Abstract
Extensive medical research showed that patients, with high protein concentration in urine, have various kinds of kidney diseases, referred to as proteinuria. Urinary protein biomarkers are useful for diagnosis of many health conditions – kidney and cardio vascular diseases, cancers, diabetes, infections. This review focuses on the instrumental quantification (electrophoresis, chromatography, immunoassays, mass spectrometry, fluorescence spectroscopy, the infrared spectroscopy, and Raman spectroscopy) of proteins (the most of all albumin) in human urine matrix. Different techniques provide unique information on what constituents of the urine are. Due to complex nature of urine, a separation step by electrophoresis or chromatography are often used for proteomics study of urine. Mass spectrometry is a powerful tool for the discovery and the analysis of biomarkers in urine, however, costs of the analysis are high, especially for quantitative analysis. Immunoassays, which often come with fluorescence detection, are major qualitative and quantitative tools in clinical analysis. While Infrared and Raman spectroscopies do not give extensive information about urine, they could become important tools for the routine clinical diagnostics of kidney problems, due to rapidness and low-cost. Thus, it is important to review all the applicable techniques and methods related to urine analysis. In this review, a brief overview of each technique's principle is introduced. Where applicable, research papers about protein determination in urine are summarized with the main figures of merits, such as the limit of detection, the detectable range, recovery and accuracy, when available.
Keywords: Urine proteomics, Biomarkers, Human serum albumin, Immunoassays, Mass spectrometry, Fluorescence spectroscopy
Graphical abstract
Highlights
-
•
Urinary protein biomarkers are useful for diagnosis of many conditions: kidney and cardio vascular diseases, cancers.
-
•
Liquid chromatography – mass spectroscopy is a powerful tool for urine proteomics, but used mostly in science.
-
•
Immunoassays are widely used in both clinical and bio-analytical laboratories.
-
•
IR and Raman spectroscopies are promising tools for diagnostics of urine due to low-cost and rapidness.
1. Introduction
1.1. Urine composition
Urine is a readily available liquid for the medical diagnosis of patients. Urine tests are non-invasive procedures that involve no pain or discomfort for patients to determine problems with kidney function since extensive medical research showed that patients, with high protein concentration in urine, have various kinds of illnesses of the kidney, referred to as proteinuria [1]. Thus the precise and simple determination of urinary concentrations of total protein is important for diagnostic purposes. Since urine is a complex matrix, it poses challenges for the analytical determination of proteins and other constituents. Among the reasons that make urine challenging for researchers to analyze are: urine matrix is complex, it consists of various inorganic and organic compounds, from low-molar mass molecules to polymers; urine could contain cells, such as blood cells, or bacteria, which changes the composition of urine in time rapidly; an analytical method for diagnosis of proteinuria should cover protein presence in urine in a wide range from 0.01 mg/ml to 10 mg/ml. Also, it is important to note that the concentration of protein in urine taken from patients can vary widely depending on dieting, exercising, and time of the day a patient urinated. It is widely accepted that analysis of urea taken throughout a day is the best representation of any illnesses in the human body if any. The so-called – the urine 24-h volume test, that measures the amount of urine the human body produces in a day. Another option is to determine creatinine to protein ratio in urea since creatinine to protein ratio is positively correlated with 24-h volume test for quantifying proteinuria [2,3]. The noninvasive collection of samples and wide range of diagnostic targets found in urine makes urinalysis well suited for point-of-care (PoC) monitoring applications, in which testing is done at the time and place of patient care [4]. Table 1 shows constituents in urine taken from the urine 24-h volume test.
Table 1.
Composition of urine (averages of selected components in 24-h collection test) [5].
| Component | Average weight (mg) |
|---|---|
| Water | 1,200,000.0 |
| Urea | 24,000.0 |
| Creatinine | 1335.0 |
| Uric acid | 505.0 |
| Albumin | 90.0 |
In healthy individuals, Tamm–Horsfall protein (also known as uromodulin) is the most abundant protein in urine (50%), followed by albumin (20%) and immunoglobulin (5%) [6]. Other constituents in the urine of either sick or healthy patients include, but not limited to: glucose; low abundant proteins, such as bilirubin, and urobilinogen; cells, such as Erythrocyte, Leukocyte, and other cells; bacteria [7].
1.2. Urine as a diagnostic tool
The urine of a healthy individual contains up to 150 mg of protein in total measured throughout a day, of which approximately 20 mg is albumin (human serum albumin) [1]. Albumin excretion of 30–300 mg a day, which is called microalbuminuria, is an early and sensitive marker of diabetic nephropathy [8], cardiovascular and renal disease [9]. The 15–30 mg/L albumin concentration is a critical value that could indicate kidney problems when it is repeatedly exceeded [10]. Though one of the main proteins in urine is albumin, there are thousands of other types of proteins. Furthermore, the removal of albumin from the urine helps to identify the low abundant proteins [11]. Marimuthu et al. by using high-resolution Fourier transform mass spectrometry were able to identify 1823 proteins in the urine of healthy subjects [12]. Using mass spectroscopy and sub-fractionating normal urine by successive steps (vesicle separation, CPLL, and solvent treatments) Santucci et al. were able to identify 3429 individual proteins [13]. All those discovered proteins could be potential biomarkers for diseases. In a review paper by Röthlisberger et al. it was stated that besides albumin, other proteins such as CD14, hh-FABP, BNP/NT-proBNP, NGAL, ORM1 are potential biomarkers of cardiovascular disease [14]. Another biomarker in human urine, Bence Jones protein (BJP), has an important diagnosis and prognosis value for multiple myeloma, cancer formed in a white blood cell called plasma cell [15]. Other types of chemical substances in urine can be important biomarkers as well, for example, urine microRNAs have the potential to be a valid marker for bladder cancer detection [16].
Proteinuria is the main clinical presentation of glomerular diseases (the glomerulus are complex capillary set that are located in the nephrons – renal cells) [17]. The glomerular diseases can be primarily when the disease caused by kidney diseases (such as glomerulonephritis), or secondary, when kidney glomerulus becomes target organ affected by different diseases such as diabetes and cardiovascular diseases, autoimmune and inflammatory disorders, amyloidosis and neoplasms, cancer and many other including genetic disorders [17,18]. The levels of proteinuria, which usually measures in clinical settings, may vary depending on severity of disease and glomerular injury, despite any cause of disease. Any detectable proteinuria in the urine between 30 and 300 mg/24h called albuminuria (or microalbuminuria), above the 300 mg/24h – proteinuria. Depending the amount of proteinuria they considered to named nephritic range proteinuria below 3.5 g/24h and nephrotic range proteinuria when protein loss excess the 3.5 g/24h (heavy proteinuria). The most of the cases, proteinuria caused by cardiovascular diseases limited with nephritic range proteinuria, whereas in renal diseases and cancer the proteinuria may reach nephrotic range proteinuria [19,20]. Since urine is in direct contact with kidneys, analysis of urine is important for diagnosis of renal diseases, such as chronic kidney disease. Several viral infections such as Epstein-Barr virus, hepatitis B and C viruses, herpes zoster, hantavirus, human immunodeficiency virus, dengue fever, COVID-19 and many others also may lead to proteinuria [21,22]. The potential mechanisms of proteinuria are related to primary affect to glomerulus and/or secondary to autoimmune response to infection [23,24]. Chronic kidney disease (CKD) is a major public health problem. Albuminuria, urinary sediment abnormality and other markers of kidney damage are criteria of CKD, according to international guidelines [25]. Fassett et al. summarized biomarkers for CKD in urine – cystatin C, β-trace protein, NGAL, KIM-1, NAG and many others [26]. A more recent article on this topic is discussed in Ref. [27].
In the United States, it was estimated in 2003 that 11% of the adult population (aged 20 or older) has chronic kidney disease [28]. In a review paper about the prevalence of chronic kidney disease (CKD) by Zhang et al., the authors studied relevant data across America, Europe, Asia, Australia [29]. They reported that the median prevalence of CKD was 7.2% in persons aged 30 years or older. In persons aged 64 years or older prevalence of CKD varied from 23.4% to 35.8%. The authors concluded that worldwide, CKD is becoming a common disease in the general population. In a more recent paper, Wei et al. studied data on kidney damage among elderly people in Wuhan, China [30]. The age-standardized prevalence of kidney damage decreased renal function and proteinuria was 17.2, 13.5, and 5.3%. In the US, patients aged 66 or older have a mortality rate of 111.2 per 1000 due to CKD, compared to 45.2 per 100 due to non-CKD reasons as of 2014 [31]. It remains among the few growing causes of mortality which made CKD the 13th leading cause of death in 2013 [32].
Other renal diseases include diabetic nephropathy, autosomal dominal polycystic kidney disease, paediatric renal disease, acute kidney injury, and renal transplant rejection [33]. Besides renal diseases, urine can also be a useful source of information related to cancers and non-renal diseases. Theodorescu et al. found biomarkers in urine related to urothelial carcinoma, also called transitional cell carcinoma, by the means of capillary electrophoresis coupled with mass spectroscopy [34]. It is the most common type of bladder cancer. Bhasin et al. developed a bioresistor device to detect bladder cancer marker DJ-1 in urine at concenctration of 10 pM in 1 min [35]. Zhang et al. in their review summarized information about cancer biomarkers in urine [36]. In another work related to non-renal disease, Zimmerli et al. investigated urine samples of patients with coronary artery disease [37]. Rossing et al. identified potential biomarkers for diabetes [38]. Raja et al. investigated into the potential of albuminuria as a biomarker of diabetic complications [39]. Moreover, researchers could diagnose patients with viruses from their urine samples. Yang et al. developed an immunoassay cassette with a handheld reader for HIV urine testing in point-of-care diagnostics [40]. Robles et al. analyzed viruses present in urine from patients with interstitial cystitis [41]. Other viruses in urine described in literature include – Zika virus [42], hepatitis C [43], human papiloma virus [44]. Niedrig et al. wrote a review about usefulness of saliva and urine for the diagnosis of emerging viruses [45]. They concluded that it is important to perform an investigation using non-invasive approaches, such as urine can provide, for the diagnostic of emerging viral diseases.
1.3. Clinical urine tests
Urinalysis is an abbreviation for clinical urine tests for diagnostic purposes. A urinalysis (UA) is one of the most common methods of medical diagnosis. There are three basic components to urinalysis [46]:
-
•
Gross/physical examination targets parameters that can be measured or quantified with the naked eye (or other senses), including volume, color, transparency, odor, and specific gravity.
-
•
Microscopic examination. The numbers and types of cells and/or material such as urinary casts can yield a great detail of information and may suggest a specific diagnosis.
-
•
Chemical examination of urine measures quantitatively and qualitatively for pH, blood, nitrite, protein, glucose, ketones, bilirubin, urobilinogen, ascorbic acid.
Conditions for storage of urine samples is an important consideration for chemical analysis, since undesired changes in unpreserved urine occur, such as a decrease of concentration of glucose due to consumption of it by cells or bacteria; a decrease of concentration of bilirubin due to photo-oxidation and etc [47]. The most common form of preservation of urine samples is refrigeration, also chemicals can be utilized too. Remer et al. suggest that high long-term stability and measurement validity for numerous clinical chemistry parameters stored at −22 °C without the addition of preservative for human urine can be achieved [48]. Specifically for free-light chains monoclonal immunoglobulin, Pieri et al. suggest to store urine sample at +4 °C, if analysis to be made within 72 h, otherwise for longer periods, the refrigeration should be done at −80 °C [49]. Moyle et al. found that collection and storage of canine urine samples in clean homopolymer polypropylene (HP), propylene copolymer (PC), or glass containers at 24 °C for 4 h, 4 °C for 12 h or −20 °C for 72 h is unlikely to result in clinically relevant decreases in measured protein to creatine values [50].
Historically, qualitative or semi-quantitative screening tests for urine protein relied on protein precipitation techniques. Proteins denature upon exposure to extremes of pH or temperature, and the most visible evidence of this is a decrease in solubility. In clinical laboratories, sulfosalicylic acid at room temperature may be used to detect urine protein [47]. This protein precipitation method detects all proteins—albumin and globulins. The analytical signal here is turbidity, which is the cloudiness or haziness of a fluid. This method can not detect protein below 0.05 mg/ml. Modern commercial reagent strips are available for routine protein screening use change in color due to acid-base reaction or formation of complexes between urine constituents and reagent strips material. Some of those reactions are described and compared in Ref. [51]: absorbance at 280 nm, which monitors tyrosine and tryptophan in protein (UV absorbance); the method of Lowry et al. based on complex formation between tyrosine, tryptophan or cysteine and heavy metals using Folin phenol reagent in the presence of copper ions under alkaline conditions (Lowry method) [52]; the Bradford method based on protein dye-binding using Coomassie brilliant blue (CBB method) [53]; the method of Smith et al. based on the formation of a purple complex between bicinchoninic acid and copper ions under alkaline conditions (BCA method) [54]; the method of Watanabe et al. based on protein-dye binding using pyrogallol redmolybdate reagent (PRM method) [55].
To date, different diagnostics methods in clinical settings can be used to ascertain urinary protein and albumin. Urine dipstick test, technique with acidic buffer precipitation and immuno-electrophoresis are commonly used diagnostic methods for urinary total protein quantification [56]. The protein-specific dipstick and immunochemical techniques are the fastest and cheapest way to determine proteins in the urine for diagnostic purposes. Modern automated urine analyzers are based on similar principles and are widely used in clinical routine tests [57]. A urine analyzer is a device used in the clinical setting to perform automatic urine testing. The units can detect and quantify a number of analytes including bilirubin, protein, glucose, and red blood cells. Many models contain urine strip readers, a type of reflectance photometer that can process several hundred strips per hour. Some analyzers can perform up to 240 tests an hour. Prices of urine analysis are relatively low, but they depend on a location/country and they are available from the most clinical laboratories The high-performance liquid chromatography mostly used for scientific purpose due to its relatively high cost, though relatively inexpensive versions of mass spectrometers are now available [58].
Prices of urine analysis are relatively low, but they depend on a location/country and they are available from the most clinical laboratories The high-performance liquid chromatography mostly used for scientific purpose due to its relatively high cost, though relatively inexpensive versions of mass spectrometers are now available [58].
1.4. Focus of this review
Several review papers are focused on urine proteomics. A review by Albalat et al. focuses on urinary proteins as potential biomarkers for mainly urine diseases, and capillary electrophoresis coupled mass spectroscopy as an instrumental method [33]. The review also gives information on biomarkers on non-renal diseases obtained from the urine. Kalantari et al. state that mass spectrometry as a detection technique is the most common [59]. The paper discusses technical aspects of urinary proteomics, proteomic technologies, and their advantage and disadvantages. As of 2015, several recent experiments are presented there which applied urinary proteome for biomarker discovery in renal diseases including diabetic nephropathy, immunoglobulin A (IgA) nephropathy, focal segmental glomerulosclerosis, lupus nephritis, membranous nephropathy, and acute kidney injury. Decramer et al. in their review paper focus on mass spectrometry-based urinary protein and peptide profiling [60]. The advantages and disadvantages of different mass spectroscopy methods are compared. Applications of urinary proteome analysis to urogenital and non-urogenital diseases are also discussed there.
This review, besides covering mass spectrometry, holds chapters discussing fluorescence spectroscopy, immunoassay, infrared, and Raman spectroscopy that was not given much attention compared from the reviews on urinalysis mentioned in the previous paragraph. This review also covers separation techniques – electrophoresis and chromatography. For each technique, a brief overview of the technique's principle is introduced. Where applicable, research papers about protein determination in urine are summarized with the main figures of merits, such as the limit of detection, the detectable range, recovery, and accuracy. Particularly, electrophoresis, chromatography, immunoassay, and fluorescence spectroscopy chapters are followed by summary tables with analytical parameters taken from individual research papers. It should be noted that not all analytical parameters are given in every paper, for example, sometimes recovery is unknown, and thus is not included in the following tables. Since the volumes used for all techniques are 100–1000 times smaller than a typical urine probe available from one patient for analysis, usually 50–100 mL, urine volume is not introduced in the tables. For instance for proteomics study, a typical sample size for high performance liquid chromatography (HPLC) ranges from few microliters to 0.1 mL [61]. A commercial immunoassay test kits developed by Valle et al. require 10 μL of urine [62]. This review ends with a paragraph about urinary protein biomarkers. Most of those biomarkers were identified by mass spectrometry or immunoassay methods. A summary table of experimental papers on urinary protein biomarkers with related diseases or health conditions is given.
2. Separation techniques
2.1. Electrophoresis
Electrophoresis is an electrokinetic process, with a long history of development, which separates charged particles (macro-molecules, such as proteins) in a fluid using a field of electrical charge [63]. Particles could be separated based on their charge or their mass. The electrophoretic method is a widely used method in a modern research laboratory, but not so much in a clinical laboratory. Electrophoresis is commonly used for proteins, peptides and nucleic acid analysis [64]. It could be applied as a qualitative or quantitative analytical technique on its own if a reference substance is given, or it could be used as a separation technique for a sample to be further examined by other techniques, such as mass spectroscopy, fluorescence spectroscopy and etc.
Aguzzie et al. successfully used the electrophoretic technique to detect Bence-Jones protein (BJP) in human urine with the limit of detection LOD of 1 mg/L to BJP, and LOD of 3 mg/L to other types of protein [65]. These results were obtained by immunofixation using Dako antisera on cellulose nitrate followed by further staining with gold. The authors claim that their technique does not require particular skill and is cheap. Such low levels of LOD allowed them to use unconcentrated urine samples, thereby eliminating the problem of the loss of low molecular weight proteins. Usually, though it is often necessary to concentrate the urine before electrophoresis because of the low urinary protein level [6]. This is a time-consuming process. Machii et al. used cellulose acetate membrane electrophoresis followed by colloidal silver staining on the urine of healthy subjects with success [6]. However, when they used silver staining on an agarose gel, urinary protein in healthy subjects was not detected. They estimated that urine has to be concentrated 10 times, furthermore, the pattern of the urinary fraction will change after the concentration procedure is conducted. Giovannoli et al. developed a noncompetitive capillary electrophoresis immunoassay with laser-induced fluorescence detection was used to determine human serum albumin (HSA) in buffer and urine [66]. The injection of an excess of labeled antibody off-line incubated with the analyte allows the surface capture of the free antibody and consequently the immunocomplex detection. In buffer, authors were able to achieve the limit of detection LOD of 0.60 mg/L, with the quantitative range up to 6.6 mg/L. Recovery in the urine matrix was dependent upon sample dilution and HSA added and was as low as 91 ± 8%.
