Abstract
Therapeutic innovation for human papilloma virus-related (HPV+) head and neck squamous cell carcinomas (HNSCCs) is impaired by inadequate preclinical models and absence of accurate biomarkers. This study establishes the first well-characterized panel of patient-derived xenografts (PDXs) and organoids from HPV+ HNSCCs while determining fidelity of the models to the distinguishing genetic features of this cancer type. Despite low engraftment rates, whole exome sequencing showed that PDXs retain multiple distinguishing features of HPV+ HNSCC lost in existing cell lines, including PIK3CA mutations, TRAF3 deletion, and absence of EGFR amplifications. Engrafted HPV+ tumors frequently contained NOTCH1 mutations, thus providing new models for a negatively prognostic alteration in this disease. Genotype-phenotype associations in the models were then tested for prediction of tumor progression and survival in published clinical cohorts. Observation of high tumor mutational burdens (TMBs) in the faster-growing models facilitated identification of a novel association between TMB and local progression in both HPV+ and HPV− patients that was prognostic in HPV− cases. In addition, reduced E7 and p16INK4A levels found in a PDX from an outlier case with lethal outcome led to detection of similar profiles among recurrent HPV+ HNSCCs. Transcriptional data from the Cancer Genome Atlas was used to demonstrate that the lower E2F target gene expression predicted by reduced E7 levels has potential as a biomarker of disease recurrence risk. Our findings bridge a critical gap in preclinical models for HPV+ HNSCCs and simultaneously reveal novel potential applications of quantifying mutational burden and viral oncogene functions for biomarker development.
Keywords: head and neck cancer, HPV, patient-derived xenografts, outcomes, biomarkers
INTRODUCTION
The human papilloma virus-related (HPV+) subtype of head and neck squamous cell carcinoma (HNSCC) occurs in a younger patient demographic than other HNSCCs and is rising in incidence1. A major therapeutic objective for these patients is to decrease the severe treatment-related morbidities that persist after radiation and cytotoxic chemotherapy2 while maintaining their generally favorable survival outcomes3. However, attempting to reduce treatment toxicity for this disease through therapy de-escalation carries significant risk. For instance, treatment failures increased in two recent phase III trials evaluating therapeutic efficacy of cetuximab as an alternative to standard cisplatin4, 5. Improving clinical trial design for this disease is presently impeded by the limited risk stratification offered by 8th edition AJCC staging6 and absence of molecular biomarkers. These factors are barriers both to identifying the optimal HPV+ HNSCC patients for therapy de-escalation and to developing distinct, more effective treatments for high risk cases. Among molecular prognostic features that could potentially guide treatment7-10, NOTCH1 loss of function is thus far the only mutation shown to predict poor outcomes in a published cohort of HPV+ HNSCCs10.
A second obstacle to therapeutic innovation for HPV+ HNSCCs is the paucity of experimental models that accurately recapitulate their biology in humans. Poor growth of these tumors ex vivo delayed the creation of human cancer cell lines from them, and the cell lines that are now available lack multiple genetic traits that distinguish HPV+ HNSCCs from their HPV− counterparts11. Artifacts in existing HPV+ cell lines include loss of the canonical PIK3CA activating mutations and TRAF3 deletions that are more abundant in the HPV+ subtype of HNSCC. Most HPV+ HNSCC cell lines also acquire EGFR amplifications, which are uncommon in HPV+ HNSCCs, as well as copy gains in the 3q region spanning TP63, SOX2, and PIK3CA11, 12. Infrequent surgical treatment of HPV+ HNSCCs in the past and their intrinsically poor engraftment rates in immune-deficient mice13 have impeded development of patient-derived xenograft (PDX) models. As a result, current preclinical models are mostly derived from cell lines that have limited ability to assess strategies to reduce treatment toxicity for low risk cases or evaluate intensified approaches for high risk cases.
The improved molecular fidelity of PDXs and organoids over cancer cell lines is well established14, 15 and justified our pursuit of such models despite potential pitfalls of generating them from HPV+ HNSCCs. In particular, the <25% stable engraftment rate observed by us13 raised the possibility that any models that were created would also contain major molecular selection biases. Although such engraftment biases may prevent PDXs from capturing some tumor genotypes, PDXs can also enrich for other clinically relevant molecular subgroups, including those with more aggressive behavior15, 16. For instance, rapid growth of early stage HPV− HNSCCs upon engraftment to mice was recently shown to be predictive of recurrence and mortality for that disease subtype17. These observations informed our intensive effort to generate a limited number of PDXs from HPV+ HNSCCs in order to provide preclinical models for aggressive tumor phenotypes and identify molecular traits that could aid risk stratification in the clinic.
This study aimed to characterize the largest panel of HPV+ HNSCC PDXs and organoids described to date and leverage these models to advance molecular risk stratification. The panel contained 9 HPV+ and 11 HPV− PDXs, whose comparative analysis using whole exome sequence (WES) served to evaluate the HPV+ models for retention of genetic traits specific to HPV+ HNSCCs, including those not captured by HPV+ cell lines. Cancer-related mutations in the PDXs were assessed for enrichment relative to their expected frequencies derived from the Cancer Genome Atlas (TCGA) to identify potentially aggressive genotypes that promote engraftment. A previously unknown association between high mutational burden and efficient growth of HPV+ patient-derived models was further tested for ability to predict tumor progression and outcomes in patients. In addition, distinctive molecular features found in a PDX from a rapidly lethal HPV+ case served to identify a gene expression pattern with potential for clinical prognostic utility.
MATERIALS AND METHODS
Patient derived xenografts and organoids
PDXs were established, passaged, and cryopreserved as described by us13 using NOD/SCI D/I L-2Rγ−/− mice under University of Pennsylvania IRB protocol #417200 and Wistar Institute IACUC protocol #112652. Tumors were harvested for sequencing when they reached a volume of 1 cm3. Tumor volumes were calculated from width2*length/2. PDX growth rates were estimated by the slope from a linear regression model for the logarithm of tumor volume as a function of time for each replicate; for each PDX there were two passages with 1-3 replicates for each passage. Each PDX-specific growth rate was calculated as the inverse-variance weighted sum of growth rates for each of its replicates. PDXs were dissociated as described13 and resuspended in Matrigel (600 cells/μL) for organoid culture. 50μL was plated in 24-well format with 500μL of organoid media. Organoid media was comprised of DMEM/F12+ (Gibco, Gaithersburg, MD) supplemented with N2, B27, 50 ng/mL recombinant human EGF (Thermo Fisher Scientific, Waltham, MA), 0.1 mM N-acetyl-L-cysteine (Sigma, St. Louis, MO), 2% Noggin/R-Spondin–conditioned media, 50 μg/mL Gentamicin (Invitrogen, Carlsbad, CA), Glutamax, 10 mM HEPES (Gibco), and 10 μM Y27632 (SelleckChem, Houston, TX).
Sequencing and Analysis
Whole exome sequencing of PDXs was performed from snap-frozen tissue using the Illumina HiSeq (2x150bp) platform and is summarized in Supporting Information Table S1. Genomic DNA extraction was performed with Qiagen QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) per the manufacturer’s recommendation. DNA was quantified with Qubit 2.0 DNA HS Assay (Thermo) and quality assessed by Tapestation genomic DNA assay (Agilent Technologies, Santa Clara, CA). Library preparation was performed using KAPA Hyper Prep kit (KAPA Biosystems, Wilmington, MA) per manufacturer’s instructions. Exome capture was performed with IDT xGen Exome Research Panel v1.0 (IDT, Skokie, IL). Library quality and quantity were assessed with Qubit 2.0 DNA HS Assay (Thermo), Tapestation High Sensitivity D1000 Assay (Agilent), and QuantStudio® 5 System (Applied Biosystems, Foster City, CA).
