Abstract
Rational/Objective
Computed tomography (CT) is the clinical gold-standard for high-resolution 3D visualization of cortical bone structures. However, ionizing radiation is of concern, particularly for pediatric patients. This study evaluates the feasibility of producing 3D human skull renderings using a novel bone-selective magnetic resonance imaging (MRI) technique.
Materials/Methods
A dual-radiofrequency (RF) pulse, dual-echo, 3D ultrashort echo time (UTE) sequence was applied for scanning of a cadaver skull and five healthy adult subjects. Scans were each completed within 6 minutes. Semi-automatic segmentation of bone voxels was performed using ITK-SNAP software, leading to 3D renderings of the skulls. For comparison, thin-slice head CT scans were performed. Mimics software was used to measure eight anatomic distances from 3D renderings. Lin’s Concordance Correlation test was applied to assess agreement between measurements from MR-based and CT-based 3D skull renderings.
Results
The 3D rendered MR images depict most craniofacial features (e.g., zygomatic arch), although some voxels were erroneously included or excluded in the renderings. MR-based measurements differed from CT-based measurements by mean percent difference ranging from 2.3%−5.0%. Lin’s Concordance Correlation Coefficients for MR-based vs CT-based measurements ranged from 0.998–1.000.
Conclusion
The proposed dual-RF dual-echo 3D UTE imaging technique produces high-resolution bone-specific images within a clinically feasible imaging time, leading to clear visualization of craniofacial skeletal structures. Concordance coefficients suggest good reliability of the method compared to CT. The method is currently limited by time and manual input necessary for segmentation correction. Further investigation is needed for more accurate 3D renderings and for scanning of pediatric patients.
Keywords: MRI, CT, craniofacial
Introduction
Multiple studies have raised concern of long-term health risks from computed tomography (CT) associated ionizing radiation exposure, especially in infants and young children who may require multiple scans in a lifetime1–10. Yet CT evaluation offers the advantage of allowing the generation of 3D reconstructions with high spatial resolution. Exploring the potential of magnetic resonance (MR) imaging for high resolution imaging of craniofacial or other bony structures would have appeal in reducing pediatric exposure to radiation. This technique would also have appeal in allowing the acquisition of skeletal and soft tissue images in a single scan session. This would be particularly useful for common indications, such as head trauma, knee or shoulder pain, where the etiology is unclear.
Conventional MR imaging is a non-radiative modality for imaging the head, but is primarily used to evaluate soft tissues with high water/proton content. Bone is difficult to visualize due to its very short T2 relaxation times (~300μs – 3ms) and relatively low proton density (~20% by volume), making it nearly indistinguishable from air. Images obtained using routine pulse sequences are unreliable for identifying cranial sutures in infants11. Furthermore, the poor differentiation between air and bone makes it difficult to create 3D reconstructions of the skull.
Eley et al developed a protocol for 3D gradient-echo MR imaging performed at very low flip angle to obtain images with proton-density-weighted soft-tissue contrast12. 3D surface-rendered images created after eliminating soft-tissue signal were found to be comparable to CT for anatomical measurements of skull models12, and for the identification of normal and prematurely fused cranial sutures13. While the Eley method allowed discrimination between bone and soft-tissue, it encountered problems with separating the bone signals from the signals of surrounding structures containing air (e.g. sinuses). This was because bone appeared at near background intensity (i.e. ‘black’) due to very short T2 relaxation times and relatively low proton density. Processing of images to produce 3D models required time-intensive manual segmentation due to ambiguous pixel values.
Recent developments in solid-state MRI by ultrashort echo time (UTE)14,15 or zero echo time (ZTE)16,17 methods have enabled detection and quantification of very short T2 (ranging from tens of microseconds to a few milliseconds) species of the human body such as cortical bone water18–20 and phosphorus in bone mineral21,22. Wiesinger et al developed a protocol using ZTE which allowed high resolution imaging of the skull, with good contrast between air, soft-tissue, and bone23.
In both UTE and ZTE imaging, signal reception begins within tens of microseconds after excitation on modern clinical scanners. This allows the capture of signals from submillisecond T2 signals, such as those from protons in water bound to the bone’s organic matrix. However, components with long T2 signals (i.e. water in soft tissue or residing in the bone pore spaces) would exhibit high signal intensities and obscure the short T2 signals.