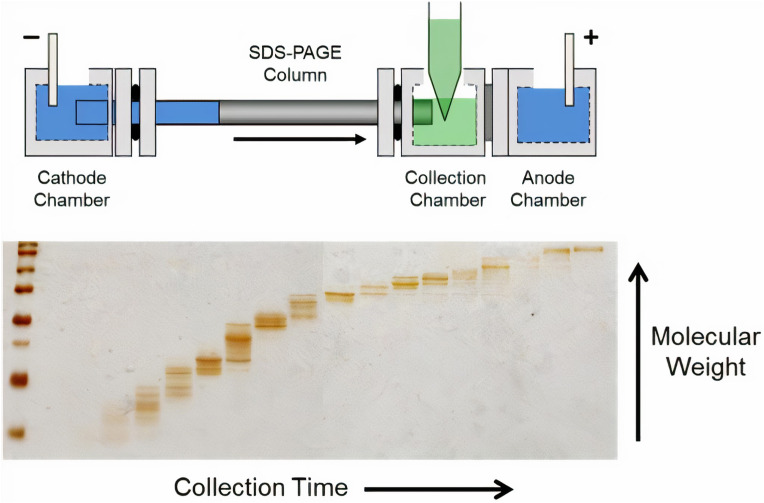
One of the variations of the electrophoretic method is called gel electrophoresis, where a gel is used as an anticonvective medium. Particularly polyacrylamide gel PAGE is used for separating proteins due to the uniform pore size provided by the polyacrylamide. Jia et al. used PAGE gel electrophoresis with sodium dodecyl sulfate (SDS-PAGE) to show a good correlation (R2 = 0.825) with the pyrogallol red-molybdate (PRM) assay technique used in clinical laboratories for urine samples [67]. Sodium dodecyl sulfate (SDS) coats proteins with a negative charge so that proteins are separated due to their size, not charge. The authors estimated that their method can detect protein as low as 5 mg/L. Tran et al. showed that the use of tube gel electrophoresis for protein separation was further expanded with the invention of gel-eluted liquid fraction entrapment electrophoresis (GELFrEE) [68]; shown in Fig. 1 .
Fig. 1.
Diagram of the GELFrEE device [68]. A gel column is utilized to achieve electrophoretic separation of proteins, analogous to SDS–PAGE, which are then eluted into the liquid-phase for manual collection. The fractionation can then be visualized by running a portion of the fractions on a SDS–PAGE gel. Reprinted from Refs. [95].
Bessonova et al. in their review paper compared capillary electrophoresis with five on-line preconcentration techniques, including field-amplified sample stacking (FASS), head-column field-amplified sample stacking (HC-FASS), stacking with a polymer solution, dynamic pH junction and large-volume sample stacking (LVSS) with reversed polarity for various analytes including protein in the urine [69]. The use of these preconcentration techniques makes it possible to increase sensitivity in the determination of analytes by a factor of 100 or higher. In another work by Bessonova et al., they found that the lowest LOD was obtained by the LVSS approach, and it was 15 mg/L with UV-detection [70].
Electrophoresis is often used to be followed by another method, such as mass spectroscopy. Wittke et al. used the on-line coupling of capillary electrophoresis (CE) with electrospray mass spectroscopy to distinguish the difference in polypeptide patterns in the urine of healthy individuals and patients with kidney disease [71]. Kiprijanovska et al. used two dimensional get electrophoresis coupled with mass spectroscopy (2D PAGE – MS) for mapping and identification purposes of the urine proteome [72]. For the same purposes, Nakayama et al. used cellulose acetate membrane electrophoresis coupled with liquid chromatography mass spectroscopy LC-MS [73].
2.2. Chromatography
Another widely used separation technique in chemistry is chromatography. In chromatography, a sample is dissolved in a fluid called the mobile phase, which carries it through a structure holding another material called the stationary phase. The various constituents of the mixture travel at different speeds, causing them to separate due to differential affinities (strength of adhesion, as an example) of the various components of the analyte towards the stationary and mobile phase results. Then resulting constituents can be further analyzed by various techniques, such as mass spectroscopy, UV-spectroscopy, fluorescence detection, etc. Depending on the mobile phase methods in chromatography could be classified as gas-chromatography (GC) [74], and liquid-chromatography (LC) [75]. Typically, proteomic sample size ranges from few microliters to 0.1 mL [61.
Ishida et al. quantitatively determined human serum albumin (HSA) in human plasma and urine by utilizing post-column high-performance liquid chromatography (HPLC) with fluorescence detection [76]. Particularly, they used 8-anilino-1-naphthalenesulfonic acid as a fluorophore to bind HSA. The sample size was 1 ml for urine, and the retention time was around 20 min. A linear relationship was observed between fluorescence peak and the amounts of HSA in both plasma and urine, as for the latter matrix, a linear relationship was observed up to 400 mg/L, with the correlation coefficient being 0.998. The LOD for HSA was 0.2 mg/L, while the recovery was 94.5 ± 3.8%. The same authors also compared their method with the immunological nephelometric method and obtained the correlation coefficient of 0.990 [77]. Often researchers compare their method with the immunoassay technique that is used in clinical practice. Brinkman et al. in their experiments using the immunonephelometric approach and HPLC determined that HPLC reveals higher values of HSA, especially in the lower concentration range, resulting in a higher prevalence of microalbuminuria [78]. Nephelometry only measures immunoreactive albumin, whereas HPLC measures both immunoreactive and immunoreactive intact albumin, as was found by Comper et al. with the HPLC method [79]. The authors also compared HPLC with radioimmunoassay (RIA), immunonephelometry (IN), and two different methods of immunoturbidimetry (IT).
Owen et al. used a new urinary albumin assay that uses size-exclusion high-performance liquid chromatography with UV-detection [80]. The limit of detection (LOD) of the assay based on a signal/noise ratio of 5 was 3.4 mg/L. The linearity of the HPLC assay was tested in a range of albumin concentrations from 4.3 to 240 mg/L. The maximum deviation from a mean recovery of 100% was 4.6% at a concentration of 66 mg/L. Total imprecision was less than 10% from 16 to 206 mg/L. They also conducted experiments to compare their approach with immunoturbidimetric (ITA) assay, which showed positive proportional bias, which decreased with increasing concentrations of albumin. Singh et al. used liquid chromatography with quadrupole time-of-flight mass spectrometry (Q-TOF, LC-MS) method for the quantification of urinary albumin using a15N-isotopically labeled bovine serum albumin as an internal standard in human urine matrix [81]. The limit of detection (LOD) was 4.84 mg/L, while the limit of quantification was 10.5 mg/L. Multiple calibration curves ranging from 4 to 625 mg/L were linear and were reproducible with the coefficient of determination (R2) of 0.999.
Micellar electrokinetic capillary chromatography (MEKC) is a hybrid method that combines chromatographic and electrophoretic separation principles, extends the applicability of capillary electrophoretic methods to neutral analytes [82]. In micellar electrokinetic capillary chromatography, surfactants are added to the buffer solution in a concentration above their critical micellar concentrations, consequently, micelles are formed; micelles that undergo electrophoretic migration like any other charged particle. Glavac et al. utilized MEKC combined with UV-detection of urine samples of patients diagnosed with proteinuria without any sample pre-treatment before analysis [83]. They were able to simultaneously identify albumin (HSA), haemoglobin (HGB), myoglobin (MYO). For all three analytes, linear correlations were obtained with the correlation coefficient (R2) being not less than 0.99. Particularly, for HSA, the limit of detection (LOD) was 0.115 mg/L, the limit of quantification (LOQ) was 0.383 mg/L, a linear range between 1 and 300 mg/L and the recovery was 101.9 ± 4.5%. In another work by Wu et al., a MEKC method combining field‐amplified sample injection (FASI) with UV-detection has been developed for the analysis of albumin (HSA) and transferrin (TRF) in human urine [84]. Field-amplified stacking or injection is an on-column (on-line) preconcentration technique which is based on the different conductivity between two solution plugs. For TRF, the authors obtained the following analytical parameters of the method: LOD 0.31 mg/L, linear range 1.00–100 mg/L with R2 0.994. For HSA: LOD 0.14 mg/L, linear range 0.50–100 mg/L with R2 0.999. The recoveries of TRF and HSA were 91.1–101.7%.
Jaffuel et al. performed Aquaporin-2 (AQP2) nephrotoxicity biomarker measurement by using liquid chromatography–multiple reaction monitoring cubed mass spectrometry assay (MRM mode in LC-MS/MS) [85]. AQPs regulate numerous downstream effector signaling molecules that promote cancer development and progression [86]. In numerous cancer types, AQP expression has shown a correlation with tumor stage and prognosis. Linearity is observed within the concentration range 0.5·10−3-50·10−3 mg/L, intra and inter-assay precision ranged from 9 to 35% at the lower limit of quantification (LLOQ), and accuracy from 94 to 114%.
Overall, due to complex composition of biofluids, including the urine, liquid chromatog-raphy in particular HPLC, which is frequently coupled with MS for detection, remains a common method in bioanalytical laboratories to separate complex mixtures. A review by Novakova et al. states that in order to decrease time for an analysis without compromising resolution and separation efficiency, three main approaches in HPLC exist – the use of monolith columns, LC at high pressures and temperatures, and HPLC at ultra-high pressures [87]. By employing those approaches, time for analysis of biofluids could be done within few minutes up to half an hourhe Table 2 below shows a summary table of quantitative methods for application of electrophoresis and chromatography for protein determination in the human urine matrix.
Table 2.
Summary table of quantitative methods for application of electrophoresis and chromatography for protein determination in the human urine matrix (unless otherwise stated in the method section). CV (coefficient of variation) parameters are taken from maximum readings, for Recovery – the range is given.
| Method | Analyte | The limit of detection, LOD | Range | Analytical parameters | Experimental parameters | Reference |
|---|---|---|---|---|---|---|
| Electrophoretic with immunofixation | BJP Bence Jones Proteins | 1 mg/L | NA | NA | Migration time 33 min and 43 min depending on buffer Voltage 220V | 4 Aguzzi et al., 1993 [65] |
| Electrophoretic with immunofixation | Transferrin, retinol binding protein, b2-microglobulin | 3 mg/L (for all) | NA | NA | Migration time 33 min and 43 min depending on buffer Voltage 220V | 5 Aguzzi et al., 1993 [65] |
| HPLC high performance liquid chromatography with fluorescence detection | HSA | 0.2 mg/L | 0.2–400 mg/L (linear, R2 = 0.998) | Recovery 94.5 ± 3.8% | Retention time for HSA 16.0 min Fluorescence enhancement factor 1286 TSK gel G2000 SWXL column | 6 Ishida et al., 1996 [76] |
| HPLC with UV-detection | HSA | 3.4 mg/L | 4.3–240 mg/L (linear) | Recovery 100 ± 4.6% at 66 mg/L | Zorbax Bio Series GF-250 column | 7 Owen et al., 2005 [80] |
| CZE capillary zone electrophoresis with preconcentration techniques | HSA | 15 mg/L | 15–1000 mg/L (linear, R2 = 0.9840) | Sensitivity enhancement factor by large volume sample stacking (LVSS) 67 | Voltage 15/25 kV | 8 Bessonova et al., 2007 [70] |
| Capillary electrophoresis immunoassay with fluorescence detection (the buffer matrix) | HSA | 0.6 mg/L | 0.6–6.67 mg/L (detectable) | Recovery 91–160% | Voltage 12–15 kV | 9 Giovannoli et al., 2007 [66] |
| Q-TOF, LC-MS using a N-15 isotope | HSA | 4.84 mg/L | 4–625 mg/L (linear, R2 = 0.999) | LOQ 10.5 mg/L CV intra assay 12.6% CV inter assay 12.2% | Cone voltage for MS 40–80 V | 10 Singh et al., 2007 [81] |
| FASI-MEKC field amplified sample injection MEKC | HSA | 0.14 mg/L | 0.50–100 mg/L (linear, R2 = 0.999) | Recovery 91.1–101.7% | Retention time for HSA around 10–12 min Voltage for MEKC 20 kV Sample injection volatage 10 kV | 11 Wu et al., 2009 [84] |
| FASI-MEKC | TRF transferrin | 0.31 mg/L | 1.00–100 mg/L (linear, R2 = 0.994) | Recovery 91.1–101.7% | Retention time for TRF around 10–12 min Voltage for MEKC 20 kV Sample injection voltage 10 kV | 12 Wu et al., 2009 [84] |
| LC–MS/MS (MRM mode) | AQP2 aquaporin-2 | 5·10−4 mg/L | 5·10−4 -5·10−2 mg/L (linear) | Accuracy from 94 to 114% CV intra assay 9% CV inter assay 35% | Retention time around 5 min a Symmetry™ C18 column | 13 Jaffuel et al., 2013 [85] |
| Reversed-phase HPLC with fluorescence detection | Tamm–Horsfall protein | 0.35 mg/L | 4.5–90.0 mg/L (linear, R2 = 0.999) | Recovery 100.0–104.2% CV intraday 1.73–2.77% CV interday 2.50–5.35% | COSMOSIL 5C18-MS-II Column | 14 Akimoto et al., 2015 [88] |
| SDS-PAGE sodium dodecyl sulfate–polyacrylamide gel electrophoresis | HSA | 5 mg/L | NA | CV 21.5% | Electrophoresis time 5 min Staining time 15 min | 15 Jia et al., 2019 [67] |
| Paper-based analytical device - ion concentration polarization PAD-ICP | HSA | 10 mg/L | 50–350 mg/L (linear, R2 = 0.994) 10–500 mg/L (detectable) | Recovery 93–108% CV 11% | 16 Gao et al., 2019 [89] |
3. Detection techniques
3.1. Mass spectrometry
Mass spectrometry is a powerful analytical technique that measures the mass-to-charge ratio of ions. Firstly, molecules in a sample must be ionized, which causes a large molecule to fragment into smaller ions; then those ions are separated by a magnetic field depending on their mass to charge ratio. The results are typically presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Usually, mass spectrometry is utilized after pre-treatment of a sample by electrophoresis or chromatography.
Mass spectrometry is a widely used technique for proteomics studies. The proteome is the entire set of proteins that is produced or modified by an organism. Research on urine proteomics is important for the identification purposes of reliable biomarkers that help the diagnosis of disease [90]. In a review by Kalantari et al. on urine proteomics is stated that common techniques for urinary proteome analysis are two-dimensional gel electrophoresis followed by mass spectrometry (2DE-MS), liquid chromatography coupled to mass spectrometry (LC-MS), surface-enhanced laser desorption/ionization coupled to mass spectrometry (SELDI-TOF), and capillary electrophoresis coupled to mass spectrometry (CE-MS) [59]. By using mass spectroscopy protein constituents of urine were identified. Also reviews by Albalat et al. and Decramer et al. focus on mass spectrometry-based urinary protein and peptide profiling [33,60]. The advantages and disadvantages of different mass spectrometry methods are compared. As was mentioned in the introductory part of this review, Marimuthu et al. by using high-resolution Fourier transform mass spectrometry were able to identify 1823 proteins in the urine of healthy subjects [12]. Santuccie et al. by using mass spectrometry and sub-fractionating normal urine by successive steps (vesicle separation, CPLL, and solvent treatments) were able to identify 3429 individual proteins [13].
Two ionization techniques in mass spectrometry are commonly used for protein studies: electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) [91]. In the ESI approach, ions are produced in an electrospray in which a high voltage is applied to a liquid to create an aerosol. ESI is different from other ionization processes (e.g. MALDI) since it may produce multiple-charged ions, effectively extending the mass range of the analyzer to accommodate the kDa-MDa orders of the magnitude observed in proteins and their associated polypeptide fragments [92]. In MALDI, a sample is dispersed in a certain solid matrix (e.g. aromatic compounds), which then irradiated by the laser that causes electronic excitation of molecules in the sample matrix [93]. Both methods ESI and MALDI produce relatively high molecular ions due to mild fragmentation of those approaches. It should be noted a specific method of MS applied to biomolecules, which is called tandem mass spectrometry, also known as MS/MS. In MS/MS two or more mass analyzers are coupled together using an additional reaction step to increase their abilities to analyze samples. In general, protein identification in mass spectroscopy is performed using either bottom-up or top-down approaches. Bottom-up protein identification methods rely on enzymatic digestion to break down proteins into smaller peptides that are easier to ionize and fragment, resulting in higher sequence coverage [94]. However, in top-down methods, intact proteins are injected into the mass spectrometer and subjected to fragmentation without pre-treatment [95]. Aside from better tracking of protein modifications, an advantage of top-down protein sequencing is that it complements imaging experiments by enabling mass measurement of the intact protein that relates more directly to the MALDI IMS generated signals [96].
Analysis of a mass-spectrum, a plot of intensity as a function of the mass-to-charge ratio, could be a challenge itself, especially for big molecules like proteins, since they can be divided in many fragments resulting in a complex spectrum. Usually, the mass-spectrum is compared by software with the existing database. Several computationally feasible approaches have emerged for performing protein inference from such data [97].
Mass spectrometry in itself does not allow for quantitative analysis due to the different physicochemical properties of different peptides and proteins [[98], [99], [100]]. However quantification in MS is possible if an internal standard is used, for such there are approaches based on labeling with stable isotopes (such as N-15, O-18), involving artificial labeling of peptides and proteins. On the other hand, there are label-free approaches, in which the samples retain their native isotope composition and are compared between separate measurements. In general, they could be classified into two categories: methods that involve comparing peptide signals at the level of LC-MS analysis; methods that involve counting the number of identified peptides or acquired fragment spectra. Also, it should be noted that label-based methods are relatively laborious while label-free methods are less accurate [98]. An example of quantitative protein determination, in this case – albumin, in the urine matrix by MS methods is described in a paper by Singh et al. [81]. They used N-15 isotopically labeled albumin as an internal standard. The authors achieved the limit of detection (LOD) of 4.84 mg/L, the limit of quantification of 10.5 mg/L with the linear range being from 4 to 625 mg/L, R2 = 0.999. Chen et al. used isotope-labeled peptides as internal standards for quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for the discovery of potential bladder cancer biomarkers [101]. Bachmann et al. experimentally compared routine measurements of urinary albumin to isotope dilution tandem mass spectrometry (IDMS) [102]. They reported that median differences between the largest positive and negative biases versus IDMS were 45%, 37%, and 42% in the concentration intervals of 12–30 mg/L, 31–200 mg/L, and 201–1064 mg/L, respectively. Lieske et al. reported their current status on a developing reference method for quantification of urinary albumin by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) [103]. They used three different labeled peptides as internal standards to cover the range of clinical values (5–1270 mg/L). Recovery was between 97% and 108% with an average of 103%. In conclusion, quantitative mass spectroscopy assays for peptides and proteins are difficult to implement, and the future will see standardization of MS validation and quality control requirements.
Overall, mass spectrometry coupled with liquid chromatography is a powerful tool for the discovery and the analysis of biomarkers of diseases from biofluid samples, which includes proteomics study of urine as well [104]. Many of urinary proteome biomarkers were discovered by this technique. However, quantitative protein determination represents a challenge by following this approach [105]. Thus, mass spectroscopy could be used for biomarker discovery but not for clinical applications [60]. Another consideration for an analytical technique is the cost of analysis. Practical usage of LC-MS is limited by factors such as instrument size, cost, ease of operation, though, low cost compact mass spectroscopy detectors coupled with cinematographic separation are being developed, their costs equal to around $50000 [58].
