Abstract
In cancer cells, a gain of stemness may have profound implications for tumor initiation, aggressiveness, and clinical outcome. However, the molecular mechanisms underlying the self-renewal maintenance of cancer stem-like cells (CSCs) remain elusive. Here, based on analysis of transcriptome sequencing, we identified a long noncoding RNA (lncRNA) named HotairM1, which is weakly expressed in human colorectal carcinoma and uveal melanoma, and a much lower expression in corresponding CSCs. Our results showed that HotairM1 depletion could promote CSC self-renewal and tumor propagation. Mechanistically, HotairM1 recruit EZH2 and SUZ12 to the promoter of its target gene HOXA1, leading to histone H3K27 trimethylation and epigenetic silencing of HOXA1. The silence of HOXA1 subsequently induces the H3K27 acetylation at the enhancer site of Nanog gene to upregulate its expression. The enrichment of Nanog could further inhibit HOXA1 expression, forming a reciprocal regulation loop augmenting the stemness maintaining effect. In summary, our results revealed a lncRNA-based regulatory loop that sustains self-renewal of CSCs, which highlights the critical role of HotairM1 in CSC development through the HOXA1-Nanog signaling loop.
Keywords: HotairM1, HOXA1-Nanog regulation loop, self-renewal, cancer stem cell
Graphical Abstract
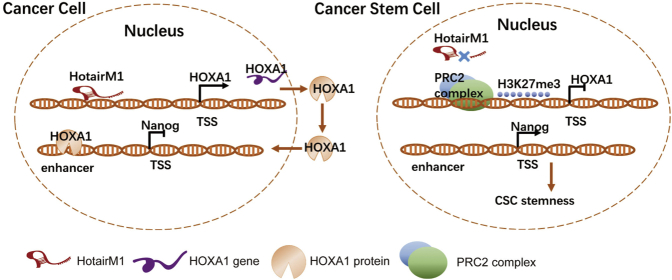
Knockdown of lncRNA HotairM1 could promote self-renewal of CSCs and subsequent tumor proliferation and propagation through inhibiting neighbor gene HOXA1 expression. HOXA1 would repress stemness maintenance of CSC through inhibiting Nanog expression.
Introduction
Cancer stem cells (CSCs), which represent a small and distinct population within the tumor, are characterized by bearing stemness markers1 and are believed to be responsible for tumor initiation, propagation, metastasis, and recurrence.2 Previous studies have confirmed the existence of CSCs in both hematologic tumors and solid tumors, as well as the important prognostic role of CSCs in patients with cancers such as breast cancer, colon cancer, melanoma, lung cancer, liver cancer, pancreatic cancer, and ovarian cancer.3, 4, 5, 6 CSCs have several properties, such as self-renewal, asymmetric division, and resistance to traditional radiation or chemotherapy,7 which are the prime sources of tumor recurrence and metastasis, and it is the reason why therapies targeting bulk cancer cells alone may not be sufficient to improve patient outcome. Thus, CSC-targeted therapy, which nowadays usually focuses on promoting differentiation or inhibiting self-renewal,8,9 does provide a promising opportunity to cure tumor cells, especially those that are responsible for recurrence and treatment failure.10,11 It is imperative to improve understanding of fundamental biology of CSCs and vital underlying mechanisms in maintaining stemness.
Long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nt with limited or no coding potential. lncRNAs play important roles in many biological processes and regulate gene expression through acing as cis-regulatory or trans-regulatory elements via diverse mechanisms.12, 13, 14, 15 Recent studies demonstrated that lncRNAs also take part in the self-renewal regulation of CSCs. Wang et al.16 found that lncTCF7 was required for liver CSC self-renewal and tumor propagation by recruiting the SWI/SNF complex to the promoter of TCF7 to regulate its expression, leading to Wnt signaling activation. Zhu et al.17 found that lnc-β-Catm could sustain the stability of β-catenin, leading to the activation of the Wnt signaling pathway, which was positively correlated with liver cancer severity and prognosis. Besides liver CSCs, lncRNAs also play roles in other kinds of tumors. Hu et al.18 found that the oncogenic lncRNA FAL1 interacts with the epigenetic repressor BMI1, modulating the transcription of a number of genes associated with the prognosis of ovarian cancer. Tan et al.19 demonstrated that lncRNA EGFR-AS1 mediated epidermal growth factor receptor (EGFR) addiction and modulated treatment response in squamous cell carcinoma. Emerging lines of evidence have shown that noncoding RNAs are closely associated with the development and progression of various CSCs. However, whether there was a relatively common lncRNA involved in the stemness maintenance of different kinds of CSCs still remains to be detected.
In our present study, through transcriptome sequencing analysis of colorectal carcinoma stem cells (CCSCs) and non-CCSCs, a lncRNA called HOX antisense intergenic RNA myeloid 1 (HotairM1) was confirmed. HotairM1 was reported to be involved in neuronal differentiation and impacts the prognosis of acute myeloid leukemia patients.20 In solid tumors, HotairM1 has been shown to function as both an oncogene suppressor and a tumor suppressor. For instance, HotairM1 promotes malignance in glioblastoma, lung cancer, and pancreatic ductal adenocarcinoma,21,22 while it inhibits tumorigenesis in colorectal cancer, head and neck tumors, gastric cancer, and hepatocellular carcinoma.23, 24, 25 However, the prognostic value of HotairM1 in uveal melanoma remains unknown. In our study, we found that HotairM1 depletion promoted stemness maintenance of CSCs and tumor propagation in uveal melanoma cells and colorectal carcinoma cells through the HOXA-Nanog regulation loop. HOXA1 is a member of the family of homeobox (HOX) genes, and it is a gene positively regulated by HotairM1.26 Our findings elucidated the tumor-suppressive role of HotairM1 in colorectal carcinoma and uveal melanoma and unveiled its molecular mechanism underlying tumor stemness maintenance, which might suggest a biomarker in predicting tumor prognosis and a novel therapeutic strategy for melanoma.
Results
HotairM1 Is Expressed at Low Levels in CSCs and Is a Positive Prognostic Factor for Tumor Patients
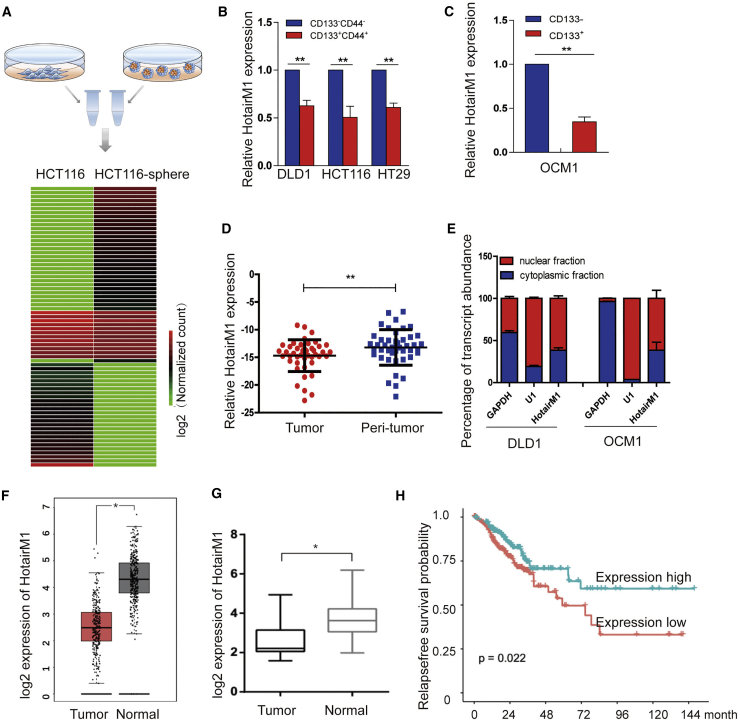
To study the potential lncRNAs that play roles in stemness maintenance in tumors, we first enriched cancer stem-like cells in tumors using an oncosphere formation assay. Cells enriched in serum-free medium (called CSCs herein) showed significantly enhanced oncosphere formation ability compared with attached cells (called non-CSCs herein), and stem-related gene expressions were also confirmed changed, as shown in Figure S1. We conducted RNA sequencing analysis using CSCs and non-CSCs to identify lncRNAs involved in CSCs (Figure 1A). According to the analysis, several significantly changed lncRNAs were detected (Figure S2), and one of the most downregulated lncRNAs called HotairM1 was identified.
