Abstract
Retention time is the most common and widely used criterion to report the separation of glycans using Liquid Chromatography (LC), but it varies widely across different columns, instruments and laboratories. This variation is problematic when inter-laboratory data is compared. Furthermore, it influences reproducibility and hampers efficient data interpretation. In our endeavor to overcome this variance, we propose the use of the Glucose Unit Index (GUI) on C18 and PGC column-based separation of reduced and permethylated glycans. GUI has previously been utilized for retention time normalization of native and labeled glycans. We evaluated this method with reduced and permethylated glycans derived from model glycoproteins fetuin and ribonuclease B (RNase B), and then implemented it to human blood serum to generate C18 and PGC column-based isomeric glycan libraries. GUI values for glycan compositions were calculated with respect to the glucose units derived from dextrin, which was employed as an elution standard. The GUI values were validated on three different LC systems (UltiMate 3000 Nano UHPLC systems) in two laboratories to ensure the reliability and reproducibility of the method. Applicability on real samples was demonstrated using human breast cancer cell lines. A total of 116 permethylated N-glycans separated on a C18 column and 134 glycans separated on a PGC column were compiled in a library. Overall, the established GUI method and the demonstration of reproducible inter- and intra-laboratory GUI values would aid the future development of automated glycan and isomeric glycan identification methods.
Keywords: Glucose unit index, Permethylated glycans, LC-MS/MS, C18 column, Porous graphitic carbon
Introduction
Glycans are the most common molecules attached to proteins during post translational modification.1, 2 Glycans are responsible for a variety of biological processes, such as protein stability,3 cell-cell recognition,4 cell signaling,5, 6 immune response,7 and host-pathogen interaction.8 Any aberration in their pattern may lead to structural alterations in proteins which can be an indication of many diseases and cancer prognosis. Thus, glycans serve as crucial biomarkers for studying various diseases, such as alzheimer’s disease,9 immune disorders,10 cardiovascular diseases,11 and cancers.6, 12 Moreover, the significance of isomeric glycan expressions in cancer cells has been highlighted, demanding more rigorous and reliable identification of glycan isomers.13
Liquid Chromatography-Mass Spectrometry (LC-MS) is one of the most widely used techniques for glycan analysis due to its high sensitivity and abundance of structural information.1, 14–19 Depending on the derivatization methods, different stationary phases such as hydrophilic interaction liquid chromatography (HILIC), porous graphitized carbon (PGC) columns, and C18 columns are typically used to perform the separation of glycans in conjunction with mass spectrometric detection and fragmentation to precisely identify and quantitate glycan structures.1, 16, 19–25 This separation accommodates not only the determination of glycan structures according to their retention times along with MS/MS, but also provides reliable quantitation by reducing competitive ionization.17, 26 Nevertheless, the lack of reproducibility of these techniques across different instruments and laboratories is a major issue that averts the development of automated tools for glycan identification and quantification.
There are several labeling and derivatization techniques available for glycans, such as 2-aminobenzamide (2-AB), procainamide, aminoxyTMT, RapiFluor-MS (RFMS) labeling, reduction and reduction with permethylation.27 Permethylation is a derivatization method in which all the existing hydrogens in a glycan molecule are converted to methyl groups by iodomethane. Permethylation offers several advantages over the native and reducing end labeled glycans.27 Due to the increased hydrophobicity, permethylated glycans can be easily separated by reversed-phase chromatography.29 Moreover, the technique improves ionization efficiency because of the increased positive ion affinity of glycans, which induces high sensitivity.30 Permethylation also benefits from the fact that it prevents fucose migration/rearrangement in protonated ions31, 32 and sialic acid loss33, which can occur during the gas phase of MS analysis due to weak glycosidic bonds and may lead to possible misinterpretation of structural identification. Zhou et al. demonstrated enhanced MS intensity for permethylated glycans. For example, intensity for sialylated glycans has been demonstrated to be more than 100 times higher than 2-AB derivatized and reduced native glycans.27
C18 and PGC columns have been instrumental in the isomeric separation of glycans. C18 has been widely used for reversed-phase separation.15, 16, 24 The reversed-phase LC analysis of premethylated glycans usually demonstrates poor isomeric separation, which was improved using a PGC column.25 Isomeric separation of permethylated glycans using PGC chromatography was first reported by Costello et al. in which they demonstrated partial isomeric separation of glycans.34 More recently, Zhou et al. reported isomeric separation of permethylated N-glycans using PGC at 75ºC to facilitate both positional and linkage N-glycan isomers.19, 25 Unlike native glycans, permethylated glycans, owing to structural stability, permit the determination of glycan isomers at high confidence in conjunction with PGC-LC-MS.
The glucose unit index (GUI), first introduced by Guile et al.35, has been employed in LC-MS based glycan analysis for retention time normalization of glycans and has been used extensively for sequencing and prediction of unknown glycan structures present in samples.36–38 The elution positions of glycans were determined by comparison with the elution positions of standard dextran. This helped determine the contribution of individual monosaccharides to the Glucose Unit (GU) values of glycan structures, thereby assisting with unknown structure predictions. The index has also been applied to determine alterations in glycosylation patterns related to various diseases.39, 40 This method of assigning elution positions is more advantageous than retention time alone because GUI serves as a calibration of the retention time. Previously, use of GUI has been demonstrated successfully with PA and 2-AB labeled glycans, where it was employed for retention time normalization and structure prediction of labeled glycans.28, 37,41–43 GlycoBase, a database used for assigning HPLC elution positions of 2-AB labeled glycans in terms of their GU values, is an outcome of such efforts which aim to automate the process of glycan characterization.44 Recently, Ashwood et al. presented a PGC-LC-MS based approach for sample-specific glycotyping as another step toward the development of automated glycan identification and quantitation processes.45 They utilized a hydrolyzed dextran ladder as an internal standard for GU determination and skyline software to reduce retention time and peak area technical variations in analyses employing a PGC column.
However, a systematic investigation of using GUI to normalize the retention time shift across different columns, instruments, and laboratories for permethylated glycans has not been made. Despite the ability of LC-MS to efficiently analyze permethylated glycans, the retention time shift across different instruments and laboratories is still a significant challenge. In addition, the use of GUI for permethylated glycans in both glycomic profiling and isomeric separation needs to be developed to obtain better and more reproducible interpretations of glycomics data. To overcome this retention time shift issue, we propose the use of GUI for permethylated glycans to calibrate the retention time. In this study, we report the utilization of permethylated GUI for both C18 and PGC columns to facilitate reliable identification of reduced and permethylated N-glycans and their isomers, derived from model glycoproteins as well as human blood serum, by GUI values. Also, to further validate the method on original samples, application of the method has been shown to characterize permethylated glycans derived from breast cancer cell lines.
