Abstract
Connectomics has demonstrated that synaptic networks and their topologies are precise and directly correlate with physiology and behavior. The next extension of connectomics is pathoconnectomics: to map neural network synaptology and circuit topologies corrupted by neurological disease in order to identify robust targets for therapeutics. In this report, we characterize a pathoconnectome of early retinal degeneration. This pathoconnectome was generated using serial section transmission electron microscopy to achieve an ultrastructural connectome with 2.18nm/px resolution for accurate identification of all chemical and gap junctional synapses. We observe aberrant connectivity in the rod-network pathway and novel synaptic connections deriving from neurite sprouting. These observations reveal principles of neuron responses to the loss of network components and can be extended to other neurodegenerative diseases.
Keywords: Connectomics, Pathoconnectomics, Retinal Degeneration, Retinitis Pigmentosa, Neurodegeneration, Abnormal Ribbon Morphology, Rod Pathway, GABAergic Amacrine Cells, Neurodegeneration
1. Introduction
1.1. Connectomics
The precise organization of neurons and their connections in brain, retina, and spinal cord defines network topologies underlying healthy nervous system processing (Marc, et al., 2013). This has been demonstrated in numerous neural networks from spinal circuits (Catela, et al., 2015), to C. Elegans (White, et al., 1986), to the cerebral cortex (Lee, et al., 2016), where distinct classes of neurons participate in synaptic networks to create precise network topologies. The study of all of the synaptic connections (ideally both chemical and electric) between all cells in a given neural network is termed connectomics. In 2011, we published the first retinal connectome, Retinal Connectome 1 (RC1): an ultrastructural connectome of rabbit retina generated to elucidate the neural networks in retina contributing to visual processing (Anderson, et al., 2011a). Many papers have detailed the specificity of retinal network motifs (Diamond, 2017, Thoreson and Dacey, 2019, Thoreson and Witkovsky, 1999, Tsukamoto and Omi, 2015, 2014, 2013), including those deriving from RC1 (Anderson, et al., 2011a, Lauritzen, et al., 2013, Lauritzen, et al., 2019, Marc, et al., 2014, Marc, et al., 2018, Sigulinsky, et al., 2020), but there are many more motifs yet to discover, particularly those involving inhibitory networks and glial interactions (Pfeiffer, et al., 2019).
1.2. Why the Retina
The retina is an ideal candidate for connectomics approaches. It is compact and highly ordered, with a series of neuronal cell soma layers and plexiform or interconnecting layers containing repeating network motifs (Masland, 2012). Understanding how retinal neural circuits are organized, may in fact reveal general principles that neural circuits use in other nervous system tissues. Annotation and analysis of retinal connectomes is delivering a more complete understanding of the organization of neural network topology in retina than we have from any other region of the mammalian CNS.
The retina is unique in its accessibility, as gross histology, functional, and behavioral assays can be conducted non-invasively, down to cellular resolution in living animals using optical coherence tomography (OCT) (Lujan, et al., 2015) and electroretinogram (ERG) (Creel, 2019) to track the progression of disease. This accessibility is important for understanding the basic network topologies of healthy retinas in addition to dynamic processes such as development (Johnson, et al., 2017, Tien, et al., 2017), and how topologies are altered through aging (Samuel, et al., 2011), and disease.
1.3. Neural Networks in Degeneration
Disruption of neural network organization leads to deficits or failures in network function (Chew, et al., 2017, Day, et al., 2006, delEtoile and Adeli, 2017). In a simple, static network, this is intuitive: Neuron A is presynaptic to neuron B, neuron B is presynaptic to neuron C, the loss of neuron A or B will lead to a lack of input to neuron C (Bargmann, et al., 1993). These simple neural network graphs rapidly gain complexity with the addition of more neurons and associated synapses. Therefore, understanding neural networks and how changes in them contribute to the failure of appropriate network processing in neurological disease or retinal degeneration is critical for understanding the progression of these diseases (Agosta, et al., 2010, Day, et al., 2006, delEtoile and Adeli, 2017, Guo, et al., 2015, Lacor, et al., 2007). Furthermore, creating targeted therapeutic interventions to stop, reverse, modify, or interact with degenerate neural networks will depend upon understanding precisely which targets are involved in networks that are altered in disease.
1.4. Retinal Degenerative Disease: Retinitis Pigmentosa and Age-Related Macular Degeneration
Progressive neurodegenerative disease seen in retinal degenerative diseases like retinitis pigmentosa (RP) (Jones, et al., 2016a) or Age-Related Macular degeneration (AMD) (Jones, et al., 2016b) have many causes, but the final remodeling pathways converge on photoreceptor cell death, retinal remodeling, and often blindness. RP is a retinal disease with hallmarks of photoreceptor degeneration and subsequent neural retinal deafferentation, affecting approximately 1 in 4000 individuals (Hamel, 2006). Although over 100 genes are associated with this disease, the sequence of degeneration and clinical findings in rod dominated RP are: rod photoreceptors degenerate causing neural retinal deafferentation, loss of night and peripheral vision, followed secondarily by the degeneration of cone photoreceptors and total or near total vision loss (Hartong, et al., 2006). In the retina, these negative plastic changes and cell death revise the circuit topology through a process called retinal remodeling (Jones and Marc, 2005, Marc, et al., 2003) that has been observed in not only human RP, but also all models examined to date of RP, as well as human AMD. For further review of photoreceptor degenerations and retinal remodeling, see: (Jones and Marc, 2005).
This report examines the ultrastructural changes in circuitry of the retina of a model of RP, resulting in rewiring, which is the specific process of changes in network topology that occur through neurite outgrowth (Fariss, et al., 2000, Lewis, et al., 1998, Strettoi and Pignatelli, 2000), formation of new synapses (Peng, et al., 2000), and loss of existing connections (Linberg, et al., 2006).
1.5. Pathoconnectomics
Recent work has demonstrated that retina behaves similarly to CNS with respect to late stage disease with many of the same features observed in retina as in other neuropathies of the brain (Pfeiffer, et al., 2020b). This, combined with the features that made the retina ideal for the initial mapping of neuronal networks, makes it the best candidate for mapping neuronal network disruptions that occur as a result of neurological disease. We are evaluating these disruptions by creating a series of connectomes of pathological tissue at advancing stages of degeneration: Pathoconnectomes. This is the first publication to our knowledge, noting the creation of pathoconnectomics approaches, or pathological connectomes of disease tissues to demonstrate how wiring is altered in models of neurodegeneration. It is our hypothesis that network changes associated with retinal degenerative disease, found in retinal pathoconectomics will expose underlying principles of negative plasticity that are generalizable across the CNS.
Our goal is to detail all of the network topologies in retina disrupted by photoreceptor degeneration. The purpose of this is two-fold: 1) To create network diagrams for better targeting of therapeutic interventions for vision loss. 2) To uncover the basic principles of neural network responses to altered or missing input. In this report, we make a first step towards accomplishing these goals through the introduction of the structure, features, and initial findings from Retinal Pathoconnectome 1 (RPC1). RPC1 is an open-access, serial-section transmission electron microscopy (ssTEM) pathoconnectome volume, generated from a rabbit model of autosomal-dominant cone-sparing RP.
2. Material and Methods
2.1. Model System
The retina used to generate RPC1 is from a heterozygous transgenic (Tg) P347L rabbit, which was originally generated in the Kondo Laboratory (Kondo, et al., 2009). The Tg P347L rabbit has been extensively characterized and follows a pattern of progression similar to that of human cone-sparing RP (Jones, et al., 2011, Ueno, et al., 2019). The rabbit was 10 months old at the time of enucleation and extensive photoreceptor degeneration had begun, though the central retina still had numerous rods intact. The retina used in the control connectome RC1, was from a 13-month-old Dutch belted rabbit. The characteristics of RC1 volume have been described previously (Anderson, et al., 2011a). All animal experiments were conducted according to the ARVO Statement for the use of animals in Opthalimic and Vision Research, with the approval of the Institutional Animal Care and Use Committee (IACUC) of the University of Utah.
2.2. Tissue Preparation
Tissues are fixed in a mixed aldehyde solution (1% formaldehyde, 2.5% glutaraldehyde), dehydrated in graded alcohols, then embedded in epoxy resins. Tissues are then serially sectioned at 70nm and prepared for TEM capture or CMP analysis by placing the section on a formvar grid or 12-well slide, respectively.