Figure 1.
HotairM1 Is Expressed at Low Levels in CSCs
(A) Workflow depicting the isolation of CSCs from the sphere formation assay using serum-free medium and differentially expressed lncRNAs from transcriptome analysis. (B) HotairM1 was detected in CD44+CD133+ colorectal cancer cells by quantitative real-time PCR analysis. Data are indicated as the mean ± SD. Data are representative of at least three independent experiments. (C) HotairM1 was detected in CD133+ OCM1 cells by quantitative real-time PCR analysis. Data are indicated as the mean ± SD. Data are representative of at least three independent experiments. (D) HotairM1 was detected in tumor tissues and paired adjacent tumor tissues (n = 37). (E) Fractionation of tumor cells followed by quantitative real-time PCR. U1 RNA served as a positive control for nuclear gene expression. GAPDH RNA served as a positive control for cytoplasmic gene expression. N, nuclear fraction; C, cytoplasmic fraction. Data are indicated as the mean ± SD. Data are representative of at least three independent experiments. (F) HotairM1 expression in colon adenocarcinoma (COAD) was analyzed using the GEPIA database (number of tumor cells, 275; number of normal cells, 349). (G) HotairM1 expression was analyzed using the GEO database (GEO: GSE104836). (H) Kaplan-Meier survival analysis of HotairM1 using GEO: GSE104836. The cutoff value was set according to the median value of HotairM1 normalized expression. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
The HotairM1 expression level was significantly lower in self-renewing spheroids compared with that in attached cells. To detect the expression of HotairM1 in different kinds of tumor CSCs, colorectal cancer cells and uveal melanoma cells were selected. Many studies have demonstrated that the small subset of CD133+CD44+ cells in colorectal cancer cells could represent the characteristics of CSCs.27 For uveal melanoma, the most commonly used stem cell marker, CD133, was selected as sorting criterion, because no universally known stem cell markers have been confirmed until now.28 Thus, a flow cytometry assay was performed to sort CD133+CD44+ cells from the HCT116 and DLD1 cell lines and CD133+ uveal melanoma cells, presumptively CSCs. Reduced HotairM1 expression was detected in CD133+CD44+ colorectal cancer cells (Figure 1B) and CD133+ uveal melanoma cells (Figure 1C). In addition, HotairM1 was also lowly expressed in some other kinds of tumors and cell lines (Figures S3 and S4), confirming the aforementioned results. Moreover, HotairM1 was significantly low in CRC tissues compared with adjacent peri-tumor tissues (Figure 1D). We further examined the localization of HotairM1 and found that HotairM1 distributed both in the nuclei and cytoplasm of cells and mainly in the nucleus, as evidenced through cellular fractionation assays (Figure 1E). To confirm HotairM1 expression in tumor tissues, we queried the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/)29 and GEO database (GEO: GSE104836) (https://www.ncbi.nlm.nih.gov/geo/). Notably, low expression of HotairM1 was confirmed in both datasets (Figures 1F and 1G). A Kaplan-Meier survival analysis was performed to investigate the effect of the HotairM1 expression level on patient relapse-free survival probability. According to the dataset for GEO: GSE104836, lower expression of HotairM1 was associated with a lower relapse-free survival probability, indicating a poorer prognosis (Figure 1H). Altogether, HotairM1 was expressed at low levels in tumors and much lower in CSCs, which was also a positive prognostic factor for CRC patients.
HotairM1 Depletion Promotes CSC Self-Renewal and Facilitates Tumor Propagation
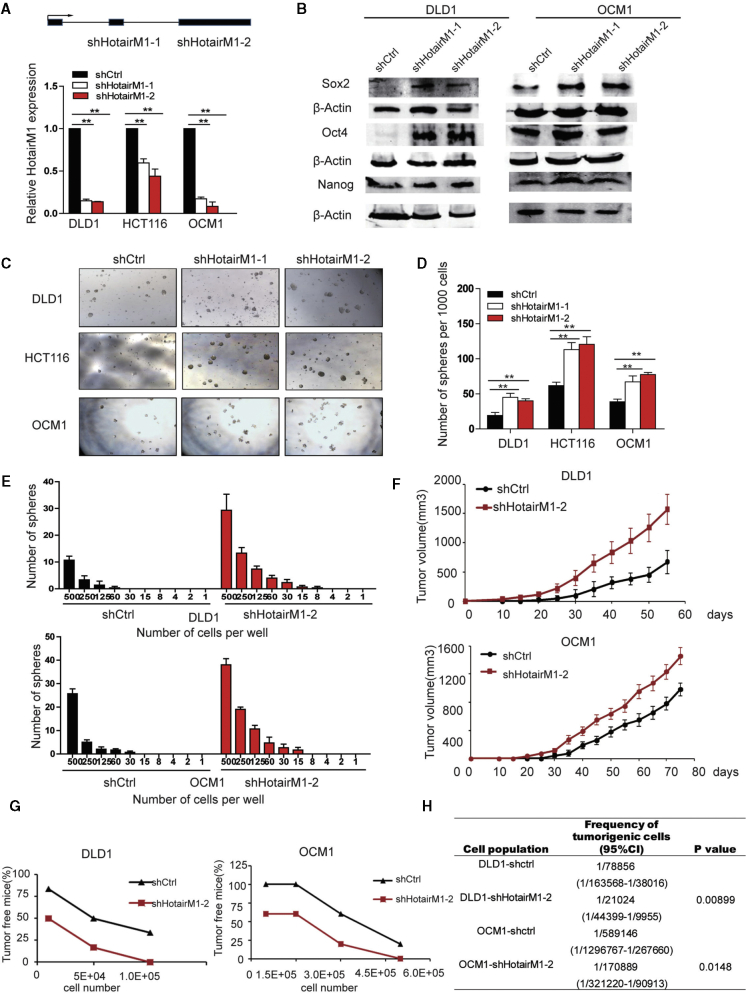
To investigate the role of HotairM1 in CSC self-renewal, HotairM1 was significantly silenced in either exon 2 or exon 3 in tumor cells using lentivirus-mediated short hairpin RNAs (shRNA1 and shRNA2, hereinafter collectively referred as shHotairM1) (Figure 2A). Notably, HotairM1 knockdown cell showed elevated expression of the pluripotent factors Oct4, Sox2, and Nanog compared with scramble cells (shCtrl) (Figure 2B). Moreover, HotairM1 depletion enhanced sphere formation ability (Figure 2C), and the quantitative analysis also confirmed that the number of spheres formed by shHotairM1 cells per site was much more than that formed by shCtrl cells (Figure 2D). In addition, HotairM1 depletion obviously increased the number of spheres formed per dish from different diluted numbers of cells through the in vitro limiting gradient dilution assay (Figure 2E).
Figure 2.
