Abstract
Peroxiredoxin 6 (PRDX6) has been associated with tumor progression and cancer metastasis. Its acting on phospholipid hydroperoxides and its phospholipase-A2 activity are unique among the peroxiredoxin family and add complexity to its action mechanisms. As a first step towards the study of PRDX6 involvement in cancer, we have constructed a human hepatocarcinoma HepG2PRDX6-/- cell line using the CRISPR/Cas9 technique and have characterized the cellular response to lack of PRDX6.
Applying quantitative global and redox proteomics, flow cytometry, in vivo extracellular flow analysis, Western blot and electron microscopy, we have detected diminished respiratory capacity, downregulation of mitochondrial proteins and altered mitochondrial morphology. Autophagic vesicles were abundant while the unfolded protein response (UPR), HIF1A and NRF2 transcription factors were not activated, despite increased levels of p62/SQSTM1 and reactive oxygen species (ROS). Insulin receptor (INSR), 3-phosphoinositide-dependent protein kinase 1 (PDPK1), uptake of glucose and hexokinase-2 (HK2) decreased markedly while nucleotide biosynthesis, lipogenesis and synthesis of long chain polyunsaturated fatty acids (LC-PUFA) increased. 254 Cys-peptides belonging to 202 proteins underwent significant redox changes. PRDX6 knockout had an antiproliferative effect due to cell cycle arrest at G2/M transition, without signs of apoptosis.
Loss of PLA2 may affect the levels of specific lipids altering lipid signaling pathways, while loss of peroxidase activity could induce redox changes at critical sensitive cysteine residues in key proteins. Oxidation of specific cysteines in Proliferating Cell Nuclear Antigen (PCNA) could interfere with entry into mitosis. The GSH/Glutaredoxin system was downregulated likely contributing to these redox changes. Altogether the data demonstrate that loss of PRDX6 slows down cell division and alters metabolism and mitochondrial function, so that cell survival depends on glycolysis to lactate for ATP production and on AMPK-independent autophagy to obtain building blocks for biosynthesis. PRDX6 is an important link in the chain of elements connecting redox homeostasis and proliferation.
Keywords: Peroxiredoxin 6, CRISPR-Cas9, Proteomics, Redox proteome, Cell cycle, Mitochondria, Lipokines, NRF2, PCNA, Glucose metabolism, Carbohydrate metabolism
Highlights
-
•
A CRISPR-Cas9 based PRDX6 KO human cell line is characterized for the first time.
-
•
Loss of PRDX6 causes mitochondrial dysfunction, autophagy and slow growth rate.
-
•
Glucose uptake and HK2 decrease; nucleotide biosynthesis and lipogenesis increase.
-
•
Oxidation of PCNA Cys residues could be responsible for cell cycle arrest at G2/M.
1. Introduction
Peroxiredoxin-6 (PRDX6), the “non-selenium glutathione peroxidase”, is the only member of “1-Cys” human peroxiredoxin family (Prdx). In contrast to the other members of the Prdx family, PRDX6 does not possess a “resolving Cys” and uses glutathione as a primary physiological reductant together with Pi isoform of glutathione-S-transferase (GST-π) instead of thioredoxin [[1], [2], [3]]. PRDX6 also differs from the rest of Prdx for its multiple functions as it also presents Ca2+-independent phospholipase A2 (iPLA2) activity [4] and lysophosphatidylcholine acyl transferase (LPCAT) activity [5], being the only Prdx acting on phospholipid hydroperoxides. PRDX6 is located in the cytoplasm but after its phosphorylation can be targeted to acidic organelles [6]. It can also translocate to damaged mitochondrial membranes to regulate mitophagy by suppressing ROS [7] and to the plasma membrane to activate NADPH oxidase (NOX1, NOX2) activity [8].
PRDX1 and PRDX6 are the only members of the Prdx family with a binding site for the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), the master regulator of the cellular antioxidant response, in their gene promoters [9]. Besides, PRDX6 stands out as the only member associated with phospholipid turnover and cell membrane repair [10].
PRDX6 has an important role in cell physiology and pathology [11,12] and both peroxidase and PLA2 activities have been associated with tumor progression. The role of PRDX6 in cancer is controversial. Elevated levels of PRDX6 have been found in a broad variety of human cancers and its overexpression promotes invasion and metastasis in lung cancer [13] and growth of lung tumors in mice through activation of JAK2/STAT3 signaling [14]. However, PRDX6 has a reducing or promoting effect on tumors depending on the state of tumor development in mice skin cells [15]. PRDX6 has also been related to pathogenesis of inflammatory, metabolic and neurodegenerative diseases as well as ocular damage and male infertility [12]. A relationship has been documented between PRDX6 and insulin signaling pathways with influence on glucose homeostasis, lipid metabolism and inflammation [11].
Several functions of PRDX6 linked to its PLA2 activity have been associated with lipid signaling, with each of the PLA2 reaction products acting as lipokines. These can be the liberated sn2-fatty acid, be it arachidonic acid (AA) [13,[16], [17], [18]] or a hydroxy fatty acid (HFA) [19] or lysophosphatidic acid (LPA) from the remaining moiety [20]. A relationship between PRDX6 PLA2 activity and the transcription factor nuclear factor erythroid 2-related factor 2 (NEF2L2 or NRF2) has been documented in several studies [18,19]. NRF2 is one of the main regulators of cellular redox status whose activity increases under oxidative stress conditions with far-reaching actions on intermediary metabolism and adaptive responses by upregulating the repair and degradation of damaged macromolecules and recently established as a driver of cancer progression, metastasis and resistance to chemotherapy [21].
PRDX6 has been related to cell cycle arrest although with apparent contradictory results. On one hand, induction of cell cycle arrest at G2/M transition in HeLa cells provoked by high concentrations of H2O2 ran parallel to Cys47 hyperoxidation and enhanced PLA2 activity of PRDX6 [22]. On the other hand, cell cycle arrest was induced by PRDX6 silencing with specific siRNA in HepG2 cells [23].
A connection exists between H2O2 concentration and the transition through specific stages during cell cycle progression [24], although the exact underlying mechanisms are not fully elucidated. The intracellular levels of H2O2 increase as the cell cycle progresses reaching the highest levels in G2-M phases [25]. NADPH oxidases (NOX), particularly NOX1 and NOX4 may be the source of H2O2 [26] and H2O2 generated by NOX in response to growth factors are required to activate the extracellular signal-regulated kinase (ERK) and p38 which increase the expression of cyclin D to promote the transition from G0 to G1 phase. Several key proteins involved in different phases of the cell cycle with redox sensitive residues at their active sites, like Cdc14B, PP1, PP2, Cdc25C, could be the targets [27,28] and it has been proposed that Prdx1, Prdx2 and Prdx3 play important roles to maintain the redox state of those target proteins thus protecting the cell against an undesired progression through the cell cycle [29,30]. More specifically and pertinent to the study reported herein, PRDX6 has been described to activate NOX1 oxidase activity through physical interaction with NOX1 components and through its PLA2 activity [31] as occurred also with NOX2 [20]. Tumor suppressor protein p53, a transcriptional regulator which is rapidly induced in response to damaged DNA, is also redox regulated by reversible oxidation of specific Cys residues [32]. High levels of H2O2 induce the activation of p53 by the protein kinase ATM [33] which leads to cell cycle arrest at G1/S or G2/M transition [24,25].
In a previous study we have detected evidences of cell cycle arrest in HepG2 cells where PRDX6 was down-regulated by treatment with specific siRNA [23]. To dig into the mechanisms of PRDX6 involvement in the regulation of cancer cell growth we have constructed a stable HepG2 cell line devoid of PRDX6 and have carried out experiments to analyze the cellular response in terms of proteomic changes, alteration of redox status, mitochondrial functions, signaling pathways and cell cycle progression. Some results support but others dissent from those of the previous silencing study, adding relevant new data to confirm PRDX6 as an important link in the chain of elements connecting cell redox homeostasis and proliferation.
2. Materials and methods
2.1. Materials and reagents
All reagents were of analytical grade and were purchased from Sigma (St. Louis, Missouri, USA) unless otherwise specified. HepG2 cell line used in this work was obtained from the ATCC LGC Standards Company (Teddington, UK). Cell culture dishes and flasks were from TPP (Switzerland). ECL was from GE Healthcare (Wauwatosa, Wisconsin, USA). Antibodies against PRDX6 (#ab133348) were from Abcam (Cambridge, UK). Antibodies against CD95 (sc715) from Santa Cruz Biotechnology Inc. (Dallas, TX, USA), caspase 3 (#96628), and 8 (#97465) were from Cell Signaling Technology (Leiden, The nederlands). MJ33 and antibodies against Actin (A-2066) were from Sigma.
2.2. Cell growth conditions
Cells were grown in EMEM Medium (Eagle Minimum Essential Medium), pH 7.4, supplemented with 10% fetal bovine serum, 2.2 g/L NaHCO3, 1 mM sodium pyruvate, 100 U/L penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin, and the corresponding selective zeocin antibiotic in 5% CO2 atmosphere at 37 °C.
2.3. Construction of a human cell line of hepatoblastoma (HepG2) without peroxiredoxin 6 using the CRISPR/Cas9 methodology (HepG2PRDX6-/-)
The CRISPR-Cas9 technique was applied to the human hepatocarcinoma HepG2 cell line, transfecting with a specific gRNA that was designed using the website of “Thermo Fisher TrueGuide Synthetic gRNA” so that the double-stranded cut was directed towards the third coding region of human prdx6 gene (Fig. 1A). 20,000 cells/cm2 were cultured in 24-well multiplates. When the cells reached 60–70% confluence they were transfected with 7.5 pmol of Cas9 Nuclease (“TrueCut™ Cas9 Protein v2”, ThemoFisher), 1.5 μL of lipofectamine (“Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent”, ThermoFisher) and 15 pmol of gRNA-prdx6. After 48 h the cells were detached, and a portion was lysed to calculate the efficiency using “GeneArtA™ Genomic Cleavage Detection Kit”(TermoFisher). The rest of the cells was used to carry out a “limit dilution” in 96-well plates, until a single clone was isolated. To provide the necessary growth and survival factors, the culture medium in these plates was complemented with 10% of filtered medium where HepG2 cells had grown to approximately 50% confluence. Each isolated clone was analyzed by Western blot to check for the absence of PRDX6.
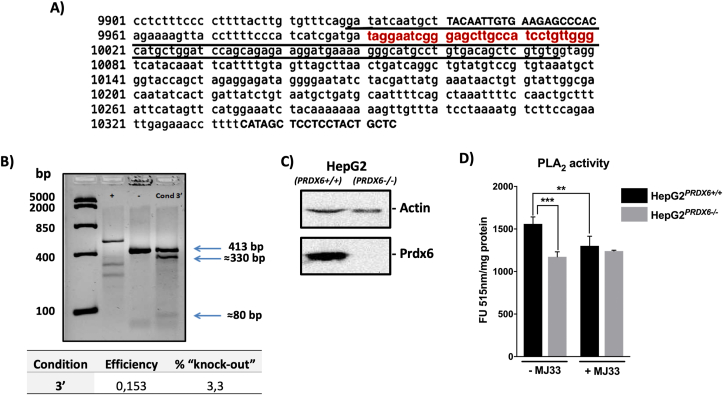
Fig. 1.
