SUMMARY
Pseudomonas aeruginosa and Burkholderia cepacia complex (Bcc) species are opportunistic lung pathogens of cystic fibrosis (CF) patients. While P. aeruginosa can initiate long-term infections in younger CF patients, Bcc infections only arise in teenagers and adults. Both P. aeruginosa and Bcc use type VI secretion systems (T6SSs) to mediate interbacterial competition. Here, we show P. aeruginosa isolates from teenage/adult CF patients, but not those from young CF patients, are outcompeted by the epidemic Bcc isolate Burkholderia cenocepacia strain AU1054 in a T6SS-dependent manner. The genomes of susceptible P. aeruginosa isolates harbor T6SS-abrogating mutations, the repair of which, in some cases, rendered the isolates resistant. Moreover, seven of eight Bcc strains outcompeted P. aeruginosa strains isolated from the same patients. Our findings suggest certain mutations that arise as P. aeruginosa adapts to the CF lung abrogate T6SS activity, making P. aeruginosa and its human host susceptible to potentially fatal Bcc superinfection.
Graphical Abstract
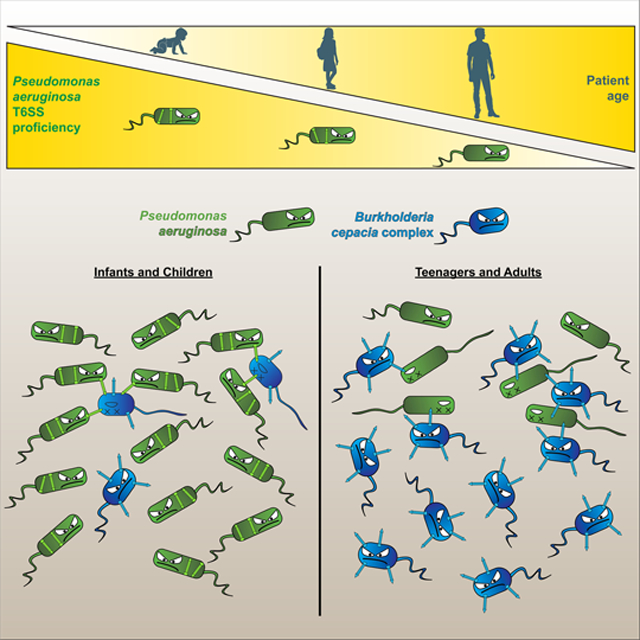
eTOC BLURB
Pseudomonas aeruginosa infects cystic fibrosis (CF) patients of all ages. Burkholderia cepacia complex (Bcc) infections are restricted to teenage/adult CF patients. Perault et al. demonstrate Bcc pathogens, using the type VI secretion system (T6SS), only outcompete host-adapted P. aeruginosa isolates, which become T6SS-deficient during evolution within the CF respiratory tract.
INTRODUCTION
The respiratory tracts of individuals suffering from cystic fibrosis (CF) are hospitable environments for microorganisms, and thus CF patients harbor complex, dynamic microbial communities in their airways that can include opportunistic pathogens (Carmody et al., 2015; Filkins and O’Toole, 2015; Lipuma, 2010; J. Zhao et al., 2012). Pseudomonas aeruginosa and certain members of the Burkholderia cepacia complex (Bcc), a taxonomic group containing at least 17 Burkholderia spp. (Salsgiver et al., 2016), cause devastating infections in CF patients, with Bcc pathogens often causing superinfections in P. aeruginosa-infected patients (Govan and Deretic, 1996; Lipuma, 2010; Mahenthiralingam et al., 2005). While P. aeruginosa infects young CF patients and is the most common opportunistic CF pathogen by early adulthood, Bcc infections are less common and, for unknown reasons, limited to older CF patients, typically teenagers and adults (Cystic Fibrosis Foundation, 2019). Unlike other CF pathogens, Bcc strains are more frequently associated with person-to-person spread (Biddick et al., 2003; Chen et al., 2001; Govan et al., 1993) and can progress to a fatal necrotizing pneumonia and bacteremia termed “cepacia syndrome” (Isles et al., 1984; Lipuma, 2010). While P. aeruginosa and Bcc do not colocalize within the lungs of CF patients (P. aeruginosa is predominantly located in the airway lumen and Bcc within phagocytes), the P. aeruginosa burden in co-infected patients tends to be lower than in patients infected by P. aeruginosa alone (Schwab et al., 2014).
Interbacterial interactions likely occur within the polymicrobial CF respiratory tract and may influence disease progression (Bisht et al., 2020; Filkins and O’Toole, 2015; O’Brien and Fothergill, 2017; Peters et al., 2012). Interbacterial competition is hypothesized to be one of the strongest determinants of ecology and evolution within polymicrobial communities (Foster and Bell, 2012). A prevalent and well-understood mechanism of interbacterial competition is that mediated by type VI secretion systems (T6SSs) (Alteri and Mobley, 2016; Russell et al., 2014), which are predicted to be present in ~25% of Gram-negative bacteria (Boyer et al., 2009). T6SSs use a bacteriophage-like mechanism to deliver effector proteins directly into target bacterial or eukaryotic cells (Basler et al., 2012; Hachani et al., 2016; Hood et al., 2010; Pukatzki et al., 2007). Antibacterial T6SS effectors disrupt diverse biological processes within target cells, and cognate immunity proteins protect T6SS-producing cells from autotoxicity (Ahmad et al., 2019; Russell et al., 2014; Ting et al., 2018). Type VI secretion (T6S) has been studied in the Bcc pathogen Burkholderia cenocepacia strain J2315 (BcJ2315), which produces a T6SS that is important for infection of macrophages and influences the host immune response (Aubert et al., 2015; 2016; Hunt et al., 2004; Rosales-Reyes et al., 2012). Recent bioinformatic analysis has revealed T6SS-encoding genes throughout the Bcc, with one system (referred to as T6SS-1) predominating; however, several species encode multiple T6SSs (Spiewak et al., 2019). The B. cenocepacia strain H111 T6SS was shown to have modest antibacterial activity (Spiewak et al., 2019). Whether T6SSs in other Bcc pathogens have antibacterial activity is unknown.
P. aeruginosa produces three separate T6SSs (the H1-, H2-, and H3-T6SSs), and while both the H1- and H2-T6SSs are antibacterial weapons, the H1-T6SS is the stronger mediator of interbacterial competition (Allsopp et al., 2017; Hood et al., 2010; Russell et al., 2011). The P. aeruginosa H1-T6SS is under intricate regulation at both the post-transcriptional and post-translational level. Phosphorelay through the GacSA two-component system induces T6SS protein production via the regulatory small RNAs (sRNAs) RsmY and RsmZ, which relieve RsmA-mediated repression of translation of transcripts encoding T6SS proteins (Goodman et al., 2004; 2009; Lapouge et al., 2008; Moscoso et al., 2011; Ventre et al., 2006). Moreover, a threonine phosphorylation pathway regulates T6SS assembly and function via signal transduction through the membrane-associated TagQRST proteins, Fha1, and the kinase and phosphatase PpkA and PppA, respectively (Basler et al., 2013; Casabona et al., 2013; Hsu et al., 2009; Mougous et al., 2007).
P. aeruginosa undergoes dramatic evolution within the CF respiratory tract to transition to a chronic infection lifestyle (Folkesson et al., 2012; Winstanley et al., 2016), with mutations in evolved strains often occurring in gacS/gacA and T6SS structural genes (Bartell et al., 2019; Kordes et al., 2019; Marvig et al., 2015). Our characterization of T6S in the Bcc led us to hypothesize host adaptation by P. aeruginosa may open the door to subsequent Bcc infections if the resident P. aeruginosa community loses T6SS activity. Here, we describe experiments conducted to test this hypothesis.
RESULTS
The BcAU1054 T6SS mediates interbacterial competition
We selected Burkholderia cenocepacia strain AU1054 (BcAU1054), which was isolated from the bloodstream of a CF patient, for our studies (Chen et al., 2001; Grigoriev et al., 2012). The BcAU1054 genome encodes a predicted T6SS on chromosome 1 (BCEN_RS13060, tssM, through BCEN_RS13170, tssL) (Figure 1A). Presumably due to errors during the sequencing of this strain’s genome (assembly GCA_000014085.1), multiple genes in this region were annotated as pseudogenes. We PCR amplified and sequenced these regions and found that each gene is actually intact (Table S1). Compared to the T6SS-encoding cluster of B. cenocepacia strain J2315 (BcJ2315), the BcAU1054 T6SS gene cluster contains an additional region (BCEN_RS13075 (tai1) through BCEN_RS13110 (vgrG1)) that includes two predicted effector-immunity (E-I)-encoding gene pairs (tae1-tai1 and tle1-tli1) (Figure S1). Immediately 3’ to the BcAU1054 core cluster (with the 5’ to 3’ direction corresponding to the sequence numbering in Figure 1A) is an additional region containing vgrG2, the predicted E-I-encoding pair tne1-tni1, and several genes predicted to be involved in phage or other mobile genetic elements.
Figure 1. The BcAU1054 T6SS mediates interbacterial competition.
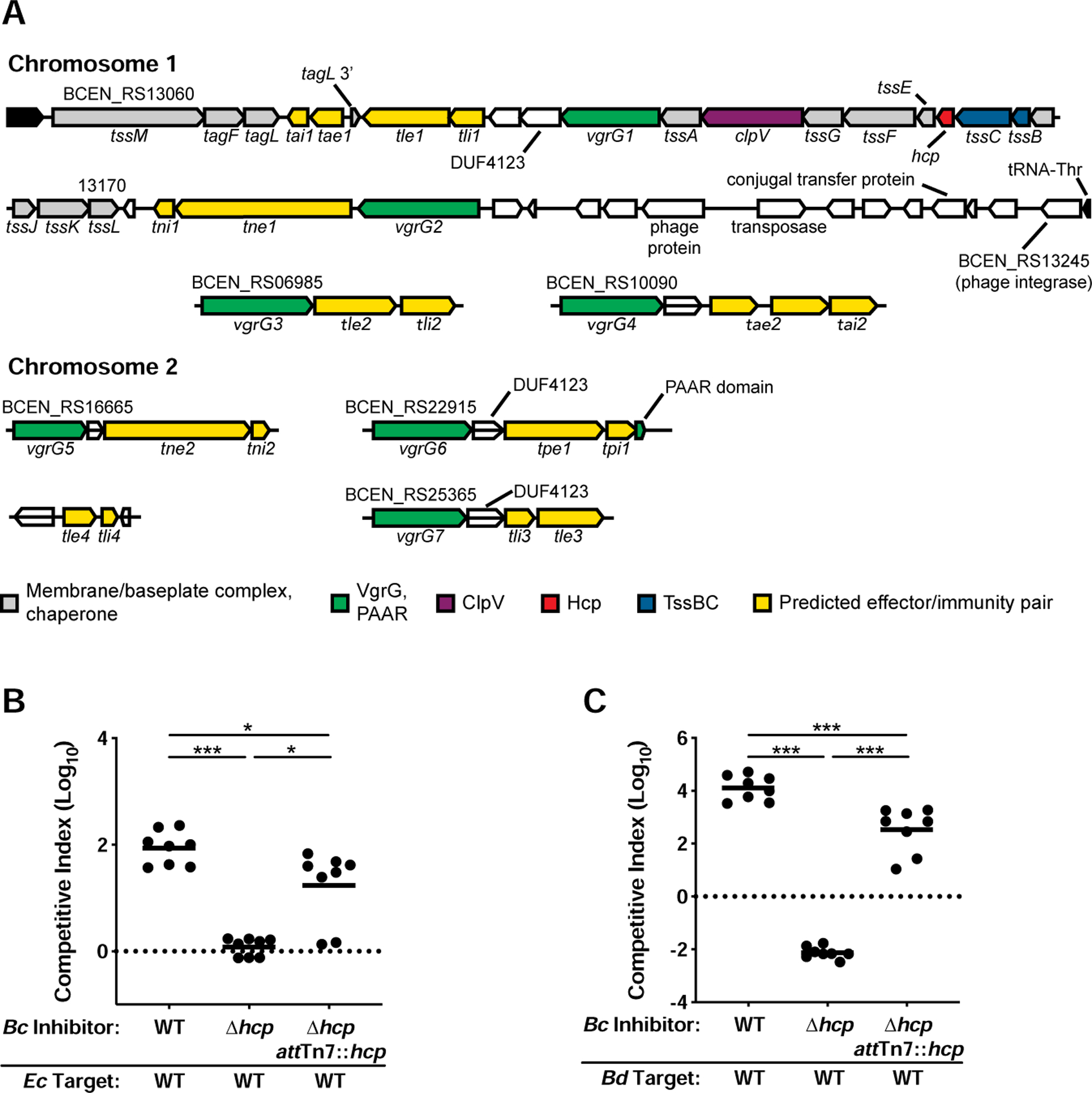
(A) Core cluster (top two lines) and accessory genes encoding the BcAU1054 T6SS and predicted effector-immunity (E-I) pairs (see Table S2). The legend indicates function of protein products. White ORFs encode DUF4123 T6SS adapter proteins, or proteins not known to be associated with the T6SS. (B and C) Competition experiments between inhibitor BcAU1054 and target E. coli (B) and B. dolosa (C). WT, Δhcp, and Δhcp attTn7::hcp BcAU1054 inhibitor strains used in each. Circles represent individual cocultures from two biological replicates, each with four technical replicates. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. *P<0.05, **P<0.0005, Mann-Whitney test.