3.2. Immunoassay
Immunoassay is a widely used technique in clinical and research laboratories to determine qualitatively and quantitatively macromolecules (usually proteins) generated from organisms [106]. Immunoassay is based on a reaction between an antigen and an antibody, which is found in the adaptive immune system of vertebrates, that includes humans [107]. The reaction is carried out in-vitro (not in a living organism), and the result of it is the formation of a complex antibody-antigen formed by spatial complementary, like lock and key, thus such interactions are highly selective. Antibody-antigen complexes are bounded by weak forces such as electrostatic forces, hydrogen bonds, hydrophobic interactions, and van der Waals forces; which means that such bindings are reversible. The specific piece of the antigen to which an antibody binds is called the epitope. Usually, in analytical papers, the analyte is the antigen while the reagent is the antibody. Immunoassay methods have been widely used in many areas of pharmaceutical analysis such as diagnosis of diseases, therapeutic drug monitoring, clinical pharmacokinetics, and bioequivalence studies in drug discovery and pharmaceutical industries [108]. Conventional immuno-chemical based urinary albumin assays for diabetic patients as well as nondiabetic patients with renal disease, including those who may suffer hypertension and cardiovascular disease, now amount to 100 million assays per year worldwide as of 2004 [79]. The widespread immunoassay methods in the pharmaceutical analysis is attributed to their inherent specificity, high-throughput, and high sensitivity for the analysis of a wide range of analytes in biological samples as well the relatively low cost of the instruments, tools, or the reagents [109].
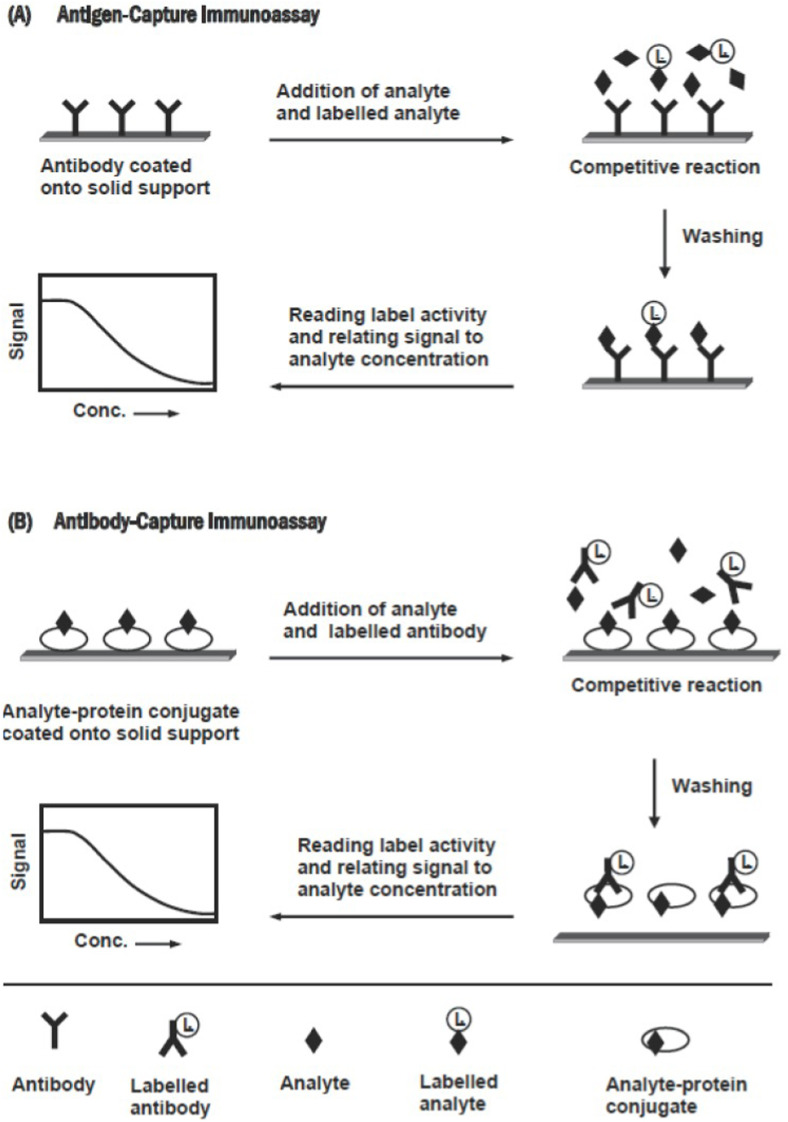
Based on signal detection immunoassays could be classified as follows:
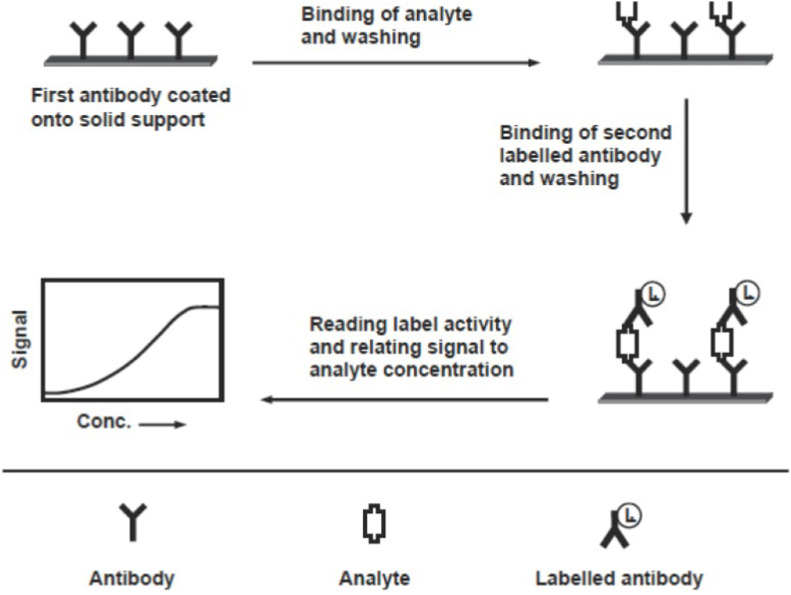
Immunoassay experiments could be classified into types – competitive and non-competitive [106,109]. The competitive immunoassay relies on the competition between the antigen of interest (the analyte) and a constant amount of a similar but labeled antigen for a limited amount of specific antibody. When these immunoanalytical reagents are mixed and incubated, the analyte is bound to the antibody-forming an immune complex. This complex is separated from the unbound reagent fraction by physical or chemical separation technique, though sometimes separation is not required. Based on whether the separation step is or is not required, immunoassay methods can be classified into a heterogeneous or a homogeneous assay, respectively. The analysis is achieved by measuring the label activity in either of the bound or free fraction. On the other hand, the non-competitive assay, also called immunometric, uses an excess of labeled specific antibody toward the analyte of interest. It requires two antibodies that bind to non-overlapping epitopes on the analyte molecule. One of the two antibodies is bound to the solid phase, and the second one is labeled and used for detection. Labels commonly used in immunoassays include radioisotopes, fluorophores, and enzymes. For both types, the schematic diagrams are given below (for the competitive – Fig. 2 , for the non-competitive – Fig. 3 ), reprinted from Ref. [109].
-
•
Radioimmunoassay RIA. Detection is based on radioactive labeling of antigen with unstable isotopes such as I-125, C-14, H-3.
-
•
Enzyme immunoassay EIA with sub-categories: ELISA – Enzyme-linked immunosorbent assay; EMIT – Enzyme-multiplied immunoassay technique; MEIA – Microparticle enzyme immunoassay.
-
•
Fluoroimmunoassay FIA
-
•
Chemiluminescence immunoassay CLIA
-
•
Immunonephelometry IN. Detection is based on an antigen-antibody complex scattering light.
-
•
Immunoturbidimetry IT. Detection is based on an antigen-antibody complex blocking light thus increasing the turbidity of the sample.
Fig. 2.
Schematic diagram for the competitive immunoassays. Reprinted from Ref. [109].
Fig. 3.
Schematic diagram for the non-competitive immunoassay. Reprinted from Ref. [109].
As it was mentioned in the introductory part, the concentration of albumin higher than 2 mg/L could be indicative of potential kidney diseases [10]. Subsequently for many immunoassays methods for albumin determination in urine the limit of detection (LOD) of magnitude around 1 mg/L would be enough for clinical purposes, thus priority should be given to the parameters such as specificity, accuracy, costs, etc. However for low abundant proteins achieving lower LOD is important [110]. In 1980–1990 common immunoassays for urinary proteins for clinical purposes were developed. Comper et al. compared four immunoassays (RIA, IN, two IT) and high-performance liquid chromatography (HPLC) [79]. The correlation coefficients calculated were high (>0.85) which is indicative of a strong linear relationship between all assays studied. While the correlation was high, the slopes of the regression line were significantly different from 1, it means that certain methods tend to underestimate or overestimate. For example, the authors found that the Beckman immunonephelometry assay consistently gives values approximately threefold lower than the Dade-Behring immunoturbidimetry assay. There have been numerous other studies that have found similar levels of variation between different immunoassays and other techniques, such as in the paper by Shaikh et al. [111]. The authors claim that theirs LC-MS and HPLC assays both performed poorly at concentrations below 20 mg/L, while immunoturbidimetric assay underestimated albumin compared with LC-MS. Hoofnagle et al. state that immunoassays have four common problems of which a researcher should be aware [112]. Assay performed with one set of reagents does not measure the same concentration as another which results in patients having measurements performed on the same platform for continuity of care. The other three problems results in falsely low or high results depending on the analyte concentration, antibodies used. While immunoassays methods have their shortcomings they are still widely used in clinical practice. Modern immunoassays are actively being developed and some of their analytical parameters are given in the summary Table 3 .
Table 3.
Summary table of quantitative methods for application of immunoassay for protein determination in the human urine matrix (unless otherwise stated in the method section). CV (coefficient of variation) parameters are taken from maximum readings, for Recovery – the range is given.
| Method | Reagent for detection (if any) | Analyte | The limit of detection, LOD | Range | Analytical and experimental parameters | Reference |
|---|---|---|---|---|---|---|
| Fluoroimmunoassay | Fluorescein isothiocyanate | HSA | 0.5 mg/L | 0.5–20 mg/L (linear R2 = 0.99) | Total time for experiment 4–6 h Recovery 105 ± 7% | 19 Chavers et al., 1984 [113] |
| Immunoturbidimetry | No reagent | HSA | 4 mg/L | 4–35.5 mg/L (linear) | Time so screen 15 min Recovery 95%–104% | 20 Lloyd et al., 1987 [114] |
| Immunonephelometry | No reagent | HSA | 0.34 mg/L | 0.34–43.0 mg/L (detectable) | Automated version 240 samples an hour CV (intraassay) 6% CV (interassay) 9% | 21 Marre et al., 1987 [115] |
| Immunoturbidimetry | No reagent | HSA | 2 mg/L | 2–260 mg/L (detectable) | CV (interassay) 4.8% | 22 Bakker et al., 1988 [116] |
| Chemiluminescence | Acridinium ester | HSA | 0.01 mg/L | 0.3–10 mg/L (detectable) | Recovery 90–106% | 23 Horton et al., 1989 [117] |
| Radioimmunoassay | Na125I | HSA | 0.015 mg/L | 1.18–14.96 mg/L (detectable) | Recovery 90–106% | 24 Horton et al., 1989 [117] |
| Time resolved Fluorescence Resonance Energy Transfer | Europium chelate (donor)/cyanine 5 (acceptor) | HSA | 5.5 mg/L | 10–320 mg/L (detectable) | Experiment time 12 min Recovery 103–122% CV (within-run) 6.9–10% CV (between-run) 7.5–13% | 25 Qin et al., 2003 [118] |
| Chemiluminescence enzyme-linked immunosorbent assay based on Avidin–biotin | 4-methoxy4-(3-phosphatephenyl)-spiro-(1,2-dioxetane-3,2 -adamantane) (AMPPD) to detect enzyme activity | HSA | 0.089 mg/L | 0.15–15 mg/L (linear R2 = 0.9902) | Low usage of antibody CV (intra-assay) 10.7% CV (inter-assay) 15% Recovery 112% | 26 Zhao et al., 2005 [119] |
| Chemiluminescence enzyme-linked immunosorbent assay based on fluorescein-isothiocyanate (FITC) | 4-methoxy4-(3-phosphatephenyl)-spiro-(1,2-dioxetane-3,2 -adamantane) (AMPPD) to detect enzyme activity | HSA | 0.089 mg/L | 0.15–15 mg/L (linear R2 = 0.9906 | CV (intra-assay) 10.5% CV (inter-assay) 11.9% Recovery 109% | 27 Zhao et al., 2005 [119] |
| Magnetic two-site immunoassay | Dextran-coated nanoscaled to detect magnetic permeability | HSA | 5 mg/L | 0–400 mg/L (linear) | Experiment time 6.5 min CV 11% | 28 Lu et al., 2006 [120] |
| Capillary electrophoresis immunoassay with spectrophotometric detection | Hydroxylamine | HSA | 0.60 mg/L | 0–6.67 mg/L (detectable) | Voltage 12 kV Recovey 91–160% | 29 Giovannoli et al., 2007 [66] |
| Electrochemical immunosensor | Fe3O4/Au colloid -modified gold electrode | NMP22 Nuclear Matrix Protein 22 | 5·10−4 mg/L | 1.2·10−3-0.2 mg/L (linear, R2 = 0.9932) | CV 3.9% | 30 Ning et al., 2007 [121] |
| Resonance scattering (RS) spectral immunoassay | Cu2O for RS | HSA | 7.2·10−6 mg/L | 1.4·10−5–4.3·10−4 mg/L | CV 4.5% Recovery 92.1–106.8% | 31 Jiang et al., 2009 [122] |
| Fluoroimmunoassay, a quantum-dot (QS) optical immunosensor (the buffer matrix) | CdSe/ZnS QS | HSA | 0.032 mg/L | 0.2–200 mg/L (detectable) | Low cost immunosensor. | 32 Tu et al., 2012 [123] |
| Time-resolved fluorescence resonance energy transfer immunoassay | Europium based oligonucleotides (donor)/oligonucleotide(acceptor) | HSA | 0.0039 mg/L | 0.0039–0.5 mg/L (linear) 0.0039–1 mg/L (detectable) | Experiment time 40 min Tested in serum, salive, urine CV 3% | 33 Wang et al., 2012 [124] |
| Chemiluminescence lateral flow immunoassay | Luminol/enhancer/H2O2 with silicon photosensor | HSA | 2.5 mg/L | 2.5–850 mg/L (detectable) | CV 20% Recovery 85–112% | 34 Zangheri et al., 2016 [125] |
| Amperometric immunosensor (the buffer matrix) | Carbon nanotubes/gold nanoparticles screenprinted electrodes | p53 protein | 6.1·10−4 mg/L | 8.4·10−4-0.437 mg/L (linear) | CV 10% Recovery 84–144% | 35 Giannetto et al., 2017 [126] |
| Piezoelectric immunosensor, label-free immunoassay | Quartz crystal coated with a silver electrode | HSA | 0.0095 mg/L | 0.1–100 mg/L | Diagnostic sensitivity 98.7% Diagnostic specificity 100% | 36 Theansun et al., 2017 [127] |
| Time-resolved fluorescence immunoassay | Europium (III) and samarium (III) | cystatin-C | 1.26 10−3 mg/L | 0.42 10−3-0.9563 mg/L (linear, R2 = 0.9991) | Recovery 99.36% CV 3.2–5.9% (inter) CV 3.4–6.3% (intra) | 37 Liu et al., 2017 [128] |
| Time-resolved fluorescence immunoassay | Europium (III) and samarium (III) | β2-microglobulin | 2.13 10−3 mg/L | 0.86 10−3-0.9754 mg/L (linear, R2 = 0.9998) | Recovery 100.18% CV 3.0–6.8% (inter) CV 3.1–8.4% (intra) | 38 Liu et al., 2017 [128] |
| Chemiluminescence immunoassay | Magnetic microparticles using carboxylic acid groups, acridinium | monomeric laminin-γ2 | 1.0 10−5 mg/L | 1.0 10−5-0.02 mg/L (detectable) | Recovery 82.3–96.2% | 39 Nakagawa et al., 2017 [129] |
| Fluoroimmunoassay, optical microchips | Streptavidin-fusion based variants | HSA | 0.65 mg/L | Limit of quantification 2 mg/L | 40 Semeradtova et al., 2018 [130] | |
| Electrochemical ELISA | Gold sensor chips with a hair comb design | CFHR1 complement factor H-related 1 | 1.29 10−3 mg/L | 1 10−3-0.01 mg/L (linear) | 41 Arya et al., 2018 [131] | |
| Electrochemical ELISA | Gold sensor chips with a hair comb design | NUMA1 nuclear mitotic apparatus protein 1 | 0.97 10−3 mg/L | 1 10−3-0.1 mg/L (linear) | 42 Arya et al., 2018 [131] | |
| Electrochemical immunosensor | Gold nanoparticles-platinum nanoparticles-metal organic frameworks | NMP22 nuclear matrix protein 22 | 1.7·10−6 mg/L | 5·10−6-5·10−4 mg/L (linear, R2 = 0.9833) 5·10−4-0.02 mg/L (linear, R2 = 0.9909) | CV 1.93% Recovery 96%–106% | 43 Zhao et al., 2019 [110] |
| Fluoroimmunoassay, graphene oxide-mediated fluorescence quenching aptasensor | 87-nucleotide ssDNA as aptamer | HSA | 0.05 mg/L | 0.1–14.0 mg/L (detectable) | Cheap 0.3$ per reaction | 44 Chawjiraphan et al., 2020 [132] |
| Enzyme fluroimmunoassay, sandwich-type ultramicroELISA | 4-methylumbelliferil phosphate/diethanolamine-Hcl/sodium azide | HSA | 1.44·10−3 mg/L | 1.44·10−3-0.2 mg/L (detectable) | CV 3.98–4.35% (intra) CV 7.59–8.92% (inter) Recovery 94.26–98.50% Time 1.5 h | 45 Valle et al., 2020 [62] |
3.3. Fluorescence spectroscopy
Fluorescence is a luminescence phenomenon in which a compound emits light after absorption of electromagnetic irradiation [133]. In most cases, the emitted light has a longer wavelength, and therefore lower energy, than the absorbed radiation. Often in chemical analysis, a sample is irradiated in UV or visible range, and as a result, fluorescence occurs in the visible and near-infrared region respectively. Fluorescent materials cease to glow nearly immediately when the radiation source stops, unlike phosphorescent materials, which continue to emit light for some time after. Fluorescence spectroscopy is one of the most common techniques with applications in geology, chemistry, medicine, and astronomy [133]. Because of the sensitivity that the method affords, fluorescent molecule concentrations as low as 1 part per trillion can be measured [134]. Fluorescence spectroscopy can be used on its own, and also it is often used in conjunction with high-performance liquid chromatography as a detector. Often fluorescence detection is used in immunoassays to either label antigen or antibody.