Construction and characterization of a stable HepG2PRDX6-/-cell line. A) Sequence of prdx6 gene with the exon coding region underlined, the gRNA region complementary to the prdx6 gene in red color and the sequence of the commercial primers in capital letters. B) Three bands are detected in the fourth lane, corresponding to the original amplified region with these primers (413 bp) and two bands resulting from the cut by Cas9 nuclease (330 and 80 bp). Efficiency and probability of obtaining a knockout was calculated. C) Analysis of knockout clone for PRDX6 protein by Western blot. D) The PLA2 activity of the constructed HepG2PRDX6-/- cell line compared to the standard HepG2 cell line; specific activity in arbitrary fluorescence units per mg protein in the standard assay ± the PRDX6 specific inhibitor MJ33, was determined. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.4. Cell proliferation and viability, nuclear area, apoptosis and cell cycle
Cell proliferation was analyzed using a colorimetric ELISA (Roche Applied Science, Penzberg, Germany). 20,000 cells/cm2 were cultured in 96-well multiplates. After 24 h cells were incubated with 10 μM BrdU labelling solution for 6 h at 37 °C following the protocol recommended by the manufacturer. Total number of cells and cell viability in a HepG2 cell suspension were quantified using the trypan blue dye exclusion method. To measure the nuclear area, 20,000 cells/cm2 were cultured on a coverslip in a 24-well plate. After 48 h cells were fixed in methanol and permeabilized with 0.2% Triton-X100 solution in PBS and were stained with DAPI. The area of cell nuclei was measured on the DAPI pictures using the open source software “ImageJ” [34]. Apoptosis was determined by Western blot analysis of CD95 and caspase-3 and -8 and by flow cytometry. BrdU incorporation into DNA was also determined by flow cytometric analysis: 1 × 106 cells were plated in 60 cm2 dish and after 24 h a 3-h pulse with BrdU (10 mg mL−1) was carried out. BrdU incorporation was determined using the APC BrdU Flow Kit (Becton Dickinson Biosciences, Franklin Lakes, NJ, USA). The proportions of cell cycle phases were also determined by flow cytometric analyses of 7-AAD-stained HepG2 cells. The cytometer used was BD LSRFortessa SORP (BD Biosciences) equipped with 4 lasers and allowing for the simultaneous analysis of up to 20 parameters plus dispersion FSC and SSC. The data were processed with the software BD FACSDiva v8.0.1 (BD Biosciences).
2.5. Transmission electron microscopy
Cells were detached, collected by centrifugation and fixed in 2.5% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4) for 30 min, post fixed in 1% osmium tetraoxide in the same buffer for 30 min, dehydrated in graded ethanol, washed with propylene oxide, embedded in Epon, and then sectioned on an ultramicrotome at 90 nm thickness. Thin sections were stained with 5% uranyl acetate and 5% lead citrate and then examined on a JEM1400 (Japan) transmission electron microscope at 80 kV.
2.6. Seahorse extracellular flux analysis of mitochondrial respiration
Agilent Seahorse XF Cell Mito Stress Test was applied to HepG2PRDX6+/+ and HepG2PRDX6-/- cells and oxygen consumption rate (OCR) determined using Agilent Seahorse XF24 Analyzer (Agilent Seahorse Bioscience, Santa Clara, CA, USA). 48 h before the assay, cells were seeded at 40,000 cells per well in a Seahorse 24-well XF Cell Culture microplate in 250 μL of culture medium and were allowed to adhere for 24 h in 5% CO2 atmosphere at a 37 °C. In addition, the Seahorse XF Sensor Cartridge was hydrated for around 24 h with 500 μL of Seahorse XF Calibrant Solution in a non-CO2 atmosphere at 37 °C to remove CO2 from the media that would interfere with measurements. After that, cells were washed three times with XF Assay Medium supplemented with 10 mM glucose, 2 mM sodium pyruvate and 2 mM glutamine, pH 7.4, and then maintained in XF assay media at 37 °C in a non-CO2 incubator for 1 h. Mitochondrial function of the cells was analyzed by sequential injections of the modulators oligomycin (1 μM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 1 μM) and rotenone (0.5 μM), dissolved in pre-warmed XF Assay Medium and loaded into the designated injection ports of the hydrated sensor cartridge. The loaded XF Sensor Cartridge with a XF Utility Plate was placed into the XF24 Analyzer and calibrated. After calibration, the XF Utility Plate with the calibration fluid was replaced with the plate containing cells. Measurement cycles were performed at the beginning and after addition of each modulator and the parameters basal, ATP production-linked, maximal, proton leak-linked OCR, spare respiratory capacity and non-mitochondrial respiration determined following manufacturer's guidelines. Data were normalized to protein concentration determined at the end of the assay.
2.7. Measurement of enzymatic activities, glucose, lactate, ROS and protein
PLA2 activity in HepG2 cells was assayed using the Red/Green BODIPY based EnzChek Phospholipase A2 Assay (Invitrogen) (pH 7.0, in the absence of calcium) in 96-well plates [35]. Cells were sonicated in PLA2 buffer (50 mM Tris-HCl, 1 mM EGTA, pH 7.0, protease and phosphatase inhibitors), and centrifugated at 15,000×g for 15 min. The supernatant was collected and 50 μg protein was preincubated for 10 min at 37 °C in the absence or presence of MJ33 (10 μM) [36]. The substrate, 50 μL of Red/Green BODIPY, was added and samples were analyzed for 1 h in a Microplate Reader (Varioskan, Thermo Scientific) at excitation 470 nm, emission 515 nm. Glucose and lactate concentration in the culture medium were determined by standard enzymatic colorimetric assays at 505 nm (Labkit, Chemelex, S.A., Spain). ROS were measured by the DCFDA/H2DCFDA method using a commercial ROS Assay Kit (Canvax Biotech S.L., Córdoba, Spain). Protein concentration in the samples was determined by the Bradford method (Bio-Rad) using BSA as standard.
2.8. Global and redox proteome analysis
The experiments were routinely carried out at 20,000 cells/cm2 and reduced cysteines were blocked with “light” Iodoacetamide (IAM) while reversibly oxidized Cys were labelled with “heavy” IAM (Iodoacetamide-13C2, 2d2) incorporating Δmass of 4 Da. The whole methodological strategy as well as the proteolytic digestion was as described previously [23]. Four biological replicas of each sample were analyzed. Proteomics analyses were performed at the Proteomics Facility (SCAI) at the University of Córdoba. The mass spectrometer used was the Thermo Orbitrap Fusion (Q-OT-qIT, Thermo Scientific) equipped with a nano-UHLC Ultimate 3000 (Dionex-Thermo Scientific). The setting of the instrument was as described before [37].
“Label-free” quantification was performed using the MaxQuant (v1.5.7.0) free software [38] configuring “light” IAM and “heavy” IAM and methionine oxidation as variable modifications. No imputation of missing values was implemented. The intensity values were transformed into logarithm base 2 and only the conditions with 3 or more values per identification were considered and analyzed through the Student's T-test. The criteria for considering a significantly expressed protein were that it was identified and quantified using at least two unique peptides and it showed a fold change of at least 1.5 and a q < 0.05 value. MS2 spectra were searched with SEQUEST engine against a database of Uniprot_Human_Oct2018 (www.uniprot.org). The Cys-containing peptides independently labelled with light and heavy IAM modifications were quantified using the open software Skyline [39]. The reduced/oxidized ratio for each specific Cys residue was calculated in each sample in order to define the redox proteome. The intensities of misscleavaged peptides were taken into account for the calculations.
2.9. SDS-PAGE and Western blotting
SDS-PAGE was performed in 12% Criterium XT Precast Gels (Bio-Rad) for detection of specific proteins (PRDX6, Grx1, Caspases 3 and 8, CD95 and Actin). After electrophoresis, proteins were transferred to a nitrocellulose membrane in a semi-dry electrophoretic transfer system (Bio-Rad) for 45 min at 400 mA. Transfer and protein load were checked by staining with Ponceau reagent. The membranes were incubated overnight at 4 °C with the corresponding dilutions of primary antibodies: 1:500, CD95; 1:2000, PRDX6 and caspase-3 and -8; and 1:4000, Actin. They were then washed with TBS-T and incubated with the corresponding peroxidase-conjugated secondary antibody (anti-rabbit, anti-goat or anti-mouse) used at 1:8000 dilution. The chemiluminescent signal induced by ECL reagent (GE Healthcare) was detected in a ChemiDoc image analyzer (Bio-Rad) and quantified by densitometry with Quantity-One 1-D analysis software (Bio-Rad) using Actin as reference for loading normalization.
2.10. Statistics
Where appropriate, results are expressed as mean ± SD of at least three independent experiments. Statistical analysis of the data were performed by Mann-Whitney test, unpaired Student's T-test or two-way ANOVA, depending on the experiment design, as detailed in the figure legends, with the threshold for statistically significant differences set at p adjust <0.05 value, except for the proteomics data that were statistically analyzed using the method of Storey [40] using the R-package and values with q < 0.05 were considered significant.
3. Results
3.1. Construction and characterization of a HepG2 cell line knockout for PRDX6 (HepG2PRDX6-/-)
Cells were transfected following the CRISPR-Cas9 technique as has been described in M&M and the efficiency was calculated, getting a value of 0.153, which meant a probability of obtaining knockout cells of 3.3% (Fig. 1B). The cells, preserved after transfection, were used to carry out a limit dilution in 96-well plates, to isolate one single clone per well. In this way, a considerable number of clones, 157, were able to grow. They were transferred to 24-well plates for later analysis by Western blotting. Only one clone was knocked out for PRDX6 which did not react with specific antibodies against PRDX6 but did with anti-Actin (Fig. 1C). Analysis of the PRDX6 gene of knockout clone by sequencing and incubation of the amplified specific region of 413 bp (Fig. 1A) with gRNA and Cas9, both demonstrated that the mutation was biallelic and the two alleles were repaired differently (Suppl. Fig. 1). Collectively, we concluded that the monoclonal HepG2PRDX6-/- cell line constructed, knockout for human PRDX6, was biallelic and heterozygous.
The phospholipase A2 activity (iPLA2) was markedly lower in HepG2PRDX6-/- cells than in HepG2PRDX6+/+ (Fig. 1D). The PRDX6 peroxidase activity was not determined because it cannot be discriminated from other peroxidases in crude extracts with the currently available assays. The LPCAT activity of PRDX6 was not assayed either but the quantitative proteomic analysis (see below) showed that the enzyme LPCAT1, which was not detected in normal HepG2 cells (Suppl. file 1) was highly expressed in PRDX6 knockout cells, likely as a compensatory response mechanism.
3.2. Differential quantitative proteomic analysis of HepG2PRDX6-/- and HepG2PRDX6+/+ cells
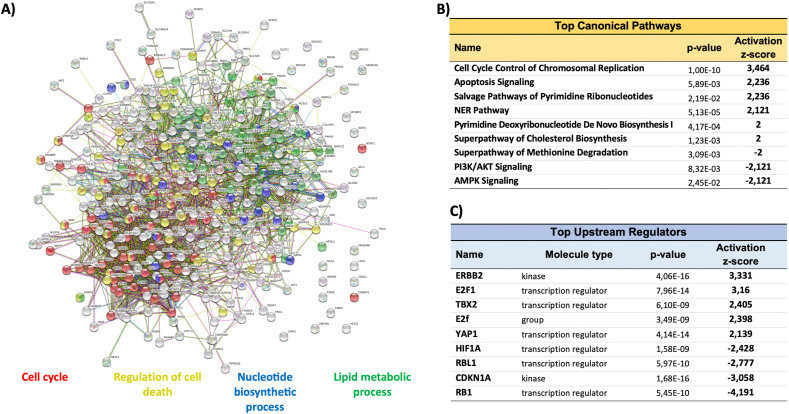
A quantitative proteomic analysis was carried out to get a perception of the effects of eliminating PRDX6 at a global scale. 2843 proteins were identified of which 378 identified with ≥2 unique peptides, fold change ≥1.5 or ≤0.67 and q-value<0.05 were considered differentially expressed (Supplementary file 1). Systems analysis with STRING showed four GO terms predominantly affected: Cell Cycle, Nucleotide Biosynthetic Process, Lipid Metabolic Process and Regulation of Cell Death (Fig. 2A). The main canonical pathway activated by the lack of PRDX6 according to IPA analysis was “Cell Cycle Control of Chromosomal Replication” while “PI3K/AKT signaling” and “AMPK signaling” were inactivated (Fig. 2B). IPA also predicted the activation of several upstream regulators like ERBB2, E2F1 and YAP1, whereas RB1, CDKN1A and HIF1A were inactivated (Fig. 2C). It is worth noting that there was not a compensatory response regarding antioxidant systems: among the peroxiredoxin family of proteins, the levels of Prdx1, Prdx2 and Prdx4 did not change while those of the mitochondrial members Prdx3 and Prdx5 and peroxiredoxin-like 2A (PXL2A/FAM213A/PAMM) decreased significantly as well as did Grx1 (Suppl. file 1).