To identify additional genes potentially encoding E-I pairs, we first analyzed genes located near the seven annotated vgrG genes (Figure 1A). VgrG proteins, along with PAAR-repeat proteins, form the puncturing tip of the T6SS needle and typically associate with effectors encoded by nearby genes (Pukatzki et al., 2007; Russell et al., 2014; Shneider et al., 2013). Based on previous nomenclature (Russell et al., 2014), we named predicted cell membrane-degrading effectors Tle for T6SS lipase effector, nucleic acid-degrading effectors Tne for T6SS nuclease effector, cell wall-degrading effectors Tae for T6SS amidase effector, and used Tpe for the T6SS pore-forming effector. We named cognate immunity proteins Tli, Tni, Tai, and Tpi. Three of the predicted E-I-encoding gene pairs (tle1-tli1, tle3-tli3, and tpe1-tpi1) are near ORFs encoding proteins with the domain of unknown function (DUF) 4123, which is a conserved chaperone domain for T6SS effectors (Liang et al., 2015) (Figure 1A). Protein secondary structure analysis using Phyre2 (Kelley et al., 2015) and HHpred (Zimmermann et al., 2018) predicted antibacterial enzymatic activities for all potential effectors (Table S2). A duplication of the 3’ end of tagL flanking tae1-tai1 (Figure 1A) suggests these predicted E-I-encoding genes inserted into the BcAU1054 T6SS core cluster via a transposon, and Tae1 is predicted to have a glycosyl hydrolase domain. We identified the tle4-tli4 gene pair by searching for DUFs shared among E-I-encoding genes, as DUF3304 is only present in the BcAU1054 genome within tli1, tli3, and tli4 (Table S2).
To determine if the BcAU1054 T6SS mediates interbacterial competition, we generated an unmarked, in-frame deletion mutation in hcp, which encodes the inner tube protein of the T6S apparatus (the BcAU1054 genome only has one hcp gene). Over 5 h coculture, BcAU1054 outcompeted Escherichia coli DH5α by ~2 logs, whereas BcAU1054 Δhcp had no competitive advantage (Figure 1B). BcAU1054 outcompeted another Bcc pathogen, Burkholderia dolosa strain AU0158 (BdAU0158), by ~4 logs over 5 h, and BcAU1054 Δhcp was outcompeted by BdAU0158 by ~2 logs (Figure 1C). Ectopic expression of hcp from the genomic attTn7 site partially restored the ability of BcAU1054 Δhcp to outcompete E. coli DH5α and BdAU0158 (Figures 1B and 1C). Growth rate differences did not determine competitive fitness as BcAU1054 and BcAU1054 Δhcp had similar growth rates (Figure S2). The BcAU1054 T6SS, therefore, is a potent weapon capable of killing competitor bacteria.
At least five BcAU1054 T6SS effectors mediate interbacterial competition
To determine which predicted effectors are involved in T6SS-mediated interbacterial competition by BcAU1054, we generated nine mutants, each containing an unmarked, in-frame deletion mutation in one of the predicted E-I-encoding gene pairs. We screened these mutants by engineering them to produce green fluorescent protein (GFP) and coculturing them, individually, with either wild-type (WT) or Δhcp BcAU1054 strains for ~20 h, and then measuring GFP fluorescence intensity and the OD600 of the cocultures. For four of the mutants (Δtle1Δtli1, Δtne1Δtni1, Δtne2Δtni2, and Δtpe1Δtpi1), the GFP/OD600 values for cocultures with WT BcAU1054 were about half of what they were for cocultures with BcAU1054 Δhcp, indicating these mutants were outcompeted in a T6SS-dependent manner, presumably because they lack functional immunity proteins (Figure S3). We then cocultured each of these four mutants with WT and Δhcp BcAU1054 strains and measured competition quantitatively. In each case, the E-I deletion mutant was outcompeted by its parental strain in a T6SS-dependent manner (Figure 2A). Ectopic expression of the cognate immunity gene in each mutant rescued it from T6SS-mediated killing by the parental strain (Figure 2A), providing evidence that Tle1-Tli1, Tne1-Tni1, Tne2-Tni2, and Tpe1-Tpi1 are true antibacterial E-I pairs associated with the BcAU1054 T6SS.
Figure 2. At least five BcAU1054 T6SS effectors mediate interbacterial competition.
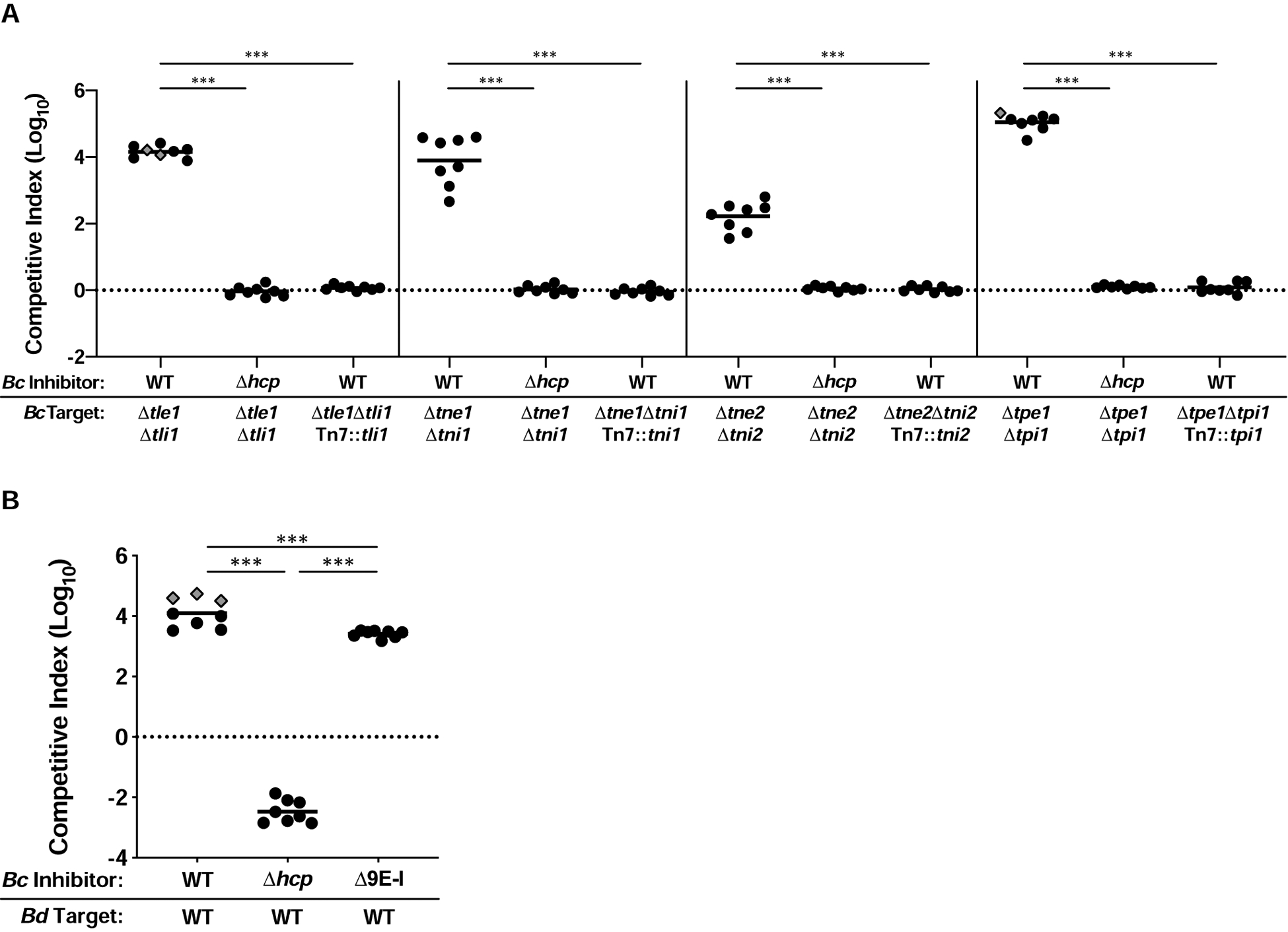
(A) Competition experiments between WT and Δhcp BcAU1054 inhibitor strains and BcAU1054 mutants lacking E-I-encoding genes, including mutants complemented with cognate immunity genes. (B) Competition experiments between WT, Δhcp, and Δ9E-I BcAU1054 inhibitor and BdAU0158 target strains. For (A and B), circles/diamonds represent individual cocultures from two biological replicates, each with four technical replicates. Grey-filled diamonds represent competitions from which no target cells were recovered. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. ***P<0.0005, Mann-Whitney test.
To determine if there are additional E-I-encoding gene pairs in BcAU1054, we generated a mutant lacking all nine predicted E-I-encoding gene pairs (BcAU1054 Δ9E-I) and assessed its ability to outcompete target bacteria. This nonuple mutant outcompeted BdAU0158 by ~3.5 logs (slightly less than WT BcAU1054) (Figure 2B), indicating at least one more effector delivered by the BcAU1054 T6SS exists.
Susceptibility of P. aeruginosa CF isolates to the BcAU1054 T6SS correlates with patient age
We next sought to determine whether the BcAU1054 T6SS targets the prevalent CF pathogen P. aeruginosa. In cocultures with P. aeruginosa reference strain PAO1, PAO1 outcompeted both WT and Δhcp strains of BcAU1054, though showed a slightly (~0.5-log) greater ability to outcompete T6SS-active than T6SS-inactive BcAU1054 (Figure 3A). These results are consistent with two theories on the regulation of H1-T6SS activity by PAO1: T6SS-dueling (Basler et al., 2013), in which the PAO1 H1-T6SS only deploys following antagonism by a neighboring cell, and the P. aeruginosa response to antagonism (PARA) (LeRoux et al., 2015), in which PAO1 activates aggressive behaviors, like T6SS activity, following detection of kin cell lysates. The ability of PAO1 to outcompete BcAU1054 was dependent on the H1-T6SS, as a transposon insertion in vipA1, which encodes a protein necessary for H1-T6SS activity (Basler et al., 2013), caused PAO1 to be outcompeted by BcAU1054 (Figure 3A).
