Abstract
BACKGROUND & AIMS:
Liquid biopsies, or blood samples, can be analyzed to detect circulating tumor cells (CTCs), cell-free DNA (cfDNA), and extracellular vesicles, which might identify patients with hepatocellular carcinoma (HCC) or help determine their prognoses. We performed a systematic review of studies of analyses of liquid biopsies from patients with HCC and their comparisons with other biomarkers.
METHODS:
We performed a systematic review of original studies published before December 1, 2019. We included studies that compared liquid biopsies alone and in combination with other biomarkers for the detection of HCC, performed multivariate analyses of the accuracy of liquid biopsy analysis in determining patient prognoses, or evaluated the utility of liquid biopsy analysis in monitoring treatment response.
RESULTS:
Our final analysis included 112 studies: 67 on detection, 46 on determining prognosis, and 25 on treatment monitoring or selection. Ten studies evaluated assays that characterized cfDNA for detection of HCC in combination with measurement of α-fetoprotein (AFP)—these studies found that the combined measurement of cfDNA and AFP more accurately identified patients with HCC than measurement of AFP alone. Six studies evaluated assays for extracellular vesicles and 2 studies evaluated assays for CTC in detection of HCC, with and without other biomarkers—most of these studies found that detection of CTCs or extracellular vesicles with AFP more accurately identified patients with HCC than measurement of AFP alone. Detection of CTCs before surgery was associated with HCC recurrence after resection in 13 of 14 studies; cfDNA and extracellular vesicles have been studied less frequently as prognostic factors. Changes in CTC numbers before vs after treatment more accurately identify patients with HCC recurrence than pretreatment counts alone, and measurements of cfDNA can identify patients with disease recurrence or progression before changes can be detected by imaging. We found little evidence that analyses of liquid biopsies can aid in the selection of treatment for HCC. Quality assessment showed risk of bias in studies of HCC detection and determination of prognosis.
CONCLUSIONS:
In a systematic review of 112 studies of the accuracy of liquid biopsy analysis, we found that assays for CTCs and cfDNA might aid in determining patient prognoses and monitoring HCC, and assays for cfDNA might aid in HCC detection, but there is a risk of bias in these studies. Studies must be standardized before we can assess the clinical utility of liquid biopsy analysis in the detection and management of patients with HCC.
Keywords: Liver Cancer, Outcome, Therapy, Prediction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and the third leading cause of cancer death worldwide.1 HCC is one of the few cancers with increasing incidence in the United States and this is driven primarily by the increase in nonalcoholic fatty liver disease and the peak in hepatitis C–related complications.2–4 Prognosis after HCC diagnosis remains poor, with a median survival of fewer than 2 years.5
There are several challenges in HCC management. First, current surveillance strategies using ultrasound and α-fetoprotein (AFP) are inadequate, with a sensitivity of 63% and a specificity of 84%, for the detection of early stage HCC.6 Although other biomarkers such as AFP-L3 and des-γ-carboxyprothrombin have shown promise for early detection, they have not been validated sufficiently for routine clinical use.7,8 Second, we lack effective therapies and biomarkers to guide personalized selection of locoregional and systemic therapies for patients with advanced-stage HCC.9,10 A major contributor to the lack of progress in developing more effective and precise therapies for HCC is the lack of available tissue to study tumor mutations and biology because HCC can be diagnosed reliably and treated based on imaging alone and routine diagnostic biopsy is not currently recommended by guidelines.11–13 Only recently has deep sequencing of human HCC tissue identified molecular subtypes with distinct prognoses.14–16
A liquid biopsy, using circulating tumor-derived markers, may address limitations in both early detection and lack of molecular data on HCC. One example is circulating tumor cells (CTCs), the presence of which represents an intermediate stage between localized disease and distant metastasis.17 CTCs can be detected in virtually all major solid tumors in the setting of metastatic disease as well as some early and intermediate-stage cancers.17–19 Deep sequencing of CTCs can identify mutations, some of which are distinct from those detected in primary tumor samples, as has been shown in multiple myeloma20 and prostate cancer,21 and may provide more comprehensive information, compared with a needle biopsy, as a result of molecular heterogeneity of tumors within the same patient and sometimes within the same tumor. Another approach is the detection of cell-free DNA (cfDNA), released into the circulation from dead cancer cells and/or secretion from viable cancer cells.22 cfDNA can be quantified and characterized for integrity, and also can be sequenced to detect mutations, methylation, and insertions–deletions.22 cfDNA mutational profiles have been investigated for the detection and prognosis of HCC, and for identification of clinically actionable mutations.23–25 A third example is extracellular vesicles (EVs), which are formed by budding of lysosomes, cell membranes, or apoptotic bodies, which subsequently can be released into the circulation.26 EVs affect cell–cell communication and have been investigated as both targets of therapy and as therapeutic modalities themselves.27 Furthermore, EV presence/concentration and analysis of EV contents have been studied as circulating cancer biomarkers.28 Liquid biopsy has not been studied extensively for the early detection of other cancers,29 however, given inadequacies in early detection of HCC and an identifiable high-risk population to target for surveillance (ie, patients with cirrhosis), liquid biopsy could fill an important gap in HCC detection.
The role of liquid biopsy in HCC detection and prognosis has been reviewed recently.30–32 One important clinical question yet to be addressed is whether liquid biopsy outperforms or offers incremental value to existing biomarkers, most notably AFP. Another question not addressed is whether liquid biopsy is a useful method for monitoring patients receiving HCC therapy. Thus, we conducted a systematic review of the role of liquid biopsy for HCC detection, prognostication and/or prediction of response to therapy, monitoring for recurrence, and treatment selection, focusing on the comparison of liquid biopsy with existing biomarkers used in clinical practice.
Methods
Literature Search
We performed a systematic review of Medline/PubMed, EMBASE, and the Cochrane library through December 1, 2019, with no start date restriction. Search terms are detailed in Supplementary Table 1. We also screened all articles referenced in these selected studies and in several recent review articles for eligibility.30–32 There were no language restrictions; English abstracts of non-English language articles were screened and, when applicable, full-text studies were reviewed by authors fluent in their respective languages. (Chinese was the only non-English language for which full text review was required.)
Inclusion criteria were as follows: studies of patients diagnosed with HCC in which CTCs, cfDNA, or EVs were evaluated. Studies were required to report associations between liquid biopsy and 1 of the following: (1) detection, comparing circulating markers in patients with vs those without HCC, or sensitivity of the markers for detecting HCC; (2) prognosis/prediction, evaluating the effect of levels/presence of circulating markers on clinically relevant outcomes and/or response to therapy; (3) monitoring of residual disease during or after treatment; or (4) choice of HCC therapy. Finally, detection studies had to report either a comparison between liquid biopsy and existing biomarkers or the incremental value of liquid biopsy when combined with existing biomarkers (eg, AFP), and prognostic studies had to report the effect of liquid biopsy on prognosis in multivariable analysis.
Abstracts were reviewed independently by 2 authors (V.L.C. and D.X.), with discrepancies resolved by a third author (N.D.P.).
Data Extraction
We independently abstracted the required information from eligible studies using standardized forms developed by the investigators. We collected information on inclusion/exclusion criteria and country/countries of studies. For controls, we determined whether they had chronic liver disease (CLD) or not, and, if they did, we extracted the etiology of underlying liver disease. For participants with HCC, we abstracted this information plus HCC stage and cancer treatment modalities.
Clinical End Points
For the detection component of this study, the primary outcome of interest was the diagnostic accuracy of the liquid biopsy in distinguishing HCC patients from participants without HCC. We separately analyzed the comparisons of the following: HCC vs CLD and HCC vs healthy controls. For prognosis, outcomes of interest were mortality/overall survival, and tumor progression or recurrence.
A meta-analysis was not conducted owing to the heterogeneity in which components of liquid biopsy were evaluated.
Quality Assessment
Quality assessment was performed using the QUality Assessment of Diagnostic Accuracy Studies-2 tool for diagnostic studies33 and the QUality In Prognosis Studies tool for prognostic studies.34
Results
A Preferred Reporting Items for Systematic Reviews and Meta-Analyses flowchart illustrating study selection is shown in Figure 1.35 The database search identified 2225 studies, of which 79 were included in the systematic review. We also reviewed all studies referenced in the selected studies, yielding 33 additional studies for inclusion in the systematic review. In total, 112 studies were included in this systematic review: 67 on detection, 46 on prediction/prognostication, and 25 on treatment monitoring/selection. Ten studies investigated both detection and prognosis; 3 studies investigated both detection and treatment monitoring/selection; 7 studies investigated both prognosis and treatment monitoring/selection; and 3 studies investigated detection, prognosis, and treatment monitoring/selection. We identified only 3 studies on treatment selection, which was not adequate for a systematic review and therefore this application is not discussed further in this article.36–38
Figure 1.
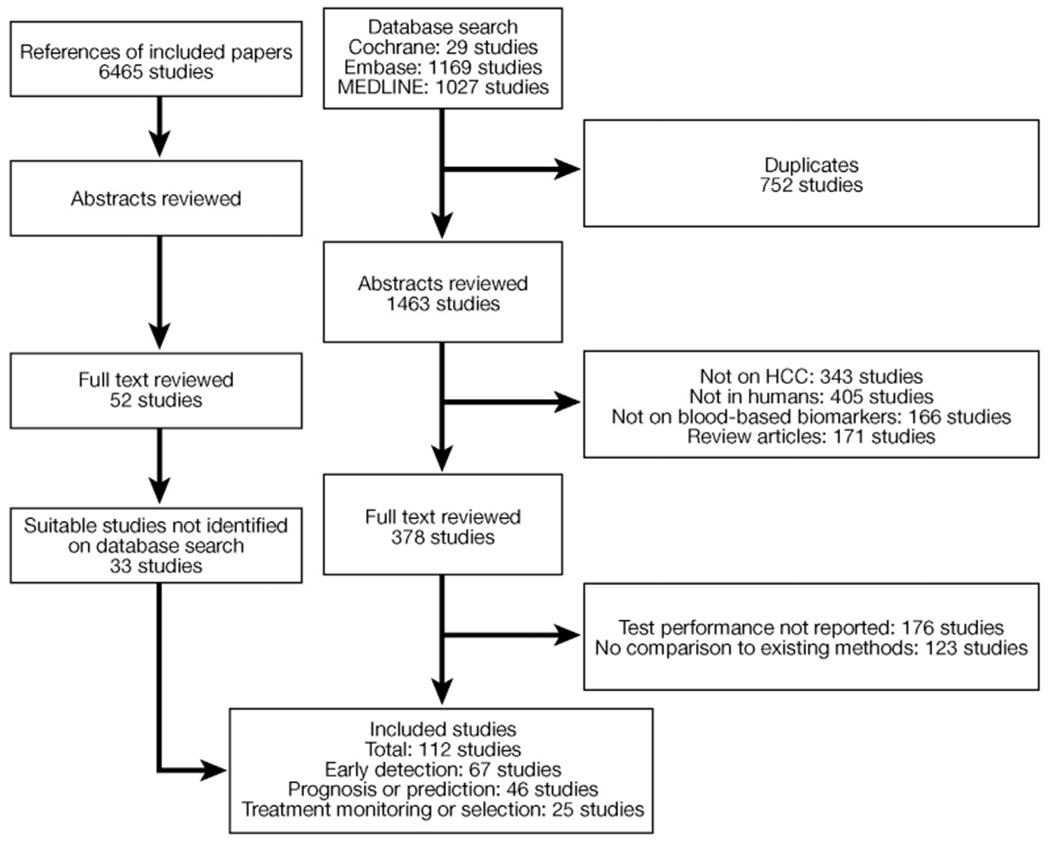
Preferred Reporting Items for Systematic Reviews and Meta-Analyses flowchart illustrating study selection. HCC, hepatocellular carcinoma.
Detection
We evaluated the utility of liquid biopsy for HCC detection based on either performance vs AFP, performance in AFP-positive vs AFP-negative patients, or a combination of liquid biopsy and AFP vs AFP or liquid biopsy alone.
Circulating tumor cells.
Seventeen studies investigated the utility of CTCs for HCC detection (Table 1).39–55 The most commonly used method to detect CTCs was positive selection for epithelial markers such as cytokeratins, epithelial cell adhesion molecule, or asialoglycoprotein receptor (8 studies). Another method used was Canpatrol (2 studies), which used filtration followed by cell staining to identify both epithelial and mesenchymal phenotypes. In general, these studies showed that CTC levels were superior to AFP in distinguishing HCC from controls (3 of 4 studies reporting test characteristics for CTCs vs AFP), that CTC levels were increased in AFP-negative HCC patients (11 of 13 studies), or that a combination of CTCs and AFP was superior to AFP alone in differentiating HCC from controls (2 of 2 studies).
Table 1.
