Abstract
Processing of animal carcasses and other animal wastes in rendering plants is a significant source of antibiotic resistant microorganisms. The main goal of this study was to investigate the resistance to 18 antibacterial agents including β-lactams, fluoroquinolones, colistin and virulence factors (iss, tsh, cvaC, iutA, papC, kps and ibeA genes) in 88 Escherichia coli strains isolated from a rendering plant over 1 year period. ESBL (Extended-spectrum beta-lactamases) and plasmid-mediated Amp were screened by interpretative reading of MIC. ESBL phenotype was detected in 20.4% of samples and high level of resistance to fluoroquinolone was found in 27.2% of strains. Cephalosporinase CTX-M1, cephamycinase CMY-2, integrase 1 and transposon 3 genes were detected by PCR. Furthermore, there were found three CMY-2 producing E. coli with O25b-ST131, resistant to the high level of enrofloxacin and containing the gene encoding the ferric aerobactin receptor (iutA). One enrofloxacin resistant E. coli strain possessed iss, ibeA, kps and papC virulence genes also with CMY-2, integrase1 and Tn3. ST131 E. coli with CMY-2 has a zoonotic potential and presents a serious health risk to humans.
Subject terms: Antimicrobials, Natural hazards, Occupational health, Public health
Introduction
Escherichia coli is regarded as a representative indicator of antimicrobial resistance of Gram-negative bacteria.
Risk of transmission of β-lactam resistance to nosocomial E. coli, especially the increasing resistance to 3rd and 4th generation cephalosporins and carbapenems became a serious issue worldwide1.
AmpC β-lactamases are enzymes commonly isolated from Enterobacteriaceae and a few other cephalosporin resistant bacteria, encoded on the chromosomes. They mediate resistance to cephalosporins (cefoxitin, cefazolin, cephalothin), most penicillins and β-lactamase inhibitor combinations2. Jacoby2 described induction or overexpression of chromosomally-located, species-specific AmpC genes and the acquisition of plasmid-encoded AmpC genes (e.g. blaCMY-like, blaACC-like, blaDHA-like) in E. coli.
Resistance of E. coli to cephalosporins and fluoroquinolones is of considerable concern, as the prevalence of horizontally acquired resistance mechanisms has increased significantly in the past 20 years. High-level resistance to fluoroquinolones is associated with DNA gyrase and topoisomerase mutations although intermediate resistance or enhancement of clinical resistance is possible by chromosomal efflux pump upregulation and/or plasmid-borne genes encoding target site protection (qnr), efflux (qepA), or modification of fluoroquinolones by acetylase (aac(6′)-Ib-cr)3.
Escherichia coli sequence type 131 (ST131) is a worldwide pandemic clone responsible for the most part of community-onset antimicrobial-resistant infections. A high prevalence of the clone (∼ 30–60%) has been identified amongst fluoroquinolone-resistant E. coli. Moreover, it potentially possesses a variety of β-lactamase genes; they most often include CTX-M family β-lactamases, and, less frequently, TEM, SHV and CMY β-lactamases4.
ST131 E. coli is widely disseminated within the antibiotic-resistant community and was associated with hospital-onset of E. coli infection in the UK. It has been identified among ESBL-producing isolates in seagulls and rats, but in Spain the prevalence of the clone amongst poultry and on pig farms was low4.
The E. coli ST131 clones cause many multidrug nosocomial infections worldwide (cystitis pyelonephritis, abdominal, soft tissue infections, meningitis, osteoarticular infection, myositis, septic shock and other) and probably can produce biofilms5.
Processing of food animal carcasses and secondary raw materials of animal origin in rendering plants is a significant source of microorganisms that contributes to the risk of pollution of the environment. A large body of information is available about antibiotic resistance of microorganisms in municipal wastewater treatment plants that can act as reservoirs and environmental suppliers of antibiotic resistance. They may serve as reservoirs for ESBLs and AmpC producing Enterobacteriaceae6–8.
However, there are still some gaps in our knowledge about the role of rendering plants in the spread of antibiotic resistance.
The aim of the study was to determine the resistance to 18 antibacterial agents including β-lactams, fluoroquinolones, colistin and virulence factors (iss, tsh, cvaC, iutA, papC, kps and ibeA genes) in E. coli isolated from surface swabs collected in the processing space and from waste water produced by the investigated rendering plant.