Figure 3. Susceptibility of P. aeruginosa CF isolates to the BcAU1054 T6SS correlates with patient age.
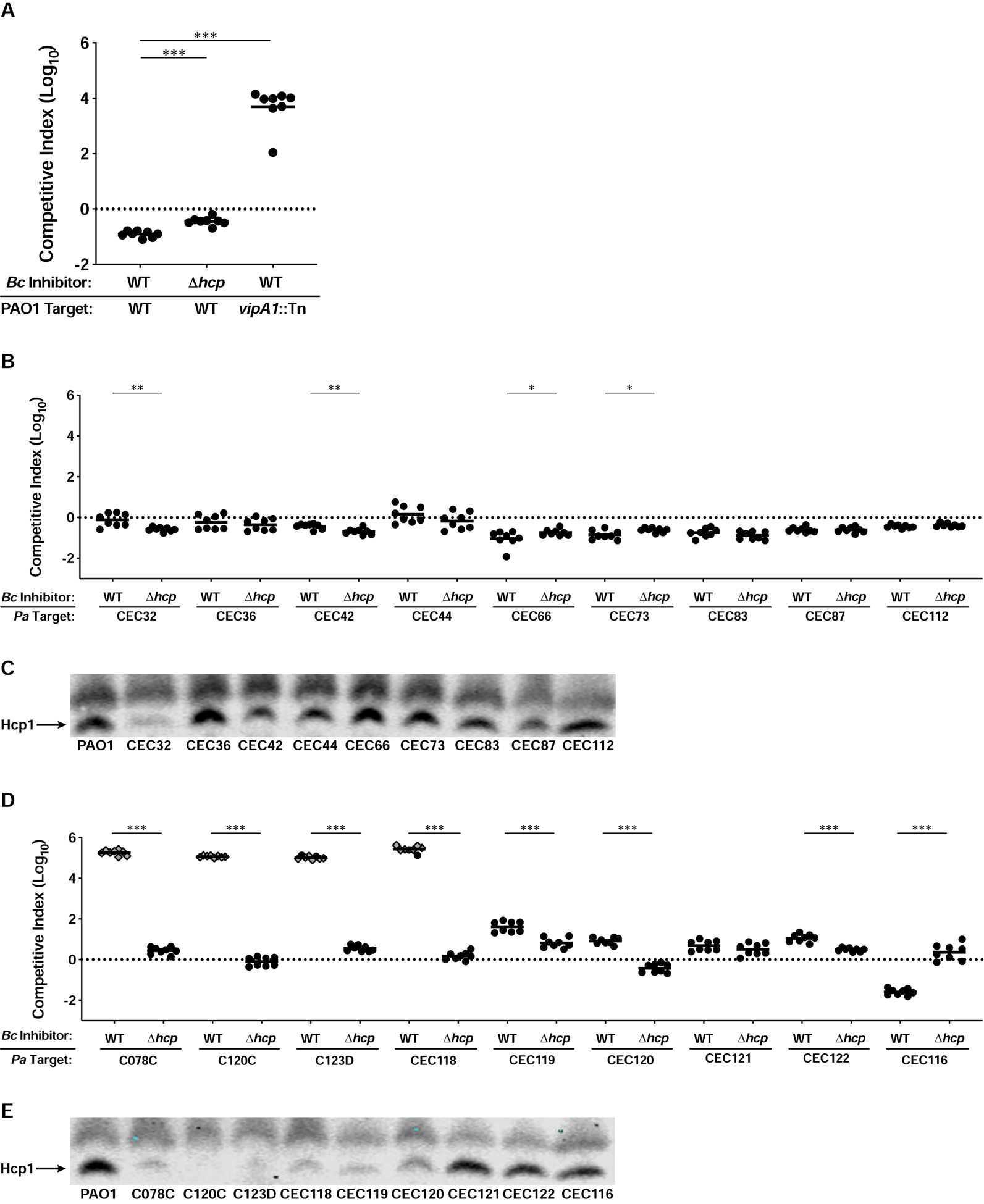
(A) Competition experiments between WT and Δhcp BcAU1054 inhibitor strains and WT and vipA1::Tn PAO1 target strains. (B) Competition experiments between WT and Δhcp BcAU1054 inhibitor strains and P. aeruginosa infant/child CF isolate targets. (C) Immunoblot for Hcp1 production by PAO1 and P. aeruginosa infant/child CF isolates. (D) Competition experiments between WT and Δhcp BcAU1054 inhibitor strains and P. aeruginosa teenage/adult CF isolate targets. (E) Immunoblot for Hcp1 production by PAO1 and P. aeruginosa teenage/adult CF isolates. For (A, B, and D), circles/diamonds represent individual cocultures from two biological replicates, each with four technical replicates. Grey-filled diamonds represent competitions from which no target cells were recovered. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. *P<0.05, **P<0.005, ***P<0.0005, Mann-Whitney test. For (C and E), non-specific band above Hcp1 serves as loading control. Blots are representative of at least two experiments per strain.
PAO1 was originally isolated from a wound infection and has undergone decades of laboratory passage and diversification (Chandler et al., 2019; Holloway, 1955; Holloway and Morgan, 1986; Klockgether et al., 2010). To investigate T6SS-mediated interactions between BcAU1054 and P. aeruginosa strains relevant to CF infection, we used collections of P. aeruginosa strains isolated from CF patients (Burns et al., 2001; Rosenfeld et al., 2001). BcAU1054 did not outcompete any of the P. aeruginosa strains isolated from infants or young children (≤three years old) and was often slightly outcompeted by these strains (Figure 3B). By contrast, BcAU1054 had the striking ability to outcompete nearly half of the P. aeruginosa strains isolated from teenagers and adults (11–31 years old) in a T6SS-dependent manner, oftentimes efficiently enough to prevent recovery of any P. aeruginosa from the cocultures (Figure 3D). We also determined if the nonuple E-I deletion mutant of BcAU1054 could outcompete the susceptible P. aeruginosa strains. Although BcAU1054 Δ9E-I was strongly outcompeted by PAO1, it retained a competitive advantage against C078C, C120C, C123D, and CEC118 (Figure S4). C120C was less susceptible to killing by BcAU1054 Δ9E-I than were C078C, C123D, and CEC118 (Figure S4), suggesting C120C is less sensitive to the unidentified effector(s) associated with the BcAU1054 T6SS, and BcAU1054 T6SS effectors exhibit target strain-specific variability in toxicity. The ability of BcAU1054 to kill P. aeruginosa from older CF patients correlates with the clinical presentation of Bcc infections, which arise only in teenagers and adults (Cystic Fibrosis Foundation, 2019).
Host-adapted P. aeruginosa isolates that are sensitive to the BcAU1054 T6SS harbor T6SS-abrogating mutations
We sequenced the genomes of the P. aeruginosa clinical isolates used for the experiments described above. Of the four T6SS-susceptible isolates (C078C, C120C, C123D, and CEC118), three contain mutations in gacS or gacA, which encode a two-component system required for T6SS protein production (Table 1) (Goodman et al., 2004; Marden et al., 2013; Moscoso et al., 2011). C078C contains a missense mutation in gacS (gacSG1715A), resulting in the variant protein GacSG572D. C123D contains a genomic deletion spanning the gacS gene and the nearby pirRSA genes, which encode a siderophore iron-acquisition system (Ghysels et al., 2005). C120C contains a premature stop codon in gacA (gacAC349T). The C078C genome also contains a premature stop codon in pppA (pppAG111A). PppA is a post-translational regulator of H1-T6SS activity (Mougous et al., 2007), and is required for efficient T6SS-mediated competition (Basler et al., 2013). CEC118 has a small deletion in fha1 (fha1Δ404–424) resulting in the loss of seven amino acid residues from Fha1, another post-translational regulator of H1-T6SS activity (Mougous et al., 2007).
Table 1.
Competition sensitivity, Hcp1 production, and putative T6SS-abrogating mutations of P. aeruginosa CF isolates used in this study.
| P. aeruginosa CF Isolate | Patient age (in years) at isolation | Susceptibility to BcAU1054 T6SS, or to B. cenocepacia paired isolate | Hcp-1 production | Putative T6SS- abrogating mutation(s) | RetS substitutions in gacS/gacA mutants |
|---|---|---|---|---|---|
| Infant/Child Isolates | |||||
| CEC32 | 1 | − | + | N/A | N/A |
| CEC36 | <1 | − | +++ | N/A | N/A |
| CEC42 | 2 | − | +++ | N/A | N/A |
| CEC44 | 2 | − | +++ | N/A | N/A |
| CEC66 | 1 | − | +++ | N/A | N/A |
| CEC73 | 3 | − | +++ | N/A | N/A |
| CEC83 | 1 | − | +++ | N/A | N/A |
| CEC87 | <1 | − | +++ | N/A | N/A |
| CEC112 | 3 | − | +++ | N/A | N/A |
| Teenage/Adult Isolates | |||||
| C078C | 31 | +++ | + | gacSG1715A (GacSG572D), pppAG111A (PppATRUNC) | RetSA46v, RetSR144H |
| C120C | 12 | +++ | − | gacAC349T (GacATRUNC) | RetSA46v, RetSL856Q |
| C123D | 19 | +++ | − | RetSA46V | |
| CEC118 | 17 | +++ | + | fha1Δ404–424 (Fha1Δ134–140) | N/A |
| CEC119 | 25 | + | + | N/D | N/A |
| CEC120 | 11 | + | + | N/D | N/A |
| CEC121 | 19 | − | +++ | N/A | N/A |
| CEC122 | 18 | +/− | +++ | N/A | N/A |
| CEC116 | 11 | − | +++ | N/A | N/A |
| Co-Infection Isolates 1 | |||||
| PaAU4391 | 39 | +++ | +++ | fha1Δ405–425 (Fha1Δ135–141) | N/A |
| PaAU5159 | 26 | ++ | +++ | N/D | N/A |
| PaAU7618 | 33 | + | + | gacAG175A (GacAG59S) | N/D |
| PaAU10617 | 17 | − | − | N/D | N/A |
| PaAU19694 | 36 | +++ | + | gacAC162A (GacAD54E) | N/D |
| PaAU22775 | 38 | + | − | N/D | N/A |
| PaAU23781 | 39 | + | − | gacSG1568A (GacSTRUNC) | N/D |
| PaAU29744 | 33 | +++ | +++ | N/D | N/A |
To investigate if the ability of BcAU1054 to outcompete P. aeruginosa strains isolated from teenage/adult CF patients correlates with a loss of H1-T6SS activity in the P. aeruginosa strains, we assessed production of Hcp1, the major subunit protein of the H1-T6SS inner tube, during growth on agar. Every P. aeruginosa isolate that was outcompeted by the BcAU1054 T6SS showed either negligible or diminished Hcp1 production compared to PAO1 (Figures 3D and 3E). CEC121, CEC122, and CEC116, which were not outcompeted by the BcAU1054 T6SS, produced Hcp1 at levels similar to PAO1 (Figure 3E). Of the isolates producing diminished levels of Hcp1, two (C078C and CEC118) were strongly outcompeted by BcAU1054 and contain pppA or fha1 mutations that likely abrogate T6SS activity independent of Hcp1 production. By contrast, mutations in genes encoding known post-translational T6SS regulators were not detected within CEC119 and CEC120, possibly explaining why these isolates were not strongly outcompeted by BcAU1054. Every P. aeruginosa strain isolated from an infant or young child except CEC32 produced Hcp1 at or near levels similar to PAO1 (Figure 3C), which correlates with their resistance to being outcompeted by the BcAU1054 T6SS (Figure 3B).