2.3. Image Acquisition
Transmission electron microscopy (TEM) provides the high resolution required for connectomics approaches to visualize the network components, along with the quantitative aspects of computational molecular phenotyping (CMP) to molecularly label cell identity (Anderson, et al., 2011a). These approaches have been previously used to create a healthy rabbit connectome RC1: a 250μm in diameter volume of retina spanning from the apical inner nuclear layer (INL) to the basal ganglion cell layer (GCL), which serves as a ground truth for normal retinal circuit topology. RPC1 was constructed in identical fashion. In electron microscopy, adequate resolution is required to definitively identify all network components (2.18nm/px is the lowest resolution in which gap junctions can be positively identified) and the identification of pre- and post-synaptic structures is crucial to the accurate identification of networks. Therefore, our routine TEM acquisitions are acquired at 2.18 nm/pixel, and sections can be re-imaged at 0.27 nm/pixel, with a goniometric tilt as necessary to confirm certain circuit identities like gap junctions (Kolb and Famiglietti, 1974, Marc, et al., 2018, Sigulinsky, et al., 2020).
2.4. Volume Assembly and Annotation
Serial TEM images were acquired on a JEOL JEM-1400 electron microscope with a 16-Mpixel Gatan camera, while CMP images were acquired using a Leica light-microscope affixed with 8-bit CCD camera. RPC1 was assembled using the custom Nornir tools developed for volume assembly. Nornir combines individual images acquired from transmission electron microscopy or from a light-microscope, and registers individual image tiles into assembled mosaics before using automated registration to align adjacent sections throughout the volume, with minimal manual correction (Anderson, et al., 2009, Marc, et al., 2012). Following volume assembly, navigation and annotation of the dataset is performed in the Viking software environment (Anderson, et al., 2011b). Annotation of every chemical or electrical synapse type is made to avoid false positives of connectivity and minimize missed structures. For further description of synapse identification metrics used in this study, see supplementary Fig S1–S3.
2.5. Computational Molecular Phenotyping (CMP)
Along with ultrastructural data, molecular tagging of cellular identity is fundamental to understanding contributors to neuronal networks. CMP allows for cell classification through quantitatively evaluating the combinations of small molecules, which are stoichiometrically trapped during fixation (Marc and Cameron, 2002, Marc, et al., 1995, Ottersen, 1989), and detected with glutaraldehyde-tolerant IgGs. The concentrations of the probed amino acids and proteins are used in the identification of cells beyond their morphology and synaptology (Table 1). For more in depth information on CMP technologies and usage see (Marc, et al., 1995, Pfeiffer, et al., 2020a). Small molecule IgGs to GABA, L-glutamate, L-glutamine, glycine, and taurine, in addition to IgGs targeting glial fibrillary acidic protein (GFAP) were used in the classification of cells in the RPC1 volume.
Table 1.
Immunocytochemistry Reagents:
| Reagent | RRID | Source | Dilution |
|---|---|---|---|
| anti-L-glutamate IgG | AB_2532055 | Signature Immunologics | 1:50 |
| anti-glycine IgG | AB_2532057 | Signature Immunologics | 1:50 |
| anti-L-glutamine IgG | AB_2532059 | Signature Immunologics | 1:50 |
| anti-taurine IgG | AB_2532060 | Signature Immunologics | 1:50 |
| anti-GABA IgG | AB_2532061 | Signature Immunologics | 1:50 |
| anti-GFAP | AB_10013382 | Dako | 1:200 |
2.6. Viking Software
Viking annotations encode information about the size, structure, and location of all annotated structures within connectomes and pathoconnectomes, facilitating database-base queries of parameters including statistics of encoded structures (Anderson, et al., 2011b, Marc, et al., 2012). These databases are publicly accessible at www.connectomes.utah.edu and http://websvc1.connectomes.utah.edu/RPC1/OData.
2.7. Figure Assembly
Figures were assembled using screenshots from the RPC1 volume in Viking (http://connectomes.utah.edu/, RRID:SCR_005986), 3D renderings of cells were generated in VikingView (DOI: 10.5281/zenodo.3267451, https://zenodo.org/record/3267451#.XSUW1OhKguU), and whole volume renderings were generated in Blender (http://www.blender.org/; RRID: SCR_008606). Final figure assembly was done in Photoshop 6.0.
3. Results
3.1. Retinal pathoconnectome 1 (RPC1)
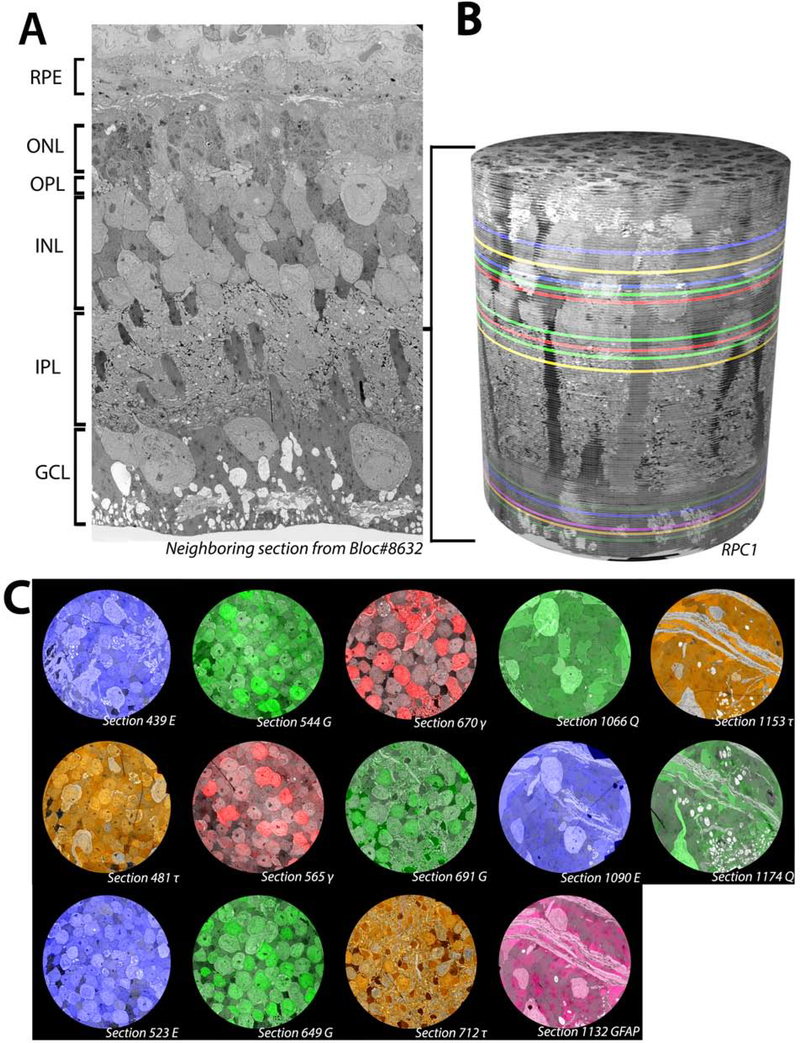
RPC1 is composed of 946 serial TEM sections with 14 intercalated IgG probes generated against small molecules important in metabolism (Fig 1). In this volume, 1 out of 30 sections cut were placed on slides for Computational Molecular Phenotyping (CMP) probing, while the other sections were placed on formvar grids for ssTEM imaging. The RPC1 volume is narrower in diameter (70μm) than our control RC1 volume (250μm), allowing for more rapid capture and assembly, while still large enough to capture repeated rod-network motifs (Lauritzen, et al., 2019). RPC1 is the first pathoconnectome volume in a series of 3, each with increasingly progressive photoreceptor degeneration (with eventual neurodegeneration in later timepoints) to chronicle landmarks in rewiring as the disease progresses. RPC1 derives from a 10-month-old Tg P347L rabbit retina, model of RP (see methods). In the selected region of retina (peri-visual streak), photoreceptor degeneration had initiated and the outer nuclear layer (ONL) was reduced in thickness by approximately 50% (Fig 1A). Though rod photoreceptors were the most heavily degenerated neuronal class, many rods remain morphologically intact. The remaining rod photoreceptors, along with the inner retinal cell population, allowed for connectomics-based evaluation of early-stage RP rewiring, corresponding to a clinical point where human patients would still have vision, though would likely be suffering from scotopic deficits as well as adaptation deficits. In total, 321 neuronal and glial somas have been identified within the volume and assigned to broad classes in RPC1. The breakdown of cells is as follows: 113 photoreceptors, 5 horizontal cells, 81 amacrine cells, 79 bipolar cells, 4 ganglion cells, at least 36 Müller glia, and 4 microglia. Examination of these cell classes has revealed altered network topologies associated with rewiring in early photoreceptor degeneration, prior to complete rod degeneration or inner retinal neuronal loss.
Figure 1.
