Abstract
Discovery of new anti-tuberculosis (TB) drugs is a time-consuming process due to the slow-growing nature of Mycobacterium tuberculosis (Mtb). A requirement of biosafety level 3 (BSL-3) facility for performing research associated with Mtb is another limitation for the development of TB drug discovery. In our screening of BSL-1 Mycobacterium spp. against a battery of TB drugs, M. smegmatis (ATCC607) exhibits good agreement with its drug susceptibility against the TB drugs under a low-nutrient culture medium (0.5% Tween80 in Middlebrook 7H9 broth). M. smegmatis (ATCC607) enters its dormant form in 14 days under a nutrient-deficient condition (a PBS buffer), and shows resistance to a majority of TB drugs, but shows susceptibility to amikacin, capreomycin, ethambutol, and rifampicin (with high concentrations) whose activities against non-replicating (or dormant) Mtb were previously validated.
Keywords: Mycobacterium tuberculosis surrogate, Mycobacterium smegmatis, TB drugs, non-replicating Mycobacterium smegmatis
INTRODUCTION
Since the emergence of multidrug-resistant Mycobacterium tuberculosis (Mtb), the importance of discovery of new tuberculosis (TB) drugs has been documented in a number of scientific publications [1–5]. Public sector research agencies and nonprofit organizations play an important role in the development of TB drugs due to the fact that pharmaceutical companies have shied away from drug discoveries of neglected diseases including TB [6, 7]. A limited number of academic institutions have supported biosafety level 3 (BSL-3) laboratory and biocontainment facilities to perform TB researches in the U.S. In addition to inconvenient accessibility for other researchers to the required facility, the pathogenicity, and a slow-growing nature of Mtb might discourage scientists from TB drug discovery. Particularly, a whole cell-based assay frequency of TB drug leads using Mtb strains is one of the factors that leads to a slow process of medicinal chemistry [1, 8–17]. Applying non-pathogenic Mycobacterium smegmatis strains to TB drug discovery has shown a limited number of successes in the past [18–21]. Under rich-nutrient culture media (or recommended culture conditions), a majority of M. smegmatis strains are not susceptible to the TB drugs (e.g., rifampicin, INH, and ethambutol) [20]. In the screening of M. smegmatis strains susceptible to the TB drugs, it was found that M. smegmatis (ATCC 607) was effectively killed by the representative TB drugs (e.g., rifampicin, INH, ethambutol, and amikacin) in 2–4 days under a low-nutrient culture condition (0.5% Tween 80 in Middlebrook 7H9 broth). To the best of our knowledge, it is the first observation that an M. smegmatis strain shows similar drug susceptibility to Mtb H37Rv strain. Herein, we report 1) drug susceptibility of M. smegmatis (ATCC607) against 20 antibacterial agents including the 1st and 2nd line, and investigational TB drugs, 2) mechanistic studies of antibacterial effects of the representative TB drugs against M. smegmatis (ATCC607), and 3) responses of some TB drugs against non-replicating M. smegmatis (ATCC 607).
MATERIALS AND METHODS
General / Chemicals and Reagents
All chemicals and antibiotics were purchased from commercial sources and used without further purification unless otherwise noted. Difco Middlebrook 7H10 agar, Middlebrook 7H9 broth, 44 Brain Heart Infusion Agar/Broth, Tween 80, ADC and OADC enrichment were purchased from Fisher Scientific. Resazurin (Alamar blue) was purchased from Sigma-Aldrich. Note: Middlebrook 7H9 contains following ingredients (g•L−1): ammonium sulfate (0.50), disodium phosphate (2.50), monopotassium phosphate (KH2PO4, 1.00), sodium citrate (0.10), magnesium sulfate (0.05), calcium chloride (0.0005), zinc sulfate (0.001), copper sulfate (0.001), ferric ammonium citrate (0.04), L-glutamic acid (0.50), pyridoxine (0.001), biotin (0.0005). Middlebrook 7H10 contains following ingredients (g•L−1): ammonium sulfate (0.50), disodium phosphate (1.50), monopotassium phosphate (KH2PO4, 1.50), sodium citrate (0.40), magnesium sulfate (0.025), calcium chloride (0.0005), zinc sulfate (0.001), copper sulfate (0.001), ferric ammonium citrate (0.04), L-glutamic acid (0.50), pyridoxine hydrochloride (0.001), biotin (0.0005), malachite green (0.00025), agar (15.00).
Bacterial and macrophage strains
Mycobacterium smegmatis (ATCC607) was purchased from ATCC. Mycobacterium tuberculosis H37Rv was acquired from BEI Resources (NIAID).
MIC assays
Log phase bacterial culture
All liquid bacterial culturing was performed with a conical flask with an air filter. A single colony of a bacterial strain (M. tuberculosis) was grown on a Difco Middlebrook 7H10 nutrient agar (enriched with 10% OADC and 0.4% glycerol). Seed cultures and larger cultures of M. tuberculosis were obtained using Middlebrook 7H9 broth enriched with 10% OADC and 0.4% glycerol. M. smegmatis (ATCC607) was cultured on a 0.5% Tween 80 Middlebrook 7H10 nutrient agar (0.4% glycerol). Seed cultures and larger cultures of M. smegmatis (ATCC607) were obtained using 0.5% Tween 80 in Middlebrook 7H9 (0.4% glycerol). The culture flasks were incubated for 3–4 days for M. smegmatis (ATCC607), and for 10–12 days for M. tuberculosis H37Rv in a shaking incubator at 37 °C with a shaking speed of 200 rpm and cultured to mid-log phase (optical density - 0.5). The optical density was monitored at 600 nm using a 96 well microplate reader.
