/8 and the cytosolic RNA sensor, the retinoic acid-inducible gene I (RIG-I) and protein 5 associated with melanoma differentiation (MDA5). These virus-specific RNA structures culminate in the oligomerization of these receptors and the activation of transcription factors, mainly interferon regulatory factors (IRFs) and nuclear factor kB (NF-kB) [43].
Transcriptional activation of IRFs and NF-kB results in two general antiviral programs. The first is related to cellular antiviral defenses which are mediated by the transcriptional induction of type I and III interferons (IFN-I and IFN-III, respectively) and subsequent positive regulation of IFN-stimulated genes (ISGs). The second antiviral response involves the recruitment and coordination of specific leukocytes through the secretion of pro-inflammatory chemokines and cytokines [44,45]. If properly located and early activated both systems can limit CoV infection [46].
Thus, viral pathogens develop mechanisms to escape immune recognition and suppress the functions of IFNs and ISGs. CoVs can interfere with any of the following steps of innate antiviral immunity: (1) innate detection, (2) IFN production, (3) IFN signaling and (4) effective ISG function [5]. Several studies have shown that structural and non-structural proteins in SARS-CoV are responsible for suppressing IFN release in vitro and in vivo [5,11,47] and that SARS-CoV-2 acts in a similar way, as it inhibits IFN-I and IFN-III production in infected cell lines, primary human bronchial cells as well as in a ferret model [48]. In fact, patients with severe COVID-19 demonstrate remarkably impaired IFN-I signatures compared to mild or moderate cases [11].
In addition to blocking IFN signaling, CoVs also promote the activation of other inflammatory pathways. SARS-CoV-2, through non-structural proteins NSP9 and NSP10, induces the production of IL-6 and IL-8, potentially by inhibiting NKRF, an endogenous NF-kB repressor. Collectively, these processes lead to a progressive increase in viral load and hypercytokinaemia, also called “cytokine storm”, observed in patients with COVID-19 [48].
After infecting lung cells such as type II pneumocytes, SARS-CoV-2 destroys these cells through the pyroptosis process, thus triggering a local immune response. In most cases, this process is able to resolve the infection. However, in some cases, a dysfunctional immune response occurs, which can cause the severe lung damage and develop into a systemic pathology [49].
It is noted that the immune dysfunction is mediated by a polarized response by T helper 1 (TH1) cells which produce pro-inflammatory cytokines such as IL-6, IFNγ, MCP-1 and IP-10 [50]; these cytokines are also elevated in the blood of patients affected by the disease. The secretion of these cytokines and chemokines attracts immune cells mainly monocytes and T lymphocytes from the blood to the infection sites which in turn will produce more cytokines, making the process of cell chemoattraction consecutive; this amplification loop results in an exacerbated production of cytokines such as IL-2, IL-7, IL-10, granulocyte colony stimulating factor (G-CSF), IP-10, MCP-1, macrophage inflammatory protein 1α (MIP1α) and tumor necrosis factor α (TNF-α), in addition to those mentioned above [49,51]. These events increased vascular permeability, fluid and protein leakage as well as reduced gas exchange [43,52].
The histopathological characteristics of COVID-19-related pneumonitis includes epithelial changes with diffuse alveolar damage, pneumocyte denudation and atypia, vascular changes including microvascular damage and thrombi, deposits of intra-alveolar fibrin with the presence of hyaline membrane associated with insufficient surfactant production [53].
One of the main cytokines to play an important role in the pathophysiology of COVID-19 is IL-6. Elevated levels of IL-6 are related to an increased inflammatory process, cardiovascular damage, triggering an excessive release of several cytokines which has been called a cytokine storm, and worsening the clinical condition of patients [51,54].
The cytokine IL-6 activates numerous cells that express the glycoprotein receptor (gp130), the membrane-bound IL-6 receptor and a soluble form of the IL-6 receptor that interacts with gp130 and thus causes the activation of signaling JAK/STAT, which in turn stimulates the production of IL-6 [43]. This cytokine can further activate macrophages to secrete MCP-1, increase expression of cell adhesion molecules as well as stimulate the proliferation and migration of vascular smooth muscle cells [24]. Thus, the abnormal increase in IL-6 levels may be implicated at least in part in the occurrence cardiovascular diseases (e.g. coronary atherosclerosis, inflammation in the vascular system resulting in diffuse microangiopathy with thrombosis) observed in COVID-19 patients [34,55].