One method to enhance visualization of short T2 signals is to acquire two or more images at different TE’s, and filter out long-T2 components during post-processing. Dual-echo subtraction has been shown to be an efficient method for optimal short T2 contrast24–26. A recent method further enhances discrimination of bone signals by also varying the duration of radiofrequency (RF) pulses27. Bone water proton magnetization exhibits a substantial level of signal decay during the application of longer RF pulses due to its very short T2 relaxation time, while soft-tissue retains nearly the same level of signal regardless of the RF pulse lengths. This permits an alternative means to differentiate between bone and soft tissues.
A rapid bone MRI method involving a 3D DUal-RAdiofrequency aNd Dual-Echo (DURANDE) UTE pulse sequence was developed for obtaining 3D renderings of the human skull. The development and technical details of this rapid pulse sequence are described in detail in a prior publication28.
The clinical translatability of the bone-selective MR method was explored, with comparison to the current gold-standard of thin-slice CT imaging.
Methods
Imaging technique
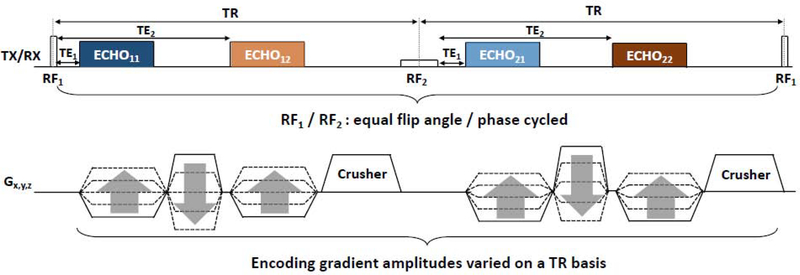
Figure 1 shows the diagram of a dual-RF, dual-echo 3D UTE pulse sequence. Two RF pulses differing in duration and amplitude are alternately applied in successive repetition time (TR) along the pulse train. Within each TR, two echoes are acquired. Acquisition of the first echo starts at the ramp-up of the encoding gradient (TE1), allowing for capture of signals with very short lifetimes (bone), while that of the second starts after a longer delay (TE2). In total, four echoes are obtained: ECHO11 (RF1TE1), ECHO12 (RF1TE2), ECHO21 (RF2TE1), and ECHO22 (RF2TE2). During reconstruction, ECHO11 is combined with ECHO21 (Image 1) and ECHO12 is combined with ECHO22 (Image 2). Image 2 is then subtracted from Image 1 to yield the final bone-selective image.
Figure 1. Pulse sequence diagram.
Diagram of the dual-RF, dual-echo 3D UTE pulse sequence, in which RF1 (short ~ 40μs) and RF2 (long ~ 520μs) are alternately played out and four independent signals are produced: ECHO11, ECHO12, ECHO21, and ECHO22.
Imaging parameters: TR/TE1/TE2 = 7/0.06/2.46 ms, RF1/RF2 durations = 40/520 μs, flip angle = 12° (for both RFs), matrix size = 256, field-of-view = 280mm3, voxel size = 1.1 mm isotropic, number of radial spokes = 25,000, and scan time = 6 min.
Cadaver skull imaging
Imaging protocol
The outer-lying soft tissue and brain tissue were removed from a fresh, disarticulated human cadaver skull (Science Care) prior to imaging. The pulse sequence described above was applied at 3 T field strength (Siemens Prisma, Erlangen, Germany). The skull was placed in a direct horizontal position conventionally used for imaging of the head. For comparison, a CT scan (Siemens Somatom Definition, Erlangen, Germany) was also performed of the human cadaver skull with 1 mm slice thickness.
3D rendering
Semi-automatic segmentation of bone voxels was performed using the classification feature of ITK-SNAP29. The user draws examples of tissue classes in the image, using a paint brush tool to label each class example with a corresponding color. A machine learning algorithm uses these examples to assign classifications to the rest of the image. In this study, the user drew examples of bone tissue, soft tissue and air. After segmentation, the 3D model of the skull was generated using the ITK-SNAP software. The initial 3D model was reviewed for errors in segmentation. For example, an air voxel may be erroneously classified by the software as a bone voxel. Manual correction was required in all segmentations. Manual correction required the user to use the paint brush tool to re-label the erroneous voxels and repeat the machine learning algorithm to re-classify the rest of the image. The final 3D model exported as an STL file.
Segmentation of the CT scan was performed using preset bone CT thresholds on the Mimics software (Materialise®, Ghent, Belgium), the standard protocol at the senior author’s institution. After segmentation, the 3D model was automatically generated using the Mimics software and exported as an STL file.