Studies on Use of Circulating Tumor Cells for Hepatocellular Carcinoma Detection
| Study | CTC definition | HCC patients | Controls | Comparator, AFP cut off value, ng/mL | Findings: sensitivity/specificity, AUC |
|---|---|---|---|---|---|
| Bahnassy et al,39 2014 | CD45(−) and either CK19, CD90, or CD133(+) | N = 70 Stage: 74% (late stage) Treatment: NR |
33 CLD, 30 healthy | AFP ratio | CTCs had poorer test characteristics than AFP ratio; HCC vs CLD: CK19(+) CTCs: 87.1%/82.5% CD90(+) CTCs: 82.5%/89.6% CD133(+) CTCs: 40.0%/6.3% AFP ratio: 95.7%/90.5% |
| Bhan et al,40 2018 | CD45(−) and hydrodynamics, followed by HCC score based on gene expression | N = 54 Stage: 39% within Milan criteria Treatment: 40% ablation 30% TACE, 28% radiation therapy, 21% resection, 19% sorafenib, 9% liver transplant, 13% othera |
39 CLD, 10 healthy | No cut-off value provided | HCC score outperformed AFP in HCC vs CLD HCC score: 85%/95% AFP >20 ng/mL: 55%/100% |
| Cheng et al,41 2019 | CanPatrol | N = 113 Stage: 65% BCLC 0/A Treatment: NR |
57 CLD | 400 | CTCs outperformed and provided incremental benefit to AFP AFP: 44.3%/89.5%, AUC, 0.67 All CTCs (>1/5 mL): 72.6%/61.4%, AUC, 0.77 All CTCs or AFP: AUC, 0.82 |
| Fang et al,42 2014 | CellSearch | N = 42 Stage: 48% >7 cm maximal tumor size, 45% >3 tumors Treatment: 100% TACE |
10 CLD, 10 healthy | 400 | CTCs: 74%/100% Sensitivity 89% among patients with high AFP and 61% with low AFP (P = .08) |
| Guo et al,43 2007 | CD45(−) EpCAM(+) then AFP mRNA | N = 44 Stage/treatment: NR |
7 healthy | 20 | AFP mRNA: sensitivity, 72.7%; overall, 50% among AFP <20 ng/mL, and 86.7% among AFP >1000 ng/mL (P < .05) |
| Guo et al,45 2014 | Two methods: CellSearch and quantitative PCR for EpCAM in CD45-cells | N = 222 Stage: NR Treatment: 53% resection, 25% TACE, 22% radiotherapy |
49 CLD, 71 healthy | No cut-off value provided | Note: cohort may overlap with Guo et al44 2018 EpCAM-mRNA(+) CTCs: 42.6%/96.7%, AUC, 0.70 EpCAM-mRNA(+) CTCs plus AFP: 73.0%/93.4%, AUC, 0.86 |
| Guo et al,44 2018 | PCR score: EpCAM, CD133, CD90, CK19 | N = 395 Training: 66% BCLC 0/A, 98% resection, 2% TACE Validation: 48% BCLC 0/A, 67% resection, 33% TACE |
301 CLD, 210 healthy | 20 | Note: cohort may overlap with Guo et al45 2014 PCR score: Overall: 72.5%/95.0%, AUC, 0.88 AFP low: 77.7%/95.0%, AUC, 0.89 AUC based on stage: 0.92 (stage 0), 0.86 (stage A), 0.91 (stage B) and 0.86 (stage C) AFP alone: 57.0%/90.0%, AUC, 0.77 |
| Kalinich et al,46 2017 | PCR score: expression of AFP, AHSG, ALB, APOH, FABP1, FGB, FGG, RBP4, and TF | N = 63 Stage: 15/25/13/46% BCLC 0/A/B/C+D Treatment: 66% ablation, 40% TACE, 28% resection, 23% liver transplant, 19% radiation therapy, 18% sorafenib, 9% SIRT, 15% othera |
26 CLD, 31 healthy | 100 | 15 patients with both PCR score and AFP: 4 PCR score (+) 1 AFP (+) 5 with both assays (+) 5 with both assays (−) 6 patients within Milan criteria: 2 PCR score (+) and 0 AFP (+) |
| Kelley et al,47 2015 | CellSearch | N = 20 Stage: 100% BCLC C Treatment: NR |
10 CLD | 400 | AFP ≤400 ng/mL: sensitivity, 90% AFP <400 ng/mL: sensitivity, 10% (P = .008) |
| Liu et al,48 2013 | CD45(−) ICAM-1(+) | N = 60 Stage: 72% maximal tumor size >5 cm, 12% multifocal tumors Treatment: 100% surgical |
N/A | 20 | High levels of CTCs in 56% of AFP(+) and 33% of AFP(−) patients (P = .14) |
| Sun et al,49 2013 | CellSearch | N = 123 Stage: 82/18% BCLC 0+A/B+C Treatment: 100% surgery |
5 CLD, 10 healthy | 400 | ≥2 CTCs/7.5 mL: Overall: 41.5%/100% High AFP: sensitivity, 54.7% Low AFP: sensitivity, 31.4% (P = .009) |
| Takahashi et al,50 2016 | Microcavity and CD45(−) EpCAM(+) CK(+) | N = 19 Stage: mixed Treatment: NR |
11 CLD | 4 | CTCs: sensitivity, 47.3% overall With high AFP, higher numbers of CTC detected (91.9 ± 50.1 vs 3.9 ± 2.1; P < .05) |
| Xu et al,51 2011 | ASGPR(+) | N = 85 Stage: 38/22/32/8% TNM I/II/III/IV Treatment: NR |
37 CLD, 20 healthy | 20 or 100 | CTCs: 81 %/100% No significant differences in CTC levels based on either AFP cut-off value |
| Xue et al,52 2018 | Two methods: CellSearch and either CD45(−) CK(+) DAPI(+) hybridization signal for CEP8 ≥2 or CD45(−) CK(−) DAPI(+) and hybridization signal for CEP8 >2 | N = 30 Stage: 80/20 BCLC 0+A/B+C Treatment: 100% liver transplant |
N/A | 400 | CTCs measured by hybridization in: Overall cohort: 70%/100% Low AFP: sensitivity, 90% High AFP: sensitivity, 30% (P = .002) |
| Yao et al,53 2005 | CD45(−) EpCAM(+) then AFP mRNA | N = 49 Stage/treatment: NR |
36 CLD, 18 healthy | 20 |
AFP mRNA Overall: 72.1%/66.7% Low AFP: sensitivity, 75.0% High AFP: sensitivity, 71.0% (P > .05) |
| Yin et al,54 2018 | CanPatrol | N = 80 Stage: 11/31/45/13% TNM I/II/III/IV Treatment: 51% surgery, 23% TACE, 26% no treatment |
10 healthy | 20 | Overall cohort: any CTCs 77.5%/100%, Twist (+) CTCs 67.5%/100% Low AFP: sensitivity, 35.3% or 17.7% for any CTCs or Twist (+) CTCs, respectively (P < .001) High AFP: sensitivity, 88.9% or 71.8% for any CTCs or Twist (+) CTCs, respectively (P < .001) |
| Zhou et al,55 2016 | CD45(−) EpCAM-mRNA(+) | N = 49 Stage: NR Treatment: 100% resection |
N/A | 400 | Any CTCs: Overall: 34.6%/100% Low AFP: sensitivity, 28.2% High AFP: sensitivity, 60% (P = .06) |
NOTE. Test characteristics are reported either as sensitivity (%)/specificity (%), area under the receiver operating characteristic curve; sensitivity (%)/specificity (%); or as individual parameters.
AFP, α-fetoprotein; ASGPR, asialoglycoprotein receptor; AUC, area under the receiver operating characteristic curve; BCLC, Barcelona Clinic Liver Cancer; CD, cluster of differentiation; CEP8,___; CK, cytokeratin; CLD, chronic liver disease; CTC, circulating tumor cell; DAPI, 4’,6-diamidino-2-phenylindole (nuclear stain); EpCAM, epithelial cell adhesion molecule; HCC, hepatocellular carcinoma; ICAM, intercellular adhesion molecule; mRNA, messenger RNA; NR, not reported; TACE, transarterial chemoembolization.
Patients may have received more than 1 therapy type.
Most of the included studies compared the sensitivity of liquid biopsy for HCC detection in individuals with AFP levels above vs below a specific cut-off value, which varied from 4 to 400 ng/mL (most commonly 20 ng/mL).42,43,46–55 A large study found that a CTC-derived polymerase chain reaction (PCR) score (quantifying expression of cancer-related genes in the blood) was increased in 125 of 171 (73%) patients with an AFP level less than 20 ng/mL.44 One study using CanPatrol, a detection method that is not biased toward epithelial vs mesenchymal CTC phenotypes, reported an overall sensitivity of 78%, with lower sensitivity among patients with an AFP level less than 20 ng/mL (N = 17) than those with an increased AFP level (N = 63): 35% vs 89%, respectively (P < .001).54Another study using Cell- Search, which only identifies epithelial cells, found that CTCs were detected in 1 of 10 patients with an AFP level less than 400 ng/mL compared with 9 of 10 patients with an AFP level of 400 ng/mL or greater (P = .008).47 A third study found that CD45(−) epithelial cell adhesion molecule (+) cells were detected in 6 of 12 patients with an AFP level less than 20 ng/mL vs 13 of 15 with an AFP level greater than 1000 ng/mL (P < .05).43 Other studies found that CTC levels were not statistically significantly different in patients with HCC and increased vs normal AFP levels.42,46,51,53,55
Two studies evaluated whether CTCs provided incremental value to AFP alone for identifying HCC patients. One study using CanPatrol for the detection of CTCs in 113 HCC vs 57 CLD patients reported the area under the receiver operating characteristic curves (AUCs) for identifying HCC patients were 0.67 for AFP at a cut-off value of 400 ng/mL, 0.77 for CTCs, and 0.82 for a combination of CTCs and AFP (sensitivity not reported).41 Another study found that CTCs, defined as the presence of EpCAM–messenger RNA, had a sensitivity of 42.6% and an AUC of 0.70 for differentiating HCC from controls that comprised CLD patients and healthy controls, while AFP (cut-off value, 400 ng/mL) had a sensitivity of 39.5% (AUC not reported),45 and a combination of CTCs and AFP had a sensitivity of 73.0% and an AUC of 0.86. No studies reported that CTCs added no incremental benefit to AFP; however, the risk of publication bias exists.
Three studies compared AFP and CTCs in differentiating HCC from controls. A large study compared 395 HCC patients with 301 CLD and 210 healthy controls and found that a CTC-derived PCR score showed a sensitivity of 72.5%, a specificity of 95.0%, and an AUC of 0.88 compared with 57.0%, 90.0%, and 0.77 for AFP at a cut-off value of 20 ng/mL.44 This score performed well in patients with early stage HCC, with an AUC of 0.92 among patients with Barcelona Clinic Liver Cancer (BCLC) stage 0, and 0.86 in patients with BCLC stage A disease. Another study also found that a CTC-derived PCR score showed higher sensitivity than AFP (cut-off value, 20 ng/mL) in differentiating HCC vs CLD (85% vs 55%).40 In contrast, 1 study found that CTCs (defined as CD45[−] plus either cytokeratin 19, CD90, or CD133[+]) had inferior sensitivity and specificity compared with the AFP ratio (undefined) in distinguishing HCC vs CLD.39
Cell-free DNA.
Several components of cfDNA may differ in patients with vs without cancer, such as total amount of cfDNA, cfDNA mutations (especially in candidate genes), or cfDNA methylation. Thirty-nine studies on cfDNA and HCC detection met inclusion criteria.56–94 Ten studies compared diagnostic test characteristics of cfDNA properties with AFP; of these, 9 found cfDNA to be superior,56,57,62,63,71,72,80,81,92 although 1 did not not.82 Sixteen studies evaluated the sensitivity of cfDNA for the diagnosis of HCC among patients with normal AFP levels, results were variable, ranging from 15% to 100%, in part owing to heterogeneity in which components of cfDNA were measured.57,59,61,69,73,77–79,84,86–91,94 Ten studies compared scores incorporating various components of cfDNA (ranging from amount of cfDNA to methylation of or mutation in specific genes) and AFP, and found that the combination had superior test characteristics than AFP level alone.60,64–68,74,76,83,85,93 Thirteen of 14 studies comparing the combination of cfDNA and AFP with cfDNA alone found that the combination had superior sensitivity and/or specificity,60,64–68,70,75,76,83,85,89,93 however, the incremental value of AFP was not observed in a recent large study in the United States.74
Extracellular vesicles.
Eleven studies on EVs in HCC detection were included (Supplementary Table 2).95–105 Generally, EV properties added to AFP in detection capability, but the value of EVs for HCC detection is less clear than CTCs or cfDNA, in part related to greater heterogeneity in which properties of EVs were studied.