Usually, analyte molecules do not emit strong luminescence for their analytical detection. Hence an analyte usually modified with a fluorophore, or a fluorescent probe, in other words. The fluorophore (or a fluorescent probe) is a fluorescent chemical compound that can re-emit light upon light excitation. A suitable fluorescent probe should have the following characteristics: (i) is conveniently excitable, without simultaneous excitation of the biological matrix, and detectable with conventional instrumentation; (ii) is bright, that is, possesses a high molar absorption coefficient at the excitation wavelength and a high fluorescence quantum yield, (iii) is soluble in relevant buffers, cell culture media or body fluids, (iv) is sufficiently stable under relevant conditions, (v) has functional groups for site-specific labeling, (vi) has reported data about its photophysics, and (vi) is available in reproducible quality [135]. Most common fluorophores are usually organic molecules, that typically contain several combined aromatic groups or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to a macromolecule, serving as a marker (or dye, or tag, or reporter) for affine or bioactive reagents (antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, i.e., fluorescent imaging and spectroscopy. Some examples of widely used fluorophores are fluorescein, and by its amine reactive isothiocyanate derivative fluorescein isothiocyanate, methylene blue. Over the last two decades, the role of semiconductor quantum dots (QD) and other types of nanoparticles have grown considerably in bionanalysis and bioimaging. Petryayeva et al. conclude that QDs as fluorophores and for other bioimaging purposes offer nontrivial advantages, such as the brightness needed for sensitive detection, the photostability needed for tracking dynamic processes, the multiplexing capability needed to elucidate complex systems or the nanoscale interface needed for biomolecular engineering of novel probes and biosensors [136]. The fluorescent labeling in immunoassays is the common practice (see page 20).
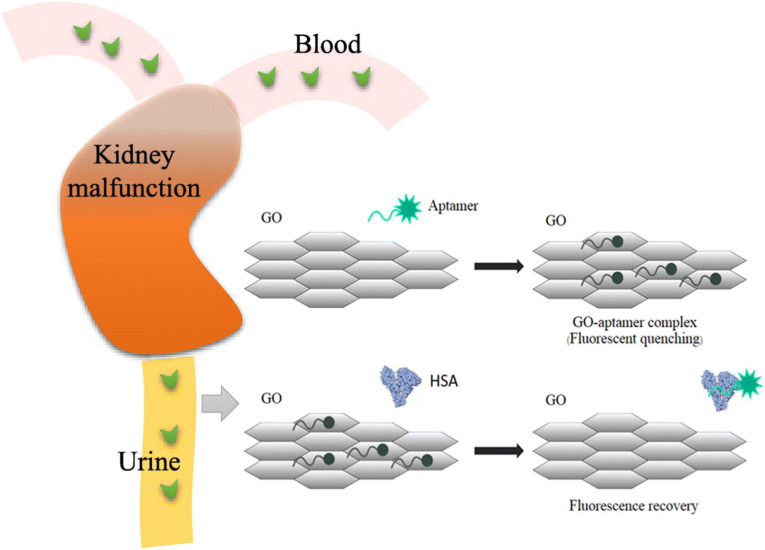
A method of fluorescence, where two light-sensitive molecules, that transfer energy between them, is called fluorescence resonance energy transfer. Fluorescence resonance energy transfer (FRET) involves the transfer of energy from a fluorescent donor molecule via a nonradiative dipole-dipole interaction to an acceptor molecule. The emission of the energy transfer-excited acceptor can be distinguished from donor emission by the use of a longer-wavelength filter (spatial resolution). The fluorescent lanthanide chelates of europium and terbium are attractive FRET donors. The use of a long-lifetime donor fluorophore and a short-lifetime acceptor fluorophore together with pulsed-laser excitation and time-resolved detection (temporal resolution) are effective in reducing the background signal. Qin et al. used time-resolved FRET for the determination of urinary albumin [118]. Particularly, they used, in the competitive homogeneous assay, an albumin-specific monoclonal antibody labeled with a stable fluorescent europium chelate as donor and an albumin labeled with cyanine 5 (Cy5) as acceptor. The limit of detection was 5.5 mg/L with the detectable range being 10–320 mg/L. Another interesting approach to fluorescence spectroscopy is the fluorescent quenching. Chawjiraphan et al. developed a method to determine HSA in urine and serum by the graphene oxide-mediated (GO) fluorescence quenching aptasensor [132]. When albumin was added to the complex GO with the fluorescence-labeled aptamer, the aptamer detached from the complex to bind albumin, which resulted by an increase in fluorescence intensity, as shown in Fig. 4 . The limit of detection was 0.05 mg/L and the detection range is 0.1–14.0 mg/L. Other quantitative works using the fluorescence spectroscopy are summarized in Table 4 .
Fig. 4.
Schematic of graphene oxide-mediated fluorescence quenching aptasensor for the detection of albuminuria in urine and HSA in human serum. When albumin was added to the complex GO with the fluorescence-labeled aptamer, the aptamer detached from the complex to bind albumin, which resulted by an increase in fluorescence intensity. Reprinted from Refs. [132].
Table 4.
Summary table of quantitative methods for application of fluorescence spectroscopy for protein determination in the human urine matrix (unless otherwise stated in the method section). CV(coefficient of variation) parameters are taken from maximum readings, for Recovery – the range is given.
| Method | Fluorophore | Analyte | The limit of detection, LOD | Range | Analytical and experiment parameters | Reference |
|---|---|---|---|---|---|---|
| Fluoroimmunoassay FIA | Fluorescein isothiocyanate (FITC) | HSA | 0.5 mg/L | 0.5–20 mg/L (linear R2 = 0.99) | Total time for experiment 4–6 h Recovery 105 ± 7% | 47 Chavers et al., 1984 [113] |
| HPLC high performance liquid chromatography with fluorescence detection | 8-anilino-1-naphtalenesulfonic acid | HSA | 0.2 mg/L | 0.2–400 mg/L (linear, R2 0.998) Recovery 94.5 ± 3.8% | Retention time for HSA 16.0 min TSK gel G2000 SWXL column | 48 Ishida et al., 1996 [76] |
| Time resolved Fluorescence Resonance Energy Transfer | Europium chelate (donor)/cyanine 5 (acceptor) | HSA | 5.5 mg/L | 10–320 mg/L (detectable) | Experiment time 12 min Recovery 103–122% CV (within-run) 6.9–10% CV (between-run) 7.5–13% | 49 Qin et al., 2003 [118] |
| Spectrofluorimetry | 4-dimethylamino-2,5-dihydroxychalcone (DMADHC) | HSA | 0.5 mg/L | 1–95 mg/L (linear, R2 = 0.99) | CV 1.15% Recovery 95.6–103.0% | 50 Xu et al., 2005 [137] |
| Chemiluminescence enzyme-linked immunosorbent assay based on fluorescein-isothiocyanate FITC | 4-methoxy4-(3-phosphatephenyl)-spiro-(1,2-dioxetane-3,2 -adamantane) (AMPPD) to detect enzyme activity | HSA | 0.089 mg/L | 0.15–15 mg/L (linear R2 = 0.9906 | CV (intra-assay) 10.5% CV (inter-assay) 11.9% Recovery 109% | 51 Zhao et al., 2005 [119] |
| Capillary electrophoresis immunoassay with fluorescence detection (the buffer matrix) | Fluorescein 5-isothiocyanate (FITC) | HSA | 0.6 mg/L | 0.6–6.67 mg/L (quantification range) | Voltage 12–15 kV Recovery 91–160% | 52 Giovannoli et al., 2007 [66] |
| Synchronous fluorescence determination | triphenylmethane acid dye methyl blue | HSA | 0.03 mg/L | 0.03–266.0 mg/L (linear, R2 = 0.9974) 266.0–665.0 mg/L (linear, R2 = 0.9911) |
Recovery 99.0–103.3% CV 2.3% | 53 Hou et al., 2007 [138] |
| Synchronous fluorescence determination | 5-Aminosalicylic acid (5-ASA) | HSA | 0.552 mg/L | 1.60–414 mg/L (linear) | Recovery 100.6–103.7% CV 1.26% | 54 Cui et al., 2008 [139] |
| Spectrofluorimetry | CdS (core)/SiO2 (nanoparticles) | BSA | 0.18 mg/L | 0.6–30 mg/L (linear, 0.9998) | Recovery 94–105% CV 2.1% | 55 Zhu et al., 2009 [140] |
| Synchronous fluorescence determination (the buffer matrix) | Methylen blue | BSA | 0.0089 mg/L | 0.08–40 mg/L (linear, R2 = 0.998) | CV 1.7% | 56 Liu et al., 2010 [141] |
| Spectrofluorimetry (the human plasma) | Terbium-danofloxacin (Tb3+-Dano) | HSA | 5.4 mg/L | 13.3–66.5 mg/L (linear, R2 = 0.9899) | CV (intra-assay) 6.44% CV (inter-assay) 8.08% Recovery 94.3–97.8% | 57 Ramezani et al., 2012 [142] |
| Spectrofluorimetry immunoassay (the buffer matrix) | CdSe/ZnS quantum dots | HSA | 0.032 mg/L | 0.2–200 mg/L (detectable) | NA | 58 Tu et al., 2012 [123] |
| Time-resolved fluorescence resonance energy transfer immunoassay | Europium based oligonucleotides (donor)/oligonucleotide(acceptor) | HSA | 0.0039 mg/L | 0.0039–0.5 mg/L (linear) 0.0039–1 mg/L (detectable) | Experiment time 40 min Tested in serum, saliva, urine CV 3% | 59 Wang et al., 2012 [124] |
| Constant-energy synchronous fluorescence | No fluorescent label | HSA | 0.007 mg/L | 0.1–220 mg/L (linear) | CV 2.2% | 60 Madrakian et al., 2015 [143] |
| Spectrofluorimetry in near infrared region | Based on hemicyanine dye | HSA | 1.74 mg/L | 27–2750 mg/L (linear, R2 = 0.9994) | Recovery 107.23–108.79% CV 1.00% | 61 Li et al., 2016 [144] |
| Spectrofluorimetry | poly(thymine) (poly T)-templated copper nanoparticles (CuNPs) | HSA | 5.45 mg/L | 10–166 mg/L (linear, R2 = 0.9976) | Recovery 97–101% | 62 Chen et al., 2017 [145] |
| Spectrofluorimetry (the human serum matrix) | CuInZnS | HSA | 3 mg/L | 5.0–6650 mg/L | Recovery 87.57–94.39% CV 1.96% | 63 Gui et al., 2017 [146] |
| Spectrofluorimetry in near infrared region | Dicyanomethylene-4H-chromene-derived | HSA | 1.26 mg/L | 1.26–232 mg/L (detectable) | Good agreement with a conventional methods | 64 Rajasekhar et al., 2017 [147] |
| Time-resolved fluorescence immunoassay | Europium (III) and samarium (III) | cystatin-C | 1.26 10−3 mg/L | 0.42 10−3-0.9563 mg/L (linear, R2 = 0.9991) | Recovery 99.36% CV 3.2–5.9% (inter) CV 3.4–6.3% (intra) | 65 Liu et al., 2017 [128] |
| Time-resolved fluorescence immunoassay | Europium (III) and samarium (III) | β2-microglobulin | 2.13 10−3 mg/L | 0.86 10−3-0.9754 mg/L (linear, R2 = 0.9998) | Recovery 100.18% CV 3.0–6.8% (inter) CV 3.1–8.4% (intra) | 66 Liu et al., 2017 [128] |
| Fluoroimmunoassay, optical microchips | Streptavidin-fusion based variants | HSA | 0.65 mg/L | NA | Limit of quantification 2 mg/L | 67 Semeradtova et al., 2018 [130] |
| Fluoroimmunoassay, graphene oxide-mediated fluorescence quenching aptasensor | 87-nucleotide ssDNA as aptamer | HSA | 0.05 mg/L | 0.1–14.0 mg/L (detectable) | Cheap 0.3$ per reaction | 68 Chawjiraphan et al., 2020 [132] |
| Enzyme fluroimmunoassay, sandwich-type ultramicroELISA | 4-methylumbelliferil phosphate/diethanolamine-Hcl/sodium azide | HSA | 1.44·10−3 mg/L | 1.44·10−3-0.2 mg/L (detectable) | CV 3.98–4.35% (intra) CV 7.59–8.92% (inter) Recovery 94.26–98.50% Time 1.5 h | 69 Valle et al., 2020 [62] |
3.4. The Infrared and Raman spectroscopies
The absorption of infrared radiation IR excites vibrational transitions of molecules [148]. The infrared spectral region covers wavelengths from 780 nm to 1000 μm which can be further subdivided into the near-infrared region from 780 nm to 2500 nm, the mid-infrared region from 2500 nm to 50 μm and the far-infrared region from 50 μm to 1000 μm. Since vibrational frequency and probability of absorption depend on the strength and polarity of the vibrating bonds, they are influenced by intra- and intermolecular effects [149]. The approximate position of an infrared absorption band is determined by the vibrating masses and the type of bond (single, double, triple), the exact position by electron-withdrawing or donating effects of the intra- and intermolecular environment and by coupling with other vibrations. Information that can be derived from the infrared spectrum:
-
•
Chemical structure of the vibrating group
-
•
Chemical properties of neighbouring groups in a molecule
-
•
Redox state
-
•
Bond parameters
-
•
Hydrogen bonding
-
•
Electric fields
-
•
Conformational freedom
Basically, all polar bonds contribute to the infrared absorption so there is no need to specifically label biomolecules to detect them. However, the infrared spectrum of biomolecules is challenging to analyze since they are complex with many overlapping bands. Certain mathematical and statistical procedures have to be utilized to analyze the resulting spectrum. The IR spectroscopy is often used to identify organic structures because functional groups give rise to characteristic bands both in terms of intensity and position (frequency). The IR spectra of organic molecules are often interpreted as having two regions: functional group region (>1500 cm−1 = 6.7 μm), and fingerprint region (<1500 cm−1). In the fingerprint region, there are many troughs that form an intricate pattern that can be used as a fingerprint to determine the compound. Modern infrared spectrometers are usually Fourier transform infrared (FTIR) spectrometers [149]. The FTIR spectrometer works differently than the dispersive spectrometer where a monochromatic light beam is absorbed by a sample. This technique shines a beam containing many frequencies of light at once and measures how much of that beam is absorbed by the sample. Next, the beam is modified to contain a different combination of frequencies, giving a second data point. This process is rapidly repeated many times over a short timespan. Afterward, a computer takes all this data and works backward to infer what the absorption is at each wavelength by a Fourier transform.
Another analytical technique that relies on molecular vibrations is Raman spectroscopy [150]. Raman spectroscopy is a powerful technique to determine vibrational modes of molecules to provide a structural fingerprint, like infrared spectroscopy, by which molecules can be identified. Infrared spectroscopy typically yields similar information because certain molecular vibrations can occur in both techniques but some only detectable in either, which makes them complimentary as well. Also, transitions that have large Raman intensities often have weak IR intensities and vice versa. Raman spectroscopy relies upon the inelastic scattering of photons. The laser light interacts with molecular vibrations, phonons or other excitations in the system, resulting in the energy of the laser photons being shifted up or down. The shift in energy gives information about the vibrational modes in the system. Raman spectra for biomolecules is hard to analyze due to overlapping bands, the same issue as in the IR spectroscopy. Certain mathematical procedures must be employed on Raman spectra to analyze a sample.
Despite difficulties in the analysis of large biomolecules such as proteins, the IR, and Raman spectroscopy make their advances in this regard. For the protein analysis with IR, the MIR region plays an important role since it includes bands that predominantly arise from three conformationally sensitive vibrations arising from the peptide backbone—namely, amide I, amide II, and amide III [151]. Amide group vibrations of the backbone have attracted much attention in protein IR spectroscopy, as they are native to all proteins and inform on secondary conformation and solvation. Such bands include the amide I region, between 1600 and 1700 cm-1, primarily resulting from C O stretching vibration, the amide II band, related to CN stretch and NH in-plane bending, the amide III vibration, associated with CN stretching, NH bending, and CO in-plane bending, and, finally, the amide A (NH stretch) band. Conventional IR and Raman methods often lack enough sensitivity. Surface-Enhanced Infrared Absorption Spectroscopy (SEIRAS) [152], and Surface-Enhanced Raman spectroscopy SERS [[153], [154], [155]] improve sensitivity to overcome this problem. Both SEIRAS and SERS utilize the surface plasmon effect from the interaction of light with metallic nanoparticles. SERS has an enhancement factor (of the order of 106–1012) compared to the traditional Raman signal [156]. For SEIRA, the surface-enhancement is comparatively modest (~101–103) [157]. Although the enhancement factor of SEIRAS is smaller than that of SERS, the cross-section for IR absorption is several orders of magnitude higher than the corresponding Raman cross-section. Despite the modest enhancement factor, the SEIRAS technique may be sufficient for many applications [152].
It should be emphasized again that the IR and Raman spectra are rich in data, and mathematical interpretations of those along with the right calibration procedure are important for analysis. Jessen et al. developed a method for simultaneous determination of glucose, triglycerides, urea, cholesterol, albumin and total protein in human plasma by Fourier transform infrared spectroscopy [158]. They put their samples of 40 μL without any reagent treatment or preconcentration into water-thermostated CaF cells to measure transmission. The spectra were recorded between 3100 and 950 cm−1. In the analysis, the first derivative of the original spectra was used. The first derivative was computed by using the Savitzky–Golay algorithm with 9 smoothing points. For each sample, two IR spectra were recorded and the mean spectrum was subsequently corrected by the subtraction of a water blank spectrum. Since the IR spectra are complex, a calibration procedure is important. The authors, used between 144 and 169 patient samples for each analyte to cover a wide range of concentrations. Jessen et al. compared their results between the FTIR method by other reference methods, which were mainly based on immunoassays. For each analyte, the FTIR spectra are processed by a stepwise selection of wavenumbers giving the optimal least-squares correlation between the measured IR signal, and the analyte concentration measured by the reference method. The correlation coefficients between the FTIR and the reference method were 0.87<R2 < 1.00, and specifically for albumin it was R2 = 0.92. For albumin, the calibration range was 20–60 g/L. The authors concluded that the developed FTIR methods use a simple and robust technology to achieve stable and accurate results that meet international quality criteria for the measurement of glucose, triglycerides, urea, cholesterol, albumin, and total protein in human plasma.