HotairM1 Is Required for the Self-Renewal Maintenance of CSCs
(A) HotairM1 was silenced in tumor cells by two independent shRNAs. HotairM1 silencing efficiency was determined using quantitative real-time PCR. Data are indicated as the mean ± SD. (B) Pluripotent transcription factors (Oct4, Sox2, and Nanog) were analyzed in HotairM1-depleted cells using western blot. (C) Onco-sphere formation assay using serum-free medium was performed to detect the sphere-forming capacity. Representative images of tumor spheres at a dose of 1,000 cells per well are shown. (D) Statistical analysis of spheres per 1,000 cells was performed. Data are indicated as the mean ± SD. (E) In vitro limiting dilution assay using different numbers of cells was performed. Spheres formed from different doses of cells were statistically analyzed. (F) HotairM1-silenced or shCtrl tumor cells were subcutaneously injected into BALB/c nude mice for observation of tumor growth. The results are shown as the mean ± SD. (G) HotairM1-silenced tumor cells and shCtrl cells were injected into mice in gradient doses of cells. The number of tumors formed in each mouse was calculated and analyzed. (H) Tumorigenic cell frequency in HotairM1-silenced and shCtrl tumor cells was analyzed with a limiting dilution assay (http://bioinf.wehi.edu.au/software/elda/). CI, confidence interval. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
Next, we explored whether the tumor behavior could be significantly altered by HotairM1 in vivo. Stably silenced HotairM1 and scramble cells were injected into nude mice. We found that HotairM1 knockdown cells showed a much more obvious tumor growth advantage over the shCtrl group (Figure 2F), including enhanced xenografted tumor growth ability, a bigger tumor size, and increased tumor weight. Representative hematoxylin and eosin (H&E) staining is shown in Figure S5. Additionally, in vivo limiting dilution assay showed a higher tumor formation rate in the HotairM1 depletion group compared with shCtrl cells (Figure 2G). Extreme limiting dilution analysis (ELDA) displayed a higher CSC frequency in comparison with shCtrl cells (Figure 2H), indicating that HotairM1 depletion noticeably promoted the expansion of CSCs.
HotairM1 Depletion Promotes Tumor Cell Proliferation and Migration
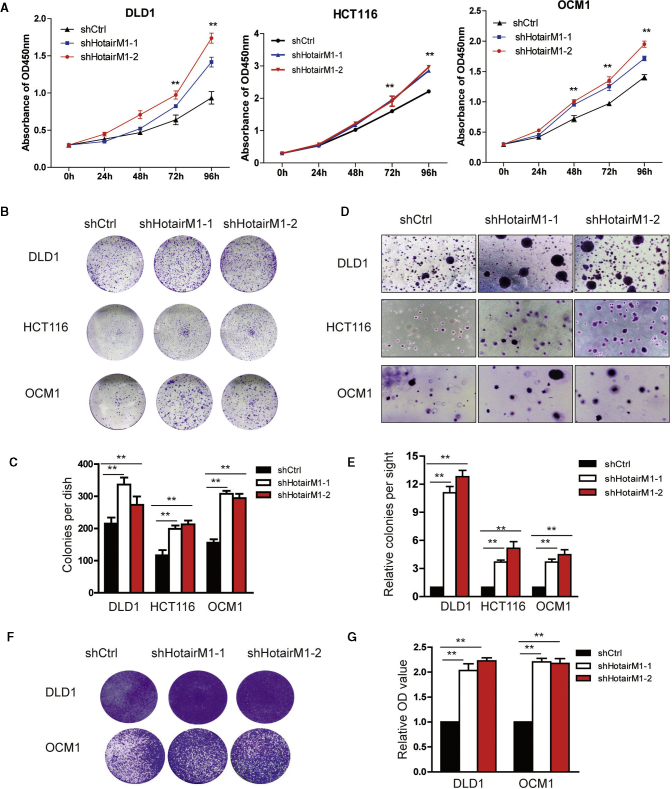
Next, we examined the effects of HotairM1 knockdown on tumor cell proliferation and migration. In DLD1, HCT116, and OCM1 cells, cell proliferation is significantly increased in HotairM1 depletion cells (Figure 3A). Besides, the reduction of HotairM1 could promote the formation of colonies (Figure 3B), and quantitative analysis also confirmed the significant change (Figure 3C). A soft agar assay showed that HotairM1 knockdown increased the number of spheres, and statistical analysis confirmed the changes (Figures 3D and 3E). Then, tumor migration ability was also detected. Transwell assay showed that shHotairM1 cells significantly increased migration ability, and quantitative analysis confirmed the changes (Figures 3F and 3G). Collectively, all these results showed that tumor cell proliferation and migration were significantly increased mainly because of the increasing ratio of CSCs in tumors.
Figure 3.
HotairM1 Depletion Facilitates Tumor Cell Proliferation and Migration
(A) The proliferation ability of HotairM1-knockdown and shCtrl tumor cells was analyzed using the CCK8 assay. The optical density 450 nm (OD450) value was recorded, and the growth index was analyzed. (B) Images of colonies formed from the plate cloning assay are shown. (C) Colonies formed in the well were calculated and analyzed. Data are indicated as the mean ± SD. (D) Tumorigenesis ability was determined using a soft agar assay. Representative images of stained colonies formed in the upper agar are shown. (E) The colony number in the upper agar was counted, and the relative colonies per site were calculated. Data are indicated as the means ± SD. (F) Migration ability of shHotairM1 and shCtrl tumor cells was analyzed through a Transwell assay. Images of cells on the outside of the Transwell are shown. (G) The OD630 absorbance values of stained migrated cells were obtained to calculate the migration rate. Data are indicated as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
HotairM1 Overexpression Impairs Self-Renewal, Proliferation, and Tumorigenesis of CSCs
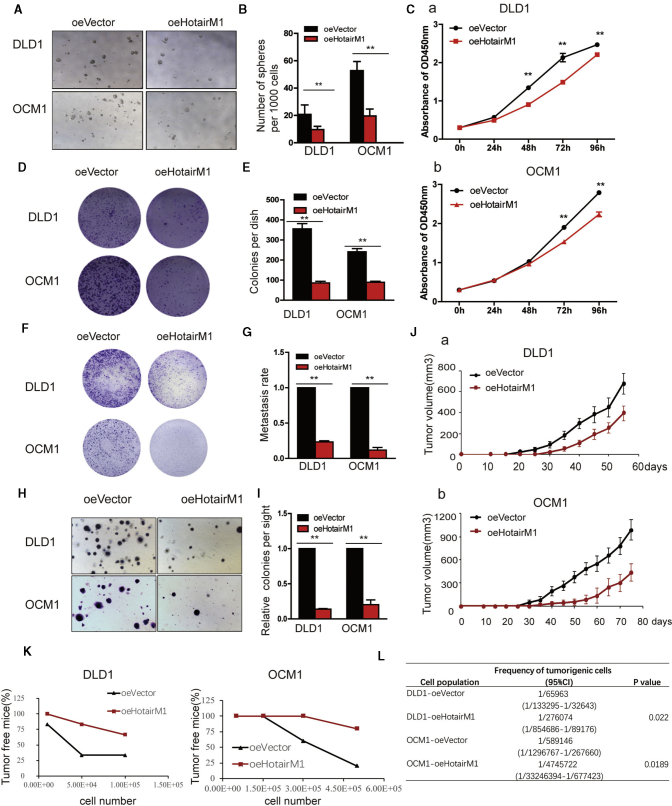
We next overexpressed HotairM1 in tumor cells and established HotairM1 stably overexpressing cell lines. HotairM1 overexpression dramatically inhibited sphere formation both in volume and number (Figures 4A and 4B). A cell counting Kit-8 (CCK8) assay showed that HotairM1 overexpression significantly inhibited tumor cell growth (Figure 4C). The colonies formed in plates were reduced in the overexpression group (Figures 4D and 4E). Similar results were also observed in a Transwell assay. HotairM1 overexpression significantly reduced the migrating number of tumor cells (Figures 4F and 4G). In addition, colonies formed in soft agar were suppressed in the overexpression group (Figures 4H and 4I). To investigate the ability of HotairM1 to suppress tumor formation in vivo, we established a xenograft model using overexpressed cells. We found that HotairM1 overexpression notably repressed tumor progression (Figure 4J), and representative H&E staining is shown in Figure S6. Consistently, HotairM1 overexpression significantly reduced tumor-initiating capacity (Figure 4K), and corresponding tumorigenic cell frequency was downregulated in the overexpression group (Figure 4L). Taken together, these data indicate that HotairM1 plays a critical role in the self-renewal maintenance of CSCs and further tumorigenesis.