Biometric accuracy assessment
Eight anatomic distances were measured in both CT- and MRI-based 3D renderings of the human cadaver skull. The STL files of the 3D renderings were uploaded to 3-Matic (Materialise®, Ghent, Belgium) software and anatomic distances were measured using the ruler tool. These distances were compared with those directly measured on the cadaver skull, with calipers (resolution 1 mm). Each distance was measured 20 times by a single assessor and the mean value calculated.
From AP view
Maximum craniocaudal aperture of the right orbit.
Maximum craniocaudal aperture of the left orbit.
Height of mandible from menton to inferior alveolar point
Maximum height of piriform aperture.
Distance between lateral most aspect of zygomatic arches
Maximum maxilla width along superior alveolar ridge
From axial view
Maximum cranial length
Maximum cranial width
These anatomic measures were selected as they are useful for pre-operative planning for craniofacial procedures, including during virtual surgical planning sessions that utilize 3D CT scan renderings.
Lin’s Concordance Correlation test was applied to assess agreement between mean measurements obtained from MR-based and CT based 3D skull renderings, cadaver and MR-based rendering, and cadaver and CT-based rendering.
This experiment was repeated after a two week time interval to provide a second sample. Between scan sessions, the skull was stored in a −34C freezer designated for fresh cadaver specimens.
Healthy Adult Subject Skull Imaging
IRB approval was obtained to recruit male and female adult volunteers age 18 or older, who are not receiving care as a patient, to complete a head CT scan and a bone-selective MRI scan of the head. The same imaging protocol described for the cadaver skull imaging was used. Subjects with craniofacial anomalies, prior craniofacial surgery, implants made from ferromagnetic materials, or needing sedation were ineligible for the recruitment. No contrast was used for any subject. Subjects did not undergo a full conventional head MRI scan.
The CT scan followed the senior author’s institution craniofacial imaging protocol, with a 0.75mm slice thickness but lower dose radiation. A single scanner (Siemens Somatom Definition, Erlangen, Germany) was used for all scans.
3D rendering of the skull from MR scans and CT scans, as well as comparison of craniometric measurements, were performed as previously described.
Results
Cadaver Skull Imaging
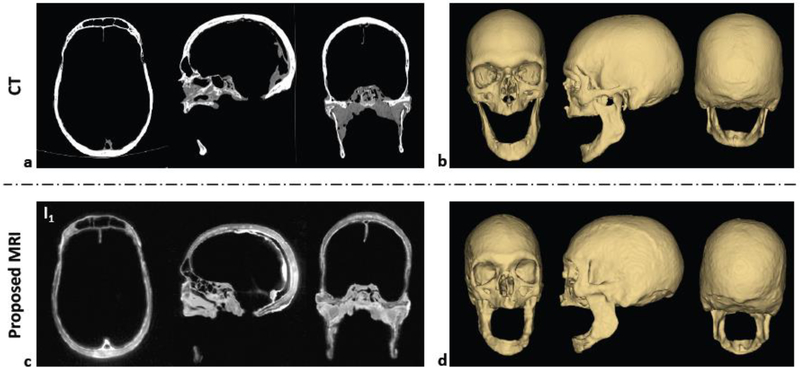
Figure 2 compares cadaver skull images obtained from CT and the proposed MR method, along with corresponding 3D renderings. The subtraction of Image 2 (ECHO12 combined with ECHO22) from Image 1 (ECHO11 combined with ECHO21) yielded an image with enhanced bone contrast. Compared to CT, the 3D rendered images maintain most features over the entire head (e.g., zygomatic arch), except for the appearance of some artifacts in the mandibular region.
Figure 2. Cadaver skull images.
Comparison of ex vivo cadaver human skull images between CT (a, b) and the proposed MRI method (c, d). Magnitude images in three orthogonal planes (a, c) are shown along with 3D renderings for three different views (b, d).
Table 1 presents the mean anatomic measurements obtained from MR-based and CT-based 3D skull renderings, as well as direct measurements from the cadaver skull.
Table 1.
Mean anatomic distances, measured directly from cadaver skull, and from MR and CT imaging.
| Mean Measurement (cm±SD) | Modality | ||
|---|---|---|---|
| MR | CT | Cadaver | |
| Cranial length | 19.9±0.2 | 19.4±0.1 | 18.6±0.1 |
| Cranial width | 13.9±0.1 | 13.9±0.1 | 13.2±0.1 |
| L orbit height | 3.7±0.1 | 3.5±0.1 | 3.5±0.1 |
| R orbit height | 3.6±0.1 | 3.4±0.1 | 3.4±0.1 |
| Piriform aperture | 3.3±0.1 | 3.7±0.0 | 3.6±0.1 |
| Inter-zygomatic arch width | 12.5±0.1 | 12.2±0.1 | 12.3 0.1 |
| Mandibular height | 2.6±0.1 | 2.7±0.1 | 27±0.1 |
| Maxilla width | 5.0±0.1 | 5.0±0.1 | 4.9±0.1 |
Table 2 presents the mean absolute and percent differences when comparing the three modalities.