Whole exome sequencing resulted in 21-250 million total raw reads with 150 base pair median length in all PDX samples. Read quality was assessed and ≥86 percent reads with Phred scores ≥30 were observed in all samples. Target coverage of 98-99.8 percent was obtained with ≥10X depth in all whole exome datasets. Percent duplicate paired reads ranged between 9 and 24 percent across samples.
DNA for targeted sequencing was obtained from snap-frozen or formalin-fixed tissue, and NOTCH1 and PIK3CA exons were sequenced using an Ion AmpliSeq Custom Panel (Thermo). DNA was isolated using the PureLink Genomic DNA Kit (Invitrogen) for snap frozen primary specimens and QIAamp DNA FFPE Tissue kit (Qiagen) for FFPE primary specimens. The Qubit Broad Range dsDNA kit (Thermo) and the NGS FFPE QC kit (Agilent, Santa Clara, CA) assays were used to determine DNA quantity and quality, respectively.
Deletions and damaging mutations (frameshift, stopgain, stoploss, missense with damaging R-SVM score) were predicted to confer loss of function, while amplifications and non-damaging missense mutations in the COSMIC v70 database were considered gain-of-function. For principal component analysis (PCA), damaging mutations were weighted by allele frequency. Non-transformed continuous scores for each gene were used to calculate principal components.
Mutational variant calling
For whole exome sequencing, mutational variants were processed through the bcbio pipeline and mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA). GATK HaplotypeCaller recalibrated the mapped reads based on quality and realigned them around indel regions. Low complexity regions (LCRs) were removed and mutational variants were called when identified by two out of four variant callers: Freebayes, Vardict, MuTect2, or VarScan. Variants were limited to exomic mutations with allele frequencies ≥4% and minimum read depths ≥10. For targeted sequencing, Ion Torrent’s software was used for alignment, and its default parameters were employed for somatic variant calling. Minimum allele frequency of 2%, a minimum depth of 100x were used as cut-offs to acquire the list of mutations. In the absence of matched normal controls for both whole exome and targeted sequencing, somatic variants were identified if their population frequency was <1% based on the 1000 Genome Project, ESP6500, ExAC, and CG46 databases. Only nonsynonymous, frameshift, stoploss, or stopgain mutations were called. Nonsynonymous mutations were further filtered by either a damaging radial support vector machine (R-SVM) score or presence in the COSMIC v70 database. Finally, mutations that passed all other filtering criteria but were found in three or more PDXs were discarded due to potentially being missed by the population frequency analysis.
Copy number analysis
Copy number alterations (CNAs) were called using the CNVkit software toolkit and mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA). Each sample was first median-centered and corrected for read-depth bias. In the absence of matched normal controls, the threshold for calling amplifications or deletions was based on the scaled log2 copy ratios of each CNA segment relative to a generated ground reference. The ground reference was calculated by taking a weighted average (Tukey’s biweight location) of the log2 converted read-depth value of each bin across all samples. Thresholds for deletions and amplifications were log2 values of <−0.25 and >0.2, respectively.
TCGA data analysis
The cBioPortal18 was used to analyze genomic and clinical information from the Head and Neck Squamous Cell Carcinoma (TCGA, Provisional)19, Esophageal Carcinoma (TCGA, Provisional)20 and Cervical Squamous Cell Carcinoma (TCGA, PanCancer Atlas)21 data sets. Samples from disease types other than squamous cell carcinoma were removed from analysis. Additional patient data for the TCGA HNSCC cohort including clinical follow-up were downloaded from the Genomic Data Commons (project TCGA-HNSCC). The HPV+ oropharyngeal HNSCCs in TCGA were identified based on expression of viral E6/E722. Overall and disease-free survival data were updated for TCGA HPV+ cases using follow-up tables V1.0 and V4.8, and cases with persistent disease were assigned disease-free survival (DFS) of 1 day. HPV+ TCGA tumors were restaged clinically by the 8th edition AJCC manual. Full clinical annotation of the 53 HPV+ oropharyngeal HNSCCs in TCGA is provided in Supporting Information Table S2. Normalized mRNA expression for TCGA samples was downloaded from the Broad GDAC Firebrowse repository and ln(x+1)-transformed for clustering. Unsupervised hierarchical clustering and heatmap visualization were performed with ClustVis23 using correlation distance and average linkage.
Real-time quantitative PCR (qPCR)
CDKN2A copy number was estimated by qPCR as described24. Briefly, PCR reactions were carried out in a volume of 20 μl, using 10 μl of 2X PowerSYBR Green PCR Master Mix (Applied Biosystems). The thermal cycling conditions were: 95°C for 10 min followed by 40 cycles at 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. The primers used were: CDKN2A forward: 5’-GGCTGGCTGGTCACCAGA-3’; CDKN2A reverse: 5’- CGCCCGCACCTCCTCTAC-3’; GAPDH promoter forward: 5’-TACTAGCGGTTTTACGGGCG-3’; GAPDH promoter reverse: 5’-TCGAACAGGAGCAGAGAGCGA-3’. Reverse transcriptase qPCR was performed as previously described13. Primers for reverse transcriptase qPCR were: HPV16 E7 forward: 5’-CCGGACAGAGCCCATTACAA-3’; HPV16 E7 reverse: 5’-CGAATGTCTACGTGTGTGCTTTG-3; HPV16 E6 forward: 5’-TCAGGACCCACAGGAGCG-3’; HPV16 E6 reverse: 5’-CCTCACGTCGCAGTAACTGTTG-3; E2 forward: 5’-TGGAAACACATGCGCCTAGAA-3’ E2 reverse: 5’- GTTGCAGTTCAATTGCTTGTAATGC-3’; p16 forward: 5’-AGCATGGAGCCTTCGGCTGA-3’; p16 reverse: 5’-CCATCATCATGACCTGGATCG-3’.
Western Blotting and Immunohistochemistry
PDX tissues were lysed in RIPA buffer (Thermo) and protein quantified by BCA assay (Thermo). 50ug protein was separated on 10% ECL gels (GE, Pittsburgh, PA) and transferred to nitrocellulose using the Trans-Blot® System (Bio-Rad, Hercules, CA). Antibodies are listed in Supporting Information Table S3.
Statistical analysis
Mutational frequencies among cohorts were compared by Fisher’s exact test. Differences in mutation frequency and TMB were evaluated using Student’s t-test assuming unequal variances. Randomization testing to compare p16 expression between groups was based on 100,000 permutations using the Welch t-statistic. Significance testing of Pearson correlations was performed using Student’s t-test. Survival was compared by log-rank test.
Data Availability
The raw data has been deposited in the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) and can be accessed through the accession PRJNA597082.
RESULTS
PDXs retain genetic hallmarks of HPV+ HNSCC and avoid artifacts seen in HPV+ cell lines.