In brief the pathophysiology of SARS-CoV-2 infection is characterized by an imbalance between the host late antiviral response and a dysfunction of the immune system which results in an exacerbated inflammatory response strongly implicated in multiple organ associated to the imbalance between the enzymes of the renin angiotensin system [48,56] (Fig. 1 ).
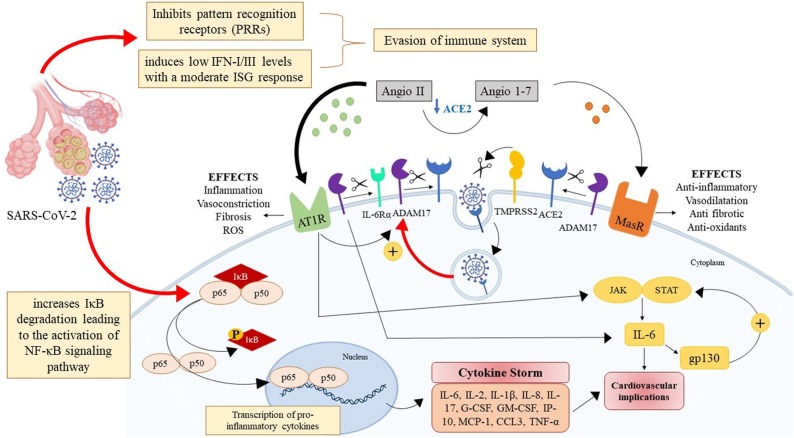
Fig. 1.
Hypothetical mechanism by which SARS-CoV-2 successfully evade the immune system while maintaining an inflammatory feedback loop. Following infection coronaviruses remain highly pathogenic at least in part due to the various viral mechanisms allowing them to evade and suppress the IFN response. The CoVs interfere with several processes in innate antiviral immunity. The SARS-CoV-2 can inhibit pattern recognition receptors (PPRs) and/or reduce IFN-I/III levels. ADAM17 (a desintegrin and metalloproteinase 17)-mediated proteolytic cleavage of ACE2 is upregulated by endocytosed SARS-CoV-2 viral particle, that results in over-production of angiotensin II (Angio II) by the related enzyme ACE. The elevate angiotensin II levels increase the activity of angiotensin 1 receptors (AT1R) as found in conditions like fibrosis and inflammation as well as increase reactive oxygen species (ROS) formation and vasoconstriction. In turn, increased angiotensin II enhances IL-6 production via JAK/STAT pathway, thus establishing a positive feedback loop. Moreover, the angiotensin II/AT1 receptor axis activates ADAM17 that cleaves and inactivates ACE2, enhancing angiotensin II levels. In addition, ADAM17 induction also cleaves the membrane form of IL-6Rα to the soluble form (sIL-6Rα), followed by the gp130-mediated activation of STAT, further amplifying the IL-6 signal. The SARS-CoV-2 can also activate NF-κB signaling pathway through increased IκB degradation, a process that leads to the transcription of several pro-inflammatory cytokines that results in what is called a cytokine storm.
3. Renin-angiotensin and kallikrein-kinin systems
The angiotensin-converting enzyme 2 (ACE2) is part of two systems that physiologically regulate a wide variety of effects [57]. The kallikrein-kinin system role in physiological control was first described when it was observed that hypotension and muscle relaxation were caused by a factor later called bradykinin [58] released from the action of trypsin and snake venoms on the so-called bradykininogens. Later, it was also discovered that the so-called bradykinin-enhancing peptides were found in the venom of snakes of the Bothrops jararaca species and had the effect of inhibiting the degradation of bradykinin [59,60].
ACE and ACE2 are fundamental enzymes of the renin-angiotensin system (RAS) involved in hydroelectrolytic control and blood pressure regulation [61]. The system keeps the levels of vasoconstrictor and vasodilator agents in balance. A drop in blood pressure causes the juxtaglomerular cells of the kidney to increase renin production and release that will cleave angiotensinogen produced by the liver into angiotensin I. Once in circulation, angiotensin I can be cleaved by two different enzymes, the angiotensin converting enzyme 1 (ACE or kininase II) which generates the angiotensin II and by a neutral endopeptidase to release angiotensin 1–7. The angiotensin converting enzyme 2 (ACE 2) (propylendopeptidase) cleaves angiotensin II into angiotensin 1–7 [62].