Six studies evaluated the combination measurement of EVs and AFP compared with AFP alone.95,96,98,102–104 A large study (200 HCC patients, with 200 CLD and 200 healthy controls) found that AUCs of exosomal levels of 3 long noncoding RNAs were 0.96 and 0.53 in the training and validation cohorts, respectively, and 0.97 and 0.87 for the combination of these RNAs and AFP.98 Another study found that exosomal miR-122, miR-148a, and AFP together showed an AUC of 0.947 for distinguishing HCC vs cirrhotic controls, vs 0.665 for AFP alone.102
Five studies compared EV properties with AFP without evaluating incremental value compared with AFP.97,99–101,104 A large study with 71 HCC patients and 131 controls (37 of whom had liver disease) found that a machine learning–based score of EV long noncoding RNAs outperformed AFP for differentiating HCC vs benign liver lesions, with an AUC of 0.946 vs 0.834 (P = .037).97 The utility of EVs may depend on disease etiology: 1 study reported that EV miR-212 had a higher AUC than AFP alone (0.886 vs 0.849) among HBV-related HCC, but not among non–HBV-related HCC (0.793 vs 0.840); P values were not provided.105
Quality assessment.
Results of the QUality Assessment of Diagnostic Accuracy Studies-2 analysis are shown in Supplementary Tables 3, 4, 5, and 6. The detection of HCC usually was reliable, with most studies using either cross-sectional imaging or biopsy, and in most cases the HCC patients were representative of the general HCC population. However, most studies did not specify how patients were selected (ie, consecutive patients vs convenience sampling). Furthermore, the HCC diagnosis in nearly all studies was established before the diagnostic liquid biopsy and most studies did not mention blinding, raising the possibility of bias. In addition, many studies did not separately report results for early stage HCC, or for comparisons with cirrhosis controls.
Prognosis
Liquid biopsy has the potential to serve as a prognostic biomarker or as a predictive biomarker for response to therapy. For this section, we included only studies that reported liquid biopsy properties in multivariate analysis to identify the incremental value of liquid biopsy in prognostication beyond conventional factors such as tumor stage.
Circulating tumor cells.
Twenty-one studies investigated CTCs as prognostic markers (Table 3).36,44,45,48,49,55,106–120 Most studies (15 studies) focused on patients undergoing resection.44,48,49,55,106–110,113–116,118,120 In this subpopulation, 13 of 14 studies found a significant association between the presence of CTC preoperatively and tumor recurrence or disease-free survival,44,48,49,55,106–109,113–116,118,120 and 2 of 5 studies found a significant association between the presence of CTCs and overall survival.48,107,109,117,120 The remaining studies included populations undergoing other therapies.45,111,112,119 In all of these studies, the presence of CTC pretreatment was associated with a poorer prognosis.
Table 3.
Studies On Circulating Tumor Cell as Predictive or Prognostic Markers
| Study | CTC definition | HCC patients | Outcome, adjusted analysis |
|---|---|---|---|
| Dong et al,106 2015 | CellSearch | N = 156 Stage: NR Treatment: 88% resection |
Any CTCs: increased recurrence (P = .001) Consistent lack of CTCs: greater time to relapse (P = .015) Adjusted for NR |
| Fan et al,107 2011 | CD44(+) CD90(+) CD45(−) | N = 82 Stage: 5%/34%/34%/27% TNM I/II/III/IV Treatment: 100% resection |
CTCs associated with poorer: Median recurrence-free survival (6.0 vs >46.5 mo) 2-year recurrence-free survival (22.7% vs 64.2%) 2-year overall survival (58.5% vs 94.1%); P < .001 for all Adjusted for tumor size, tumor number, TNM stage, or blood transfusions |
| Fu et al,108 2019 | CanPatrol | N = 25 Stage: NR Treatment: 100% liver transplant |
Number of interstitial CTCs associated with recurrence post-transplant Adjusted for NR |
| Guo et al,45 2014 | Two methods: CellSearch or quantitative PCR for EpCAM in CD45(−) cells | N = 299 Stage: NR Treatment: 53% resection, 25% TACE, 22% radiotherapy |
EpCAM(+) mRNA pretreatment associated with poorer outcomes: Surgery: recurrence HR, 2.71 (1.52–4.86) Adjusted: satellite lesions TACE: progression HR, 3.75 (1.41–9.97) Unadjusted Radiotherapy: progression HR, 5.07 (1.39–18.47) Unadjusted |
| Guo et al,44 2018 | PCR score: EpCAM, CD133, CD90, CK19 | N = 395 Training: Stage: 66% BCLC 0/A Treatment: 98% resection, 2% TACE Validation: Stage: 48% BCLC 0/A Treatment: 67% resection, 33% TACE |
Increased recurrence with increased PCR score: Training: HR, 2.69 (1.62–4.48) Adjusted for tumor size and vascular invasion Validation: HR, 3.13 (1.36–7.19) Adjusted for tumor encapsulation, tumor size, satellite lesions, and vascular invasion Associations were significant among BCLC stage 0/A cancers and with AFP ≤20 ng/mL |
| Ha et al,109 2019 | Tapered slit filter | N = 105 Stage: 19%/81% BCLC 0/A Treatment: 100% resection |
Increase in CTC count after surgery: Increased recurrence: HR, 2.28 (1.06–4.90), adjusted for HBV etiology, tumor size, AST, ALT, and satellite lesions No difference in overall survival Presence of preoperative CTCs: no association with recurrence Presence of postoperative CTCs: no association with recurrence |
| Hamaoka et al,110 2019 | Glypican-3(+) | N = 85 Stage: NR Treatment: 100% resection |
Presence of CTCs associated with higher risk of microscopic portal vein invasion Adjusted for AFP, AFP-L3, multifocal disease, prothrombin time, and albumin |
| Li et al,36 2016 | Density-based, CD45(−), pan-CK(+), and either pAkt1/2/3 or pERK1/2(+) | N = 59 Stage: NR Treatment: 100% sorafenib |
High proportion of pERK (+) pAkt (−) CTCs: Superior progression-free survival: HR, 9.39 (3.24–27.19) Adjusted for Child-Pugh class, TNM stage, and presence of pERK(−) pAkt(+) CTCs |
| Liu et al,48 2013 | CD45(−) ICAM-1(+) | N = 60 Stage: 72% maximal tumor size >5 cm, 12% multifocal tumors Treatment: 100% resection |
High proportion of ICAM-1 cells associated with: Poorer disease-free survival (HR, 7.15; 2.99–17.05), adjusted for AFP, tumor size, tumor number, portal vein tumor thrombus, and ascites No association with overall survival (HR, 2.28; 0.95–7.82), adjusted: portal vein tumor thrombus, ascites, and prealbumin |
| Nel et al,111 2014 | CD45(−) and either EpCAM, panCK, or CD133(+) | N = 10 Stage: NR Treatment: 100% SIRT |
Low CD133+ cell ratio associated with increased progression (P = .02) Adjusted for AFP (P = .02) |
| Ogle et al,112 2016 | CD45(−), size | N = 69 Stage: NR Treatment: 39% arterial therapy, 13% sorafenib, 6% liver transplant, 4% resection, 28% supportive care only |
Presence of CTCs at any time (N = 69): Poorer overall survival: HR, 2.34 (1.01–5.43) Adjusted for tumor size, portal vein thrombus, and extrahepatic disease Decreased time to progression (P = .006) Presence of CTCs post-treatment (N = 40): Poorer overall survival: HR, 6.16 (1.71–22.33) Adjusted for tumor size, portal vein thrombus, and extrahepatic disease Decreased time to progression (P = .002) |
| Qi et al,113 2018 | CanPatrol | N = 112 Stage: 39/21/30 BCLC 0+A/B/C Treatment: 100% resection |
CTCs associated with HCC recurrence: CTC count: HR, 1.04 (1.03–1.05)a Mesenchymal CTC percentage: HR, 1.02 (1.01–1.03)a Mesenchymal > epithelial CTC percentage: HR, 1.03 (1.01–1.03)a Mesenchymal = epithelial CTC percentage: HR, 1.00 (0.99–1.03) Mesenchymal < epithelial CTC percentage: HR, 1.00 (0.99–1.01) Epithelial CTC percentage: HR, 1.00 (0.99–1.01) |
| Schulze et al,114 2017 | CellSearch | N = 57 Stage: NR Treatment: 100% resection |
CTCs associated with poorer: Recurrence: HR, 2.3 (P = .027) Recurrence-free survival: 5.0 ± 1.5 vs 12.0 ± 2.5 mo (P = .039) Adjusted for incomplete resection |
| Sun et al,49 2013 | CellSearch | N = 123 Stage: 82%/18% BCLC 0+A/B+C Treatment: 100% resection |
Presence of CTCs associated with: Increased recurrence: HR, 5.20 (2.65–10.21) Adjusted for AFP, tumor size, tumor encapsulation, satellite lesions, and vascular invasion |
| Sun et al,115 2018 | CellSearch | N = 73 Stage: 77%/23% BCLC 0+A/B+C Treatment: 100% resection |
Presence of CTCs from different vascular sites associated with: Intrahepatic recurrence: Peripheral vein: HR, 0.77 (0.14–5.19) Peripheral artery: HR, 2.54 (0.87–7.42) Peripheral vein CTC or clusters: HR, 3.48 (1.40–8.61) Adjusted for CTC presence in the above locations, AFP, vascular invasion Lung metastasis: Hepatic vein CTC: HR, 0.59 (0.04–9.54) Intrahepatic inferior vena cava CTC: HR, 0.67 (0.10–4.40) Hepatic vein CTC or clusters: HR, 42.20 (3.73–477.80) Adjusted for CTC presence in the above locations |
| von Feldon et al,116 2017 | CellSearch | N = 57 Stage: 46%/32%/21% T1/T2/T3, vascular invasion in 38% Treatment: 100% resection |
CTCs associated with increased recurrence: HR, 3.1 (1.0–9.4) Adjusted for resection margin |
| Vona et al,117 2004 | Filter (diameter > 25 mm) | N = 44 Stage: no extrahepatic metastasis, 39% multinodular, 39% maximum tumor ≤3 cm, 45% portal vein thrombus Treatment: NR |
Presence of CTCs or clusters associated with poorer overall survival (HR, NR; P = .02). NS after adjustment for eligibility for surgery, Child–Pugh class, unifocal disease, or portal vein thrombus |
| Wang et al,118 2018 | CanPatrol | N = 62 Stage: 37%/63% BCLC 0+A/B+C Treatment: 100% resection |
CTCs associated with recurrence: Total CTCs: unadjusted HR, 2.95 (1.18–7.35), NS after adjustment Mesenchymal CTCs: unadjusted HR, 4.74 (2.04–11.01), adjusted HR, 3.45 (1.39–8.56) Mixed CTCs: unadjusted HR, 2.94 (1.31–6.59), NS after adjustment Adjusted for tumor size, portal vein tumor thrombus, AFP, Edmondson stage, alkaline phosphatase, and the 3 categories of CTCs |
| Wu et al,119 2019 | CD45(−) and abnormal chromosome 8 amplification by FISH | N = 155 Stage: 38%/14%/48% BCLC A/B/C, 34%/35%/28%/4% TNM I/II/III/IV Treatment: 100% TACE |
Pre-TACE CTCs associated with poorer overall survival: HR, 2.84 (1.41–5.73) Adjusted for ECOG score and serum Dickkopf-1 concentration |
| Yu et al,120 2018 | CellSearch | N = 139 Stage: 40%/60% BCLC 0+A/B+C Treatment: 100% resection |
4 categories: (1) persistent (+), (2) preoperatively (+) but postoperatively (−), (3) preoperatively (−) but postoperatively (+), (4) persistently (−). For a 1-point increase in category: Disease-free survival: HR, 0.53 (0.41–0.68) Overall survival: HR, 0.48 (0.36–0.66) Adjusted: multifocal vs unifocal disease, tumor size >5 cm, macroscopic tumor thrombosis, and microscopic vascular invasion |
| Zhou et al,55 2016 | EpCAM mRNA(+) | N = 49 Stage: 90% BCLC 0/A Treatment: 100% resection |
High EpCAM mRNA increased recurrence: HR, 6.67 (1.94–22.88) Adjusted for satellite lesion presence and Treg/CD4+ T-cell proportion |
AFP, α-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BCLC, Barcelona Clinic Liver Cancer; CD, cluster of differentiation; CK, cytokeratin; CTC, circulating tumor cell; ECOG,____; EpCAM, epithelial cell adhesion molecule; FISH, fluorescence in situ hybridization; HR, hazard ratio; ICAM, intercellular adhesion molecule; mRNA, messenger RNA; pAkt,_____; panCK,_____; PCR, polymerase chain reaction; pERK,_____; NR, not reported; SIRT, selective internal radiation therapy; TACE, transarterial chemoembolization; Treg, regulatory T cell.
Significant after adjustment for AFP, cirrhosis, tumor size, BCLC stage, number of nodes, microvascular invasion, portal vein thrombus, and tumor capsule.
Cell-free DNA.
Fifteen studies on cfDNA met inclusion criteria (Table 4).37,56,58,63,73,77,86,92,121–127 Five studies focused on the total amount of cfDNA,37,63,125–127 5 studies focused on methylation either in single or multiple loci,73,77,86,92,123 and 5 studies focused on mutations or copy number variation.37,56,58,122,124 In general, a greater amount of cfDNA was associated with poorer overall survival, and tumor progression or recurrence.126 Two studies reported that RASSF1A methylation was associated with decreased overall survival.73,123 No other cfDNA properties were evaluated in more than 1 study.