Material and methods
A rendering plant is a processing operation where materials of animal origin are recycled. The following processes are involved: unloading of raw material brought for processing, its sorting, primary processing and sampling, sterilisation, separation of fat and feed meals of animal origin, pressing, processing of feed meals, and processing of animal fat. The operation premises are divided to a section used for common processing of materials of categories I and II and (high risk—meat-bone meal have to be burned at a temperature of 850 °C) a separate section for processing of category III materials (lower risk—meat-bone meal can be used for production of pet granules).
Destruction (crushing of material to 50 mm particles) and sterilisation of animal by-products using the temperature of 133 °C and pressure of 3 bars during 20 min ensure high level of sanitization and limit the risk of spread of microorganisms to the environment. The above mentioned parameters are critical for adequate processing of raw materials entering the rendering plant9. In the dryer the solid portion is separated from the liquid one (water) and the dried meat-bone meal is pressed during which process the fat is separated from the meat-bone mash. The technological procedure in the rendering plant includes processing of the wastewater and installation of a biological air washer—the components important for reducing the hygiene-epidemiological risks.
Sampling
Over 1 year period (10 times) swabbing procedure was used to obtain samples from surfaces in the processing section of the investigated rendering plant and additional samples were collected from raw wastewater in rendering plant. Altogether 88 samples were obtained and examined. Each sample from surface swabs and waste water was inoculated and multiplied in Buffered peptone water (Oxoid, Basingstoke, United Kingdom) and then were sub-cultured on Mac Conkey agar (Oxoid) at 37 °C overnight10.
Identification of E. coli
The suspect E. coli colonies from McConkey agar were identified by a matrix-assisted laser desorption/ionization (MALDI-TOF) biotyper (Bruker Daltonics, Leipzig, Germany). Bacterial extracts for mass spectrometry measurements were prepared as recommended by the manufacturer of the MS instrument. For MALDI-TOF analysis, one colony was spotted onto a ground steel target (Bruker Daltonik GmbH, Leipzig, Germany) and air dried for 15 min.
Each sample spot was overlaid with 2 μl of matrix solution (saturated solution of α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile with 2.5% trifluoroacetic acid), and again air dried for 15 min. To identify the relevant microorganisms, the raw spectra obtained for each isolate were imported into a BioTyper software, version 2.0 (Bruker Daltonik GmbH, Leipzig, Germany), and analysed without any user intervention10.
Antibiotic susceptibility
Eighty eight isolates of E. coli (one sample-one strain) were analysed for their antibiotic susceptibility and for the presence of ESBLs, pAmpC and for the high level fluoroquinolone resistance.
Minimal inhibitory concentrations (MIC) were determined according to VET01-S211 and EUCAST12, by a Miditech system (Bratislava, Slovakia) with interpretative reading of MIC13. The antibiotics used in the presented study were as follows: ampicillin (AMP), ampicillin and sulbactam (A + IB), ceftazidime (CAZ), ceftazidime with clavulanic acid (CAC), ceftriaxon (CTR), ceftiofur (CFF), cefquinome (CFQ), ertapenem (ETP), gentamicin (GEN), streptomycin (STM), nalidixic acid (NAL), ciprofloxacin (CIP), enrofloxacin (ENR), chloramphenicol (CMP), florfenicol (FLO), tetracycline (TET), cotrimoxazol (COT), colistin (COL).
Phenotypic confirmation of mechanisms of ESBLs and pAmpC to the β-lactams (CTR, CAZ, CAC) was carried out by reading the MIC levels12,14.
Phenotypic interpretation of chromosomal quinolone-resistance mechanisms was based on modification of the method by Kmet and Kmetova15. High-level resistance MIC for CIP (≥ 4 mg/L) and ENR (≥ 16 mg/L) involved three mutations in QRDR (gyrA and parC).
Genes of antibiotic resistance
ESBL genes for cefotaximases CTX-M16, plasmid ampicillinase/cephamycinase CIT17, plasmid quinolone resistance genes: oqxA, oqxB18, qepA19, qnrA, qnrB, qnrS and aac(6′)IbCr20, integrase 1 (Int1)21, Tn3—transposon22 were determined by PCR. DNA sequencing of the PCR products from cefotaximases (CTX-M1) and ampicilinases with primers CIT (CMY-2) was carried out. The O25b-ST131 clone was also detected by PCR23.