Experimental Materials
Fetuin, ribonuclease B (RNase B), and pooled human blood serum (HBS) from Sigma Aldrich were used. Two breast cancer cell lines, MDA-MB-231 (231) and HTB22, were purchased from American Type Culture Collection (ATCC). MDA-MB-231BR (231BR) was received as a gift from Dr. Paul Lockman (Texas Tech Health Sciences Center, School of Pharmacy, Amarillo, TX). The media for cell lines were purchased from ATCC. Trypsin-EDTA 1X (0.25% trypsin/2.21 Mm EDTA) was obtained from Dow Corning (Auburn, MI). Trypsin/Lys-C mix (mass spectrometry grade) was purchased from Promega (Madison, WI). Dextrin (derived from maize starch), Formic acid (FA), iodoacetamide (IAA), ammonium bicarbonate, dithiothreitol (DTT), sodium deoxycholate (SDC), and ammonium bicarbonate (ABC) were purchased from Sigma Aldrich (St. Louis, MO). HPLC grade water was purchased from Avantor Performance Materials (Center Valley, PA). HPLC grade acetonitrile (ACN), methanol, and ethyl alcohol were purchased from Fisher Scientific (Fair Lawn, NJ). Borane-ammonia complex (97% purity), dimethyl sulfoxide (DMSO), iodomethane, sodium hydroxide beads, and formic acid were ordered from Sigma Aldrich. 10XG7 buffer (0.5M phosphate buffer saline) and PNGase F were obtained from New England Biolabs.
Sample Preparation
Standard Proteins and Dextrin
20 μg each of fetuin and RNase B were treated with G7 buffer (final concentration 20 mM) and denatured in a 90°C water bath for 30 minutes. Samples were cooled to room temperature and 100U of PNGase F (500U/μl stock diluted 10X and 2 μl added) were added to each sample, followed by an 18 hour incubation at 37°C.19 After digesting with PNGase F, de-N-glycosylated proteins were removed by precipitating with 90% ethanol at 20ºC for 1 hour. Solution was then subjected to centrifugation and supernatant was collected and dried. The reducing ends of the purified glycans were then reduced by the addition of 10 μl of borane-ammonia complex (97% purity, 10 μg/μl) to the dried solution. Samples were incubated in a 60ºC water bath for 1 hour. After incubation, methanol was added to the reduced glycan samples to remove borane in the form of borate. Samples were then dried in a centrifugal vacuum concentrator to remove the borate. This methanol washing step was repeated three times until all borate had been removed. Reduced samples were then permethylated through the solid-phase permethylation method previously reported.46–49 Briefly, dried samples were resuspended in 1.2 μl of water and 30 μl of DMSO. Then, 20 μl of iodomethane was added. DMSO-soaked sodium hydroxide beads were packed in spin columns and washed once with 200 μl of DMSO using a centrifuge at 1800 rpm for 2 min. Samples were then loaded to the columns and incubated for 25 minutes at room temperature. Then, another 20 μl of iodomethane was added to the samples and incubated for additional 15 minutes at room temperature. After permethylation, permethylated glycans were collected by centrifugation at 1800 rpm for 2 min. Next, 30 μl of acetonitrile was added to the columns and eluents were collected by centrifugation. Eluents were then dried and prepared for LC-MS/MS analysis by resuspending in 20% ACN and 0.1% FA. Each injection comprised of 1 μg of initial protein material, resuspended in 6 μl of 20% ACN and 0.1% FA
Dextrin standard was directly reduced and permethylated following the procedures described above. 1 μg of dextrin was injected before each sample as an external standard. The dextrin was also used to spike the regular samples and used as an internal standard. For this purpose, 2 μg dextrin was added to 20 μg of initial protein, before sample reduction. Reduction, and permethylation were then performed as mentioned previously
Human Blood Serum
10 μl of human blood serum was treated with 90 μl of G7 buffer (20 mM). Samples were kept in a 90ºC water bath for 30 minutes to denature the proteins. After cooling the sample to room temperature, 600U of PNGase F (1.2μl of 500U/μl stock) were added to the samples before they were incubated at 37ºC for 18 hours. The proteins were then precipitated by adding 90% ethanol. For reduction and permethylation, the same procedures were followed as described previously for the standard proteins and dextrin. For internal standard, 1 μg dextrin was spiked into initial 10 μl serum, before reduction and permethylation. For each injection, released N-glycans from 1 μl of the initial serum sample were resuspended in 6 μl of 20% ACN and 0.1% FA.
Cell Culture, Harvesting, and Sample Preparation
Cell line sample 231 was cultured in a RPMI-1640 medium containing 10% fetal bovine serum and 2% penicillin-streptomycin solution. Culturing for 231BR was done in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum and 2% penicillin-streptomycin solution. HTB22 was grown in Eagles’s Minimum Essential Medium (EMEM) containing 10% fetal bovine serum and 2% penicillin-streptomycin solution.
Frozen cells were thawed in a 37ºC water bath. They were then inoculated in 75 cm2 flasks. Cells were incubated at 37ºC for 4–8 days. The cells were fed every 2–4 days before reaching a cell confluence of 90%. After this, cells were washed with a 10 ml aliquot of phosphate-buffered saline (PBS) twice and detached by 2.2 ml trypsin-EDTA solution. Fresh medium was incubated at 37ºC for 5 minutes and was added to the cell solution to neutralize trypsin. Cell suspension was then seeded in three 175 cm2 flasks and incubated at 37ºC for 7 days. When the cells reached a confluence of 80%, they were washed and again detached as described previously. 10 ml of fresh medium was then added, and the cells were harvested by centrifuging at 500X g for 5 minutes. Collected cell pellets were washed twice with PBS to remove medium. The harvested cells were stored in −20ºC.
1/5th of the total cells in the 175 cm2 flask were suspended in 100 μl of water. Cells were lysed by bead beater and were ultrasonicated for at least 1 hour with ice. 100 μl of 5% SDC was added to the sample and vortexed. 2 μl of the sample was taken out for protein assay. 500 mM PBS was then added to the cell solution to bring the final concentration of PBS to 20 mM. Samples were then denatured in a 90ºC water bath for 30 minutes. 500U (1 μl) of PNGase F were added after the samples were cooled down to room temperature and were then incubated at 37ºC for 18 hours. Formic acid was added to the samples (2 μl FA to 200 μl sample) and vortexed thoroughly to remove SDC. The samples were then centrifuged for 1000 rpm for 2 minutes and again for 10 minutes at full speed. Supernatant was transferred to a new tube and dried. 90% ethanol was added to the dried samples and they were stored in −20ºC for 1 hour to precipitate de-N-glycosylated proteins. Centrifugation was then performed at maximum speed for 10 minutes and the supernatant was again transferred to a new tube and dried. The samples were then resuspended in 250 μl of 5% ACN containing 0.1% TFA. Activated charcoal microspin columns were washed three times with 200 μl of 85% ACN containing 0.1% TFA, and then three times with 200 μl of 5% ACN containing 0.1% TFA. Samples were then loaded on the columns and allowed to pass through by centrifuging at 1000 rpm for 2 minutes. The flow through was again loaded on the same column and allowed to pass through the column twice. The microspin columns were then washed two times with 200 μl of 5% ACN containing 0.1% TFA. They were then transferred to new tubes and glycans were eluted by adding 200 μl of 50% ACN containing 0.1% TFA two times and centrifuging.50 Reduction and permethylation was performed as described above.