Table 2.
Summary of mean absolute differences and mean absolute percent differences between the three measurement modalities.
| Comparison | Mean Difference (cm±SD) | Mean Percent Difference (%±SD) |
|---|---|---|
| MR vs CT | 0.2± 0.2 | 4.5±3.5 |
| MR vs Cadaver | 0.4±0.4 | 5.5±2.6 |
| CT vs Cadaver | 0.2±0.3 | 2.0±1.9 |
Table 3 presents the Lin’s Concordance Correlation Coefficients when comparing the three modalities, for both the initial scan (Sample 1) and the scan completed two weeks later (Sample 2).
Table 3.
Summary of Sample 1 and Sample 2 Lin‟s Concordance Correlation Coefficients
| Comparison | Sample 1 Lin’s Concordance Correlation Coefficient | 95% CI | Sample 2 Lin’s Concordance Correlation Coefficient | 95% CI |
|---|---|---|---|---|
| MR vs CT | 0.999 | 0.997–1.000 | 0.992 | 0.986–0.999 |
| MR vs Cadaver | 0.996 | 0.991–1.000 | 0.989 | 0.980–0.999 |
| CT vs Cadaver | 0.998 | 0.995–1.000 | 0.999 | 0.997–1.001 |
Healthy Adult Subject Skull Imaging
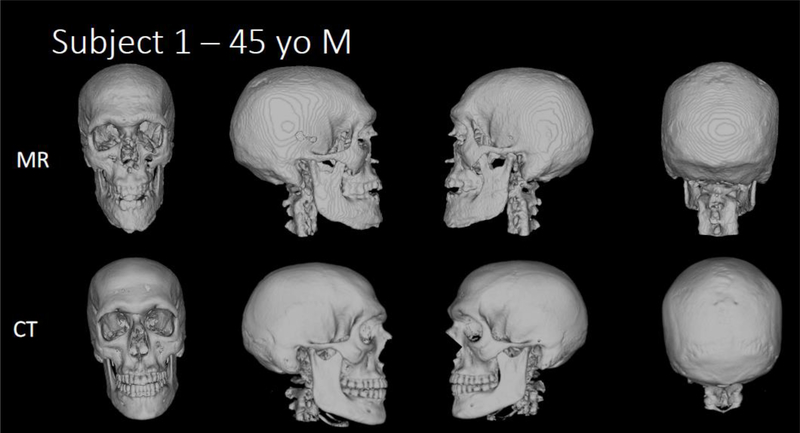
Five healthy adult subjects were recruited for this study. Figures 3a–e compare the 3D renderings of the MR and CT scans of Subjects 1–5. Table 4 summarizes the mean percent differences and Lin’s Concordance correlation coefficients for the five subjects.
Figure 3a-e b. Human subject skull images.
3D renderings of the MR and CT scans of Subjects 1–5.
Table 4.
Mean percent differences and Lin’s Concordance Correlation Coefficients of craniometric measurements for Subjects 1–5.
| Subject | Mean percent difference (%±SD) | Lin’s Concordance Correlation Coefficient | 95% CI |
|---|---|---|---|
| 1 (45yo M) | 2.8±3.2 | 0.999 | 0.998–1.000 |
| 2 (26yo F) | 2.9±3.9 | 1.000 | 0.999–1.000 |
| 3 (27yo M) | 5.0±4.9 | 0.998 | 0.995–1.001 |
| 4 (27yo F) | 2.3±1.7 | 1.000 | 0.999–1.000 |
| 5 (35yo M) | 3.2±4.0 | 0.998 | 0.996–−1.001 |
Discussion
DURANDE UTE, in combination with the bone-selective image reconstruction, enables high-resolution (~1.1 mm) skull imaging of the whole head in clinically feasible imaging time of six minutes. The dual-RF based UTE bone imaging method enhances differentiation of cortical bone from long T2 species (such as soft tissue). The resolution and differentiation of the cortical bone enabled semi-automatic segmentation of MR images and subsequent 3D rendering of the skull. Craniometric measurement comparisons suggested high concordance (concordance coefficient >0.990) of the bone-selective MR method in comparison to the current clinical standard of thin-slice 3D head CT.