Virulence factors
Screening of E. coli isolates for ExPEC (Extra intestinal Pathogenic E. coli) virulence genes was carried out by PCR amplification of the following: iutA—ferric aerobactin receptor; cvaC—colicin V and kpsII—capsular polysialic acid virulence factor24; iss—increased serum survival25; tsh—temperature sensitive haemaglutinin26; papC—P fimbrial adhesin27; and ibeA—invasive factor of E. coli strains responsible for neonatal meningitis in humans28.
Ethics approval and consent to participate
This study does not qualify for review by the University of veterinary medicine and pharmacy Ethics Board.
Results
Antimicrobial susceptibility profiles
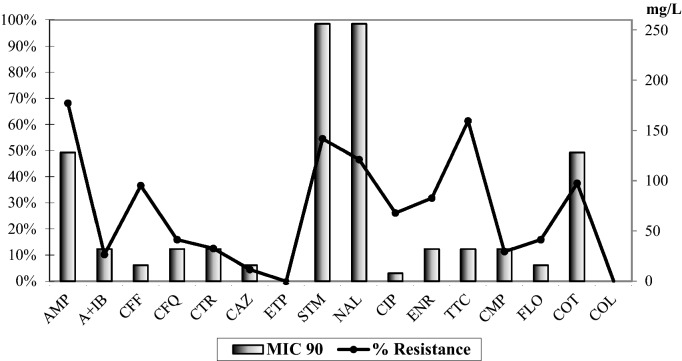
A modified microdilution method with the VetMIC panel was used to detected antimicrobial resistance in 88 E. coli strains (Fig. 1).
Figure 1.
Antibiotic resistance and MIC90 of 88 strains of E. coli isolated from rendering plant.
The highest incidence of beta-lactam resistance was observed for ampicillin (68%), followed by cephalosporins—veterinary ceftiofur (36% and MIC90 16 mg/L) and cefquinome (15.9% and MIC90 32 mg/L), ceftriaxone (12.5% and MIC90 32 mg/L) and ceftazidime (4.5% and MIC90 16 mg/L). Were detected also high resistance to streptomycin (54.5%), tetracyclin (61.4%), florfenicol (15.9%), chloramphenicol (11.39%) and cotrimoxazol (37.5%). All strains were susceptible to ertapenem with MIC 90 0.06 mg/L and colistin. A ratio of ceftazidime MIC to ceftazidime-clavulanic acid MIC greater than 8 indicates the presence of ESBL. From among all 88 investigated E. coli strains the ESBL phenotype was confirmed in 18 strains (20.4%). Three phenotypic confirmed ESBL strains from 18 strains which did not present CTX-M or CMY-2 genes are not in the Table 1. However, CMY-2 is related to AmpC.
Table 1.
Genotyping and antibiotic resistance in 15 selected E. coli isolated from rendering plant.
| No | ESBL/AmpC | Mobilome | Virulence | Antibiotic resistance | ST131 |
|---|---|---|---|---|---|
| 1 | CTX-M1, CMY-2 | Int1, Tn3 | cvaC, iutA | CTR,TTC, STM, FLO, COT | |
| 111 | CTX-M1, CMY-2 | Int1, Tn3 | cvaC, iutA | CTR, TTC, STM, FLO, COT | |
| 2 | CMY-2 | Int1, Tn3 | cvaC, iutA | ENR | ST131 |
| 3 | CMY-2 | Int1, Tn3 | iutA | TTC, ENR,COT | ST131 |
| 51 | CMY-2 | Int1, Tn3 | iutA | TTC, STM, ENR, COT | ST131 |
| 21 | CMY-2 | Int1, Tn3 | iss, ibeA, kps, papC | CTR, TTC, STM, ENR, COT | |
| 71 | CMY-2 | Int1, Tn3 | ND | TTC, FLO | |
| O3 | CMY-2 | Int1 | iss | TTC, STM, ENR | |
| P1 | CMY-2 | Int1 | iss, papC | STM, GEN, ENR,CMP | |
| K1 | CMY-2 | Tn3 | iss | TTC, ENR,COT | |
| P2 | Tn3 | papC | TTC, ENR, FLO, COT | ||
| O7 | Tn3 | papC | CTR, TTC, STM, NAL | ||
| C51 | Int1 | iss, papC, | CTR, TTC, ENR, CMP, COT | ||
| 4C | Int1 | iss, cvaC, iutA, papC | CTR, TTC, STM,ENR,CMP | ||
| C52 | Int1 | iss, iutA, tsh | CTR, TTC, STM, ENR, CMP, COT |
iutA ferric aerobactin receptor; cvaC colicin V; kps capsular polysialic acid virulence factor; iss increased serum survival; tsh temperature sensitive haemaglutinin; papC P fimbrial adhesin; ibeA invasive factor of E. coli; ND not defined.