Fig. 2.
Systems analysis of the differential proteomics analysis. The set of statistically significant differentially expressed proteins with Fold Change ≥1.5 or ≤0.67 (q ≤ 0.05) in HepG2PRDX6-/- compared to HepG2PRDX6+/+ was analyzed, A) with the STRING algorithm for clustering in terms of “GO Biological Process” and network plotted with the four main processes colored red, blue, green and yellow and other processes presented in grey color, and B) with IPA for canonical pathways enrichment. Only those pathways above the significance activation threshold z-score ≥2 and ≤-2, are shown. C) Predicted top upstream regulators involved; filter set for complex, group, kinase, phosphatase, transcription regulator and chemical-endogenous mammals; significance above activation threshold, z-score ≥2 and ≤-2. Statistical significance is indicated by the p-value. Only top upstream regulators with p-values <1.0E-08 are included in this list. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Altogether, this systems analysis reveals a strong effect of PRDX6 knockout on cell cycle progression and on metabolism. These and other effects will be considered in more detail below.
3.3. The absence of PRDX6 has an impact on the redox proteome
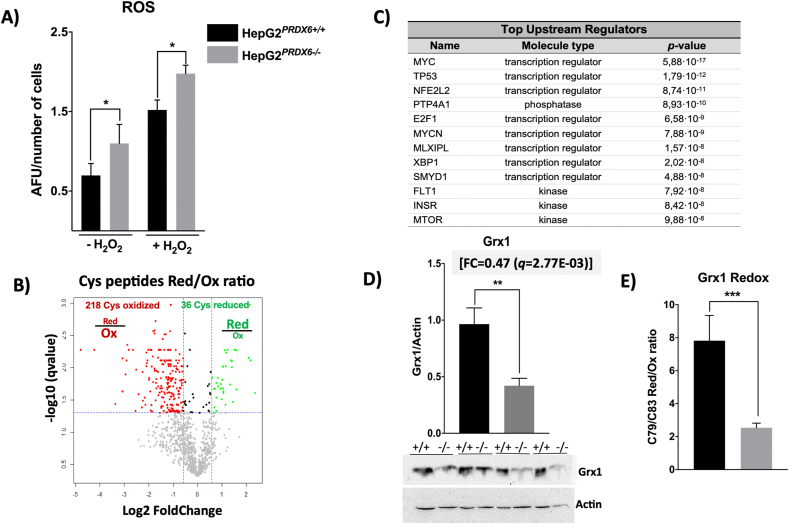
According to the canonical role of peroxiredoxins as part of the antioxidant cellular defenses, an increase in ROS should be expected when PRDX6 is eliminated, with consequences on redox homeostasis. The levels of ROS determined by the DCFDA fluorescence assay were actually higher in HepG2PRDX6-/- cells (Fig. 3A). To check the impact on protein thiols we analyzed the Redox Proteome following our already tested methodology. We detected a total of 2158 Cys-labelled peptides of which 1112 could be quantified and 254, belonging to 202 proteins, showed a significant change ≥1.5 or ≤0.67 (q ≤ 0.05) in their reduced/oxidized ratio upon PRDX6 knockout (Supplementary file 2). Among these peptides, 218 were more oxidized and 36 more reduced (Fig. 3B). IPA and DAVID systems analysis of the set of redox modified proteins revealed a widespread impact on metabolism and signaling pathways. The five more significant KEGG pathways were “Carbon metabolism”, “Glycolysis/gluconeogenesis”, “Biosynthesis of amino acids”, “Metabolic pathways”, “Fatty acid degradation” (Suppl. file 2). Among the predicted up-stream regulators with the highest overlapping score (p-value ≤ 1.00E-07) were MYC, TP53, E2F1 related to cell cycle regulation, NFE2L2 (NRF2), involved in the response to oxidative stress, XBP1, MLXIPL (ChREBP) and INSR, related to UPR and lipid and carbohydrate homeostasis (Fig. 3C).
Fig. 3.
ROS and Grx1 levels and systems analysis of the differential redox proteome of HepG2PRDX6-/-cell line. A) ROS levels in basal and 100 μM H2O2 treated cells expressed as arbitrary fluorescence units in the DCFDA assay normalized for number of cells (n = 4). B) Volcano plot of the change the in reduced/oxidized ratio of quantified Cys-peptides in PRDX6-knockout HepG2 cells; dotted lines indicate the significance thresholds, q-value<0.05 and fold change ≥1.5 or ≤0.67. C) IPA systems analysis of the redox proteome showing the upstream regulators (filter set as in Fig. 2) predicted to be affected by redox changes in their target proteins, ordered by increasing p-value. D) Grx1 levels determined by Western blot (n = 4). The fold change (FC) determined by the quantitative proteomic analysis (n = 4) is shown on top of the graph; membrane images in which WT (+/+) and KO (−/−) paired samples from different experiments (n = 4) were run in the same gel; membranes were cut at appropriate height according to expected migration of Grx1 (≈12 kDa) to reveal with specific antibody. Loading in each lane was normalized to actin. E) Redox change in Cys79/Cys83 of Grx1 in PRDX6 knockout cells; the reduced/oxidized ratio in shown. Statistical significance was assessed by Student's t-test and is shown with a number of asterisks inversely proportional to the p-value (*** ≤0.001, **≤0.005, *≤0.05).
Compared to the redox changes in enzymes of carbon metabolism produced when PRDX6 was silenced with specific siRNA [23], those of cytosolic proteins like HK2-Cys368 and PHGDH-Cys281 behaved similarly, but those of mitochondrial proteins like CLIC1-Cys191 and PCK2-Cys55/63 responded the opposite way and were more oxidized in PRDX6 knockout cells (Suppl. file 2). Special attention was paid to Grx1 given its functional relationship to 1-Cys PRDXs [41]: from the global proteome and by Western blot, Grx1 was down-regulated (Fig. 3D) and its two “extra” cysteines, C79/C83, were more oxidized (FC = 0.32; q = 7,0E-03) (Fig. 3E) in PRDX6 knockout cells. Reciprocally, the “extra” Cys91 of PRDX6 was also more oxidized in siRNA-Grx1 treated HepG2 cells [37] although these cysteine residues are not essential for the activity of either protein [1,42], despite their redox sensitivity. C91 of PRDX6 was prone to glutathionylation [37] and C79/C83 of Grx1 can form a disulfide bond that implies drastic structural rearrangements [43] that may be potential sites for regulation [44].
Proliferating cell nuclear antigen (PCNA) and Protein arginine N-methyltransferase 1 (PRMT1), two proteins involved in cell cycle progression and regulation underwent oxidative and reductive changes, respectively (Suppl. file 2).
The scope of these redox changes will be expounded on and discussed in context below.
3.4. PRDX6 elimination leads to mitochondrial dysfunction, ER stress and metabolic alterations
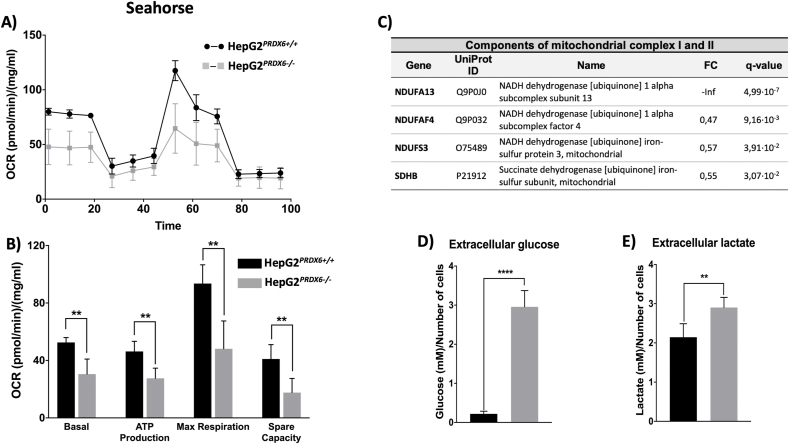
The reported connection between PRDX6 and mitochondrial clearance and the increased accumulation of LC3B and p62 induced by PRDX6 depletion [7] together with our discovery that SQSTM1-p62 was markedly increased (Fold change = 1.9; q = 1,1E-03) in HepG2 cells upon PRDX6 knockout (see Suppl. file 1), prompted us to check the mitochondrial functional state. In vivo extracellular flow analysis (Seahorse) showed nearly 50% decrease in basal and maximal respiration as well as in respiratory capacity reserve in HepG2 cells lacking PRDX6 (Fig. 4A and B). A drastic decrease in the levels of several critical components of the electron transport chain involved in assembly and catalysis of Complex I and Complex II (Fig. 4C) should explain the observed mitochondrial respiratory dysfunctionality. Mitochondrial appearance was prominently different in HepG2PRDX6-/- cells (Fig. 5).
Fig. 4.
Mitochondrial functionality of HepG2PRDX6-/-cell line. A) Representative recording of oxygen consumption rate (OCR) during the extracellular flow analysis (“Seahorse”) following the protocol for “Mitochondrial Function”; differences between WT and Prdx6 KO cells are highly significant despite a drop of OCR after addition of FCCP. B) Histogram plots of the calculated basal and maximum respiratory rate, ATP production rate and respiratory capacity reserve, normalized for protein content (n = 4). C) Table with mitochondrial protein components of Complexes I and II that were down-regulated in HepG2PRDX6−/- according to the quantitative proteomic analysis. UniProt ID, Protein name, fold change and statistical score are shown; “-inf” indicates that the protein was detected in normal HepG2 cells but not in PRDX6 knockout cells. D) Extracellular glucose and E) lactate concentrations determined by standard methods normalized for number of cells (n = 4). (**** p-value ≤ 0.0001; ** p-value ≤ 0.005).
Fig. 5.