Determination of minimum inhibitory concentration (MIC)
The antibiotics were dissolved in DMSO or water (a final concentration of 1 mg per 100 μL). This concentration was used as the stock solution for all studies. Bacterial cultures at 0.2 optical density, were treated with serial dilutions of inhibitors in aerobic conditions and incubated at 37 °C for 4 and 14 days for M. smegmatis and M. tuberculosis, respectively. Alamar blue (2%, 20 μL) was added and incubated in a static incubator at 37 °C for 4–12 h. The lowest concentration at which the color of Alamar blue was completely retained as blue was read as the MIC (Pink = Growth, Blue = No growth). The absorbance measurements were also performed using a Biotek Synergy XT, 96 well plate reader at 570 nm and 600 nm.
Generation of drug-resistant M. smegmatis (ATCC607) strains
Drug-resistant M. smegmatis (ATCC607) strains against rifampicin (RIF), ethambutol (EMB), isoniazid (INH), capreomycin (CAP), and amikacin (AMK) were generated according to the same procedure. M. smegmatis (ATCC607) (100 μL of 1×107 CFU•mL−1) was plated on 0.5% Tween 80 Middlebrook 7H10 nutrient agar plate (55 cm2) containing antibiotics (minimum bactericidal concentration (MBC)). The colonies grown on the antibiotic-containing agar plate were collected and suspended in a PBS buffer (~1×107 CFU•mL−1), and 100 μL of the bacterial suspension was plated on the agar plate containing antibiotics (1.5xMBC). This process was repeated until the cells acquire >5 times higher MIC level than the wild type. The concentrations of antibiotic were gradually increased (2.0x, 2.5x, 3.0x, 3.5x, 4.0x, 5.0x, 7.0x, 8.0x, 9.0x, 10.0x, and 20xMBC). The isolated resistant cells were confirmed by the MIC assay with the generated resistant strain.
Genetic analyses of M. smegmatis (ATCC607) resistant strains
The chromosomal DNAs from the resistant mutants, including RIFR-M. smegmatis, EMBR-M. smegmatis, CapR-M. smegmatis, and AMKR-M. smegmatis, INHR-M. smegmatis and their parental control M. smegmatis (ATCC607), were isolated using a NucleoSpin Tissue kit (Macherey-Nagel). The target genes of these antibiotics, including embB, rpoB, embB, rrs, inhA, katG, and ahpC, were amplified using the purified genomic DNA as a template by PCR using high fidelity DNA polymerase (BioLabs) and gene-specific primers listed in Table S1 (supporting information). The PCR products were purified using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) or ExoSAP-IT Express PCR Cleanup reagents (ThermoFisher Scientific) and sequenced. The DNA sequences of these target genes were blasted against their corresponding DNA sequences of M. smegmatis in the NIH Genome database.
Formation of non-replicating M. smegmatis (ATCC607)
The starter culture of M. smegmatis (ATCC607) was obtained from a single colony by incubation in 0.5% tween 80-Middlebrook 7H9 medium at 37 °C for 4 days. The seed culture (1 mL) was inoculated into 0.5% Tween 80 Middlebrook 7H9 (50–75 mL). This culture was incubated in a shaking incubator (200 rpm, 37 °C) for 48–72 h. A stationary phase (OD ~1.0) culture was kept in a static incubator (37 °C) for 30, 60, or 150 days.
Formation of non-replicating M. smegmatis (ATCC607) under a nutrient-deficient condition
The starter culture of M. smegmatis (ATCC607) was obtained from a single colony by incubation in 0.5% tween 80-Middlebrook 7H9 medium (35 mL) at 37 °C for 4 days. The culture was centrifuged (4,500 rpm for 10 min.) and suspended in phosphate-buffered saline (PBS, pH 7.2) for 14 days (37 °C)
Antimycobacterial activity of TB drugs against non-replicating M. smegmatis (ATCC607)
Non-replicating M. smegmatis (ATCC607) generated via the procedure described above was kept in the presence of each drug (5xMBC or 20xMBC) (total volume 200 μL) for 5–15 days at 37 °C. The cultures were diluted 4,000 times (10×20×20), and 100 μL of the diluted cell culture was plated on 0.5% Tween 80 Middlebrook 7H10 nutrient agar plate. Colony-forming units (CFUs) of survival M. smegmatis cells on agar plates were counted after incubation at 37 °C for 4 days.
RESULT
Susceptibility of M. smegmatis (ATCC607) against representative TB drugs
The MIC values of representative TB drugs, and positive- and negative-controls against M. smegmatis (ATCC607) were examined under a nutrient-deficient condition (0.5% Tween 80-Middlebrook 7H9 broth). These data are summarized in Table 1. Table 1 also includes their MIC values (reported in literature/databases or obtained in our lab as noted) [22–31], drug target(s), and drug-resistant mechanism(s) against M. tuberculosis (Mtb). A majority of FDA-approved TB drugs (isoniazid, rifampicin, ethambutol, bedaquline, aminoglycosides, streptomycin, capreomycin, ethionamide, clofazimine, and cycloserine) showed good correlation in the MIC values between M. smegmatis (ATCC607) strain and Mtb H37Rv. Drug susceptibility of rifampicin (RIF) against M. smegmatis was 16–19 times higher MIC value than that against Mtb. On the other hand, the fluoroquinolones (moxifloxacin and ciprofloxacin) exhibited 5–10 times lower MIC values against M. smegmatis than those against Mtb. Pyrazinamide is known to show pH-dependent susceptibility against Mtb in vitro [32, 33]. At pH 6.6 it killed M. smegmatis efficiently with much lower MIC value (0.097–1.6 μg/mL) than that against Mtb. The MIC value of tunicamycin, a MraY/WecA inhibitor [34, 35], against M. smegmatis exhibited equal to that against Mtb. Glycopeptide antibiotics, vancomycin, and ristocetin A [8], had 3–10 times better susceptibility to M. smegmatis than Mtb. The negative-controls showed a good agreement in their susceptibility: colistin did not show antibacterial activity against M. smegmatis. Under the aerobic conditions, metronidazole was not effective in inhibiting the growth of M. smegmatis [36]. In these susceptibility tests, all TB drugs showed susceptibility against M. smegmatis under a slow growth condition, and a majority of TB drugs showed good or meaningful correlations for their MIC values except pyrazinamide.