Resistance to enrofloxacin was detected in 31.8% (MIC90 32 mg/L) of strains, to ciprofloxacin in 26.1% (MIC90 8.0 mg/L) and to nalidixic acid in 46.5% (MIC90 256 mg/L) of strains. Plasmid-mediated quinolone resistance (qnrS) and eflux genes oqxA, oqxB and genes qepA, gnrA, qnrB, qnrS, aac(6′)IbCr were not detected. High levels of MIC 90 for enrofloxacin and for ciprofloxacin indicate the presence of chromosomal fluoroquinolone resistance15.
Multidrug resistance was defined as resistance to at least three non-related antibiotics. Multidrug resistant E. coli isolated from rendering plant occurred in 15 strains of E. coli (17%), which were selected for PCR analysis (with the exception of E. coli strain No. 2).
The strains with the most interesting combination of properties were selected in the Table 1.
PCR detection of integrons, gene casset, virulence factors, CTX-M and CMY-2
We confirmed an integron-mediated antibiotic resistance in the selected isolates. Class 1 integron cassettes were confirmed in 12 E. coli strains, of them the transposons Tn3 gene and six virulence genes cvaC, iutA, iss, ibeA, kps, papC were detected in 7 isolates.
The genes cvaC, iutA, iss, papC, were the most frequently detected virulence genes detected in E. coli strains. Genes of CTX-M1, CMY-2, integrase 1, transposons Tn3, cvaC and iutA were detected in two strains of E. coli.
Three multidrug-resistant clones O25b-ST131 with CMY-2 and enrofloxacin resistance with Int1, Tn 3, cvaC and iutA were detected.
Discussion
Critical control points in rendering plants with regard to high level contamination, bioaerosol production and the risk for the environment involve unloading of the raw material and wastewater treatment. In the unloading section of the rendering plant, the raw material is dumped from collecting containers of the transport vehicles to destructors. This is associated with potential aerosolisation of liquids, such as blood, intestinal contents and similar.
Very similar drugs (beta lactams, penicillin, ampicillin, cloxacillin, tetracyclines, sulphonamides and potentiated sulphonamides, cephalosporins, and fluoroquinolones) have been used in both human medicine and agriculture production29.
Antimicrobials used in poultry production have the potential to accumulate in poultry feathers and during the rendering process are not completely destroyed. Poultry feathers can be recycled to a feather meal and used as a fertilizer and animal feed, thereby providing a potential pathway for re-entry of drugs into the human food supply30.
Hofacre et al.31 found that a high percentage of feed samples for poultry containing meat and bone meal from rendering plant were contaminated by bacteria resistant to amoxicillin, ampicillin, cephalothin or clavulanic acid. Some samples contained bacteria resistant to kanamycin, trimethoprim/sulfamethoxazole or ciprofloxacin. The presence of mobile genetic elements mediate multi-drug resistance was proved in many of the isolated bacteria.
Higher than 30% prevalence among the 3rd-generation cephalosporin-resistant E. coli was detected mostly in poultry production32.
In our study we observed the highest incidence of beta-lactams resistance to ampicillin (68%), followed by cephalosporins—veterinary ceftiofur (36%), cefquinom (15.9%) and ceftriaxone (12.5%) while the beta-lactam resistance to ceftazidime was detected only in 4.5% of all strains.
ESBLs and AmpC beta-lactamases are usually responsible for the mediation of resistance to 3rd-generation cephalosporins in E. coli33.