Table 4.
Studies on Cell-Free DNA as Predictive or Prognostic Markers
| Study | cfDNA property | Cancer stage/treatment | Outcome, adjusted analysis |
|---|---|---|---|
| An et al,56 2019 | Any mutation | N = 26 Stage: 77%/23% TNM I/II+III Treatment: 100% resection |
Presence of cfDNA mutations postresection: shorter disease-free survival (median, 8.3 mo vs unreached) Adjusted for portal vein tumor thrombus |
| Cai et al,57 2019 | Fraction of single-nucleotide or copy number variants | N = 34 Stage: NR Treatment: 100% resection |
Presence of ctDNA (mutated cfDNA) postoperatively: Decreased relapse-free survival Decreased overall survival (P = .001 for both) Combining ctDNA and des-carboxyprothrombin further increased predictive power |
| Chelis et al,121 2013 | SOX17 promoter methylation | N = 87 Stage: 6%/17%/77% BCLC A/B/C Treatment: NR |
Methylated vs nonmethylated: overall survival 5 vs 14 mo, P = .006 Adjusted for performance status (P = .04) |
| El-Shazly et al,63 2010 | Total amount, integrity | N = 25 Stage/treatment: NR |
Overall survival: cfDNA amount: HR, 0.54 (0.20–1.60) cfDNA integrity: HR, 1.86 (1.20–2.88) Adjusted for tumor size, TNM stage, vascular invasion, nodal involvement, metastatic disease |
| Howell et al,122 2017 | CSMD3 mutations | N = 51 Stage/treatment: NR |
CSMD3 mutations, increased mortality: HR, 4.56 (1.17–17.69) Adjusted for age, Child–Pugh score, BCLC stage |
| Huang et al,123 2011 | APC or RASSF1A methylation | N = 72 Stage: 24%/76% UICC I+II/III+IV Treatment: NR |
Overall survival poorer with RASSF1A or APC methylation: RASSF1A methylation: HR, 3.26 (1.48–7.21), adjusted for age, sex, tumor size, TNM stage, α-fetoprotein APC methylation: poorer on unadjusted analysis, NS after adjustment |
| Jiao et al,124 2018 | TERT mutations | N = 218 Stage: 39%/22%/33% TNM I/II/III Treatment: NR |
Presence of TERT mutations: All HCC patients, decreased overall survival (P = .006) but NS after adjustment for tumor stage (P = .19) HCC patients with cirrhosis: trend toward significance after adjustment for tumor stage (P = .051) |
| Kanekiyo et al,73 2015 | Methylation of RASSF1A, CCND2, CFTR, SPINT2, SRD5A2, and/or BASP1 | N = 125 Stage: 46%/54% TNM I+II/III+IV Treatment: surgical |
Methylation of ≥3 of 6 genes: Disease-free survival: HR, 2.18, P < .0001 Overall survival: HR, 4.20, P < .001 Adjusted for tumor size, tumor differentiation, venous invasion stage, cirrhosis, AFP, DCP, methylation of all 6 individual genes |
| Li et al,77 2018 | Methylation of IFGBP7 | N = 155 Stage: 63%/37% TNM I+II/III/IV Treatment: 100% resection |
Methylation of IFGBP7 associated with: Increased recurrence: HR, 4.99 (1.51–16.47), adjusted for tumor size, vascular invasion, TNM stage Poorer overall survival: HR, 3.86 (2.07–7.20), adjusted for tumor size, vascular invasion, TNM stage, early tumor recurrence |
| Oh et al,37 2019 | Total amount, genomic instability, and VEGFA amplification | N = 151 Stage: 97% BCLC C Treatment: 100% sorafenib |
Higher amount of cfDNA associated with: Shorter time to progression: HR, 1.17 (1.20–2.44), P = .002, adjusted for AFP Shorter overall survival: HR, 3.50 (2.36–5.20), P < .0001, adjusted for macroscopic vascular invasion and AFP Genomic instability associated with: Shorter time to progression: HR, 2.09 (1.46–3.00), P < .0001, adjusted for AFP Shorter overall survival: HR, 3.35 (2.24–5.01), P < .0001), adjusted for macroscopic vascular invasion and AFP |
| Ono et al,125 2015 | Total amount | N = 46 Stage: 24%/39%/33%/4% T1/T2/T3/T4, all N0/M0 Treatment: 100% surgical (resection or liver transplant) |
Presence of cfDNA associated with: Increased recurrence (P = .01), unadjusted Increased extrahepatic metastasis (P = .04), unadjusted Similar overall survival (P = .07), unadjusted Increased microscopic vascular invasion: HR, 6.10 (1.11–33.33), adjusted for AFP, DCP, tumor size Among patients with serial cfDNA measurements, cfDNA levels correlated with clinical course (eg, decreased after therapy and increased with recurrence) |
| Park et al,126 2018 | Total amount | N = 55 Stage: 23%/23%/27%/27% TNM I/II/III/IV Treatment: 100% radiotherapy |
cfDNA levels decreased after successful but not unsuccessful therapy Higher post-therapy cfDNA was associated with: Similar overall survival (P = .15) Similar progression-free survival (P = .26) Increased intrahepatic failure: HR, 2.41 (1.06–5.46), adjusted for cirrhosis, multifocal tumors, TNM stage Decreased local control: HR, 1.96 (0.57–6.81), adjusted for cirrhosis, multifocal tumors, TNM stage |
| Tangkijvanich et al,86 2007 | LINE-1 hypomethylation | N = 85 Stage: 82% >5 cm, 36% multiple tumors Treatment: NR |
LINE-1 hypomethylation associated with poorer overall survival: HR, 1.74 (1.09–2.79) Adjusted for age, sex, AFP, hepatitis B virus etiology, Child–Pugh class, tumor size, tumor number, venous invasion, extrahepatic metastasis, CLIP score, and HCC therapy |
| Tokuhisa et al,127 2007 | Total amount | N = 87 Stage: 46%/44%/10% TNM I/II/III Treatment: 100% resection |
High cfDNA associated with: Poorer overall survival: HR, 3.4 (1.5–7.6), adjusted for tumor size Increased distant metastasis: HR, 4.5 (1.3–14.9), adjusted for tumor grade Similar disease-free survival (P = .19) |
| Xu et al,92 2017 | Methylation of 8 genes: SH3PXD2A, C11orf9, PPFIA1, chromosome 17:78, SERPINB5, NOTCH3, GRHL2, and TMEM8B | N = 1049 Stage: 16/16/52/12 TNM I/II/III/IV Treatment: NR |
Overall survival based on high risk score: Training set (n = 680): HR, 2.41 (1.90–3.03) Validation set (n = 369): HR, 1.55 (1.25–1.92) Adjusted for AFP, TNM stage, sex, age |
AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer; cfDNA, cell-free DNA; CLIP, Cancer of the Liver Italian Program; ctDNA, circulating tumor DNA; DCP, des-carboxyprothrombin; HR, hazard ratio; NR, not reported; TACE, transarterial chemoembolization; UICC, Union for International Cancer Control.
Properties of cfDNA were associated with overall survival in multivariable analysis in most studies,37,58,73,77,86,92,121–123,127 although 3 studies did not confirm such associations.63,124,125
Extracellular vesicles.
Ten studies on EVs met the inclusion criteria (Supplementary Table 6).95,128–136 Most studies focused on exosomal microRNAs,128–135 although 2 studies also investigated long noncoding RNAs130,136 and another study evaluated RAB11A messenger RNA.95 Six of these studies focused on patients with early stage disease and/or undergoing surgical resection.95,131,132,134–136 The only microRNA included in more than 1 study was miR-21, and all 3 studies found an association with increased disease progression and poorer disease-free survival.130,133,135
Quality assessment.
Results of the QUality In Prognosis Studies tool to assess the quality of prognostic studies are shown in Supplementary Tables 7, 8, and 9. Although measured outcomes consistently were objective (ie, based on survival or imaging), there was concern for risk of bias owing to lack of prespecified cut-off values and nonconsecutive patient selection.
Monitoring on Therapy
Thirteen studies used CTCs to monitor the response to therapy,44–46,52,108,109,113,119,120,137–140 12 evaluated cfDNA,58,66,125,126,141–148 and none evaluated EVs.
Most studies on CTCs evaluated changes in CTC concentrations before vs after therapy, most commonly resection or locoregional therapy. Typically, patients who persistently had increased CTC counts after therapy or who initially had low/undetectable CTC counts who then increased posttherapy had a poorer prognosis than patients with persistently negative CTC or whose CTC concentrations decreased after therapy.44,45,52,108,109,113,119,120,139,140 One study showed longitudinal measurements of a CTC-derived PCR score detected recurrence more accurately than AFP.46 Another study found no association between preresection CTC count and overall or progression-free survival, but an increase in CTC count after resection was associated with poorer overall survival and progression-free survival.140
Few studies examined the effect of post-treatment cfDNA properties on prognosis, and those that did failed to show an association between post-treatment cfDNA and progression-free survival or overall survival.126,141 Several studies evaluated dynamic changes in cfDNA after therapy58,143,144,146,148 and found that cfDNA tended to increase at or before the time of disease recurrence/progression more reliably than did AFP. Indeed, 2 studies reported changes in cfDNA mutational profiles preceded radiographic documentation of tumor relapse/progression by up to 8 weeks.143,146 It should be noted that all of the studies evaluating cfDNA for monitoring of tumor response were very small (N < 20), and results of some are available only in abstract form.
Discussion
In this systematic review, we identified 112 studies investigating CTCs, EVs, or cfDNA as biomarkers for detection, prognostication, and monitoring for treatment response in HCC. We found that liquid biopsy has value in early HCC detection and prognosis beyond existing diagnostic tests and tumor staging, respectively. In addition, liquid biopsy may be useful for monitoring response to therapy. Overall, CTCs, EVs, and cfDNA are appealing targets of biomarker development.
For detection, cfDNA methylation scores appear to have the most favorable test characteristics of all liquid biopsy tools included in this systematic review. In particular, their sensitivity for early stage HCC compares favorably with that of AFP and ultrasound, with large studies on early detection in more than 2000 patients showing more than 75% sensitivity and more than 90% specificity for TNM stage I or BCLC stage 0 disease in phase 2 biomarker studies (case-–control comparisons).74,92,149 In contrast, the presence of CTCs has high specificity (>90%) but generally only modest sensitivity (~60%), whereas EVs have highly variable diagnostic accuracy. We note that CTC and EV gene expression have shown greater promise for early HCC detection46,97,98; however, these findings require external validation. In contrast, cfDNA mutations generally have low sensitivity for early stage HCC and CTC mutational profiles have not been well studied in HCC. Validation of cfDNA methylation scores in more robust phase 2 studies in which early stage HCC is compared with appropriate controls and is shown to perform well in diverse etiologies of liver disease are required before larger phase 3 and 4 studies are appropriate.149
As prognostic/predictive biomarkers, CTCs have been the most extensively evaluated liquid biopsy type with numerous studies consistently showing their utility in predicting recurrence after curative therapy beyond that of conventional metrics such as cancer stage. However, whether the prognostic value of CTCs also applies to patients receiving local ablative therapies or arterial-based locoregional therapies is unclear, and the value of CTCs in predicting outcomes in patients receiving noncurative therapy is even less certain. The literature on cfDNA (especially methylation scores) and EVs and prognosis is comparatively limited and less consistent in terms of which outcomes and populations have been studied. That does not necessarily imply that they are less prognostically important however: for example, circulating tumor DNA has higher sensitivity than CTCs at detecting minimal residual disease in several different tumor types.150 How these differences in sensitivity may translate into utility as predictive markers in HCC is not known.
Finally, we note the potential utility of a liquid biopsy for monitoring treatment response. Several small studies have shown that CTCs or cfDNA may better predict tumor recurrence or progression than AFP and changes in CTCs or cfDNA may precede changes on imaging. We caution that the studies reporting these findings are small and many are available only in abstract form. In addition, the question arises regarding how to manage abnormal liquid biopsy findings in the absence of imaging evidence of progression/recurrence. A trial in patients with metastatic breast cancer receiving first-line chemotherapy found no benefit to changing therapy based on concerning CTC trends.151 Larger studies will be required to confirm the utility of liquid biopsy in monitoring tumor progression/recurrence and whether it can be used to guide clinical management of HCC.
Although liquid biopsy is a promising tool, we identified several limitations in the literature. First, many studies did not include appropriate study populations. For early detection, the relevant comparison is between early stage HCC and patients at risk of HCC (ie, those with cirrhosis or selected patients with chronic hepatitis B). However, many diagnostic studies included CLD patients without cirrhosis or advanced fibrosis, or even healthy controls for comparisons. Furthermore, results for early stage HCC were not always reported separately. Finally, most studies have not separately examined the accuracy of liquid biopsy for detection of HCC owing to viral vs nonviral etiologies.