Table 1.
Comparison of MIC values of representative TB drugs, positive- and negative-controls against M. smegmatis (ATCC607) and M. tuberculosis H37Rv.
 | ||||
|---|---|---|---|---|
| Molecule | Molecular target | Primary resistant mechanism in Mtb | M. smegmatis MIC μg·mL−1 a,b | M. tuberculosis H37Rv MIC μg·mL−1 |
| Isoniazid (INH) | InhA | katG, ahpC mutations | 0.012–0.78 | 0.02–0.2c |
| Rifampicin (RIF) | β-subunit of RNA polymerase | rpoB mutations | 0.97–1.6 | 0.05–0.1c |
| Ethambutol (EMB) | Arabinosyl transferase | embB mutations | 0.3–0.5 | 1.0–5.0c |
| Pyrazinamide | fatty acid synthase (FAS) I | pncA mutation | 0.012–0.78 (pH 6.6) | 50–400(pH dependent)b |
| Bedaquiline | ATP synthase | aipE mutation, efflux pumps | 0.024–0.048 | 0.03–0.10c |
| Amikacin (AMK) | 16S ribosome RNA | 16S ribosome mutations | 1.0–1.6 | 2.0–4.0c |
| Kanamycin | 16S ribosome RNA | 16S ribosome mutations | 0.39–1.6 | 0.04–0.10c |
| Streptomycin | 16S ribosome RNA | 16S ribosome mutations | 1.56–6.3 | 0.50–2.0c |
| Capreomycin (CAP) | 16S ribosome RNA | 16S ribosome mutations | 0.78–1.6 | 2.0–4.0c |
| Ethionamide | InhA | ethA mutations | 3.1–6.3 | 0.5–2.0c |
| Moxifloxacin | DNA gyrase | gyrA/gyrB mutations | 0.048–0.39 | 0.25–2.0c |
| Ciprofloxacin | DNA gyrase | gyrA/gyrB mutations | 0.048–0.39 | 0.25–2.0c |
| Cycloserine | Alanine racemase (Alr) and D-alanine:D-alanine ligase (Ddl) | unknown | 6.3–13 | 16–25c |
| Linezolid | Ribosomal L3 protein, 23S ribosomal RNA | rplC T460C | 0.39–1.56 | 0.25–0.50c |
| Clofazimine | Bacterial membrane | unknown | 0.048–0.19 | 0.13–0.20c |
| Tunicamycin | MraY, WecA | unknown | 6.25–12.5 | 6.3–12.5a,c |
| Vancomycin | Cell wall biosynthesis | unknown | 3.1–12.5 | 6.25–25.0a,c |
| Ristocetin A | Cell wall biosynthesis | unknown | 0.19–0.39 | 0.5–3.9c |
| Colistin | Bacterial membrane | unknown | >50 | >50a,c |
| Metronidazole | A prodrug that is activated by a nitroreductase enzyme to reactive species (effective only hypoxic conditions) | unknown | >50 | >50a,c |
All MIC data were generated in this studies. Microplate Alamar (Risazurin) blue assays were applied (see Experimental). All experiments were triplicated.
Selected MIC data performed in an enriched medium were summarized in Supporting Information (for a comparison).
The MIC data were cited from databases and/or literatures.
Determination of M. smegmatis’ mechanisms of resistance against representative TB drugs
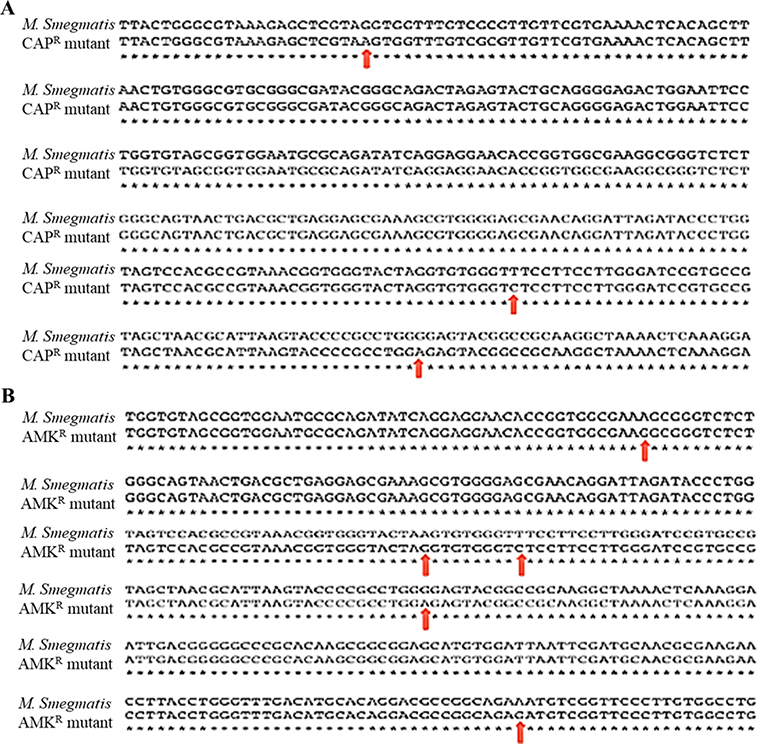
We have generated drug-resistant strains of M. smegmatis (ATCC607) against rifampicin (RIF), ethambutol (EMB), isoniazid (INH), capreomycin (CAP), and amikacin (AMK). Their MIC values were 5-times or higher than those of the wild-type strain. It has been studied that RIF, EMB, CAP and AMK exhibit antibacterial activity by targeting RpoB (bacterial RNA polymerase) [37, 38], EmbB (embB gene encoding arabinosyltransferase activity) [39–41] and 16S ribosome RNA (encoded by the rrs gene) [42, 43], respectively. INH targets InhA, and drug-resistant Mtb strains have mutations in the katG (Mtb catalase-peroxidase) and/or ahpC (alkyl hydroperoxide reductase C) gene [38, 44–46]. To understand whether the resistance of M. smegmatis to these antibiotics has resulted from the mutations of their target genes identified in the mutant Mtb cells, we isolated the chromosomal DNAs from the resistant mutants, including RIFR-M. smegmatis, EMBR-M. smegmatis, CAPR-M. smegmatis, and AMKR-M. smegmatis (intermediate resistant), and their parental control M. smegmatis (ATCC607). Except for embB, the other target genes, including rpoB, rrs, inhA, katG, and ahpC, were obtained by PCR and they were sequenced. The DNA sequences of these target genes were blasted against their corresponding DNA sequences in the NIH Genome database [47]. The DNA sequence alignment revealed E401Q, E462Q, A638G, A653G, and G656S mutations in the protein sequence of RpoB in the RIFR-M. smegmatis mutant, compared with their parental