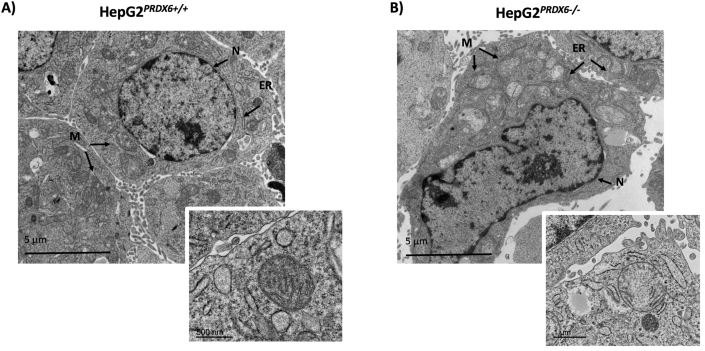
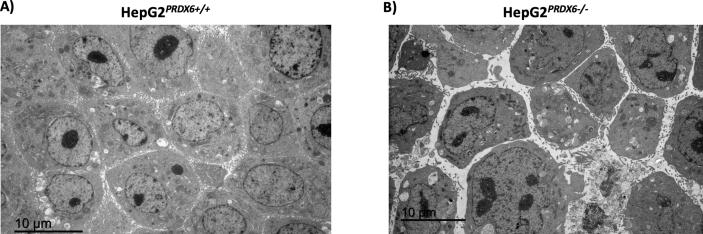
Electron micrographs of HepG2PRDX6+/+and HepG2PRDX6-/-cells. A) Transmission electron microscopy (TEM) image of HepG2 showing normal hepatocytic organelles; nucleus (N), abundant mitochondria with cristae (M), rough and smooth endoplasmic reticulum (ER) are highlighted. B)Idem for HepG2 lacking PRDX6 showing alterations in cellular and nuclear morphology as well as in hepatocytic organelles; mitochondria with less cristae and dilated endoplasmic reticulum cisternae are indicated. Insets showing an amplified selection from each type of cell have been included to show the different appearance of mitochondria; scale bars are indicated.
The levels of extracellular glucose were much higher and those of Insulin receptor (INSR) and key glycolytic enzymes (HK2, PFKL, ALDOC), lower in HepG2PRDX6−/- cells (Fig. 4D; Suppl. file 1) confirming a decreased rate of glucose catabolism in the absence of PRDX6 as observed previously when the cells were treated with specific siRNA-PRDX6 [23]. However, the levels of extracellular lactate were higher in HepG2PRDX6−/- cells (Fig. 4E), and the levels of several components of the Pyruvate Dehydrogenase Complex (PDHK, DLAT) were lower, indicating that despite the lower consumption of glucose, glycolysis to lactate was its prevailing metabolic fate under conditions of decreased mitochondrial respiratory capacity.
Electron microscopy showed that HepG2 cells lacking PRDX6 have much less functional mitochondria than the wild type. They had a rarefied matrix and less cristae than normal HepG2 cells (Fig. 5). A decreased level (fold change = 0.67; q = 4.20E-04) of mitochondrial Lon protease (LONP1), the major component of the mitochondrial protein quality control system under oxidative stress conditions [45], could have caused accumulation of misfolded proteins thus explaining this appearance. ER stress was enhanced in KO cells as revealed by dilated ER cisternae (Fig. 5) and by down-regulation of HSPA5/BiP (FC = 0.63; q = 4.1E-03; Suppl. file 1), one of the main regulators of the unfolded protein response (URP) [46]. Autophagic vesicles were more abundant in KO cells Suppl. Fig. 2, a survival strategy under the stress caused by accumulation of damaged proteins and by starvation induced by the lack of PRDX6.
A number of other archetypal mitochondrial proteins were also down-regulated, including four members of the pyruvate and 2-oxoglutarate dehydrogenase complexes (DLAT, DLST, PDHX, OGDH), succinate dehydrogenase (SDHB), PEP carboxykinase (PCK2) and pyruvate carboxylase (PC) and antioxidant enzymes like PRDX3 and PRDX5 (Suppl. file 1).
Altogether, our results obtained by different experimental approaches, Seahorse, proteomics and electron microscopy, support a role for PRDX6 in regulating mitochondrial biogenesis and function, and in the control of metabolism.
3.5. NRF2 is not fully active despite the increased levels of ROS in HepG2 cells lacking PRDX6
Under the conditions of increased levels of ROS in HepG2PRDX6−/- cells the canonical cellular response should imply activation of NRF2 so that antioxidant defenses under the control of ARE promoter be induced. Two markers of NRF2 activation, NQO1 and GSTM3 were only slightly up-regulated, but the proteomic analysis showed prominently decreased levels of many other positively regulated markers and increased levels of negatively regulated markers [21,47] (Table 1). As mentioned above, the three cytosolic members of the peroxiredoxin family did not change while the two mitochondrial members and peroxiredoxin-like 2A decreased significantly. These data indicate that the activity of NRFF2 is lower in cells lacking PRDX6. The presence of a TPA-responsive element (TRE) within the ARE at the NQO1 promotor allows for different transcription factors to compete for binding to both these elements [48] what would explain slightly induced levels of NQO1 even if NRF2 is not fully active.
Table 1.
Changes in protein levels of NFR2 targets in HepG2PRDX6−/- cells.
| Gene |
UniProt ID |
Name |
FC |
q-value |
|---|---|---|---|---|
| Positively regulated by Nrf2 | ||||
| AKR1C2 | P52895 | Aldo-keto reductase family 1 member C2 | -Inf | 1,86·10−10 |
| AKR1C3 | P42330 | Aldo-keto reductase family 1 member C3 | 0,64 | 5,47·10−2 |
| MGST2 | Q99735 | Microsomal glutathione S-transferase 2 | 0,46 | 9,91·10−3 |
| GCLC | P48506 | Glutamate cysteine ligase catalytic subunit | 0,67 | 8,15·10−3 |
| GCLM | P48507 | Glutamate cysteine ligase regulatory subunit | 1,15 | 4,94·10−1 |
| TXNRD1 | Q16881 | Thioredoxin reductase 1, cytoplasmic | 0,73 | 2,97·10−1 |
| GSTM3 | P21266 | Glutathione S-transferase Mu 3 | 1,51 | 4,51·10−2 |
|
NQO1 |
P15559 |
NAD(P)H dehydrogenase [quinone] 1 |
1,34 |
4,13·10−2 |
|
Negatively regulated by Nrf2 |
||||
| FADS2 | O95864 | Fatty acid desaturase 2 | +Inf | 4,70·10−13 |
| FASN | P49327 | Fatty acid synthase | 1,44 | 9,75·10−4 |
| HMGCS1 | Q01581 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 3,45 | 5,58·10−4 |
| CYB5A | P00167 | Cytochrome b5 | 1,68 | 2,47·10−2 |
| RBP1 | P09455 | Retinol-binding protein 1 | 3,74 | 1,65·10−3 |
3.6. Loss of PRDX6 has an antiproliferative effect but does not induce apoptotic cell death
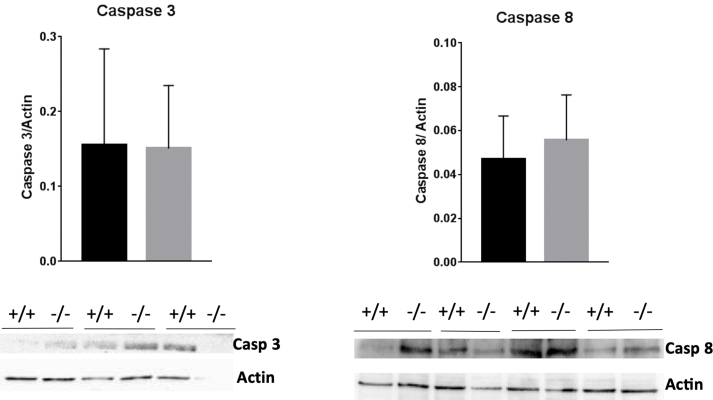
Proliferation measured as BrdU incorporation into DNA and the number of cells counted two, three and four days after seeding were significantly lower in knockout HepG2PRDX6−/- cells compared with normal HepG2PDRX+/+ cells (Fig. 6A and B). The decrease in the number of cells is not justified by cell death since the viability did not decrease significantly (data not shown) and no signs of increased cell death due to apoptosis was observed by electron microscopy (Fig. 5) nor by flow cytometry (Fig. 7A and B). Coherently, the levels of caspases 3 and 8, as determined by Western blot (Fig. figs3), and by proteomics (Suppl. file 1) did not change significantly. Moreover, there was a significant decrease in CD95 protein levels (Fig. 6C) as we had also observed previously in HepG2 cells with low levels of PRDX6 [23]. This receptor has been traditionally related to apoptosis but nowadays its involvement in other multiple non-apoptotic functions is well known and stimulation of CD95 in cancer cells can turn this death receptor into tumor promoting receptor [49].
Fig. 6.
Growth characteristics of HepG2PRDX6−/-cell line. A) Cell count. B) Proliferation rate. Determinations were done after 24 h of cell culture in 4 different experiments (n = 4). C) CD95 levels in control and knockout cells determined by Western blot; membrane images in which WT (+/+) and KO (−/−) paired samples from different experiments (n = 4) were run in the same gel; membranes were cut at appropriate height according to expected migration of CD95 (≈38 kDa) to reveal with specific antibody. Loading in each lane was normalized to actin. Statistical significance was assessed by unpaired Student's t-test. Number of asterisks inversely proportional to the p-value (**< 0.005, * < 0.05).
Fig. 7.
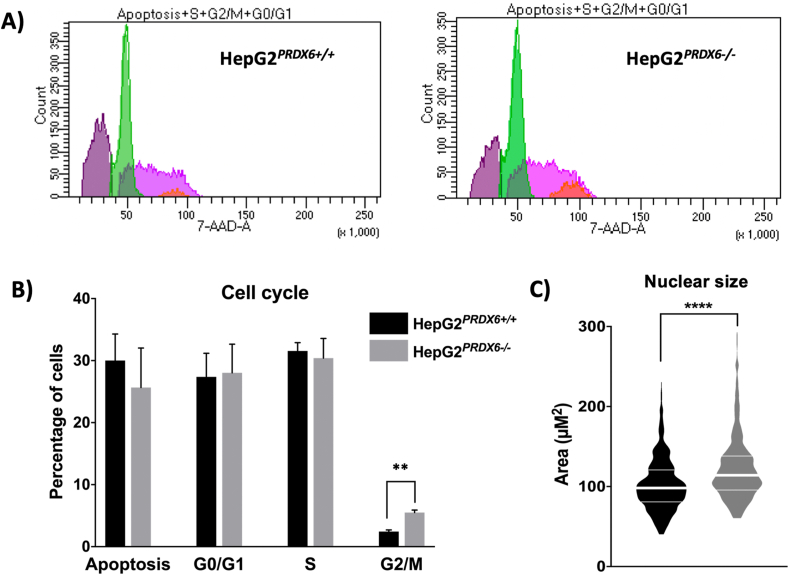
Cell cycle parameters of HepG2(PRDX6−/-)cell line. Flow cytometry analysis showing the distribution of cells along the different cell cycle phases and those undergoing apoptosis in normal and PRDX6 knockout HepG2 cells. A) Recordings of cytometer fluorescence event counts of one representative experiment (purple, apoptotic cells; green, G0/G1 phases; pink, S phase; orange, G2/M phases). B) Cell cycle phase distribution of the cells (n = 4; multiple t-test, ** p-value ≤ 0.005). C) Size of the nuclei determined as described in Materials and Methods section 2.4 (n > 500; unpaired, non-parametric Mann-Whitney test; Median and quartiles are shown, **** p-value ≤ 0.0001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Loss of PRDX6 promotes cell cycle arrest at G2/M phase
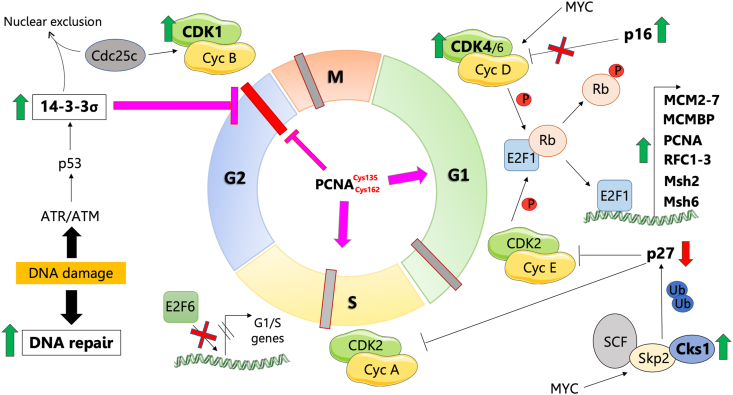
As described above in subsection 2 and Fig. 2, PRDX6 knockout had a clear effect on the cell cycle since a number of cell cycle proteins were differentially expressed (Scheme 1; Table 2). Increased levels of Cyclin-dependent kinase 4 (CDK4), Cyclin-dependent kinase 1 (CDK1), Cyclin-dependent kinases regulatory subunit 1 (CSK1B) and Proliferating cell nuclear antigen (PCNA) in HepG2PRDX6−/- would favor the G1/S transition, a likely result of mitogenic signals activation by oxidative stress associated with PRDX6 deficiency. Proteins related to MCM complex, DNA replication and DNA repair and involved in G1/S progression, such as all 6 replication-related proteins (MCM2-7), Mini-Chromosome Maintenance Complex-Binding Protein (MCMBP), Replication Factor C (RFC1-3), DNA ligase 1 (LIG1) and DNA Mismatch Repair Proteins 2 and 6 (Msh2 and Msh6) were also increased in cells devoid of PRDX6. Even Thymidylate Synthase (TYMS) and Ribonucleoside-Diphosphate Reductase subunit M2 (RRM2), two enzymes responsible for the synthesis of deoxyribonucleotides were also prominently elevated in HepG2PRDX6−/- cells. In addition, E2F1-3 upstream regulators appeared activated and RB1 inactivated according to IPA (Fig. 2C), which agrees with cell cycle progression beyond G1/S. However, proteins that favor cell cycle arrest at G1/S by inhibiting CDK4/6, such as CDKN2A/p16 were also increased in knockout cells, although according to IPA analysis, CDKN1A and CDKN2A are inactive (Fig. 2C and Suppl. file 1). Our results also agree with entry of the cells into S phase since the levels of the inhibitor of CDK2/Cyclin complex, cyclin-dependent kinase inhibitor 1B (CDKN1B/p27kip1) decreased in PRDX6 knockout cells while cyclin-dependent kinases regulatory subunit 1 (CSK1B) and S-phase kinase-associated protein 1 (SKP1) which are involved in polyubiquitination of p27kip1 increased.