ACE2 is a membrane-bound monocarboxypeptidase having great similarities to ACE (sometimes called ACE1) which is a dipeptidylcarboxypeptidase [62]. ACE levels is far higher than the level of ACE2. ACE2 can be constitutively found in the lungs and other tissues and the angiotensin-(1–7) that it generates is a vasodilating agent. Angiotensin-(1–7) can be further metabolized into angiotensin-(1–5) by the ACE [63]. However, ACE2 does not cleave bradykinin [64,65] but cleaves the BK metabolite desArg9-BK which is the most potent B1 receptor agonist and inflammatory mediator [61].
Both angiotensin II and angiotensin (1–7) have important and opposite biological effects, resulting from the activation of specific receptors. Angiotensin II is capable of activating the receptors called AT 1 found in several organs. Its activation leads to vasoconstriction, increased release of antidiuretic hormone (ADH), aldosterone, water and Na+ reabsorption, remodeling, cell proliferation, thrombosis and inflammation (increased recruitment of lymphocytes, PMN and expression of adhesion molecules). The activation of the MAS receptor (also called Receptor 3) by Angiotensin (1–7) causes vasodilation, apoptosis, reduction in cell proliferation [66] and fibrosis [67]. It also reduces cytokine production (IL1β, IL6, TNF, IL12), inhibits the intracellular signaling including NFkB, MCP1, p38 MAPK, CCL2, JNK, ERK ½, and act as an antifibrogen, cardioprotector, nephroprotector as well as protecting against lung injuries [[68], [69], [70], [71]] (Fig. 2 ).
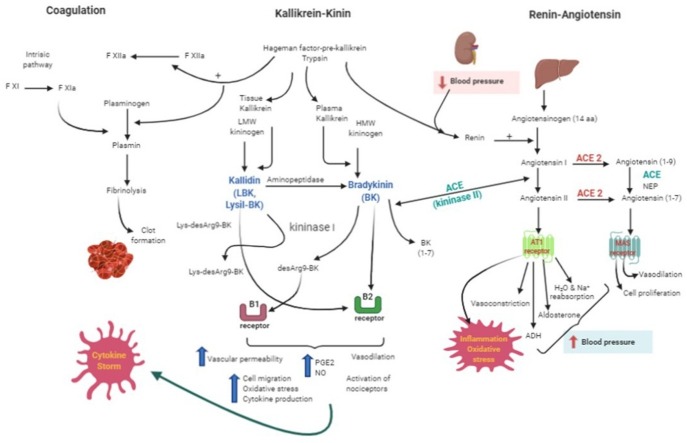
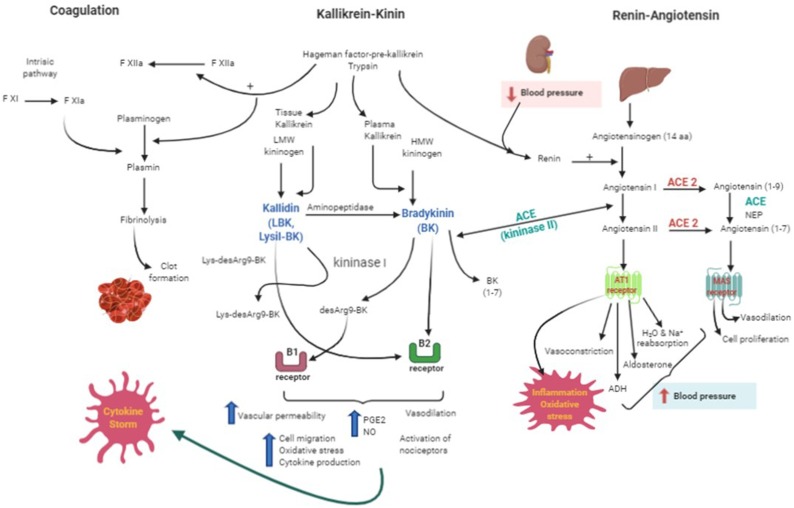
Fig. 2.