Restoration of H1-T6SS protein production can rescue host-adapted P. aeruginosa from T6SS-mediated elimination by BcAU1054
Phosphorelay through the GacSA two-component system activates production of the sRNAs RsmY and RsmZ, which are required for T6SS protein production by P. aeruginosa (Goodman et al., 2009; 2004; Lapouge et al., 2008; Moscoso et al., 2011; Ventre et al., 2006). To determine if lack of gacS/gacA function is responsible for susceptibility of the P. aeruginosa strains with mutations in these genes, we introduced a plasmid (pJN-rsmZ) into C120C, C123D, and C078C to express rsmZ (induced by arabinose) independent of the GacSA phosphorelay (Intile et al., 2014; Janssen et al., 2018). The vector backbone (pJN105) served as a negative control. In competitions against BcAU1054 on agar containing 0.1% arabinose, C120C pJN-rsmZ was rescued from T6SS-mediated elimination by BcAU1054, while C120C pJN105 was not (Figure 4A). Consistent with this result, C120C pJN-rsmZ, but not C120C pJN105, produced Hcp1 when grown under inducing conditions (Figure 4D). pJN-rsmZ did not promote Hcp1 production in C123D or C078C (Figure 4D), and these strains were still strongly outcompeted by the BcAU1054 T6SS (Figures 4B and 4C). C123D and C078C may harbor additional, unidentified mutations that render them T6SS-deficient, or the ectopic rsmZ strategy may simply not work in these strains. Because C078C also contains a premature stop codon in pppA, we delivered the WT pppA gene under control of a constitutive promoter to the attTn7 site, and also introduced pJN-rsmZ, into this strain, but expression of these genes failed to rescue C078C from T6SS-mediated elimination by BcAU1054 (Figures 4E and 4F). Lastly, we delivered the WT fha1 gene to the attTn7 site of CEC118, as this isolate has a truncated fha1, but constitutive expression of full-length fha1 did not rescue CEC118 from being outcompeted by BcAU1054 (Figure 4G). It is not surprising that constitutive expression of full-length pppA and full-length fha1 did not rescue C078C and CEC118, respectively, as both isolates were defective for Hcp1 production (Figure 3E). It is also possible the natively-produced truncated PppA and Fha1 variants act as dominant negatives in these strains.
Figure 4. Restoration of H1-T6SS protein production can rescue host-adapted P. aeruginosa from T6SS-mediated elimination by BcAU1054.
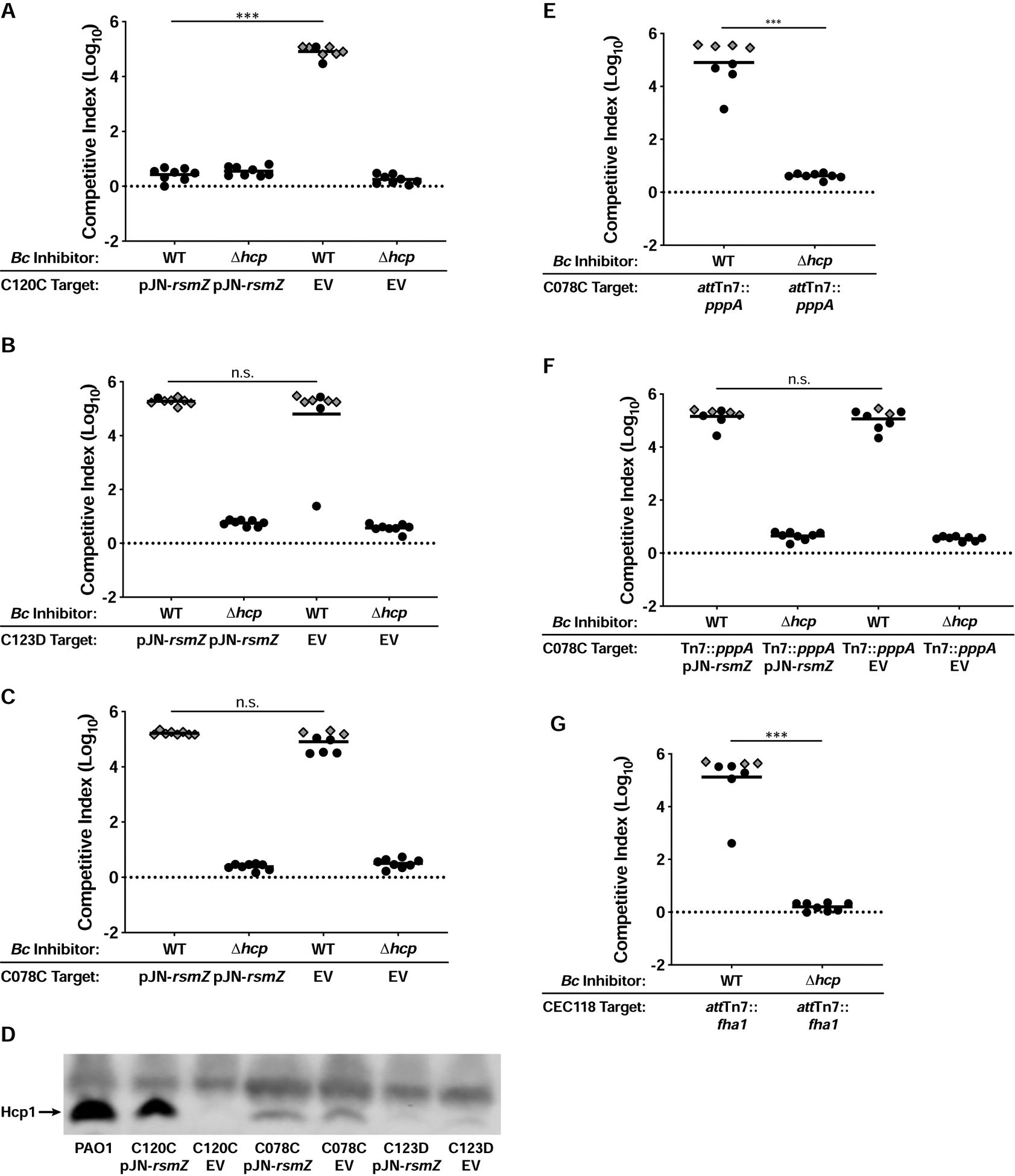
(A, B, and C) Competition experiments between WT and Δhcp BcAU1054 inhibitors and P. aeruginosa teenage/adult CF isolates C120C (A), C123D (B), and C078C (C) harboring the pJN-rsmZ and pJN105 (EV) plasmids. Competitions conducted on agar containing 0.1% arabinose. (D) Immunoblot for Hcp1 production by C120C, C078C, and C123D harboring pJN-rsmZ and pJN105 (EV) during growth on agar containing 0.1% arabinose, and PAO1 for comparison. Non-specific band above Hcp1 serves as loading control. The blot is representative of at least two experiments per strain. (E) Competition experiments between WT and Δhcp BcAU1054 inhibitor and C078C attTn7::pppA target strains. (F) Competition experiments between WT and Δhcp BcAU1054 inhibitor and C078C attTn7::pppA target strains harboring pJN-rsmZ and pJN105 (EV). Competitions conducted on agar containing 0.1% arabinose. (G) Competition experiments between WT and Δhcp BcAU1054 inhibitor and CEC118 attTn7::fha1 target strains. For (A, B, C, E, F, and G), circles/diamonds represent individual cocultures from two biological replicates, each with four technical replicates. Grey-filled diamonds represent competitions from which no target cells were recovered. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. n.s.=not significant, ***P<0.0005, Mann-Whitney test.
Additional Bcc pathogens kill host-adapted P. aeruginosa in a T6SS-dependent manner
To investigate whether T6SS-mediated killing of host-adapted P. aeruginosa is a common feature of Bcc strains, we used Burkholderia multivorans strain CGD2M (BmCGD2M) and BdAU0158, which encode predicted T6SS-1 systems. BmCGD2M and BdAU0158 encode one and two additional predicted T6SSs, respectively. We generated plasmid disruption mutations in the tssC1 genes of these strains’ T6SS-1 clusters (BmCGD2M tssC1::pAP82 and BdAU0158 tssC1::pAP83), and competed these mutants and the parental strains against P. aeruginosa strains PAO1 and C078C. Both BmCGD2M and BdAU0158 strongly outcompeted C078C, but not PAO1, and they did so in a T6SS-dependent manner (Figure 5). These data suggest T6S may provide many Bcc pathogens a competitive advantage against host-adapted P. aeruginosa.
Figure 5. Additional Bcc pathogens kill host-adapted P. aeruginosa in a T6SS-dependent manner.

(A) Competition experiments between WT and tssC1::pAP82 BmCGD2M inhibitor strains and PAO1 and C078C target strains. (B) Competition experiments between WT and tssC1::pAP83 BdAU0158 inhibitor strains and PAO1 and C078C target strains. For (A and B), circles/diamonds represent individual cocultures from two biological replicates, each with four technical replicates. Grey-filled diamonds represent competitions from which no target cells were recovered. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. *P<0.05, ***P<0.0005, Mann-Whitney test.
B. cenocepacia isolates from CF patients with concurrent P. aeruginosa infections outcompete their paired P. aeruginosa isolates under T6SS-permissive conditions
Our data, together with data from other groups (Bartell et al., 2019; Kordes et al., 2019; Marvig et al., 2015), suggest T6SS-abrogating mutations can accumulate as P. aeruginosa evolves within the CF respiratory tract, and patients harboring T6SS-null P. aeruginosa may be susceptible to Bcc superinfections. To explore this hypothesis further, we acquired eight B. cenocepacia-P. aeruginosa co-infection pairs, each isolated from a separate concurrently-infected CF patient. During coculture on agar, seven of eight B. cenocepacia isolates outcompeted their paired P. aeruginosa isolate by as little as ~1 log (BcAU7523 vs. PaAU7618) or as great as ~4 logs (BcAU29704 vs. PaAU29744) (Figure 6A). Since genetic manipulation of recent human isolates is often not possible, we took advantage of the fact that growth in shaking liquid cultures is non-permissive for T6SS-mediated competition (Hood et al., 2010; Majerczyk et al., 2016; Russell et al., 2011; Speare et al., 2020), likely because cells are not in contact long enough to allow T6SS effector delivery to target cells. The competitive advantages of B. cenocepacia isolates over their paired P. aeruginosa isolates dropped dramatically during shaking liquid growth compared to growth on agar (Figure 6B). In two cases (PaAU7618 and PaAU19694), liquid growth provided P. aeruginosa a competitive advantage over its paired B. cenocepacia isolate (Figure 6B).
Figure 6. B. cenocepacia isolates from CF patients with concurrent P. aeruginosa infections outcompete their paired P. aeruginosa isolates under T6SS-permissive conditions.