Second, prognostic studies were highly inconsistent in which outcomes were reported, which raises concern for selective reporting and limits meta-analyses. There is a risk of bias based on nonconsecutive patient selection and ad hoc or post hoc cut-off values, for example, in analysis of cfDNA methylation levels or CTC concentration. These limitations can be avoided in future studies by using prespecified cut-off values and outcome reporting. We propose minimal reporting parameters of overall survival and (depending on stage and treatment type) either progression-free survival or tumor recurrence for future studies on liquid biopsy for prognostication.
A third, more fundamental limitation is that there has been wide variation in how liquid biopsy is defined. CTCs are the best-standardized of the 3 types of liquid biopsy we analyzed in this study and there is already a Food and Drug Administration–cleared CTC detection method for use in prostate, breast, and colorectal cancers.18 However, there are a number of alternate methods of CTC detection, some of which focus on circulating cells with epithelial phenotypes while others focus on cell size or morphology rather than cell surface markers, which make direct comparisons across methodologies difficult.17 Similarly, multiple methylation scores have been developed from cfDNA, each of which includes different loci.89,92 EV studies largely have focused on microRNAs, and, again, there is little consistency in which microRNAs are included in analyses. Analysis of human CTCs, EVs, and cfDNA is an evolving field, and exploratory research is critical to developing a fundamental understanding of these entities. Establishment of optimal methodologies and standards in liquid biopsy technologies is required before robust validation studies and clinical utilization.
In conclusion, liquid biopsy has the potential as a biomarker for HCC detection, prognostication, and monitoring.152 Further^research using standardized definition and methods of detection, quantification, and characterizing of CTCs, cfDNA, and EVs; as well as standardized study design, patient selection, and outcome reporting; followed by validation of promising biomarkers will be required before liquid biopsy can be recommended in the detection and clinical management of HCC.
Supplementary Material
Table 2.
Studies on Use of Cell-Free DNA for Hepatocellular Carcinoma Detection
| Study | cfDNA property | HCC patients | Control | Comparator, AFP cut-off value, ng/mL | Findings: sensitivity/specificity, AUC |
|---|---|---|---|---|---|
| Amount or integrity | |||||
| Chen et al,60 2013 | Total amount | N = 39 Stage: multifocal disease in 13% Treatment: NR |
45 healthy | NS | Sensitivity: cfDNA: 56.4% AFP: 53.8% cfDNA + AFP: 71.8% (P < .05 for 3-way comparison) cfDNA + AFP + α-L-fucosidase vs cfDNA + AFP: 89.7% (P < .05) |
| El-Shazly et al,63 2009 | Total amount, integrity | N = 25 Stage: 12%/32%/48%/8% TNM I/II/III/IV Treatment: NR |
25 CLD, 15 healthy | 20 | HCC vs CLD comparison: Total DNA amount: 72%/68%, AUC, 0.57 DNA integrity: 88%/92%, AUC, 0.75 |
| Huang et al,68 2012 | Total amount | N = 72 Stage: 24%/76% TNM e I+II/III+IV Treatment: NR |
37 CLD, 41 healthy | 400 | Total cfDNA amount: HCC vs CLD: 60%/78%, AUC, 0.71 HCC vs healthy: 90%/90%, AUC, 0.95 Total DNA + AFP (HCC vs healthy): 95%/94%, AUC, 0.97 |
| Huang et al,66 2016 | cfDNA integrity | N = 53 Stage: 24%/76% TNM I+M/III+IV Treatment: 100% resection |
15 CLD, 22 healthy | 20 | cfDNA integrity: 43.4%/100%/0.71 AFP: 50.9%/100%/0.61 cfDNA integrity + AFP: 79.2%/100%/0.85 |
| Iizuka et al,71 2006 | Total amount | N = 52 Stage: 46%/37%/17% TNM I/II/III Treatment: 100% resection |
30 CLD, 16 healthy | AFP: 10.2 DCP: 29.5 ng/mL |
AFP: 69.2%/72.7% DCP: 73.1%/75.0% cfDNA amount: 69%/93% (P < .05 vs both AFP and DCP) |
| Marchio et al,80 2018 | Total amount, TP53 R249S mutation by digital droplet PCR | N = 149 Stage/treatment: NR | 164 CLD, 49 healthy | 10 | AUC: Proportion of droplets with TP53 R249S: 0.83 AFP: 0.81 (P > .05 vs TP53 R249S droplets) cfDNA amount: 0.59 |
| Piciocchi et al,82 2013 | Total amount | N = 66 Stage: 59% within Milan criteria Treatment: NR |
76 CLD | 14 | cfDNA: 91%/43%, AUC, 0.69 AFP: 45%/83%, AUC, 0.64 (P > .05) |
| Ren et al,84 2006 | Total amount, chromosome 8p allelic imbalance (D8S258 or D8S264) | N = 79 Stage: 62%/38% TNM I+II/III+IV Treatment: NR |
20 CLD, 20 healthy | 20 | Total amount: HCC vs healthy, 52%/95%, AUC, 0.80 Allelic imbalance at D8S258: sensitivity, 57% for TNM stage III/IV and 22% for stage I/II High cfDNA concentration + allelic imbalance: abnormal in 8 of 24 with low AFP |
| Yan et al,93 2018 | Total amount | N = 24 Stage: 58%/33%/8% BCLC A/B/C Treatment: NR |
62 CLD | 80.5 | cfDNA amount: 62.5%/93.6%, AUC, 0.82 AFP alone: AUC, 0.67 cfDNA amount + AFP + age: 87%/100%, AUC, 0.98 (P < .05) |
| Mutations | |||||
| An et al,56 2019 | cfDNA mutations | N = 26 Stage: 77%/23% TNM I/II+III Treatment: 100% resection |
20 CLD | NS | AUC of different tests: cfDNA concentration: 0.917 Mutation number: 0.878 ctDNA concentration (cfDNA concentration times variant allele frequency): 0.871 Maximal variant allele frequency: 0.802 AFP: 0.783 |
| Cai et al,57 2019 | Fraction of single-nucleotide or copy number variants | N = 34 Stage: NR Treatment: 100% resection |
N/A | NR | ctDNA: sensitivity, 100% AFP: sensitivity, 56% AFP-L3%: sensitivity, 50% DCP: sensitivity, 82% |
| Igetei et al,69 2008 | TP53 R249S mutation | N = 85 Stage/treatment: NR |
77 healthy | 400 | cfDNA TP53 R249S mutation: Overall: 7.6%/100% Patients with HCC and AFP measurements: 16.7% overall, 20% without increased AFP (P > .05) |
| Liao et al,78 2016 | TERT, CTNNB1, or TP53 mutation | N = 41 Stage: 42% ≥5 cm, 27% multiple tumors, 61% vascular invasion Treatment: 100% resection |
10 healthy | 20 | cfDNA mutations: sensitivity, 23% vs 13% in high vs low AFP (P = .70) Specificity, 90% |
| Qu et al,83 2019 | Integrated hepatitis B virus DNA and mutations in TP53, CTNNB1, AXIN1, and TERT promoter Age, sex, AFP, and DCP also included |
Training: N = 65 Validation: N = 24 |
Training: 70 CLD Validation: 307 CLD |
None | Training cohort: patients with (+) AFP or ultrasound 93%/93%, AUC, 0.93 Both the cfDNA and protein markers contributed predictive power Validation cohort: 331 patients with (−) AFP and ultrasound screening sensitivity, 100%, specificity only 4 of 24 |
| Xiong et al,90 2019 | Mutations in TP53, ARID1A, FLCN, SETD2, PTEN, BUB1B, CTNNB1, JAK1, AXIN1, EPS15, or CACNA2D4 | N = 37 Stage: NR Treatment: 100% resection |
6 healthy | 400 | cfDNA mutations: Overall: 65%/100% Low AFP: 73%/100% High AFP: 53%/100% |
| Xu et al,91 2015 | Copy number variation of 1q, 7q, or 19q forward strand or 1p, 9q, or 14q reverse strand | N = 31 Stage: mean maximum tumor size, 5.7 cm (SD, 3.1 cm) Treatment: 100% resection |
8 CLD | 10 | Copy number variation score: All HCC: 83.9%/100% HCC <5 cm maximum tumor size: 68.8%/100% HCC <3 cm maximum tumor size: 57.1%/100% Low AFP: sensitivity, 7 of 10 |
| Methylation/epigenetics | |||||
| Cai et al,57 2019 | 5hmC modifications in cfDNA | N = 1204 Stage: 12%/36%/25%/13% BCLC 0/A/B/C Treatment: NR |
392 CLD, 958 healthy | 20 | Early stage HCC vs CLD: 5hmC-based score: 82.7%/67.4%, AUC, 0.87 in training set, 0.85 in validation set AFP: 44.8%/76.1%, AUC, 0.79 in training set, 0.69 in validation set |
| Chan et al,59 2008 | RASSF1A methylation | N = 63 Stage: NS Treatment: 100% resection |
63 CLD, 50 healthy | 20 |
RASSF1A methylation detected in: 93% HCC (50% among normal AFP) 58% CLD 8% healthy controls |
| Chu et al,61 2004 | P16 methylation | N = 46 Stage/treatment: NR |
23 CLD | 20 |
P16 methylation: Overall cohort: 48%/83% Normal AFP HCC: sensitivity, 50% |
| Dou et al,62 2016 | CDH1, DNMT3b, or ESR1 promoter methylation | N = 183 Stage/treatment: NR |
173 CLD, 50 healthy | NS | Methylation frequency: HCC: CDH1 31%, DNMT3b 41%, ESR1 30% CLD: <10% for all 3 genes Healthy controls: 0% AUC for methylation of any gene, 0.75 (0.70–0.80) AUC for AFP, 0.62 (0.55–0.68) |
| Han et al,64 2014 | TGR5 promoter methylation | N = 160 TNM stage: 59%/41% I+II/III+IV Treatment: NR |
88 CLD, 45 healthy | 200 | HCC vs CLD: TGR5 methylation + AFP: 68.1%/78.4% AFP alone: 30.6%/92.1% TGR5 alone: 48.1%/86.3% |
| Hu et al,65 2017 | UBE2Q1 methylation | N = 80 TNM stage: 43%/57% I+II/III+IV Treatment: NR |
80 CLD, 20 healthy | 200 |
UBE2Q1 methylation + AFP: 53.8%/87.5%, AUC, 0.76 UBE2Q1 methylation alone: 66%/58%, AUC, 0.62 AFP alone: 53.8%/87.5%, AUC, 0.67 |
| Huang et al,67 2014 | INK4A methylation | N = 66 Stage: 24%/23%/27%/26% TNM I/II/III/IV Treatment: NR |
43 CLD | 200 |
INK4A methylation + AFP: sensitivity, 80.3% (P < .05 vs AFP alone) AFP alone: sensitivity, 45.5% INK4A methylation alone: sensitivity, 74.2% |
| Iizuka et al,70 2011 | SPINT2 and SRD5A2 methylation | N = 220 Stage: 15%/34%/30%/21% TNM I/II/III/IV Treatment: NR |
202 CLD | AFP: 20 DCP: 40 mAU/mL |
Methylation of SPINT2 and SRD5A2, AFP, and DCP: 82%/82% AUC, 0.72 for ≤5 cm HCC and 0.89 for >5 cm HCC AFP alone: 57.4%/85.7% DCP alone: 60.2%/89.3% |
| Ji et al,72 2014 | MT1M and MT1G methylation | N = 121 Stage: 53%/47% TNM I+II/III+IV Treatment: 100% noncurative |
37 CLD, 31 healthy | 20 |
MT1M methylation: 49% HCC, 5% CLD, 7% healthy controls MT1G methylation: 70% HCC, 16% CLD, 13% healthy controls MT1M or MT1G methylation: HCC vs CLD: 90%/81%, AUC, 0.86 HCC vs healthy: 91%/84%, AUC, NR AFP alone: HCC vs healthy: 56.0%/62.1% |
| Kanekiyo et al,73 2015 | RASSF1A, CCND2, CFTR, SPINT2, SRD5A2, and/or BASP1 methylation | N = 125 Stage: 46%/54% TNM I+II/III+IV Treatment: 100% resection |
N/A | AFP: 20 DCP: 40 ng/mL |
Serum methylation score: Positive in 41% vs 48% of high vs low AFP Positive in 42% vs 46% of high vs low DCP (P > .05 for both) |
| Kisiel et al,74 2018 | Methylation score: HOXA1, EMX1, ECE1, AK055957, PFKP, CLEC11A | N = 116 Stage: 4%/44%/15%/30%/7% BCLC 0/A/B/C/D Treatment: NR |
80 CLD, 98 healthy | 10 | Methylation score: HCC vs cirrhosis: 95%/86%, AUC, 0.93–no improvement after adding AFP HCC vs healthy: 95%/95% Sensitivity based on cancer stage: 75% (BCLC stage 0), 93% (stage A/B), and 100% (stage C/D) |
| Kuo et al,75 2014 | HOXA9 methylation | N = 40 Stage/treatment: NR |
34 healthy | 10 |
HOXA9 methylation: 73%/97% HOXA9 methylation + AFP: 95%/97% |
| Li et al,76 2014 | IGFBP7 promoter methylation | N = 136 Stage: 51%/49% TNM I+II/III+IV Treatment: NR |
46 CLD, 35 healthy | 20 |
IGFBP7 promoter methylation + AFP: 85%/41% (P < .05 for both sensitivity and specificity vs AFP) IGFBP7 methylation alone: 65%/83% AFP alone: 57%/52% |
| Li et al,77 2018 | IGFBP7 promoter methylation | N = 155 Stage: 63%/37% TNM I+II/III+IV Treatment: 100% resection |
60 CLD, 20 healthy | 200 | Serum IGFBP7 promoter methylation: sensitivity, 69% vs 67% in high vs low AFP (P = .81) |
| Lu et al,79 2017 | Methylation score: APC, COX2, RASSF1A, miR-203 | N = 203 Stage: 89%/11% TNM I+II/III+IV Treatment: 100% resection |
104 CLD, 50 healthy | 20 | HCC vs controls: 84%/83%, AUC, 0.87 Sensitivity, 75% of patients with low AFP |
| Oussalah et al,81 2018 | SEPT9 methylation | Derivation cohort (France): N = 51, BCLC 25%/39%/31%/2% A/B/C/D Validation cohort (Germany): N = 47, BCLC stage 39%/22%/15%/24% A/B/C/D |
Derivation cohort: 135 CLD Validation cohort: 56 CLD |
NS |