C18-LC Conditions
Separations were carried out using three different UltiMate 3000 Nano UHPLC systems (Thermo Scientific™, San Jose, CA) for inter- and intra-laboratory comparison studies. The optimum temperature for the oven was kept at 55ºC.51 Mobile phase A was 98% water, 2% ACN, and 0.1% FA and mobile phase B was 100% ACN and 0.1% FA. The gradient started at 20% for mobile phase B and was then increased to 42% in 11 minutes. After 48 minutes of sample run, it reached 55% and was then escalated to 90% in next 1 minute. The organic phase remained at 90% until 54 minutes of total sample run and was then decreased to 20% for the last 6 minutes of the sample run.
PGC-LC Conditions
For isomeric investigation of GUI for reduced and permethylated glycans, the separation was performed with a HyperCarb PGC column (75 μm × 150 mm, 5 μm particle size, Thermo Scientific™, Pittsburgh, PA) using the same HPLC systems mentioned above. The oven temperature was 75ºC, an optimized temperature that was reported in our previous publication.51 Both of the mobile phases were the same as discussed in the previous section. Mobile phase B was kept at 20% for the initial 10 minutes. After 20 minutes, the organic phase was 50%, which was then increased to 95% at 60 minutes. It was maintained at the same concentration until 87 minutes, before being decreased to 20% at 88 minutes. The concentration was maintained at 20% until the end of the sample run at 90 minutes.
MS Conditions
LTQ Orbitrap Velos (Thermo Scientific) and Exactive (Thermo Scientific) equipments were utilized in this study. The LTQ Orbitrap Velos mass spectrometers in labs 1 and 2 were set to the positive ion mode with 1.6 kV ESI voltage. Full MS was performed at 100,000 resolution, and the scan range was 200–2000 m/z. MS2 was acquired by CID and HCD. Normalized collision energy (CE) was 30% for CID and 45% for HCD. The activation Q was 0.25 and the injection time was 10 ms. The repeat count of dynamic exclusion was 2, repeat duration was 30 s, exclusion list size was 50, and exclusion duration was 60 s. Data-dependent acquisition mode (DDA) was applied, which allowed for the selection of the top 4 most intense ions from full MS for the subsequent CID and HCD. The precursor ion selection window was set to 1.50, the intensity threshold for MS2 was 5000 counts, and singly charged ions were set to be excluded for MS2.
The Exactive in lab 1 was operated in positive mode; mass range was 200–2000 m/z; resolution was 100,000; ESI voltage was 1.6 kV; capillary voltage was 60; capillary lens voltage was 120 kV; and skimmer voltage was 60 kV.
Data Analysis
Xcalibur 2.2 SP1 software was used to analyze the LC-MS data. The EIC of full MS data was used to determine glycan composition, with a mass tolerance of 10 ppm. Boxcar 7 point smoothing was enabled for the peaks. For data acquired from Exactive, the identification of the glycans was achieved through full MS and the normalized retention times were compared to dataset from the two Orbitrap Velos included in the study, whose compositions were identified by MS2. The retention times for each glycan from the samples as well as for oligosaccharide units from the dextrin ladder were identified. The retention times of oligosaccharide units derived from dextrin ladders were used to create initial standard curves of retention time versus GUI values with third order polynomial fitting using OriginLab® software. The standard curves were then used to calculate GUI from retention time data for reduced and permethylated glycans derived from model glycoprotein samples. Later, the standard dextrin was spiked in the reduced and permethylated glycomic samples derived from pooled HBS as an internal standard to establish GUI, as our results demonstrated that it does not inhibit the ionization efficiency of the permethylated glycans.
Results and Discussions
The investigation of GUI values was conducted on two different columns, C18 and PGC. Additionally, three different LC-systems were included in the study to normalize the GUI values across multiple instruments and laboratories. Dextrin, a linear oligoglucose consisting of a mixture of glucose oligomers was reduced and permethylated to establish a standard curve of GUI values. The wide range and distribution of oligoglucose provided a glucose ladder for the determination of GUI values for permethylated glycans from our samples. The investigation of GUI on two different columns served the purpose of establishing GUI ranges for characterization of permethylated glycans and their isomers. Retention times for reduced and permethylated glycans on LC were compared to the retention times of reduced and permethylated standard glucose units from the dextrin. These comparisons were done on different columns, instruments, and laboratories to acquire system and laboratory independent normalized elution values for permethylated glycans. A representation of a dextrin ladder subjected to separation on three different LC systems is shown in the Figure S1. The figure demonstrates the retention times of GUI 2 to 10 on different systems. Additionally, separation of dextrin spiked in to the HBS samples is also shown in the figure.
Investigation of GUI for Reduced and Permethylated Glycans on a C18 Column
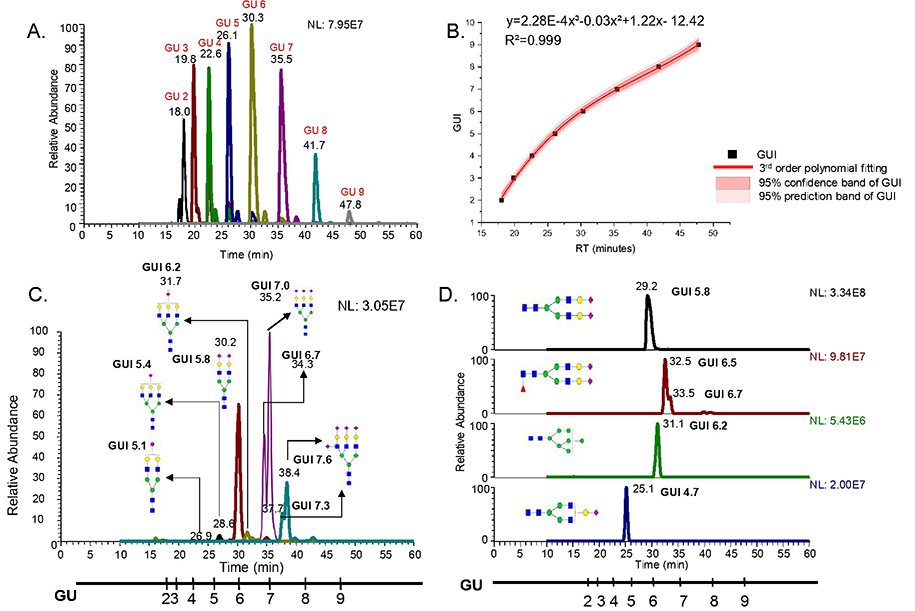
To acquire retention times of the permethylated dextrin glucose units, the dextrin was subjected to C18-LC-MS. Glucose units falling between retention times 15 min to 55 min, were extracted in order to cover the glycans eluted within this range. A chromatogram depicting the retention times of different glucose units separated on a C18 column using LC-1 is shown in Figure 1A. These standard glucose units were detected under the same gradient used for biological glycomic samples. A standard plot of glucose units against their retention times was then constructed with third order polynomial fitting, which was found to be the best fit for the standard units. This standard plot served to calculate the GUI values of the glycans extracted from the samples. The standard curve constructed from dextrin on a C18 column is shown in Figure 1B. The coefficient of determination (R2) of more than 0.999 obtained from the curve denotes the high accuracy of the correlation between retention time and GUI.
Figure 1.