Overview of RPC1. (A) Vertical section through tissue directly adjacent to the tissue processed for the RPC1 volume. (B) 3D composite volume of RPC1. Pseudocolored sections illustrate the locations of CMP sections for small molecules indicated in 1C. (C) Overlaid CMP sections from RPC1 on their adjacent TEM sections.
3.2. Rod-network pathologies
Rod photoreceptors are the first cells in this heterogeneous neuronal network to degenerate. Therefore, the first network we sought to characterize in RPC1 was the rod-mediated network. The literature of retinal networks is vast, so only a simplified description of the rod-network is provided below along with a graphical description (Fig 2). For more comprehensive descriptions of additional retinal networks see: (Diamond, 2017, Lauritzen, et al., 2019, Marc, et al., 2014, Marc, et al., 2018, Thoreson and Dacey, 2019).
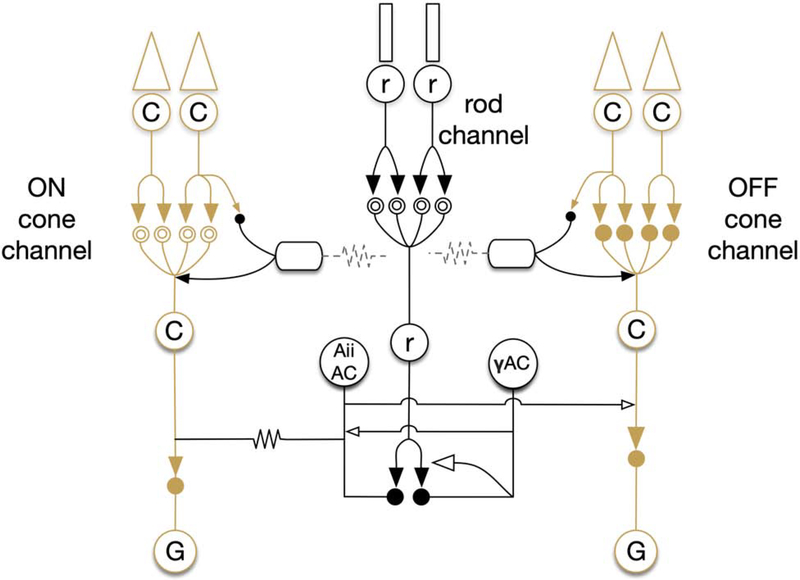
Figure 2:
Simple diagram of bipolar cell network. Solid arrows indicate glutamatergic synapses. Open arrows indicate inhibitory synapses (GABA or glycine). Solid circles indicate ionotropic glutamate receptors, while mGluR6 is indicated by the double open circles. Gap junctions are indicated by zig-zag lines.
Rod photoreceptors (rods) are specialized neurons responsible for detection of photons in low-light (scotopic) conditions. Following photon capture, rods initiate an enzymatic cascade ultimately leading to a depolarization. This depolarization causes a decrease in glutamate release into the synaptic cleft, signaling to postsynaptic rod bipolar cell (RodBCs) dendrites, via the first synapse in vision located in the outer plexiform layer (OPL). Axons of RodBCs branch and synapse in the deepest layer of the inner plexiform layer (IPL), close to, but not connecting with ganglion cells in the ganglion cell layer (GCL). RodBCs are presynaptic to at least 2 classes of amacrine cells: GABAergic amacrine cells (γACs) and Aii glycinergic amacrine cells (Aii GACs). Aii GACs are chemically presynaptic to OFF cone bipolar cells (OFF CBC) using the neurotransmitter glycine, causing inhibition. However, in the ON-layer, Aii GACs make gap junctions with ON cone bipolar cells (ON CBCs), sharing their polarization state through these gap junctions with the ON CBCs. ON CBCs in turn synapse with ganglion cells that ultimately project out of the retina, and into the brain via the optic nerve, thereby passing the rod signal out of the retina. In healthy retina, RodBCs are never presynaptic to ganglion cells and therefore “piggyback” on the ON CBC pathway using the Aii GAC as described above. This rod pathway is highly conserved and observed in all mammalian retinas.
3.2.1. RodBC Outer Plexiform Layer Pathology
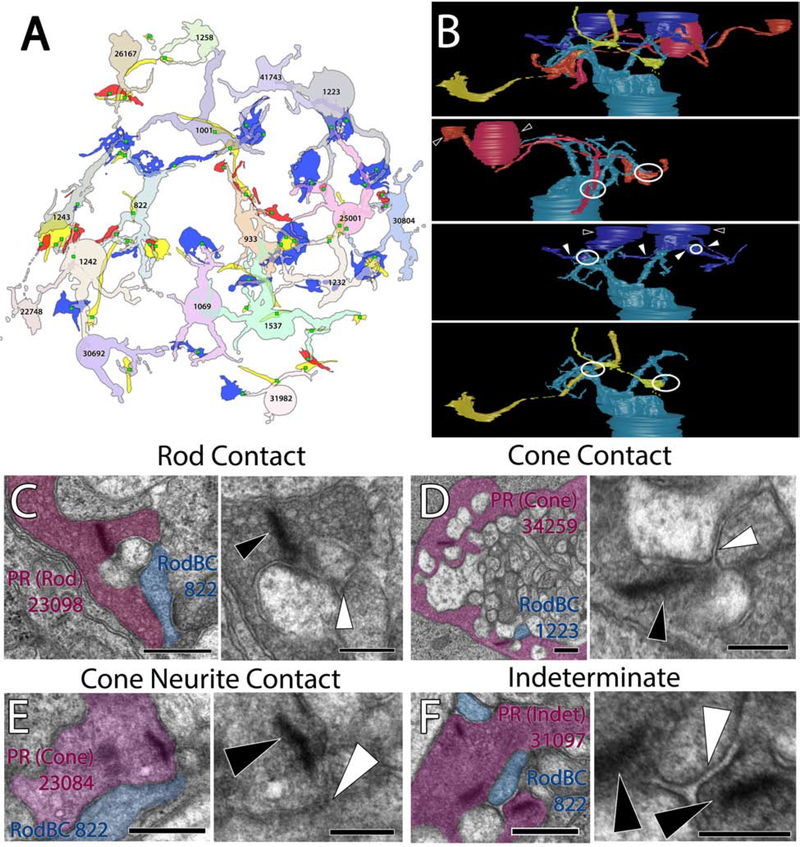
RPC1 contains 17 RodBCs, 8 of which have axonal and dendritic arbors contained entirely within the volume. RodBCs are traditionally thought to be exclusively postsynaptic to rod photoreceptors in the healthy retina. In contrast, recent studies have proposed that RodBCs may be post-synaptic to cones in addition to rods in healthy retina. In evaluating these studies, we find the resolution and lack of clear post-synaptic densities to the cone pedicles make it difficult to determine whether the RodBCs are truly synapsing with the cone pedicle or merely traversing in close proximity (Behrens, et al., 2016, Pang, et al., 2018). Because our RC1 volume does not include an OPL, we could not directly assess whether these contacts exist using our identification metrics, though the serial TEM reconstruction of RodBCs in a healthy mouse by Tsukamoto and Omi failed to identify any cone inputs to these cells (Tsukamoto and Omi, 2013). In photoreceptor degeneration retinal diseases however, it is widely accepted that RodBCs retract their rod-contacting dendrites as rods die and extend new neurites towards cone pedicles making peripheral contacts (Peng, et al., 2000). In evaluating the OPL of RPC1, we found this process initiates prior to complete rod degeneration. RodBCs maintained contact with existing rods, while many simultaneously demonstrated aberrant contacts with cones (Fig 3). We observed 10 RodBCs post-synaptic to cone photoreceptors, with 4 of those cone-contacting RodBCs maintaining at least 1 post-synaptic density to a rod axon terminal (spherule) (Fig 3A). Though most cone-contacts are with traditional cone axon terminals (pedicles) (Fig 3B), we found one instance of a sprout off the primary pedicle with a secondary terminal presynaptic to RodBC 822 (Fig 3C+D). We found an additional 7 RodBCs with densities opposing non-traditional structures containing ribbons (fig 3E). In these cases, we found 1 RodBC opposing axonal ribbons and no terminal, and 6 opposing ribbon terminals inconsistent with morphology typically associated with rod spherules or cone pedicles and could not be conclusively morphologically classified as a rod or cone terminal. Because 9 of the RodBC dendritic arbors evaluated are not entirely contained within the volume, the number of cone contacts made by RodBCs in RPC1 may be greater than the currently tabulated 58.8% (10/17). These results confirm previous light microscopy work indicating RodBCs extend neurites to cone pedicles as the rod photoreceptors degenerate, and expands upon this by determining RodBCs extend neurites towards cone pedicles prior to becoming fully deafferented from rod spherules.