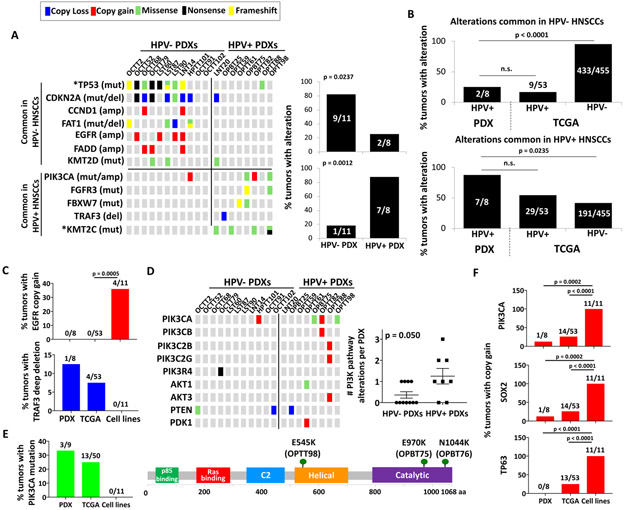
Oropharyngeal primary HNSCCs or lymph node metastases that were positive for p16INK4A by immunohistochemistry (IHC) were used to generate 9 stable HPV+ PDXs (Supporting Information Table S4). Whole exome sequencing (WES) was completed for 8 HPV+ PDXs, and targeted sequencing was performed selectively on a 9th HPV+ PDX that failed to provide full WES due to excess mouse DNA. Sequenced models all contained HPV-16 DNA and expressed HPV-16 transcripts by qPCR (Supporting Information Figure S1). WES profiles of the HPV+ PDXs were compared to those of the 53 HPV+ oropharyngeal HNSCCs in TCGA (Supporting Information Table S2)22. WES simultaneously performed on 11 PDXs from HPV− HNSCCs (Supporting Information Table S4) provided controls to help highlight distinguishing features of HPV+ HNSCCs preserved in the models. A list of genetic differences between HPV− and HPV+ HNSCCs was curated to include alterations that differed significantly in frequency in direct comparison of HPV+ to HPV− HNSCCs within a study or have been reported to be highly enriched in one subtype but not the other. (Supporting Information Table S5)10,19,25-33. These differences were broadly preserved between the two groups of PDXs (Figure 1A, left). The alterations less common in HPV+ cases were markedly under-represented in HPV+ PDXs, and the more common alterations were significantly over-represented (Figure 1A, right). High fidelity of the HPV+ and HPV-neg PDXs to their respective tumors of origin is further illustrated by comparing the same list of alterations between the PDXs and the HNSCCs in TCGA (Figure 1B). Key distinguishing features of HPV+ disease retained by the models included a paucity of TP53 and CDKN2A alterations10, 19,25-31 and multiple KMT2C mutants27, 33. The HPV+ PDXs also avoided EGFR copy gains, which are rare in human HPV+ HNSCCs but are typically acquired by their cell lines, and captured a characteristic TRAF3 deep deletion not found in HPV+ cell lines11 (Figure 1C). Furthermore, the HPV+ PDXs retained the increase in alterations predicted to activate the PI3-kinase pathway that are a hallmark of HPV+ HNSCC (Figure 1D). PIK3CA hotspot mutations, which are lost in HPV+ cell lines11, 12, were identified in 3 HPV+ PDXs, including a canonical activating E545K helical domain mutant (Figure 1E, Supporting Information Table S6). The two other mutants (E970K and N1044K) were in the catalytic domain and are previously described as hotspots in a population-scale cohort of tumor samples34, with N1044K identified by targeted sequencing of the HPV+ PDX that failed WES. Finally, the 3q amplicon spanning PIK3CA, TP63, and SOX2 that appears de novo during HNSCC cell line generation11, 12 was not enriched the HPV+ PDXs (Figure 1F). Together, these findings supported the superior fidelity of the PDXs over cell lines in representing key genetic traits of HPV+ HNSCC and thus their distinct utility as HPV+ cancer models.
Figure 1. PDXs retain genetic hallmarks of HPV+ HNSCC and avoid artifacts seen in HPV+ cell lines.
A. Frequencies in PDXs of mutations and CNAs that differ between HPV− and HPV+ HNSCCs individually (left) and in aggregate (right). Unadjusted p-values determined by Fisher’s exact test. *p<0.025 B. Frequencies of alterations common in HPV− (top) and HPV+ (bottom) HNSCCs compared between HPV+ PDXs and HPV− or HPV+ HNSCCs in TCGA. C. Frequencies of EGFR and TRAF3 CNAs in HPV+ PDXs, TCGA cases, and a panel of 11 HPV+ HNSCC cell lines (USMCC47, USMCC104, USMCC105, VUMCSCC4149, VUMCSCC4755, VUMCSCC4975, UPCISCC90, UPCISCC152, UPCISCC154, UDSCC2, 93VU147T) previously sequenced by Cheng et al.11. p-values determined by Fisher’s exact test. D. Mutations and CNAs in proximal components of the PI3K/Akt pathway compared between HPV− and HPV+ PDXs (left). Number of altered PI3K pathway genes per tumor in HPV− vs. HPV+ PDXs (right). p-value determined by two-tailed Student’s t-test assuming unequal variances. E. Frequencies of PIK3CA mutations in HPV+ PDXs, TCGA cases, and cell lines (left). Positions of PIK3CA mutations in HPV+ PDXs (right). F. Frequencies of 3q gene copy gains in HPV+ PDXs, TCGA cases, and cell lines. p-values determined by Fisher’s exact test.
HPV+ PDX generation efficiently captures Notch pathway mutations.
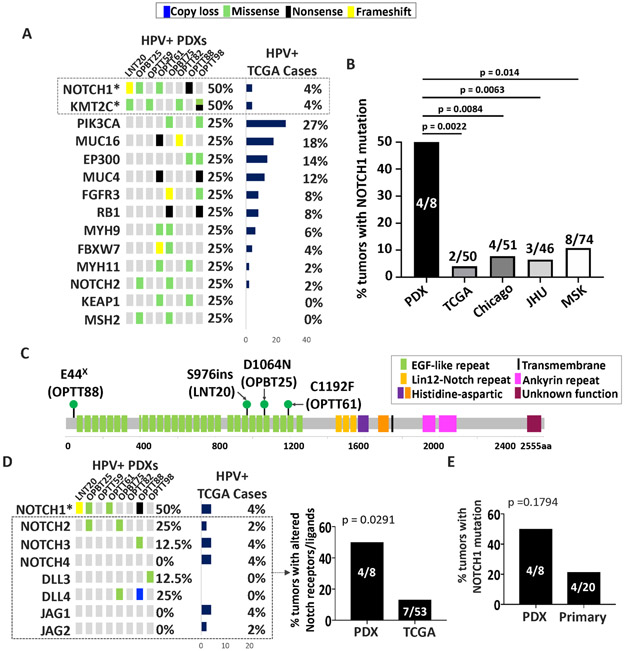
Despite overall fidelity of HNSCC PDXs to the genetic landscape of HPV+ tumors, low engraftment rates in immunodeficient mice13 predicted molecular selection biases in the PDXs. To evaluate for effects that might provide models for more aggressive tumor subtypes, mutations in genes in the Cancer Gene Census that occurred in at least two HPV+ PDXs were compared in frequency to their counterparts in TCGA (Figure 2A). KMT2C mutations were increased in the PDXs relative to TCGA but are found at high frequency in other HPV+ HNSCC series27, 33. By contrast, significantly more NOTCH1 mutants with predicted loss of function were found in the PDXs relative to TCGA and the three other large published cohorts (Figure 2B). The mutations occurred predominantly in the N-terminal EGF ligand-binding domain (Figure 2C), like those described to be negatively prognostic10. These mutations were accompanied by an increase in alterations across the other Notch receptors and ligands in aggregate (Figure 2D). A similar increase was not evident in the HPV− PDXs (Supporting Information Figure S2), despite comparable background frequencies of Notch pathway mutations in HPV+ and HPV− HNSCCs19. To evaluate whether the increase in HPV+ models arose through selective engraftment, targeted sequencing was used to quantify NOTCH1 mutants in the population of HPV+ tumors from which engraftment was attempted. A trend toward more NOTCH1 mutations in tumors that stably engrafted was observed (Figure 2E). Of note, no associations were observed between lymph node metastasis and either NOTCH1 mutation status or engraftment success (Supporting Information Tables S7-8). Taken together, these findings demonstrate that PDXs efficiently capture alterations predicted to inactivate the Notch pathway in HPV+ HNSCCs. A paucity of similar changes in HPV+ cell lines11, 12 establishes the PDXs as compelling models for this class of molecular alteration, which likely plays a major role in HPV+ HNSCC biology35 and may have negative prognostic significance10.