A. Chromatogram showing dextrin separated on C18 column B. Standard dextrin plot showing third order polynomial fitting C. Permethylated glycans derived from fetuin, separated on C18 column D. Permethylated glycans derived from human blood serum, separated on C18 column
After the establishment of a standard fitting curve, reduced and permethylated N-glycans derived from fetuin and RNase B were employed for the initial investigation and establishment of GUI values. Glycans obtained from fetuin consisting of sialylated structures, and RNase B consisting of oligomannose structures, served as ideal candidates for initial study. Figure 1C shows the separation of permethylated glycans derived from fetuin, using a C18 column. The scale shown parallel to the retention times axis illustrates the normalized retention time of the reduced and permethylated dextrin in terms of GUI. This scale helped determine the GUI values for individual permethylated glycans derived from fetuin, as shown in the figure. Since there was a need to include structurally diverse and more complex glycans, the method was also applied to pooled HBS samples. A total of 116 permethylated glycans were identified from human blood serum on a C18 column and their GUI values were calculated using the standard curve generated from dextrin. Some of the glycan structures obtained from human blood serum with corresponding GUI values are presented with respect to the GUI scale in Figure 1D. In both figures, individual GUI values for reduced and permethylated glycans derived from the samples are shown. Another striking finding in these results was the retention time shift for individual reduced and permethylated glycans across different samples, which did not fluctuate with the GUI value. As an example, HexNAc4Hex5NeuAc2 (shown in both Figure 1C and Figure 1D) shows a retention time shift of 1 minute between two samples run on the same system and on the same day. Despite this retention time fluctuation, the GUI for the structure in both samples was the same, demonstrating the importance of retention time normalization using GUI.
A great advantage of using GUI was that we were able to obtain normalized retention time values for the investigated compositions between model glycoproteins and pooled HBS samples. This proved that the GUI values were independent of the sample being investigated and confirmed the robustness of the method for use with a C18 column. Thus, the GUI from permethylated dextrin is well-suited for glycomic retention time normalization using C18-LC-MS.
Investigation of GUI for Reduced and Permethylated Glycans on a PGC Column
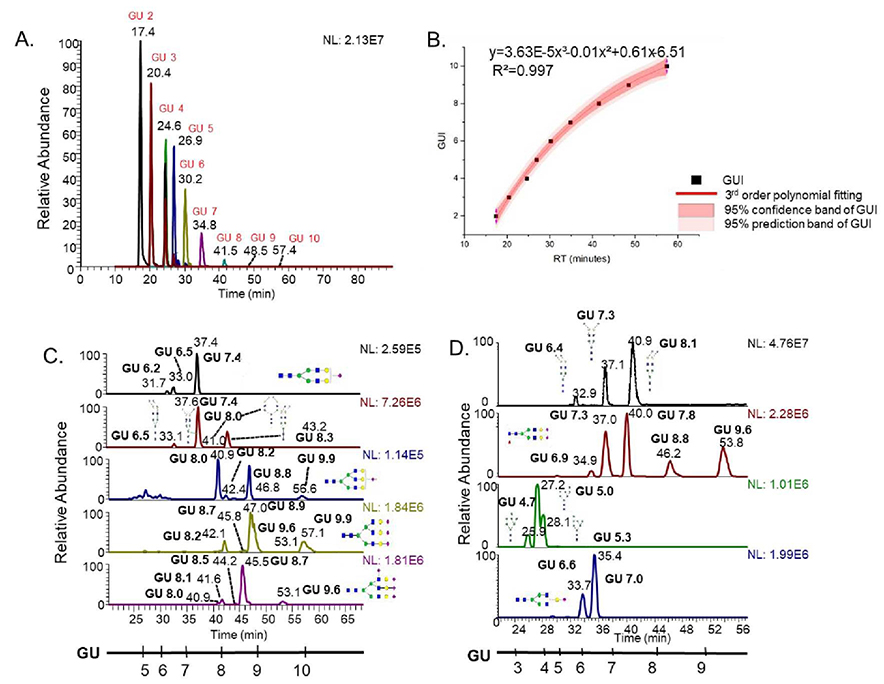
A GUI calibration curve for PGC-LC-MS was established using permethylated dextrin in the same manner as for a C18 column. Figure 2A depicts the extracted chromatogram of different glucose unit peaks detected on a PGC column. Glucose units 2 to 10 were observed, which covered most of the permethylated isomeric glycan peaks in PGC-LC-MS analysis. Again, third order polynomial fitting was found to be the best fit for dextrin retention times versus the GUI standard curve. Figure 2B represents the fitting curve of the PGC glucose unit plots. The R2 was greater than 0.999 using a third order polynomial function, signifying a high degree of correlation between GUI and retention time on a PGC column. Similar to the C18 GUI curve, the PGC GUI curve generated a high fitting accuracy and was applied to normalize the retention times of permethylated isomeric glycans using PGC-LC-MS. Figure 2C shows the application of GUI on fetuin derived permethylated glycans separated on a PGC column. Multiple peaks denoting different isomers for individual glycan compositions are visible on the chromatogram. The GUI scale, shown parallel to the retention time axis, denotes the normalized retention times for reduced and permethylated oligosaccharide units derived from dextrin. Here, GUI values for reduced and permethylated glycan isomers were calculated the same way as that of glycans separated on C18 column. The analysis was repeated with pooled HBS. Figure 2D illustrates the application of GUI on permethylated glycan isomers derived from HBS. The figure shows different permethylated glycan isomers derived from pooled HBS with corresponding structures and GUI values. The GUI scale is again shown parallel to the retention time, describing the elution pattern of the standard dextrin on a PGC column. As before, the reproducible GUI values for individual reduced and permethylated glycan isomers were obtained from the model glycoproteins and pooled HBS.
Figure 2.
A. Chromatogram showing dextrin separated on PGC column B. Standard dextrin plot showing third order polynomial fitting C. Permethylated glycans derived from fetuin, separated on PGC column D. Permethylated glycans derived from human blood serum, separated on PGC column
It must be emphasized again that GUI was able to attain system independent, reproducible elution values with an average standard deviation of 0.2, despite retention time shifts ranging between 0.5 to 3.0 minutes for reduced and permethylated glycans derived from different samples. We were able to attain normalized GUI values on a PGC column, regardless of the sample investigated, thus demonstrating the robustness of this method on a PGC column. Therefore, GUI can be successfully employed for retention time normalization of reduced and permethylated glycans on a PGC column.
Interestingly, the retention time gaps between glucose units were not uniform across the C18 and PGC GUI curves. For the C18 column, it could be because the separation was performed using a multistep gradient that consists of multiple linear gradients (as presented in the methods section). The non-linear gradient caused the different elution powers for various glucose units, resulting in inconsistent retention time changes among glucose units. Nevertheless, PGC separation, which follows a linear elution gradient, also shows a non-linear elution pattern for permethylated glucose units. However, since we used the same gradient for both the standard curve and subsequent biological samples and found a high accuracy of fitting, this phenomenon did not affect the use of GUI on different glycomic samples. This can also be corroborated with the C18 and PGC based separations discussed above, where successful normalization of retention time in terms of GUI was attained for reduced and permethylated glycans and glycan isomers across different samples.