control (Figure 2A). One of important mechanisms in M. smegmatis against RIF is its ADP-ribosylation by mono ADP-ribosyltransferase encoded by arr (MSMEG_1221). In this experiment, we cannot rule out the possibility of mutation in the arr gene in RIFR-M. smegmatis mutant [48]. A544G, A562T, E591Q, and R598P mutations were identified in the protein sequence of KatG, whereas no mutation of inhA occurred in the INHR-M. smegmatis mutant compared to its parental control (Figure 2B). Three nucleotide mutations of A564G, C818T, and A869G were identified in 16S rRNA gene (rrs) of the CapR-M. smegmatis strain (Figure 3A). Five nucleotide mutations of G710A, G809A, C818T, A869G, and G997A were revealed in the rrs of the AMKR-mutant (Figure 3B). These results observed in RIF-, INH-, CAP-, and AMK-resistant M. smegmatis strains suggested that the mutations in the same target genes identified in the Mtb mutants contribute to resistance to the corresponding antibiotics.
Figure 2.
The amino acid alignments of the bacterial RNA polymerase subunit protein B (RpoB) between RifampicinR (RifR) mutant and parental control M. smegmatis (A) and KatG protein between IsoniazidR (INHR) mutant and parental control M. smegmatis (B). The red arrow represents the site mutation in RpoB or KatG.
Figure 3.
16S rRNA gene rrs sequence alignment between: (A) CapreomycinR (CAPR) mutant and its parental control M. smegmatis (ATCC607), and (B) AmikacinR (AMKR) mutant and its parental control M. smegmatis (ATCC607).
Response of TB drugs against non-replicating M. smegmatis (ATCC607)
Mtb can persist many years within host tissues [49, 50]. Subpopulations of Mtb to enter a dormant phase lead to the long-term treatment adherence for TB and recurrence of TB. In vitro studies using the dormant forms of Mtb have demonstrated that non-replicating Mtb cells show resistance to a majority of TB drugs [1, 51]. We have cultured M. smegmatis (ATCC607) strain for up to 150 days in medium containing 0.5% Tween 80 as the source of the primary carbons [52]. Bactericidal activity of amikacin (AMK), capreomycin (CAP), rifampicin (RIF), isoniazid (INH), and ethambutol (EMB) were examined against M. smegmatis (ATCC607) cultured for 30 days, 60 days, and 150 days. AMK killed non-replicating forms (incubation periods of 30, 60, and 150 days) of M. smegmatis in 5 days at 5-times the minimum bactericidal concentration (5xMBC) (Table 2) [53]. Similarly, susceptibility of the other TB drugs (CAP, RIF, INH, and EMB) showed no apparent difference against non-replicating M. smegmatis cultured for 30, 60, and 150 days. CAP was also determined to be an effective agent for killing non-replicating M. smegmatis at 5xMBC; over 2-log (99%) reductions of colony-forming unit (CFU) were observed in 5 days [54]. No CFU was counted for non-replicating M. smegmatis treated with CAP (5xMBC) for 15 days. RIF showed efficacy against non-replicating M. smegmatis in a concentration-dependent manner: a 25–30% reduction of the CFU with the drug concentration at 5xMBC for 5 days, and over 90% reduction at 20xMIC for 5 days [55, 56]. 15 Days of treatment of RIF against non-replicating M. smegmatis did not significantly show the CFU reduction. INH, a negative control, was not effective in killing non-replicating M. smegmatis at 20xMBC for 5 and 15 days, respectively [57, 58]. EMB was not efficacious in killing non-replicating M. smegmatis at 20xMBC for 5 days. However, it showed bactericidal activity against non-replicating M. smegmatis in a time-dependent manner: 15 Days treatment of EMB showed no countable colony [59]. The same susceptibility testing was performed with non-replicating M. smegmatis generated under a nutrient-deficient condition (stored in a PBS buffer for 14 days). AMK, CAP, RIF, and EMB displayed an equal or very similar trend of antimycobacterial activities to those observed against non-replicating M. smegmatis formed via 30–150 days incubation in the growth medium.
Table 2.