Figure 2. HPV+ PDX generation efficiently captures Notch pathway mutations.
A. Genes in Cancer Gene Census mutated in two or more HPV+ PDXs compared to the same genes in HPV+ TCGA HNSCCs. Unadjusted p-values: *p<0.005 B. Frequencies of NOTCH1 mutations in HPV+ PDXs compared with cases in the HPV+ TCGA, Chicago 27, JHU 33, and MSK 10 patient cohorts. p-values were determined by Fisher’s exact test. C. Positions of NOTCH1 mutations in HPV+ PDXs. D. Mutations and CNAs in NOTCH receptors and ligands in HPV+ PDXs with predicted loss of function (left). Unadjusted p-values: *p<0.005 Frequency of mutations and/or CNAs in NOTCH receptor (excluding NOTCH1) or NOTCH ligand genes in HPV+ PDXs vs. TCGA (right). E. Percent of tumors with mutant NOTCH1 in HPV+ PDXs and HPV+ human HNSCCs from which engraftment failed. p-values were determined by Fisher’s exact test.
High mutational burden is associated with efficient growth of HPV+ PDXs and organoids.
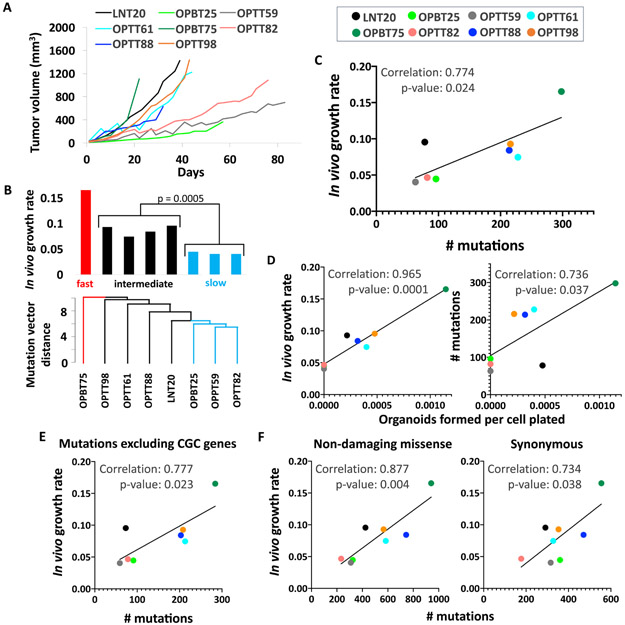
To identify genetic features that might impact HPV+ HNSCC progression, growth properties of the HPV+ PDXs were evaluated for relationships with their mutational profiles. PDX growth curves (Figure 3A) were quantified by averaging the regression coefficients from two consecutive passages, and growth rates were expressed as slopes weighted by the variability among three to six replicates (Supporting Information Table S9). The relationship between mutation profile and growth rate was evaluated using principal component analysis (PCA), and a relatedness dendrogram was created based on the Euclidean distances among allele-frequency weighted mutation vectors (Figure 3B, bottom). The dendrogram segregated fast, intermediate, and slow-growing PDXs (Figure 3B, top), whose PCA grouping is also illustrated on a projection plot (Supporting Information Figure S3). Although the small PDX sample size was ill-suited to identifying growth associations with individual genes, a significant positive association was apparent between growth rate and the total number of called mutations, quantified here as tumor mutational burden (TMB) (Figure 3C). Furthermore, creation of in vitro organoid models from the PDXs revealed associations between organoid growth efficiency (Supporting Information Table S10) and both in vivo growth rate (Figure 3D, left) and TMB (right, Supporting Information Figure S4). These in vitro proliferative features indicated the PDX growth rates to be tumor-autonomous features and not artifacts of the mouse milieu. Thus, the Cancer Gene Census gene list was used to interrogate whether mutations in the subset of these cancer genes account for the growth rate differences. Surprisingly, subtracting mutations in known oncogenes and tumor suppressors from the TMB count failed to diminish the strength of the growth association (Figure 3E). This finding was consistent with alterations outside the known cancer genome driving PDX growth or simply an accumulation of passenger mutations in rapidly growing models. The latter possibility was supported by the growth association also being present for point substitutions excluded by the mutation-calling pipeline, including missense mutations without damaging R-SVM scores (Figure 3F, left) and synonymous mutations (right). Collectively, these findings revealed that faster growing HPV+ models harbor more passenger mutations and indicated potential utility for TMB as a predictor of tumor-autonomous HPV+ HNSCC progression.
Figure 3. High mutational burden is associated with efficient growth of HPV+ PDXs and organoids.
A. Representative in vivo growth curves of HPV+ PDXs during the first passage after initial engraftment B. Dendrogram illustrating Euclidean distances among allele frequency-weighted mutation vectors in HPV+ PDXs (bottom). Growth rates of HPV+ PDXs are ordered by mutation vector distances (top). p-value was determined by Student’s t-test assuming unequal variances. C. In vivo PDX growth rates vs. total mutations per PDX. D. Organoid formation efficiency by PDX cells vs. in vivo growth rate (left) and number of mutations (right). E. PDX growth rates vs. number of mutations excluding all genes in Cancer Gene Census. F. PDX growth rates vs. substitutions excluded by mutation calling pipeline: missense mutations without damaging R-SVM scores (left) and synonymous substitutions (right). Pearson correlation coefficients and p-values based on two-tailed Student t-distribution are shown for each plot.
High TMB is associated with HPV+ and HPV− tumor progression and is prognostic in early HPV− cases.