In the cadaver skull study, comparison of eight anatomic distance measurements obtained from MR and CT images yielded a mean absolute difference of 2mm and percent difference of 4.5%. The concordance coefficients of 0.999 (Sample 1) and 0.992 (Sample 2) correspond to a substantial strength of agreement between MR and CT30. These results suggest good reliability of the MR method when compared to CT. However, CT had a greater concordance to direct cadaver measurements (Sample 1: 0.999, Sample 2: 0.999) than MR (Sample 1: 0.996, Sample 2: 0.989). We note that MR-based measurements had decreased correlation with CT-based and cadaver measurements in Sample 2 compared to in Sample 1. This may be because the freezing and thawing of the skull decreased the bone water content, yielding weaker bone signals and lower MR image quality. Due to resource limitations, we were only able to scan one cadaver at two time points. We acknowledge that scanning the cadaver head multiple times would allow more thorough evaluation of the precision of the method.
The proposed MR sequence also produced bone-specified images of healthy adult subject skulls, with sufficiently high resolution for 3D rendering. Eight anatomic distance measurements obtained from MR and CT images yielded mean percent differences ranging from 2.3–5.0%, and concordance coefficients ranging from 0.998 to 1.000, corresponding to a substantial strength of agreement. These results suggest that the method has good reliability for adult skull imaging when compared to CT. Notably, the method was reliable for imaging of human adult subject skulls despite the presence of significantly more soft tissue than the pre-stripped human cadaver skull. Our data is currently limited to that of two cadaver skull scans and five adult subjects. Recruitment of additional subjects would allow the analysis to be performed across subjects with a concordance coefficient for each anatomic measure, and this will occur in the next phase of the study.
Lambdoid sutures can be observed in MR-based 3D renderings of all five adult subject skulls, most prominently in Subject 4 (Figure 3d, column 4). However for all subjects, the sutures are more defined in the CT-based 3D renderings. The ability to visualize the sutures has clinical significance particularly for the diagnosis of craniosynostosis in infants and young children. These scans suggest that the bone-selective MR sequence and subsequent processing method still needs to be optimized to meet the current gold standard for visualization of cranial sutures (thin-slice head CT scans). The lower quality of 3D MR renderings is likely in part due to the lower in-plane resolution of MRI (1.1mm) compared to CT (0.30–0.51mm)
Segmentation of MR images was performed in a semi-automatic fashion with ITK-SNAP. This required the user first to identify bone vs air vs soft tissue voxels in order to train the machine learning algorithm. In both the cadaver skull and adult skull studies, errors in segmentation required manual correction. It was difficult to differentiate air, soft tissue and bone voxels in some areas (around zygomatic arch, sinuses and facial bones). These were areas where the bone was particularly thin, and where there was significant interface between air, soft tissue and bone structures. In several of the adult human subjects, the coronoid process on either or both sides appeared to extend above the zygomatic arch in the MR-based 3D renderings. This is likely because of the similar voxel intensities of the bone and fibrous capsule, making classification using ITK-SNAP software difficult. Thus, during segmentation, some voxels were erroneously included or excluded in the renderings of the bony skull by the ITK-SNAP software. Some obvious errors could be manually corrected by the user by re-classifying the voxels. Due to the limitation of the software the authors are unable to provide the exact number of voxels requiring reclassification per subject, and acknowledge that this would be a valuable way to quantify error. Furthermore, more subtle errors remained, especially in areas where the bone was very thin, causing partial volume mixing. Thus, semi-automatic segmentation of MR scans can be time-intensive (requiring up to 20 minutes per subject) and user-dependent. These are notable limitations of the proposed method, especially when compared to the automatic segmentation capability for CT scans.
Another source of error arises from the anatomical points used for measuring distances. These may have differed between MR-based renderings, CT-based renderings and the cadaver skull. For example, a difference found in the piriform aperture height may well be due to an actual discrepancy in the piriform aperture size across the different skull models. Or, the difference may be because the two anatomical points chosen to measure the height on the MR-based skull model did not correspond exactly to those chosen for the CT-based skull or the actual cadaver skull. Each anatomic distance was measured twenty times and the mean value was calculated in order to account for the random error when selecting anatomical points to measure distances. However, a systematic error of measuring height from non-corresponding anatomical points may have occurred. For example, the user may have consistently used anatomical points on the MR-based skull model that were lateral to the anatomical points used on the CT-based skull model. Calculating mean values from repeated measurements would not have accounted for this systematic error.