Ma et al. synthesized thiourea-functionalized silica nanoparticles and used them for the preconcentration of albumin in urine [159]. The adsorbent with the analyte was then used for near-infrared diffuse reflectance spectroscopy measurement directly and the partial least squares model was established for quantitative prediction. Forty samples were taken as a calibration set for establishing the PLS model and 17 samples were used for validation of the method. The principle of the PLS method is the combination of different signal processing methods and wavenumber regions. The wavenumber regions that produced the best results in cross-validation were selected. The correlation coefficient and the root mean squared error of cross-validation is 0.9986 and 0.43, respectively. The residual predictive deviation value of the model is as high as 18.8. The recoveries of the 17 validation samples in the concentration range of 3.39–24.39 mg/L are between 95.9 and 113.1%. In another work by Hall et al., the authors performed multiple linear least-squares (MLR) and an enhanced partial least-squares (PLS) using this time the second-derivative of the absorbance data to determine analytes, including protein, in unmodified human serum [160]. Pezanniti et al. used a polynomial based spectral smoothing method to the urine matrix [161]. In general, all the mentioned works requires no specific reagents, they are fast and are able to quantify multiple analytes with one spectrum.
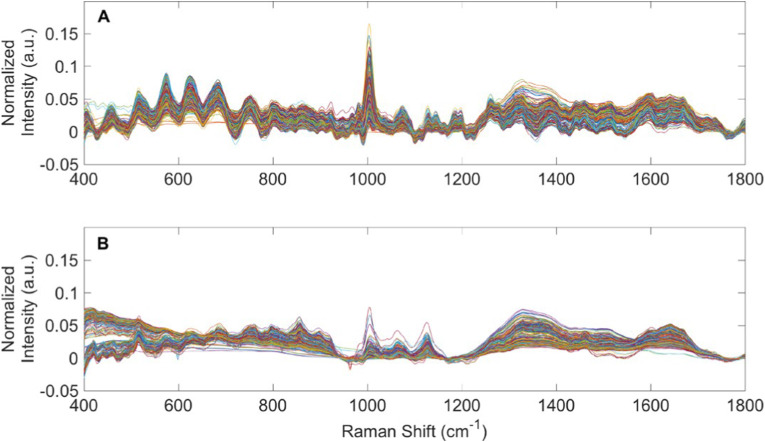
Premasiri et al. investigated urine by Raman spectroscopy [5]. The result was that since urea is in high concentration, normal Raman spectroscopy is sufficient for analysis of the analyte. However, all other components are in low concentrations requiring the use of SERS. Bispo et al. correlated the amount of urea, creatinine, and glucose in urine from patients with diabetes mellitus and hypertension with the risk of developing renal lesions by means of Raman spectroscopy and principal component analysis [162]. They took Raman spectra of control subjects and patients with illnesses, and then the spectra were submitted to principal component analysis (PCA) followed by discriminant analysis. The essence of PCA is to yield an orthogonal basis in which different individual dimensions of the data are uncorrelated. The discriminating model showed a better overall classification rate of 70%. The same approach was utilized by Almeida et al. to estimate concentrations of urea and creatinine in the human serum of normal and dialysis patients [163]. PCA showed high discrimination between dialysis and normality (95% accuracy). Senger et al. analyzed by the Raman spectroscopy three types of patients: patients receiving peritoneal dialysis (PD) therapy for end-stage kidney disease; patients receiving peritoneal dialysis (PD) therapy for end-stage kidney disease; urine from healthy human volunteers, as shown in Fig. 5 [164]. The authors used the set of mathematical procedures on spectra which included spectral processing (e.g., truncation, baselining, and vector normalization); principal component analysis (PCA); statistical analyses (ANOVA and pairwise comparisons); discriminant analysis of principal components (DAPC); and testing DAPC models using a leave-one-out build/test validation procedure. Their approach was able to identify “unknown” urine specimens as from PD patients or healthy human volunteers with better than 96% accuracy (with better than 97% sensitivity and 94% specificity).
Fig. 5.
Raman spectra of PD patient urine and spent dialysate.(A) Averaged, baselined, and vector normalized Raman spectra from 362 urine specimens obtained from patients receiving PD therapy for ESKD. (B) Averaged, baselined, and vector normalized Raman spectra from 395 spent PD dialysate specimens. Reprinted from Ref. [164].
Chamuah et al. used a blu-ray DVD as a SERS substrate for the detection of albumin, creatinine, and urea in urine [165]. The minimum concentration of albumin, creatinine, and urea which can be measured by Raman spectrometer is 0.1 mg/L, 0.2 mg/L, and 0.6 mg/L respectively. The authors investigated correlations between the SERS signal taken from their signature Raman peaks (albumin 1208 cm−1, creatinine 1444 cm−1, urea 1018 cm−1) and the concentration of the respective substance. The linear correlation coefficients (R2) were 0.956 for albumin, 0.976 for creatinine, 0.982 for urea. Dou et al. determined quantitatively concentration of glucose, acetone, urea, and creatinine in the human urine [166]. Their detection limits were 410 mg/L, 400 mg/L, 49 mg/L, and 15 mg/L, respectively. Good correlations were obtained between the concentrations and the Raman intensities with the R values of 0.924 (glucose), 0.987 (acetone), 0.962 (urea), and 0.923 (creatinine), respectively. The authors showed that the excitation wavelength is an important factor to reduce fluorescence of urine, which reduces the signal to noise ratio. The laser with a wavelength of 750 nm performed better than 600 nm, 650 nm, 700 nm. Gaipov et al. have begun to study patients with elevated protein content in urine vs control group (normal protein content) using combination of hybrid Brillouin-Raman spectroscopy and SERS [167].
Chamuah et al. assigned the signature Raman peaks to the corresponding vibrations in analytes [165]:
-
1.
For albumin – peaks at 1208 cm−1 and 1370 cm−1 are attributed to SO2 symmetric stretch and CH bond deformation of albumin.
-
2.
For creatinine – the Raman signature peaks are observed at 888 cm−1, 958 cm−1, and 1444 cm-1 which could be due to C O stretching, C–C stretching and CH3 anisometric deformation respectively.
-
3.
For urea – a very strong Raman peak at 1018 cm−1 which corresponds to C–N stretching mode.
Throughout is important consideration for laboratory tests. Zhu et al. performed quantitative SERS detection of creatinine in urine, and then compared with a clinically validated enzymatic “creatinine kit” [168]. They wrote that SERS detection process could be completed within 2 min compared with 11 min for the creatinine kit, indicating the practicality of the quantitative SERS technique as high-throughput platform for relevant clinical and forensic analysis.
4. Urinary protein biomarkers
Human urine contains thousands of individual proteins [12,13]. Excessive presence of some of them are associated with certain diseases related not only to renal diseases, but also to cancer, diabetes, and infections. Those molecules that are associated with a certain biological condition are called biomarkers. Most of those biomarkers in human urine were identified by comparing patients with healthy individuals by the means of chromatography or electrophoretic separation followed by mass spectroscopy, or by the immunoassay. Literature review of urinary protein biomarkers with related diseases is summarized in Table 5 , corresponding diagnostic sensitivity and diagnostic specificity are given [169]. Some of the listed research papers used Western blot as a technique [170]. Due to the usage of antibodies, this technique was counted as the immunoassay in Table 5. Most of the methods mentioned followed established procedures. To get a general idea of those experimental methods, refer to the previous paragraphs about separation and detection techniques, and Table 2, Table 3, Table 4
Table 5.
Protein biomarkers in urine. Sensitivity is the percentage of individuals who have a given disorder who are identified by the method as positive for the disorder. Specificity is the percentage of individuals who do not have a given condition who are identified by the method as negative for the condition.
| Method | Urinary Protein Biomarker | Disease | Sensitivity/Specificity | Reference |
|---|---|---|---|---|
| LC-MRM/MS | Prothrombin | Bladder cancer | 71.1%/75% | Chen et al., 2012 [101] |
| SELDI-TOF-MS | urinary glycopeptides m/z 1201 and 1449 | Endometrial cancer, ovarian cancer, cervical cancer | 100%/91.67% | Ak et al., 2016 [175] |
| LC–MS/MS | S100-A9 and Granulins | Hepatocellular carcinoma | NA | Huang et al., 2015 [176] |
| LC-MS/MS | leucine-rich alpha-2-glycoprotein | Ovarian cancer | NA | Smith et al., 2014 [177] |
| MS, labelfree quantitative (LFQ) mass spectrometry, parallel reaction monitoring | LYPD1, LYVE1, PTMA and SCGB1A1 | Ovarian cancer | 92%/78% | Sandow et al., 2018 [178] |
| GeLC-MS/MS analysis, immunoassay – ELISA | LYVE-1, REG1A, and TFF1 | Pancreatic cancer | 76.9%/86.8% | Radon et al., 2015 [171] |
| LC-MS/MS, reaction monitoring, Western Blot | FABP5 | Prostate cancer | 60%/100% | Fujita et al., 2017 [172] |
| LC-MS, immunoassay – ELISA (commercial kits), Western Blot | Flotilin-2, PARK7 | Prostate cancer | 68%/93% (immunoassay) | Wang et al., 2017 [173] |
| CE-TOF-MS | CKD273 | Chronic kidney disease | 33%/83% | Pontillo et al., 2017 [179] |
| CE-MS | mucin-1 | Chronic kidney disease | NA | Zhang et al., 2017 [180] |
| 2D DIGE-MS (2D gel electropheris) | preliminary protein profiling for potential biomarkers | Influenza Virus | NA | Prescott et al., 2010 [181] |
| LC-TOF-MS, immunoassay – ELISA (commercial kits) | α-fetoprotein | Hepatitis B | 90.1%/95.4% | Zhan et al., 2019 [174] |
| Immunoassay | VEGF, IL-8, MMP-9, MMP-7, survivin and Cyfra 21.1 | Bladder cancer | NA | Gogalic et al., 2015 [182] |
| Immunoassay – Western Blot (commercial kits) | FGFR3 and Cyclin D3 | Bladder cancer | 73%/90% | Blanca et al., 2016 [183] |
| Chemiluminescence immunoassay (self made) | monomeric laminin-γ2 | Bladder cancer | 72%/92% | Nakagawa et al., 2017 [129] |
| Immunoassay – ELISA (commercial kits) | uCyr61 and uTFF3 | Colorectal cancer | 75.5%/69.8% | Shimura et al., 2019 [184] |
| Radioimmunoassay125I (commercial kits) | TGFα and AFP | Hepatocellular carcinoma | 86.7%/NA | Tsai et al., 1997 [185] |
| Immunoassay – Western blot | AQP1 and PLIN2 | Kidney cancer | NA | Morrissey et al., 2015 [186] |
| Multiplex immunoassay (commercial kits) | HE4, CEA, and TTR | Ovarian cancer | 93.7%/70.6% | Lee et al., 2019 [187] |
| 2DE – Western blot (commercial kits) | clusterin, leucine-rich alpha-2-glycoprotein | Ovarian cancer | NA | Mu et al., 2013 [188] |
| Multiplex immunoassay (commercial kits) | HSA, KIM-1, MCP-1 | Chronic kidney disease | NA | Zhang et al., 2018 [189] |
| Immunoassay – ELISA (commercial kits) | EGF | Chronic kidney disease | NA | Ju et al., 2015 [190] |
| Immunoassay – ELISA (commercial kits) | EGF, MCP-1 | Diabetic kidney disease | NA | Wu et al., 2020 [191] |
| Immunoassay | HSA, KIM-1, NGAL and MCP-1 | Diabetic kidney disease | 60%/NA | Nowak et al., 2018 [192] |
| Immunoassay – ELISA (commercial kits) | MCP-1, EGF | Diabetic kidney disease | 75.7%/73.9% | Satirapoj et al., 2018 [193] |
| Immunoassay – ELISA (commercial kits) | AQP5 | Diabetic nephropathy | 85%/88% | Lu et al., 2016 [194] |
| Immunoassay – ELISA (commercial kits) | nonalbumin protein-to-creatinine ratio | Diabetic nephropathy | 92.3%/81.7% | Kim et al., 2017 [195] |
| Solid phase immunoassay (commercial kits) | NGAL, urinary type-IV collagen | Diabetic nephropathy | NA | Gaipov et al., 2019 [196] |
| Immunoassay – ELISA (commercial kits) | β2MG, MCP-1 | Autosomal dominant polycystic kidney disease | NA | Messchendorp et al., 2018 [197] |
| Immunoassay – ELISA (commercial kits) | HGF | Cardiovascular disease | NA | von Scholten et al., 2016 [198] |
| Immunoassay – ELISA (commercial kits) | EGF, MCP-1 | Glomerulonephritis | NA | Chanrat et al., 2018 [199] |
| Immunoassay – ELISA (commercial kits), immunonephelometry | NGAL, KIM-1, Cystatin-C | Acute kidney injury | NA | Dubin et al., 2018 [200] |
| Multiplexed immunoassay | L.infantum biomarkers: Li-isd1, Li-txn1, and Li-ntf2 L.donovani biomarkers: Ld-mao1, Ld-ppi1 |
Visceral leishmaniasis | 82.2%/100% | Abeijon et al., 2019 [201] |
| Fluorescence emission spectroscopy, synchronous fluorescence excitation spectra | NADH, FAD | Kidney cancer | 90%/90% | Atif et al., 2018 [202] |
Below are some observations and conclusions regarding Table 5 about the content of the listed research papers:
-
•
Urinary protein analysis are useful for not only renal diseases. Urine proteomics can be used for detection of at least 15 conditions, including 8 kinds of cancer; 5 kinds of renal complications; and at least 3 different infections. The list of health complications that can be diagnosed from urine analysis is bigger if non-protein substances are included, such as peptides, nucleic acids, low molar mass metabolites, and etc; that can be found in reviews [14,26,27,36].
-
•New potential urinary protein biomarkers are found primarily by mass spectrometry nowadays. However, further extensive validation on clinically representative population is required. Often it is achieved through the immunoassay, if the appropriate method is developed with a sufficient analytical sensitivity, since the immunoassay methods are expressive, low-cost and quantitative compared to the chromatography coupled with mass spectrometry.
-
◦In total 34 references are included in Table 5; 12 of them utilized mass spectrometry, 24 utilized the immunoassay, and 1 paper – fluorescence spectroscopy.
- ◦
-
◦All of the mentioned work (except [129]) in the immunoassay followed manufacturer's protocol, or if a custom immunoassay was used, other established procedure from their own or previous works.
-
◦ELISA-based immunoassay was the most popular method accounting for 13 references.
-
◦
-
•
Diagnostic sensitivity and specificity increased if a panel of several biomarkers was considered simultaneously. Though, the panel should be carefully chosen as not all of the components provide increase in diagnostic sensitivity and specificity.
5. Conclusion
Urine is a readily available liquid for the medical diagnosis of patients. Urine tests are non-invasive procedures that involve no pain or discomfort for patients to determine problems with kidney function since extensive medical research showed that patients, with high protein concentration in urine, have various kinds of illnesses of the kidney, referred to as proteinuria [1]. Thus the precise and simple determination of urinary concentrations of total protein is important for diagnostic purposes. In this review, we focused on instrumental determination of abundant proteins in urine by electrophoresis, chromatography, mass spectrometry, immunoassay, fluorescence, IR, and Raman spectroscopy. The mentioned techniques are not mutually exclusive, for example, high performance liquid chromatography is usually a separation step followed by mass spectrometry, another example is labelling with fluorophores in immunoassay. Each technique has its own strength to analyze urine for specific information, hence, it is hard to compare them directly, and to find a common denominator for comparison. Overall, number of cited experimental papers that were covered by this review in Table 2, Table 3, Table 4, Table 5: electrophoresis and chromatography – 22 in total and 8 related to HSA detection; mass spectrometry – 17 in total and 5 related to HSA; immunoassay – 47 in total and 17 related to HSA; fluorescence spectroscopy – 22 in total and 19 related to HSA; IR and Raman spectroscopy – 10 in total; all topics – 101 papers. Thus, almost half of the cited literature by the means of immunoassays, which is in a good correlation that immunoassays are widespread in clinics and science.
The limit of detection is important parameter for evaluating an analytical method. In order to develop a method, the limit of detection should take into account which amount of analyte has clinical significance. The 15–30 mg/L albumin concentration is a critical value that could indicate kidney problems when it is repeatedly exceeded [10]. For the corresponding articles discussed above, the limit of detection for electrophoresis, chromatography, mass spectroscopy, immunoassay, fluorescence exceeded 2 mg/L for albumin, which is enough for the clinical purpose. For some methods, explained in Ref. [117,119,122,123,127,132,143], the limit of detection does not exceed 0.1 mg/L for albumin in the urine matrix. For low abundant proteins the limit of detection must be obviously much lower. For example, in the article for Aquaporin-2 determination by Jaffuel et al. the limit of detection was 5·10−4 mg/L [85]. In another work by Zhao et al., the LOD for Nuclear matrix protein 22 was 1.7·10−6 mg/L [110]. For the IR and Raman spectroscopy methods the LOD might not be so relevant factor. The IR and Raman methods is better characterized by how accurate a particular method can distinguish whether a spectra belongs to a healthy subject or not, as evidenced from works [158,164].
Electrophoresis and chromatography are powerful separation techniques which are accompanied by further detection, usually UV–vis spectrometry or fluorescence. Since the human urine matrix is very complex consisting of inorganic and organic compounds, those techniques are useful for urine analysis [7]. Though certain methods might require preconcentration techniques to enhance sensitivity [69]. High-performance liquid chromatography with mass spectroscopy detection is a particularly powerful tool for proteomics studies, as it was able to identify 1823 proteins in urine [12]. Mass spectroscopy in itself does not allow for quantitative analysis due to the different physicochemical properties of different peptides and proteins [[98], [99], [100]]. However quantification in MS is possible if an internal standard is used, for such there are approaches based on labeling with stable isotopes (such as N-15, O-18), involving artificial labeling of peptides and proteins, as in work by Singh et al., specifically in the urine matrix [81]. The immunoassay is widely used technique both in clinical practice and in research papers for urine analysis. Though different immunoassay methods can overestimate or underestimate an analyte concentration [79]. They can hurt patients due to inaccurate results [112]. The IR and Raman spectroscopy have advantage of easy preparation of samples to analyze. They require no specific reagents, or no reagents at all, no preconcentration or dilution, as evidenced from Refs. [158,162,165]. They are rapid. On the other hand, due to overlapping bands of large biomolecules, and the complexity of urine components – hard to analyze their spectra that require mathematical analysis.
Overall, there is no "silver bullet" modern quantification method for the analysis of proteins in urine that has been reported and validated so far. However, at least for the routine clinical diagnostics of kidney problems, applications of relatively direct methods (such as SERS or IR) spectroscopy may expand as instrumentation, substrates (at least SERS substrates), software and computing power for data analysis (peak deconvolution) become progressively less expensive.