Scheme 1.
Cell cycle state in PRDX6 knockout HepG2 cells. The cell cycle is shown in the middle with a different color for each phase and the phase transitions and checkpoints indicated by intersecting thick short lines; proteins differentially expressed in HepG2PRDX6−/- cells as determined by quantitative proteomics are represented; processes and regulators predicted to be activated or inhibited by systems analysis have also been included; green and red vertical arrows indicate up- and down-regulated proteins, respectively; red crosses indicate inhibition; purple arrows and blunt lines indicate positive or negative action, respectively. Read subsection 3.7. in the main text for a full explanation.
Table 2.
Differential cell cycle related proteins in HepG2PRDX6−/- cells.
| Gene | UniProt ID | Name | FC | q-value |
|---|---|---|---|---|
| TYMS | P04818 | Thymidylate synthase | +inf | 4,70·10−13 |
| RRM2 | P31350 | Ribonucleoside-diphosphate reductase subunit M2 | +inf | 1,45·10−12 |
|
CDKN2A; CDKN2B |
P42771; P42772 |
Cyclin-dependent kinase inhibitor 2A/4B | +inf | 2,56·10−9 |
| CKS1B | P61024 | Cyclin-dependent kinases regulatory subunit 1 | +inf | 3,84·10−10 |
| CDK4 | P11802 | Cyclin-dependent kinase 4 | +inf | 4,92·10−11 |
| CDK5RAP1 | Q96SZ6 | CDK5 regulatory subunit-associated protein 1 | +inf | 3,84·10−10 |
| TOP2A | P11388 | DNA topoisomerase 2-alpha | +inf | 2,28·10−8 |
| CETN2 | P41208 | Centrin-2 | +inf | 1,51·10−9 |
| LIG1 | P18858 | DNA ligase 1 | +inf | 2,25·10−10 |
| CDK1 | P06493 | Cyclin-dependent kinase 1 | 10,40 | 3,81·10−4 |
| MCM2 | P49736 | DNA replication licensing factor MCM2 | 2,99 | 6,89·10−4 |
| MCM3 | P25205 | DNA replication licensing factor MCM3 | 3,28 | 1,04·10−4 |
| MCM4 | P33991 | DNA replication licensing factor MCM4 | 2,82 | 7,71·10−3 |
| MCM5 | P33992 | DNA replication licensing factor MCM5 | 3,18 | 9,75·10−4 |
| MCM6 | Q14566 | DNA replication licensing factor MCM6 | 2,81 | 4,45·10−4 |
| MCM7 | P33993 | DNA replication licensing factor MCM7 | 2,70 | 1,58·10−4 |
| PCNA | P12004 | Proliferating cell nuclear antigen | 1,82 | 2,00·10−3 |
| RPA1 | P27694 | Replication protein A 70 kDa DNA-binding subunit | 1,51 | 1,94·10−2 |
| MCMBP | Q9BTE3 | Mini-chromosome maintenance complex-binding protein | 2,12 | 9,23·10−3 |
| RFC1 | P35251 | Replication factor C subunit 1 | 2,70 | 3,32·10−5 |
| RFC2 | P35250 | Replication factor C subunit 2 | 1,93 | 6,45·10−3 |
| RFC3 | P40938 | Replication factor C subunit 3 | 1,74 | 3,07·10−2 |
| MSH2 | P43246 | DNA mismatch repair protein Msh2 | 1,57 | 4,22·10−2 |
| MSH6 | P52701 | DNA mismatch repair protein Msh6 | 1,63 | 1,22·10−3 |
| SKP1 | P63208 | S-phase kinase-associated protein 1 | 1,52 | 8,05·10−3 |
| SFN | P31947 | 14-3-3 protein sigma | 2,19 | 2,81·10−3 |
| CUL5 | Q93034 | Cullin-5 | -inf | 4,40·10−10 |
| CDKN1B | P46527 | Cyclin-dependent kinase inhibitor 1B | -inf | 1,86·10−10 |
Centrin-2 (CETN2) and DNA topoisomerase 2-alpha (TOP2A), two proteins necessary for G2/M transition and mitosis, were also increased in HepG2PRDX6−/- cells. Regarding p53, the lower expression of its repressor cullin-5 (CUL5) and higher expression of its activator 14-3-3σ (SFN) in HepG2PRDX6−/- is worth noting.
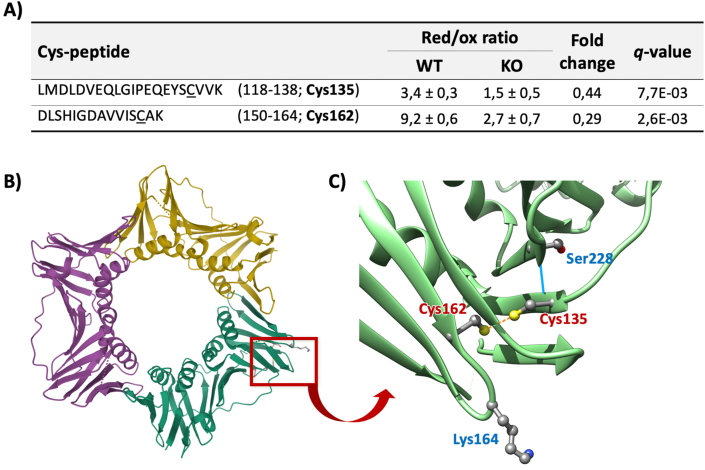
Besides these important changes in the levels of cell cycle related proteins, we have also detected meaningful redox changes in some of these proteins. Among them, PCNA deserves special attention as it has been described to induce cell cycle progression through G1/S and arrest at G2/M [50]. It was induced (Table 2) in HepG2PRDX6−/- and notably, residues Cys135 and Cys162, underwent a 3-fold decrease in their reduced/oxidized ratio (Suppl. file 2; Fig. 8A). Whether this redox change affects the activity of PCNA remains to be demonstrated, but, interestingly, they are located in two different β-pleated sheets in the region of the protein near the binding site for most PCNA interacting proteins and they are within reach (3.45 Å) in the 3D structure. Actually, they form a disulfide bond in some of the structures resolved so far (Fig. 8B and C).
Fig. 8.
Redox changes of PCNA Cys135-Cys162 in HepG2PRDX6−/-cell line. A) Cys-peptides of PCNA undergoing significant changes in their reduced/oxidized ratios in PRDX6 knockout HepG2 cells. The sequence of the peptides, the values and the fold change (FC) of their red/ox ratios and the statistical scores are shown. B) Structure of the protein highlighting its ring-shape (PDB 3VKX (Punchihewa et al., 2012)); the red square highlights a region of the protein involved in interaction with its partner regulatory proteins. C) inset: detail of the residues clustered around in the red square, twisted 180°, showing Cys135 and Cys162 close enough (3.06 Å) to form a disulfide bond (orange dotted line); Ser228, essential for interactions with regulatory proteins (Baple et al., 2014), is connected to Cys135 by a hydrogen bond (blue line); and Lys164, site for ubiquitination and sumoylation; Image made using UCSFChimera (Pettersen et al., 2004). Baple et al., 2014. J. Clin. Invest. 124:3137–3146. https://doi.org/10.1172/JCI74593. Pettersen et al., 2004. J. Comput. Chem. 25:1605–1612. https://doi.org/10.1002/jcc.20084. Punchihewa et al., 2012. J. Biol. Chem. 287:14289–14300. https://doi.org/10.1074/jbc.M112.353201. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
These proteomics data clearly show that lack of PRDX6 affects the cell cycle, and flow cytometric analysis provided evidence that the cells were arrested at G2/M phase (Fig. 7A and B). Moreover, we observed a significant increase in nuclear size both by staining with DAPI (Fig. 7C) and by transmission electron microscopy (Fig. 5 and Fig. figs2) in HepG2PRDX6−/-, which is consistent with arrest at G2/M.
4. Discussion
Through the CRISPR/Cas9 technique we have obtained a stable monoclonal hepatocarcinoma cell line, HepG2, knockout for PRDX6. The process of CRISPR-Cas9-mediated gene disruption through non-homologous end joining (NHEJ) is caused by “indels”, which consist of insertions or deletions of few nucleotides. It is a cell survival response to be able to bind to broken DNA chains, where the “indels” must not be multiples of three, so that the corresponding gene produces a non-functional protein. In this case the mutation was biallelic and heterozygous. We demonstrated that the two alleles were repaired differently: one allele had a deletion of one nucleotide, and the other allele had an elimination of 11 nucleotides. Loss of PLA2 activity and increased levels of ROS as a consequence of PRDX6 elimination were experimentally confirmed.
4.1. Role of NRF2
The increased levels of ROS indicate that the cell response to lack of PRDX6 does not involve the expected activation of antioxidant defense mechanisms. ROS should have oxidized redox sensitive Cys in KEAP1 to set NRF2 free from ubiquitination and ready to move to the nucleus to activate ARE promoters for antioxidant defense [21]. However, this does not seem to be the case, indicating that either oxidative equivalents do not reach KEAP1 cysteines when PRDX6 is absent or that NRF2 is being down-regulated by a KEAP1-independent mechanism. Trx-dependent redox regulation of Nrf2 in the nucleus through Cys514, a critical redox sensitive Cys residue conserved in the b-zip family of transcription factors, could be involved, but the levels and redox status of the Trx system are not affected in knockout cells (Suppl. files 1 and 2). However, redox regulation of NRF2 nuclear translocation [51] could be affected by the marked decrease observed in the Grx/GSH system in PRDX6 knockout cells. S349-phosphorylated p62/SQSTM1 activates NRF2 by binding to and inactivating KEAP1 [52]. Although p62/SQSTM1 is highly induced in HepG2PRDX6−/- cells, this NRF2 activating mechanism does not prevail in these cells either because p62 is not phosphorylated or because other NRF2 inactivating mechanisms are functioning.