To improve segmentation accuracy and automation, additional adult subjects will be recruited in the next phase of the study. The scans will be used as a dataset for multi-atlas segmentation (MAS). MAS utilizes a dataset of training images (“atlases”, previously manually labeled by an expert) to automatically label objects in a target image31,32.
Until this point, the sequence has not been applied to younger pediatric skulls. Recruiting patients of ages ranging from infancy to early adolescence would provide valuable information regarding how potential changes in bone structure in younger pediatric skulls may affect image acquisition. By 6 months, the single cortical layer in the newborn skull already begins to differentiate to the mature skull structure of a cortical layer sandwiched between cancellous bony layers33. Furthermore, the actual bone mineral content of infant skulls does not differ significantly from that of mature skulls34. Thus, it is not anticipated that the signal produced from school-age pediatric skulls will be different enough to necessitate changes in the image acquisition protocol. However, this assumption may be wrong. The smaller head size of younger pediatric patients may also contribute to lower relative resolution at a given voxel size. Adjustments to the pulse sequence or bone segmentation algorithm may be required.
The scans of adult human subjects allowed for the acquisition of a high quality image with little to no motion artifact. Very young patients (infants to age 6) would be sedated and would presumably also have little motion artifact. But motion artifact is more likely for scans of younger children who do not need sedation but may find it difficult to hold still for extended period of time. While the scan time (6 minutes) is relatively short compared to conventional MRI, it is possible that some children around ages 7–11 will still exhibit small movements while in the scanner, decreasing quality of images acquired. Therefore, recruiting pediatric patients from a wider age range would provide valuable information regarding the clinical translatability of the MR imaging method. To address the issue of undesired subject movement during a scan, a method to correct the issue retrospectively will be investigated. Briefly, multiple low-resolution subset images are obtained by using a slightly modified acquisition view order35. Then, motion-related inconsistencies among those images are corrected in each subset k-space (an image acquisition space representing spatial frequencies of the MR image). Finally, all k-space data are combined to produce a single high-resolution motion-corrected image.
Advances in CT 3D craniofacial rendering has decreased the effective dose of radiation (0.2–2mSV)36, and MR imaging time does often require more frequent sedation/anesthesia, which is also of increasing concern amongst pediatric practitioners. However, it would be valuable to have another radiologic option for bone imaging, in particular for young patients who would also require sedation for CT, or who are already clinically indicated for MRI of the brain. These include pediatric patients requiring evaluation for trauma (including non-accidental trauma) or for lytic skull lesions in suspected cases of Langerhans Cell Histiocytosis.
Lastly, the new MR technique could be applied to imaging of extremities, and would be particularly useful for evaluation of knee and shoulder problems. An additional area of further research is the comparison of CT and MR for detection of fractures. The study would include the generation of fractures in a cadaver skull, which is then imaged with CT and MRI. The ability of CT versus MRI to detect fractures of varying degrees (e.g. 2 mm displaced fracture, vs. 1 mm, vs. non-displaced) could be compared.
Conclusion
The results of this study suggest the feasibility of the presented bone-selective MRI technique for whole-skull imaging at high spatial resolution in a clinically practical scan time. Further investigation is required to optimize post-processing for more realistic 3D renderings and more accurate anatomic measurements, as well as for optimizing for imaging pediatric patients.
Acknowledgements
Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number TL1TR001880. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- CT
Computed tomography
- MR
Magnetic Resonance
- RF
Radiofrequency
- UTE
Ultrashort echo time
- ZTE
Zero echo time
Footnotes
Conflicts of Interest
The authors report no relevant financial disclosures related to this current work.
Disclosure: None of the authors has a financial interest in any of the products, devices, or drugs mentioned in this manuscript.