Figure 4.
HotairM1 Overexpression Impairs the Tumorigenic Capacity of CSCs
(A) HotairM1-overexpressing tumor cells were established, and onco-sphere formation ability was assessed. Representative images of spheres formed per well at a dose of 1,000 cells per well are shown. oeVector, overexpression of empty vector. oeHotairM1, overexpression of HotairM1. (B) The number of spheres per well was counted and analyzed. (C) The proliferation ability of HotairM1-overexpressing and oeVector tumor cells was analyzed using the CCK8 assay, and absorbance of OD450 was analyzed. (D) Images of colonies formed from the plate cloning assay are shown. (E) Colonies formed in the dish were counted and analyzed. (F) Images of migrating oeHotairM1 and oeVector tumor cells were analyzed through Transwell assay. (G) OD630 absorbance values of stained migrated cells were calculated. (H) Tumorigenesis ability was determined using a soft agar assay. Representative images of stained colonies formed in the upper agar are shown. (I) The colony number in the upper agar was counted, and the relative colonies per site were calculated. (J) HotairM1-overexpressing and oeVector cells derived from tumor cells were diluted and subcutaneously implanted into BALB/c nude mice. Tumors were observed over 3 months. (K) HotairM1-overexpressing tumor cells and oeVector cells were injected into mice in gradient doses. The number of tumors formed in each mouse was calculated and analyzed. (L) Tumorigenic cell frequency in HotairM1-overexpressing and oeVector cells was determined. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
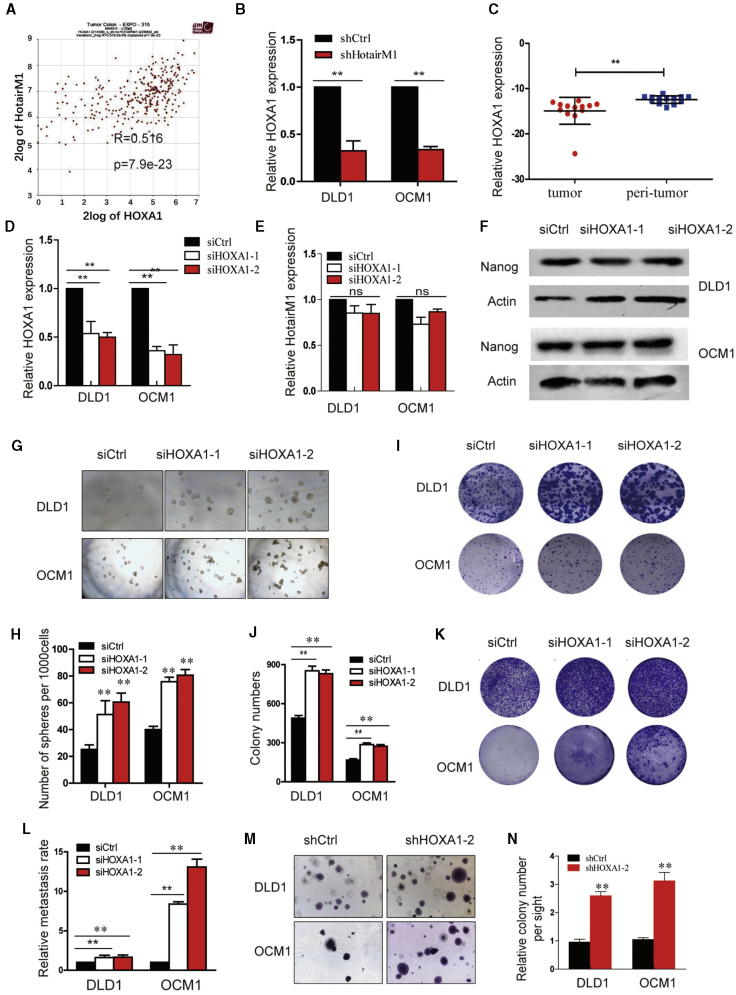
HOXA1 Is a Downstream Target of HotairM1 in CSCs
To understand the regulatory role of HotairM1 in gene expression, we first analyzed the location of HotairM1. HotairM1 is located at chromosome 7 (chr7), next to HOXA1, and HOXA1 and HotairM1 were positively correlated (Pearson correlation coefficient r = 0.516, p < 0.001) (Figure 5A). HotairM1 knockdown significantly reduced the expression of HOXA1 (Figure 5B). In addition, we also detected the expression of HOXA1 in tumor tissues, and HOXA1 was lowly expressed in tumors, which was consistent with previous results (Figure 5C). To determine whether HotairM1 functions upstream of HOXA1 in the regulation of CSC self-renewal, we silenced HOXA1 expression (Figure 5D). However, the silence of HOXA1 did not affect the expression of HotairM1, which suggested that HOXA1 may be a downstream target of HotairM1 (Figure 5E).
Figure 5.
HOXA1 Is a Downstream Target of HotairM1 in CSCs
(A) The correlation between HOXA1 and HotairM1 determined through TCGA analysis. (B) HOXA1 expression after HotairM1 was stably knocked down. (C) Expression of HOXA1 was detected in tumor tissues and paired adjacent peri-tumor tissues. (D) HOXA1 was silenced in tumor cells by two independent siRNAs, and silencing efficiency was detected using real-time PCR. (E) HotairM1 expression after HOXA1 was silenced was detected. (F) Western blot of Nanog expression in DLD1 cells and OCM1 cells transfected with HOXA1 siRNAs. (G) Sphere formation assay was performed to detect the sphere-forming capacities of HOXA1-silenced cells and control cells. Representative images of tumor spheres at a dose of 1,000 cells per well are shown. (H) Statistical analysis of spheres per 1,000 cells was performed. (I) Images of colonies formed from the plate-cloning assay are shown. (J) Colonies formed in the dish were calculated and analyzed. (K) Migration abilities of HOXA1-silenced and control tumor cells were analyzed through Transwell assay. Images of cells on the outside of the Transwell are shown. (L) The OD630 absorbance values of stained migrated cells were obtained to calculate the migrating rate. (M) Tumorigenesis ability was determined using a soft agar assay. Representative images of stained colonies formed in the upper agar are shown. (N) The colony number in the upper agar was counted, and the relative colonies per site were calculated. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
To determine the function of HOXA1 in CSC self-renewal, we silenced HOXA1, and western blot assay showed that Nanog expression was significantly increased (Figure 5F). Through the non-serum oncosphere formation ability, both the volume and the number of spheres formed by HOXA1-depleted cells were greater than the control group, as shown in Figures 5G and 5H. Regarding the influence of HOXA1 on cell proliferation and migration, CCK8 assay (Figure S7) and plate colony formation assay confirmed the increased proliferation ability (Figures 5I and 5J), and Transwell assays showed a stronger migration ability of HOXA1-depleted cells (Figures 5K and 5L). Besides, the colonies formed by HOXA1 knockdown cells showed a much larger volume and number than those formed by the scramble cells in soft agar assay (Figures 5M and 5N). These results suggested that HOXA1 functions as a downstream target of HotairM1 and plays important roles in the maintenance of CSC stemness and further tumor cell proliferation and tumorigenesis.