Figure 3:
RodBC dendritic contacts. (A) Representative rod photoreceptor synapse of RodBC 822 postsynaptic to rod. (B) Representative cone photoreceptor synapse between a cone photoreceptor and RodBC 1223. (C) 3D rendering of cone photoreceptor 23084 main terminal and 2 neurite processes. (D) Representative photoreceptor synapse between a cone neurite ribbon and RodBC 822 (E) Representative photoreceptor synapses between an indeterminate photoreceptor and RodBC 822. Black arrowheads indicate ribbon densities, while white arrowheads indicate associated post-synaptic densities. Viking Annotation and Pseudocolored Images Scale bars 500nm. Higher Magnification Scale Bars 250nm
3.2.2. RodBC Inner Plexiform Layer Pathology
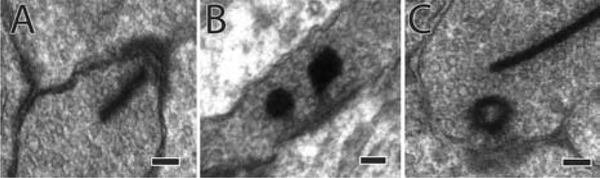
We then compared the inner retinal network of RPC1 with the previously described IPL network topologies of RC1 to determine the potential network impacts of changes in RodBC inputs. RodBCs are typically only associated with chemical synapses within the IPL. Specifically, RodBCs in the healthy retina are found presynaptic via glutamatergic ribbon synapses to Aii GACs and yACs. We found the total ribbon synapses made by complete RodBCs of RPC1 (n=8) were similar to representative RodBCs (n=5) from RC1. RPC1 ribbons: 34.125 ± 4.29 (mean ± standard deviation) and RC1 ribbons: 34.4 ± 2.5 (p=0.8, unpaired Student’s-t-test). Although the numbers of ribbon synapses were not altered, some ribbon synapses of bipolar cells, including RodBCs, were morphologically altered. Rather than the typical “bar” shaped ribbons (Fig 4A), a subset of ribbons (n=16) had a spherical morphology (Fig 4B+C) commonly associated with immature or disassembling ribbon synapses (Adly, et al., 1999, Spiwoks-Becker, et al., 2004, Strettoi, et al., 2002). In addition to their presynaptic signaling, RodBCs are also heavily postsynaptic to inhibitory synapses from yACs (Lauritzen, et al., 2019). The prevalence of postsynaptic densities was also similar between RPC1: 69.625 ± 14.050 and RC1 62.8 ± 13.498 (p=0.406, unpaired Student’s-t-test). These results suggest that chemical synapse numbers are not substantially changed at this early stage of photoreceptor degeneration.
Figure 4:
Spherical Ribbons in RodBCs of RPC1. (A) Example normal ribbon made by RodBCs. (B) 2 example spherical ribbons. (C) The hollow conformation of spherical synapses. Scale bars: 100nm
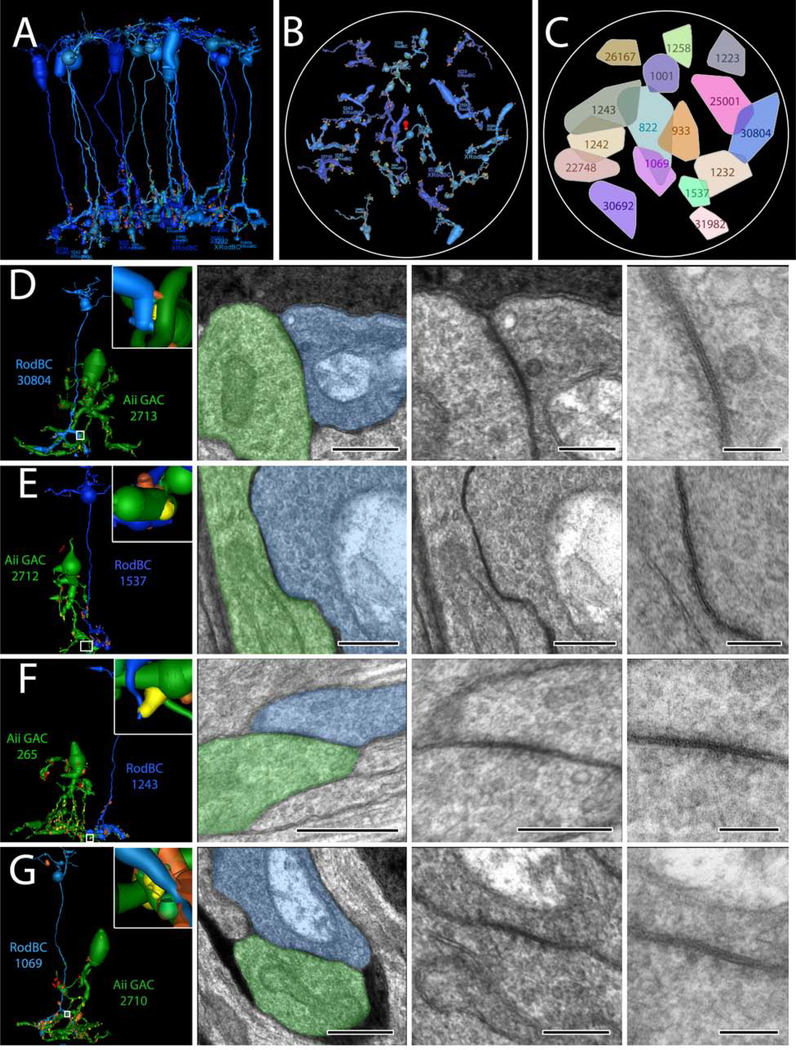
Chemical synapses between RodBCs, Aii GACs, and yACs appear to be largely intact in the early degenerate retina and occur in similar frequency to that observed in healthy retina. However, we observed the emergence of gap junctions involving RodBCs of RPC1. In the adult healthy mammalian retina, RodBCs do not make gap junctions. This is confirmed in our RC1 volume, where none of the 104 RodBC axonal arbors are found to contain a single gap junction (Sigulinsky, et al., 2020). In contrast, all 17 RodBCs found within RPC1 formed at least 1 gap junction. In total, we have identified 50 candidate gap junctions, 18 of which were confirmed using high magnification (0.43nm/pixel) and goniometric tilt (Fig 5), which revealed the confirmatory pentalaminar structure associated with gap junctions. At least one gap junction per RodBC was confirmed using high magnification with the exception of RodBC 26167, which is largely off volume and recapture was not possible of its only identified candidate gap junction. Next, we identified gap junctionally coupled partners of these RodBCs. No gap junctions were found between paired RodBCs in RPC1, despite substantial overlap of arbors in some areas (Fig 5C). Of the 50 gap junctions, 48 (96%) were positively identified with Aii GACs. In all cases of gap junctions with Aii GACs in RPC1, the individual RodBC was also heavily presynaptic to the same Aii GAC via glutamatergic ribbon synapses, often within the same varicosity as the gap junction. Of the gap junctions not made with a confirmed Aii GAC, 1 was with an unidentified partner, which could not be conclusively identified due to being near the volume edge. The other non-Aii GAC partner was an ON CBC. These data demonstrate that RodBCs, early in the retinal degenerative process, alter their inner retina synaptology through the formation of gap junctions, especially with Aii GACs, though chemical synaptology remains intact. There also appears to be preferred class-specific partnering via gap junctions that may reveal the presence of cell-cell markers or targets for therapeutic intervention. What implications this has for network performance or stability is currently unclear, but is an area of future modeling work.
Figure 5:
RodBC gap junctions with Aii GACs (A) 3D rendering of all 16 RodBCs from RPC1. (B) RodBC axonal arbor fields in RPC1. (C) Convex Hulls of all RodBCs in RPC1 (D-E) Gap junctions between RodBCs and Aii GACs. Left panel is 3D rendering with inset higher magnification image of specific gap junction annotations. White boxed image corresponds to white box in adjacent left panel. * indicate ribbons and # indicates gap junctions arising in same varicosity. 100nm scale images are 25k recaptures of gap junction structures.
3.3. GABAergic Amacrine Cell Neurite Sprouting
There are 31 identified and annotated GABAergic amacrine cell (γAC) somas contained within the RPC1 volume. In the healthy retina, γACs (with the exception of some starburst amacrine cells) have a soma that is predominately positioned within the INL just apical to the IPL, with processes extending solely into the IPL. Within the IPL yACs make conventional inhibitory chemical synapses with multiple partners, and some classes are also known to make extensive gap junctions with other yACs and certain classes of ganglion cells (Marc, et al., 2018). All 107 γACs contained within RC1 are consistent with this description.