Similar ESBL phenotypes with high level fluoroquinolones resistance in animal E. coli isolated from a Slovak poultry slaughterhouse was described by Gregova et al.10. Aggregated European Community data for E. coli isolates from broilers showed that over 50% of isolates were resistant to ciprofloxacin34. Moreover, the fluoroquinolones-resistant E. coli typically exhibited clinically significant elevations in MIC values35. E. coli isolates resistant to fluoroquinolones are often resistant to other antibiotic groups and genes of virulence36.
The CMY-2-producing E. coli O25b-ST131 represent a clonal lineage that differs from the CTX-M-15-producing ST131-O25b cluster. ST131-O25b strains with the presence of ESBL-type CTX-M-15 and resistance to fluoroquinolones have been reported worldwide. They are frequently a cause of infections, particularly of the urinary tract of humans. In human patients in Europe, approximately 1% of the 3rd generation cephalosporin-resistant E. coli produce CMY-2. However, recent studies in Asia showed higher rates and an increasing trend among the 3rd-generation cephalosporin-resistant E. coli isolates has been reported33.
Recently, ten multi-resistant strains of E. coli that harboured CMY-2 were observed with increasing tendency in the European livestock production. However, ST131 isolates with CMY-2 production have been reported rarely37,38.
We also detected multidrug-resistant clones O25b-ST131 with CMY-2 and enrofloxacin resistance with Int1, Tn 3, cvaC and iutA.
CMY-2-producing E. coli isolates were also detected in products from meat, livestock animals and human patients. The predominant way of transmission of blaCMY-2 genes between animals and humans is the horizontal transfer of temporarily stable blaCMY-2-carrying IncK2 and IncI1 plasmids33. This suggests a zoonotic potential of the blaCMY-2 genes and their transmission by horizontal transfer and clonal spread along the food production chain33,37.
We also detected resistant strains (CTR, TTC, STM, FLO, COT) with genes, such as blaCTX-M-1, blaCMY-2 together with virulence factors cvaC, iutA and mobile elements (Int1, Tn3).
Extraintestinal virulence genes encoding adhesins, iron capture systems, toxins, and protectins have been correlated with successful colonization of gut in humans and animals39,40.
Our study of E. coli strains from wastewater showed that virulence genes cvaC, iutA, iss, papC were the most frequently detected in them. In some E. coli samples, we detected genes kps, tsh, papC, ibeA.
Similarly, examination of meat from healthy broilers from Slovakia conducted by Drugdova et al.41 showed presence of antimicrobial-resistant E. coli strains with virulence factors (most frequently iutA, iss, cvaC, tsh and papC) related to avian pathogenic or human uropathogenic E. coli.
The study conducted in Canada42 revealed high prevalence of many virulence genes (ompT, traT, uidA, vat, hemF, iss and cvaC), including the genes responsible for adhesion, fimH and kpsMT KII, in ExPEC isolates from frozen poultry meat.
Chicken meat and eggshells also harbour E. coli strains containing genes of virulence papA, papC, sfa, foc, afa, dra, kpsM II and iutA43.
Bok et al.44 observed that virulence genes (fimH, papAH, iutA, iroN, ompT, traT, and iss) were more frequently identified in isolates from piglets than from sows. E. coli from piglets constituted a substantial reservoir of extraintestinal virulence genes and could increase the potential risk of extraintestinal infections. According this study44 the mobile genetic elements transmitted via horizontal gene transfer play an important role in the evolution of E. coli resistance. Most ExPEC virulence genes are clustered together on mobile genetic elements, usually on pathogenicity islands (PAI) or virulence plasmids, exhibiting a unique organization.
Cunha et al.45 characterized APEC strains from different poultry farms in Brazil, that harboured a number of virulence factors such as sfa, usp (100% each), pap (85%), kpsMTII (66%), hlyA (52%), cnf1 (22%), ibeA (4%), iss (37%), tsh, ompT, and hlyF (8% each), and cvi/cva (0%).
Ten out of 13 tetracycline resistant strains carried the Int1 gene, and 6 of them the iutA gene. Five from among 8 streptomycin-resistant strains carried iutA and Int1 genes, which indicate a horizontal transfer of resistant genes between bacteria. The high number of isolates resistant to streptomycin, tetracycline and cotrimoxazol can be spread by same mobile genetic elements.