SEPT9 methylation: 85%/91%, AUC, 0.96 AFP alone: AUC, 0.85 (P = .002 vs SEPT9 methylation) |
| Sun et al,85 2013 | TFPI2 methylation | N = 43 Stage: 40%/19%/33%/9% TNM I/II/III/IV Treatment: NR |
24 CLD, 26 healthy | 400 |
TFPI2 methylation or AFP alone: similar sensitivity (47% vs 54%) TFPI2 methylation + AFP: sensitivity, 61% (P < .05) |
| Tangkijvanich et al,86 2007 | LINE-1 hypomethylation | N = 85 Stage: 82% >5 cm, 36% multiple tumors Treatment: various |
93 CLD, 30 healthy | 400 | LINE-1 hypomethylation: sensitivity in high vs low AFP (61.7% vs 52.6%), P > .05 |
| Wang et al,87 2006 | GSTP1 methylation | N = 32 Stage: NR Treatment: 100% resection |
8 CLD | N/A | Methylated cfDNA GSTP: sensitivity, 50% (61% among patients with tissue GSTP1 methylation), specificity, 100% Methylation detected in 4 of 9 patients with low AFP |
| Wei et al,88 2018 | SOCS3 promoter methylation | N = 119 Stage: 34% tumor >5 cm, 17% >1 tumor, 17% portal vein involvement Treatment: 100% resection |
157 CLD, 50 healthy | 400 |
SOCS3 cfDNA methylation: Overall: 28.6%/95% High vs low AFP (52.9% vs 10.3%), P < .001 |
| Wen et al,89 2015 | Methylation score: RGS10, ST8SIA6, RUNX2, VIM, CACNA1C, TBX2, SOX9 (5’ end), NEDD4L (intron), ALX3, ZNF683 (3’ end), KCNQ4 (i), ERG, PTPN18 (intron), SYN2, LINC00682 (3’ end), CPLX1 (intron), FLJ42709, UBD (3’ end), SNX10 (3’ end), TRPS1 (intron) | N = 36 Stage: 36%/25%/22%/17% TNM I/II/III/IV Treatment: NR |
17 CLD, 38 healthy | 20 | Two cfDNA methylation scores, either score positive: Training set: 93%/91% Validation set: 100%/80% Combined cohort: 94%/89% Sensitivity, 100% in patients with low AFP (N = 10) |
| Xu et al,92 2017 | Methylation score: cg10428836, cg26668608, cg25754195, cg05205842, cg11606215, cg24067911, cg18196829, cg23211949, cg17213048, cg25459300 |
N = 1098 Stage: 16%/16%/52%/12% TNM I/II/III/IV Treatment: NR |
835 healthy | 25 | cfDNA levels: Training set: 85.7%/94.3%, AUC, 0.97 Validation set: 83.3%/90.5%, AUC, 0.94 AFP: AUC, 0.82 (P < .05 vs cfDNA) |
| Yeo et al,94 2005 | RASSF1A methylation | N = 40 Stage: 30% ≥5 cm Treatment: 100% resection |
10 healthy | 20 |
RASSF1A methylation Overall: 43%/100% Low AFP: sensitivity, 36% |
NOTE. Test characteristics are reported either as sensitivity (%)/specificity (%), area under the receiver operating characteristic curve; sensitivity (%)/specificity (%); or as individual parameters.
5hmC, 5-hydroxymethylcytosine; AFP, α-fetoprotein; AUC, area under the receiver operating characteristic curve; BCLC, Barcelona Clinic Liver Cancer; CLD, chronic liver disease; cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; DCP, des-carboxyprothrombin; HCC, hepatocellular carcinoma; NR, not reported; PCR, polymerase chain reaction.
Table 5.
Studies on Liquid Biopsy to Monitor Disease or Guide Treatment Selection
| Study | Liquid biopsy | HCC patients | Finding |
|---|---|---|---|
| Circulating tumor cells | |||
| Chen et al,137 2019 | CanPatrol | N = 113 Stage: 35%/65% BCLC 0+A/B+C Treatment: 100% resection |
No significant changes postresection in total CTC count or CTC subtype counts |
| Fu et al,108 2019 | CanPatrol | N = 25 Stage: NR Treatment: 100% liver transplant |
19 of 22 patients with pretransplant CTC had lower numbers of CTCs post-transplant CTC counts increased in 6 of 8 with post-transplant recurrence |
| Guo et al,45 2014 | PCR for EpCAM in CD45(−) cells | N = 77 Stage: NR Treatment: 45% resection, 55% TACE or radiotherapy |
Surgery: EpCAM expression Decreased postresection: 48.6% vs 17.1% positive preresection vs postresection Persistent expression or initially negative then positive expression higher recurrence: 50%-75% vs 7.7%-12.5% TACE/radiotherapy: EpCAM expression Decreased post-treatment: 50.0% vs 10.0% positive pretreatment vs post-treatment All patients with progressive disease had increased EpCAM expression post-treatment (P < .01) |
| Guo et al,44 2018 | PCR score: EpCAM, CD133, CD90, CK19 | N = 60 Stage: NR Treatment: 100% resection |
PCR scores decreased postresection: 70.0%-31.7% positive Persistently positive scores and scores that became positive postresection were associated with higher recurrence |
| Ha et al,109 2019 | Tapered slit filter | N = 105 Stage: 19%/81% BCLC stage 0/A Treatment: 100% resection |
Patients with AFP <200 ng/mL: increase in postoperative CTCs associated with: Poorer overall survival (P = .047) Poorer recurrence-free survival (P < .001) Similar findings in subgroup with HCC and cirrhosis |
| Kalinich et al,46 2017 | PCR score | N = 5 Stage: 80% BCLC C/D Treatment: NR |
PCR scores post-therapy correlated with recurrence |
| Lee et al,138 2017 | CellSearch | N = 10 Stage: NR Treatment: 100% SBRT |
Objective response at 3 months after SBRT correlated with percentage change in CTC (r = 0.703, P = .023), but not change in AFP |
| Qi et al,113 2018 | CanPatrol | N = 112 Stage: 39%/21%/30% BCLC stage 0+A/B/C Treatment: 100% resection |
8–10 days after resection, total CTC counts decreased whereas the proportion of mesenchymal-type CTCs increased 8 patients with recurrence had increased CTC count 2 months post-resection Detected earlier than the recurrence |
| Wang et al,139 2018 | CanPatrol | N = 47 Stage: 30%/70% T1+2/T3+4 Treatment: 100% liver transplant |
CTCs measured either pretransplant and 1 month post-transplant (N = 20) or 1 and 3 months post-transplant (N = 27) Trends in number of all CTCs or specific CTC subgroups was not associated with recurrence post-transplant in either group |
| Wu et al,119 2019 | CD45(−) and abnormal chromosome 8 amplification by FISH | N = 155 Stage: 38%/14%/48% BCLC stage A/B/C, 34%/35%/28%/4% TNM stage I/II/III/IV Treatment: 100% TACE |
CTCs measured before TACE and 1 and 4 weeks after TACE Responders to TACE: numbers of CTCs decreased 1 and 4 weeks post-TACE Nonresponders to TACE: CTC counts higher at week 4 than pre-TACE; pre-TACE CTC counts also were higher in nonresponders |
| Xue et al,52 2018 | Two methods: CellSearch and either CD45(−) CK(+) DAPI(+) hybridization signal for CEP8 ≥2 or CD45(−) CK(−) DAPI(+) and hybridization signal for CEP8 >2 | N = 23 (subset of larger study) Stage: NR Treatment: 100% liver transplant |
iFISH CTC counts were measured 3 months post-transplant Compared with pretransplant levels, decreased in 65.2%, did not change in 21.7%, and increased in 13.0% of patients 1 patient with a marked increase in CTC count from 2/7.5 to 12/7.5 mL had extrahepatic recurrence 4 months later Outcomes in the remaining patients were not reported |
| Ye et al,140 2018 | CanPatrol | N = 42 Stage: 88%/12% TNM stage I+II/III+IV, 81%/19% BCLC stage A+B/C+D Treatment: 100% resection |
Preoperative CTC counts: not associated with overall survival or progression-free survival Postoperative CTC count >5: Poorer progression-free survival: HR, 6.89 (1.64–29.00) No association with overall survival: HR, 15.65 (0.80–304.64) Increase in CTC counts postoperatively: Poorer progression-free survival: HR, 39.58 (4.22–371.64) All HRs were adjusted for tumor stage, number of tumors, tumor size, age, and sex |
| Yu et al,120 2018 | CellSearch | N = 139 Stage: 40%/60% BCLC stage 0+A/B+C Treatment: 100% resection |
CTC counts measured 1 day before and 3 days after surgery CTC counts increased postoperatively in 42%, were unchanged in 33%, and decreased in 33% Patients with persistently increased CTC counts had the poorest overall survival and disease-free survival |
| Cell-free DNA | |||
| Alunni-Fabroni et al,141 2019 | cfDNA amount | N = 13 Stage: 23%/31%/46% BCLC stage A/B/C Treatment: TARE plus sorafenib (77%), RFA + sorafenib (15%), RFA alone (8%) |
cfDNA was measured after locoregional therapy, and every 8 weeks thereafter cfDNA levels at 16 and 24 weeks postintervention trended toward an association with poorer overall survival (P = .057 and .095, respectively) 3 patients with reported data: cfDNA concentration trends corresponded to disease recurrence and provided information beyond AFP trends alone |
| Cai et al,57 2019 | Fraction of single-nucleotide or copy number variants | N = 34 Stage: NR Treatment: 100% resection |
Four patients with data reported: all had initial reduction in SNV/CNV fraction after initial surgery Three patients with recurrence: cfDNA amount and SNV/CNV fraction dynamics changed dynamically with recurrent cancer |
| Chen et al,142 2018 | cfDNA sequencing | N = 11 Stage: NR Treatment: 100% resection |
1 patient: CHD4 A1789T and NCOA2 A2777T allele frequency corresponded to tumor volume Another patient, FOXA1 T254G and ABL1 T642G allele frequency corresponded to tumor burden |
| Du et al,143 2018 | Mutations in cfDNA NRAS, BRAF, PIK3CA, KRAS, ARID1A, AXIN1, ARID2, TERT, TP53, and CTNNB1 | N = 3 Stage: advanced stage Treatment: TACE |
Mutational burden identified relapse weeks before changes in CT or AFP |
| Evans et al,144 2019 | cfDNA amount | N = 18 Stage: NR Treatment: TACE |
All patients, cfDNA levels decreased after TACE and then increased at time of progression True in both AFP-positive and AFP-negative tumors |
| Higuera et al,145 2019 | cfDNA mutations | N = 12 Stage/treatment: NR |
Two patients undergoing resection, follow-up samples >20 months later showed no mutations One patient, a TERT mutation with 8% frequency was found 10 months pre-HCC diagnosis |
| Huang et al,66 2016 | DNA integrity | N = 13 (subset of larger study) Stage: 24%/76% TNM stage I+II/III+IV Treatment: 100% resection |
cfDNA integrity increased postresection in 11 of 13 patients with longitudinal measurements (P = .0003) NR whether this correlated to long-term outcomes |
| Li et al,146 2019 | cfDNA mutations | N = 3 Stage: 33%/67% BCLC A/C Treatment: TACE |
All 3 patients had week 4 CT with treatment response while week 10 CT showed disease recurrence Two patients: cfDNA mutational burden remained stable at week 1 but increased at week 4 after TACE; AFP continued to decrease during this time Third patient: mutational burden was lower at week 1 then increased at week 4; AFP increased at both weeks 1 and 4 |
| Ono et al,125 2015 | Total amount | N = 46 Stage: 24%/39%/33%/4% T1/T2/T3/T4, all N0/M0 Treatment: 100% surgical (resection vs liver transplant) |
Among patients with serial cfDNA measurements, cfDNA levels correlated with clinical course (eg, decreased after therapy and increased with recurrence) |
| Park et al,126 2018 | Total amount | N = 55 Stage: 23%/23%/27%/27% TNM stage I/II/III/IV Treatment: radiotherapy |
cfDNA levels decreased after successful but not unsuccessful radiotherapy |
| Wong et al,147 2003 | p16INK4a methylation | N = 10 Stage: NR Treatment: 100% resection |
Plasma p16INK4a median methylation decreased in 5 of 8 patients after resection (P = .07) Buffy coat p16INK4a median methylation decreased in 10 of 10 patients after resection (P = .01) |
| Yang et al,148 2019 | cfDNA mutations | N = 12 Stage: NR Treatment: 100% sorafenib |
Dynamic changes in cfDNA mutations in 19 genes were associated with therapeutic efficacy |
AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer; CD, cluster of differentiation; cfDNA, cell-free DNA; CK, cytokeratin; CNV,____; CT, computed tomography; CTC, circulating tumor cell; EpCAM, epithelial cell adhesion molecule; FISH, fluorescence in situ hybridization; HR, hazard ratio; iFISH,____; NR, not reported; PCR, polymerase chain reaction; RFA,____; SBRT,____; SNV,____; TACE, transarterial chemo-embolization. TARE, transarterial radioembolization.