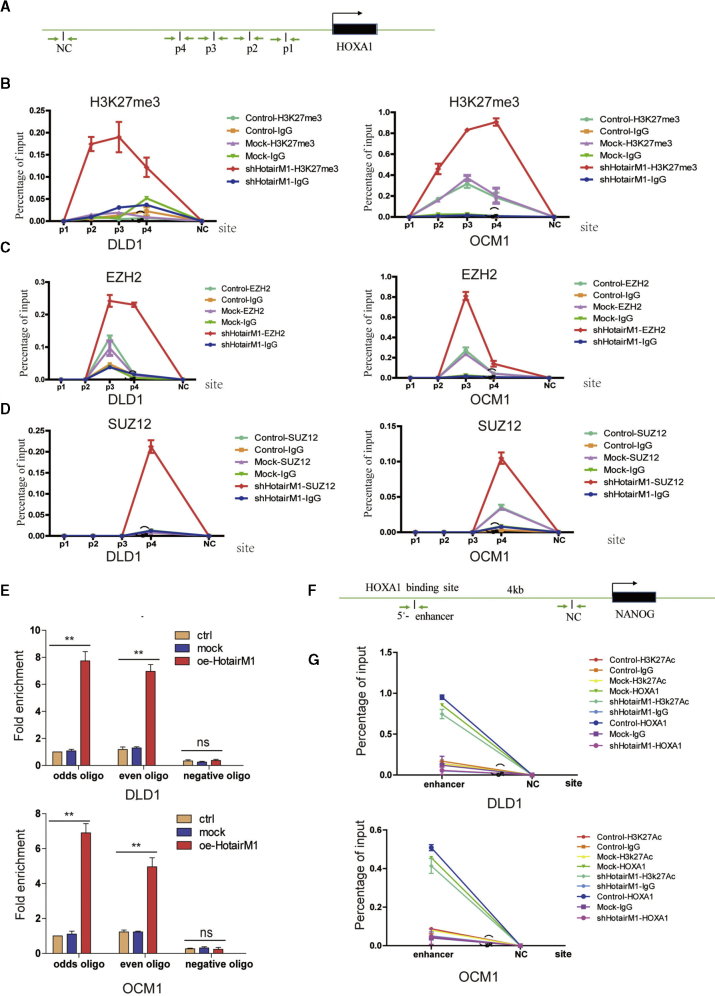
HotairM1 Regulates HOXA1 Expression by Activating Histone H3K27 Methylation
To elucidate the mechanism underlying the suppressive role of HotairM1 in CSC expansion, we first analyzed the potential regulation via a nuclear, chromosome-related mechanism, since HotairM1 mostly located in the nucleus. To explore this possibility, we first detected whether histone methylation status, a common epigenetic modification, was changed in the HOXA1 promoter. Several sites in the HOXA1 promoter (Figure 6A) were designed, and H3K27 methylation in HotairM1 depletion cells was detected through chromatin immunoprecipitation (ChIP) assay. Tri-methylation of lysine 27 on histone 3(H3K27me3) occurs at sites p2, p3, and p4 (Figure 6B). A similar positive trend of H3K27 methylation was found in shHotairM1 OCM1 cells. Taken together, these results demonstrated that HotairM1 inhibits HOXA1 expression by promoting H3K27 methylation at the HOXA1 promoter. Polycomb repressive complex 2 (PRC2) is the most recognized histone methyltransferase that mediates H3K27 methylation, consisting of three important components: EED, SUZ12, and EZH2.30 ChIP was performed to detect the binding of these proteins to the HOXA1 promoter. EZH2 binds at the p3 and p4 sites in HotairM1-depleted DLD1 cells, while EZH2 binds at the p3 site in OCM1 cells (Figure 6C). SUZ12 binds at the p4 site in both shHotairM1 DLD1 cells and shHotairM1 OCM1 cells (Figure 6D). These results confirmed that the PRC2 complex could bind to the HOXA1 promoter and mediate H3K27 methylation alteration, causing the repression of HOXA1 in HotairM1-depleted cells. What role did HotairM1 play during this process? Chromatin oligonucleotide precipitation (ChOP) assay was performed, and HotairM1 was detected to bind to the HOXA1 promoter, whereas this interaction was not observed in shCtrl cells, as shown in Figure 6E. Taken together, these results demonstrated that HotairM1 influenced PRC2 occupancy of the promoter of the HOXA1 gene. When HotairM1 was depleted, the binding at the HOXA1 promoter was abrogated, and this region could be competitively occupied by PRC2 complex mediating the H3K27 tri-methylation.
Figure 6.
HotairM1 Triggers HOXA1 Expression to Inhibit Nanog Expression
(A) Schematic of sites in HOXA1 promoter detected using the ChOP assay and ChIP assay. (B) Real-time PCR examination of histone H3K27me3 changes in HOXA1 promoter when HotairM1 was knocked down through the ChIP assay. IgG was used as the negative control. The value obtained for untreated uveal melanoma cells was set as 1. Data are indicated as the mean ± SD. (C) Real-time PCR examination of EZH2 changes in HOXA1 promoter when HotairM1 was knocked down using ChIP assay. IgG was used as the negative control. Data are shown as the mean ± SD. (D) Real-time PCR examination of SUZ12 changes in HOXA1 promoter when HotairM1 was knocked down using ChIP assay. IgG was used as the negative control. Data are indicated as the mean ± SD. (E) Real-time PCR examination of the binding of HotairM1 to the HOXA1 promoter through the ChOP assay. The HotairM1 oligo indicates the biotinylated antisense oligonucleotides against the HotairM1 lncRNA. Negative oligo indicates the scramble oligonucleotides, which were used as a negative control in the ChOP assay. The value obtained for untreated uveal melanoma cells was set to 1. Data are indicated as the mean ± SD. (F) Schematic of sites in Nanog enhancer detected using ChIP assay. Data are indicated as the mean ± SD. (G) Real-time PCR examination of the changes of HOXA1, H2K27Ac, and IgG in the Nanog enhancer site through ChIP assay. Data are indicated as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01, by two-tailed Student’s t test.
HotairM1 Triggers HOXA1 Expression to Inhibit Nanog Expression
HotairM1 depletion suppressed the expression of HOXA1 but increased expression of pluripotency markers Oct4, Sox2, and Nanog. Whether the expression of these genes was dependent on HOXA1 is unknown. Considering that HOXA1 and these factors are all located in the nucleus, we explored epigenetic modulation using a ChIP assay. As shown in Figure 6F, we analyzed a 3-kb locus upstream of the transcription start sites (TSSs) of the Nanog gene. We found that HotairM1 depletion abrogated the binding capacity of HOXA1 at the Nanog enhancer site and induced the acetylation of H3K27, upregulating Nanog expression in shHotairM1 cells (Figure 6G). However, no correlation of HOXA1 with Oct4 or Sox2 was identified in our study. Besides, the effect of Nanog on HOXA1 was also explored in our research. As shown in Figure S8, Nanog could conversely bind at the HOXA1 enhancer site and inhibit the acetylation of H3K27, thus further inhibiting HOXA1 expression, forming an inter-regulation loop. The inhibitory effect of HOXA1 was enhanced through two ways, including HotairM1 inhibition and the Nanog-HOXA1 inter-regulation loop (Figure 7).
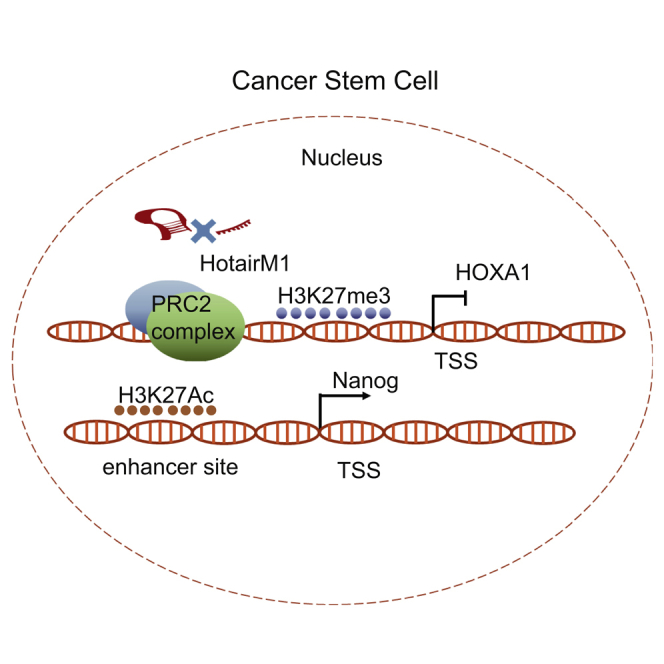
Figure 7.