The interplay between renin-angiotensin, kallikrein-kinin and coagulation system activation during COVID infection. Renin cleaves the angiotensinogen into angiotensin I that can be converted to angiotensin II by the angiotensin converting enzyme (ACE or kininase II). Both angiotensin I and II can also be cleaved by angiotensin converting enzyme II (ACE 2) into angiotensin (1-9) and angiotensin (1-7) respectively. Angiotensin II will activate receptors (AT1) which leads to oxidative stress, inflammation, increase in blood pressure due to a vasoconstriction whereas the angiotensin (1-7) will produce vasodilation and potentiates the effects of bradykinin). Lysyl-bradykinin (LBK) and bradykinin (BK) produced after plasma kallikrein cleavage of low or high molecular weight kininogen (LMW or HMW kininogen) respectively, will: 1) directly activate the B2 receptors; 2) be cleave by the kininase I leading to the metabolites lys-desArg9BK and desArg9BK (B1 receptor agonists). LBK can also be converted to BK through an aminopeptidase action. Activation of B1 or B2 receptors will culminate in inflammatory events exacerbated by the cytokine storm. In the coagulation cascade, the Hageman factor can activate factor XII and convert plasminogen into plasmin which leads to fibrinolysis and clot formation. ACE = angiotensin converting enzyme; ACE2=angiotensin converting enzyme 2; NPE = neutral endopeptidase; HMW kininogen = high molecular weight kininogen; LMW kininogen = low molecular weight kininogen; LBK = lysil-bradykinin; BK = bradykinin. Created with BioRender.com.
ACE also has a fundamental role in the Kallikrein Kinin System (KKS). Kinins are produced from the action of a group of serine proteases called kallikreins on protein precursors named the kininogens. The kinin activation/synthesis pathway originates with the action of Hageman factor and/or trypsin in the plasma and tissue kallikreins, each cleaving the high and low molecular weight kininogen giving rise to bradykinin (BK) and kallidin (lysyl-BK), respectively ([72,73]). Once formed, Lys-Bk can still be converted to BK by the action of an aminopeptidase. Other carboxypeptidases (M and N) remove arginine from the C-terminal and give rise to desArg9-BK (DABK) and Lys-desArg9-BK (LDABK), respectively from BK and Lys-BK [74,75].
The interconnection of the kallikrein-kinin and renin-angiotensin systems is done by angiotensin-converting enzymes present in both systems. In plasma the half-life of kinins is in the order of seconds [60] being cleaved mainly by the angiotensin-converting enzyme 1 (ACE or kininase II) which removes the carboxyl terminal of BK and generates BK (1–7), an inactive peptide. This metabolite can be rapidly cleaved by ACE generating BK (1–5) [76,77]. Other aminopeptidases and carboxypeptidases can also degrade bradykinin.
Once synthesized, both bradykinin and lysyl-BK can activate B2 receptors constitutively expressed in different organs of the body and whose activation causes an increase in vascular permeability, vasodilation and activation of nociceptors. They can also activate the B1 receptors which are expressed in high concentrations in a diversity of tissues in inflammatory conditions. However, the binding and stimulation of B1R by BK is dependent on its conversion to DesArg9-BK by Kininase I, a carboxypeptidase N or M. In fact, BK has low affinity for the native and cloned B1R [78,79]. Although both receptors can be activated by either BK and lysyl-BK, the metabolite resulting from BK cleavage, DesArg9-BK is the most specific and potent agonist of the type 1 receptor [75] (Fig. 2).
3.1. Renin-angiotensin and kallikrein systems in COVID-19
Like other SARS−COV, SARS−COV2 is able to enter the host cell. Its entry results from the induction of a TNF-α converting enzyme (TACE) that is dependent on the shedding of the ectodomain of ACE2 with its consequent downregulation, an effect that facilitates the entry of the virus into the host cell. Once bound, ACE2 is hijacked and internalized by the cell. In addition, cleavage of its extracellular domain is also observed [80]. Downregulation of the activity and availability of ACE2 cause an imbalance in the renin-angiotensin system, an accumulation of angiotensin II and a reduction in levels of angiotensin (1–7) that favors the binding of angiotensin II on the AT1 receptor and the effects of its activation; this is one of the hypotheses presented to explain the cases of severe respiratory syndrome that occurs in some SARS-CoV-2 infections.
Nicolau et al. [57] suggested that the inhibition of the ACE2 pathway reduces the degradation of angiotensin II and DesArg9-BK. Together, this triad of factors can cause an increase in the levels of angiotensin II, a reduction in the synthesis of angiotensin (1–7) and an increase in the availability of DesArg9-BK. The activity of TMPRSS2 (a kallikrein-like effect) on plasma kininogen could also lead to an increase in the production of BK and DesArg9-BK [81]. The cysteine protease present in the virus has also the same action on kininogen which suggests that these enzymes may become candidates for the production of kinins during infection by the virus [81,82].