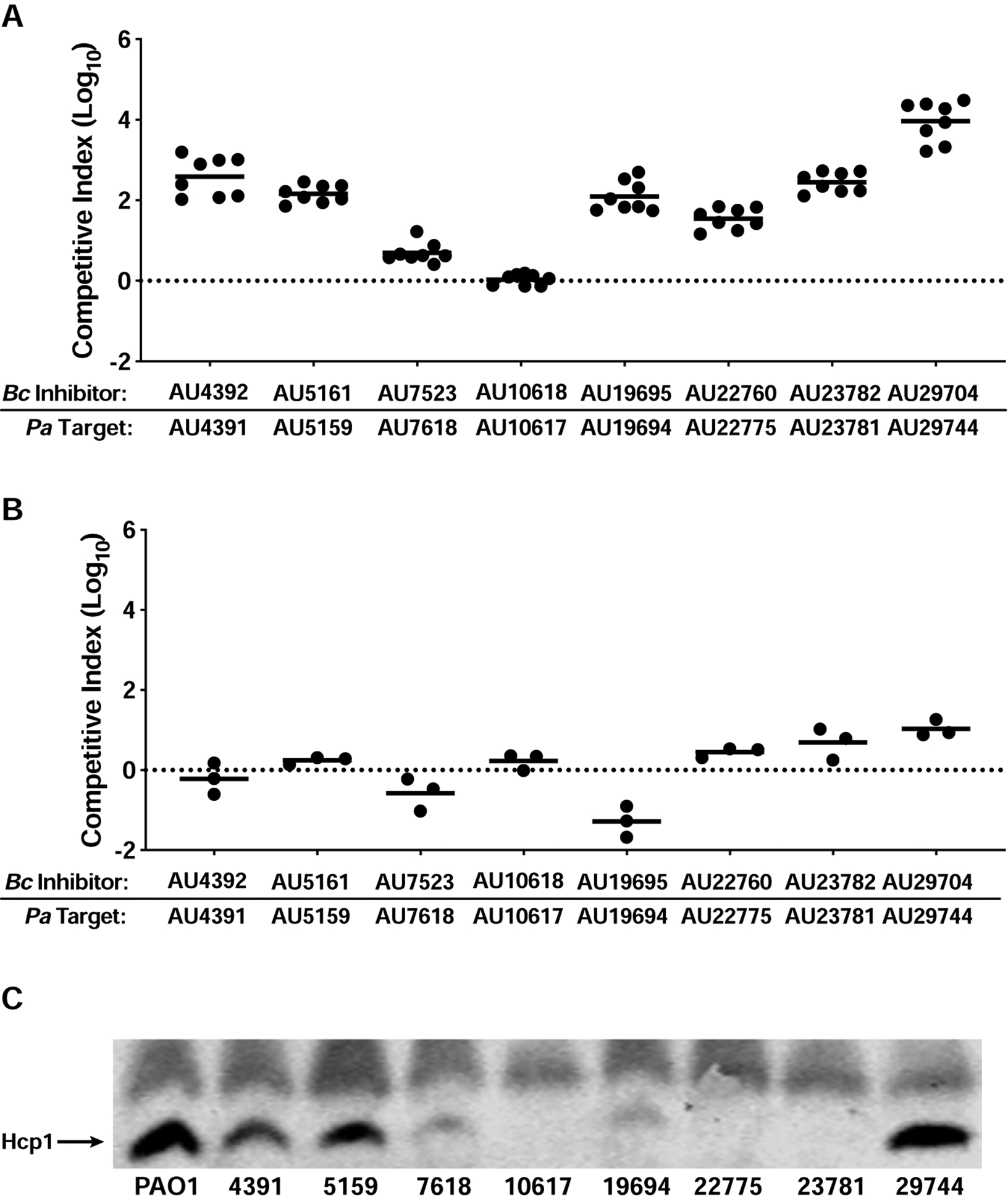
(A and B) Competition experiments between B. cenocepacia-P. aeruginosa co-infection isolate pairs on agar (A) and in shaking liquid culture (B). In (A), circles represent individual cocultures from two biological replicates, each with four technical replicates. In (B), circles represent individual cocultures from three biological replicates. Solid horizontal lines represent mean log10 C.I. values. Dotted horizontal lines (log10 C.I. = 0) indicate no competitive advantage for either strain. (C) Immunoblot for Hcp1 production by P. aeruginosa co-infection isolates, and PAO1 for comparison. Non-specific band above Hcp1 serves as loading control. The blot is representative of at least two experiments per strain.
To identify potential genetic explanations for the competitive disadvantages of the P. aeruginosa co-infection isolates, we PCR-amplified the genes involved in H1-T6SS production/activity that are mutated in the P. aeruginosa strains isolated from adults for which we have whole-genome sequence information (Table 1) and sequenced these PCR products. Three isolates contain mutations in gacS or gacA (PaAU7618 contains a gacAG175A mutation resulting in GacAG59S, PaAU19694 contains a gacAC162A mutation resulting in GacAD54E, and PaAU23781 contains a premature stop codon in gacS (gacSG1568A)) (Table 1). PaAU4391 contains a small deletion in fha1 (fha1Δ405–425) that is nearly identical to the mutation in CEC118 (fha1Δ404–424) (Table 1). Western blotting showed negligible or diminished Hcp1 production by five P. aeruginosa co-infection isolates (PaAU7618, PaAU10617, PaAU19694, PaAU22775, and PaAU23781) compared to PAO1 (Figure 6C). PaAU4391, PaAU5159, and PaAU29744 produced Hcp1 at levels similar to PAO1 (Figure 6C), though PaAU4391 has an fha1Δ405–425 mutation that may abrogate T6SS activity without affecting protein production. PaAU5159 and PaAU29744 may harbor mutations in other genes important for post-translational regulation of T6SS activity. Together, our results suggest Bcc pathogens may only be able to invade a P. aeruginosa-colonized CF respiratory tract if the P. aeruginosa population, or at least a subpopulation, has lost T6SS activity.
DISCUSSION
The underlying reasons for the propensity of Bcc pathogens to infect only older CF patients, and to cause superinfections in those colonized with P. aeruginosa (Folescu et al., 2015; McCloskey et al., 2001; Whiteford et al., 1995) are unknown. During our investigation of T6S in the Bcc, we found that none of the P. aeruginosa strains isolated from infant or child CF patients, but almost half of the strains isolated from teenage and adult CF patients, were susceptible to T6SS-mediated killing by BcAU1054. Additional Bcc pathogens (BmCGD2M and BdAU0158) also efficiently outcompeted susceptible P. aeruginosa strains via T6SS activity, and seven of eight B. cenocepacia strains from patients with concurrent P. aeruginosa infections outcompeted their paired P. aeruginosa strains under conditions promoting T6SS-mediated interactions. These data suggest that one reason Bcc pathogens are restricted to infecting older CF patients is because only in these patients are resident P. aeruginosa susceptible to T6SS-mediated competition by Bcc bacteria.
We found that differential susceptibility of P. aeruginosa strains to T6SS-mediated competition by Bcc pathogens depends on T6SS functionality in P. aeruginosa. Disruption of vipA1 to inactivate the H1-T6SS converted PAO1 from being resistant to T6SS-mediated competition by BcAU1054 to being outcompeted by four logs. We found that all of the susceptible P. aeruginosa strains isolated from teenagers or adults harbor mutations predicted to abrogate production and/or function of their T6SSs, all failed to produce substantial amounts of Hcp1, and for one strain, elimination by BcAU1054 was prevented by activating production of its T6SS proteins. Consistent with these observations, B. cenocepacia isolates strongly outcompeted their co-isolated P. aeruginosa strains when the bacteria were cocultured on a solid surface (conducive to contact-dependent interactions) and not when cocultured in liquid medium. The P. aeruginosa co-infection isolates were also typically deficient in Hcp1 production. These data indicate that, at least for the P. aeruginosa strains studied here, the main factor in determining susceptibility to T6SS-mediated competition by Bcc bacteria is whether P. aeruginosa produces a functional T6SS.
The T6S-abrogating mutations we identified in P. aeruginosa CF isolates in this study fell into two classes: those in genes encoding post-translational regulators of T6SS activity (pppA and fha1), and those in genes encoding the GacSA two-component regulatory system. Fha1 is required for the initial assembly of the T6S apparatus, whereas PppA is required for disassembly of apparatuses and recycling of T6SS proteins into new apparatuses. Mutations in pppA or fha1 are expected to prevent efficient T6SS activity without affecting production of individual T6SS components (Basler et al., 2013; Mougous et al., 2007). Consistent with this expectation, Hcp1 was detectable in PaAU4391, which contains a small deletion in fha1, but this co-infection isolate was outcompeted by its paired B. cenocepacia isolate. By contrast, mutations in gacA or gacS are expected to prevent production of the entire T6S apparatus. GacS is one of four hybrid sensor kinases that controls phosphorylation, and hence activation, of the GacA response regulator. LadS functions with GacS to activate GacA, while RetS blocks GacS activity, thereby inhibiting GacA activation (Chambonnier et al., 2016; Goodman et al., 2009). The PA1611-encoded sensor kinase promotes GacA activation by relieving RetS inhibition of GacS (Kong et al., 2013). When active, GacA induces production of two sRNAs, RsmY and RsmZ, which bind to, and prevent activity of, RsmA, a pleiotropic global regulator that impedes translation of many target genes (Brencic and Lory, 2009; Brencic et al., 2009). When not inhibited by RsmY or RsmZ, RsmA activity results in production of factors associated with acute infection (e.g., flagella, type III secretion, type IV pili) and lack of production of factors and phenotypes associated with chronic infection (e.g., exopolysaccharide production, biofilm, T6S). The RetS/PA1611/LadS/GacSA signaling pathway is therefore considered to function as a switch between acute and chronic infection modes (Balasubramanian et al., 2013; Goodman et al., 2009; 2004).
While there is evidence that the genes encoding the RetS/PA1611/LadS/GacSA signaling pathway are intact when P. aeruginosa establishes infection initially in the CF lung, mutations arise in retS within some strains over time (e.g., 11/36 clone types in the 2015 Marvig et al. study), and, at least for those studied, all retS-mutated strains acquire subsequent mutations in gacS/gacA or rsmA (Bartell et al., 2019; Marvig et al., 2015). Our data are consistent with these reports, as three out of nine teenage/adult P. aeruginosa isolates used in our study were gacS/gacA mutants and also contained nonsynonymous retS mutations, though it is unknown whether these mutations affect RetS function. Three out of eight P. aeruginosa co-infection isolates contained gacS/gacA mutations and did not produce Hcp1; their retS statuses are unknown. Thus, there appears to be a selection for lack of GacSA activity following mutation of retS within the CF respiratory tract, and we envisage this selection could be either T6S-independent or T6S-dependent; a Gac-regulated target other than T6S may drive this selection, with loss of T6S being simply a consequence of Gac inactivation, or T6S itself could be what is selected against. We and others (Kordes et al., 2019; Marvig et al., 2015) have detected mutations in genes encoding proteins specific for T6SS assembly and function in P. aeruginosa strains isolated from older CF patients, supporting the hypothesis that T6S may be disadvantageous to P. aeruginosa during chronic infection in the CF lung.
Why might P. aeruginosa lose T6SS activity in later stages of host colonization? Given the polymicrobial nature of the CF respiratory tract, it is reasonable to hypothesize that maintaining a potent antibacterial weapon like the T6SS would be beneficial. However, P. aeruginosa T6SS proteins are immunogenic (Mougous et al., 2006), and avoiding the host immune response could be equally, or more, beneficial. Additionally, production of T6SSs is energetically costly, and while T6SS-mediated competition may be worth the cost during early stages of infection, these structures may be dispensable once P. aeruginosa has established its niche. As indicated by the proportion of reads in metagenomic samples, P. aeruginosa can constitute over 90% of all bacterial cells within the airways of certain CF patients (Carmody et al., 2015; 2013; J. Zhao et al., 2012). Under these conditions, T6S-mediated interspecies competition should not be required. Loss of T6S by bacteria colonizing humans has been shown with gut resident Bacteroides spp., as T6SS-proficient Bacteroides are more prevalent in the unstable infant gut microbiota than they are in adult gut microbiota where individual Bacteroides spp. or strains predominate (Verster et al., 2017). One might expect that similar selective pressures would act on Bcc during CF infection. However, the B. cenocepacia T6SS is required for murine infection (Hunt et al., 2004), and at least some Bcc strains produce a T6SS effector (TecA) that promotes survival within macrophages (Aubert et al., 2016; 2015; Rosales-Reyes et al., 2012), suggesting there is a strong selective advantage for Bcc pathogens to remain T6SS-active while infecting the CF lung.