Schematic of HotairM1 in the Regulation of CSC Stemness Maintenance
HotairM1 in tumors recognizes and binds to the HOXA1 promoter. This binding could competitively inhibit the recruition of SUZ12 and EZH2 to this region. HOXA1 expression inhibited the acetylation of H3K27 at the Nanog enhancer site, resulting in the downregulation of the pluripotent factor Nanog (left). When HotairM1 was knocked down, SUZ12 and EZH2 were recruited to the HOXA1 promoter, promoting H3K27 trimethylation modification and leading to epigenetic silence of HOXA1. The silence of HOXA1 promoted acetylation of H3K27 at the Nanog enhancer site, resulting in the increased expression of Nanog. In the meantime, the induction of Nanog could conversely inhibit the acetylation of H3K27 at the enhancer site of HOXA1, further inhibiting HOXA1 expression, which forms a cross-regulatory loop (right).
Discussion
Recently, CSCs have been considered to be responsible for tumor metastasis and recurrence due to their characteristics, such as self-renewal and ability to initiate tumorigenesis.31 CSCs are found not only in blood tumors but also in solid tumors, such as colon cancer, melanoma, breast cancer, lung cancer, and ovarian cancer.32, 33, 34 The isolation mainly includes flow cell sorting and sphere formation,35 and the self-renewal capacities of CSCs could be detected through serial passages of colonies assay, in vitro sphere formation assay, and in vivo transplantation assay.36 Recent studies have demonstrated that lncRNAs played important roles in cell-fate programming and reprogramming, and they could emerge as key components of the address code, allowing proteins, genes, and chromosomes to be activated or deactivated.37, 38, 39 In this study, cells isolated through sphere formation using serum-free medium were detected and defined as cancer stem-like cells. Critical lncRNAs that functioned during the stemness maintenance process were identified via transcriptome sequencing analysis, and a lncRNA significantly lowly expressed in CSCs called HotairM1 was identified.
HotairM1 was first described as a myeloid-specific regulator of the HOXA genes40 and was identified as a promising molecule in leukemia and several solid tumors. Wang et al.26 found that HotairM1 could contribute to the three-dimensional chromatin organizational changes required for HOXA gene activation in tumor cells. Chen et al.41 found that HotairM1 plays pivotal roles in the degradation of the oncoprotein PML-RARA and in myeloid cell differentiation by regulating autophagy pathways in acute promyelocytic leukemia. However, little is known about the relationship between HotairM1 and self-renewal capability of CSCs. Then, the role of HotairM1 in colorectal CSCs and uveal melanoma stem cells was detected in our study. Our results demonstrated that HotairM1 knockdown promotes CSCs stemness maintenance and increases tumorigenesis through target gene HOXA1.
HOXA1 plays a critical role in early embryonic development in humans and is the most rapidly induced gene during retinoid-induced differentiation of murine embryonic stem cells (ESCs).42 Growing evidence suggests that alterations in HOXA1 genes have also been associated with tumor prognosis and progression, and it could be both an oncogene and a tumor suppressor in a variety of tumors.43 Overexpression of HOXA1 is associated with tumor progression in glioblastoma, melanoma, prostate cancer, gastric cancer, and breast cancer.44, 45, 46, 47, 48 Conversely, HOXA1 silence was associated with poor prognosis of small-cell lung cancer and giant-cell tumor of bone.49,50 However, the number of studies on HOXA1 expression during stemness maintenance in CSCs is limited.48 Our study demonstrates that HOXA1 was downregulated in CSCs, and its expression was positively correlated with HotairM1 expression. Depletion of HOXA1 could increase the frequency of stem cells in uveal melanoma and colorectal cancer and thus promote tumorigenesis. These data also suggest that overexpression of HOXA1 can be a positive prognostic indicator for patient survival. Mechanistically, the local epigenetic status of HOXA1 was controlled by HotairM1 through the RNA-DNA interactions.22 HotairM1 could bind at the promoter region of HOXA1 and impedes the binding of EZH2 and SUZ12 at this site, inducing tri-methylation of H3K27 and inhibiting HOXA1 expression. Recent studies have found that SUZ12 is necessary in mediating strong chromatin binding in a manner independent from the rest of the core PRC2 complex.30,51 Our studies also confirmed that the binding of SUZ12 contributes to the binding of methyltransferase, catalyzing trimethylation of lysine 27 on histone H3. Our study provides insight into how HotairM1 regulated CSC stemness maintenance in uveal melanoma cells and colorectal cancer cells, and it explains the precise transcriptional control of the HOXA1 gene.
HOXA1 is one of the most rapidly induced genes in ESC differentiation, playing important roles in the differentiation and pluripotency process.52 Previous studies have found that Hoxa1 has many target genes in Wnt, transforming growth factor β (TGF-β), Hedgehog, and Hippo signaling pathways through genome-wide Hoxa1 binding data in mouse ESCs.53 These major signaling pathways could maintain cells in a pluripotency state by modulating the expression of components in a core pluripotency gene regulatory network consisting of Nanog, Oct4, and Sox2.54 These genes are found to be regulated by HOXA1 in mouse ESCs.52 In our study, HOXA1 was found to be recruited to the enhancer site of Nanog and influence the expression of the Nanog through inhibiting the acetylation of H3K27. At the same time, Nanog could bind at the enhancer site of HOXA1 and inhibit the expression of HOXA1 through inhibiting H3K27 acetylation, forming a cross-regulatory interaction.
In summary, HotairM1 was expressed at markedly lower levels in CSCs, uveal melanoma tissues, and colorectal cancer tissues. Knockdown of HotairM1 significantly increased the proportion of CSCs and maintained the stemness. HotairM1 depletion could induce the tri-methylation of H3K27, silencing HOXA1 expression. Moreover, HOXA1 regulated the expression of the critical stemness-related gene Nanog by inducing the acetylation of H3K27 at the enhancer site, and Nanog could conversely downregulate HOXA1 expression. The HOXA1-Nanog inter-regulation loop augments the effect and acts as a critical pathway in the stemness maintenance of CSCs and tumor progression.
Materials and Methods
Cell Culture
The human HCT116, DLD1, HT29, OCM1, RPE, and 293T cell lines were bought from Shanghai Institute of Life Sciences Cell Resource Center and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) supplemented with 10% certified heat-inactivated fetal bovine serum (FBS; GIBCO), penicillin (100 U/mL), and streptomycin (100 mg/mL) in a humidified 5% CO2 atmosphere.
Real-Time Polymerase Chain Reaction and Quantitative Real-Time PCR
Total RNA from cells and tissues was extracted using TRIzol Reagent (GIBCO), and cDNA was synthesized using the PrimeScript RT-PCR Kit (Takara Bio). Real-time PCR was performed using the Premix Ex Taq reagent (Takara Bio), and real-time PCR was performed using the SYBR Select Master Mix (Applied Biosystems) and an ABI 7500 Real-Time PCR System (Applied Biosystems). HotairM1 and HOXA1 expression in tumor samples was analyzed using cDNA microarrays (Superchip Biotech, Shanghai, China). The primers for qPCR are listed in Table S1.