Antimycobacterial activity of representative TB drugs against non-replicating M. smegmatis (ATCC607) generated via long-term culturing and a nutrient deficient conditiona
| TB drugs | MBC (μg·mL−1)b | Drug conc. (μg·mL−1)c | Treatment time (days) | CFU·mL−1 d | |||
|---|---|---|---|---|---|---|---|
| M. smegmatis (30 days)e | M. smegmatis (60 days)e | M. smegmatis (150 days)e | M. smegmatis (14 days, nutrient def.)f | ||||
| Amikacin (AMK) | 1.6 | 7.5 (5 × MBC) | 5 | 0 | 0 | 0 | 0 |
| Amikacin (AMK) | 1.6 | 7.5 (5 × MBC) | 15 | 0 | 0 | 0 | 0 |
| Capreomycin (CAP) | 2.5 | 12.5 (5 × MBC) | 5 | 2.0 × 105 | 8.0 × 104 | 4.0 × 104 | 2.0 × 105 |
| Capreomycin (CAP) | 2.5 | 12.5 (5 × MBC) | 15 | 0 | 0 | 0 | 0 |
| Rifampicin (RIF) | 1.6 | 7.5 (5 × MBC) | 5 | 1.2 × 107 | 1.1 × 107 | 1.0 × 107 | 1.9 × 107 |
| Rifampicin (RIF) | 1.6 | 32.0 (20 × MBC) | 5 | 6.0 × 105 | 4.0 × 105 | 3.9 × 105 | 6.8 × 106 |
| Rifampicin (RIF) | 1.6 | 32.0 (20 × MBC) | 15 | 5.5 × 105 | 3.9 × 105 | 3.2 × 105 | 5.7 × 106 |
| Isoniazid (INH) | 1.0 | 20.0 (20 × MBC) | 5 | 2.8 × 107 | 2.5 × 107 | 2.4 × 107 | 2.8 × 107 |
| Isoniazid (INH) | 1.0 | 20.0 (20 × MBC) | 15 | 2.6 × 107 | 2.3 × 107 | 2.2 × 107 | 2.5 × 107 |
| Ethambutol (EMB) | 0.5 | 10.0 (20 × MBC) | 5 | 3.8 × 107 | 3.2 × 107 | 3.3 × 107 | 3.2 × 107 |
| Ethambutol (EMB) | 0.5 | 5.0 (5 × MBC) | 15 | 0 | 0 | 0 | 0 |
| No drug (control) | - | - | 5 | 4.0 × 107 | 3.9 × 107 | 3.8 × 107 | 3.6 × 107 |
Non-replicating M. smegmatis (ATCC607) was kept in the presence of each drug (5×MBC or 20×MBC) (total volume 200 μL) for 5 days at 37 °C. All experiments were triplicated.
The minimum bactericidal concentration (MBC) was determined by colony-forming unit (CFU) of M. smegmatis (ATCC607) cultured on agar plates contacting TB drug.
Drug concentrations used are 5- or 20-times the MBC of each drug.
M. smegmatis colony-forming unit (CFU) grown on the agar plate (55 cm2, 0.5 % tween 80-Middlebrook 7H11 agar base) was counted after 4 days at 37 °C.
The culture was produced by inoculation of single colony of M. smegmatis (ATCC607) into 25 mL 0.5% tween 80-Middlebrook 7H9 medium (0.4% glycerol), followed by incubation under stationary conditions at 37 °C for 30, 60, and 150 days, respectively.
The starter culture of M. smegmatis (ATCC607) was obtained from a single colony by incubation in 0.5% tween 80-Middlebrook 7H9 medium (0.4% glycerol) at 37 °C for 4 days. The culture was centrifuged (4,500 rpm for 10 min.) and suspended in PBS (pH 7.2) for 14 days (37 °C)
DISCUSSION
M. smegmatis is a useful research surrogate for pathogenic Mycobacterial species in laboratory experiments. For example, it is an excellent expression host for the production of recombinant proteins from various mycobacterial species [60]. However, under the recommended growth conditions (nutrient-rich conditions), M. smegmatis strains are resistant to many TB drugs (e.g., rifampicin and isoniazid); M. smegmatis strains displayed resistance (MIC >20 μg•mL−1) to the key drugs in the 1st-line anti-TB drugs, such as rifampicin (RIF) and isoniazid (INH) [20]. Although resistant mechanisms have not been studied thoroughly, a number of TB scientists agree that M. smegmatis has intrinsic resistance to these drugs [61, 62]. Nonetheless, low susceptibility of M. smegmatis to several TB drugs discourages many scientists to apply M. smegmatis as a surrogate to screening TB drug leads. Under a slow-growing culture condition (0.5% tween 80-Middlebrook 7H9 broth), M. smegmatis (ATCC607) displayed antimicrobial susceptibility against all TB drugs tested (Table 1). Importantly, a majority of the 1st- and 2nd-line TB drugs showed a good correlation in their MIC levels against M. smegmatis (ATCC607) to those against M. tuberculosis (Mtb) H37Rv (a laboratory strain. Pyrazinamide showed better susceptibility against M. smegmatis (ATCC607) at an initial pH of 6.6 than that against Mtb [63]. All positive- and negative-control agents showed a good correlation in the MIC values between M. smegmatis (ATCC607) and Mtb. The observed drug susceptibility agreement between M. smegmatis (ATCC607) and Mtb premises that M. smegmatis strains could be reliable surrogates for Mtb in a slow-growing culture medium. We have spent about half a year to isolate mutant strains against rifampicin (RIF), isoniazid (INH), capreomycin (CAP), amikacin (AMK), and ethambutol (EMB). These resistant strains showed 5-times or higher MIC than those against the wild-type strain. The target genes (rpoB, katG, ahpC, inhA, and rrs) for RIF, INH, Cap, and AMK were obtained successfully from their resistant strains. Analyses of the amino acid or gene sequence alignments between the wild-type strain and these drug-resistant mutants revealed that RIF, INH, CAP, and AMK-resistant M. smegmatis strains show their resistances by the mutations of the same target genes that are identified in the corresponding Mtb mutants. Although a limited number of the drug- resistant mechanistic studies have been performed, it could be concluded that M. smegmatis (ATCC607) acquires the predicted resistant mechanisms against four TB drugs (selected from two of 1st and 2nd line TB drugs). These genotypic data further support that M. smegmatis (ATCC607) is a reliable and convenient surrogate for identifying new anti-TB drug leads. However, we have not performed whole-genome sequencing of the mutants, thus, we cannot rule out the possibility that other factors or genes beside the known target genes contribute to the observed drug resistances.