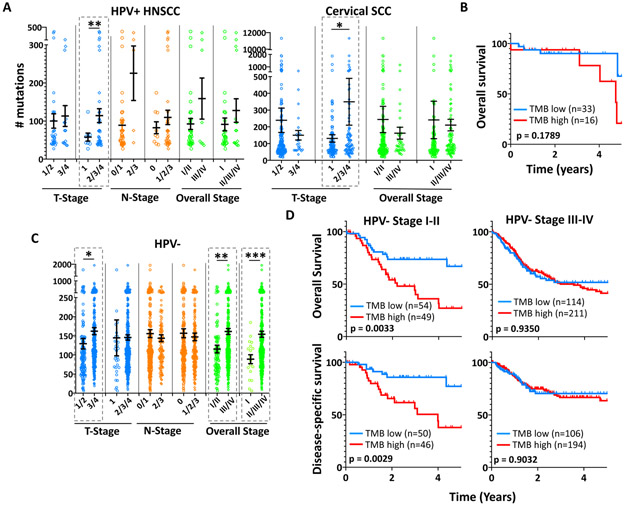
The positive association between TMB and HPV+ PDX growth efficiency prompted assessment of TMB’s utility for predicting clinical behavior. Comparison of TMBs among 8th edition TNM stage groups for HPV+ HNSCCs identified decreased TMB in cases with limited (T1) local disease (Figure 4A, left). This finding was validated using the cervical squamous cell carcinomas in TCGA as a second HPV+ cohort (Figure 4A, right). To evaluate prognostic relevance of TMB, its distribution among the HPV+ HNSCCs was scanned by ROC analysis for 5-year overall survival (OS), which identified a cut point of 103 that best segregates outcomes based on Youden’s Index (Supporting Information Table S11); however, this TMB cut-point was not predictive of OS (Figure 4B). Because local disease burden is more strongly predictive of poor outcome in HPV− cases, the same analysis was performed on the HPV− HNSCC cohort in TCGA. Here, TMB was significantly elevated in both locally advanced (T3/T4) and advanced stage (III/IV) disease (Figure 4C). Moreover, survival curves for groups defined by the optimal TMB cut-point based on Youden’s Index for this cohort (99.5, Supporting Information Table S12) demonstrated decreased disease-specific survival (DSS) and OS among early stage but not advanced stage cases with high TMB (Figure 4D). This finding was validated using TCGA data for esophageal squamous cell carcinoma (ESCC), a second HPV− malignancy with comparable risk factors, tissue origin, histology, and genetic landscape36. As observed in HNSCCs, a TMB cut point in ESCC was found that predicted OS for early but not advanced cases (Supporting Information Figure S5). Together, these results support an association between high TMB and local progression of all HNSCCs and establish a prognostic relationship between high TMB and decreased survival of early stage HPV− cases.
Figure 4. High TMB is associated with local progression in HPV+ and HPV− HNSCCs.
Differences in mean TMB among different TNM and overall stage groups of HPV+ HNSCC (left) and cervical SCC (right) TCGA cases. *p<0.05, **p<0.025 Error bars represent SEM. p-values were determined by two-tailed Student’s t-test assuming unequal variances. B. OS for HPV+ HNSCCs in TCGA divided by a TMB cutoff of 103. C. Differences in mean TMB among different TNM and overall stage groups of HPV− TCGA cases. *p<0.05, **p<0.001, ***p<0.0001 D. 5-year OS and DSS for early stage (I/II) and advanced stage (III/IV) HPV− TCGA HNSCCs segregated by a TMB of 99.5 (AUC=0.7463, p=0.0166). Staging for HPV+ HNSCC cases is updated to the 8th edition AJCC clinical stage. 6th/7th edition clinical stage is shown for HPV− cases. Survival p-values were determined by log-rank test.
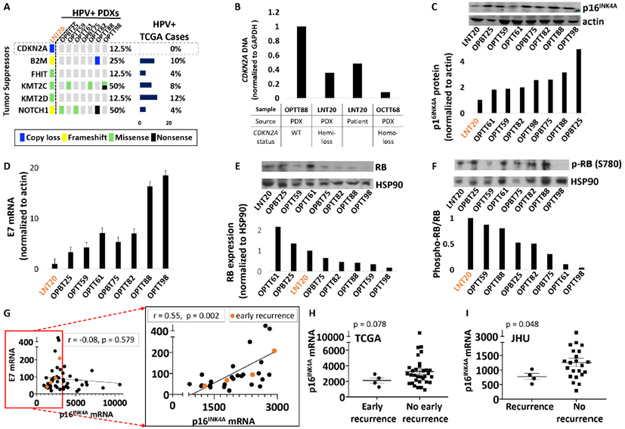
Lower p16INK4A expression in an atypical HPV+ PDX may be shared by recurrent HPV+ cases.
To aid risk stratification in HPV+ HNSCCs, we shifted focus to the molecular traits of LNT20, the only HPV+ PDX whose patient of origin suffered an early recurrence and had a lethal outcome within 6 months of completing initial therapy (Supporting Information Table S4). Among the tumor suppressors altered in this PDX, hemizygous loss of CDKN2A was the sole distinguishing feature relative to the other HPV+ PDXs and the 53 HPV+ HNSCCs in TCGA (Figure 5A). Presence of the hemizygous CDKN2A loss in both the PDX and its tumor of origin was further supported by DNA qPCR (Figure 5B). Although the PDX and original tumor retained p16INK4A overexpression by IHC, its p16INK4A mRNA (Supporting Information Figure S6) and protein levels (Figure 5C) were the lowest among the HPV+ PDXs. Because viral E7 acts upstream of p16INK4A upregulation in HPV-related cancers, lower E7 was considered as a possible additional source of reduced p16INK4A in this model. Among the PDXs, LNT20 also contained the lowest E7 transcript levels (Figure 5D), which was consistent with its relatively high levels of Rb protein (Figure 5E). Furthermore, LNT20 contained the highest ratio of phospho-Rb to Rb (Figure 5F), in keeping with the increase in CDK4/6 activity predicted by its lower p16INK4A levels. To evaluate HPV+ HNSCCs for aggressive features mimicking LNT20’s rapidly lethal phenotype, recurrences among HPV+ TCGA cases were examined for p16INK4A and E7 expression relative to the rest of the cohort. Although E7 transcript levels lacked correlation with p16INK4A across the full range of p16INK4A expression in the cohort (Figure 5G, left), a significant association was apparent at the lower levels of p16INK4A where the early recurrences (within 2 years of definitive therapy) appeared (Figure 5G, right inset). Despite absence of CDKN2A alterations in the cohort, there was a trend toward lower p16INK4A expression in the recurrences within 2 years (p=0.078, Figure 5H) and in all the recurrences (p = 0.082, Supporting Information Figure S7). This observation was strengthened using RNA sequencing data from a second cohort of HPV+ oropharyngeal HNSCCs37 (JHU), where p16INK4A levels were significantly reduced in recurrent cases (p = 0.048, Figure 5I). Taken together, these findings suggest a novel association between HPV+ HNSCC recurrence and reduced p16INK4A upregulation downstream of lower viral E7 levels. However, neither E7 nor p16INK4A transcript levels had prognostic utility alone or in combination based on Kaplan-Meier analyses (not shown).
Figure 5. Reduced p16INK4A levels in an atypical HPV+ PDX are shared by recurrent HPV+ cases.
A. Tumor suppressors mutated or deleted in LNT20 relative to the other HPV+ PDXs and TCGA cases. B. CDKN2A DNA levels in the LNT20 PDX and its tumor of origin, relative to control PDXs with WT or deleted CDKN2A. C. p16INK4A protein by western blot (top) with band densities normalized to actin (bottom) in HPV+ PDXs. D. HPV16 E7 mRNA levels normalized to actin in HPV+ PDXs. E RB protein (top) with corresponding band densities normalized to HSP90 (bottom) in HPV+ PDXs. F. phospho-RB protein (Ser780) (top) and phospho-RB to total RB ratios (bottom). G. E7 vs. p16INK4A mRNA in all TCGA HPV+ HNSCCs (left) and those with a normalized p16INK4A mRNA level of <3000 (right inset). Recurrences within 2 years (early recurrence) are highlighted in orange. Pearson correlation coefficients and p-values based on two-tailed Student t-distribution are shown for each plot. H. p16INK4A mRNA expression in TCGA HPV+ HNSCCs that recurred within 2 years (early recurrence) or were disease-free at the time of last early recurrence. Bars represent mean ± SEM. p-values were determined by randomization test (n = 100,000 permutations) with Welch’s t-statistic. I. p16INK4A expression in recurrent vs. non-recurrent HPV+ cases with matched follow-up duration in the JHU cohort. Bars represent mean ± SEM. p-values were determined by randomization test (n = 100,000 permutations) with Welch’s t-statistic.