Funding Statement
The authors of this review acknowledge funding from the Nazarbayev University Collaborative Research Program (CRP) for 2020–2022 (Funder Project Reference: 091019CRP2105).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Naderi A.S.A., Reilly R.F. Primary care approach to proteinuria. J. Am. Board Fam. Med. 2008;21:569–574. doi: 10.3122/jabfm.2008.06.070080. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y., Yang X., Zhang Y., Yue S., Mei X., Bi L., Zhai W., Ren X., Ding Y., Zhang S., Deng Z., Sun Y. Correlation of urine protein/creatinine ratios to 24-h urinary protein for quantitating proteinuria in children. Pediatr. Nephrol. 2020;35:463–468. doi: 10.1007/s00467-019-04405-5. [DOI] [PubMed] [Google Scholar]
- 3.Kamińska J., Dymicka-Piekarska V., Tomaszewska J., Matowicka-Karna J., Koper-Lenkiewicz O.M. Diagnostic utility of protein to creatinine ratio (P/C ratio) in spot urine sample within routine clinical practice. Crit. Rev. Clin. Lab Sci. 2020:1–20. doi: 10.1080/10408363.2020.1723487. [DOI] [PubMed] [Google Scholar]
- 4.Mahoney E., Kun J., Smieja M., Fang Q. Review—point-of-Care urinalysis with emerging sensing and imaging technologies. J. Electrochem. Soc. 2019;167 [Google Scholar]
- 5.Premasiri W.R., Clarke R.H., Womble M.E. Urine analysis by laser Raman spectroscopy. Laser Surg. Med. 2001;28:330–334. doi: 10.1002/lsm.1058. [DOI] [PubMed] [Google Scholar]
- 6.Machii R., Kubota R., Hiratsuka N., Sugimoto K., Masudo R., Kurihara Y., Kobayashi S., Shiba K. Urinary protein fraction in healthy subjects using cellulose acetate membrane electrophoresis followed by colloidal silver staining. J. Clin. Lab. Anal. 2004;18:231–236. doi: 10.1002/jcla.20028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simerville J.A., Maxted W.C., Pahira J.J. Urinalysis: a comprehensive review. Am. Fam. Physician. 2005;71:1153–1162. [PubMed] [Google Scholar]
- 8.Viberti G.C., Jarrett R.J., Mahmud U., Hill R.D., Argyropoulos A., Keen H. Microalbuminuria as a predictor of clinical nephropathy in insulin-dependent diabetes mellitus. Lancet. 1982;319:1430–1432. doi: 10.1016/s0140-6736(82)92450-3. [DOI] [PubMed] [Google Scholar]
- 9.Lambers Heerspink H.J., Brinkman J.W., Bakker S.J., Gansevoort R.T., de Zeeuw D. Update on microalbuminuria as a biomarker in renal and cardiovascular disease. Curr. Opin. Nephrol. Hypertens. 2006;15:631–636. doi: 10.1097/01.mnh.0000247496.54882.3f. [DOI] [PubMed] [Google Scholar]
- 10.Miller W.G., Bruns D.E., Hortin G.L., Sandberg S., Aakre K.M., McQueen M.J., Itoh Y., Lieske J.C., Seccombe D.W., Jones G., Bunk D.M., Curhan G.C., Narva A.S. Current issues in measurement and reporting of urinary albumin excretion. Clin. Chem. 2009;55:24–38. doi: 10.1373/clinchem.2008.106567. [DOI] [PubMed] [Google Scholar]
- 11.Gopalan G., Rao V., Kakkar V. An overview of urinary proteomics applications in human diseases. IJHTS. 2010;I:183–192. [Google Scholar]
- 12.Marimuthu A., O'Meally RobertN., Chaerkady R., Subbannayya Y., Nanjappa V., Kumar P., Kelkar D.S., Pinto S.M., Sharma R., Renuse S., Goel R., Christopher R., Delanghe B., Cole RobertN., Harsha H.C., Pandey A. A comprehensive map of the human urinary proteome. J. Proteome Res. 2011;10:2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santucci L., Candiano G., Petretto A., Bruschi M., Lavarello C., Inglese E., Righetti P.G., Ghiggeri G.M. From hundreds to thousands: widening the normal human Urinome (1) Journal of Proteomics. 2015;112:53–62. doi: 10.1016/j.jprot.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Röthlisberger S., Pedroza-Diaz J. Urine protein biomarkers for detection of cardiovascular disease and their use for the clinic. Expet Rev. Proteonomics. 2017;14:1091–1103. doi: 10.1080/14789450.2017.1394188. [DOI] [PubMed] [Google Scholar]
- 15.Kyle R.A., Steensma D.P. In: Multiple Myeloma. Moehler T., Goldschmidt H., editors. Springer; Berlin, Heidelberg: 2011. History of multiple myeloma; pp. 3–23. [Google Scholar]
- 16.Kutwin P., Konecki T., Borkowska E.M., Traczyk-Borszynska M., Jablonowski Z. CEJU; 2018. Urine miRNA as a Potential Biomarker for Bladder Cancer Detection – a Meta-Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll M.F., Temte J.L. Proteinuria in adults: a diagnostic approach. Am. Fam. Physician. 2000;62:1333–1340. [PubMed] [Google Scholar]
- 18.Wingo C.S., L W. Clapp, Proteinuria: potential causes and approach to evaluation. Am. J. Med. Sci. 2000;320:188–194. doi: 10.1097/00000441-200009000-00010. [DOI] [PubMed] [Google Scholar]
- 19.E B., R G., P F.-S., E M., M B. Classification of renal proteinuria: a simple algorithm. Clin. Chem. Lab. Med. 2002;40:1143–1150. doi: 10.1515/CCLM.2002.201. [DOI] [PubMed] [Google Scholar]
- 20.D'Amico G., Bazzi C. Pathophysiology of proteinuria. Kidney Int. 2003;63:809–825. doi: 10.1046/j.1523-1755.2003.00840.x. [DOI] [PubMed] [Google Scholar]
- 21.Huart J., Bouquegneau A., Lutteri L., Erpicum P., Grosch S., Resimont G., Wiesen P., Bovy C., Krzesinski J.-M., Thys M., Lambermont B., Misset B., Pottel H., Mariat C., Cavalier E., Burtey S., Jouret F., Delanaye P. 2020. Proteinuria in COVID-19: Prevalence, Characterization and Prognostic Role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaparro A.I., Mitchell C.D., Abitbol C.L., Wilkinson J.D., Baldarrago G., Lopez E., Zilleruelo G. Proteinuria in children infected with the human immunodeficiency virus. J. Pediatr. 2008;152:844–849. doi: 10.1016/j.jpeds.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Leung A.K.C., Wong A.H.C., Barg S.S.N. Proteinuria in children: evaluation and differential diagnosis. Am. Fam. Physician. 2017;95:248–254. [PubMed] [Google Scholar]
- 24.Toblli J.E., Bevione P., Di Gennaro F., Madalena L., Cao G., Angerosa M. Understanding the mechanisms of proteinuria: therapeutic implications. International Journal of Nephrology. 2012 doi: 10.1155/2012/546039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster A.C., Nagler E.V., Morton R.L., Masson P. Chronic kidney disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 26.Fassett R.G., Venuthurupalli S.K., Gobe G.C., Coombes J.S., Cooper M.A., Hoy W.E. Biomarkers in chronic kidney disease: a review. Kidney Int. 2011;80:806–821. doi: 10.1038/ki.2011.198. [DOI] [PubMed] [Google Scholar]
- 27.Bidin M.Z., Shah A.M., Stanslas J., Seong C.L.T. Blood and urine biomarkers in chronic kidney disease: an update. Clin. Chim. Acta. 2019;495:239–250. doi: 10.1016/j.cca.2019.04.069. [DOI] [PubMed] [Google Scholar]
- 28.Coresh J., Astor B.C., Greene T., Eknoyan G., Levey A.S. Prevalence of chronic kidney disease and decreased kidney function in the adult US population: third national health and nutrition examination survey. Am. J. Kidney Dis. 2003;41:1–12. doi: 10.1053/ajkd.2003.50007. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q.-L., Rothenbacher D. Prevalence of chronic kidney disease in population-based studies: systematic review. BMC Publ. Health. 2008;8:117. doi: 10.1186/1471-2458-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei H., Yan Y., Gong J., Dong J. Prevalence of kidney damage in Chinese elderly: a large-scale population-based study. BMC Nephrol. 2019;20:341. doi: 10.1186/s12882-019-1525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He K., Saran R. Chapter 3: morbidity and mortality in patients with CKD. Am. J. Kidney Dis. 2017;69:S67–S92. [Google Scholar]
- 32.Murray C. 1990–2013: a Systematic Analysis for the Global Burden of Disease Study 2013, the Lancet. vol. 385. 2015. Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death; pp. 117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albalat A., Mischak H., Mullen W. Prilozi; 2011. Urine Proteomics in Clinical Applications: Technologies, Principal Considerations and Clinical Implementation. [PubMed] [Google Scholar]
- 34.Theodorescu D., Wittke S., Ross M.M., Walden M., Conaway M., Just I., Mischak H., Frierson H.F. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 35.Bhasin A., Sanders E.C., Ziegler J.M., Briggs J.S., Drago N.P., Attar A.M., Santos A.M., True M.Y., Ogata A.F., Yoon D.V., Majumdar S., Wheat A.J., Patterson S.V., Weiss G.A., Penner R.M. Virus bioresistor (VBR) for detection of bladder cancer marker DJ-1 in urine at 10 pM in one minute. Anal. Chem. 2020;92:6654–6666. doi: 10.1021/acs.analchem.0c00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W., Zhang X.J., Chao S.Y., Chen S.J., Zhang Z.J., Zhao J., Lv Y.N., Yao J.J., Bai Y.Y. Update on urine as a biomarker in cancer: a necessary review of an old story. Expert Rev. Mol. Diagn. 2020;20:477–488. doi: 10.1080/14737159.2020.1743687. [DOI] [PubMed] [Google Scholar]
- 37.Zimmerli L.U., Schiffer E., Zürbig P., Good D.M., Kellmann M., Mouls L., Pitt A.R., Coon J.J., Schmieder R.E., Peter K.H., Mischak H., Kolch W., Delles C., Dominiczak A.F. Urinary proteomic biomarkers in coronary artery disease. Mol. Cell. Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Rossing K., Mischak H., Dakna M., Zürbig P., Novak J., Julian B.A., Good D.M., Coon J.J., Tarnow L., Rossing P. Urinary proteomics in diabetes and CKD. J. Am. Soc. Nephrol. 2008;19:1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raja P., Maxwell A.P., Brazil D.P. Cardiovasc Drugs Ther; 2020. The Potential of Albuminuria as a Biomarker of Diabetic Complications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W., Yang D., Gong S., Dong X., Liu L., Yu S., Zhang X., Ge S., Wang D., Xia N., Yu D., Qiu X. An immunoassay cassette with a handheld reader for HIV urine testing in point-of-care diagnostics. Biomed. Microdevices. 2020;22 doi: 10.1007/s10544-020-00494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robles M.T.S., Cantalupo P.G., Duray A.M., Freeland M., Murkowski M., van Bokhoven A., Stephens Shields A.J., Pipas J.M., Imperiale M.J. Analysis of viruses present in urine from patients with interstitial cystitis. Virus Gene. 2020;56:430–438. doi: 10.1007/s11262-020-01767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bui T.T., Moi M.L., Morita K., Hasebe F. Development of universal and lineage-specific primer sets for rapid detection of the Zika virus (ZIKV) in blood and urine samples using one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) Jpn. J. Infect. Dis. 2020;73:153–156. doi: 10.7883/yoken.JJID.2019.073. [DOI] [PubMed] [Google Scholar]
- 43.Lu T., Han Y., Zhang R., Zhang K., Lin G., Li J. Quantitative detection of hepatitis C virus RNA in urine of patients with chronic hepatitis C using a novel real-time PCR assay. J. Med. Virol. 2019;91:115–123. doi: 10.1002/jmv.25280. [DOI] [PubMed] [Google Scholar]
- 44.Marhash A., Al-Mayah Q., Al-Kazaaly E. Detection and phylogenetic analysis of human papilloma virus in urine from a sample of Iraqi women with vaginal discharge. J. Pure Appl. Microbiol. 2018;12:2183–2192. [Google Scholar]
- 45.Niedrig M., Patel P., El Wahed A.A., Schädler R., Yactayo S. Find the right sample: a study on the versatility of saliva and urine samples for the diagnosis of emerging viruses. BMC Infect. Dis. 2018;18:707. doi: 10.1186/s12879-018-3611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X. Urinalysis: a review of methods and procedures. Crit. Care Nurs. Clin. 2010;22:121–128. doi: 10.1016/j.ccell.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Brunzel N.A. Elsevier/Saunders; St. Louis, Mo.: 2013. Fundamentals of Urine and Body Fluid Analysis. [Google Scholar]
- 48.Remer T., Montenegro-Bethancourt G., Shi L. Long-term urine biobanking: storage stability of clinical chemical parameters under moderate freezing conditions without use of preservatives. Clin. Biochem. 2014;47:307–311. doi: 10.1016/j.clinbiochem.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Pieri M., Pignalosa S., Dinallo V., Crisanti A., Casalino P., Bernardini S., Dessi M., Rossella Z. Free light chains nephelometric assay: human urine stability in different storage conditions. Clin. Chem. Lab. Med. 2016;54:e273–e274. doi: 10.1515/cclm-2015-1174. [DOI] [PubMed] [Google Scholar]
- 50.Moyle P.S., Specht A., Hill R. Effect of common storage temperatures and container types on urine protein : creatinine ratios in urine samples of proteinuric dogs. J. Vet. Intern. Med. 2018;32:1652–1658. doi: 10.1111/jvim.15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto K., Funaba M. Comparison of various methods for the determination of total protein in urine. Clin. Chem. Lab. Med. 2006:44. doi: 10.1515/CCLM.2006.201. [DOI] [PubMed] [Google Scholar]
- 52.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 53.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 54.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe N., Kamei S., Ohkubo A., Yamanaka M., Ohsawa S., Makino K., Tokuda K. Urinary protein as measured with a pyrogallol red-molybdate complex, manually and in a Hitachi 726 automated analyzer. Clin. Chem. 1986;32:1551–1554. [PubMed] [Google Scholar]
- 56.Viswanathan G., Upadhyay A. Assessment of proteinuria. Adv. Chron. Kidney Dis. 2011;18:243–248. doi: 10.1053/j.ackd.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Oyaert M., Delanghe J.R. Semiquantitative, fully automated urine test strip analysis. J. Clin. Lab. Anal. 2019;33 doi: 10.1002/jcla.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bu X., Regalado E.L., Hamilton S.E., Welch C.J. The emergence of low-cost compact mass spectrometry detectors for chromatographic analysis. Trac. Trends Anal. Chem. 2016;82:22–34. [Google Scholar]
- 59.Kalantari S., Jafari A., Moradpoor R., Ghasemi E., Khalkhal E. Human urine proteomics: analytical techniques and clinical applications in renal diseases. International Journal of Proteomics. 2015 doi: 10.1155/2015/782798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Decramer S., de Peredo A.G., Breuil B., Mischak H., Monsarrat B., Bascands J.-L., Schanstra J.P. Urine in clinical proteomics. Mol. Cell. Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Jiang X., Feng S., Tian R., Han G., Jiang X., Ye M., Zou H. Automation of nanoflow liquid chromatography-tandem mass spectrometry for proteome analysis by using a strong cation exchange trap column. Proteomics. 2007;7:528–539. doi: 10.1002/pmic.200600661. [DOI] [PubMed] [Google Scholar]
- 62.Valle R.D., Mora J.M.C., Martín N.L.C.S., Pérez L.H., Torres M.E.L., Cisneros R.L., Ramos R.P.O., Rodríguez A.M., Álvarez I.V. An enzyme immunoassay for determining albumin in human urine samples using an ultra-microanalytical system. J. Immunoassay Immunochem. 2020;41:896–912. doi: 10.1080/15321819.2020.1807357. [DOI] [PubMed] [Google Scholar]
- 63.Righetti P.G. Electrophoresis: the march of pennies, the march of dimes. J. Chromatogr. A. 2005;1079:24–40. doi: 10.1016/j.chroma.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Kašička V. Recent developments in capillary and microchip electroseparations of peptides (2017–mid 2019) Electrophoresis. 2020;41:10–35. doi: 10.1002/elps.201900269. [DOI] [PubMed] [Google Scholar]
- 65.Aguzzi F., Gasparro C., Bergami M.R., Merlini M. High-sensitivity electrophoretic method for the detection of bence Jones protein and for the study of proteinuria in unconcentrated urines. Ann. Clin. Biochem. 1993;30:287–292. doi: 10.1177/000456329303000310. [DOI] [PubMed] [Google Scholar]
- 66.Giovannoli C., Anfossi L., Baggiani C., Giraudi G. A novel approach for a non competitive capillary electrophoresis immunoassay with laser-induced fluorescence detection for the determination of human serum albumin. J. Chromatogr. A. 2007;1155:187–192. doi: 10.1016/j.chroma.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 67.Jia J., Pan J., Xu H., Wang S., Bai B. Urine protein quantification in stacking gel by SDS-PAGE. Electrophoresis. 2019;40:487–490. doi: 10.1002/elps.201800379. [DOI] [PubMed] [Google Scholar]
- 68.Tran J.C., Doucette A.A. Gel-eluted liquid fraction entrapment Electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal. Chem. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 69.Kartsova L.A., Bessonova E.A. Preconcentration techniques in capillary electrophoresis. J. Anal. Chem. 2009;64:326–337. [Google Scholar]
- 70.Bessonova E.A., Kartsova L.A., Shmukov A.U. Electrophoretic determination of albumin in urine using on-line concentration techniques. J. Chromatogr. A. 2007;1150:332–338. doi: 10.1016/j.chroma.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 71.Wittke S., Fliser D., Haubitz M., Bartel S., Krebs R., Hausadel F., Hillmann M., Golovko I., Koester P., Haller H., Kaiser T., Mischak H., Weissinger E.M. Determination of peptides and proteins in human urine with capillary electrophoresis–mass spectrometry, a suitable tool for the establishment of new diagnostic markers. J. Chromatogr. A. 2003;1:173–181. doi: 10.1016/s0021-9673(03)00713-1. [DOI] [PubMed] [Google Scholar]
- 72.Kiprijanovska S., Stavridis S., Stankov O., Komina S., Petrusevska G., Polenakovic M., Davalieva K. Mapping and identification of the urine proteome of prostate cancer patients by 2D PAGE/MS. International Journal of Proteomics. 2014;2014 doi: 10.1155/2014/594761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakayama A., Kubota R., Sakatsume M., Suzuki H., Katayama A., Kanamori K., Shiba K., Iijima S. Cellulose acetate membrane electrophoresis based urinary proteomics for the identification of characteristic proteins: CAME-based urinary proteomics. J. Clin. Lab. Anal. 2016;30:359–367. doi: 10.1002/jcla.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zoccali M., Tranchida P.Q., Mondello L. Fast gas chromatography-mass spectrometry: a review of the last decade. Trac. Trends Anal. Chem. 2019;118:444–452. [Google Scholar]
- 75.Rigano F., Tranchida P.Q., Dugo P., Mondello L. High-performance liquid chromatography combined with electron ionization mass spectrometry: a review. Trac. Trends Anal. Chem. 2019;118:112–122. [Google Scholar]
- 76.Ishida J., Abe K., Nakamura M., Yamaguchi M. High-performance liquid chromatographic determination of human serum albumin in plasma and urine by post-column fluorescence enhancement detection using 8-Anilino-1-naphthalenesulfonic acid. Biol. Pharm. Bull. 1996;19:1391–1395. doi: 10.1248/bpb.19.1391. [DOI] [PubMed] [Google Scholar]
- 77.Lizana J., Hellsing K. Manual immunonephelometric assay of proteins, with use of polymer enhancement. Clin. Chem. 1974;20:1181–1186. [PubMed] [Google Scholar]
- 78.Brinkman J.W., Bakker S.J.L., Gansevoort R.T., Hillege H.L., Kema I.P., Gans R.O.B., De Jong P.E., De Zeeuw D. Which method for quantifying urinary albumin excretion gives what outcome? A comparison of immunonephelometry with HPLC. Kidney Int. 2004;66:S69–S75. doi: 10.1111/j.1523-1755.2004.09219.x. [DOI] [PubMed] [Google Scholar]
- 79.Comper W.D., Jerums G., Osicka T.M. Differences in urinary albumin detected by four immunoassays and high-performance liquid chromatography. Clin. Biochem. 2004;37:105–111. doi: 10.1016/j.clinbiochem.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 80.Owen W.E., Roberts W.L. Performance characteristics of an HPLC assay for urinary albumin. Am. J. Clin. Pathol. 2005;124:219–225. doi: 10.1309/F6WV-K152-5KLQ-GXR4. [DOI] [PubMed] [Google Scholar]
- 81.Singh R., Crow F.W., Babic N., Lutz W.H., Lieske J.C., Larson T.S., Kumar R. A liquid chromatography-mass spectrometry method for the quantification of urinary albumin using a novel 15N-isotopically labeled albumin internal standard. Clin. Chem. 2007;53:540–542. doi: 10.1373/clinchem.2006.078832. [DOI] [PubMed] [Google Scholar]
- 82.Hancu G., Simon B., Rusu A., Mircia E., Gyéresi Á. Principles of micellar electrokinetic capillary chromatography applied in pharmaceutical analysis. Adv. Pharmaceut. Bull. 2013;3:1–8. doi: 10.5681/apb.2013.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glavač N.K., Injac R., Kreft S. Optimization and validation of a capillary MEKC method for determination of proteins in urine. Chroma. 2009;70:1473–1478. [Google Scholar]
- 84.Wu Y.-W., Liu J.-F., Xiao T.-X., Han D.-Y., Zhang H.-L., Pan J.-C. Field-amplified sample injection for the determination of albumin and transferrin in human urines by MEKC. Electrophoresis. 2009;30:668–673. doi: 10.1002/elps.200800684. [DOI] [PubMed] [Google Scholar]
- 85.Jaffuel A., Lemoine J., Aubert C., Simon R., Léonard J.-F., Gautier J.-C., Pasquier O., Salvador A. Optimization of liquid chromatography–multiple reaction monitoring cubed mass spectrometry assay for protein quantification: application to aquaporin-2 water channel in human urine. J. Chromatogr. A. 2013;1301:122–130. doi: 10.1016/j.chroma.2013.05.068. [DOI] [PubMed] [Google Scholar]
- 86.Dajani S., Saripalli A., Sharma-Walia N. Water transport proteins–aquaporins (AQPs) in cancer biology. Oncotarget. 2018;9:36392–36405. doi: 10.18632/oncotarget.26351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nováková L., Vlčková H. A review of current trends and advances in modern bio-analytical methods: chromatography and sample preparation. Anal. Chim. Acta. 2009;656:8–35. doi: 10.1016/j.aca.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 88.Akimoto M., Hokazono E., Ota E., Tateishi T., Kayamori Y. Highly sensitive reversed-phase high-performance liquid chromatography assay for the detection of Tamm–Horsfall protein in human urine. Ann. Clin. Biochem. 2016 Jan;53(Pt. 1):75–84. doi: 10.1177/0004563215583698. [DOI] [PubMed] [Google Scholar]
- 89.Gao H., Liu J.-J., Liu Y.-Q., Wu Z.-Y. Detection of urine protein by a paper-based analytical device enhanced with ion concentration polarization effect. Microfluid. Nanofluidics. 2019;23:51. [Google Scholar]
- 90.Alharbi R.A. Proteomics approach and techniques in identification of reliable biomarkers for diseases. Saudi J. Biol. Sci. 2020;27:968–974. doi: 10.1016/j.sjbs.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicolescu T.O. Mass Spectrometry; 2017. Interpretation of Mass Spectra. [Google Scholar]
- 92.Ho C., Lam C., Chan M., Cheung R., Law L., Lit L., Ng K., Suen M., Tai H. Electrospray ionisation mass spectrometry: principles and clinical applications. Clin. Biochem. Rev. 2003;24:3–12. [PMC free article] [PubMed] [Google Scholar]
- 93.Hillenkamp F., Karas M., Beavis R.C., Chait B.T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 94.Protein Analysis by Shotgun/Bottom-Up Proteomics | Chemical Reviews, (n.d.).