Although P. aeruginosa and Bcc bacteria ultimately colonize different sites in the CF airways (Schwab et al., 2014), Bcc pathogens must traverse the lumen, where P. aeruginosa can exist in large populations, before invading host cells. Therefore, transient Bcc-P. aeruginosa interactions likely occur, and our data support the hypothesis that the outcome of these interactions depends on the T6S proficiency of the resident P. aeruginosa. P. aeruginosa populations within individual CF patients exhibit genotypic and phenotypic diversity across different regions of the respiratory tract (Jorth et al., 2015), and thus Bcc bacteria may only need to interact with a subpopulation of P. aeruginosa that has lost T6SS activity in order to initiate an infection and invade host cells. Experiments using animal models and human microbiome analyses have shown that T6SS-mediated competition occurs within mammalian intestines (M. C. Anderson et al., 2017; Sana et al., 2016; Verster et al., 2017; Wexler et al., 2016; W. Zhao et al., 2018), though whether such interactions occur in the CF respiratory tract is unknown. These questions would be better addressed with animal models of CF disease. Unfortunately, a dearth of robust, efficient animal models for chronic bacterial infections has inhibited progress in the understanding of these infections (Fisher et al., 2011; Kukavica-Ibrulj and Levesque, 2008; Semaniakou et al., 2018).
While our data are consistent with T6SS-mediated competition between Bcc pathogens and P. aeruginosa affecting susceptibility of older CF patients to the Bcc, we hypothesize additional factors may prevent Bcc infections in young patients. Staphylococcus aureus is the most prevalent pathogen of young CF patients (Cystic Fibrosis Foundation, 2019), and S. aureus colonization could preclude Bcc infection. Additionally, changes in the immune response, physiology, and/or nutritional environment of the CF respiratory tract over time could cause these tissues to be more hospitable to Bcc pathogens later in the lives of CF patients compared to those in infants and children. CF patients are often on antibiotic regimens to treat opportunistic infections, and regular use of antibiotics may promote Bcc pathogen colonization of older patients. Other unknown factors could also be at play.
In our studies, the T6SS-1 provided strong competitive advantages to three Bcc pathogens (BcAU1054, BmCGD2M, and BdAU0158) against host-adapted P. aeruginosa. Gene clusters encoding T6SS-1 are prevalent throughout the Bcc (Spiewak et al., 2019), suggesting T6SS-mediated killing of host-adapted P. aeruginosa may be a common asset of Bcc pathogens. The role of additional T6SSs in Bcc pathogens remains unknown, but it appears that interbacterial antagonism is mostly mediated by the T6SS-1, at least under the conditions used in this study. Our investigation of the BcAU1054 T6SS revealed four bona fide antibacterial E-I pairs; however, our bioinformatic prediction of E-I pair-encoding genes missed at least one gene pair, as BcAU1054 Δ9E-I maintained a strong competitive advantage against BdAU0158. The unidentified effector(s) is/are not encoded by gene(s) near vgrG genes, nor are there shared domains between the effectors we identified and the unidentified effector(s), suggesting the unidentified effector(s) may be members of an uncharacterized class of T6SS toxins. Our screening for antibacterial effectors and follow-up competition experiments were specific to intrastrain antagonism (BcAU1054 vs. BcAU1054) under one condition (LSLB agar at 37°C). The predicted E-I pairs that our screen suggested were not important for intrastrain competition may be important for interstrain/interspecies competition or competition under different conditions (e.g., temperature, salt, pH); similar conditional efficiency has been demonstrated for P. aeruginosa T6SS effectors (LaCourse et al., 2018). Supporting this hypothesis, BcAU1054 Δ9E-I outcompeted T6SS-null P. aeruginosa teenage/adult isolates to varying degrees, suggesting the additional, unidentified effector(s) have prey cell-specific activity.
There is growing appreciation for the genotypic and phenotypic diversity of P. aeruginosa within the CF respiratory tract (Folkesson et al., 2012; Jorth et al., 2015; Winstanley et al., 2016). Although reference strains are powerful tools for studying bacterial pathogens, they do not always perfectly represent the strains currently infecting humans. Our investigations illuminate differences between PAO1 and recently collected P. aeruginosa CF isolates specific to T6SS-mediated competition against Bcc pathogens, as well as demonstrate varying abilities of P. aeruginosa CF isolates to compete against BcAU1054. Our data support a model in which resident P. aeruginosa populations must evolve to lose T6SS activity in order for Bcc pathogens to colonize the CF respiratory tract. If true, not only is the Bcc T6SS an important colonization factor, but assessing the T6S potential of resident P. aeruginosa could predict susceptibility of CF patients to deadly Bcc superinfections.
STAR METHODS TEXT
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peggy A. Cotter (peggy_cotter@med.unc.edu).
Materials Availability
Bacterial strains and plasmids generated in this study are available upon request from the Lead Contact.
Data and Code Availability
Sequencing reads generated as part of this study are available at the NCBI Sequencing Read Archive: PRJNA607994 and PRJNA609958.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and growth conditions
All bacterial strains in this study were cultured at 37°C in low salt lysogeny broth (LSLB: 10 g/L tryptone, 5 g/L yeast extract, 5 g/L sodium chloride) or on LSLB agar (1.5% agar). Antibiotics to select for Burkholderia strains were used at the following concentrations, when applicable: 30 μg/mL gentamicin, 250 μg/mL kanamycin, 50 μg/mL trimethoprim, 40 μg/mL tetracycline. Antibiotics to select for P. aeruginosa strains were used at the following concentrations, when applicable: 20 or 35 μg/mL chloramphenicol, 20 μg/mL nalidixic acid, 50μg/mL trimethoprim, 75 μg/mL gentamicin, 40 μg/mL tetracycline. 20 μg/mL nalidixic acid was used to select for E. coli DH5α, when applicable. LSLB agar was supplemented with 200 μg/mL 2,6-diaminopimelic acid to support the growth of E. coli strain RHO3.
METHOD DETAILS
Genetic manipulations
E. coli strain RHO3 was used to conjugate plasmids into Burkholderia spp. and P. aeruginosa. The pEXKm5 allelic exchange vector (López et al., 2009) was used to generate unmarked, in-frame deletion mutations in BcAU1054. Briefly, ~500 nucleotides 5’ to and including the first three codons of the gene to be deleted were fused to ~500 nucleotides 3’ to and including the last three codons of the gene by overlap extension PCR and cloned into pEXKm5. Following selection of BcAU1054 merodiploids with the plasmids integrated into the chromosome, cells were grown for 4 h in YT broth (10 g/L yeast extract, 10 g/L tryptone) at 37°C with aeration, subcultured 1:1000 in fresh YT broth, and grown overnight at 37°C with aeration. After overnight growth, cells that lost the cointegrated plasmid following the second homologous recombination step were selected on YT agar (1.5% agar) containing 25% sucrose and 100 μg/mL 5-bromo-4-chloro-3-indoxyl-β-D-glucuronide (X-Gluc). Deletion mutants were screened for by PCR and verified by sequencing regions spanning the deletions.
The pUC18T-mini-Tn7T suite of plasmids (Choi et al., 2005) was used to deliver antibiotic resistance gene cassettes to the attTn7 sites of BcAU1054 and P. aeruginosa. The trimethoprim resistance-conferring plasmid pUC18T-mini-Tn7T-Tp was generated in this study by restriction digesting out dhfRII from pUC18T-mini-Tn7-Tp-PS12-mCherry (LeRoux et al., 2012) using MscI and NcoI and ligating into digested pUC18T-mini-Tn7T-Km (Choi et al., 2005) lacking nptII (the kanamycin resistance-conferring gene). pUC18-miniTn7-kan-gfp (Norris et al., 2010) was used to generate GFP-producing BcAU1054 E-I deletion mutants. Complemented BcAU1054 mutant strains (with either hcp or T6SS immunity-encoding genes) were generated by PCR amplifying the genes of interest and cloning the sequences into pUCS12Km, with genes expressed off the constitutive ribosomal S12 subunit gene promoter of Burkholderia thailandensis strain E264 (M. S. Anderson et al., 2012). BcAU1054 strains constitutively expressing lacZ were generated using pECG10 (M. S. Anderson et al., 2012). P. aeruginosa isolates C078C and CEC118 were complemented with pppA and fha1 genes from PAO1, respectively, by cloning these sequences into pUCS12Km, digesting out the genes and upstream constitutive promoter PS12, and cloning these fragments into pUC18T-mini-Tn7T-Tet (M. S. Anderson et al., 2012). For all pUC18T-mini-Tn7T-based cassette delivery to the attTn7 sites of BcAU1054 and P. aeruginosa, the transposase-encoding pTNS3 helper plasmid was used in triparental conjugation. BmCGD2M tssC1::pAP82 and BdAU0158 tssC1::pAP83 were generated by cloning ~500 internal nucleotides of the tssC1 genes into pUC18T-mini-Tn7T-Km, conjugating the plasmids into BmCGD2M and BdAU1058, and selecting for plasmid cointegrants on kanamycin.
Interbacterial competition experiments
All competition experiments were conducted for 5 h on LSLB agar at 37°C, with an ~1:1 starting cell ratio of inhibitor and target strains, unless stated otherwise. Cells were collected from overnight liquid cultures, centrifuged for 2 min at 15,000 rpm, washed in 1X phosphate buffered saline (PBS), diluted to an OD600 of 1.0, and equal volumes of inhibitor and target cells were mixed. For BcAU1054 vs. E. coli DH5α competitions, BcAU1054 1.0 OD600 cell suspensions were diluted 1:3 in 1X PBS before mixing with DH5α 1.0 OD600 cell suspensions to attain an ~1:1 starting cell ratio. 20 μL spots of mixtures were plated on LSLB agar in 24-well plates, allowed to dry, and incubated at 37°C for 5 h. Starting mixtures were also serially diluted and plated on antibiotic-containing selective media to enumerate inhibitor and target strains at the initial time point. Following 5 h, competition spots were resuspended in 1 mL 1X PBS within wells, serially diluted, and plated on antibiotic-containing selective media to separately grow inhibitor and target strains. Colony counts at the initial and 5 h time points allowed for competitive index (C.I.) calculations as follows: C.I. = (inhibitort5/targett5)/(inhibitort0/targett0). A positive log10 C.I. indicates the inhibitor strain outcompeted the target strain, a negative log10 C.I. indicates the target strain outcompeted the inhibitor strain, and a log10 C.I. of ~0 indicates neither strain had a competitive advantage. For cocultures from which no target bacteria were recovered, the target colony count was set at the limit of detection (one colony forming unit at a 10−2 dilution) and indicated by grey-filled diamonds; thus, the calculated C.I.’s for these competitions are likely underestimations.
Liquid competitions between B. cenocepacia-P. aeruginosa co-infection isolates were set up following the above protocol, except 20 μL of cell mixtures were inoculated into 1 mL LSLB and grown for 5 h at 37°C shaking at 220 rpm. For competitions between BcAU1054 and P. aeruginosa clinical isolates, BcAU1054 WT and Δhcp strains constitutively expressing lacZ were used and inocula/competitions were plated onto antibiotic-containing LSLB agar with 40 μg/mL 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-Gal) to help differentiate between P. aeruginosa and BcAU1054 colonies. Competitions between BcAU1054 and pJN-rsmZ/pJN105-harboring P. aeruginosa teenage/adult isolates were conducted on LSLB agar containing 0.1% L-arabinose.