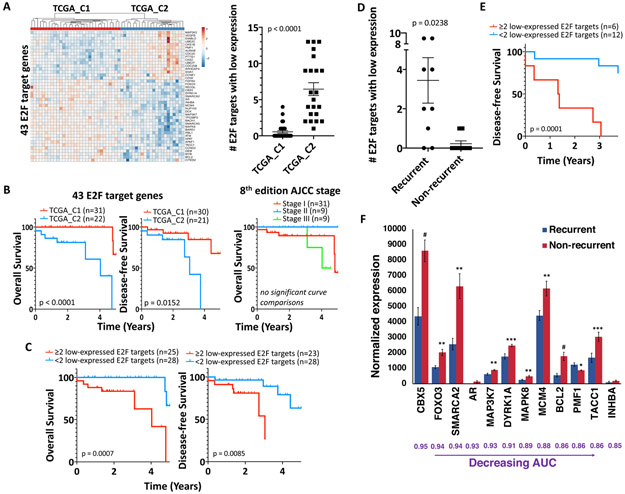
Reduced E2F target gene expression is prognostic for HPV+ HNSCCs in TCGA
Because neither E7 nor p16INK4A levels were prognostic, risk stratification was instead pursued by profiling expression of E2F target genes, which are upregulated via E7 function and have higher expression in HPV+ vs. HPV− HNSCCs38. Unsupervised hierarchical clustering of the 53 HPV+ oropharyngeal HNSCCs in TCGA using a list of 325 E2F target gene transcripts38 identified a 22-case cluster (TCGA_C2) with decreased DFS and OS (Supporting Information Figure S8). The target gene list was narrowed by excluding transcripts with area under the ROC curve (AUC) of <0.8 (Supporting Information Table S13) to define 43 transcripts explanatory of TCGA_C2 cluster (Figure 6A, left). Upregulation of the 43 E2F target genes was significantly diminished in TCGA_C2, where more genes were expressed at levels at least one standard deviation below their medians for the total HPV+ case cohort (Figure 6A, right). Furthermore, most of the 43 E2F targets that are upregulated in HPV+ vs. HPV− oropharyngeal HNSCCs in TCGA lacked upregulation in TCGA_C2 (Supporting Information Figure S9). Prediction of DFS and OS using the 43-transcript profile contrasted with the lack of prognostic utility for 8th edition AJCC staging in the same cohort (Figure 6B). To evaluate potential clinical utility of this E2F target gene profile, the HPV+ TCGA cases were classified based on the number of low-expressing E2F target genes in each sample. Log rank analysis of the 43 gene profile defined an optimal cut point of 2 or more low-expressed E2F targets to be strongly predictive of OS and DFS (Figure 6C). To further assess the 43 genes for potential utility as biomarkers, the recurrent cases were compared to all the patients with known disease-free status at the time of the latest recurrence (3.75 years), creating a sub-cohort of 9 recurrent cases and 9 controls. The recurrences had significantly more low-expressed E2F targets than controls (Figure 6D), and presence of 2 or more low-expressed genes here predicted worse DFS as well (Figure 6E). Computing ROC curves for expression of each gene identified 12 genes among the 43 with a significant AUC (range 0.85-0.95) and an FDR of <0.05, with 11 of the genes having lower expression in recurrences vs. controls (Figure 6F, Supporting Information Table S14). Taken together, these findings are evidence that HPV+ HNSCCs with poor prognosis have downregulation of several E2F target transcripts, thus offering promise for further biomarker development in cohorts containing more recurrent cases.
Figure 6. Reduced E2F target gene expression is prognostic for HPV+ HNSCCs in TCGA.
A. Expression of the 43 E2F target genes with AUC ≥0.8 in the HPV+ HNSCCs in TCGA (left). Number of E2F target genes with low expression (normalized mRNA levels at least one standard deviation below their medians for the total HPV+ case cohort) in TCGA_C1 vs. TCGA_C2 cases (right), p-value determined by two-tailed Student’s t-test assuming unequal variances. B. 5-year OS (left) and DFS (middle) for E2F target-derived clusters TCGA_C1 vs. TCGA_C2. OS of HPV+ TCGA HNSCCs segregated by 8th edition clinical AJCC stage (right). C. 5-year OS (left) and DFS (right) for HPV+ TCGA HNSCCs segregated by number of E2F targets with low expression. D. Number of E2F target genes with low expression in the 9 recurrent HPV+ TCGA cases versus the 9 non-recurrent controls. E. 3.75-year DFS for HPV+ TCGA recurrent cases and controls segregated by number of E2F targets with low expression. F. Normalized mRNA expression for E2F target genes with a significant AUC (range 0.85-0.95) in HPV+ TCGA recurrences vs. controls, p-values determined by two-tailed Student’s t-test assuming unequal variances. Survival p-values were determined by log-rank test.
DISCUSSION
This study addresses a need for improved preclinical resources to advance therapy for HPV+ HNSCC and creates new foundations for future biomarker development in this disease. The HPV+ PDXs characterized here are the first models systematically shown to retain the multiple genetic hallmarks of HPV+ HNSCC that are lost in human HPV+ cancer cell lines11. Despite potential for low PDX engraftment rates to create selection biases, the only bias identified was an over-representation of NOTCH1 mutations and other Notch pathway alterations with predicted loss of function. Possible enrichment of NOTCH1 mutations by engraftment here contributes to emerging evidence for HPV+ HNSCCs harboring these alterations having distinct biologic features and more aggressive behavior10. The models thus offer unique tools to address the biologic and therapeutic significance of attenuated Notch signaling in a tumor subtype that may have high clinical relevance. A novel association in this study between progression of the HPV+ HNSCC models and high TMB was preserved across multiple HPV+ and HPV− SCC patient cohorts, including cases in the head and neck, esophagus, and cervix. TMB’s prognostic utility was robustly evident in early stage HPV− tumors, whereas outcomes prediction for HPV+ HNSCCs was served by distinctive features of an HPV+ PDX from a lethal case. The reduced p16INK4A upregulation downstream of low E7 levels in this outlier PDX was suggested to be present in recurrent HPV+ HNSCCs in two cohorts. The decrease in E2F target gene expression predicted to accompany low E7 was prognostic in TCGA, providing a new biologic basis with potential to distinguish subgroups of HPV+ HNSCCs warranting distinct therapeutic approaches. Lack of a suitable validation set for this result highlights the need for transcriptional analyses of HPV+ HNSCCs cohorts containing more poor outcomes to establish expression classifiers with prognostic utility.
The negative prognostic potential of reduced E2F target upregulation in this study strengthens the early evidence for oncogenic HPVs creating a greater imprint upon the host transcriptional landscape in those HPV+ HNSCCs that are readily curable. Specifically, the poor prognosis HPV+ case cluster identified by us in TCGA using E2F target transcripts were nearly identical to a case cluster independently defined based on reduced dysregulation of an entirely distinct set of HPV-modulated transcripts7. Evidence that diminished E2F target expression is part of a wider decrease in viral effects upon host gene expression in poor prognosis cases holds promise for development of clinical risk stratifiers based on a limited set of host and viral proteins.