M. smegmatis (ATCC607) enters its non-replicating state within 30 days in 0.5% tween 80-Middlebrook 7H9 medium. Noticeable loss of viability of non-replicating M. smegmatis was observed during extended culturing from 30 to 150 days (see viable cells vs time curve in SI). Conveniently, M. smegmatis generated under a nutrient-deficient condition (stored in a PBS for 14 days) could predict the efficacy of TB drugs against non-replicating (or dormant) Mtb. Antimycobacterial activity of AMK, CAP, RIF, and EMB (positive-controls) was demonstrated with non-replicating M. smegmatis. INH, a negative-control, was not effective in killing non-replicating M. smegmatis. In vitro evaluation of dormant form of Mtb has not provided useful information on the effect of drugs on clinical tests. Drug susceptibility of non-replicating M. smegmatis (ATCC607) showed some correlations with in vitro data obtained with non-replicating Mtb; for example, bactericidal activity of capreomycin (CAP) against non-replicating tubercle bacilli was previously reported [64–66].
In summary, the application of a non-pathogenic M. smegmatis to a preliminary screening of library molecules and iterative medicinal chemistry should facilitate TB drug discovery programs. Under nutrient limiting conditions, certain M. smegmatis strains would display similar drug susceptibility observed against Mtb. In our screening of libraries of antibacterial molecules, we have not observed disagreement in drug susceptibility profiles between M. smegmatis (ATCC607) and Mtb H37Rv. However, the MIC values obtained with M. smegmatis should not represent absolute data but are relative data to evaluate the antibacterial activity of molecules against Mtb. We recommend evaluating new molecules with the MIC values of <6.25 mg•mL−1 against M. smegmatis (ATCC607) in bacterial growth inhibitory activity (MIC) assays against Mtb strain(s). Assay results of drug susceptibility of new antimycobacterial agents against M. smegmatis (ATCC607) and its drug-resistant strains and their MIC correlations to Mtb strains will be reported elsewhere.
Supplementary Material
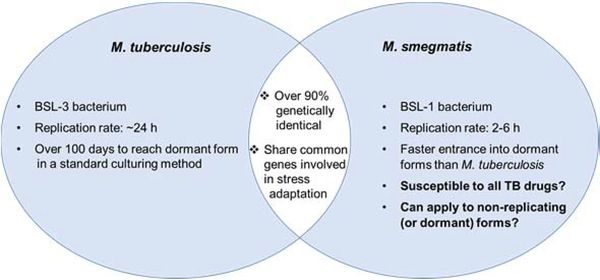
Figure 1.
Identification of M. smegmatis that is susceptible to TB drugs and beneficial of its application to TB drug discovery.
ACKNOWLEDGEMENTS
The National Institutes of Health is greatly acknowledged for financial support of this work (Grant GM114611). M. K. thanks UTRF (University of Tennessee Health Science Center) for generous financial support (Innovation award R079700292). NMR data were obtained on instruments supported by the NIH Shared Instrumentation Grant. The following reagent was obtained through BEI Resources, NIAID, NIH: Mycobacterium tuberculosis, Strain H37Rv. J. Y. thanks the support of Cystic Fibrosis Foundation Award (JI1810).
Footnotes
This article is dedicated to the memory of Dr. Isao Kitagawa, Professor Emeritus of pharmaceutical sciences at Osaka University, an inspirational scientist.
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
REFERENCES
- 1.Lauzardo M, Peloquin CA. Tuberculosis therapy for 2016 and beyond. Exp Opin Pharm. 2016;17:1859–1872. [DOI] [PubMed] [Google Scholar]
- 1.Siricilla S, Mitachi K, Wan B, Franzblau SG, Kurosu M. Discovery of a capuramycin analog that kills nonreplicating Mycobacterium tuberculosis and its synergistic effects with translocase I inhibitors J Antibiot. 2015;68:271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keri RS, Sasidhar BS, Nagaraja BM, Santos AM. Recent progress in the drug development of coumarin derivatives as potent antituberculosis agents. Eur J Med Chem. 2015;100:257–269. [DOI] [PubMed] [Google Scholar]
- 3.Muniyan R, Gurunathan J. Antimycobacterial activity of potential plant metabolites with emphasis on management of drug resistant Mycobacterium tuberculosis strains. Res J Biotech. 2017;12:75–86. [Google Scholar]
- 4.Liu J, Ren HP. Tuberculosis: current treatment and new drug development. Anti-Infect Agents Med Chem. 2006;5:331–344. [Google Scholar]
- 5.Tomioka H, Namba K. Development of antituberculous drugs: current status and future prospects. Tuberculosis. 2006;81:753–774. [PubMed] [Google Scholar]
- 6.Chatelain E, Ioset JR. Drug discovery and development for neglected diseases: the DNDi model. Drug Des Devel Ther. 2011;5:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran M A breakthrough in R&D for neglected diseases: New ways to get the drugs we need. PLoS Med. 2005;2:e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitachi K, Yun HG, Gillman CD, Skorupinska-Tudek K, Swiezewska E, Clemons WM, Kurosu M. Substrate tolerance of bacterial glycosyltransferase MurG: Novel fluorescence-based assays. ACS Infect Dis. 2019; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemieux MR, Siricilla S, Mitachi K, Eslamimehr S, Wang Y, Yang D, Pressly JD, Kong Y, Park F, Franzblau SG, Kurosu M. An antimycobacterial pleuromutilin analogue effective against dormant bacilli. Bioorg Med Chem. 2018;26:4787–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurosu M Vitamin K2 biosynthesis: drug targets for new antibacterials. Vitamin K2: Vital for Health and Wellbeing. 2017;281–295. [Google Scholar]