Conclusion
In conclusion, the present investigations illustrated the current state of antibiotic resistance of E. coli strains in the investigated rendering plant. We detected the presence of E. coli with CTX-M, cephamycinase CMY-2 genes and high level of chromosomal resistance to fluoroquinolones. Furthermore, we found three CMY-2 producing E. coli O25b-ST131, resistant to a high level of enrofloxacin with cvaC and iutA virulence factors. The CMY-2 producing E. coli isolates have a zoonotic potential and pose a serious health risk.
Considering that rendering plant is an important source of resistant bacteria, our data highlight the importance of adequate protection of the working personnel and observation of strict hygiene measures at operation premises.
Acknowledgements
Publication was supported by the University of veterinary medicine and pharmacy in Košice.
Author contributions
G.G. and V.K. designed the study. G.G. and V.K. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. G.G. and V.K. contributed substantially to the study design, data analysis and data interpretation. G.G. and V.K. contributed substantially to the writing of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the project No. APVV-16-0171 from the Slovak Research and Development Agency.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010;300(6):371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby GA. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009;22(1):161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fàbrega A, Madurga S, Giralt E, Vila J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 5.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or Ampc β-lactamases in food and food-producing animals. EFSA J. 2011;9(8):2322. doi: 10.2903/j.efsa.2011.2322. [DOI] [Google Scholar]
- 7.Cornejova T, Venglovsky J, Gregova G, Kmetova M, Kmet V. Extended spectrum beta-lactamases in Escherichia coli from municipal wastewater. Ann. Agric. Environ. Med. 2015;22:447–450. doi: 10.5604/12321966.1167710. [DOI] [PubMed] [Google Scholar]
- 8.Karkman A, Do TT, Walsh F, Virta MPJ. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018;26:220–228. doi: 10.1016/j.tim.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Regulation EC 1774/2002 laying down health rules concerning animal by-products not intended for human consumption (2002).
- 10.Gregova G, Kmetova M, Kmet V, Venglovsky J, Feher A. Antibiotic resistance of Escherichia coli isolated from a poultry slaughterhouse. Ann. Agric. Environ. Med. 2012;19:75–77. [PubMed] [Google Scholar]
- 11.CLSI document VET01-S2 Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 1–168 (Publ. Clinical and Laboratory Standards Institute, Wayne, 2013).
- 12.EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 2.0, 1–43 (2017).
- 13.Gattringer R, Niks M, Ostertág R, Schwarz K, Medvedovic H, Graninger W, et al. Evaluation of MIDITECH automated colorimetric MIC reading for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2002;49:651–659. doi: 10.1093/jac/49.4.651. [DOI] [PubMed] [Google Scholar]
- 14.Livermore DM, Winstanley TG, Shannon KP. Interpretative reading: Recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 2001;48:87–102. doi: 10.1093/jac/48.suppl_1.87. [DOI] [PubMed] [Google Scholar]
- 15.Kmet V, Kmetova M. High level of quinolone resistance in Escherichia coli from healthy chicken broilers. Folia Microbiol. 2010;55:79–82. doi: 10.1007/s12223-010-0013-x. [DOI] [PubMed] [Google Scholar]
- 16.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum ß-lactamases. J. Antimicrob. Chemother. 2005;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Perez, C., Regalado-González, C., Rodríguez-Rodríguez, C.A. & Barbosa-Rodríguez, J.R. Incorporation of antimicrobial agents in food packaging films and coatings. InAdvances in Agricultural and Food Biotechnology(ed. Guevara‐González, R.G. Torres‐Pacheco, I.) 193 – 216 (Publ. Research Signpost 2006).