Lentivirus Packaging and Generation of Stable Cell Lines
239T cells were incubated with DMEM with Lipofectamine 2000 reagent (Invitrogen) to transfect with plasmids. The supernatants containing viruses were collected and filtered through a 0.45-mm cellulose acetate filter and concentrated with an Amicon Ultra-15 Centrifugal Filter Unit (Millipore). Tumor cells were seeded in a six-well plate, and the medium was replaced with a virus-containing supernatant supplemented with 10 ng/mL polybrene (Sigma-Aldrich). Selection was performed through incubation with puromycin (InvivoGen), and colonies were selected and expanded for further analyses.
Small Interfering RNAs (siRNAs)
The siRNAs were designed and synthesized by Biotend. Cells were seeded and transfected with 125 pmol of siRNA (corresponding to the tested gene or a negative control) using Lipofectamine 2000. Six hours after transfection, the medium was replaced by fresh complete medium, and 48 h after transfection, the cells were harvested for the detection of knockdown efficiency and other experiments. Cells cultured for longer than 3 days were silenced again using siRNA. The primers for siRNA are listed in Table S1.
Western Blot Analysis
Different groups of cells were harvested and rinsed twice with phosphate-buffered saline (PBS). Cell extracts were prepared in lysis buffer, and protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. After blocking with 5% bovine serum albumin for 1 h at room temperature, the membrane was incubated with antibodies against Oct4, Sox2, Nanog (Cell Signaling Technology), and HOXA1 (Thermo Fisher Scientific) overnight. The membrane was then incubated with a secondary antibody, and the band signals were visualized and quantitatively analyzed using the Odyssey Infrared Imaging System (LI-COR, USA).
Database Analysis
To validate the potential roles of HotairM1 and HOXA1 in tumors, we searched the GEPIA (http://gepia.cancer-pku.cn/) and the GEO (https://www.ncbi.nlm.nih.gov/geo/; GEO: GSE104836) databases, which provided gene expression data, follow-up information, and correlation indexes of colorectal cancer cases. Kaplan-Meier survival analysis of HotairM1 was analyzed, and the cutoff value was set according to the median value of HotairM1 normalized expression.
Sphere Formation Assay and In Vitro Limiting Dilution Assay
Cells were cultured in sphere formation medium (SFM) composed of DMEM/F12 medium, 2% B27, 20 ng/mL EGF, and 20 ng/mL basic fibroblast growth factor (bFGF) in low-adhesion plates (Corning); spheres were collected for the subsequent experiments. Spheres were digested and dispersed, and then, 1,000 cells were counted and seeded into the wells again. Ten days after culture, spheres were counted and analyzed. For the in vitro dilution assay, cells were differentially diluted and seeded into SFM. After culturing for 10 days, spheres were collected, counted, and analyzed.
Transwell Assay
The migratory ability of the cells was evaluated using a 24-well Transwell system with polycarbonate filters (pore size, 8 mm; Millipore). The upper compartment contained 100,000 cells in medium without serum, while the lower compartment contained 10% FBS without cells. After 2 days of incubation, the cells that migrated from the upper to the lower compartment were stained with 1% crystal violet and washed using 33% acetic acid. The absorbance values of the liquid at 630 nm were determined using a microplate reader.
Colony Formation Assay
First, 0.6% agar in cell incubation medium was spread in the wells of a six-well plate, and 5,000 cells were re-suspended in 0.3% agar complete medium and seeded into the upper layer. The cells were cultured for 3–4 weeks. The colonies in the soft agar were stained with 0.05% crystal violet and photographed.
Xenograft Model and In Vivo Limiting Dilution Assay
Animal experiments were approved by the Shanghai Jiao Tong University Animal Care and Use Committee and were conducted following the animal policies of Shanghai Jiao Tong University in accordance with the guidelines established by the National Health and Family Planning Commission of China. Cells were harvested and injected subcutaneously into the flank of nude mice. The tumor volume was measured and recorded.
Different amounts of different groups of cells were injected into both sides of mice, and the number of tumors formed was recorded. The tumorigenic cell frequency was calculated using the ELDA site (http://bioinf.wehi.edu.au/software/elda/).
RNA-Sequencing Analysis
Colorectal CSCs were enriched in the sphere formation assay, and then spheres were collected. Normal colorectal carcinoma cells were collected as non-CSCs. Total RNA was extracted, and the lncRNA and mRNA expression profiles were analyzed using RNA sequencing.
Cytoplasmic and Nuclear RNA Isolation
Cytoplasmic and nuclear RNA were extracted using the Fisher BioReagents SurePrep Nuclear or Cytoplasmic RNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The RNA was reverse-transcribed to cDNA and used for real-time PCR. The primers for PCR are listed in Table S1.
ChOP
The ChOP assay was performed as previously described. Briefly, cells were fixed and crosslinked, and then the cell pellet was suspended in lysis buffer with 100 U/mL RNase (Promega). After sonication and centrifugation of nuclear lysates, the optimal sonicated chromatin was detected with 2% agarose gels. The sheared DNA complexes were immune-precipitated with biotinylated even oligonucleotides, odd oligonucleotides, and biotinylated control oligonucleotides, which were all incubated at a proper annealing temperature for 5 min and then slowly cooled to room temperature. Beads were used to capture the biotinylated DNA/RNA complexes, and after crosslink reversal and purification, the DNA samples were ready for PCR analysis. The primers for ChOP-qPCR are listed in Table S1.
ChIP
The ChIP assay was performed using the EZ-Magna ChIP A/G Kit (Millipore) according to the manufacturer’s instructions. Cells were fixed and centrifuged, and the pellets were resuspended in ChIP lysis buffer. After sonication, the sheared DNA complex of nuclear lysates was collected, and antibodies were added, including H3K27me3, SUZ12, EZH2 (Active Motif), RNA polymerase II, and immunoglobulin G (IgG) (Millipore). The mixture was incubated overnight, and magnetic beads were added to pull down the DNA-protein-antibody complexes. After elution, the samples were ready for real-time PCR analysis. The primers for ChIP-qPCR are listed in Table S1.
Statistical Analysis
Gene expression (quantitative real-time PCR and ChIP-qPCR) analyses were performed in triplicate, and the data are expressed as the mean ± SD, as described. For comparisons of relative expression, the control group was normally set at 1 or 100% when compared with the treated groups. Differences between two groups were analyzed using unpaired two-sided Student’s t test. The significance value is represented as ∗p <0.05. All statistical analyses and data graphs were generated in GraphPad Prism software.
Author Contributions
X.F. and H.Z. designed the experiments. F.Z. and F.L. wrote the paper. F.L. and Y.X. conducted the experiments. S.G. and X.X. analyzed the data.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81802919, 81802739, 81772875, and U1932135), the National Key R&D Program of China (2018YFC1106100), and the Science and Technology Commission of Shanghai (17DZ2260100 and 19JC1410200). The study sponsor had no role in the design of the study; the data collection, analysis, or interpretation; the writing of the article; or the decision to submit for publication.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.008.
Contributor Information
Feifei Zhang, Email: feifeizhang415@163.com.
He Zhang, Email: zhanghe@sjtu.edu.cn.
Xianqun Fan, Email: fanxq@sjtu.edu.cn.