ACE2 downregulation causes RAS imbalance, angiotensin II accumulation, plus angiotensin-(1–7) decrease, which shifts angiotensin II binding toward the AT1, leading to pro-inflammatory and cardiovascular injury mechanisms. It also causes imbalance of the KKS, DesArg9-BK accumulation and BKB1R activation leading to pro-inflammatory repercussions. SARS-CoV2 invasion in target cells also depends on a TMPRSS2 membrane protease necessary for the cleavage of its spike protein. Active TMPRSS2 additionally cleaves the kininogen and activates the production of bradykinin (BK). The virus itself also expresses a cysteine protease which could activate the kinin pathway by interacting with kininogen. BK via BKB2R also contributes to the proinflammatory pathway by activating nitric oxide (NO) and prostaglandins (PGs) synthesis.
BKB1R is strongly upregulated by inflammatory mediators (mainly cytokines) which increase endothelial permeability and leukocyte migration. A positive feedback loop between BKB1R and cytokines generates the “cytokines storm” that in turn leads to a sepsis-like conditions. In addition, kallikrein causes an imbalance of coagulation system by activating factor 12 (FXII) and plasmin. The two mechanisms contribute to the formation of intravascular microthrombi observed mainly in lung tissues (Fig. 2). The presence of plasmin increases cleavage of spike proteins which boosts SARS-CoV-2 virulence.
Taken together all of these events lead to a positive inflammatory feedback loop contributing and amplifying the lung damage caused by COVID-19 and a poor prognosis for the treatment of patients who develop severe illness caused by SARS-CoV-2. A better understanding of the several pathways involved in the disease can lead to more treatment options with better prognosis.
4. Drug repurposing in COVID-19 treatment
The repositioning of a drug for the treatment of new diseases has several advantages such as the reduced development time, the known side effects and the reduced financial risk when compared to a new drug development (39). Various investigators have tested a diversity of proteins from SARS-CoV-2 as potential targets for drug repurposing as they take part in virus entry and replication in cells or in important metabolic pathways and/or immune responses in the host. Other approaches included targeting potential SARS-CoV-2 Mpro (a protease essential for virus replication) inhibitors with FDA-approved antivirals, such as inhibitors of HIV-1 [e. g., lopinavir and ritonavir] and HCV [e.g., boceprevir] proteases, as well as antineoplastic [e.g. carmofur] and antibacterial [e.g., doxycycline] drugs [24,83,84]. Another important viral target is the PLpro that recognizes and hydrolyses ubiquitin from cellular proteins; this regulates the post-translational modifications of signaling molecules that trigger innate immune response of the host [85]. Inhibitors of SARS-CoV-2 PLpro include fostamatinib disodium (a tyrosine kinase inhibitor) and natural products (platycodin D), two examples of FDA-approved drugs [86]. TMPRSS2 is also a potential drug target since its inhibition or blockade could prevent virus infection. Examples of this group are serine protease inhibitors camostat mesylate and others [33].
One important condition that can be observed in several cases is the so called “cytokine storm”. A syndrome caused by an overactivation of the immune system and a huge production of cytokines affecting several organs [87]. Cytokine receptor antagonists also have therapeutic potential in the condition coined “cytokine storm”. Humanized antibodies (i.e., tocilizumab and sarilumab) could be used as an IL-6 receptor antagonist and are under clinical trial for the treatment of COVID-19 [88].
Drugs aiming at the host system such as the RAS is another valuable approach. The interaction between S protein and ACE2 makes the protein a potential drug target for the treatment of COVID-19. Different approaches were suggested including the development of a vaccine using the S protein, the administration of a soluble form of ACE2 or the administration of small molecules or antibodies that prevent the interaction ACE2-S protein [89]. Another option could be the use of drugs presenting anti-SARS-CoV effect through glycosylation of ACE2 (i.e. chloroquine) [90].
ACE inhibitors such as Captopril and angiotensin receptor antagonists such as Losartan, highly popular drugs for the treatment of hypertension and other cardiovascular diseases have raised a lot of questions in relation to the COVID-19 treatment [91]. It has been suggested that patients using these drugs could be at higher risk of COVID infection since inhibitors of RAS may increase expression of ACE2. Although these drugs do not directly inhibit/affect ACE2 activity, experimental protocols showed that they can upregulate the expression/activity of ACE2 [92,93], increasing the availability of ACE2 to SARS-CoV-2. This is still controversial. The use of one of these groups of drugs could rather protect patients from lung injury due to a reduction in the angiotensin II pathway activated by the angiotensin receptor 1 [94].