Dextrin Spiked in the Samples as Internal Standard for Calculation of GUI
Dextrin served as an ideal standard for the normalization of retention times of glycans on different LC-systems because of its ability to yield a wide range of glucose units which can be used for classification of glycans. Furthermore, the dextrin standard spiked into the samples would better normalize the retention time shift for each sample run, as it would be insusceptible to inter-injection variations. For this purpose, dextrin standard was spiked into the samples before they were reduced. Samples spiked with dextrin were then reduced and permethylated according to the procedure discussed in the methods section above. To confirm that spiked-in dextrin did not considerably decrease the ionization efficiency of the glycans present in a sample, a scatter plot of peak areas from spiked and unspiked dextrin was prepared (Figure S2). The scatter plot shows a high correlation between the peak areas from two different samples, with R2 more than 0.996 and a straight-line slope. The error bars on the data points indicate the standard deviations for triplicate runs. After comparing the peak areas, it was found that there is no considerable inhibition of ionization in dextrin-spiked samples. Further, to verify that the spiking did not introduce any shifts in GUI, a scatter plot of the GUI values from both dextrin-spiked and unspiked samples was prepared. Figure S3 depicts the reproducibility of GUI values in both dextrin-spiked and unspiked samples. Again, a high R2 value of more than 0.99 and a straight-line slope shows the reproducibility of GUI values between spiked and unspiked samples. Therefore, this method can be considered a better alternative to using dextrin as an external standard, as it would permit the calculation of GUI values more precisely with respect to the standard.
GUI Reproducibility
Reproducibility of this classification method was tested across different days, laboratories, instruments as well as gradients. The method was proved to be independent of day to day, intra-lab as well as inter-lab variations but it does depend on the type of gradient being used. Two different gradients, in addition to the regular gradients used for the study were tested for separation of human blood serum. After calculation of the GUI values, it was found that the values are unique to each gradient. Therefore, it is advised that calibration curve for GUI should be constructed when a new gradient is used.
Day-to-Day Reproducibility of GUI
The foremost criterion for a method to qualify is its reproducibility. Since we wanted to inspect the reproducibility of elution positions of individual permethylated glycans with respect to the standard, samples were analyzed on different days using this method. To determine the reproducibility of GUI values on different days, the samples were subjected to C18-LC-MS as well as PGC-LC-MS on different days on the same LC system (LC-1), with a minimum interval of one week between two consecutive analyses. The GUI values of the same sample acquired from analyses on different days were compared to investigate the variations of this method. Figure S4A shows a scatter plot depicting the reproducibility of GUI values across different days on a C18 column. With R2 of 0.998 and a straight-line slope, the reproducibility of GUI from the same sample between two days is evident. The samples were run in triplicates and the standard deviations of data points are shown as errors bars. Similarly, Figure S4B depicts the reproducibility of the same sample on a PGC column. Here, R2 of 0.973 and a straight-line slope illustrates the reproducibility of GUI between two different days on a PGC column. Again, the error bars represent the standard deviation.
Another noteworthy finding during this investigation was although retention times for individual reduced permethylated glycans fluctuated across days, there were low or no corresponding shifts in GUI values. Most of the glycans showed a retention time shift of 0.5 minutes to 1 minute on the same system on two different days, while the shift in GUI was between 0 to 0.1. The data discussed hitherto and the reproducibility of GUI values between two days depict the robustness of this method. Our results indicate that the method can be adopted for retention time normalization of reduced and permethylated glycans, irrespective of the kind of sample being investigated and the time of investigation.
Intra-Lab Reproducibility of GUI
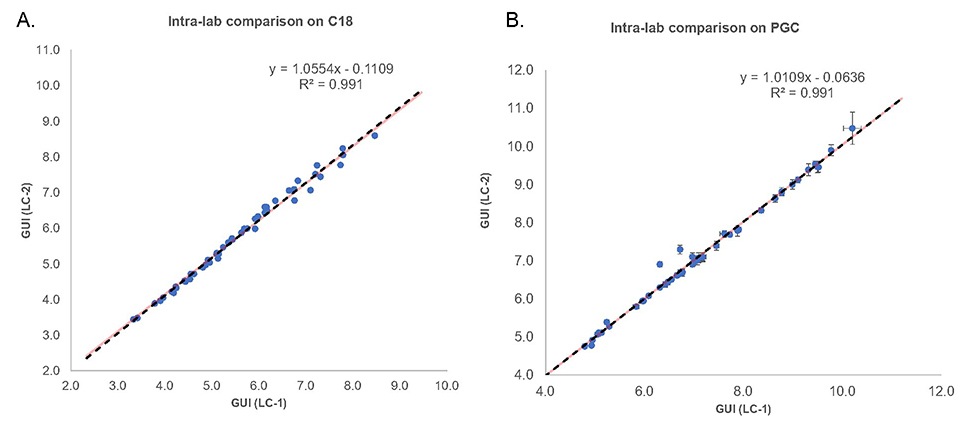
After validating the reproducibility of the method on different days, the samples were applied to a different LC system (LC-2) within the same lab. This experiment served to investigate any bias associated with the GUI values due to the LC system employed for initial study. The glycans derived from HBS samples were again spiked with dextrin, followed by reduction and permethylation. GUI values with were and were compared to the elution positions determined on LC system 1. Again, to investigate the reproducibility of these values, a scatter plot for GUI values obtained on two different systems was generated. Figure 3A shows the scatter plot that compares HBS-derived reduced and permethylated glycan GUI values from LC-1 and LC-2 on C18 column, compared against each other. The slope of the scatter plot is a straight line, as depicted by the equation, and shows a high degree of correlation between the GUI values from different instruments. The R2 value of 0.991 shows the high reproducibility of the values across the two instruments on a C18 column. Figure 3B shows the intra-laboratory reproducibility of reduced and permethylated glycans on PGC columns. Again, the R2 value of 0.991 and the straight-line slope of the scatter plot shows good reproducibility of GUI values using PGC columns on two different LC-systems. The error bars on both the plots represent standard deviations for triplicate runs. This experiment demonstrated that the data was unbiased and independent of the LC system employed for the initial study.
Figure 3.
Scatter plot showing intra-lab comparison of GUI values. Red and black dashed lines show the theoretical lines and trendlines, respectively.