What You Need to Know.
Background
Liquid biopsies, or blood samples, can be analyzed to detect circulating tumor cells, cell-free DNA, and extracellular vesicles, which might identify patients with hepatocellular carcinoma (HCC) or help determine their prognoses.
Findings
In a systematic review of 112 studies of the accuracy of liquid biopsy analysis, we found that assays for circulating tumor cells and cell-free DNA might aid in the detection or monitoring of HCC, or in determining patient prognoses. However, there was a risk of bias in these studies.
Implications for patient care
Studies of liquid biopsy analysis must be standardized before we can assess its utility in the detection and management of patients with HCC.
Acknowledgments
Funding
Supported in part by a University of Michigan Training T32 grant in Basic and Translational Digestive Sciences (National Institute of Diabetes and Digestive and Kidney Diseases 5T32DK094775) (V.L.C. and D.X.), and by National Cancer Institute grants 5U01CA230669 (A.S.L.), 5U01CA230669 (N.D.P.), and 5U01CA230694 (N.D.P.).
Abbreviations used in this paper:
- AFP
α-fetoprotein
- AUC
area under the receiver operating characteristic curve
- BCLC
Barcelona Clinic Liver Cancer
- cfDNA
cell-free DNA
- CLD
chronic liver disease
- CTC
circulating tumor cell
- EV
extracellular vesicle
- HCC
hepatocellular carcinoma
- miR
microRNA
- PCR
polymerase chain reaction
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Clinical Gastroenterology and Hepatology at www.cghjournal.org, and at https://doi.org/10.1016/j.cgh.2020.04.019.
Conflicts of interest
These authors disclose the following: Max Wicha is the founder of OncoMed; Anna Lok serves on the advisory board for Epigenomics; and Neehar Parikh is a consultant for Bristol-Myers Squibb, Exelixis, and Freenome, serves on the advisory boards of Eisai, Bayer, Exelixis, and Wako Diagnostics, and has received research grants from Bayer, Target Pharmasolutions, and Exact Sciences. The remaining authors disclose no conflicts.
References
- 1.Bertuccio P, Turati F, Carioli G, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol 2017; 67:302–309. [DOI] [PubMed] [Google Scholar]
- 2.Njei B, Rotman Y, Ditah I, et al. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology 2015; 61:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanwal F, Kramer JR, Duan Z, et al. Trends in the burden of nonalcoholic fatty liver disease in a United States cohort of veterans. Clin Gastroenterol Hepatol 2016;14:301–308 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapper EB, Parikh ND. Mortality due to cirrhosis and liver cancer in the United States, 1999-2016: observational study. BMJ 2018;362:k2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tandon P, Garcia-Tsao G. Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int 2009;29:502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology 2018;154:1706–1718.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lok AS, Sterling RK, Everhart JE, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 2010; 138:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berhane S, Toyoda H, Tada T, et al. Role of the GALAD and BALAD-2 serologic models in diagnosis of hepatocellular carcinoma and prediction of survival in patients. Clin Gastroenterol Hepatol 2016;14:875–886 e6. [DOI] [PubMed] [Google Scholar]
- 9.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–390. [DOI] [PubMed] [Google Scholar]
- 10.Cheng A-L, Kang Y-K, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol 2009;10:25–34. [DOI] [PubMed] [Google Scholar]
- 11.Tapper EB, Lok AS. Use of liver imaging and biopsy in clinical practice. N Engl J Med 2017;377:756–768. [DOI] [PubMed] [Google Scholar]
- 12.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology 2005;42:1208–1236. [DOI] [PubMed] [Google Scholar]
- 13.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008;359:1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderaro J, Couchy G, Imbeaud S, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017;67:727–738. [DOI] [PubMed] [Google Scholar]
- 16.Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavroudis D Circulating cancer cells. Ann Oncol 2010;21(Suppl 7):vii95–vii100. [DOI] [PubMed] [Google Scholar]
- 18.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897–6904. [DOI] [PubMed] [Google Scholar]
- 19.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;2004:781–791. [DOI] [PubMed] [Google Scholar]
- 20.Lohr JG, Kim S, Gould J, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med 2016;8:363ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011; 11:426–437. [DOI] [PubMed] [Google Scholar]
- 23.Wyatt AW, Azad AA, Volik SV, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2016;2:1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer 2010;116:1918–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang T, Zhai C, Su C, et al. The diagnostic value of circulating cell free DNA quantification in non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer 2016; 100:63–70. [DOI] [PubMed] [Google Scholar]
- 26.Andaloussi SEL, Mager I, Breakefield XO, et al. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347–357. [DOI] [PubMed] [Google Scholar]
- 27.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawaz M, Camussi G, Valadi H, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol 2014;11:688–701. [DOI] [PubMed] [Google Scholar]
- 29.Pantel K Blood-based analysis of circulating cell-free DNA and tumor cells for early cancer detection. PLoS Med 2016;13: e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin T, Peng H, Wu H. Clinical value of circulating liver cancer cells for the diagnosis of hepatocellular carcinoma: a metaanalysis. Biomed Rep 2013;1:731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan JL, Yang YF, Yuan CH, et al. Circulating tumor cells for predicting the prognostic of patients with hepatocellular carcinoma: a meta analysis. Cell Physiol Biochem 2015; 37:629–640. [DOI] [PubMed] [Google Scholar]
- 32.Liao W, Mao Y, Ge P, et al. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 2015;94:e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–536. [DOI] [PubMed] [Google Scholar]
- 34.Hayden JA, van der Windt DA, Cartwright JL, et al. Assessing bias in studies of prognostic factors. Ann Intern Med 2013; 158:280–286. [DOI] [PubMed] [Google Scholar]
- 35.Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ 2015;350:g7647. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Shi L, Zhang X, et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget 2016; 7:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh CR, Kong SY, Im HS, et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer 2019;19:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang X, Zhang J, et al. Microfluidic chip for isolation of viable circulating tumor cells of hepatocellular carcinoma for their culture and drug sensitivity assay. Cancer Biol Ther 2016; 17:1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahnassy AA, Zekri AR, El-Bastawisy A, et al. Circulating tumor and cancer stem cells in hepatitis C virus-associated liver disease. World J Gastroenterol 2014;20:18240–18248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhan I, Mosesso K, Goyal L, et al. Detection and analysis of circulating epithelial cells in liquid biopsies from patients with liver disease. Gastroenterology 2018;155:2016–2018.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y, Luo L, Zhang J, et al. Diagnostic value of different phenotype circulating tumor cells in hepatocellular carcinoma. J Gastrointest Surg 2019;23:2354–2361. [DOI] [PubMed] [Google Scholar]
- 42.Fang ZT, Zhang W, Wang GZ, et al. Circulating tumor cells in the central and peripheral venous compartment - assessing hematogenous dissemination after transarterial chemoembolization of hepatocellular carcinoma. Onco Targets Ther 2014; 7:1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J, Yao F, Lou Y, et al. Detecting carcinoma cells in peripheral blood of patients with hepatocellular carcinoma by immunomagnetic beads and RT-PCR. J Clin Gastroenterol 2007;41:783–788. [DOI] [PubMed] [Google Scholar]
- 44.Guo W, Sun YF, Shen MN, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res 2018;24:2203–2213. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, Yang XR, Sun YF, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res 2014;20:4794–4805. [DOI] [PubMed] [Google Scholar]
- 46.Kalinich M, Bhan I, Kwan TT, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2017; 114:1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley RK, Magbanua MJM, Butler TM, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer 2015;15:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, Li N, Yu X, et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology 2013;144:1031–1041.e10. [DOI] [PubMed] [Google Scholar]
- 49.Sun YF, Xu Y, Yang XR, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013;57:1458–1468. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K, Ofuji K, Nosaka T, et al. Development of a novel circulating tumor cells isolation system in patients with hepatocellular carcinoma using a microcavity array. Hepatology 2016;64:632A–633A.26773713 [Google Scholar]
- 51.Xu W, Cao L, Chen L, et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin Cancer Res 2011;17:3783–3793. [DOI] [PubMed] [Google Scholar]
- 52.Xue F, Shi S, Zhang Z, et al. Application of a novel liquid biopsy in patients with hepatocellular carcinoma undergoing liver transplantation. Oncol Lett 2018;15:5481–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao F, Guo JM, Xu CF, et al. Detecting AFP mRNA in peripheral blood of the patients with hepatocellular carcinoma, liver cirrhosis and hepatitis. Clin Chim Acta 2005;361:119–127. [DOI] [PubMed] [Google Scholar]
- 54.Yin LC, Luo ZC, Gao YX, et al. Twist expression in circulating hepatocellular carcinoma cells predicts metastasis and prognoses. Biomed Res Int 2018;2018:3789613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Wang B, Wu J, et al. Association of preoperative EpCAM circulating tumor cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer 2016;16:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An Y, Guan Y, Xu Y, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res 2019;11:6462–6474. [PMC free article] [PubMed] [Google Scholar]
- 57.Cai J, Chen L, Zhang Z, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019;68:2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Z, Chen G, Zeng Y, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res 2019;25:5284–5294. [DOI] [PubMed] [Google Scholar]
- 59.Chan KC, Lai PB, Mok TS, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem 2008;54:1528–1536. [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Zhang H, Zhang LN, et al. Value of circulating cell-free DNA in diagnosis of hepatocellular carcinoma. World J Gastroenterol 2013;19:3143–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chu HJ, Heo J, Seo SB, et al. Detection of aberrant p16INK4A methylation in sera of patients with liver cirrhosis and hepatocellular carcinoma. J Korean Med Sci 2004;19:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dou CY, Fan YC, Cao CJ, et al. Sera DNA methylation of CDH1, DNMT3b and ESR1 promoters as biomarker for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma. Dig Dis Sci 2016;61:1130–1138. [DOI] [PubMed] [Google Scholar]
- 63.El-Shazly SF, Eid MA, El-Sourogy HA, et al. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis C virus-related hepatocellular carcinoma. Int J Biol Markers 2010;25:79–86. [DOI] [PubMed] [Google Scholar]
- 64.Han LY, Fan YC, Mu NN, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor gpbar1 (TGR5) is a potential biomarker for hepatitis B virus associated hepatocellular carcinoma. Int J Med Sci 2014;11:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu N, Fan XP, Fan YC, et al. Hypomethylated ubiquitin-conjugating enzyme 2 Q1 (UBE2Q1) gene promoter in the serum is a promising biomarker for hepatitis B virus-associated hepatocellular carcinoma. Tohoku J Exp Med 2017;242:93–100. [DOI] [PubMed] [Google Scholar]
- 66.Huang A, Zhang X, Zhou SL, et al. Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J Cancer 2016;7:1798–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang G, Krocker JD, Kirk JL, et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med 2014;52:899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z, Hua D, Hu Y, et al. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol Oncol Res 2012;18:271–276. [DOI] [PubMed] [Google Scholar]
- 69.Igetei R, Otegbayo JA, Ndububa DA, et al. Detection of p53 codon 249 mutation in Nigerian patients with hepatocellular carcinoma using a novel evaluation of cell-free DNA. Ann Hepatol 2008;7:339–344. [PubMed] [Google Scholar]
- 70.Iizuka N, Oka M, Sakaida I, et al. Efficient detection of hepatocellular carcinoma by a hybrid blood test of epigenetic and classical protein markers. Clin Chim Acta 2011; 412:152–158. [DOI] [PubMed] [Google Scholar]
- 71.Iizuka N, Sakaida I, Moribe T, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res 2006; 26:4713–4719. [PubMed] [Google Scholar]
- 72.Ji XF, Fan YC, Gao S, et al. MT1M and MT1G promoter methylation as biomarkers for hepatocellular carcinoma. World J Gastroenterol 2014;20:4723–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanekiyo S, Iizuka N, Tsunedomi R, et al. Preoperative serum methylation signature as prognostic tool after curative hepatectomy in patients with hepatocellular carcinoma. Anticancer Res 2015;35:997–1007. [PubMed] [Google Scholar]