Supplemental Information
References
- 1.Pützer B.M., Solanki M., Herchenröder O. Advances in cancer stem cell targeting: How to strike the evil at its root. Adv. Drug Deliv. Rev. 2017;120:89–107. doi: 10.1016/j.addr.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Kreso A., Dick J.E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Delacruz A. Using circulating tumor cells as a prognostic indicator in metastatic castration-resistant prostate cancer. Clin. J. Oncol. Nurs. 2012;16:E44–E47. doi: 10.1188/12.CJON.E44-E47. [DOI] [PubMed] [Google Scholar]
- 4.Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 5.Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A.R., Poletti A. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 6.Wang W.Y., Hsu C.C., Wang T.Y., Li C.R., Hou Y.C., Chu J.M., Lee C.T., Liu M.S., Su J.J., Jian K.Y. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology. 2013;145:1110–1120. doi: 10.1053/j.gastro.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 7.Cole A.J., Fayomi A.P., Anyaeche V.I., Bai S., Buckanovich R.J. An evolving paradigm of cancer stem cell hierarchies: therapeutic implications. Theranostics. 2020;10:3083–3098. doi: 10.7150/thno.41647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saygin C., Matei D., Majeti R., Reizes O., Lathia J.D. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 2019;24:25–40. doi: 10.1016/j.stem.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Yang L., Yan W., Zhai J., Pizzo D.P., Chu P., Chin A.R., Shen M., Dong C., Ruan X. Chemotherapy Induces Breast Cancer Stemness in Association with Dysregulated Monocytosis. Clin. Cancer Res. 2018;24:2370–2382. doi: 10.1158/1078-0432.CCR-17-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Díaz-Carballo D., Saka S., Klein J., Rennkamp T., Acikelli A.H., Malak S., Jastrow H., Wennemuth G., Tempfer C., Schmitz I. A Distinct Oncogenerative Multinucleated Cancer Cell Serves as a Source of Stemness and Tumor Heterogeneity. Cancer Res. 2018;78:2318–2331. doi: 10.1158/0008-5472.CAN-17-1861. [DOI] [PubMed] [Google Scholar]
- 12.Petruk S., Sedkov Y., Riley K.M., Hodgson J., Schweisguth F., Hirose S., Jaynes J.B., Brock H.W., Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W., Alvarez-Dominguez J.R., Lodish H.F. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhu P., Wang Y., Huang G., Ye B., Liu B., Wu J., Du Y., He L., Fan Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 18.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.P. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan D.S.W., Chong F.T., Leong H.S., Toh S.Y., Lau D.P., Kwang X.L., Zhang X., Sundaram G.M., Tan G.S., Chang M.M. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017;23:1167–1175. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Weissman S.M., Newburger P.E. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–787. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian X., Ma J., Wang T., Tian J., Zhang Y., Mao L., Xu H., Wang S. Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front. Immunol. 2018;9:473. doi: 10.3389/fimmu.2018.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q., Dong C., Cui J., Wang Y., Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018;37:265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan L., Kong J., Tang J., Wu Y., Xu E., Lai M., Zhang H. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J. Cell. Mol. Med. 2016;20:2036–2044. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Mi L., Xuan Y., Gao C., Wang Y.H., Ming H.X., Liu J. LncRNA HOTAIRM1 inhibits the progression of hepatocellular carcinoma by inhibiting the Wnt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:4861–4868. doi: 10.26355/eurrev_201808_15622. [DOI] [PubMed] [Google Scholar]
- 25.Lu R., Zhao G., Yang Y., Jiang Z., Cai J., Zhang Z., Hu H. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/ PTEN axis in gastric cancer. J. Cell Biochem. 2019;120:4952–4965. doi: 10.1002/jcb.27770. [DOI] [PubMed] [Google Scholar]
- 26.Wang X.Q., Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017;45:1091–1104. doi: 10.1093/nar/gkw966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai H.T., Chiang C.T., Tseng W.K., Chao T.C., Su Y. GATA6 enhances the stemness of human colon cancer cells by creating a metabolic symbiosis through upregulating LRH-1 expression. Mol. Oncol. 2020;14:1327–1347. doi: 10.1002/1878-0261.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doherty R.E., Sisley K., Hammond D.W., Rennie I.G., Cross N.A. Phenotypic Plasticity in Uveal Melanoma Is Not Restricted to a Tumor Subpopulation and Is Unrelated to Cancer Stem Cell Characteristics. Invest. Ophthalmol. Vis. Sci. 2017;58:5387–5395. doi: 10.1167/iovs.17-22272. [DOI] [PubMed] [Google Scholar]
- 29.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laugesen A., Højfeldt J.W., Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suvà M.L., Tirosh I. The Glioma Stem Cell Model in the Era of Single-Cell Genomics. Cancer Cell. 2020;37:630–636. doi: 10.1016/j.ccell.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Haraguchi N., Ishii H., Mimori K., Tanaka F., Ohkuma M., Kim H.M., Akita H., Takiuchi D., Hatano H., Nagano H. CD13 is a therapeutic target in human liver cancer stem cells. J. Clin. Invest. 2010;120:3326–3339. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visvader J.E., Lindeman G.J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Boiko A.D. Isolation of melanoma tumor-initiating cells from surgical tissues. Methods Mol. Biol. 2013;961:253–259. doi: 10.1007/978-1-62703-227-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahmad H.F., Cheaito K., Chalhoub R.M., Hadadeh O., Monzer A., Ballout F., El-Hajj A., Mukherji D., Liu Y.N., Daoud G., Abou-Kheir W. Sphere-Formation Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front. Oncol. 2018;8:347. doi: 10.3389/fonc.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck B., Blanpain C. Unravelling cancer stem cell potential. Nat. Rev. Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 37.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Lian Z., Padden C., Gerstein M.B., Rozowsky J., Snyder M., Gingeras T.R., Kapranov P., Weissman S.M., Newburger P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z.H., Wang W.T., Huang W., Fang K., Sun Y.M., Liu S.R., Luo X.Q., Chen Y.Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Kumar B., Parrish M.E., Slaughter B.D., Unruh J.R., Gogol M., Seidel C., Paulson A., Li H., Gaudenz K., Peak A. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015;25:1229–1243. doi: 10.1101/gr.184978.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L., Hu N., Wang C., Zhao H., Gu Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17:319–329. doi: 10.1080/15384101.2017.1407893. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Brock A., Krause S., Li H., Kowalski M., Goldberg M.S., Collins J.J., Ingber D.E. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Sci. Transl. Med. 2014;6:217ra2. doi: 10.1126/scitranslmed.3007048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wardwell-Ozgo J., Dogruluk T., Gifford A., Zhang Y., Heffernan T.P., van Doorn R., Creighton C.J., Chin L., Scott K.L. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33:1017–1026. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Liu G., Shen D., Ye H., Huang J., Jiao L., Sun Y. HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncol. Rep. 2015;34:1203–1210. doi: 10.3892/or.2015.4085. [DOI] [PubMed] [Google Scholar]
- 47.Yuan C., Zhu X., Han Y., Song C., Liu C., Lu S., Zhang M., Yu F., Peng Z., Zhou C. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J. Exp. Clin. Cancer Res. 2016;35:15. doi: 10.1186/s13046-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia H., Liu Y., Wang Z., Zhang W., Qi M., Qi B., Jiang X. Long Noncoding RNA HOTAIRM1 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells Through Regulation of HOX Gene Expression. Neurotherapeutics. 2020;17:754–764. doi: 10.1007/s13311-019-00799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao F., Bai Y., Chen Z., Li Y., Luo L., Huang J., Yang J., Liao H., Guo L. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur. J. Cancer. 2014;50:1541–1554. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 50.Ni L.Y., Zhao J.D., Lu Y.H., Li W., Li B.L., Wang X.C., Meng Q.G. MicroRNA-30c suppressed giant-cell tumor of bone cell metastasis and growth via targeting HOXA1. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4819–4827. [PubMed] [Google Scholar]
- 51.Højfeldt J.W., Laugesen A., Willumsen B.M., Damhofer H., Hedehus L., Tvardovskiy A., Mohammad F., Jensen O.N., Helin K. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 2018;25:225–232. doi: 10.1038/s41594-018-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Kumar B., Parker H.J., Parrish M.E., Lange J.J., Slaughter B.D., Unruh J.R., Paulson A., Krumlauf R. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc. Natl. Acad. Sci. USA. 2017;114:5838–5845. doi: 10.1073/pnas.1610612114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Kumar B., Parker H.J., Paulson A., Parrish M.E., Zeitlinger J., Krumlauf R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev. Biol. 2017;432:151–164. doi: 10.1016/j.ydbio.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 54.Pera M.F., Tam P.P. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.