Although the reduced p16INK4A and E7 levels that prompted analysis of E2F targets lacked independent prognostic utility, they are intimately related to E2F target gene expression and might contribute to the prognostic transcriptional profile in some cases. Specifically, low E7 levels in the outlier PDX and some early recurrent TCGA cases may reflect greater reliance on host oncogenic drivers and serve as one of multiple mechanisms limiting E2F target upregulation in high-risk tumors. Similarly, the p16INK4A upregulation thought to occur downstream of E7 expression in HPV-related cancers39 may be reduced in a subset of recurrent cases in addition to the atypical PDX. In such instances, secondary events that prevent a reported addiction of HPV+ cancers to high p16INK4A levels40 may be necessary for cancer progression.
Because high p16INK4A may impair DNA repair by inhibiting homologous recombination, relative reduction in p16INK4A levels could mediate the lethal phenotype of some HPV+ HNSCCs by increasing radiation resistance. The hemizygous CDKN2A deletion in the outlier HPV+ PDX is thus particularly intriguing. Although CDKN2A mutation or loss is commonplace and negatively prognostic in HPV− HNSCCs41, these alterations are absent in HPV+ TCGA cases. However, CDKN2A alterations were reported in 10% of aggressive pulmonary metastatic recurrences from HPV+ HNSCCs42, prompting speculation that rare CDKN2A alterations in a few HPV+ untreated cases are negatively prognostic as well.
Whereas the association between TMB and PDX model growth here was not prognostic in HPV+ HNSCCs, reduced survival of early HPV− cancers with more mutations justifies further evaluation of TMB in that clinical context. Although high TMB was linked to local progression in both HNSCC subtypes, lesser ability for HPV+ local disease to impact survival might explain prognostic utility being limited to HPV− disease. TMB in the early HPV-neg HNSCCs with poor prognosis may also reflect an increase in smoking-related DNA damage43, which is not as apparent in HPV+ HNSCCs from smokers44. To date, high TMB has been extensively studied as a predictor of immunotherapy responses45 but not as a correlate of the tumor-autonomous progression quantified here in the models. Recent analysis in a cohort of HNSCCs treated non-surgically showed a marginally significant association of TMB with OS46. The association shown here narrows TMB’s prognostic to early stage HPV− disease, where it is also not limited to patients receiving chemoradiation. Prediction of tumor progression in both the milieu of an immunodeficient mouse and patients is therefore a novel finding that may lead to expanded utility for TMB as a biomarker. Progress of TMB assays toward clinical application47 thus creates added opportunities to simultaneously identify certain high-risk tumors based on TMB at presentation and escalate initial therapy for them via checkpoint inhibition.
Over-representation of NOTCH1 mutations in the HPV+ models adds to evidence that PDX generation sometimes selects for aggressive tumor genotypes15-17. Loss-of-function mutants in the Notch pathway HNSCCs indicate a tumor suppressor role48 and are similarly associated with poor outcomes in HPV− cases49. Canonical Notch signals in normal squamous epithelia promote hierarchical differentiation, and their loss enhanced tumorigenesis in a HNSCC mouse model expressing E6/E735. Furthermore, Notch activation downstream of E2F function is observed in development and served to limit tumor growth in a hepatocellular carcinoma model upon Rb loss50. Thus, silencing Notch may similarly cooperate with E7-mediated Rb degradation to drive carcinogenesis in select HPV+ HNSCCs.
There are notable limitations to the insights gained here from patient-derived models. First, the small sample size does not capture the full genetic diversity among HPV+ HNSCCs. Secondly, NOTCH1 mutation has been shown to be prognostic in only one HPV+ HNSCC cohort thus far10. Despite that study being the largest linking HPV+ HNSCC mutations to outcomes, lack of adequate validation cohorts makes the significance of the Notch pathway alterations in the PDXs less clear. Finally, the E2F target transcript profile could not be reduced to a mature gene expression classifier that is prognostic because adequate clinical cohorts with molecular annotation are presently lacking. Nevertheless, the knowledge gained from this small HPV+ PDX panel illustrates the value of pursuing patient-derived models even for tumor types that do not grow efficiently outside humans.
Supplementary Material
Novelty and Impact:
Therapeutic advancement for HPV-related oropharyngeal cancers is hindered by inadequate biomarkers and few experimental models. This study represents the first detailed characterization of patient-derived xenografts and organoids created from these tumors, which typically grow poorly outside humans. The models’ retention of genetic hallmarks lost in cell lines addresses the deficit in preclinical tools for this disease. The same analysis identified a novel basis for quantifying viral effects on host gene expression to serve biomarker development.
ACKNOWLEDGEMENTS
This work is supported by NIH R01-DE027185 (D Basu, P Gimotty), P30-DK050306 (Core Facilities), P30CA016620 (Bioinformatics Core of the Abramson Cancer Center), and P30CA010815 (Wistar Institute Cancer Center).
Abbreviations:
- CNA
copy number alteration
- DFS
disease-free survival
- DSS
disease-specific survival
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papilloma virus
- OS
overall survival
- PCA
principle component analysis
- PDX
patient-derived xenograft
- R-SVM
radial support vector machine
- SCC
squamous cell carcinoma
- TCGA
the cancer genome atlas
- TMB
tumor mutational burden
- WES
whole exome sequence
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
REFERENCES
- 1.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J Clin Oncol 2015;33: 3235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirghani H, Blanchard P. Treatment de-escalation for HPV-driven oropharyngeal cancer: Where do we stand? Clin Transl Radiat Oncol 2018;8: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ, Sturgis EM, Burtness B, Ridge JA, Ringash J, Galvin J, Yao M, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 2019;393: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, Dalby M, Mistry P, Sen M, O'Toole L, Al Booz H, Dyker K, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet 2019;393: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Würdemann N, Wagner S, Sharma SJ, Prigge ES, Reuschenbach M, Gattenlöhner S, Klussmann JP, Wittekindt C. Prognostic Impact of AJCC/UICC 8th Edition New Staging Rules in Oropharyngeal Squamous Cell Carcinoma. Front Oncol 2017;7: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleber-Netto FO, Rao X, Guo T, Xi Y, Gao M, Shen L, Erikson K, Kalu NN, Ren S, Xu G, Fisch KM, Akagi K, et al. Variations in HPV function are associated with survival in squamous cell carcinoma. JCI insight 2019;4: 10.1172/jci.insight.124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Yan B, Lou H, Shen Z, Tong F, Zhai A, Wei L, Zhang F. Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol Immunol 2018;96: 28–36. [DOI] [PubMed] [Google Scholar]
- 9.Chera BS, Kumar S, Beaty BT, Marron D, Jefferys SR, Green RL, Goldman EC, Amdur R, Sheets N, Dagan R, Hayes DN, Weiss J, et al. Rapid Clearance Profile of Plasma Circulating Tumor HPV Type 16 DNA during Chemoradiotherapy Correlates with Disease Control in HPV-Associated Oropharyngeal Cancer. Clin Cancer Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dogan S, Xu B, Middha S, Vanderbilt CM, Bowman AS, Migliacci J, Morris LGT, Seshan VE, Ganly Identification of prognostic molecular biomarkers in 157 HPV-positive and HPV-negative squamous cell carcinomas of the oropharynx. Int J Cancer 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H, Yang X, Si H, Saleh AD, Xiao W, Coupar J, Gollin SM, Ferris RL, Issaeva N, Yarbrough WG, Prince ME, Carey TE, et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell reports 2018;25: 1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalu NN, Mazumdar T, Peng S, Shen L, Sambandam V, Rao X, Xi Y, Li L, Qi Y, Gleber-Netto FO, Patel A, Wang J, et al. Genomic characterization of human papillomavirus-positive and -negative human squamous cell cancer cell lines. Oncotarget 2017;8: 86369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facompre ND, Sahu V, Montone KT, Harmeyer KM, Nakagawa H, Rustgi AK, Weinstein GS, Gimotty PA, Basu D. Barriers to generating PDX models of HPV-related head and neck cancer. Laryngoscope 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell chemical biology 2017;24: 1092. [DOI] [PubMed] [Google Scholar]