- 11.Mitachi K, Yun HG, Kurosu SM, Eslamimehr S, Lemieux MR, Klaic L, Clemons WM, Kurosu M. Novel FR-900493 analogues that inhibit the outgrowth of Clostridium difficile spores. ACS Omega. 2018;3:1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitachi K, Aleiwi BA, Schneider CM, Siricilla S, Kurosu M. Stereocontrolled total synthesis of muraymycin D1 having a dual mode of action against Mycobacterium tuberculosis. J Am Chem Soc. 2016;138:12975–12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitachi K, Siricilla S, Yang D, Kong Y, Skorupinska-Tudek K, Swiezewska E, Franzblau SG, Kurosu M. Fluorescence-based assay for polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) and identification of novel antimycobacterial WecA inhibitors. Anal Biochem. 2016;512:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siricilla S, Mitachi K, Skorupinska-Tudek K, Swiezewska E, Kurosu M. Biosynthesis of a water-soluble lipid I analogue and a convenient assay for translocase I. Anal Biochem. 2014;461:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. J Med Chem. 2012;55:3739–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurosu M, Crick DC. MenA is a promising drug target for developing novel lead molecules to combat Mycobacterium tuberculosis. Med Chem. 2009;5:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosu M, Narayanasamy P, Biswas K, Dhiman R, Crick DC. Discovery of 1,4-dihydroxy-2-naphthoate prenyltransferase inhibitors: New drug leads for multidrug-resistant Gram-positive pathogens. J Med. Chem. 2007;50:3973–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namouchi A, Cimino M, Favre-Rochex S, Charles P, Gicqul B. Phenotypic and genomic comparison of Mycobacterium aurum and surrogate model species to Mycobacterium tuberculosis: implications for drug. BMC Genomics. 2017;l18:530/1–530/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal P, Miryala S, Varshney U. Use of Mycobacterium smegmatis deficient in ADP-ribosyltransferase as surrogate for Mycobacterium tuberculosis in drug testing and mutation analysis. PLoS One. 2015;10:e0122076/1–e0122076/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaturvedi V, Dwivedi N, Tripathi RP, Sinha S. Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J Gen Appl Microbiol. 2007;53:333–337. [DOI] [PubMed] [Google Scholar]
- 21.Quan S, Venter H, Dabbs ER. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother. 1997;41:2456–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzblau SG, Witzig RS, Mclaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heifets L MIC as a quantitative measurement of the susceptibility of Mycobacterium avium strains to seven antituberculosis drugs. Antimicrob Agents Chemother. 1988;32:1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khanna A, Raj VS, Tarai B, Sood R, Pareek PK, Upadhyay DJ, Sharma P, Rattan A, Saini KS, Singh H. Emergence and molecular characterization of extensively drug-resistant Mycobacterium tuberculosis clinical isolates from the Delhi region in India. Antimicrob Agents Chemother. 2010;54:4789–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorrentino F, Gonzalez del RR, Zheng X, Matilla JP, Gomez PT, Hoyos MM, Herran MEP, Losana AM, Av-Gay Y. Development of an intracellular screen for new compounds able to inhibit Mycobacterium tuberculosis growth in human macrophages. Antimicrob Agents Chemother. 2016;60:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Yang J, Tan G, Liu H, Liu Y, Guo Y, Gao R, Wan B, Yu F. Drug resistance characteristics of Mycobacterium tuberculosis isolates from patients with tuberculosis to 12 antituberculous drugs in China. Front Cell Infect Microbiol. 2019;9:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelan J, Maitra A, Gupta A, Bhakta S, McNerney R, Coll F, Nair M, Pain A, Clark TG. The draft genome of Mycobacterium aurum, a potential model organism for investigating drugs against Mycobacterium tuberculosis and Mycobacterium leprae. Int J Mycobacteriol. 2015;4:207–216. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs MT, May RJ, Heider F, Reimers T, Sy SKB, Peloquin CA, Derendorf H. Mycobacterium tuberculosis Strains H37ra and H37rv have equivalent minimum inhibitory concentrations to most antituberculosis drugs. Int J Mycobacteriol. 2018;7:156–161. [DOI] [PubMed] [Google Scholar]
- 29.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues L, Villellas C, Bailo R, Viveiros M, Aínsa JA. Role of the mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ollinger J, Bailey MA, Moraski GC, Casey A, Florio S, Alling T, Miller MJ, Parish T. A dual read-out assay to evaluate the potency of compounds active against Mycobacterium tuberculosis. PLoS One. 2013;8:e60531/1–e60531/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchison DA, Zhang Y. Recent developments in the study of pyrazinamide: an update. Prog Resp Res. 2011;40(Antituberculosis Chemotherapy):32–43. [Google Scholar]
- 33.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52:790–795. [DOI] [PubMed] [Google Scholar]
- 34.Mitachi K, Kurosu SM, Eslamimehr S, Lemieux MR, Ishizaki Y, Clemons WM, Kurosu M. Semi-synthesis of an anticancer DPAGT1 inhibitor from a muraymycin biosynthetic intermediate. Org Lett. 2019;21:876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurosu M Structure-based drug discovery by targeting N-glycan biosynthesis, dolichyl-phosphate N-acetylglucosaminephosphotransferase. Future Med Chem. 2019;11:927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J Infect Dis. 2008;198: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu JH, Wang BW, Pan M, Zeng YN, Rego H, Javid B. Rifampicin can induce antibiotic tolerance in mycobacteria via paradoxical changes in rpoB transcription. Nature Comm. 2018;9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drobniewski FA, Wilson SM. The rapid diagnosis of isoniazid and rifampicin resistance in Mycobacterium tuberculosis - a molecular story. J Med Microbiol. 1998;47:189–196. [DOI] [PubMed] [Google Scholar]