- 18.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, et al. Prevalence of qnr, aac(6')-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 2012;56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamane K, Wachino J, Suzuki S, Arakawa Y. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 2008;52:1564–1566. doi: 10.1128/AAC.01137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 2006;50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000;44:1568–1574. doi: 10.1128/AAC.44.6.1568-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weill FW, Demartin M, Fabre L, Grimot PAD. Extended-spectrum-β-lactamase (TEM-52)-producing strains of Salmonella enterica of various serotypes isolated in France. J. Clin. Microbiol. 2004;42:3359–3362. doi: 10.1128/JCM.42.7.3359-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 2009;64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 25.Foley SL, Home SM, Giddings CW, Robinson M, Nolan LK. Iss from a virulent avian Escherichia coli. Avian Dis. 2000;44:185–191. doi: 10.2307/1592523. [DOI] [PubMed] [Google Scholar]
- 26.Dozois CM, Dho-Moulin M, Brée A, Fairbrother JM, Desautels C, Curtiss R., III Relation between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the tsh genetic region. Infect. Immun. 2000;68:4145–4154. doi: 10.1128/IAI.68.7.4145-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Bouguénec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microb. 1992;30:1189–1193. doi: 10.1128/JCM.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germon P, Chen YH, He L, Blanco JE, Brée A, Schouler SH. IbeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology. 2005;151:1179–1186. doi: 10.1099/mic.0.27809-0. [DOI] [PubMed] [Google Scholar]
- 29.McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002;34:S93–S106. doi: 10.1086/340246. [DOI] [PubMed] [Google Scholar]
- 30.Love DC, Halden RU, Davis MF, Nachman KE. Feather meal: A previously unrecognized route for reentry into the food supply of multiple pharmaceuticals and personal care products (PPCPs) Environ. Sci. Technol. 2012;46:3795–3802. doi: 10.1021/es203970e. [DOI] [PubMed] [Google Scholar]
- 31.Hofacre ChL, White DG, Maurer JJ, Morales C, Lobsinger Ch, Hudson Ch. Characterization of antibiotic-resistant bacteria in rendered animal products. Avian Dis. 2001;45:953–961. doi: 10.2307/1592874. [DOI] [PubMed] [Google Scholar]
- 32.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012;18(7):646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 33.Pietsch M, Irrgang A, Roschanski N, Brenner MG, Hamprecht A, Rieber H, et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics. 2018;19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EFSA/ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2016;14(2):4380. doi: 10.2903/j.efsa.2016.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drugdova Z, Kmet V. Prevalence of β-lactam and fluoroquinolone resistance, and virulence factors in Escherichia coli isolated from chickens in Slovakia. Biologia. 2013;68:11–17. doi: 10.2478/s11756-012-0142-6. [DOI] [Google Scholar]
- 36.Boyd BL, Maynard MJ, Morgan-Linnell SK, Horton LB, Sucgang R, Hamill RJ, et al. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 2008;53:229–234. doi: 10.1128/AAC.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schonning K, Agerso Y, et al. Host-specific patterns of genetic diversity among IncI1-Igamma and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl. Environ. Microbiol. 2016;82(15):4705–4714. doi: 10.1128/AEM.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Day MJ, Rodriguez I, van Essen-Zandbergen A, Dierikx C, Kadlec K, Schink AK, et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016;71(5):1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 39.Frömmel U, Lehmann W, Rödiger S, Böhm A, Nitschke J, Weinreich J, et al. Adhesion of human and animal Escherichia coli strains in association with their virulence-associated genes and phylogenetic origins. Appl. Environ. Microbiol. 2013;79:5814–5829. doi: 10.1128/AEM.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic from different sources: Recent reports. Gut Pathog. 2019;11:10. doi: 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drugdová Z, Kmeť V, Bujňáková D. Virulence factors in Escherichia coli isolated from chicken meat in Slovakia. J. Food. Nutr. Res. 2010;49(1):10–13. [Google Scholar]
- 42.Aslam M, Toufeer M, Narvaez Bravo C, Lai V, Rempel H, Manges A, et al. Characterization of extraintestinal pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. Int. J. Food Microbiol. 2014;177:49–56. doi: 10.1016/j.ijfoodmicro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell NM, Johnson JR, Johnston B, Curtiss R, Mellata M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 2015;81:1177–1187. doi: 10.1128/AEM.03524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bok E, Kożańska A, Mazurek-Popczyk J, Wojciech M, Baldy-Chudzik K. Extended phylogeny and extraintestinal virulence potential of commensal Escherichia coli from piglets and sows. Int. J. Environ. Res. Public Health. 2020;17(1):366. doi: 10.3390/ijerph17010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha MPV, Saidenberg AB, Moreno AM, Ferreira AJP, Vieira MAM, Gomes TAT, et al. Pandemic extra-intestinal pathogenic Escherichia coli (ExPEC) clonal group O6-B2-ST73 as a cause of avian colibacillosis in Brazil. PLoS ONE. 2017;12(6):1–11. doi: 10.1371/journal.pone.0178970. [DOI] [PMC free article] [PubMed] [Google Scholar]