Inter-Lab Reproducibility
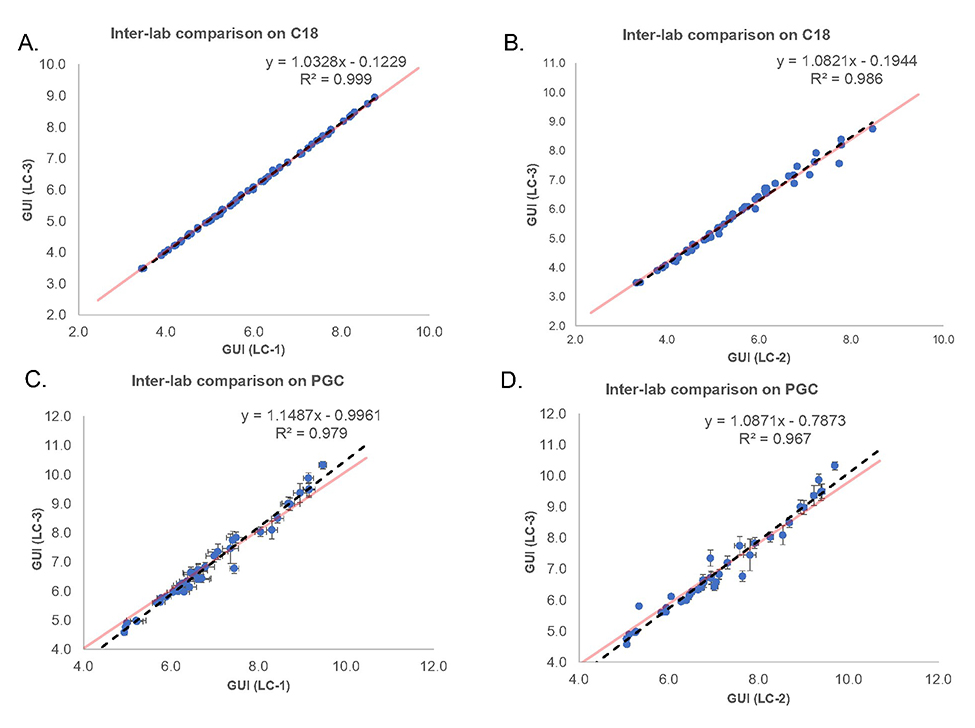
For method transfer validation, the GUI values for permethylated glycans were also investigated in a different laboratory. This was a critical part of the study, as the normalized data is expected to aid automated permethylated glycan characterization in glycomic samples in future. This is possible only if the elution positions of permethylated glycans with respect to the standard do not vary by laboratory. To confirm the reproducibility of the method in a different laboratory setting, the samples were subjected to separation on a third LC system (LC-3) in a different lab. Again, the GUI values for the glycans were calculated as described earlier. The values obtained were compared with the values from LC-1 and LC-2 using scatter plots (Figure 4). In Figure 4A, the GUI values for reduced and permethylated glycans obtained from LC-1 in laboratory-1 are compared against GUI values obtained from LC-3 in laboratory-2 on C18 columns. The R2 of nearly 1 and the straight-line slope, as evident from the equation, depict the reproducibility of the GUI values. Also, this reproducibility of GUI was attained despite the retention time shifts of 0.3 to 1.3 minutes, while the GUI shifted in the range of 0.0 to 0.3. Figure 4B depicts the inter-laboratory reproducibility of GUI values on C-18 columns between LC-2 and LC-3. Here, the GUI values for reduced and permethylated glycans obtained from LC-2 in laboratory-1 were compared to the GUI values acquired from LC-3. The retention time values vary in the range of 0.2 to 1 minute, whereas the GUI varies from 0.0 to 0.2. This reproducibility is also evident from the high degree of correlation visible in the scatter plot. Figure 4C and Figure 4D show the inter-laboratory reproducibility of GUI on PGC columns. In Figure 4C, the GUI values from LC-1 are compared to the GUI values from LC-3, and in Figure 4D the comparison is made between the GUI values obtained from LC-2 and LC-3. The standard deviation of GUI for PGC ranges between 0.2 to 0.5, higher as compared to C18. The results indicate more stable separation using C18 column compared to PGC, as demonstrated by larger standard deviation of GUI from PGC. Regardless of this, it must be noted that the GUI reproducibility is not affected by this behavior of PGC column. These results, showing strong reproducibility of GUI values across different LC-systems, validate that this method can be successfully adapted across different laboratories and instruments for the identification of permethylated glycans and their isomers on PGC columns.
Figure 4.
Scatter plot showing inter-lab comparison of GUI values. Red and black dashed lines show the theoretical lines and trendlines, respectively.
Libraries of GUI values for permethylated N-glycan compositionswere compiled for both C18 and PGC columns. We believe that these libraries will aid in the identification of permethylated glycans and glycan isomers from original samples. Table S1 shows the compilation of HBS-derived permethylated N-glycans separated on C18 columns. A total of 116 glycans were included in the C18 GUI library. The standard deviation across all the instruments and laboratories was calculated and reported for each structure. The GUI standard deviations for all the reduced and permethylated glycans present in the library range from 0.1 to 0.4; this is better than the retention time fluctuations, which vary between 0.2 to 3 minutes across different instruments. Table S2 shows the PGC GUI library, with 134 glycans and their respective standard deviation values. Again, the high retention time fluctuations that are present across different instruments and laboratories can be normalized using GUI, which varies from 0.1 to 0.5.
It should be mentioned that even though the C18 and PGC libraries compiled by us do not include all the possible structures, still they are a significant step in the direction of improved inter-laboratory data analysis. With a few exceptions, most of the isomeric structures present in the libraries were not identified, but we were successful in obtaining reproducible GUI values and demonstrating the reproducibility and robustness of the method. GUI for the assigned isomeric structures were proposed on the basis of our previous work.25 It is worth mentioning that the libraries do show GUI overlaps due to overlapping standard deviations. Thus, we propose the utilization of GUI in conjunction with the m/z values provided in the tables for the identification of permethylated glycans and glycan isomers. Also, due to the dependence of the GUI values on the gradient used, it is proposed that the values included in the library are only applicable to the glycans separated using the same gradients as mentioned in the methods section in this study. However, establishing GUI for other column dimensions could be easily achieved by using dextrin ladder
Application of GUI to Cell Line Samples
To further demonstrate the application of GUI on real samples, three different breast cancer cell lines, 231 (metastatic breast cancer cells), HTB22 (metastatic breast cancer cells) and 231BR (brain seeking metastatic breast cancer cells) were employed. Reduced and permethylated glycans derived from cell line samples were spiked with dextrin as an internal standard, following the same procedure described earlier for pooled HBS samples. The samples were separated on both C-18 and PGC columns using LC-1 and analyzed using LTQ Orbitrap Velos from Lab-1, under the same conditions utilized for standard protein and serum samples. The GUI values for glycans released from these samples were then calculated with the help of the standard dextrin plot obtained from the dextrin-spiked samples. A total of 61 permethylated N-glycan compositions were derived from the cell lines on a C18 column, of which 60 compositions were confirmed to yield reproducible GUI values according to the library. A single structure HexNAc3Hex6 showed a GUI shift of only 0.1 GU. Meanwhile, a total of 70 structures were confirmed from cell lines on a PGC column.