NRF2 could also be inactivated by a mechanism dependent on GSK-3β [53] either through PI3K/Akt pathway or by ERBB2. Inhibition of the PI3K/Akt pathway (Fig. 2B) would keep GSK-3β active to phosphorylate and inactivate NRF2. At the same time, activation of ERBB2 (Fig. 2C) would phosphorylate and activate GSK-3β, but induction of PPP2R1A (Suppl. file 1) would counteract this effect.
NRF2 negative regulation by HRD1 through down-regulation of the UPR could be operative, as occur during liver cirrhosis [54]. Actually, the pathway from UPR to NRF2 ubiquitination requires active IRE1α, which is maintained in its inactive monomeric state by binding to the endoplasmic reticulum chaperone HSPA5/BiP. This chaperone is markedly repressed in KO cells, which could stimulate HRD1 binding to and proteasomal degradation of NRF2. Finally, inhibition of NRF2 activity by RXRα [55] cannot be discarded since this pathway is activated in PRDX6 knockout cells (Suppl. file 1).
These data indicate that NRF2-dependent activation of antioxidant defenses in PRDX6 knockout cells is blocked by KEAP1-independent mechanisms.
4.2. Lipid metabolism and PLA2 activity of PRDX6
Lipogenesis, including cholesterol biosynthesis, was induced in PRDX6 knockout cells to cope with the need to synthesize new membranes prior to cell division. The prominent induction of Fatty acylCoA-6-desaturase (FADS2) and microsomal cytochrome b5, two enzymes responsible for the first steps of long chain polyunsaturated fatty acids (LC-PUFA) and eicosanoids biosynthesis, could be a compensatory cellular response to deprivation of arachidonic acid (AA) caused by lack of PRDX6. A critical regulatory component of eicosanoid biosynthesis is at the level of availability of unesterified fatty acids liberated from membrane phospholipids. In normal HepG2 cells, AA could be provided by PRDX6 PLA2 activity if acting on sn-2-arachidonate containing phospholipids. Early characterization of PRDX6 PLA2 showed high activity with AA containing phospholipids [56] and we have previously detected increased levels of 1-palmitoyl-2-arachidonoyl-GPC in PRDX6 down-regulated HepG2 cells [23]. Moreover, a relationship has been described between AA and PRDX6 tumor promoting functions [13] through activation of NOX2 or Src family kinases (SFK) [16].
The PLA2 activity of PRDX6 on phosphatidylcholine phospholipids would stimulate the production of lysophosphatidic acid (LPA) and the sequential action of its peroxidase and PLA2 activities would also liberate a hydroxy fatty acid (HFA) [19,20]. Given the importance to dissect the mechanism of action of PRDX6 in cell physiology and the roles of AA, HFA and LPA as lipokines, it would be worth defining the substrate specificity of its PLA2 activity in vivo to solve this question.
In summary, the release of lipokines would be compromised in cells devoid of PRDX6-dependent PLA2 activity. This would have consequences in signaling pathways affecting cell integrity as observed at the level of mitochondria and endoplasmic reticulum.
4.3. Mitophagy/autophagy
Prominent down regulation of respiratory complex I subunits, diminished respiratory capacity, conspicuous mitochondrial morphological changes and marked up-regulation of p62 and down-regulation of HSP5A/Bip point to mitochondrial respiratory dysfunction and ER stress upon PRDX6 knockout in HepG2 cells. Involvement of NRF2 inactivation in mitochondrial dysfunction, and particularly in mitochondrial biogenesis, has been put forward by several studies [57] and may play a role in the mitochondrial response of HepG2 cells to the absence of PRDX6. E2F1, which is activated in PRDX6 knockout cells, could be also involved as it is a transcription factor that, beyond a mere cell cycle regulator, plays a role in activating lipogenesis and nucleotide synthesis and represses mitochondrial biogenesis and oxidative phosphorylation [58]. The marked increase in TOMM34, the cytosolic cochaperone that protects mitochondrial protein precursors in an unfolded state, speaks of accumulation of mitochondrial precursors in the cytosol [59]. This would partly explain the general decrease in the levels of mitochondrial proteins observed in PRDX6 KO cells.
It is well known that a complex connection exist between mitochondria and ER so that signals from damaged mitochondria may have stimulated the production of ROS from the ER surface with the involvement of NOX complexes [60]. HepG2 cells respond to H2O2-induced oxidative stress by activation of the unfolded protein response (UPR) and autophagy for survival [61]. However, the UPR was not activated in HepG2PRDX6−/- cells despite mitochondrial damage and ER stress, as indicated by decreased levels of HSP5A/Bip, nor was apoptosis as evidenced by flow cytometry and because caspases were not induced.
The presence of oxidizable phospholipids acylated with polyunsaturated fatty acids (PUFA-PLs), as inferred from the striking induction of Δ6-desaturase (FADS2) and LPCAT, and defective lipid peroxide repair caused by lack of PRDX6, together with high levels of ROS and inactive NRF2 could lead to cell death by ferroptosis [62,63], although proteomics and microscopy data do not provide additional evidences.
Autophagy was induced instead, as revealed by the augmented presence of autophagosomes. The significant increase in HSP90AB1 and CDC37 (FC = 1.46 and 1.43, respectively; Suppl. file 1) are also indicative of mitophagy activation [64]. In this way the cells would obtain building blocks for biosynthesis to compensate for the diminished glucose uptake. The canonical mechanism of autophagy regulation involves nutrient sensing kinases mTOR and AMPK, but other independent pathways of autophagy induction have been described [65]. Since AMPK signaling was inhibited in PRDX6 knockout cells (Fig. 2B), induction by unsaturated fatty acids requiring an intact Golgi apparatus [66] or through p38/MAPK14 [67], which is upregulated in PRDX6 KO cells, could be called.
PRDX6 has been associated with regulation of glucose homeostasis and lipid metabolism through the insulin signaling pathway in muscle cells of mice [11]. Our proteomics results show that HepG2PRDX6−/- cells have lower levels of insulin receptor and PDPK1, a kinase that plays a central role activating PKB/AKT1 in insulin signaling. This is evidence for the involvement of PRDX6 in the regulation of metabolism in human hepatocarcinoma cells. These results together with a reduced uptake of glucose and the diminished flow through the glycolytic pathway (key glycolytic enzymes are decreased) cause these cells to have lost the characteristic Warburg effect of tumor cells what makes PRDX6 a good target for anticancer drugs.
4.4. Cell cycle
Our results indicate that PRDX6 deficiency in HepG2 causes a decrease in the number of cells as well as in proliferation as a consequence of cell cycle arrest at G2/M phase. Cell cycle progression is a biological process regulated by multiple redox-dependent signal transduction pathways to ensure the correct cell division and where the effect of ROS on cell fate oscillates during the cell cycle [24]. Thus, it is not surprising that the lack of PRDX6 affects cell cycle progression. We observed changes in proteins involved in G1 and S phases of the cell cycle that point to a successful progression through them with a central role of p27 degradation, p21 inactivation and E2F1 activation. However, previous reports claimed that PRDX6 silencing arrests cell cycle at G1/S by a mechanism dependent on its PLA2 activity [17,23,68]. Therefore, the arrest that we observe at G2/M should be related to its peroxidase activity, instead. A correlation has been reported between PRDX6 loss of peroxidatic activity by hyperoxidation and H2O2 induced G2/M arrest in HeLa cells [22].
The connection between PRDX6 and cell cycle could take place at the level of redox changes at key proteins and from our results, PCNA stands as a likely candidate. PCNA is necessary for the progression through transitions M/G1 and G1/S, for the execution of phase S and for S phase and G2 phase checkpoints [50]. In PRDX6 knockout HepG2 cells, PCNA seems to be operative in all these steps where it is required except for the last checkpoint dependent on ATR/Chk1. In this phase, progression through the cycle entails PCNA detachment from DNA with the help of several proteins followed by its degradation [50]. Redox changes that we found in Cys135 and Cys162 of PCNA could affect protein interactions implicated in detachment from DNA, what would constitute a new connection between the redox status and cell cycle.
There are many proteins known to interact with PCNA to accommodate its activity to the cellular demands and interestingly, one of them is PRDX6 [69,70]. A single point mutation, Ser228Ile (variant dbSNP:rs369958038), in the human PCNA gene was recently identified to cause a disease. The mutation lies near the binding site for most PCNA-interacting proteins and results in significantly decreased interaction with FEN1, LIG1 and ERCC5 [71]. Cys135 and Ser228 are located in front of each other in two adjacent strands of an antiparallel β-sheet. One of the main PCNA regulatory mechanisms consists of ubiquitination and sumoylation at Lys164 also in the vicinity of Cys135, Cys162 and Ser228 (Fig. 8C). It is tempting to speculate that formation of a C135–C162 disulfide bond could compromise the flexibility needed for the large conformational changes that dramatically transform the binding pocket for PCNA client proteins [72]. However, whether PRDX6 could be among the proteins PCNA needs to detach from DNA, either indirectly through H2O2 or NOX stimulation or even by direct interaction [69,70] remains to be demonstrated, but would constitute a new connection between the cell redox status and the cell cycle. Interestingly, Cys135 and Cys162 are not present in fungal and plant PCNA indicating that they might have been acquired by mammals through evolution as a “redox switch” to fine tune the link between the cellular redox state and progression of the cell cycle. This is an interesting starting hypothesis worth further investigation. Down-regulation of Grx1 and GCLC (Table 1 and Fig. 3C) as a consequence of NRF2 low activity would compromise the action of the GSH/Grx system allowing for the oxidation of those critical cysteine residues. The presence of Grx1 in the nuclei of many cells [[73], [74], [75]] suggest that Grx1 could play a role in this so far hypothetical cell cycle regulatory redox mechanism. The absence of the protein Peroxiredoxin-like 2A (PXL2A/Fam213A/PAMM), an antioxidant peroxiredoxin secreted to the extracellular space [76] should be taken into account as another contributor to a more oxidative environment in PRDX6 KO cells.
In summary, G2/M arrest induced by the absence of PRDX6 could take place through oxidative redox changes at critical cysteine residues of key proteins as part of the existing interplay between the redox status and progression of the cell cycle [24,25]. Oxidative changes resulting in the formation of an intramolecular disulfide bond between Cys330 and Cys377 in Cdc25c are important for binding of Cdc25c to 14-3-3 [77]. Formation of this disulfide induced by NRF2 genetic disruption led to G2/M arrest and was reverted by GSH [78]. Similarly, oxidation of key Cys residues leading to p53 activation and cell cycle arrest are well known [32,33]. Although we have not detected redox changes in Cdc25c and p53 in PRDX6 knockout cells, its functioning cannot be discarded. Besides, the data presented herein suggest the operation of a so far unknown additional redox mechanism of cell cycle regulation by oxidation of PCNA Cys135 and Cys162.