IRB:
This study has been approved by the Institutional Review Board for research involving human subjects at the Children’s Hospital of Philadelphia.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krille L, Dreger S, Schindel R, et al. Risk of cancer incidence before the age of 15~years after exposure to ionising radiation from computed tomography: results from a German cohort study. Radiat Env Bioph. 2015;54(1):1–12. doi: 10.1007/s00411-014-0580-3 [DOI] [PubMed] [Google Scholar]
- 2.Rebecca S-B, Lipson J, Marcus R, et al. Radiation Dose Associated With Common Computed Tomography Examinations and the Associated Lifetime Attributable Risk of Cancer. Arch Intern Med. 2009;169(22):2078–2086. doi: 10.1001/archinternmed.2009.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathews JD, Forsythe A V, Brady Z, et al. Cancer risk in 680 000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million Australians. Bmj. 2013;346(May21 1):f2360. doi: 10.1136/bmj.f2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miglioretti DL, Johnson E, Williams A, et al. The Use of Computed Tomography in Pediatrics and the Associated Radiation Exposure and Estimated Cancer Risk. Jama Pediatr. 2013;167(8):700–707. doi: 10.1001/jamapediatrics.2013.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhns LR, Oliver WJ, Christodoulou E, Goodsitt MM. The Predicted Increased Cancer Risk Associated With a Single Computed Tomography Examination for Calculus Detection in Pediatric Patients Compared With the Natural Cancer Incidence. Pediatr Emerg Care. 2011;27(4):345. doi: 10.1097/PEC.0b013e3182132016 [DOI] [PubMed] [Google Scholar]
- 6.Kmietowicz Z Computed tomography in childhood and adolescence is associated with small increased risk of cancer. Bmj. 2013;346(May22 16):f3348. doi: 10.1136/bmj.f3348 [DOI] [PubMed] [Google Scholar]
- 7.Fazel R, Krumholz HM, Wang Y, et al. Exposure to {Low-Dose} Ionizing Radiation from Medical Imaging Procedures. New Engl J Med. 2009;361(9):849–857. doi: 10.1056/NEJMoa0901249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de González A, Mahesh M, Kim K-P, et al. Projected Cancer Risks From Computed Tomographic Scans Performed in the United States in 2007. Arch Intern Med. 2009;169(22):2071–2077. doi: 10.1001/archinternmed.2009.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redberg RF. Cancer Risks and Radiation Exposure From Computed Tomographic Scans: How Can We Be Sure That the Benefits Outweigh the Risks? Arch Intern Med. 2009;169(22):2049–2050. doi: 10.1001/archinternmed.2009.453 [DOI] [PubMed] [Google Scholar]
- 10.Pearce MS, Salotti JA, Little MP, et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: A retrospective cohort study. Lancet. 2012;380(9840):499–505. doi: 10.1016/S0140-6736(12)60815-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eley KA, Sheerin F, Taylor N, R W-SS, Golding SJ. Identification of normal cranial sutures in infants on routine magnetic resonance imaging. 2013;24(1):317–320. [DOI] [PubMed] [Google Scholar]
- 12.Eley KA, Mcintyre AG, Watt-Smith SR, Golding SJ. “Black bone” MRI: A partial flip angle technique for radiation reduction in craniofacial imaging. Br J Radiol. 2012;85(1011):272–278. doi: 10.1259/bjr/95110289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eley KA, Watt-Smith SR, Sheerin F, Golding SJ. “Black Bone” MRI: a potential alternative to CT with three-dimensional reconstruction of the craniofacial skeleton in the diagnosis of craniosynostosis. Eur Radiol. 2014:2417–2426. doi: 10.1007/s00330-014-3286-7 [DOI] [PubMed] [Google Scholar]
- 14.Bergin CJ, Pauly JM, Macovski A. Lung parenchyma: projection reconstruction {MR} imaging. Radiology. 1991;179(3):777–781. doi: 10.1148/radiology.179.3.2027991 [DOI] [PubMed] [Google Scholar]
- 15.Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic Resonance: An Introduction to Ultrashort {TE} {(UTE)} Imaging. J Comput Assist Tomo. 2003;27(6):825. doi: 10.1097/00004728-200311000-00001 [DOI] [PubMed] [Google Scholar]
- 16.Madio D, Lowe I. Ultra-fast imaging using low flip angles and fids. Magn Reson Med. 1995;34(4):525–529. doi: 10.1002/mrm.1910340407 [DOI] [PubMed] [Google Scholar]
- 17.Weiger M, Pruessmann K, Hennel F. {MRI} with zero echo time: Hard versus sweep pulse excitation. Magn Reson Med. 2011;66(2):379–389. doi: 10.1002/mrm.22799 [DOI] [PubMed] [Google Scholar]