- 74.Kisiel JB, Dukek BA, Kanipakam R, et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology 2019; 69:1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuo CC, Lin CY, Shih YL, et al. Frequent methylation of HOXA9 gene in tumor tissues and plasma samples from human hepatocellular carcinomas. Clin Chem Lab Med 2014;52:1235–1245. [DOI] [PubMed] [Google Scholar]
- 76.Li F, Fan YC, Gao S, et al. Methylation of serum insulin-like growth factor-binding protein 7 promoter in hepatitis B virus-associated hepatocellular carcinoma. Genes Chromosomes Cancer 2014;53:90–97. [DOI] [PubMed] [Google Scholar]
- 77.Li F, Qiao CY, Gao S, et al. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis B virus-associated hepatocellular carcinoma after hepatectomy. Free Radic Res 2018; 52:455–464. [DOI] [PubMed] [Google Scholar]
- 78.Liao W, Yang H, Xu H, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016;7:40481–44090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu CY, Chen SY, Peng HL, et al. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget 2017;8:6406–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchio A, Amougou Atsama M, Bere A, et al. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med 2018;18:421–431. [DOI] [PubMed] [Google Scholar]
- 81.Oussalah A, Rischer S, Bensenane M, et al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine 2018; 30:138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piciocchi M, Cardin R, Vitale A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013;7:1050–1057. [DOI] [PubMed] [Google Scholar]
- 83.Qu C, Wang Y, Wang P, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A 2019; 116:6308–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ren N, Qin LX, Tu H, et al. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol 2006;132:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun F-K, Fan Y- C, Zhao J, et al. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci 2013;58:1010–1015. [DOI] [PubMed] [Google Scholar]
- 86.Tangkijvanich P, Hourpai N, Rattanatanyong P, et al. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta 2007;379:127–133. [DOI] [PubMed] [Google Scholar]
- 87.Wang J, Qin Y, Li B, et al. Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin Biochem 2006; 39:344–348. [DOI] [PubMed] [Google Scholar]
- 88.Wei L, Huang Y, Zhao R, et al. Detection of promoter methylation status of suppressor of cytokine signaling 3 (SOCS3) in tissue and plasma from Chinese patients with different hepatic diseases. Clin Exp Med 2018;18:79–87. [DOI] [PubMed] [Google Scholar]
- 89.Wen L, Li J, Guo H, et al. Genome-scale detection of hyper-methylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25:1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiong Y, Xie CR, Zhang S, et al. Detection of a novel panel of somatic mutations in plasma cell-free DNA and its diagnostic value in hepatocellular carcinoma. Cancer Manag Res 2019; 11:5745–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu H, Zhu X, Xu Z, et al. Non-invasive analysis of genomic copy number variation in patients with hepatocellular carcinoma by next generation DNA sequencing. J Cancer 2015;6:247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155–1161. [DOI] [PubMed] [Google Scholar]
- 93.Yan L, Chen Y, Zhou J, et al. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis 2018;67:92–97. [DOI] [PubMed] [Google Scholar]
- 94.Yeo W, Wong N, Wong WL, et al. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int 2005;25:266–272. [DOI] [PubMed] [Google Scholar]
- 95.Abd El Gwad A, Matboli M, El-Tawdi A, et al. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem 2018;119:8600–8610. [DOI] [PubMed] [Google Scholar]
- 96.Cheng R, Ban L, Tu T, et al. Utility of microvesicles as plasma biomarkers in patients with hepatocellular carcinoma. J Gastroenterol Hepatol 2015;30:5.25536461 [Google Scholar]
- 97.Li Y, Zhao J, Yu S, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem 2019;65:798–808. [DOI] [PubMed] [Google Scholar]
- 98.Lu Y, Duan Y, Xu Q, et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med 2020; 24:1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pu C, Huang H, Wang Z, et al. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol 2018;9:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang W, Li H, Zhou Y, et al. Peripheral blood microvesicles are potential biomarkers for hepatocellular carcinoma. Cancer Biomarkers 2013;13:351–357. [DOI] [PubMed] [Google Scholar]
- 101.Wang X, Kwak KJ, Yang Z, et al. Extracellular mRNA detected by molecular beacons in tethered lipoplex nanoparticles for diagnosis of human hepatocellular carcinoma. PLoS One 2018; 13:e0198552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Zhang C, Zhang P, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med 2018;7:1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu H, Chen Y, Dong X, et al. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 2018;27:710–716. [DOI] [PubMed] [Google Scholar]
- 104.Xu H, Dong X, Chen Y, et al. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med 2018;56:479–484. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Y, Xi H, Nie X, et al. Assessment of miR-212 and other biomarkers in the diagnosis and treatment of HBV-infection-related liver diseases. Curr Drug Metab 2019;20:785–798. [DOI] [PubMed] [Google Scholar]
- 106.Dong SL, Zhang BH, Chen XP. Preoperative circulating tumor cells predict impaired time to tumor recurrence in HCC patients after curative resection. HPB 2015;17:261. [Google Scholar]
- 107.Fan ST, Yang ZF, Ho DW, et al. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: a prospective study. Ann Surg 2011;254:569–576. [DOI] [PubMed] [Google Scholar]
- 108.Fu B, Yang Q, Tang H, et al. Correlation between classification of circulating tumor cells in peripheral blood and early recurrence in patients with hepatocellular carcinoma after liver transplantation. Transplantation 2019;103:311. [Google Scholar]
- 109.Ha Y, Kim TH, Shim JE, et al. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: a prospective study. Hepatol Int 2019;13:726–735. [DOI] [PubMed] [Google Scholar]
- 110.Hamaoka M, Kobayashi T, Tanaka Y, et al. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: a prospective study. PLoS One 2019;14: e0217586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nel I, Baba HA, Howner A, et al. Presence of circulating tumor cells with stem cell characteristics predicts response to SIRT in hepatocellular carcinoma. Oncol Res Treat 2014;37:53. [Google Scholar]
- 112.Ogle LF, Orr JG, Willoughby CE, et al. Imagestream detection and characterisation of circulating tumour cells - a liquid biopsy for hepatocellular carcinoma? J Hepatol 2016;65:305–313. [DOI] [PubMed] [Google Scholar]
- 113.Qi LN, Xiang BD, Wu FX, et al. Circulating tumor cells undergoing emt provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res 2018; 78:4731–4744. [DOI] [PubMed] [Google Scholar]
- 114.Schulze K, Von Felden J, Krech T, et al. EpCAM-positive circulating tumor cells as liquid biomarker for early micro-metastases and HCC recurrence risk. J Hepatol 2017;66:S457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun YF, Guo W, Xu Y, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res 2018;24:547–559. [DOI] [PubMed] [Google Scholar]
- 116.von Felden J, Schulze K, Krech T, et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget 2017;8:89978–89987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vona G, Estepa L, Beroud C, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology 2004;39:792–797. [DOI] [PubMed] [Google Scholar]
- 118.Wang Z, Luo L, Cheng Y, et al. Correlation between postoperative early recurrence of hepatocellular carcinoma and mesenchymal circulating tumor cells in peripheral blood. J Gastrointest Surg 2018;22:633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu X, Yang C, Yu H, et al. The predictive values of serum dickkopf-1 and circulating tumor cells in evaluating the efficacy of transcatheter arterial chemoembolization treatment on hepatocellular carcinoma. Medicine (Baltimore) 2019;98:e16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu JJ,Xiao W, Dong SL, et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer 2018;18:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chelis L, Balgouranidou I, Koukaki T, et al. Assessment of SOX-17 gene promoter hypermethylation in hepatocellular carcinoma. J Clin Oncol 2013;31. [Google Scholar]
- 122.Howell J, Atkinson S, Pinato DJ, et al. Identification of actionable mutations in circulating cell-free tumour DNA as a prognostic biomarker in hepatocellular carcinoma. J Hepatol 2017;66:S449. [DOI] [PubMed] [Google Scholar]
- 123.Huang ZH, Hu Y, Hua D, et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702–707. [DOI] [PubMed] [Google Scholar]
- 124.Jiao J, Watt GP, Stevenson HL, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: prevalence and risk factors. Hepatol Commun 2018;2:718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ono A, Fujimoto A, Yamamoto Y, et al. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol Gastroenterol Hepatol 2015;1:516–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park S, Lee EJ, Rim CH, et al. Plasma cell-free DNA as a predictive marker after radiotherapy for hepatocellular carcinoma. Yonsei Med J 2018;59:470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tokuhisa Y, lizuka N, Sakaida I, et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer 2007; 97:1399–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jang SY, Kim G, Hur K, et al. Circulating serum exosomal microRNA-203 as a non-invasive biomarker for predicting prognosis in hepatocellular carcinoma. J Hepatol 2017; 66:S672–S673. [Google Scholar]
- 129.Lam YF, Ng KTP, Man K. Prognostic role of exosomal miR-1290 in hepatocellular carcinoma recurrence after curative treatments. Transplantation 2018;102:41. [Google Scholar]
- 130.Lee YR, Kim G, Tak WY, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer 2019;144:1444–1452. [DOI] [PubMed] [Google Scholar]
- 131.Liu W, Hu J, Zhou K, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther 2017;10:3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi M, Jiang Y, Yang L, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 2018;119:4711–4716. [DOI] [PubMed] [Google Scholar]
- 133.Suehiro T, Miyaaki H, Kanda Y, et al. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett 2018;16:3267–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sugimachi K, Matsumura T, Hirata H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer 2015;112:532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tian XP, Wang CY, Jin XH, et al. Acidic microenvironment upregulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics 2019;9:1965–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu F, Wang CX, Tian H. Expression and clinical significance of LncRNA ZFAS1 in serum exosomes of patients with hepatocellular carcinoma. Chin J Cancer Prev Treatment 2019; 26:849–854. [Google Scholar]
- 137.Chen Y, Li S, Li W, et al. Circulating tumor cells undergoing EMT are poorly correlated with clinical stages or predictive of recurrence in hepatocellular carcinoma. Sci Rep 2019;9:7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee VH, Wong VCL, Lam CT, et al. Correlation of circulating tumor cells with response to SBRT for inoperable hepatocellular carcinoma. Int J Radiat Oncol Biol Physics 2017;99:E165. [Google Scholar]
- 139.Wang S, Zheng Y, Liu J, et al. Analysis of circulating tumor cells in patients with hepatocellular carcinoma recurrence following liver transplantation. J Invest Med 2018;66:929–934. [DOI] [PubMed] [Google Scholar]
- 140.Ye X, Li G, Han C, et al. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Manag Res 2018; 10:5639–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Alunni-Fabbroni M, Ronsch K, Huber T, et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: a translational exploratory study from the SORAMIC trial. J Transl Med 2019;17:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen G, Cai Z, Li Z, et al. Clonal evolution in long-term follow-up patients with hepatocellular carcinoma. Int J Cancer 2018; 143:2862–2870. [DOI] [PubMed] [Google Scholar]
- 143.Du X, Li Z, Zhang H. Detection of early recurrence in HCC patients after TACE treatment with plasma cfDNA. J Clin Oncol 2018;36:e16146. [Google Scholar]
- 144.Evans J, Pinato D, Ward C, et al. Longitudinal monitoring of cell-free DNA predicts for transarterial chemo-embolization failure in hepatocellular carcinoma. J Hepatol 2019;70:e362. [Google Scholar]
- 145.Higuera M, Vargas-Accarino E, Soria ME, et al. Massive sequencing of circulating DNA as a potential tool for diagnosis and follow-up in patients with hepatocellular carcinoma. J Hepatol 2019;70:e841–e842. [Google Scholar]
- 146.Li Z, Xiao D, Li X, et al. Early recurrence detected in hepatocellular carcinoma patients after transcatheter arterial chemo-embolization treatment with plasma cell-free DNA. Eur J Gastroenterol Hepatol 2019;31:885–892. [DOI] [PubMed] [Google Scholar]
- 147.Wong IH, Zhang J, Lai PB, et al. Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Cancer Res 2003;9:1047–1052. [PubMed] [Google Scholar]
- 148.Yang P, Ji G, Jiang Q, et al. Mutation profiles in circulating tumor DNA (ctDNA) to predict the efficacy of sorafenib treatment in patients with advanced hepatocellular carcinoma (HCC). J Clin Oncol 2019;37. [Google Scholar]
- 149.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001; 93:1054–1061. [DOI] [PubMed] [Google Scholar]
- 150.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Smerage JB, Barlow WE, Hortobagyi GN, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol 2014;32:3483–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Broersen LHA, van Pelt GW, Tollenaar RAEM, et al. Clinical application of circulating tumor cells in breast cancer. Cell Oncol 2014;37:9–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