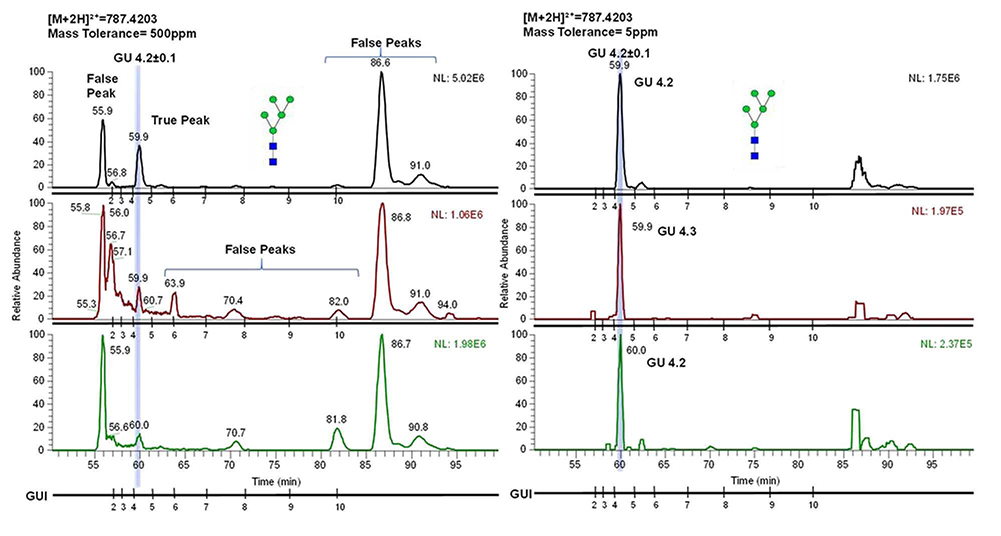
The GUI library would be further valuable by identifying glycans based on their elution positions on low mass accuracy instruments. Figure 5 shows the utilization of GUI for identification of reduced and permethylated glycans derived from breast cancer cell line 231 on a low mass accuracy instrument using C18-LC-MS. The chromatogram shown in the figure is a simulation where the mass tolerance of the chromatogram was increased to 500ppm, to show the complexity of the chromatogram as well as how it could be difficult to identify the glycan at such high mass tolerance in the absence of MS/MS data. However, the application of GUI would ease the process of assigning the true peaks in a chromatogram, because the GUI of a permethylated glycan would not fluctuate with respect to the dextrin for a given gradient.
Figure 5.
Chromatograms simultaneously showing separation of permethylated glycan HexNAC2Hex5 on instrument with low mass accuracy (mass tolerance of 500ppm) and high mass accuracy (mass tolerance of 5ppm). The figure depicts the ability to identify the glycan by determining its elution position based on GUI.
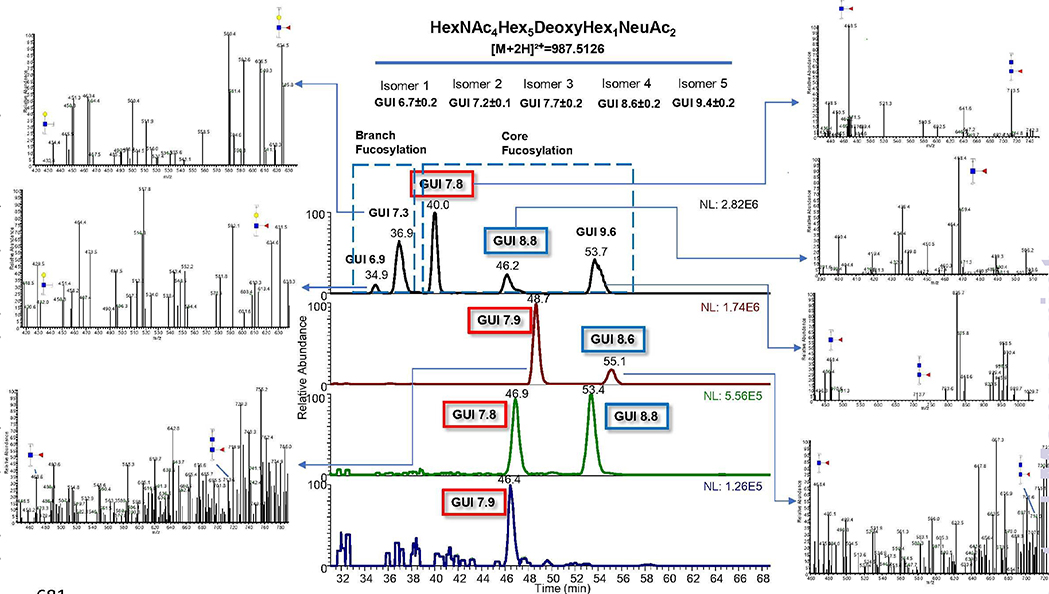
This method would also aid the identification of permethylated glycan isomers in complex samples. Figure 6 demonstrates an example where the EIC of HexNAc4Hex5DeoxyHex1NeuAc2 from HBS is compared to the three breast cancer cell lines. Five possible isomers are shown in the HBS sample, along with their GUI values and standard deviations. First two peaks represent branch fucosylation while the last three peaks represent core fucosylation. MS2 spectra showing the diagnostic ions for branch and core fucosylation are also depicted. This figure nicely illustrates how GUI could help to identify different premethylated glycan isomer expressions in various biological samples. In this case, 2 isomers each from 231 and 231BR and 1 isomer from HTB22 were identified using GUI and their values indicate that they have core fucosylation. Another noteworthy phenomenon in this example was the reproducibility of GUI regardless of the retention time shifts between HBS and the cell lines. The use of two different PGC columns for the separation of permethylated glycans derived from HBS and the cell lines led to a shift in retention times without any fluctuation of the GUI values. This also demonstrated the inter-column reproducibility of the GUI values.
Figure 6.
Chromatograms showing separation of HexNAc4Hex5DeoxyHex1NeuAc2 on PGC column. First panel depicts the separation of HBS derived structure while second, third and fourth panels depict the separation of the same structure derived from 231, 231BR and HTB22 cell lines, respectively.
Conclusion
In this study, we examined the use of dextrin as a GUI standard to normalize the retention time of permethylated N-glycans derived from standard glycoproteins and human blood serum. The obtained GUI values proved to be reproducible between inter- as well as intra-laboratory analysis. The reproducibility of GUI, regardless of fluctuations in retention time, verified the robustness of this method. Two different columns, C-18 and PGC, were employed to depict the efficacy of this method for permethylated glycan profiling as well as for permethylated glycan isomeric separation. Moreover, samples spiked with dextrin acting as internal standard were used for better retention time shift normalization. The method was shown to be valid for complex real samples by depicting the separation of reduced and permethylated glycans and glycan isomers from breast cancer cell lines. These results demonstrate a foundational example of the retention time normalization of permethylated glycans based only on GUI and m/z, independent of the LC system or even the laboratory used. Due to the low variations in GUI values as compared to retention times observed across different LC systems and across laboratories, we believe that this method of characterization, coupled to enzymatic digestion experiments and sensitive structural identification techniques, will further aid in the development of automated tools for the identification of permethylated glycans based on their GUI normalized retention times.
Supplementary Material
Acknowledgement
This work was supported by the grant from National Institutes of Health, NIH (1R01GM112490–04, 1R01GM130091–01, and 1U01CA225753–01).
Footnotes
Data availability
The raw data is available on GlycoPost under announced ID GPST000071.1 (Since the data is not public yet, we can access the uploaded data by requesting a temporary URL and Pin Code. We would be glad to provide the preview information to the reviewers anytime during the review process).
Conflict of Interest
The authors declare no conflict of interest.
This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication.