- 15.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, Roman-Roman S, Seoane J, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery 2014;4: 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon HG, Oh K, Lee J, Lee M, Kim JY, Yoo TK, Seo MW, Park AK, Ryu HS, Jung EJ, Kim N, Jeong S, et al. Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast cancer research and treatment 2015; 154: 13. [DOI] [PubMed] [Google Scholar]
- 17.Karamboulas C, Bruce JP, Hope AJ, Meens J, Huang SH, Erdmann N, Hyatt E, Pereira K, Goldstein DP, Weinreb I, Su J, O'Sullivan B, et al. Patient-Derived Xenografts for Prognostication and Personalized Treatment for Head and Neck Squamous Cell Carcinoma. Cell Rep 2018;25: 1318–31.e4. [DOI] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2: 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Network CGAR, University AWGA, Agency BC, Hospital BaWs, Institute B, University B, University CWR, Institute D-FC, University D, Centre GPC, School HM, Biology IfS, et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541: 169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173: 291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarthy A, Henderson S, Thirdborough SM, Ottensmeier CH, Su X, Lechner M, Feber A, Thomas GJ, Fenton TR. Human Papillomavirus Drives Tumor Development Throughout the Head and Neck: Improved Prognosis Is Associated With an Immune Response Largely Restricted to the Oropharynx. J Clin Oncol 2016;34: 4132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic acids research 2015;43: W566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laud K, Marian C, Avril MF, Barrois M, Chompret A, Goldstein AM, Tucker MA, Clark PA, Peters G, Chaudru V, Demenais F, Spatz A, et al. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. Journal of medical genetics 2006;43: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333: 1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, Pillay N, Forster M, Cronin MT, Lipson D, Miller VA, Brennan TA, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV− tumors. Genome medicine 2013;5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Strieker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, Agrawal N, Fakhry C, Quon H, Subramaniam RM, Zuo Z, Seiwert T, et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Annals of oncology: official journal of the European Society for Medical Oncology 2015;26: 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perdomo S, Anantharaman D, Foll M, Abedi-Ardekani B, Durand G, Reis Rosa LA, Holmila R, Le Calvez-Kelm F, Tajara EH, Wunsch-Filho V, Levi JE, Vilensky M, et al. Genomic analysis of head and neck cancer cases from two high incidence regions. PloS one 2018; 13: e0191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbison RA, Kubik M, Konnick EQ, Zhang Q, Lee SG, Park H, Zhang J, Carlson CS, Chen C, Schwartz SM, Rodriguez CP, Duvvuri U, et al. The mutational landscape of recurrent versus nonrecurrent human papillomavirus-related oropharyngeal cancer. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vossen DM, Verhagen CVM, van der Heijden M, Essers PBM, Bartelink H, Verheij M, Wessels LFA, van den Brekel MWM, Vens C. Genetic Factors Associated with a Poor Outcome in Head and Neck Cancer Patients Receiving Definitive Chemoradiotherapy. Cancers (Basel) 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols AC, Palma DA, Chow W, Tan S, Rajakumar C, Rizzo G, Fung K, Kwan K, Wehrli B, Winquist E, Koropatnick J, Mymryk JS, et al. High frequency of activating PIK3CA mutations in human papillomavirus-positive oropharyngeal cancer. JAMA Otolaryngol Head Neck Surg 2013; 139: 617–22. [DOI] [PubMed] [Google Scholar]
- 33.Haft S, Ren S, Xu G, Mark A, Fisch K, Guo TW, Khan Z, Pang J, Ando M, Liu C, Sakai A, Fukusumi T, et al. Mutation of chromatin regulators and focal hotspot alterations characterize human papillomavirus-positive oropharyngeal squamous cell carcinoma. Cancer 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, Schultz N, Taylor BS. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nature biotechnology 2016;34: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyman PE, Buehler D, Lambert PF. Loss of Function of Canonical Notch Signaling Drives Head and Neck Carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24: 6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dotto GP, Rustgi AK. Squamous Cell Cancers: A Unified Perspective on Biology and Genetics. Cancer cell 2016;29: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo T, Gaykalova DA, Considine M, Wheelan S, Pallavajjala A, Bishop JA, Westra WH, Ideker T, Koch WM, Khan Z, Fertig EJ, Califano JA. Characterization of functionally active gene fusions in human papillomavirus related oropharyngeal squamous cell carcinoma. Int J Cancer 2016;139: 373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson ME, Cantalupo PG, Pipas JM. Identification of Head and Neck Cancer Subtypes Based on Human Papillomavirus Presence and E2F-Regulated Gene Expression. mSphere 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proceedings of the National Academy of Sciences of the United States of America 2011;108: 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen WS, Bindra RS, Mo A, Hayman T, Husain Z, Contessa JN, Gaffney SG, Townsend JP, Yu JB. CDKN2A Copy Number Loss Is an Independent Prognostic Factor in HPV-Negative Head and Neck Squamous Cell Carcinoma. Frontiers in oncology 2018;8: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna GJ, Kacew A, Chau NG, Shivdasani P, Lorch JH, Uppaluri R, Haddad RI, MacConaill LE. Improved outcomes in PI3K-pathway-altered metastatic HPV oropharyngeal cancer. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desrichard A, Kuo F, Chowell D, Lee KW, Riaz N, Wong RJ, Chan TA, Morris LGT. Tobacco Smoking-Associated Alterations in the Immune Microenvironment of Squamous Cell Carcinomas. J Natl Cancer Inst 2018; 110: 1386–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grønhøj C, Jensen DH, Agander T, Kiss K, Høgdall E, Specht L, Bagger FO, Nielsen FC, von Buchwald C. Deep sequencing of human papillomavirus positive loco-regionally advanced oropharyngeal squamous cell carcinomas reveals novel mutational signature. BMC Cancer 2018; 18: 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51: 202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eder T, Hess AK, Konschak R, Stromberger C, Jöhrens K, Fleischer V, Hummel M, Balermpas P, von der Grün J, Linge A, Lohaus F, Krause M, et al. Interference of tumour mutational burden with outcome of patients with head and neck cancer treated with definitive chemoradiation: a multicentre retrospective study of the German Cancer Consortium Radiation Oncology Group. Eur J Cancer 2019; 116: 67–76. [DOI] [PubMed] [Google Scholar]
- 47.Truesdell J, Miller VA, Fabrizio D. Approach to evaluating tumor mutational burden in routine clinical practice. Transl Lung Cancer Res 2018;7: 678–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333: 1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vettore AL, Ramnarayanan K, Poore G, Lim K, Ong CK, Huang KK, Leong HS, Chong FT, Lim TK, Lim WK, Cutcutache I, McPherson JR, et al. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome medicine 2015;7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A, Vogel H, Sylvester KG, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med 2011;208: 1963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data has been deposited in the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) and can be accessed through the accession PRJNA597082.