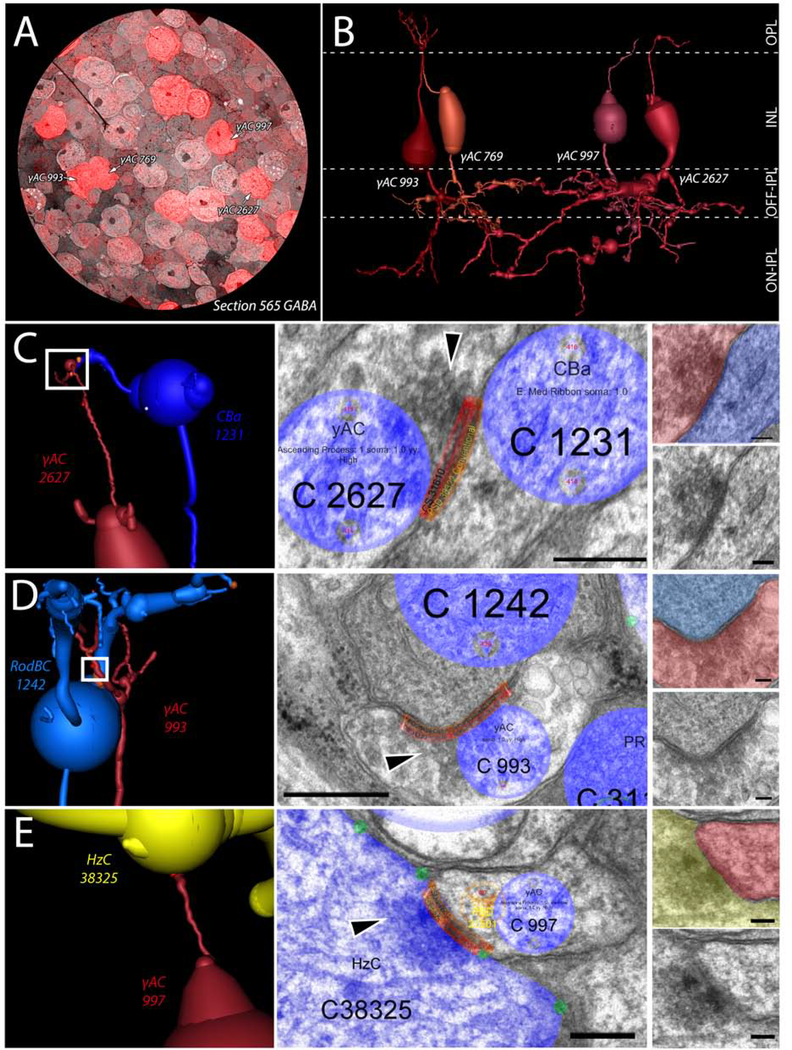
Conversely in disease, prior work has demonstrated that processes of γACs can be found extending apically towards the OPL during photoreceptor degeneration (Jones, et al., 2011). However, it was unclear whether these GABAergic processes were neurites that extended off of the normal processes below the soma before extending apically, or whether they arose from the soma itself. In RPC1, 4 of the 31 γACs were found to extend processes from the apical side of their somas into the OPL (γAC cell #s: 769, 993, 997, and 2627) (Fig 6). Not only is this confirmatory at ultrastructure of the previously described novel GABAergic processes in the OPL, but demonstrates that the extension of these processes arises from the soma, and manifests prior to complete loss of rods. We then analyzed the synaptic connections made by these aberrant apical processes and found chemical synapses in the OPL made by these γAC neurites. However, the number of synapses and their pre- and post-synaptic partners was variable. γAC 769 extended into the OPL, but made no identifiable synapses. Conversely, γAC 993 was the most complex of these apical extending processes. γAC 933 made 6 synaptic contacts in the OPL: 2 postsynaptic densities to conventional synapses originating from horizontal cells and 4 synapses in which γAC 993 was presynaptic to multiple classes of ON bipolar cells, creating a novel 2-hop inhibitory network between horizontal cells and ON bipolar cells. γAC 997 was postsynaptic to a single horizontal cell, and γAC 2627 was presynaptic to a horizontal cell and an OFF bipolar cell. These connectivities are novel in 2 distinct ways: 1) γACs in RPC1 make typologically correct contacts with bipolar cells, even when encountered in the wrong lamina. 2) γACs make aberrant synapses with horizontal cells that they never come in physical proximity with in the healthy retina, potentially affecting center-surround inhibition created by the horizontal cells. This demonstrates the ability of deviant processes that infiltrate an aberrant layer to make synapses with numerous typologically correct and incorrect partners, clearly altering the neural networks downstream of cells actively undergoing degeneration.
Figure 6:
Ascending process of GABAergic amacrine cells. (A) CMP overlay of GABA section 565 on its neighboring TEM section. Intensity of red color indicates higher levels of GABA contained within the cell. (B) 3D rendering of the 4 γACs with ascending processes in RPC1. (C) Synapse between γAC 2627 and OFF-BC 1231 in the OPL. White box indicates region of synapse shown on the right in increasing magnification. Black arrowhead indicates the presynaptic vesicle cloud. (D) Synapse between γAC 993 and RodBC 1242 in the OPL. White box indicates region of synapse shown on the right in increasing magnification. Black arrowhead indicates the presynaptic vesicle cloud. (E) Synapse between γAC 997 and HzC 38325 in the OPL. White box indicates region of synapse shown on the right in increasing magnification. Black arrowhead indicates the presynaptic vesicle cloud. Scale bars: 250nm (Viking annotated) 100nm (smaller inset).
4. Discussion
Rewiring of neural networks through plasticity mechanisms is a prominent consequence of injury or disease in the central nervous system. Multiple studies have explored the mechanisms of neurite outgrowth (Lin, et al., 2012), localized the sites of projections (Fisher, et al., 2005, Peng, et al., 2000), and identified a subset of synaptic partners (Fisher, et al., 2005, Morimoto, et al., 2004, Schmid, et al., 2016). Unfortunately, exploring this using antibody probes relies on pre-existing expectations of proteins that may be involved. In addition, even when the correct protein is predicted, many synaptic structures underlying networks are too small to resolve in light microscopy or lower resolution electron microscopy approaches. Ultrastructural pathoconnectomes captured at 2nm/pixel allows for exploration and characterization of these novel networks formed by native neurons and aberrant neurites to establish precise wiring topologies, as well as identify potential rulesets that neural tissues undergoing degeneration may adopt. These networks also identify targets for potential therapeutic intervention.
In RPC1, this region of retina still contains both cones and a substantial population of rod photoreceptors. Therefore, this region would likely still exhibit both photopic and scotopic light responses, but might have difficulty adapting between the two environments given corruption of the rod network through the introduction of gap junctions. These findings support clinical observations in patients undergoing early stages of progressive retinal degenerative diseases (Kalloniatis and Fletcher, 2004). Our observations of changes in the rod pathway and?AC connectivities are important contributing components to understanding the progression of retinal degenerative disease, and are fundamentally important to implementing therapeutic interventions. It is crucial to note that these network revisions are occurring prior to substantial inner retina neuronal cell loss, and certainly prior to the network completely failing.
RodBCs retract processes away from rod photoreceptor terminals (while maintaining contact with others) and instead contact cone pedicles in addition to apparent neurites off of cone pedicles. In some ways this could be predicted from earlier studies, but the precise connectivity required connectomic analysis. Beier and colleagues demonstrated in 2017, that following laser ablation of rod photoreceptors, RodBCs would extend new processes to rods still intact at the edge of the ablation zone (Beier, et al., 2017). These results demonstrate deafferented RodBCs do search for new photoreceptor inputs, though in this study it was concluded that there is a preferential selection for rod photoreceptor terminals when they are available. Our findings indicate that while rods are likely still the preferred input for RodBCs, they will begin making aberrant synapses with cone terminals prior to the complete loss of available rods, including with neurite sprouts off of existing cone terminals. A prior study by Fei et al., demonstrated cone neurite sprouting in late retinal degeneration following substantial cone degeneration (Fei, 2002). However, we find this process initiates early in photoreceptor degeneration, prior to the complete loss of rod photoreceptors and onset of cone degeneration. Also, in 2017 work from the Kerschensteiner group demonstrated high plasticity in RodBCs where following partial ablation of RodBCs, the surrounding RodBCs will expand their dendritic and axonal territories to fill the gap (Johnson, et al., 2017). We find no clear morphological changes in the axons of the RodBCs of RPC1, however, we also do not observe a clear decrease in RodBC packing density. It is possible this phenomenon will emerge in future pathoconnectomes of more advanced retinal degeneration as inner retinal cell death initiates. Finally, the plasticity of the RodBCs and their ability to restore some afferent input during photoreceptor degeneration is likely also related to the age of the animal. The Kerschensteiner group recently published a study describing the plasticity of cone bipolar cells in restoring afferent input at different ages (Shen, et al., 2020). This study found that of ON-cone bipolar cells most lose the plasticity to completely restore proper inputs as the animal ages. While they did not explicitly investigate RodBCs, it is likely that age of the animal plays a role in the RodBC plasticity and ability to restore the same levels of afferent input should rod degeneration be halted or therapeutically restored. Our results extend our knowledge of RodBC inputs during early retinal degeneration and indicate that early in retinal degeneration there is mixing of the rod and cone inputs through the RodBCs.