BcAU1054 T6SS E/I screen
Cocultures were set up following the same protocol as in the interbacterial competition experiments. For monocultures, cell suspensions (at an OD600 of 1.0) were mixed 1:1 with 1X PBS before plating. For cocultures and monocultures, 20 μL spots were plated on LSLB agar within 24-well plates, spots were allowed to dry, and plates were incubated at 37°C for ~20 h. Following incubation, the cultures were resuspended in 1 mL 1X PBS within wells, 100 μL were added to 96-well plates, and OD600 values and GFP fluorescence intensities (485 nm excitation, 530 nm emission) were measured on a PerkinElmer Wallac VICTOR3TM plate reader.
Hcp1 immunoblotting
P. aeruginosa strains were swabbed onto LSLB agar and grown overnight at 37°C. For pJN-rsmZ/pJN105-harboring P. aeruginosa teenage/adult isolates, strains were swabbed onto LSLB agar containing 75 μg/mL gentamicin and 0.1% L-arabinose and grown overnight at 37°C. Following overnight incubation, cells were scraped off plate, resuspended in 1 mL cold 1X PBS, centrifuged for 2 min at 15,000 rpm, washed in 1 mL cold 1X PBS, and diluted to an OD600 of 5.0. Cells were then centrifuged for 2 min at 15,000 rpm and resuspended in 200 μL 2X SDS-PAGE sample loading buffer (6X SDS-PAGE sample loading buffer: 375 mM Tris-HCl, 9% sodium dodecyl sulfate (SDS), 50% glycerol, 0.03% bromophenol blue, 1.3 M β-mercaptoethanol), boiled at 99°C for 15 min, and samples were sheared 10 times through a 26G needle. Samples were resolved on 12% SDS-PAGE gels (5 μL loaded), transferred to nitrocellulose membranes, and membranes were blocked with 5% (w/v) non-fat dry milk in 1X PBS for 1 h with rotation at room temperature (RT). Membranes were then washed three times in 1X PBS and incubated with anti-Hcp1 polyclonal peptide antibody (diluted 1:1000 in 5% (w/v) non-fat dry milk in 1X PBS+0.1% Tween®20 (PBS-T)) for 1 h with rotation at RT. Membranes were then washed three times in 1X PBS-T, incubated with IRDye® 800CW goat anti-rabbit IgG secondary antibody (diluted 1:25,000 in 5% (w/v) non-fat dry milk in 1X PBS-T) for 30 min with rotation at RT, washed three times in 1X PBS, and imaged on a LI-COR Odyssey® fluorescence imager.
Sequencing
Genomic DNA was purified from P. aeruginosa isolates C078C, C120C, and C123D using the Promega Wizard Genomic DNA Purification Kit. Paired-end TruSeq (Illumina) libraries were generated and sequenced on the Illumina MiSeq 2×150 platform at the High-Throughput Sequencing Facility at the University of North Carolina at Chapel Hill. Demultiplexed FASTQ files were mapped to the PAO1 reference genome (assembly GCA_000006765.1) using the Geneious Prime standard assembler. Sequencing reads can be accessed in BioProject PRJNA609958.
To sequence P. aeruginosa CEC isolate genomes, genomic DNA was isolated using a GenElute Bacterial Genomic DNA Kit (Sigma Aldrich, NA2110; St. Louis, MO) following kit instructions with the following exception: all DNA was eluted in 400uL of ultra-pure DEPC-treated water (ThermoFisher Scientific, Waltham, MA). Concentration of DNA preps was determined using a NanoDrop 1000 (ThermoFisher Scientific, Waltham, MA). All preps were stored at −20C. The 150bp sequencing reads from the Illumina platform were assembled using spades v.3.7.1 with careful mismatch correction and the assemblies were filtered to contain only contigs ≥500bp with ≥5X k-mer coverage. The assemblies were further examined for characteristics that would suggest the genome was of high quality (<400 contigs) and potentially P. aeruginosa. All reads and assemblies are deposited at NCBI under BioProject PRJNA607994.
Specific P. aeruginosa co-infection isolate genes were sequenced by PCR-amplifying genes of interest and submitting the PCR products for Sanger sequencing.
Bioinformatic analysis of BcAU1054 T6SS-encoding genes and effector proteins
The BcAU1054 and BcJ2315 (genome assembly GCA_000009485.1) T6SS-encoding core clusters were aligned in Geneious Prime using the Mauve plugin (Darling et al., 2004). Phyre2 (Kelley et al., 2015) and HHpred (Zimmermann et al., 2018) were used to predict the secondary structures and catalytic activities of potential BcAU1054 T6SS effector proteins.
QUANTIFICATION AND STATISTICAL ANALYSES
All statistical significance was calculated by Mann-Whitney tests using GraphPad Prism v. 8. Specific details of statistical analysis, including total number of samples and replicates, can be found in the figure legends.
Supplementary Material
Table S1. Whole genome sequencing errors in BcAU1054 T6SS genes; related to Figure 1. Correct sequences provided (determined by PCR amplification and Sanger sequencing) for two genes misannotated as pseudogenes – clpV and tne1. Sequences also provided for tni1 and tni2, which are not annotated.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-PAO1 Hcp1 polyclonal peptide antibody | Gift from Dr. John Mekalanos | N/A |
| IRDye® 800CW Goat anti-rabbit IgG Secondary Antibody | LI-COR Biosciences | 926–32211; RRID: AB_621843 |
| Bacterial and Virus Strains | ||
| All bacterial strains used in this study listed in Table S3 | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gentamicin | Gold Biotechnology | G-400–25 |
| Kanamycin | Gold Biotechnology | K-120–25 |
| Nalidixic acid | Sigma-Aldrich | N4382 |
| Trimethoprim | Gold Biotechnology | T-350–5 |
| Tetracycline | Fisher Scientific | BP912–100 |
| Chloramphenicol | Fisher Scientific | BP904–100 |
| 5-bromo-4-chloro-3-indoxyl-β-D-glucuronide (X-Gluc) | Gold Biotechnology | G1281C1 |
| 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) | Gold Biotechnology | X4281C |
| L-arabinose | Gold Biotechnology | A-300–100 |
| D-sucrose | Fisher Scientific | BP220–1 |
| 2,6-diaminopimelic acid | Alfa Aesar | AAB22391–06 |
| Critical Commercial Assays | ||
| Wizard Genomic DNA Purification Kit | Promega | A1120 |
| GenElute™ Bacterial Genomic DNA Kit | Sigma-Aldrich | NA2110 |
| Deposited Data | ||
| P. aeruginosa isolate whole genome sequences | This study | NCBI Sequence Read Archive: PRJNA607994 and PRJNA609958 |
| Recombinant DNA | ||
| All plasmids used in this study listed in Table S4 | ||
| Software and Algorithms | ||
| Geneious Prime | Geneious, Software, Newark, New Jersey, USA | http://www.geneious.com; RRID: SCR_010519 |
| Phyre2 | (Kelley et al., 2015) | http://www.sbg.bio.ic.ac.uk/~phyre2; RRID: SCR_010270 |
| HHPred | (Zimmermann et al., 2018) | https://toolkit.tuebingen.mpg.de/tools/hhpred; RRID: SCR_010276 |
| Mauve | (Darling et al., 2004) | http://gel.ahabs.wisc.edu/mauve/; RRID: SCR_012852 |
| GraphPad Prism version 8.0 for Mac | GraphPad, Software, La Jolla, California, USA | http://www.graphpad.com; RRID: SCR_022798 |
HIGHLIGHTS.
Burkholderia cepacia complex (Bcc) pathogens produce functional antibacterial T6SSs
Bcc pathogens use T6SSs to outcompete host-adapted P. aeruginosa CF isolates
Host-adapted P. aeruginosa CF isolates harbor T6SS-abrogating mutations
T6SS-mediated competition dynamics may restrict Bcc infections to older CF patients
ACKNOWLEDGEMENTS
We thank John Mekalanos for providing the α-Hcp1 peptide antibody, and John LiPuma for providing the B. cenocepacia-P. aeruginosa co-infection isolates. We thank members of the Cotter Lab for helpful discussion while designing experiments and analyzing data for this study. This work was supported by NIH awards NIHR01GM121110 to P.A.C. and U19AI110820 to D.A.R, a fellowship from the University of North Carolina at Chapel Hill Graduate School to A.I.P., a graduate stipend from the University of Maryland-Baltimore Graduate School to C.E.C., and Cystic Fibrosis Foundation award COTTER18I0 to P.A.C. Funders had no role in the planning, execution, or analysis of experiments, nor in manuscript preparation and submission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmad S, Wang B, Walker MD, Tran H-KR, Stogios PJ, Savchenko A, Grant RA, McArthur AG, Laub MT, Whitney JC, 2019. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A, 2017. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A 114, 7707–7712. doi: 10.1073/pnas.1700286114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Mobley HLT, 2016. The Versatile Type VI Secretion System. Microbiol Spectr 4, 337–356. doi: 10.1128/microbiolspec.VMBF-0026-2015 [DOI] [Google Scholar]
- Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ, 2017. Shigella sonnei Encodes a Functional T6SS Used for Interbacterial Competition and Niche Occupancy. Cell Host Microbe 21, 769–776.e3. doi: 10.1016/j.chom.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Anderson MS, Garcia EC, Cotter PA, 2012. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet 8, e1002877. doi: 10.1371/journal.pgen.1002877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert DF, Hu S, Valvano MA, 2015. Quantification of type VI secretion system activity in macrophages infected with Burkholderia cenocepacia. Microbiology (Reading, Engl.) 161, 2161–2173. doi: 10.1099/mic.0.000174 [DOI] [PubMed] [Google Scholar]
- Aubert DF, Xu H, Yang J, Shi X, Gao W, Li L, Bisaro F, Chen S, Valvano MA, Shao F, 2016. A Burkholderia Type VI Effector Deamidates Rho GTPases to Activate the Pyrin Inflammasome and Trigger Inflammation. Cell Host Microbe 19, 664–674. doi: 10.1016/j.chom.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Balasubramanian D, Schneper L, Kumari H, Mathee K, 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41, 1–20. doi: 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartell JA, Sommer LM, Haagensen JAJ, Loch A, Espinosa R, Molin S, Johansen HK, 2019. Evolutionary highways to persistent bacterial infection. Nat Commun 10, 629–13. doi: 10.1038/s41467-019-08504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ, 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ, 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick R, Spilker T, Martin A, Lipuma JJ, 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett 228, 57–62. [DOI] [PubMed] [Google Scholar]
- Bisht K, Baishya J, Wakeman CA, 2020. Pseudomonas aeruginosa polymicrobial interactions during lung infection. Curr. Opin. Microbiol 53, 1–8. doi: 10.1016/j.mib.2020.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I, 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104. doi: 10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, Lory S, 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S, 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW, 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis 183, 444–452. doi: 10.1086/318075 [DOI] [PubMed] [Google Scholar]
- Carmody LA, Zhao J, Kalikin LM, LeBar W, Simon RH, Venkataraman A, Schmidt TM, Abdo Z, Schloss PD, Lipuma JJ, 2015. The daily dynamics of cystic fibrosis airway microbiota during clinical stability and at exacerbation. Microbiome 3, 12. doi: 10.1186/s40168-015-0074-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody LA, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB, Li JZ, Lipuma JJ, 2013. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc 10, 179–187. doi: 10.1513/AnnalsATS.201211-107OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona MG, Silverman JM, Sall KM, Boyer F, Couté Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I, 2013. An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environmental Microbiology 15, 471–486. doi: 10.1111/j.1462-2920.2012.02816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambonnier G, Roux L, Redelberger D, Fadel F, Filloux A, Sivaneson M, De Bentzmann S, Bordi C, 2016. The Hybrid Histidine Kinase LadS Forms a Multicomponent Signal Transduction System with the GacS/GacA Two-Component System in Pseudomonas aeruginosa. PLoS Genet 12, e1006032. doi: 10.1371/journal.pgen.1006032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CE, Horspool AM, Hill PJ, Wozniak DJ, Schertzer JW, Rasko DA, Ernst RK, 2019. Genomic and Phenotypic Diversity among Ten Laboratory Isolates of Pseudomonas aeruginosa PAO1. J. Bacteriol 201, 917. doi: 10.1128/JB.00595-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Witzmann KA, Spilker T, Fink RJ, LiPuma JJ, 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr 139, 643–649. doi: 10.1067/mpd.2001.118430 [DOI] [PubMed] [Google Scholar]
- Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP, 2005. A Tn7-based broad-range bacterial cloning and expression system. Nature Methods 2, 443–448. doi: 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Choi K-H, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP, 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol 74, 1064–1075. doi: 10.1128/AEM.02430-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation, 2019. Cystic Fibrosis Foundation Patient Registry