- 95.Catherman A.D., Skinner O.S., Kelleher N.L. Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 2014;445:683–693. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ryan D.J., Spraggins J.M., Caprioli R.M. Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr. Opin. Chem. Biol. 2019;48:64–72. doi: 10.1016/j.cbpa.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Noble W., Serang O. A review of statistical methods for protein identification using tandem mass spectrometry. Stat. Interface. 2012;5:3–20. doi: 10.4310/sii.2012.v5.n1.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nikolov M., Schmidt C., Urlaub H. In: Quantitative Methods in Proteomics. Marcus K., editor. Humana Press; Totowa, NJ: 2012. Quantitative mass spectrometry-based proteomics: an overview; pp. 85–100. [DOI] [PubMed] [Google Scholar]
- 99.Urban P.L. Quantitative mass spectrometry: an overview. Phil. Trans. Math. Phys. Eng. Sci. 2016;374:20150382. doi: 10.1098/rsta.2015.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grebe S.K.G., Singh R.J. Clinical peptide and protein quantification by mass spectrometry (MS) Trac. Trends Anal. Chem. 2016;84:131–143. [Google Scholar]
- 101.Chen Y.-T., Chen H.-W., Domanski D., Smith D.S., Liang K.-H., Wu C.-C., Chen C.-L., Chung T., Chen M.-C., Chang Y.-S., Parker C.E., Borchers C.H., Yu J.-S. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. Journal of Proteomics. 2012;75:3529–3545. doi: 10.1016/j.jprot.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 102.Bachmann L.M., Nilsson G., Bruns D.E., McQueen M.J., Lieske J.C., Zakowski J.J., Miller W.G. State of the art for measurement of urine albumin: comparison of routine measurement procedures to isotope dilution tandem mass spectrometry. Clin. Chem. 2014;60:471–480. doi: 10.1373/clinchem.2013.210302. [DOI] [PubMed] [Google Scholar]
- 103.Lieske J.C., Bondar O., Miller W.G., Bachmann L.M., Narva A.S., Itoh Y., Zegers I., Schimmel H., Phinney K., Bunk D.M. A reference system for urinary albumin: current status. Clin. Chem. Lab. Med. 2013;51:981–989. doi: 10.1515/cclm-2012-0768. [DOI] [PubMed] [Google Scholar]
- 104.Denoroy L., Zimmer L., Renaud B., Parrot S. Ultra high performance liquid chromatography as a tool for the discovery and the analysis of biomarkers of diseases: a review. J. Chromatogr. B. 2013;927:37–53. doi: 10.1016/j.jchromb.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 105.Hawkridge A.M. Quantitative Proteomics. 2014. CHAPTER 1:practical considerations and current limitations in quantitative mass spectrometry-based proteomics; pp. 1–25. [Google Scholar]
- 106.Slagle K.M., Ghosn S.J. Immunoassays: tools for sensitive, specific, and accurate test results. Lab. Med. 1996;27:177–183. [Google Scholar]
- 107.Kapingidza A.B., Kowal K., Chruszcz M. In: Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and Other Body Fluid Proteins. Hoeger U., Harris J.R., editors. Springer International Publishing; Cham: 2020. Antigen–antibody complexes; pp. 465–497. [Google Scholar]
- 108.Findlay J.W.A., Smith W.C., Lee J.W., Nordblom G.D., Das I., DeSilva B.S., Khan M.N., Bowsher R.R. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J. Pharmaceut. Biomed. Anal. 2000;21:1249–1273. doi: 10.1016/s0731-7085(99)00244-7. [DOI] [PubMed] [Google Scholar]
- 109.Darwish I.A. Immunoassay methods and their applications in pharmaceutical analysis: basic methodology and recent advances. Int. J. Biomed. Sci. 2006;2:217–235. [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao S., Zhang Y., Ding S., Fan J., Luo Z., Liu K., Shi Q., Liu W., Zang G. A highly sensitive label-free electrochemical immunosensor based on AuNPs-PtNPs-MOFs for nuclear matrix protein 22 analysis in urine sample. J. Electroanal. Chem. 2019:33–42. [Google Scholar]
- 111.Shaikh A., Seegmiller J.C., Borland T.M., Burns B.E., Ladwig P.M., Singh R.J., Kumar R., Larson T.S., Lieske J.C. Comparison between immunoturbidimetry, size-exclusion chromatography, and LC-MS to quantify urinary albumin. Clin. Chem. 2008;54:1504–1510. doi: 10.1373/clinchem.2008.107508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoofnagle A.N., Wener M.H. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J. Immunol. Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chavers B.M., Simonson J., Michael A.F. A solid phase fluorescent immunoassay for the measurement of human urinary albumin. Kidney Int. 1984;25:576–578. doi: 10.1038/ki.1984.57. [DOI] [PubMed] [Google Scholar]
- 114.Lloyd D.R., Hindle E.J., Marples J., Gatt J.A. Urinary albumin measurement by immunoturbidimetry. Ann. Clin. Biochem.: An International Journal of Biochemistry and Laboratory Medicine. 1987;24:209–210. doi: 10.1177/000456328702400218. [DOI] [PubMed] [Google Scholar]
- 115.Marre M., Claudel J.P., Ciret P., Luis N., Suarez L., Passa P. Laser immunonephelometry for routine quantification of urinary albumin excretion. Clin. Chem. 1987;33:209–213. [PubMed] [Google Scholar]
- 116.Bakker A.J. Immunoturbidimetry of urinary albumin: prevention of adsorption of albumin; influence of other urinary constituents. Clin. Chem. 1988;34:82–86. [PubMed] [Google Scholar]
- 117.Horton J.K., Davies M., Woodhead J.S., Weeks I. A rapid and sensitive method for estimating low concentrations of albumin in human urine. Clin. Chim. Acta. 1989;186:45–51. doi: 10.1016/0009-8981(89)90202-7. [DOI] [PubMed] [Google Scholar]
- 118.Qin Q.-P., Peltola O., Pettersson K. Time-resolved fluorescence resonance energy transfer assay for point-of-care testing of urinary albumin. Clin. Chem. 2003;49:1105–1113. doi: 10.1373/49.7.1105. [DOI] [PubMed] [Google Scholar]
- 119.Zhao L., Lin J.-M., Li Z. Comparison and development of two different solid phase chemiluminescence ELISA for the determination of albumin in urine. Anal. Chim. Acta. 2005;541:197–205. [Google Scholar]
- 120.Lu M., Ibraimi F., Kriz D., Kriz K. A combination of magnetic permeability detection with nanometer-scaled superparamagnetic tracer and its application for one-step detection of human urinary albumin in undiluted urine. Biosens. Bioelectron. 2006;21:2248–2254. doi: 10.1016/j.bios.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 121.Ning G., Lu-Yan W., Wei-Min X., Tian-Hua L., Qian-Li J. Electrochemical immuno-biosensor for the rapid determination of nuclear matrix protein 22 (NMP22) antigen in urine samples by Co(III) phthlocyanine/Fe3O4/Au collide coimmobilized electrode. Chin. J. Anal. Chem. 2007;35:1553–1558. [Google Scholar]
- 122.Jiang Z., Huang Y., Liang A., Pan H., Liu Q. Resonance scattering detection of trace microalbumin using immunonanogold probe as the catalyst of Fehling reagent–glucose reaction. Biosens. Bioelectron. 2009;24:1674–1678. doi: 10.1016/j.bios.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 123.Tu M.-C., Chang Y.-T., Kang Y.-T., Chang H.-Y., Chang P., Yew T.-R. A quantum dot-based optical immunosensor for human serum albumin detection. Biosens. Bioelectron. 2012;34:286–290. doi: 10.1016/j.bios.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 124.Wang R.E., Tian L., Chang Y.-H. A homogeneous fluorescent sensor for human serum albumin. J. Pharmaceut. Biomed. Anal. 2012;63:165–169. doi: 10.1016/j.jpba.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zangheri M., Di Nardo F., Mirasoli M., Anfossi L., Nascetti A., Caputo D., De Cesare G., Guardigli M., Baggiani C., Roda A. Chemiluminescence lateral flow immunoassay cartridge with integrated amorphous silicon photosensors array for human serum albumin detection in urine samples. Anal. Bioanal. Chem. 2016;408:8869–8879. doi: 10.1007/s00216-016-9991-0. [DOI] [PubMed] [Google Scholar]
- 126.Giannetto M., Bianchi M.V., Mattarozzi M., Careri M. Competitive amperometric immunosensor for determination of p53 protein in urine with carbon nanotubes/gold nanoparticles screen-printed electrodes: a potential rapid and noninvasive screening tool for early diagnosis of urinary tract carcinoma. Anal. Chim. Acta. 2017;991:133–141. doi: 10.1016/j.aca.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 127.Theansun W., Sripratumporn J., Promptmas C. Determination of albumin in urine by a quartz crystal microbalance label-free assay. Anal. Lett. 2017;50:1912–1925. [Google Scholar]
- 128.Liu Y., Liu Y.H., Bei W.J., Wang K., Cui T.T., Li H.L., Wu D.X., Chen S.Q., Tan N., Chen J.Y. A dual-label time-resolved fluorescence immunoassay for the simultaneous determination of cystatin C and β2-microglobulin in urine. Br. J. Biomed. Sci. 2017;74:193–197. doi: 10.1080/09674845.2017.1334740. [DOI] [PubMed] [Google Scholar]
- 129.Nakagawa M., Karashima T., Kamada M., Yoshida E., Yoshimura T., Nojima M., Inoue K., Shuin T., Seiki M., Koshikawa N. Development of a fully automated chemiluminescence immunoassay for urine monomeric laminin-γ2 as a promising diagnostic tool of non-muscle invasive bladder cancer. Biomarker Research. 2017;5:29. doi: 10.1186/s40364-017-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Semeradtova A., Stofik M., Vankova L., Maly P., Stanek O., Maly J. Optical microchips based on high-affinity recombinant protein binders—human serum albumin detection in urine. Sensor. Actuator. B Chem. 2018;272:441–447. [Google Scholar]
- 131.Arya S.K., Estrela P. Electrochemical ELISA-based platform for bladder cancer protein biomarker detection in urine. Biosens. Bioelectron. 2018;117:620–627. doi: 10.1016/j.bios.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 132.Chawjiraphan W., Apiwat C., Segkhoonthod K., Treerattrakoon K., Pinpradup P., Sathirapongsasuti N., Pongprayoon P., Luksirikul P., Isarankura-Na-Ayudhya P., Japrung D. Sensitive detection of albuminuria by graphene oxide-mediated fluorescence quenching aptasensor. Spectrochim. Acta Mol. Biomol. Spectrosc. 2020;231:118128. doi: 10.1016/j.saa.2020.118128. [DOI] [PubMed] [Google Scholar]
- 133.Gomes A.J., Lunardi C.N., Rocha F.S., Patience G.S. Experimental methods in chemical engineering: fluorescence emission spectroscopy. Can. J. Chem. Eng. 2019;97:2168–2175. [Google Scholar]
- 134.Rye H.S., Dabora J.M., Quesada M.A., Mathies R.A., Glazer A.N. Fluorometric assay using dimeric dyes for double- and single-stranded DNA and RNA with picogram sensitivity. Anal. Biochem. 1993;208:144–150. doi: 10.1006/abio.1993.1020. [DOI] [PubMed] [Google Scholar]
- 135.Resch-Genger U., Grabolle M., Cavaliere-Jaricot S., Nitschke R., Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 136.Petryayeva E., Algar W.R., Medintz I.L. Applied Spectroscopy; 2013. Quantum Dots in Bioanalysis: A Review of Applications across Various Platforms for Fluorescence Spectroscopy and Imaging. [DOI] [PubMed] [Google Scholar]
- 137.Xu Z., Yang W., Dong C. Determination of human serum albumin using an intramolecular charge transfer fluorescence probe: 4′-Dimethylamino-2,5-dihydroxychalcone. Bioorg. Med. Chem. Lett. 2005;15:4091–4096. doi: 10.1016/j.bmcl.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 138.Hou X., Tong X., Dong W., Dong C., Shuang S. Synchronous fluorescence determination of human serum albumin with methyl blue as a fluorescence probe. Spectrochim. Acta Mol. Biomol. Spectrosc. 2007;66:552–556. doi: 10.1016/j.saa.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 139.Cui F., Qin L., Li F., Luo H. Synchronous fluorescence determination and molecular modeling of 5-Aminosalicylic acid (5-ASA) interacted with human serum albumin. J. Mol. Model. 2008;14:1111–1117. doi: 10.1007/s00894-008-0352-6. [DOI] [PubMed] [Google Scholar]
- 140.Zhu C., Liu M., Wang P., Cao M., Cao C. Determination of albumin using CdS/SiO 2 core/shell nanoparticles as fluorescence probes. Chin. J. Chem. 2009;27:1820–1826. [Google Scholar]
- 141.Liu X., Wu X., Yang J. Protein determination using methylene blue in a synchronous fluorescence technique. Talanta. 2010;81:760–765. doi: 10.1016/j.talanta.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 142.Ramezani A.M., Manzoori J.L., Amjadi M., Jouyban A. Spectrofluorimetric determination of human serum albumin using terbium-danofloxacin probe. Sci. World J. 2012:1–9. doi: 10.1100/2012/940541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Madrakian T., Bagheri H., Afkhami A. Determination of human albumin in serum and urine samples by constant‐energy synchronous fluorescence method. Luminescence. 2015;30:576–582. doi: 10.1002/bio.2788. [DOI] [PubMed] [Google Scholar]
- 144.Li H., Yao Q., Fan J., Du J., Wang J., Peng X. An NIR fluorescent probe of uric HSA for renal diseases warning. Dyes Pigments. 2016;133:79–85. [Google Scholar]
- 145.Chen M., Xiang X., Wu K., He H., Chen H., Ma C. A novel detection method of human serum albumin based on the poly(thymine)-templated copper nanoparticles. Sensors. 2017;17:2684. doi: 10.3390/s17112684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gui W., Chen X., Ma Q. A novel detection method of human serum albumin based on CuInZnS quantum dots-Co2+ sensing system. Anal. Bioanal. Chem. 2017;409:3871–3876. doi: 10.1007/s00216-017-0332-8. [DOI] [PubMed] [Google Scholar]
- 147.Rajasekhar K., Achar C.J., Govindaraju T. A red-NIR emissive probe for the selective detection of albumin in urine samples and live cells. Org. Biomol. Chem. 2017;15:1584–1588. doi: 10.1039/c6ob02760a. [DOI] [PubMed] [Google Scholar]
- 148.Stuart B. Infrared Spectroscopy: Fundamentals and Applications. John Wiley & Sons, Ltd; 2005. Introduction; pp. 1–13. [Google Scholar]
- 149.Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 150.Yang D., Ying Y. Applications of Raman spectroscopy in agricultural products and food analysis: a review. Appl. Spectrosc. Rev. 2011;46:539–560. [Google Scholar]
- 151.López-Lorente Á.I., Mizaikoff B. Mid-infrared spectroscopy for protein analysis: potential and challenges. Anal. Bioanal. Chem. 2016;408:2875–2889. doi: 10.1007/s00216-016-9375-5. [DOI] [PubMed] [Google Scholar]
- 152.Glassford S.E., Byrne B., Kazarian S.G. Recent applications of ATR FTIR spectroscopy and imaging to proteins. Biochim. Biophys. Acta Protein Proteonomics. 2013;1834:2849–2858. doi: 10.1016/j.bbapap.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 153.Lane L.A., Qian X., Nie S. SERS nanoparticles in medicine: from label-free detection to spectroscopic tagging. Chem. Rev. 2015;115:10489–10529. doi: 10.1021/acs.chemrev.5b00265. [DOI] [PubMed] [Google Scholar]
- 154.Pilot R., Signorini R., Durante C., Orian L., Bhamidipati M., Fabris L. A review on surface-enhanced Raman scattering. Biosensors. 2019;9:57. doi: 10.3390/bios9020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharma B., Frontiera R.R., Henry A.-I., Ringe E., Van Duyne R.P. SERS: materials, applications, and the future. Mater. Today. 2012;15:16–25. [Google Scholar]