Although the numbers of ribbons and gross morphology of the RodBC axons were similar between RodBCs of RPC1 and RC1, the spherical ribbon synapses were unique to RPC1, and their altered morphology origins could be complex. With respect to retinal degeneration, Strettoi et al., described ribbons with similar morphology in the RodBCs of postnatal day 30 (pnd30) rd1 mice (Strettoi, et al., 2002). At this time point, photoreceptors are largely degenerated and RodBCs never fully develop due to the rapid onset and progression of photoreceptor degeneration in this model. Based on this and morphological similarities between ribbons in pnd30 rd1 mice, and developing ribbons, Strettoi and colleagues proposed that the spherical ribbons were due to lack of complete RodBC development in this model. In the P347L model, lack of complete development is not a confounding feature, the Tg P347L rabbit retina fully develops and does not initiate degeneration until approximately 3 months of age (Jones, et al., 2011, Kondo, et al., 2009). Spherical ribbons, however are not isolated to the developing retina. Vollrath and colleagues demonstrated the presence of spherical synaptic bodies in photoreceptor terminals appearing coordinated with time of day and illumination of the photoreceptors and are associated with the shortening and lengthening of photoreceptor terminal ribbons. A greater number of spherical bodies near ribbon synapse sites appear in the middle of the subjective day, and are largely abolished during subjective night, and in continuous darkness (Adly, et al., 1999, Spiwoks-Becker, et al., 2004). In addition, Schmitz, confirmed the findings of Vollrath and colleagues while adding the hypothesis that decreasing levels of intracellular calcium may be contributing to the the disassembly of photoreceptor ribbon synapses during high illuminance (Schmitz, 2014). Combined, these studies suggest a number of mechanisms that may be contributing to observations of spherical ribbons in degenerate retina. We have previously hypothesized that features of photoreceptor degeneration, leading to less bipolar cell input, combined with features of remodeling such as the decrease of mGluR6 expression (Jones, et al., 2011, Marc, et al., 2007, Strettoi and Pignatelli, 2000) may be leading to changes in calcium levels of inner retinal neurons along with altered gene expression (Jones, et al., 2012, Marc, 2010, Marc, et al., 2007). Extending this hypothesis, we propose that the observed spherical ribbons may be a result of decreased intracellular calcium in the bipolar cells, occurring by a similar mechanism to that observed in photoreceptors.
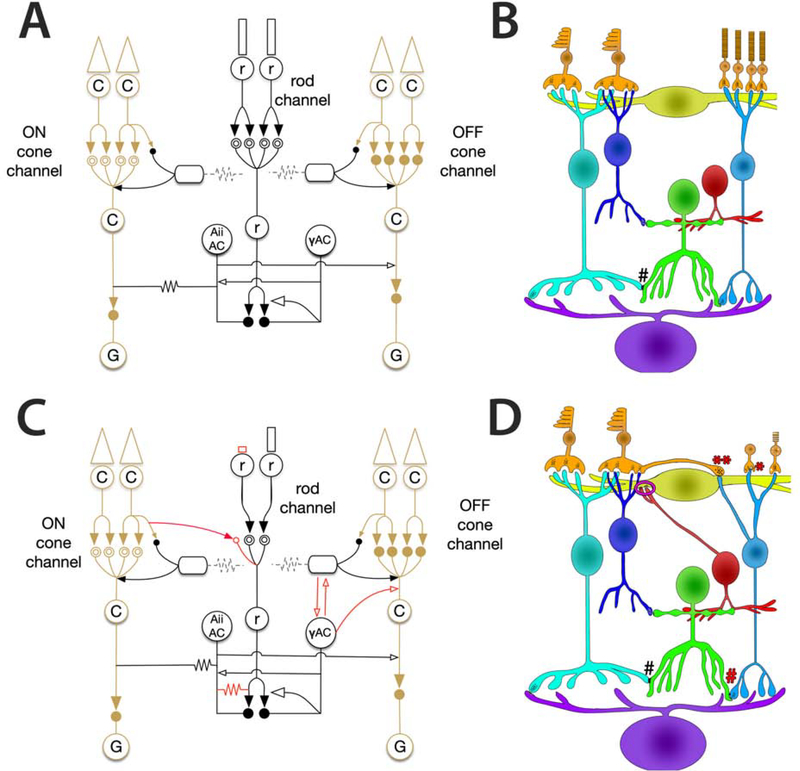
Central to connectomics analyses, specificity in synaptic partners is critical to the function of a neural network. Many complex feedforward and feedback loops have emerged as underlying properties long observed in physiological studies. For example, psychophysical studies of vision prompted many hypotheses of crossover inhibition between networks (Buck, 2014, 2004, Stabell and Stabell, 2002), which were later specifically defined using connectomics (Lauritzen, et al., 2019). The present pathoconnectome reveals the early onset of network-level changes involved in the breakdown of primary visual pathways within the retina (Fig 7). The emergence of gap junctional coupling between RodBCs and Aii GACs contributes to the deterioration of the rod-network prior to the complete degeneration of the rod photoreceptors and likely compromises the spatial and temporal resolution found in the intact network.
Figure 7:
Altered network found in RPC1. (A) Phase 0 (Healthy) bipolar cell network. (B) Phase 1 corrupted network. Solid arrows indicate glutamatergic synapses. Open arrows indicate inhibitory synapses (GABA or glycine). Solid circles indicate ionotropic glutamate receptors, while mGluR6 is indicated by the double open circles. Gap junctions are indicated by zig-zag lines. Red lines indicate aberrant connectivities observed in the RPC1 volume.
Furthermore, though the vertical pathway of rod photoreceptors to RodBCs is unsurprisingly altered, we find the extent of changes in the horizontal pathway especially interesting. Apical neurites from γACs, not only extend and traverse the OPL, but also make anatomically confirmed synapses with bipolar cells in addition to horizontal cells. Both horizontal and amacrine cells are integral to creating the center-surround opponency essential for complex vision. The novel electrical synapses formed by the emergence of gap junctions between RodBCs and Aii GACs, creates a new feedback loop with RodBCs rather than the unidirectional RodBC to Aii GAC pathway observed in healthy retina. In addition, the sprouting of horizontal and amacrine cells in various models of photoreceptor degeneration has been well documented (Fisher, et al., 1995, Jones, et al., 2003, Strettoi and Pignatelli, 2000), in this report we find this is occurring substantially before rod photoreceptors are completely degenerated. The new synaptic contacts involving ascending processes from γACs, provides a new, likely corruptive node in OPL lateral inhibition networks. At a minimum, this creates a new corrupted network that could likely diminish the center surround opponency critical for precise visual function. This will likely manifest clinically with diminished visual performance prior to the widespread neuronal loss characteristic of late-stage retinal degeneration (Jones, et al., 2003, Pfeiffer, et al., 2020b). It is also possible, depending on neuron and neurite physiology, that these processes now provide a novel feedback loop between the IPL and OPL. These results identify the neurons involved in early neurite outgrowth and defines which cells are postsynaptic partners to these aberrant processes, leading to further questions about cell to cell recognition in plasticity and how it can might be controlled experimentally and therapeutically. In the future, computational modeling of these altered networks is necessary to provide greater insight into the effects of these network changes on visual processing.
4.1. Pathoconnectomes provide a map for therapeutics
In conclusion, this study introduces the first ultrastructural pathoconnectome and demonstrates the applicability of pathoconnectomics for understanding rewiring in progressive retinal degenerative disease. Retinal remodeling exhibits the same components as CNS negative plasticity in disease, as well as similar proteomic findings observed in neurodegenerative diseases like Alzheimer’s, Parkinson’s and others (Pfeiffer, et al., 2020b). Therefore, beyond its direct applicability to retinal disease, we believe information provided by pathoconnectomics is likely to demonstrate fundamental rules in how neural networks are altered, that are applicable to disorders of the CNS as synaptic partners degenerate. These mechanisms of network changes, including those yet to be seen in pathoconnectomes of more advanced degeneration, will inform us about the progression, what ultimately causes networks to fail, and describe future targets for therapeutic intervention based on key principles of network plasticity.