References
- 1.Dong X, Huang Y, Cho BG, Zhong J, Gautam S, Peng W, Williamson SD, Banazadeh A, Torres-Ulloa KY and Mechref Y, Electrophoresis, 2018, 39, 3063–3081. [DOI] [PubMed] [Google Scholar]
- 2.Zhu R, Zacharias L, Wooding KM, Peng W and Mechref Y, Methods Enzymol, 2017, 585, 397–429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solá RJ and Griebenow K, J. Pharm. Sci, 2009, 98, 1223–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtsubo K and Marth JD, Cell, 2006, 126, 855–867. [DOI] [PubMed] [Google Scholar]
- 5.de Vreede G, Morrison HA, Houser AM, Boileau RM, Andersen D, Colombani J and Bilder D, Dev. Cell, 2018, 45, 595–605. e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzeng S-F, Tsai C-H, Chao T-K, Chou Y-C, Yang Y-C, Tsai M-H, Cha T-L and Hsiao P-W, FASEBJ, 2018, 32, 6869–6882. [Google Scholar]
- 7.Rudd PM, Wormald MR, Stanfield RL, Huang M, Mattsson N, Speir JA, DiGennaro JA, Fetrow JS, Dwek RA and Wilson IA, J Mol Biol, 1999, 293, 351–366. [DOI] [PubMed] [Google Scholar]
- 8.Tan FY, Tang CM and Exley RM, Trends Biochem Sci, 2015, 40, 342–350. [DOI] [PubMed] [Google Scholar]
- 9.Cho BG, Veillon L and Mechref Y, J Proteome Res, 2019,18, 3770–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Yu X, Ichikawa M, Lyons JJ, Datta S, Lamborn IT, Jing H, Kim ES, Biancalana M and Wolfe LA, J Allergy Clin Immunol, 2014,133, 1400–1409. el405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudelj I and Lauc G, Curr. Cardiovasc. Risk Rep, 2018,12, 16. [Google Scholar]
- 12.Adamczyk B, Tharmalingam T and Rudd PM, Biochim Biophys Acta, 2012,1820, 1347–1353. [DOI] [PubMed] [Google Scholar]
- 13.Alley WR and Novotny MV, J Proteome Res, 2010, 9, 3062–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X, Peng W, Yu C-Y, Zhou S, Donohoo KB, Tang H and Mechref Y, Anal Chem, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X, Zhou S and Mechref Y, Electrophoresis, 2016, 37, 1532–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Zhou S, Zhu J, Lubman DM and Mechref Y, Electrophoresis, 2017, 38, 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veillon L, Huang Y, Peng W, Dong X, Cho BG and Mechref Y, Electrophoresis, 2017, 38, 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Hoi KM, Stockmann H, Wan C, Sim LC, Shi Jie Tay NHBK, Poo CH, Woen S, Yang Y and Zhang P, Biotechnol J, 2018, 13, 1700185. [DOI] [PubMed] [Google Scholar]
- 19.Zhou S, Dong X, Veillon L, Huang Y and Mechref Y, Anal Bioanal Chem, 2017, 409, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alagesan K, Everest-Dass A and Kolarich D, in Glycobiophysics, Springer, 2018, pp. 77–99. [DOI] [PubMed] [Google Scholar]
- 21.Desantos-Garcia JL, Khalil SI, Hussein A, Hu Y and Mechref Y, Electrophoresis, 2011, 32, 3516–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Nie Y, Boyes B and Orlando R, J Biomol Tech, 2016, 27, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qing G, Yan J, He X, Li X and Liang X, TrAC, 2019. [Google Scholar]
- 24.Yu A, Zhao J, Peng W, Banazadeh A, Williamson SD, Goli M, Huang Y and Mechref Y, Electrophoresis, 2018, 39, 3104–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou S, Huang Y, Dong X, Peng W, Veillon L, Kitagawa DAS, Aquino AJA and Mechref Y, Anal Chem, 2017, 89, 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou S, Wooding KM and Mechref Y, Methods Mol Biol, 2017, 1503, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S, Veillon L, Dong X, Huang Y and Mechref Y, Analyst, 2017, 142, 4446–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM and Wuhrer M, Anal Bioanal Chem, 2010, 397, 3457–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vreeker GC and Wuhrer M, Anal Bioanal Chem, 2017, 409, 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang P, Mechref Y and Novotny MV, Rapid Commun Mass Spectrom, 2008, 22, 721–734. [DOI] [PubMed] [Google Scholar]
- 31.Harvey DJ, Mattu TS, Wormald MR, Royle L, Dwek RA and Rudd PM, Anal Chem, 2002, 74, 734–740. [DOI] [PubMed] [Google Scholar]
- 32.Wuhrer M, Koeleman CA, Hokke CH and Deelder AM, Rapid Commun Mass Spectrom, 2006, 20, 1747–1754. [DOI] [PubMed] [Google Scholar]
- 33.Varki A, Trends Mol Med, 2008, 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costello CE, Contado-Miller JM and Cipollo JF, J Am Soc Mass Spectrom, 2007, 18, 1799–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guile GR, Rudd PM, Wing DR, Prime SB and Dwek RA, Anal Biochem, 1996, 240, 210–226. [DOI] [PubMed] [Google Scholar]
- 36.Guttman A, Electrophoresis, 1997, 18, 1136–1141. [DOI] [PubMed] [Google Scholar]
- 37.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim Y-G, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM and Dwek RA, Anal Biochem, 2008, 376, 1–12. [DOI] [PubMed] [Google Scholar]
- 38.Rudd PM, Colominas C, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF and Dwek RA, Proteomics, 2001, 1, 285–294. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd KO, Burchell J, Kudryashov V, Yin BW and Taylor-Papadimitriou J, J Biol Chem, 1996, 271, 33325–33334. [DOI] [PubMed] [Google Scholar]
- 40.Callewaert N, Schollen E, Vanhecke A, Jaeken J, Matthijs G and Contreras R, Glycobiology, 2003, 13, 367–375. [DOI] [PubMed] [Google Scholar]
- 41.Van den Steen PE, Van Aelst I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, Moestrup SK, Fry S, Royle L, Wormald MR, Wallis R, Rudd PM, Dwek RA and Opdenakker G, J Biol Chem, 2006, 281, 18626–18637. [DOI] [PubMed] [Google Scholar]
- 42.Fabini G, Freilinger A, Altmann F and Wilson IB, J Biol Chem, 2001, 276, 28058–28067. [DOI] [PubMed] [Google Scholar]
- 43.Natsuka S and Hase S, in Glycoanalysis Protocols, ed. Hounsell EF, Humana Press, Totowa, NJ, 1998, vol. 76, pp. 101–113. [Google Scholar]
- 44.Campbell MP, Royle L, Radcliffe CM, Dwek RA and Rudd PM, Bioinformatics, 2008, 24, 1214–1216. [DOI] [PubMed] [Google Scholar]
- 45.Ashwood C, Pratt B, MacLean BX, Gundry RL and Packer NH, Analyst, 2019, 144, 3601–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang P, Mechref Y, Klouckova I and Novotny MV, Rapid Commun Mass Spectrom, 2005, 19, 3421–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu R, Zhou S, Peng W, Huang Y, Mirzaei P, Donohoo K and Mechref Y, J Proteome Res, 2018, 17, 2668–2678. [DOI] [PubMed] [Google Scholar]
- 48.Banazadeh A, Peng W, Veillon L and Mechref Y, J Am Soc Mass Spectrom, 2018, 29, 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong J, Banazadeh A, Peng W and Mechref Y, Electrophoresis, 2018, 39, 3087–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benktander JD, Gizaw ST, Gaunitz S and Novotny MV, J Am Soc Mass Spectrom, 2018, 29, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou S, Hu Y and Mechref Y, Electrophoresis, 2016, 37, 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.