- 18.Techawiboonwong A, Song H, Wehrli F. In vivo {MRI} of submillisecond T2 species with two-dimensional and three-dimensional radial sequences and applications to the measurement of cortical bone water. Nmr Biomed. 2008;21(1):59–70. doi: 10.1002/nbm.1179 [DOI] [PubMed] [Google Scholar]
- 19.Du J, Carl M, Bydder M, Takahashi A, Chung C, Bydder G. Qualitative and quantitative ultrashort echo time {(UTE)} imaging of cortical bone. J Magn Reson. 2010;207(2):304–311. doi: 10.1016/j.jmr.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Hrovat MI, Ackerman JL, et al. Bone matrix imaged in vivo by water-and fat-suppressed proton projection {MRI} {(WASPI)} of animal and human subjects. J Magn Reson Imaging. 2010;31(4):954–963. doi: 10.1002/jmri.22130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Chesler D, Glimcher M, et al. Multinuclear solid-state three-dimensional {MRI} of bone and synthetic calcium phosphates. Proc Natl Acad Sci. 1999;96(4):1574–1578. doi: 10.1073/pnas.96.4.1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seifert A, Li C, Rajapakse C, et al. Bone mineral {31P} and matrix-bound water densities measured by solid-state {31P} and {1H} {MRI}. Nmr Biomed. 2014;27(7):739–748. doi: 10.1002/nbm.3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesinger F, Sacolick LI, Menini A, et al. Zero TE MR bone imaging in the head. Magn Reson Med. 2016;75(1):107–114. doi: 10.1002/mrm.25545 [DOI] [PubMed] [Google Scholar]
- 24.Du J, Diaz E, Carl M, Bae W, Chung C, Bydder G. Ultrashort echo time imaging with bicomponent analysis. Magn Reson Med. 2012;67(3):645–649. doi: 10.1002/mrm.23047 [DOI] [PubMed] [Google Scholar]
- 25.Li C, Magland JF, Rad H, Song H, Wehrli FW. Comparison of optimized soft-tissue suppression schemes for ultrashort echo time {MRI}. Magn Reson Med. 2012;68(3):680–689. doi: 10.1002/mrm.23267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmer J, Blume U, Börnert P. Selective {3D} ultrashort {TE} imaging: comparison of “dual-echo” acquisition and magnetization preparation for improving {short-T2} contrast. Magn Reson Mater Phys Biol Med. 2007;20(2):83. doi: 10.1007/s10334-007-0070-6 [DOI] [PubMed] [Google Scholar]
- 27.Johnson EM, Vyas U, Ghanouni P, Pauly K, Pauly JM. Improved cortical bone specificity in {UTE} {MR} Imaging. Magn Reson Med. 2017;77(2):684–695. doi: 10.1002/mrm.26160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Zhao X, Song HK, Zhang R, Bartlett SP, Wehrli FW. Rapid dual-RF, dual-echo, 3D ultrashort echo time craniofacial imaging: A feasibility study. Magn Reson Med. 2019. doi: 10.1002/mrm.27625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yushkevich PA, Gao Y, Gerig G. {ITK-SNAP:} An interactive tool for semi-automatic segmentation of multi-modality biomedical images. Conf Proc {IEEE} Eng Med Biol Soc. 2016;2016:3342–3345. doi: 10.1109/EMBC.2016.7591443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin LI, McBride G, Bland JM, Altman DG. A proposal for strength-of-agreement criteria for Lin’s Concordance Correlation Coefficient. NIWA Client Rep. 2005;45(1):307–310. doi: 10.2307/2532051 [DOI] [Google Scholar]
- 31.Wang H, Suh J, Das S, Pluta J, Craige C, Yushkevich P. {Multi-Atlas} Segmentation with Joint Label Fusion. Ieee T Pattern Anal. 2013;35(3):611–623. doi: 10.1109/TPAMI.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iglesias J, Sabuncu M. Multi-atlas segmentation of biomedical images: A survey. Med Image Anal. 2015;24(1):205–219. doi: 10.1016/j.media.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margulies SS, Thibault KL. Infant Skull and Suture Properties: Measurements and Implications for Mechanisms of Pediatric Brain Injury. J Biomech Eng. 2000;122(4):364–371. doi: 10.1115/1.1287160 [DOI] [PubMed] [Google Scholar]
- 34.Kriewall TJ K MG, Tsai A. Bending properties and ash content of fetal cranial bone. J Biomech. 1981;14(2):73–79. doi: 10.1016/0021-9290(81)90166-4 [DOI] [PubMed] [Google Scholar]
- 35.Chan RW, Ramsay EA, Cunningham CH, Plewes DB. Temporal stability of adaptive 3D radial MRI using multidimensional golden means. Magn Reson Med. 2009. doi: 10.1002/mrm.21837 [DOI] [PubMed] [Google Scholar]
- 36.Kim HJ, Roh HG, Lee IW. Craniosynostosis: Updates in radiologic diagnosis. J Korean Neurosurg Soc. 2016. doi: 10.3340/jkns.2016.59.3.219 [DOI] [PMC free article] [PubMed] [Google Scholar]