Supplementary Material
Highlights.
The first ultrastructural pathoconnectome of a degenerating neural network
Rewiring of retinal networks occurs prior to complete loss of afferent input
Neurites extended by retinal neurons synapse with expected and novel partners
Novel gap junctions are formed by rod bipolar cells early retinal degeneration
Acknowledgements
This work was supported by the National Institutes of Health [RO1 EY015128(BWJ), RO1 EY028927(BWJ), P30 EY014800(Core), T32 EY024234(RLP)]; and an Unrestricted Research Grant from Research to Prevent Blindness, New York, NY to the Department of Ophthalmology & Visual Sciences, University of Utah
Abbreviations
- RodBC
Rod bipolar cell
- γAC
GABAergic amacrine cell
- Aii GAC
Aii glycinergic amacrine cell
- HzC
Horizontal cell
- RPC1
Retinal pathoconnectome 1
- RC1
Retinal connectome 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adly MA, Spiwoks-Becker I and Vollrath L, 1999. Ultrastructural changes of photoreceptor synaptic ribbons in relation to time of day and illumination. Invest Ophthalmol Vis Sci. 40 (10), 2165–72. [PubMed] [Google Scholar]
- Agosta F, Rocca MA, Pagani E, Absinta M, Magnani G, Marcone A, Falautano M, Comi G, Gorno-Tempini ML and Filippi M, 2010. Sensorimotor network rewiring in mild cognitive impairment and alzheimer's disease. Hum Brain Mapp. 31 (4), 515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Jones BW, Watt CB, Shaw MV, Yang JH, Demill D, Lauritzen JS, Lin Y, Rapp KD, Mastronarde D, Koshevoy P, Grimm B, Tasdizen T, Whitaker R and Marc RE, 2011a. Exploring the retinal connectome. Mol Vis. 17), 355–79. [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Jones BW, Yang JH, Shaw MV, Watt CB, Koshevoy P, Spaltenstein J, Jurrus E U VK, Whitaker RT, Mastronarde D, Tasdizen T and Marc RE, 2009. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 7 (3), e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Mohammed S, Grimm B, Jones BW, Koshevoy P, Tasdizen T, Whitaker R and Marc RE, 2011b. The viking viewer for connectomics: Scalable multi-user annotation and summarization of large volume data sets. J Microsc. 241 (1), 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E and Horvitz HR, 1993. Odorant-selective genes and neurons mediate olfaction in c. Elegans. Cell. 74 (3), 515–27. [DOI] [PubMed] [Google Scholar]
- Behrens C, Schubert T, Haverkamp S, Euler T and Berens P, 2016. Connectivity map of bipolar cells and photoreceptors in the mouse retina. Elife. 5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier C, Hovhannisyan A, Weiser S, Kung J, Lee S, Lee DY, Huie P, Dalal R, Palanker D and Sher A, 2017. Deafferented adult rod bipolar cells create new synapses with photoreceptors to restore vision. J Neurosci. 37 (17), 4635–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S, 2004. Rod-cone interactions in human vision in:Chalupa LM and Werner J, Book Rod-cone interactions in human vision. MIT Press, Cambridge, MA, 863–878. [Google Scholar]
- Buck S, 2014. The interaction of rod and cone signals: Pathways and psychophysics in:Werner J and Chalupa LM, Book The interaction of rod and cone signals: Pathways and psychophysics. MIT press, Cambridge, MA, 485–497. [Google Scholar]
- Catela C, Shin MM and Dasen JS, 2015. Assembly and function of spinal circuits for motor control. Annu Rev Cell Dev Biol. 31), 669–98. [DOI] [PubMed] [Google Scholar]
- Chew YL, Walker DS, Towlson EK, Vertes PE, Yan G, Barabasi AL and Schafer WR, 2017. Recordings of caenorhabditis elegans locomotor behaviour following targeted ablation of single motorneurons. Sci Data. 4), 170156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creel DJ, 2019. Electroretinograms. Handb Clin Neurol 160), 481–493. [DOI] [PubMed] [Google Scholar]
- Day M, Wang Z, Ding J, An X, Ingham CA, Shering AF, Wokosin D, Ilijic E, Sun Z, Sampson AR, Mugnaini E, Deutch AY, Sesack SR, Arbuthnott GW and Surmeier DJ, 2006. Selective elimination of glutamatergic synapses on striatopallidal neurons in parkinson disease models. Nat Neurosci. 9 (2), 251–9. [DOI] [PubMed] [Google Scholar]
- delEtoile J and Adeli H, 2017. Graph theory and brain connectivity in alzheimer's disease. Neuroscientist. 23 (6), 616–626. [DOI] [PubMed] [Google Scholar]
- Diamond JS, 2017. Inhibitory interneurons in the retina: Types, circuitry, and function. Annu Rev Vis Sci. 3), 1–24. [DOI] [PubMed] [Google Scholar]
- Fariss RN, Li ZY and Milam AH, 2000. Abnormalities in rod photoreceptors, amacrine cells, and horizontal cells in human retinas with retinitis pigmentosa. Am J Ophthalmol. 129 (2), 215–23. [DOI] [PubMed] [Google Scholar]
- Fei Y, 2002. Cone neurite sprouting: An early onset abnormality of the cone photoreceptors in the retinal degeneration mouse. Mol Vis. 8), 306–14. [PubMed] [Google Scholar]
- Fisher SK, Lewis GP, Linberg KA, Barawid E and Verardo MR, 1995. Cellular remodeling in mammalian retina induced by retinal detachment in:Kolb H, Fernandez E and Nelson R, Book Cellular remodeling in mammalian retina induced by retinal detachment. Salt Lake City (UT). [PubMed] [Google Scholar]
- Fisher SK, Lewis GP, Linberg KA and Verardo MR, 2005. Cellular remodeling in mammalian retina: Results from studies of experimental retinal detachment. Prog Retin Eye Res. 24 (3), 395–431. [DOI] [PubMed] [Google Scholar]
- Guo L, Xiong H, Kim JI, Wu YW, Lalchandani RR, Cui Y, Shu Y, Xu T and Ding JB, 2015. Dynamic rewiring of neural circuits in the motor cortex in mouse models of parkinson's disease. Nat Neurosci. 18 (9), 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel C, 2006. Retinitis pigmentosa. Orphanet J Rare Dis. 1), 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL and Dryja TP, 2006. Retinitis pigmentosa. The Lancet. 368 (9549), 1795–1809. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Tien NW, Shen N, Pearson JT, Soto F and Kerschensteiner D, 2017. Homeostatic plasticity shapes the visual system's first synapse. Nat Commun. 8 (1), 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Kondo M, Terasaki H, Lin Y, McCall M and Marc RE, 2012. Retinal remodeling. Jpn J Ophthalmol. 56 (4), 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Kondo M, Terasaki H, Watt CB, Rapp K, Anderson J, Lin Y, Shaw MV, Yang JH and Marc RE, 2011. Retinal remodeling in the tg p347l rabbit, a large-eye model of retinal degeneration. J Comp Neurol. 519 (14), 2713–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW and Marc RE, 2005. Retinal remodeling during retinal degeneration. Exp Eye Res. 81 (2), 123–37. [DOI] [PubMed] [Google Scholar]
- Jones BW, Pfeiffer RL, Ferrell WD, Watt CB, Marmor M and Marc RE, 2016a. Retinal remodeling in human retinitis pigmentosa. Exp Eye Res. 150), 149–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Pfeiffer RL, Ferrell WD, Watt CB, Tucker J and Marc RE, 2016b. Retinal remodeling and metabolic alterations in human amd. Front Cell Neurosci. 10), 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Watt CB, Frederick JM, Baehr W, Chen CK, Levine EM, Milam AH, Lavail MM and Marc RE, 2003. Retinal remodeling triggered by photoreceptor degenerations. J Comp Neurol. 464 (1), 1–16. [DOI] [PubMed] [Google Scholar]
- Kalloniatis M and Fletcher EL, 2004. Retinitis pigmentosa: Understanding the clinical presentation, mechanisms and treatment options. Clin Exp Optom. 87 (2), 65–80. [DOI] [PubMed] [Google Scholar]
- Kolb H and Famiglietti EV, 1974. Rod and cone pathways in the inner plexiform layer of cat retina. Science. 186 (4158), 47–9. [DOI] [PubMed] [Google Scholar]
- Kondo M, Sakai T, Komeima K, Kurimoto Y, Ueno S, Nishizawa Y, Usukura J, Fujikado T, Tano Y and Terasaki H, 2009. Generation of a transgenic rabbit model of retinal degeneration. Invest Ophthalmol Vis Sci. 50 (3), 1371–7. [DOI] [PubMed] [Google Scholar]
- Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, Viola KL and Klein WL, 2007. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in alzheimer's disease. J Neurosci. 27 (4), 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen JS, Anderson JR, Jones BW, Watt CB, Mohammed S, Hoang JV and Marc RE, 2013. On cone bipolar cell axonal synapses in the off inner plexiform layer of the rabbit retina. J Comp Neurol. 521 (5), 977–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen JS, Sigulinsky CL, Anderson JR, Kalloniatis M, Nelson NT, Emrich DP, Rapp C, McCarthy N, Kerzner E, Meyer M, Jones BW and Marc RE, 2019. Rod-cone crossover connectome of mammalian bipolar cells. J Comp Neurol. 527 (1), 87–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K and Reid RC, 2016. Anatomy and function of an excitatory network in the visual cortex. Nature. 532 (7599), 370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GP, Linberg KA and Fisher SK, 1998. Neurite outgrowth from bipolar and horizontal cells after experimental retinal detachment. Invest Ophthalmol Vis Sci. 39 (2), 424–34. [PubMed] [Google Scholar]
- Lin Y, Jones BW, Liu A, Tucker JF, Rapp K, Luo L, Baehr W, Bernstein PS, Watt CB, Yang JH, Shaw MV and Marc RE, 2012. Retinoid receptors trigger neuritogenesis in retinal degenerations. FASEB J. 26 (1), 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linberg KA, Lewis GP, Matsumoto B and Fisher SK, 2006. Immunocytochemical evidence that rod-connected horizontal cell axon terminals remodel in response to experimental retinal detachment in the cat. Mol Vis. 12), 1674–86. [PubMed] [Google Scholar]
- Lujan BJ, Roorda A, Croskrey JA, Dubis AM, Cooper RF, Bayabo JK, Duncan JL, Antony BJ and Carroll J, 2015. Directional optical coherence tomography provides accurate outer nuclear layer and henle fiber layer measurements. Retina. 35 (8), 1511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, 2010. Injury and repair: Retinal remodeling in:Dartt D, Book Injury and repair: Retinal remodeling. Elsevier, 414–420. [Google Scholar]
- Marc RE, Anderson JR, Jones BW, Sigulinsky CL and Lauritzen JS, 2014. The aii amacrine cell connectome: A dense network hub. Front Neural Circuits. 8), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE and Cameron D, 2002. A molecular phenotype atlas of the zebrafish retina. J Neurocytol. 30 (7), 593–654. [DOI] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Anderson JR, Kinard K, Marshak DW, Wilson JH, Wensel T and Lucas RJ, 2007. Neural reprogramming in retinal degeneration. Invest Ophthalmol Vis Sci. 48 (7), 3364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Lauritzen JS, Watt CB and Anderson JR, 2012. Building retinal connectomes. Curr Opin Neurobiol. 22 (4), 568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Watt CB, Anderson JR, Sigulinsky C and Lauritzen S, 2013. Retinal connectomics: Towards complete, accurate networks. Prog Retin Eye Res 37), 141–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Watt CB and Strettoi E, 2003. Neural remodeling in retinal degeneration. Prog Retin Eye Res. 22 (5), 607–55. [DOI] [PubMed] [Google Scholar]
- Marc RE, Murry RF and Basinger SF, 1995. Pattern recognition of amino acid signatures in retinal neurons. J Neurosci. 15 (7 Pt 2), 5106–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Sigulinsky CL, Pfeiffer RL, Emrich D, Anderson JR and Jones BW, 2018. Heterocellular coupling between amacrine cells and ganglion cells. Front Neural Circuits 12), 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH, 2012. The neuronal organization of the retina. Neuron. 76 (2), 266–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Fahnestock M and Racine RJ, 2004. Kindling and status epilepticus models of epilepsy: Rewiring the brain. Prog Neurobiol. 73 (1), 1–60. [DOI] [PubMed] [Google Scholar]
- Ottersen OP, 1989. Quantitative electron microscopic immunocytochemistry of neuroactive amino acids. Anat Embryol (Berl). 180 (1), 1–15. [DOI] [PubMed] [Google Scholar]
- Pang JJ, Yang Z, Jacoby RA and Wu SM, 2018. Cone synapses in mammalian retinal rod bipolar cells. J Comp Neurol. 526 (12), 1896–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YW, Hao Y, Petters RM and Wong F, 2000. Ectopic synaptogenesis in the mammalian retina caused by rod photoreceptor-specific mutations. Nat Neurosci. 3 (11), 1121–7. [DOI] [PubMed] [Google Scholar]
- Pfeiffer RL, Anderson JR, Emrich DP, Dahal J, Sigulinsky CL, Morrison HAB, Yang JH, Watt CB, Rapp KD, Kondo M, Terasaki H, Garcia JC, Marc RE and Jones BW, 2019. Pathoconnectome analysis of muller cells in early retinal remodeling. Adv Exp Med Biol. 1185), 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RL, Marc RE and Jones BW, 2020a. Muller cell metabolic signatures: Evolutionary conservation and disruption in disease. Trends Endocrinol Metab. 31 (4), 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RL, Marc RE and Jones BW, 2020b. Persistent remodeling and neurodegeneration in late-stage retinal degeneration. Prog Retin Eye Res. 74), 100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Zhang Y, Meister M and Sanes JR, 2011. Age-related alterations in neurons of the mouse retina. J Neurosci. 31 (44), 16033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid LC, Mittag M, Poll S, Steffen J, Wagner J, Geis HR, Schwarz I, Schmidt B, Schwarz MK, Remy S and Fuhrmann M, 2016. Dysfunction of somatostatin-positive interneurons associated with memory deficits in an alzheimer's disease model. Neuron. 92 (1), 114–125. [DOI] [PubMed] [Google Scholar]
- Schmitz F, 2014. Presynaptic [ca(2+)] and gcaps: Aspects on the structure and function of photoreceptor ribbon synapses. Front Mol Neurosci 7), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Wang B, Soto F and Kerschensteiner D, 2020. Homeostatic plasticity shapes the retinal response to photoreceptor degeneration. Curr Biol.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigulinsky CL, Anderson JR, Kerzner E, Rapp CN, Pfeiffer RL, Rodman TM, Emrich DP, Rapp KD, Nelson NT, Lauritzen JS, Meyer M, Marc RE and Jones BW, 2020. Network architecture of gap junctional coupling among parallel processing channels in the mammalian retina. J Neurosci). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiwoks-Becker I, Glas M, Lasarzik I and Vollrath L, 2004. Mouse photoreceptor synaptic ribbons lose and regain material in response to illumination changes. Eur J Neurosci. 19 (6), 1559–71. [DOI] [PubMed] [Google Scholar]
- Stabell B and Stabell U, 2002. Effects of rod activity on color perception with light adaptation. J Opt Soc Am A Opt Image Sci Vis. 19 (7), 1249–58. [DOI] [PubMed] [Google Scholar]
- Strettoi E and Pignatelli V, 2000. Modifications of retinal neurons in a mouse model of retinitis pigmentosa. Proc Natl Acad Sci U S A. 97 (20), 11020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strettoi E, Porciatti V, Falsini B, Pignatelli V and Rossi C, 2002. Morphological and functional abnormalities in the inner retina of the rd/rd mouse. J Neurosci. 22 (13), 5492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreson WB and Dacey DM, 2019. Diverse cell types, circuits, and mechanisms for color vision in the vertebrate retina. Physiol Rev. 99 (3), 1527–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreson WB and Witkovsky P, 1999. Glutamate receptors and circuits in the vertebrate retina. Prog Retin Eye Res. 18 (6), 765–810. [DOI] [PubMed] [Google Scholar]
- Tien NW, Soto F and Kerschensteiner D, 2017. Homeostatic plasticity shapes cell-type-specific wiring in the retina. Neuron. 94 (3), 656–665 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y and Omi N, 2013. Functional allocation of synaptic contacts in microcircuits from rods via rod bipolar to aii amacrine cells in the mouse retina. J Comp Neurol. 521 (15), 3541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y and Omi N, 2014. Some off bipolar cell types make contact with both rods and cones in macaque and mouse retinas. Front Neuroanat. 8), 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y and Omi N, 2015. Off bipolar cells in macaque retina: Type-specific connectivity in the outer and inner synaptic layers. Front Neuroanat 9), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Kominami T, Okado S, Inooka D, Kondo M and Terasaki H, 2019. Course of loss of photoreceptor function and progressive muller cell gliosis in rhodopsin p347l transgenic rabbits. Exp Eye Res. 184), 192–200. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN and Brenner S, 1986. The structure of the nervous system of the nematode caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 314 (1165), 1–340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.