5. Conclusions
Loss of PRDX6 deprives the cell of its lipid peroxidase and iPLA2 activities with main consequences on mitochondrial function and integrity, ER stress and cell cycle arrest at G2/M phase in cultured HepG2 cells. Activation of NRF2, the main regulator of the cellular redox status and driver of cancer progression, is inhibited in a KEAP1-independent way, so that the antioxidant defenses are not set in motion. Insulin receptor is downregulated, and uptake of glucose and respiratory capacity decrease drastically. Effects on signaling pathways could be due to altered release of lipokines from phospholipids due to absence of PRDX6 PLA2 activity. The cell displays a survival response that involves activation of the autophagy program and stimulation of biosynthetic metabolic pathways that correlate with activation of E2F1 transcription factor. There are changes in the redox proteome with as many as 202 proteins significatively affected at cysteine residues and the Grx/GSH system is weakened. Oxidation of Cys135 and Cys162 of PCNA, likely forming a disulfide bond, emerges as a new redox mechanism that adds up to other known regulatory links between the cell redox status and the cell cycle responsible for arrest at G2/M phase.
These findings support a role for PRDX6 in regulating mitochondrial biogenesis and function, in the control of metabolism and in cell proliferation in cultured cells. Further studies should confirm these effects in vivo.
Author contributions
MJLG, DJL, AFGJ and BCH performed the construction and characterization of the knockout monoclonal cell line. MJLG, DJL and RMT performed the experiments of cell proliferation, viability and nuclear area. JP, RMT and DJL analyzed the activity of PRDX6 and the levels of ROS. MJLG, DJL, AFGJ, BCH, RMT, JP and RRA performed Western blots and glucose and lactate determinations. RRA, DJL and CAP performed the Seahorse experiments. RRA, MJLG and DJL performed the flow cytometric analysis. DJL and CAP analyzed electron micrographs. MJLG and JAB analyzed proteomic results. JAB and CAP coordinated the cooperation and experimental design. All authors contributed to writing the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
Funding: This research has been financed by grants from the Spanish Ministry of Economy and Competitiveness (BFU2016-80006-P) and the Andalusian Government (Consejería de Economía, Innovación, Ciencia y Empleo, BIO-0216). BC-H and RMT have been financed by Programa de Empleo Joven, FEDER/Junta de Andalucía, EJ17-BIO216 and EJI-17-BIO216, respectively. D.J.L. is recipient of a predoctoral fellowship Mod. 6.2–2018 from the University of Córdoba.
Computer resources provided by the PAB (Andalusian Bioinformatics Platform) center located at the University of Málaga and technical support by the staff of the Microscopy and Proteomics facilities, SCAI, University of Cordoba, and by the staff of UCAIB-Flow Cytometry Unit, IMIBIC, are acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101737.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1 demonstrates that the mutation of knockout PRDX6 clone was biallelic and the two alleles were repaired differently. Supplementary figure 2 shows by transmission electron micrographs the morphological differences between HepG2PRDX6+/+ and HepG2PRDX6−/- cells. Supplementary Figure 3 shows Western blots of caspases 3 and 8. Supplementary file 1 and 2, are Excel spreadsheets containing global and redox proteomics quantitative data, respectively, in normal and Prdx6 knockout HepG2 cells.
figs1.
figs2.
figs3.
References
- 1.Chen J.W., Dodia C., Feinstein S.I., Jain M.K., Fisher A.B. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J. Biol. Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 2.Pedrajas J.R., McDonagh B., Hernandez-Torres F., Miranda-Vizuete A., Gonzalez-Ojeda R., Martinez-Galisteo E., Padilla C.A., Barcena J.A. Glutathione is the resolving thiol for thioredoxin peroxidase activity of 1-cys peroxiredoxin without being consumed during the catalytic cycle. Antioxidants Redox Signal. 2016;24:115–128. doi: 10.1089/ars.2015.6366. [DOI] [PubMed] [Google Scholar]
- 3.Rhee S.G. Overview on peroxiredoxin. Mol. Cell. 2016;39 doi: 10.14348/molcells.2016.2368. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher A.B. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A activities. Antioxidants Redox Signal. 2011;15:831–844. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher A.B., Dodia C., Sorokina E.M., Li H., Zhou S., Raabe T., Feinstein S.I. A novel lysophosphatidylcholine acyl transferase activity is expressed by peroxiredoxin 6. J. Lipid Res. 2016;57:587–596. doi: 10.1194/jlr.M064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorokina E.M., Feinstein S.I., Zhou S., Fisher A.B. Intracellular targeting of peroxiredoxin 6 to lysosomal organelles requires MAPK activity and binding to 14-3-3$ε$ Am. J. Physiol. Cell Physiol. 2011;300:C1430–C1441. doi: 10.1152/ajpcell.00285.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S., Zhang X., Zheng L., Li Z., Zhao X., Lai W., Shen H., Lv J., Yang G., Wang Q., Ji J. Peroxiredoxin 6 is a crucial factor in the initial step of mitochondrial clearance and is upstream of the PINK1-parkin pathway. Antioxidants Redox Signal. 2016;24:486–501. doi: 10.1089/ars.2015.6336. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=26560306&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 8.Ambruso D.R., Ellison M.A., Thurman G.W., Leto T.L. Peroxiredoxin 6 translocates to the plasma membrane during neutrophil activation and is required for optimal NADPH oxidase activity. Biochim. Biophys. Acta Mol. Cell Res. 2012;1823:306–315. doi: 10.1016/j.bbamcr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Hawkes H.J.K., Karlenius T.C., Tonissen K.F. Regulation of the human thioredoxin gene promoter and its key substrates: a study of functional and putative regulatory elements. Biochim. Biophys. Acta Gen. Subj. 2014;1840:303–314. doi: 10.1016/j.bbagen.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Fisher A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 2017;617:68–83. doi: 10.1016/j.abb.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacifici F., Arriga R., Sorice G.P., Capuani B., Scioli M.G., Pastore D., Donadel G., Bellia A., Caratelli S., Coppola A., Ferrelli F., Federici M., Sconocchia G., Tesauro M., Sbraccia P., Della-Morte D., Giaccari A., Orlandi A., Lauro D. Peroxiredoxin 6, a novel player in the pathogenesis of diabetes. Diabetes. 2014;63:3210–3220. doi: 10.2337/db14-0144. [DOI] [PubMed] [Google Scholar]
- 12.Arevalo J.A., Vázquez-Medina J.P. The role of peroxiredoxin 6 in cell signaling. Antioxidants. 2018;7 doi: 10.3390/antiox7120172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho J.-N., Lee S.B., Lee S.-S., Yoon S.H., Kang G.Y., Hwang S.-G., Um H.-D. Phospholipase A 2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Mol. Canc. Therapeut. 2010;9:825–832. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- 14.Yun H.-M., Park K.-R., Park M.H., Kim D.H., Jo M.R., Kim J.Y., Kim E.-C., Yoon D.Y., Han S.B., Hong J.T. PRDX6 promotes tumor development via the JAK2/STAT3 pathway in a urethane-induced lung tumor model. Free Radic. Biol. Med. 2015;80:136–144. doi: 10.1016/j.freeradbiomed.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Rolfs F., Huber M., Gruber F., Böhm F., Pfister H.J., Bochkov V.N., Tschachler E., Dummer R., Hohl D., Schäfer M., Werner S. Dual role of the antioxidant enzyme peroxiredoxin 6 in skin carcinogenesis. Canc. Res. 2013;73:3460–3469. doi: 10.1158/0008-5472.CAN-12-4369. http://cancerres.aacrjournals.org/cgi/doi/10.1158/0008-5472.CAN-12-4369 [DOI] [PubMed] [Google Scholar]
- 16.Schmitt A., Schmitz W., Hufnagel A., Schartl M., Meierjohann S. Peroxiredoxin 6 triggers melanoma cell growth by increasing arachidonic acid-dependent lipid signalling. Biochem. J. 2015;471:267–279. doi: 10.1042/BJ20141204. [DOI] [PubMed] [Google Scholar]
- 17.Kim S.Y., Chun E., Lee K.Y. Phospholipase A2of peroxiredoxin 6 has a critical role in tumor necrosis factor-induced apoptosis. Cell Death Differ. 2011;18:1573–1583. doi: 10.1038/cdd.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii T. Close teamwork between Nrf2 and peroxiredoxins 1 and 6 for the regulation of prostaglandin D2 and E2 production in macrophages in acute inflammation. Free Radic. Biol. Med. 2015 doi: 10.1016/j.freeradbiomed.2015.04.034. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=25968070&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 19.Kuda O., Brezinova M., Silhavy J., Landa V., Zidek V., Dodia C., Kreuchwig F., Vrbacky M., Balas L., Durand T., Hübner N., Fisher A.B., Kopecky J., Pravenec M. Diabetes; 2018. Nrf2-mediated antioxidant defense and peroxiredoxin 6 are linked to biosynthesis of palmitic acid ester of 9-hydroxystearic acid.http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=29549163&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vázquez-Medina J.P., Dodia C., Weng L., Mesaros C., Blair I.A., Feinstein S.I., Chatterjee S., Fisher A.B. The phospholipase A 2 activity of peroxiredoxin 6 modulates NADPH oxidase 2 activation via lysophosphatidic acid receptor signaling in the pulmonary endothelium and alveolar macrophages. Faseb. J. 2016;30:2885–2898. doi: 10.1096/fj.201500146R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Kim S.Y., Jo H.-Y.Y., Kim M.H., Cha Y.Y., Choi S.W., Shim J.-H.H., Kim T.J., Lee K.-Y.Y., So Y.K., Jo H.-Y.Y., Mi H.K., Cha Y.Y., Sung W.C., Shim J.-H.H., Tae J.K., Lee K.-Y.Y. H2O2-dependent hyperoxidation of peroxiredoxin 6 (Prdx6) plays a role in cellular toxicity via up-regulation of iPLA2 activity. J. Biol. Chem. 2008;283:33563–33568. doi: 10.1074/jbc.M806578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Grueso M.J., Tarradas Valero R.M., Carmona-Hidalgo B., Lagal D.J., Perinado J., McDonagh B., Requejo-Aguilar R., Bárcena J.A., Padilla C.A. Peroxiredoxin 6 down-regulation induces metabolic remodeling and cell cycle arrest in HepG2 cells. Antioxidants. 2019;8 doi: 10.3390/antiox8110505. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=31652719&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burhans W.C., Heintz N.H. The cell cycle is a redox cycle: linking phase-specific targets to cell fate. Free Radic. Biol. Med. 2009;47:1282–1293. doi: 10.1016/j.freeradbiomed.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Sarsour E.H., Kumar M.G., Chaudhuri L., Kalen A.L., Goswami P.C. Redox control of the cell cycle in health and disease. Antioxidants Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaura M., Mitsushita J., Furuta S., Kiniwa Y., Ashida A., Goto Y., Shang W.H., Kubodera M., Kato M., Takata M., Saida T., Kamata T. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2-M cell cycle progression. Canc. Res. 2009;69:2647–2654. doi: 10.1158/0008-5472.CAN-08-3745. [DOI] [PubMed] [Google Scholar]
- 27.Lim J.M., Lee K.S., Woo H.A., Kang D., Rhee S.G. Control of the pericentrosomal H2O2 level by peroxiredoxin I is critical for mitotic progression. J. Cell Biol. 2015;210:23–33. doi: 10.1083/jcb.201412068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusnak F., Reiter T. Sensing electrons: protein phosphatase redox regulation. Trends Biochem. Sci. 2000;25:527–529. doi: 10.1016/S0968-0004(00)01659-5. [DOI] [PubMed] [Google Scholar]