- 39.Moure R, Espanol M, Tudo G, Vicente E, Coll P, Gonzalez-Martin J, Mick V, Salvado M, Alcaide, F. Characterization of the embB gene in Mycobacterium tuberculosis isolates from Barcelona and rapid detection of main mutations related to ethambutol resistance using a low-density DNA array. J Antimicrob Chemother. 2014;69:947–954. [DOI] [PubMed] [Google Scholar]
- 40.Hazbon MH, Bobadilla del VM, Guerrero MI, Varma-Basil M, Filliol I, Cavatore M, Colangeli R, Safi H, Billman-Jacobe H, Lavender C, Fyfe J, Garcia-Garcia L, Davidow A, Brimacombe M, Leon CI, Porras T, Bose M, Chaves F, Eisenach KD, Sifuentes-Osornio J, LA Ponce de, Cave MD, Alland D. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: A novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob Agents Chemother. 2005;49:3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM, Jacobs WR. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med. 1997;3:567–570. [DOI] [PubMed] [Google Scholar]
- 42.Sirgel FA, Tait M, Warren RM, Streicher EM, Boettger EC van Helden PD, Gey van Pittius NC, Coetzee G, Hoosain EY, Chabula-Nxiweni M, Hayes C, Victor TC, Trollip A. Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb Drug Resist. 2012;18:193–197. [DOI] [PubMed] [Google Scholar]
- 43.Fu LM, Shinnick TM. Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. J Infect. 2007;54:277–284. [DOI] [PubMed] [Google Scholar]
- 44.Ho YM, Sun YJ, Wong SY, Lee ASG. Contribution of dfrA and inhA mutations to the detection of isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2009;53:4010–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62:1220–1227. [DOI] [PubMed] [Google Scholar]
- 46.Vilchèze C, Jacobs WR. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol. 2007;61:35–50. [DOI] [PubMed] [Google Scholar]
- 47. www.ncbi.nlm.nih.gov.
- 48.Imai T, Watanabe K, Mikami Y, Yazawa K, Ando A, Nagata., Morisaki N, Hashimoto Y, Furihata K, Dabbs ER. Identification and characterization of a new intermediate in the ribosylative inactivation pathway of rifampin by Mycobacterium smegmatis. Microb Drug Resist. 1999;5:259–264. [DOI] [PubMed] [Google Scholar]
- 49.Kurosu M Cell wall biosynthesis and latency during tuberculosis infections In: Cirillo J, Kong Y. (eds) Tuberculosis Host-Pathogen Interactions. Springer, Cham: 2019;pp1–21. [Google Scholar]
- 50.Mazurek J, Ignatowicz L, Kallenius, Gunilla; Svenson, Stefan B.; Pawlowski, Andrzej; Hamasur, Beston. Divergent effects of mycobacterial cell wall glycolipids on maturation and function of human monocyte-derived dendritic cells. PLoS One. 2012;7:e42515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64: 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anuchin AM, Mulyukin AL, Suzina NE, Duda VI, El-Registan GI, Kaprelyants AS. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiol. 2009;155:1071–1079. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Hashizume H, Tomishige T, Nakamura I, Matsuba M, Fujiwara M, Kitamoto R, Hanaki E, Ohba Y, Matsumoto M. Delamanid kills dormant mycobacteria in vitro and in a guinea pig model of tuberculosis. Antimicrob Agents Chemother. 2017;61:e02402/1–e02402/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heifets L, Simon J, Pham V. Capreomycin is active against non-replicating M. tuberculosis. Annal Clin Microbiol Antimicrob. 2005;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fattorini L, Piccaro G, Mustazzolu A, Giannoni F. Targeting dormant bacilli to fight tuberculosis. Mediterr J Hematol Infect Dis. 2013;5:e2013072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munsiff SS, Kambili CA, Shama D. Rifapentine for the treatment of pulmonary tuberculosis. Clin Infect Dis. 2006; 43:1468–1475. [DOI] [PubMed] [Google Scholar]
- 57.Raghunandanan S, Jose L, Kumar RA. Dormant Mycobacterium tuberculosis converts isoniazid to the active drug in a Wayne’s model of dormancy. J Antibiot. 2018;71:939–949. [DOI] [PubMed] [Google Scholar]
- 58.Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chem.2008;61:323–331. [DOI] [PubMed] [Google Scholar]
- 59.de Steenwinkel JEM, de Knegt GJ, ten Kate MT, van Belkum A, Verbrugh HA, Kremer K, van Soolingen D, Bakker-Woudenberg IAJM. Time-kill kinetics of anti-tuberculosis drugs, and emergence of resistance, in relation to metabolic activity of Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65:2582–2589. [DOI] [PubMed] [Google Scholar]
- 60.Issar S Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16: 463–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung GAC, Aktar ZJS, Duncan K. High-throughput screen for detecting antimycobacterial agents. Antimicrob Agents Chemother. 1995;39:2235–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quan S, Venter H, Dabbs ER. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother. 1997;41:2456–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salfinger M, Heifets LB. Determination of pyrazinamide MICs for Mycobacterium tuberculosis at different pHs by the radiometric method. Antimicrob Agents Chemother. 1988;32:1002–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heifets L, Simon J, Pham V. Capreomycin is active against non-replicating M. tuberculosis. Annal Clin Microb Antimicrob. 2005;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filippini P, Iona E, Piccaro G, Peyron P, Neyrolles O, Fattorini L. Activity of drug combinations against dormant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2010;54: 2712–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iacobino A, Piccaro G, Giannoni F, Mustazzolu A, Fattorini L. Mycobacterium tuberculosis is selectively killed by rifampin and rifapentine in hypoxia at neutral pH. Antimicrob Agents Chemother. 2017;61: e02296–16/1–e02296–16/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.