Taken together it is clear that there are many possible drugs already used for the treatment of various diseases that can be repositioned for the treatment of COVID-19. It is not yet possible to identify only one drug, but based on the present research effort, efficient and low-cost treatment options should be unraveled. The efforts require a better understanding the angiotensin and kinin systems in patients infected with COVID-19.
Acknowledgments
We would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do RIo de Janeiro(FAPERJ) for grant and fellowship supports.
References
- 1.Zhenming J., Yao Z., Yuan S., Bing Z., Haofeng Z., Yan W., Yan Z., Chen Z., Tianyu T., Xiaoyu D., Yinkai D., Jing Y., Xiaobao Y., Xiuna Y., Kailin Y., Xiang L., Luke G., Gengfu X., Leike Z., Haitao Y., Zihe R. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 2.Huang M.M.X., Fengxiang W., Liang H., Lijuan W.M.M., Chen M.M.K. Epidemiology and clinical characteristics of COVID-19. Arch. Iran. Med. 2020;23:268–271. doi: 10.34172/aim.2020.09. [DOI] [PubMed] [Google Scholar]
- 3.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., Bolling C., Dijkstra G., Voors A.A., Osterhaus A.D.M.S., van der Voort P.H.J., Mulder D.J., van Goor H. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park A., Iwasaki A. Type I and type III interferons – induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020;12:9–14. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y. Unveiling the origin and transmission of 2019-nCoV. Trends Microbiol. 2020;28:239–240. doi: 10.1016/j.tim.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worldometer . 2020. COVID-19 Coronavirus Pandemic.www.worldometers.info/coronavirus [Google Scholar]
- 9.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet. 2020;20:30113–30114. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L., Risson E., Saffern M., Salomé B., Selvan M.E., Spindler M.P., Tan J., van der Heide V., Gregory J.K., Alexandropoulos K., Bhardwaj N., Brown B.D., Greenbaum B., Gümüş Z.H., Homann D., Horowitz A., Kamphorst A.O., Lafaille M.A.C., Mehandru S., Merad M., Samstein R.M. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao W., Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30:367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B., Tian E.K., He B., Tian L., Han R., Wang S., Xiang Q., Zhang S., Arnaout T.E., Cheng W. Overview of lethal human coronaviruses. Signal Transd. Target Ther. 2020;5:89–104. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Báez-Santos Y.M., St. John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang L., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA Ribose 29-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2020;2 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha S.K., Prasad S.K., Islam M.A., Gurav S.S., Patil R.B., AlFaris N.A., Aldayel T.S., AlKehayez N.M., Wabaidur S.M., Shakya A. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARSCoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J. Biomolecul. Struc. Dyn. 2020;2020 doi: 10.1080/07391102.2020.1779132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabaan A.A., Ahmed S.H.A., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Aldana D.K.B., Morales A.J.R. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Le Infezioni Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 20.Feng W., Zong W., Wang F., Ju S. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review. Mol. Cancer. 2020;19:100. doi: 10.1186/s12943-020-01218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letko M., Marzi A., Munster M. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B beta coronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimoto F.K. The proteins of severe acute respiratory syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P.P., Blet A., Smyth D., Li H. The science underlying COVID-19: implications for the cardiovascular system. Circ. Res. 2020;142:68–78. doi: 10.1161/CIRCULATIONAHA.120.047549. [DOI] [PubMed] [Google Scholar]
- 25.Lam T.T.Y., Ja N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y.M., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 26.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 27.Letko M., Seifert S.N., Olival K.J., Plowright R.K., Munster V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020;18:461–471. doi: 10.1038/s41579-020-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls A.C., Park Y.C., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARSCoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 cite. iScience. 2020;23 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64:1–7. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang J., Wan Y., Costela-Ruiz C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Weber H.K., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and hostACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K., Chen W., Zhou Y., Lian J., Zhang Z., Du P. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxivorg. 2020 doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.South A.M., Tomlinson L., Edmonston D., Hiremath S., Sparks M.A. Controversies of renin–angiotensin system inhibition during the COVID-19 pandemic. Nat. Rev. Nephrol. 2020;16:305–307. doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z., Mi L., Xu J., Yu J., Wang X., Jiang J., Xing J., Shang P., Qian A., Li Y., Shaw P.X., Wang J., Duan S., Ding J., Fan C., Zhang Y., Yang Y., Yu X., Feng Q., Li B., Yao X., Zhang Z., Li L., Xue X., Zhu P. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavírus. J. Infec. Dis. 2020;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020;16:434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P. Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and imune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Basic Transl. Allergy Immunol. 2020 doi: 10.1111/ALL.14429. Accepted article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavezzi A., Troiani E., Corrao S. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020;10:24–30. doi: 10.4081/cp.2020.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transd. Targ. Ther. 2020;5:84–94. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kindler E., Thiel V. SARS-CoV and IFN: too little, too late. Cell Host Microbe. 2016;19:139–141. doi: 10.1016/j.chom.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sallenave J.M., Guillot L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in Covid-19: key therapeutic targets? Front. Immunol. 2020;11:1–9. doi: 10.3389/fimmu.2020.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type i interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia X., Guo J., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Iu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;29:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dashraath P., Wong J.L.J., Lim M.X.K., Lim L.M., Li S., Biswas A., Choolani M., Mattar C., Su L.L. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am. J. Obst. Gynecol. 2020;222:521–531. doi: 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polak S.B., Gool I.C.V., Cohen D., von der Thüsen J.H., van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod. Pathol. 2020 doi: 10.1038/s41379-020-0603-3. Accepted paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo W., Yu H., Gou J., Li X., Sun Y., Li J., Liu L. 2020. Clinical Pathology of Critical Patient with Novel Coronavirus Pneumonia (COVID-19) Preprints 2020. 2020020407. [Google Scholar]
- 55.Murakami M., Kamimura D., Hirano H. Pleiotropy and Specificity: insights from the interleukin 6 family of cytokines. Immun. Rev. 2019;50:812–831. doi: 10.1016/j.immuni.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 56.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.G., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicolau L.A.D., Magalhães P.J.C., Vale M.L. What would Sérgio Ferreira say to your physician in this war against COVID-19: how about kallikrein/kinin system? Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rocha e Silva M., Beraldo W.T., Rosenfeld G. Bradykinin, a hypotensive and smooth muscle stimulating factor released from plasma globulin by snake venoms and by trypsin. Am. J. Physiol. 1949;156:261–273. doi: 10.1152/ajplegacy.1949.156.2.261. [DOI] [PubMed] [Google Scholar]
- 59.Ferreira S.H., Rocha e Silva M. Potentiation of bradykinin and eledoisin by BPF (bradykinin potentiating factor) from Bothrops jararaca venom. Experientia. 1965;21:347–349. doi: 10.1007/BF02144709. [DOI] [PubMed] [Google Scholar]
- 60.Ferreira S.H., Vane J.R. The disappearance of bradykinin and eledoisin in the circulation and vascular beds of the cat. Br. J. Pharmacol. Chemother. 1967;30:417–424. doi: 10.1111/j.1476-5381.1967.tb02148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan N., Woolf B., Robinson K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 62.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 63.Chappell M.C. Biochemical evaluation og renin-angiotensin system: the good, bad, and absolute? Am. J. Physiol. Heart Circ. Physiol. 2016;310:H137–H152. doi: 10.1152/ajpheart.00618.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1–7 axis of the renin–angiotensin system in heart failure. Circ. Res. 2016;118:1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costa-Neto C.M., Duarte I.A., Lima V., Maria A.G., Prando E.C., Rodriguez D.Y., Santos D.A., Souza P.P.C., Parreiras-e-Silva L.T. Non-canonical signaling and roles of the vasoactive peptides angiotensins and kinins. Clin. Sci. 2014;126:753–774. doi: 10.1042/CS20130414. [DOI] [PubMed] [Google Scholar]