- 29.Heo S., Kim S., Kang D. The role of hydrogen peroxide and peroxiredoxins throughout the cell cycle. Antioxidants. 2020;9:280. doi: 10.3390/antiox9040280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phalen T.J., Weirather K., Deming P.B., Anathy V., Howe A.K., Van Der Vliet A., Jönsson T.J., Poole L.B., Heintz N.H. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J. Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon J., Wang A., Burke D.J., Boudreau H.E., Lekstrom K.J., Korzeniowska A., Sugamata R., Kim Y.-S., Yi L., Ersoy I., Jaeger S., Palaniappan K., Ambruso D.R., Jackson S.H., Leto T.L. Peroxiredoxin 6 (Prdx6) supports NADPH oxidase1 (Nox1)-based superoxide generation and cell migration. Free Radic. Biol. Med. 2016;96:99–115. doi: 10.1016/j.freeradbiomed.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maillet A., Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxidants Redox Signal. 2012;16:1285–1294. doi: 10.1089/ars.2011.4434. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330(80):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 34.Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 2017;18:529. doi: 10.1186/s12859-017-1934-z. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schattauer S.S., Land B.B., Reichard K.L., Abraham A.D., Burgeno L.M., Kuhar J.R., Phillips P.E.M., Ong S.E., Chavkin C. Peroxiredoxin 6 mediates Gαi protein-coupled receptor inactivation by cJun kinase. Nat. Commun. 2017 doi: 10.1038/s41467-017-00791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y., Feinstein S.I., Manevich Y., Chowdhury I., Pak J.H., Kazi A., Dodia C., Speicher D.W., Fisher A.B. Mitogen-activated protein kinase-mediated phosphorylation of peroxiredoxin 6 regulates its phospholipase A2 activity. Biochem. J. 2009;679:669–679. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Grueso M.J., González-Ojeda R., Requejo-Aguilar R., McDonagh B., Fuentes-Almagro C.A., Muntané J., Bárcena J.A., Padilla C.A. Thioredoxin and glutaredoxin regulate metabolism through different multiplex thiol switches. Redox Biol. 2019;21 doi: 10.1016/j.redox.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 39.Maclean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., Maccoss M.J. Skyline : an open source document editor for creating and analyzing targeted proteomics. experiments. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. http://www.pnas.org/cgi/doi/10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedrajas J.R., Padilla C.A., McDonagh B., Bárcena J.A. Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in saccharomyces cerevisiae. Antioxidants Redox Signal. 2010;13:249–258. doi: 10.1089/ars.2009.2950. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y., c Jao S., Nanduri S., Starke D.W., Mieyal J.J., Qin J. Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry. 1998;37:17145–17156. doi: 10.1021/bi9806504. http://pubs.acs.org/doi/abs/10.1021/bi9806504 [DOI] [PubMed] [Google Scholar]
- 43.Hashemy S.I., Johansson C., Berndt C., Lillig C.H., Holmgren A. Oxidation and S-nitrosylation of cysteines in human cytosolic and mitochondrial glutaredoxins: effects on structure and activity. J. Biol. Chem. 2007;282:14428–14436. doi: 10.1074/jbc.M700927200. http://www.jbc.org/cgi/doi/10.1074/jbc.M700927200 [DOI] [PubMed] [Google Scholar]
- 44.Sun C., Berardi M.J., Bushweller J.H. The NMR solution structure of human glutaredoxin in the fully reduced form. J. Mol. Biol. 1998;280:687–701. doi: 10.1006/jmbi.1998.1913. [DOI] [PubMed] [Google Scholar]
- 45.Voos W., Pollecker K. The mitochondrial lon protease: novel functions off the beaten track? Biomolecules. 2020;10 doi: 10.3390/biom10020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerezo M., Rocchi S. New anti-cancer molecules targeting HSPA5/BIP to induce endoplasmic reticulum stress, autophagy and apoptosis. Autophagy. 2017;13:216–217. doi: 10.1080/15548627.2016.1246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLeod A.K., Acosta-Jimenez L., Coates P.J., McMahon M., Carey F.A., Honda T., Henderson C.J., Wolf C.R. Aldo-keto reductases are biomarkers of NRF2 activity and are co-ordinately overexpressed in non-small cell lung cancer. Br. J. Canc. 2016;115:1530–1539. doi: 10.1038/bjc.2016.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghunath A., Sundarraj K., Nagarajan R., Arfuso F., Bian J., Kumar A.P., Sethi G., Perumal E. Antioxidant response elements: discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297–314. doi: 10.1016/j.redox.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter M.E., Hadji A., Murmann A.E., Brockway S., Putzbach W., Pattanayak A., Ceppi P. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 2015;22:549–559. doi: 10.1038/cdd.2015.3. http://www.nature.com/articles/cdd20153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maga G., Hübscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 51.Hansen J.M., Watson W.H., Jones D.P. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol. Sci. 2004;82:308–317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 52.Ichimura Y., Komatsu M. Activation of p62/SQSTM1-keap1-nuclear factor erythroid 2-related factor 2 pathway in cancer. Front. Oncol. 2018;8:1–8. doi: 10.3389/fonc.2018.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/-TrCP promotes glycogen Synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P.K., Chapman E., Fang D., Zhang D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Li Y., Luo L., Hayes J.D., Wang X.J., Tang X. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Canc. Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. http://cancerres.aacrjournals.org/cgi/doi/10.1158/0008-5472.CAN-12-3386 [DOI] [PubMed] [Google Scholar]
- 56.Akiba S., Dodia C., Chen X., Fisher A.B. Characterization of acidic Ca2+-independent phospholipase A2 of bovine lung. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998;120:393–404. doi: 10.1016/s0305-0491(98)10046-9. http://linkinghub.elsevier.com/retrieve/pii/S0305049198100469 [DOI] [PubMed] [Google Scholar]
- 57.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways. Front. Genet. 2019;10:1–12. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denechaud P.D., Fajas L., Giralt A. E2F1, a novel regulator of metabolism. Front. Endocrinol. (Lausanne) 2017;8:1–8. doi: 10.3389/fendo.2017.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bykov Y.S., Rapaport D., Herrmann J.M., Schuldiner M. Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem. Sci. 2020;xx:1–18. doi: 10.1016/j.tibs.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Leadsham J.E., Sanders G., Giannaki S., Bastow E.L., Hutton R., Naeimi W.R., Breitenbach M., Gourlay C.W. Loss of cytochrome c oxidase promotes ras-dependent ros production from the er resident nadph oxidase, yno1p, in yeast. Cell Metabol. 2013;18:279–286. doi: 10.1016/j.cmet.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Zhiming W.U., Wang H., Fang S., Xu C. Roles of endoplasmic reticulum stress and autophagy on H2O2-induced oxidative stress injury in HepG2 cells. Mol. Med. Rep. 2018;18:4163–4174. doi: 10.3892/mmr.2018.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon S.J., Stockwell B.R. The hallmarks of ferroptosis. Annu. Rev. Cell Biol. 2019;3:35–54. https://www.annualreviews.org/doi/10.1146/annurev-cancerbio-030518-055844 [Google Scholar]
- 63.Lu B., Chen X., Hong Y., Zhu H., He Q., Yang B., Ying M., Cao J. Identification of PRDX6 as a regulator of ferroptosis. Acta Pharmacol. Sin. 2019;1–9 doi: 10.1038/s41401-019-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joo J.H., Dorsey F.C., Joshi A., Hennessy-Walters K.M., Rose K.L., McCastlain K., Zhang J., Iyengar R., Jung C.H., Suen D.F., Steeves M.A., Yang C.Y., Prater S.M., Kim D.H., Thompson C.B., Youle R.J., Ney P.A., Cleveland J.L., Kundu M. Hsp90-Cdc37 chaperone complex regulates Ulk1- and atg13-mediated mitophagy. Mol. Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corona Velazquez A.F., Jackson W.T. So many roads: the multifaceted regulation of autophagy induction. Mol. Cell Biol. 2018;38:1–14. doi: 10.1128/mcb.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niso‐Santano M., Malik S.A., Pietrocola F., Bravo‐San Pedro J.M., Mariño G., Cianfanelli V., Ben‐Younès A., Troncoso R., Markaki M., Sica V., Izzo V., Chaba K., Bauvy C., Dupont N., Kepp O., Rockenfeller P., Wolinski H., Madeo F., Lavandero S., Codogno P., Harper F., Pierron G., Tavernarakis N., Cecconi F., Maiuri M.C., Galluzzi L., Kroemer G. Unsaturated fatty acids induce non‐canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moruno-Manchón J.F., Peŕez-Jiménez E., Knecht E. Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway. Biochem. J. 2013;449:497–506. doi: 10.1042/BJ20121122. [DOI] [PubMed] [Google Scholar]
- 68.Huang W.S., Huang C.Y., Hsieh M.C., Kuo Y.H., Tung S.Y., Shen C.H., Hsieh Y.Y., Teng C.C., Lee K.C., Lee K.F., Kuo H.C. Expression of PRDX6 correlates with migration and invasiveness of colorectal cancer cells. Cell. Physiol. Biochem. 2019;51:2616–2630. doi: 10.1159/000495934. [DOI] [PubMed] [Google Scholar]
- 69.Naryzhny S.N., Lee H. Proliferating cell nuclear antigen in the cytoplasm interacts with components of glycolysis and cancer. FEBS Lett. 2010;584:4292–4298. doi: 10.1016/j.febslet.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 70.Cooper S.E., Hodimont E., Green C.M. A fluorescent bimolecular complementation screen reveals MAF1, RNF7 and SETD3 as PCNA-associated proteins in human cells. Cell Cycle. 2015;14:2509—2519. doi: 10.1080/15384101.2015.1053667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baple E.L., Chambers H., Cross H.E., Fawcett H., Nakazawa Y., Chioza B.A., V Harlalka G., Mansour S., Sreekantan-Nair A., Patton M.A., Muggenthaler M., Rich P., Wagner K., Coblentz R., Stein C.K., Last J.I., Taylor A.M.R., Jackson A.P., Ogi T., Lehmann A.R., Green C.M., Crosby A.H. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J. Clin. Invest. 2014;124:3137–3146. doi: 10.1172/JCI74593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duffy C.M., Hilbert B.J., Kelch B.A. A disease-causing variant in PCNA disrupts a promiscuous protein binding site. J. Mol. Biol. 2016;428:1023–1040. doi: 10.1016/j.jmb.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 73.Padilla C.A., Martínez-Galisteo E., López-Barea J., Holmgren A., Bárcena J.A. Immunolocalization of thioredoxin and glutaredoxin in mammalian hypophysis. Mol. Cell. Endocrinol. 1992;85:1–12. doi: 10.1016/0303-7207(92)90119-Q. [DOI] [PubMed] [Google Scholar]
- 74.Rozell B., Barcena J.A., Martinez-Galisteo E., Padilla C.A., Holmgren A. Immunochemical characterization and tissue distribution of glutaredoxin (thioltransferase) from calf. Eur. J. Cell Biol. 1993;62 [PubMed] [Google Scholar]
- 75.García-Pardo L., Granados M.D., Gaytán F., Padilla C.A., Martínez-Galisteo E., Morales C., Sánchez-Criado J.E., Bárcena J.A. Immunolocalization of glutaredoxin in the human corpus luteum. Mol. Hum. Reprod. 1999;5:914–919. doi: 10.1093/molehr/5.10.914. [DOI] [PubMed] [Google Scholar]
- 76.Xu Y., Morse L.R., Da Silva R.A.B., Odgren P.R., Sasaki H., Stashenko P., Battaglino R.A. PAMM: a redox regulatory protein that modulates osteoclast differentiation. Antioxidants Redox Signal. 2010;13:27–37. doi: 10.1089/ars.2009.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savitsky P.A., Finkel T. Redox regulation of Cdc25C. J. Biol. Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 78.Reddy N.M., Kleeberger S.R., Bream J.H., Fallon P.G., Kensler T.W., Yamamoto M., Reddy S.P. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.