- Darling ACE, Mau B, Blattner FR, Perna NT, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14, 1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkins LM, O’Toole GA, 2015. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS Pathogens 11, e1005258. doi: 10.1371/journal.ppat.1005258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JT, Zhang Y, Engelhardt JF, 2011. Comparative biology of cystic fibrosis animal models. Methods Mol. Biol 742, 311–334. doi: 10.1007/978-1-61779-120-8_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folescu TW, da Costa CH, Cohen RWF, da Conceição Neto OC, Albano RM, Marques EA, 2015. Burkholderia cepacia complex: clinical course in cystic fibrosis patients. BMC Pulm Med 15, 158. doi: 10.1186/s12890-015-0148-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S, 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol 10, 841–851. doi: 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- Foster KR, Bell T, 2012. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol 22, 1845–1850. doi: 10.1016/j.cub.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Ghysels B, Ochsner U, Möllman U, Heinisch L, Vasil M, Cornelis P, Matthijs S, 2005. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett 246, 167–174. doi: 10.1016/j.femsle.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S, 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754. doi: 10.1016/j.devcel.2004.08.020 [DOI] [PubMed] [Google Scholar]
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S, 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23, 249–259. doi: 10.1101/gad.1739009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Brown PH, Maddison J, Doherty CJ, Nelson JW, Dodd M, Greening AP, Webb AK, 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342, 15–19. doi: 10.1016/0140-6736(93)91881-l [DOI] [PubMed] [Google Scholar]
- Govan JR, Deretic V, 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev 60, 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I, 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40, D26–32. doi: 10.1093/nar/gkr947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A, Wood TE, Filloux A, 2016. Type VI secretion and anti-host effectors. Curr. Opin. Microbiol 29, 81–93. doi: 10.1016/j.mib.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Held K, Ramage E, Jacobs M, Gallagher L, Manoil C, 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol 194, 6387–6389. doi: 10.1128/JB.01479-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW, 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol 13, 572–581. doi: 10.1099/00221287-13-3-572 [DOI] [PubMed] [Google Scholar]
- Holloway BW, Morgan AF, 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol 40, 79–105. doi: 10.1146/annurev.mi.40.100186.000455 [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD, 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Schwarz S, Mougous JD, 2009. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol. Microbiol 72, 1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt TA, Kooi C, Sokol PA, Valvano MA, 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun 72, 4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL, 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol 196, 357–366. doi: 10.1128/JB.01199-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H, 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr 104, 206–210. [DOI] [PubMed] [Google Scholar]
- Janssen KH, Diaz MR, Gode CJ, Wolfgang MC, Yahr TL, 2018. RsmV, a Small Noncoding Regulatory RNA in Pseudomonas aeruginosa That Sequesters RsmA and RsmF from Target mRNAs. J. Bacteriol 200, 3159. doi: 10.1128/JB.00277-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK, 2015. Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs. Cell Host Microbe 18, 307–319. doi: 10.1016/j.chom.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE, 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schöck U, Pohl TM, Wiehlmann L, Tümmler B, 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol 192, 1113–1121. doi: 10.1128/JB.01515-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Chen L, Zhao J, Shen T, Surette MG, Shen L, Duan K, 2013. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol 88, 784–797. doi: 10.1111/mmi.12223 [DOI] [PubMed] [Google Scholar]
- Kordes A, Preusse M, Willger SD, Braubach P, Jonigk D, Haverich A, Warnecke G, Häussler S, 2019. Genetically diverse Pseudomonas aeruginosa populations display similar transcriptomic profiles in a cystic fibrosis explanted lung. Nat Commun 10, 3397–10. doi: 10.1038/s41467-019-11414-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukavica-Ibrulj I, Levesque RC, 2008. Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab. Anim 42, 389–412. doi: 10.1258/la.2007.06014e [DOI] [PubMed] [Google Scholar]
- LaCourse KD, Peterson SB, Kulasekara HD, Radey MC, Kim J, Mougous JD, 2018. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat Microbiol 3, 440–446. doi: 10.1038/s41564-018-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH-T, Haas D, 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x [DOI] [PubMed] [Google Scholar]
- LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD, 2012. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc. Natl. Acad. Sci. U.S.A 109, 19804–19809. doi: 10.1073/pnas.1213963109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD, 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4, 465. doi: 10.7554/eLife.05701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Moore R, Wilton M, Wong MJQ, Lam L, Dong TG, 2015. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. U.S.A 112, 9106–9111. doi: 10.1073/pnas.1505317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma JJ, 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev 23, 299–323. doi: 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CM, Rholl DA, Trunck LA, Schweizer HP, 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl. Environ. Microbiol 75, 6496–6503. doi: 10.1128/AEM.01669-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB, 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol 3, 144–156. doi: 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- Majerczyk C, Schneider E, Greenberg EP, 2016. Quorum sensing control of Type VI secretion factors restricts the proliferation of quorum-sensing mutants. Elife 5, 317. doi: 10.7554/eLife.14712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC, 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A 110, 15055–15060. doi: 10.1073/pnas.1307217110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig RL, Sommer LM, Molin S, Johansen HK, 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet 47, 57–64. doi: 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- McCloskey M, McCaughan J, Redmond AO, Elborn JS, 2001. Clinical outcome after acquisition of Burkholderia cepacia in patients with cystic fibrosis. Ir J Med Sci 170, 28–31. doi: 10.1007/bf03167716 [DOI] [PubMed] [Google Scholar]
- Miyata ST, Unterweger D, Rudko SP, Pukatzki S, 2013. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathogens 9, e1003752. doi: 10.1371/journal.ppat.1003752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A, 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environmental Microbiology 13, 3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ, 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ, 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol 9, 797–803. doi: 10.1038/ncb1605 [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C, 1999. Broad-host-range expression vectors that carry the L- arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227, 197–203. doi: 10.1016/s0378-1119(98)00601-5 [DOI] [PubMed] [Google Scholar]
- Norris MH, Kang Y, Wilcox B, Hoang TT, 2010. Stable, site-specific fluorescent tagging constructs optimized for Burkholderia species. Appl. Environ. Microbiol 76, 7635–7640. doi: 10.1128/AEM.01188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S, Fothergill JL, 2017. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett 364, 850. doi: 10.1093/femsle/fnx128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME, 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev 25, 193–213. doi: 10.1128/CMR.00013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ, 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A 104, 15508–15513. doi: 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP, Gode CJ, Lachiewicz AM, Wolfgang MC, Conlon BP, 2017. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15, e2003981. doi: 10.1371/journal.pbio.2003981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Reyes R, Skeldon AM, Aubert DF, Valvano MA, 2012. The Type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cell. Microbiol 14, 255–273. doi: 10.1111/j.1462-5822.2011.01716.x [DOI] [PubMed] [Google Scholar]
- Rosenfeld M, Gibson RL, McNamara S, Emerson J, Burns JL, Castile R, Hiatt P, McCoy K, Wilson CB, Inglis A, Smith A, Martin TR, Ramsey BW, 2001. Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr. Pulmonol 32, 356–366. doi: 10.1002/ppul.1144 [DOI] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD, 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD, 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD, 2014. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol 12, 137–148. doi: 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD, 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549. doi: 10.1016/j.chom.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsgiver EL, Fink AK, Knapp EA, Lipuma JJ, Olivier KN, Marshall BC, Saiman L, 2016. Changing Epidemiology of the Respiratory Bacteriology of Patients With Cystic Fibrosis. Chest 149, 390–400. doi: 10.1378/chest.15-0676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM, 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A 113, E5044–51. doi: 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U, Abdullah LH, Perlmutt OS, Albert D, Davis CW, Arnold RR, Yankaskas JR, Gilligan P, Neubauer H, Randell SH, Boucher RC, 2014. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect. Immun 82, 4729–4745. doi: 10.1128/IAI.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaniakou A, Croll RP, Chappe V, 2018. Animal Models in the Pathophysiology of Cystic Fibrosis. Front Pharmacol 9, 1475. doi: 10.3389/fphar.2018.01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG, 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speare L, Smith S, Salvato F, Kleiner M, Septer AN, 2020. Environmental Viscosity Modulates Interbacterial Killing during Habitat Transition. MBio 11, 19520. doi: 10.1128/mBio.03060-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiewak HL, Shastri S, Zhang L, Schwager S, Eberl L, Vergunst AC, Thomas MS, 2019. Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. Microbiologyopen 8, e774. doi: 10.1002/mbo3.774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S-Y, Bosch DE, Mangiameli SM, Radey MC, Huang S, Park Y-J, Kelly KA, Filip SK, Goo YA, Eng JK, Allaire M, Veesler D, Wiggins PA, Peterson SB, Mougous JD, 2018. Bifunctional Immunity Proteins Protect Bacteria against FtsZ- Targeting ADP-Ribosylating Toxins. Cell 175, 1380–1392.e14. doi: 10.1016/j.cell.2018.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A, 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U.S.A 103, 171–176. doi: 10.1073/pnas.0507407103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E, 2017. The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 22, 411–419.e4. doi: 10.1016/j.chom.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler AG, Bao Y, Whitney JC, Bobay L-M, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL, 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A 113, 3639–3644. doi: 10.1073/pnas.1525637113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford ML, Wilkinson JD, McColl JH, Conlon FM, Michie JR, Evans TJ, Paton JY, 1995. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 50, 1194–1198. doi: 10.1136/thx.50.11.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley C, O’Brien S, Brockhurst MA, 2016. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol 24, 327–337. doi: 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, Lipuma JJ, 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. U.S.A 109, 5809–5814. doi: 10.1073/pnas.1120577109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Caro F, Robins W, Mekalanos JJ, 2018. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213. doi: 10.1126/science.aap8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V, 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol 430, 2237–2243. doi: 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Whole genome sequencing errors in BcAU1054 T6SS genes; related to Figure 1. Correct sequences provided (determined by PCR amplification and Sanger sequencing) for two genes misannotated as pseudogenes – clpV and tne1. Sequences also provided for tni1 and tni2, which are not annotated.
Data Availability Statement
Sequencing reads generated as part of this study are available at the NCBI Sequencing Read Archive: PRJNA607994 and PRJNA609958.