- 67.Chappell M.C., Al Zayadneh E.M. Angiotensin-(1-7) and the regulation of anti-fibrotic signaling pathways. J. Cell Signal. 2017;2:1–9. doi: 10.4172/2576-1471.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santos R.A.S., Campagnole-Santos M.J., Andrade S.P. Angiotensin-(1-7): an update. Regul. Pept. 2000;91:45–62. doi: 10.1016/s0167-0115(00)00138-5. [DOI] [PubMed] [Google Scholar]
- 69.Ferrario C.M. Does angiotensin-(1-7) contribute to cardiac adaptation and preservation of endothelial function in heart failure? Circulation. 2002;105:1523–1525. doi: 10.1161/01.cir.0000013787.10609.dc. [DOI] [PubMed] [Google Scholar]
- 70.Campbell D.J. The renin-angiotensin and the kallikrein-kinin systems. Int. J. Bioch. Cell Biol. 2003;35:784–791. doi: 10.1016/s1357-2725(02)00262-5. [DOI] [PubMed] [Google Scholar]
- 71.Simões e Silva A.C., Silveira K.D., Ferreira A.J., Teixeira M.M. ACE2, angiotensin‐(1‐7) and M as receptor axis in inflammation and fibrosis. Br. J. Pharmacol. 2013;169:477–492. doi: 10.1111/bph.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poud D., Kaplan A.P. Kinin formation: mechanisms and role in inflammatory disorders. Annu. Rev. Immunol. 1988;6:49–83. doi: 10.1146/annurev.iy.06.040188.000405. [DOI] [PubMed] [Google Scholar]
- 73.Bhoola K.D., Figueroa C.D., Worthy K. Bioregulation of kinins: kallikreins, kininogens, and kininases. Pharmacol. Rev. 1991;44:1–80. [PubMed] [Google Scholar]
- 74.Hall J.M. Bradykinin receptors: pharmacological properties and biological roles. Pharmacol. Ther. 1992;56:131–190. doi: 10.1016/0163-7258(92)90016-s. [DOI] [PubMed] [Google Scholar]
- 75.Marceau F. Kinin B1 receptor: a review. Immunopharmacol. 1995;30:1–26. doi: 10.1016/0162-3109(95)00011-h. [DOI] [PubMed] [Google Scholar]
- 76.Jaspard E., Wei L., Alhenc-Gelas F. Differences in the properties and enzymatic specificities of the two active sites of angiotensin I-converting enzyme (kininase II), Studies with bradykinin and other natural peptides. J. Biol. Chem. 1993;268:9496–9503. [PubMed] [Google Scholar]
- 77.Kuhr F., Lowry J., Zhang Y., Brovkovych V., Skidgel R.A. Differential regulation of inducible and endothelial nitric oxide synthase by kinin B1 and B2 receptors. Neuropeptides. 2010;44:145–154. doi: 10.1016/j.npep.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Regoli D., Allogho S.N., Rizzi A., Gobeil F.J. Bradykinin receptors and their antagonists. Eur. J. Pharmacol. 1998;348:1–10. doi: 10.1016/s0014-2999(98)00165-4. [DOI] [PubMed] [Google Scholar]
- 79.Leeb-Lundberg L.M.F., Marceau F., Müller-Esterl W., Pettibone D.J., Zuraw B.L. International union of pharmacology. XLV. Classification of the kinin receptor family: from molecular mechanisms to pathophysiological consequences. Pharmacol. Rev. 2005;57:27–77. doi: 10.1124/pr.57.1.2. [DOI] [PubMed] [Google Scholar]
- 80.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2013;88:1293–12307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lalmanach G., Naudin C., Lecaille F., Fritz H. Kininogens: more than cysteine protease inhibitors and kinin precursors. Biochimie. 2010;92:1568–1579. doi: 10.1016/j.biochi.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 82.Solowiej J., Thomson J.A., Ryan K., Luo C., He M., Lou J., Murray B.W. Steady-state and presteady-state kinetic evaluation of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLproCysteine protease: development of an ion-pair model for catalysis. Biochemistry. 2008;47:2617–2630. doi: 10.1021/bi702107v. [DOI] [PubMed] [Google Scholar]
- 83.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12:1–17. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARSCoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang M.L., Dai F.H., Wang Q.W., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S., Agoramoorthy G. COVID-19: consider IL-6 receptor antagonist for the therapy of cytokine storm syndrome in SARS-CoV-2 infected patients. J. Med. Virol. 2020 doi: 10.1002/jmv.26078. [Internet]. 2020 [cited 2020 Aug 8]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for Coronavirus disease 2019 (COVID-19) JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 90.Devaux C.A., Rolain J.-M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Azushima K., Morisawa N., Tamura K., Nishiyama A. Recent research advances in renin-angiotensin-aldosterone system receptors. Curr. Hypertens. Rep. 2020;22:22. doi: 10.1007/s11906-020-1028-6. [DOI] [PubMed] [Google Scholar]
- 92.Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., Diz D.I., Gallagher P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 93.Soler M.J., Ye M., Wysocki j., William J., Lloveras J., Battle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am. J. Physiol. Renal Physiol. 2009;296:F398–F405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- 94.Gurwitz D. Angiotensin receptor blockers as tentative SARSCoV2 therapeutics. Drug Dev. Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]