- 156.Nie S., Emory S.R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 157.Ataka K., Heberle J. Biochemical applications of surface-enhanced infrared absorption spectroscopy. Anal. Bioanal. Chem. 2007;388:47–54. doi: 10.1007/s00216-006-1071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jessen T.E., Höskuldsson A.T., Bjerrum P.J., Verder H., Sørensen L., Bratholm P.S., Christensen B., Jensen L.S., Jensen M.A.B. Simultaneous determination of glucose, triglycerides, urea, cholesterol, albumin and total protein in human plasma by Fourier transform infrared spectroscopy: direct clinical biochemistry without reagents. Clin. Biochem. 2014;47:1306–1312. doi: 10.1016/j.clinbiochem.2014.05.064. [DOI] [PubMed] [Google Scholar]
- 159.Ma X.-X., Wang C.-C., Cai W.-S., Shao X.-G. Quantification of albumin in urine using preconcentration and near-infrared diffuse reflectance spectroscopy. Chin. Chem. Lett. 2016;27:1597–1601. [Google Scholar]
- 160.Hall J.W., Pollard A. Near-infrared spectroscopic determination of serum total proteins, albumin, globulins, and urea. Clin. Biochem. 1993;26:483–490. doi: 10.1016/0009-9120(93)80013-k. [DOI] [PubMed] [Google Scholar]
- 161.Pezzaniti J.L., Jeng T.-W., McDowell L., Oosta G.M. Preliminary investigation of near-infrared spectroscopic measurements of urea, creatinine, glucose, protein, and ketone in urine. Clin. Biochem. 2001;34:239–246. doi: 10.1016/s0009-9120(01)00198-9. [DOI] [PubMed] [Google Scholar]
- 162.Bispo J.A.M., de Sousa Vieira E.E., Silveira L., Fernandes A.B. Correlating the amount of urea, creatinine, and glucose in urine from patients with diabetes mellitus and hypertension with the risk of developing renal lesions by means of Raman spectroscopy and principal component analysis. J. Biomed. Optic. 2013;18 doi: 10.1117/1.JBO.18.8.087004. [DOI] [PubMed] [Google Scholar]
- 163.de Almeida M.L., Saatkamp C.J., Fernandes A.B., Pinheiro A.L.B., Silveira L. Estimating the concentration of urea and creatinine in the human serum of normal and dialysis patients through Raman spectroscopy. Laser Med. Sci. 2016;31:1415–1423. doi: 10.1007/s10103-016-2003-y. [DOI] [PubMed] [Google Scholar]
- 164.Senger R.S., Sullivan M., Gouldin A., Lundgren S., Merrifield K., Steen C., Baker E., Vu T., Agnor B., Martinez G., Coogan H., Carswell W., Kavuru V., Karageorge L., Dev D., Du P., Sklar A., Pirkle J., Guelich S., Orlando G., et al. Spectral characteristics of urine from patients with end-stage kidney disease analyzed using Raman Chemometric Urinalysis (Rametrix) PloS One. 2020;15 doi: 10.1371/journal.pone.0227281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chamuah N., Saikia A., Joseph A.M., Nath P. Blu-ray DVD as SERS substrate for reliable detection of albumin, creatinine and urea in urine. Sensor. Actuator. B Chem. 2019;285:108–115. [Google Scholar]
- 166.Dou X., Yamaguchi Y., Yamamoto H., Doi S., Ozaki Y. Quantitative analysis of metabolites in urine using a highly precise, compact near-infrared Raman spectrometer. Vib. Spectrosc. 1996;13:83–89. [Google Scholar]
- 167.Gaipov A., Utegulov Z., Bukasov R., Turebekov D., Tarlykov P., Markhametova Z., Nurekeyev Z., Kunushpayeva Z., Sultangaziyev A. Development and validation of hybrid Brillouin-Raman spectroscopy for non-contact assessment of mechano-chemical properties of urine proteins as biomarkers of kidney diseases. BMC Nephrol. 2020;21:229. doi: 10.1186/s12882-020-01890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhu W., Wen B.-Y., Jie L.-J., Tian X.-D., Yang Z.-L., Radjenovic P.M., Luo S.-Y., Tian Z.-Q., Li J.-F. Rapid and low-cost quantitative detection of creatinine in human urine with a portable Raman spectrometer. Biosens. Bioelectron. 2020;154:112067. doi: 10.1016/j.bios.2020.112067. [DOI] [PubMed] [Google Scholar]
- 169.Saah A.J., Hoover D.R. “Sensitivity” and “specificity” reconsidered: the meaning of these terms in analytical and diagnostic settings. Ann. Intern. Med. 1997;126:91–94. doi: 10.7326/0003-4819-126-1-199701010-00026. [DOI] [PubMed] [Google Scholar]
- 170.Mahmood T., Yang P.-C. Western blot: technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012;4:429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Radon T.P., Massat N.J., Jones R., Alrawashdeh W., Dumartin L., Ennis D., Duffy S.W., Kocher H.M., Pereira S.P., Guarner posthumous L., Murta-Nascimento C., Real F.X., Malats N., Neoptolemos J., Costello E., Greenhalf W., Lemoine N.R., Crnogorac-Jurcevic T. Identification of a three-biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin. Canc. Res. 2015;21:3512–3521. doi: 10.1158/1078-0432.CCR-14-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fujita K., Kume H., Matsuzaki K., Kawashima A., Ujike T., Nagahara A., Uemura M., Miyagawa Y., Tomonaga T., Nonomura N. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017;7:42961. doi: 10.1038/srep42961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang L., Skotland T., Berge V., Sandvig K., Llorente A. Exosomal proteins as prostate cancer biomarkers in urine: from mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharmaceut. Sci. 2017;98:80–85. doi: 10.1016/j.ejps.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 174.Zhan Z., Guan Y., Mew K., Zeng W., Peng M., Hu P., Yang Y., Lu Y., Ren H. Urine α-fetoprotein and orosomucoid 1 as biomarkers of hepatitis B virus-associated hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 2019;318:G305–G312. doi: 10.1152/ajpgi.00267.2019. [DOI] [PubMed] [Google Scholar]
- 175.Ak M., Bk L., N A., Oh H., As S. Application of SELDI-TOF in N-glycopeptides profiling of the urine from patients with endometrial, ovarian and cervical cancer. Arch. Physiol. Biochem. 2016;122:111–116. doi: 10.3109/13813455.2016.1151441. [DOI] [PubMed] [Google Scholar]
- 176.Huang C.-H., Kuo C.-J., Liang S.-S., Chi S.-W., Hsi E., Chen C.-C., Lee K.-T., Chiou S.-H. Onco-proteogenomics identifies urinary S100A9 and GRN as potential combinatorial biomarkers for early diagnosis of hepatocellular carcinoma. BBA Clin. 2015;3:205–213. doi: 10.1016/j.bbacli.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Smith C.R., Batruch I., Bauça J.M., Kosanam H., Ridley J., Bernardini M.Q., Leung F., Diamandis E.P., Kulasingam V. Deciphering the peptidome of urine from ovarian cancer patients and healthy controls. Clin. Proteonomics. 2014;11:23. doi: 10.1186/1559-0275-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sandow J.J., Rainczuk A., Infusini G., Makanji M., Bilandzic M., Wilson A.L., Fairweather N., Stanton P.G., Garama D., Gough D., Jobling T.W., Webb A.I., Stephens A.N. Discovery and validation of novel protein biomarkers in ovarian cancer patient urine. Proteonomics Clin. Appl. 2018;12 doi: 10.1002/prca.201700135. [DOI] [PubMed] [Google Scholar]
- 179.Pontillo C., Zhang Z.-Y., Schanstra J.P., Jacobs L., Zürbig P., Thijs L., Ramírez-Torres A., Heerspink H.J.L., Lindhardt M., Klein R., Orchard T., Porta M., Bilous R.W., Charturvedi N., Rossing P., Vlahou A., Schepers E., Glorieux G., Mullen W., Delles C., et al. Prediction of chronic kidney disease stage 3 by CKD273, a urinary proteomic biomarker. Kidney International Reports. 2017;2:1066–1075. doi: 10.1016/j.ekir.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang Z.-Y., Ravassa S., Pejchinovski M., Yang W.-Y., Zürbig P., López B., Wei F.-F., Thijs L., Jacobs L., González A., Voigt J.-U., Verhamme P., Kuznetsova T., Díez J., Mischak H., Staessen J.A. A urinary fragment of mucin-1 subunit α is a novel biomarker associated with renal dysfunction in the general population. Kidney International Reports. 2017;2:811–820. doi: 10.1016/j.ekir.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Prescott M.A., Pastey M.K. Biomarker Insights; 2010. Identification of Unique Blood and Urine Biomarkers in Influenza Virus and Staphylococcus aureus Co-infection: A Preliminary Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gogalic S., Sauer U., Doppler S., Preininger C. Bladder cancer biomarker array to detect aberrant levels of proteins in urine. Analyst. 2015;140:724–735. doi: 10.1039/c4an01432d. [DOI] [PubMed] [Google Scholar]
- 183.Blanca A., Requena M.J., Alvarez J., Cheng L., Montironi R., Raspollini M.R., Reymundo C., Lopez-Beltran A. FGFR3 and Cyclin D3 as urine biomarkers of bladder cancer recurrence. Biomarkers Med. 2016;10:243–253. doi: 10.2217/bmm.15.120. [DOI] [PubMed] [Google Scholar]
- 184.Shimura T., Iwasaki H., Kitagawa M., Ebi M., Yamada T., Yamada T., Katano T., Nisie H., Okamoto Y., Ozeki K., Mizoshita T., Kataoka H. Urinary cysteine-rich protein 61 and trefoil factor 3 as diagnostic biomarkers for colorectal cancer. Transl Oncol. 2019;12:539–544. doi: 10.1016/j.tranon.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tsai J.F., Jeng J.E., Chuang L.Y., Yang M.L., Ho M.S., Chang W.Y., Hsieh M.Y., Lin Z.Y., Tsai J.H. Clinical evaluation of urinary transforming growth factor-beta1 and serum alpha-fetoprotein as tumour markers of hepatocellular carcinoma. Br. J. Canc. 1997;75:1460–1466. doi: 10.1038/bjc.1997.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Morrissey J.J., Mobley J., Figenshau R.S., Vetter J., Bhayani S., Kharasch E.D. Urine aquaporin 1 and perilipin 2 differentiate renal carcinomas from other imaged renal masses and bladder and prostate cancer. Mayo Clin. Proc. 2015;90:35–42. doi: 10.1016/j.mayocp.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee S.-W., Lee H.-Y., Bang H.J., Song H.-J., Kong S.W., Kim Y.-M. An improved prediction model for ovarian cancer using urinary biomarkers and a novel validation strategy. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mu A.K.-W., Lim B.-K., Hashim O.H., Shuib A.S. Identification of O-glycosylated proteins that are aberrantly excreted in the urine of patients with early stage ovarian cancer. Int. J. Mol. Sci. 2013;14:7923–7931. doi: 10.3390/ijms14047923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang W.R., Craven T.E., Malhotra R., Cheung A.K., Chonchol M., Drawz P., Sarnak M.J., Parikh C.R., Shlipak M.G., Ix J.H. Kidney damage biomarkers and incident chronic kidney disease during blood pressure reduction. Ann. Intern. Med. 2018;169:610–618. doi: 10.7326/M18-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ju W., Nair V., Smith S., Zhu L., Shedden K., Song P.X.K., Mariani L.H., Eichinger F.H., Berthier C.C., Randolph A., Lai J.Y.-C., Zhou Y., Hawkins J.J., Bitzer M., Sampson M.G., Thier M., Solier C., Duran-Pacheco G.C., Duchateau-Nguyen G., Essioux L., et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci. Transl. Med. 2015;7:316ra193. doi: 10.1126/scitranslmed.aac7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu L., Li X.-Q., Chang D.-Y., Zhang H., Li J.-J., Wu S.-L., Zhang L.-X., Chen M., Zhao M.-H. Associations of urinary epidermal growth factor and monocyte chemotactic protein-1 with kidney involvement in patients with diabetic kidney disease. Nephrol. Dial. Transplant. 2020;35:291–297. doi: 10.1093/ndt/gfy314. [DOI] [PubMed] [Google Scholar]
- 192.Nowak N., Skupien J., Smiles A.M., Yamanouchi M., Niewczas M.A., Galecki A.T., Duffin K.L., Breyer M.D., Pullen N., Bonventre J.V., Krolewski A.S. Markers of early progressive renal decline in type 2 diabetes suggest different implications for etiological studies and prognostic tests development. Kidney Int. 2018;93:1198–1206. doi: 10.1016/j.kint.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Satirapoj B., Dispan R., Radinahamed P., Kitiyakara C. Urinary epidermal growth factor, monocyte chemoattractant protein-1 or their ratio as predictors for rapid loss of renal function in type 2 diabetic patients with diabetic kidney disease. BMC Nephrol. 2018;19:246. doi: 10.1186/s12882-018-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lu Y., Chen L., Zhao B., Xiao Z., Meng T., Zhou Q., Zhang W. Urine AQP5 is a potential novel biomarker of diabetic nephropathy. J. Diabetes Complicat. 2016;30:819–825. doi: 10.1016/j.jdiacomp.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kim J.H., Kim S.S., Kim I.J., Lee M.J., Jeon Y.K., Kim B.H., Song S.H., Kim Y.K. Nonalbumin proteinuria is a simple and practical predictor of the progression of early-stage type 2 diabetic nephropathy. J. Diabetes Complicat. 2017;31:395–399. doi: 10.1016/j.jdiacomp.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 196.Gaipov A., Taubaldiyeva Z., Askarov M., Turebekov Z., Kozina L., Myngbay A., Ulyanova O., Tuganbekova S. Infusion of autologous bone marrow derived mononuclear stem cells potentially reduces urinary markers in diabetic nephropathy. J. Nephrol. 2019;32:65–73. doi: 10.1007/s40620-018-0548-5. [DOI] [PubMed] [Google Scholar]
- 197.Messchendorp A.L., Meijer E., Boertien W.E., Engels G.E., Casteleijn N.F., Spithoven E.M., Losekoot M., Burgerhof J.G.M., Peters D.J.M., Gansevoort R.T. Urinary biomarkers to identify autosomal dominant polycystic kidney disease patients with a high likelihood of disease progression. Kidney International Reports. 2018;3:291–301. doi: 10.1016/j.ekir.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.von Scholten B.J., Reinhard H., Hansen T.W., Oellgaard J., Parving H.-H., Jacobsen P.K., Rossing P. Urinary biomarkers are associated with incident cardiovascular disease, all-cause mortality and deterioration of kidney function in type 2 diabetic patients with microalbuminuria. Diabetologia. 2016;59:1549–1557. doi: 10.1007/s00125-016-3937-0. [DOI] [PubMed] [Google Scholar]
- 199.Chanrat E., Worawichawong S., Radinahamed P., Sathirapongsasuti N., Nongnuch A., Assanatham M., Udomsubpayakul U., Kitiyakara C. Urine epidermal growth factor, monocyte chemoattractant protein-1 or their ratio as predictors of complete remission in primary glomerulonephritis. Cytokine. 2018;104:1–7. doi: 10.1016/j.cyto.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 200.Dubin R.F., Judd S., Scherzer R., Shlipak M., Warnock D.G., Cushman M., Sarnak M., Parikh C., Bennett M., Powe N., Peralta C.A. Urinary tubular injury biomarkers are associated with ESRD and death in the REGARDS study. Kidney International Reports. 2018;3:1183–1192. doi: 10.1016/j.ekir.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Abeijon C., Alves F., Monnerat S., Wasunna M., Mbui J., Viana A.G., Bueno L.L., Siqueira W.F., Carvalho S.G., Agrawal N., Fujiwara R., Sundar S., Campos-Neto A. Development of a multiplexed assay for detection of leishmania donovani and leishmania infantum protein biomarkers in urine samples of patients with visceral leishmaniasis. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.02076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Atif M., AlSalhi M.S., Devanesan S., Masilamani V., Farhat K., Rabah D. A study for the detection of kidney cancer using fluorescence emission spectra and synchronous fluorescence excitation spectra of blood and urine. Photodiagnosis Photodyn. Ther. 2018;23:40–44. doi: 10.1016/j.pdpdt.2018.05.012. [DOI] [PubMed] [Google Scholar]