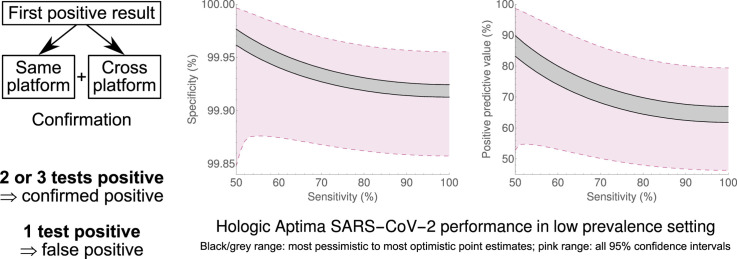
Abstract
Objectives
When the prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is low, many positive test results are false positives. Confirmatory testing reduces overdiagnosis and nosocomial infection and enables real-world estimates of test specificity and positive predictive value. This study estimates these parameters to evaluate the impact of confirmatory testing and to improve clinical diagnosis, epidemiological estimation and interpretation of vaccine trials.
Methods
Over 1 month we took all respiratory samples from our laboratory with a patient's first detection of SARS-CoV-2 RNA (Hologic Aptima SARS-CoV-2 assay or in-house RT-PCR platform), and repeated testing using two platforms. Samples were categorized by source, and by whether clinical details suggested COVID-19 or corroborative testing from another laboratory. We estimated specificity and positive predictive value using approaches based on maximum likelihood.
Results
Of 19 597 samples, SARS-CoV-2 RNA was detected in 107; 52 corresponded to first-time detection (0.27% of tests on samples without previous detection). Further testing detected SARS-CoV-2 RNA once or more (‘confirmed’) in 29 samples (56%), and failed to detect SARS-CoV-2 RNA (‘not confirmed’) in 23 (44%). Depending upon assumed parameters, point estimates for specificity and positive predictive value were 99.91–99.98% and 61.8–89.8% respectively using the Hologic Aptima SARS-CoV-2 assay, and 97.4–99.1% and 20.1–73.8% respectively using an in-house assay.
Conclusions
Nucleic acid amplification testing for SARS-CoV-2 is highly specific. Nevertheless, when prevalence is low a significant proportion of initially positive results fail to confirm, and confirmatory testing substantially reduces the detection of false positives. Omitting additional testing in samples with higher prior detection probabilities focuses testing where it is clinically impactful and minimizes delay.
Keywords: COVID-19 diagnostic testing, COVID-19 pandemic, Nucleic acid amplification techniques, SARS-CoV-2, Sensitivity and specificity
Graphical abstract
Introduction
The coronavirus disease 19 (COVID-19) pandemic [1,2] continues to cause morbidity and mortality [[3], [4], [5], [6]]. In some regions, following seasonal changes and infection control measures, incidence and prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have decreased [7], whilst testing capacity has increased [[8], [9], [10]]. As much testing capacity remains operational, in such settings many being tested will be infection-free.
The commonest clinical laboratory testing employed to detect SARS-CoV-2 is nucleic acid amplification testing (NAAT). NAAT techniques detect SARS-CoV-2 RNA using amplification steps that bind specific complementary primers to nucleic acid; probes assay the amplification progress. With well-designed primers and probes, NAAT is highly sensitive and specific. However, in practice, issues such as inadequate clinical sampling, sample degradation, and reaction inhibition affect sensitivity, and issues such as non-specific probe breakdown, amplicon contamination and sample error can affect specificity even in well-run laboratories. Initial technical validations of new diagnostic assays often only partially reflect clinical workflows. They often test limited numbers of samples, and so are underpowered to detect small proportions of false positives or false negatives. In low-prevalence, high-throughput settings, false-positive results will occur regularly, despite high specificity, causing unnecessary community isolation and contact tracing, and nosocomial infection if inpatients with false-positive tests are cohorted with infectious patients. However, the low incidence of positive results makes feasible confirmatory testing to reduce false-positive rates. This additionally enables us to understand predictors that should prompt re-evaluation of results, allowing laboratories to prepare for when an increased incidence of positive results makes confirmatory testing on all initially positive samples impracticable. Beyond these clinical benefits, this work allows improved epidemiological estimation of infection rates, which will guide policy and allow more accurate statistical analysis of infection rates in study settings (e.g. vaccine trials).
The Cambridge Clinical Microbiology and Public Health Laboratory is a regional clinical laboratory in the East of England. Since early 2020 it has undertaken SARS-CoV-2 NAAT on many inpatient and community samples. On 22nd June 2020, following decreasing local positive test result incidences, the laboratory began same- and cross-platform confirmatory testing for patients with positive results but without previous positive tests. By combining highly specific tests in a way that minimizes repetition of errors, the specificity of reported results could be increased further, reducing inconvenience and harm to the few uninfected people with initially positive results. We report the results from testing undertaken during the first month after policy implementation.
Methods
Patient sampling, initial preparation and testing
Samples were received within the routine diagnostic service. Upper-airway samples were collected into viral transport medium following Public Health England guidelines [11]. Lower-airway samples (sputum, tracheal aspirate or bronchoalveolar lavage washings) were transported in universal containers and underwent mucolysis with Mucolyse Sputum Digestant (Pro-Lab Diagnostics, Richmond Hill, Canada). Initial testing used one of two platforms: reverse transcription polymerase chain reaction (RT-PCR) against one or two targets using an in-house assay, or transcription-mediated amplification against two targets using the Aptima SARS-CoV-2 assay on the Panther System (Hologic, Marlborough, United States). Assays are detailed in the web-only Supplementary Material (Methods).
Confirmation of first positive samples
All samples testing positive for SARS-CoV-2 were identified. Samples from patients with a preceding positive SARS-CoV-2 NAAT were reported positive without confirmation, reflecting the higher prior probability of true-positive results, and the lower probability of significant adverse impact from false positives. Only preceding positive results from samples tested by our laboratory, or our local hospital's point-of-care laboratory [12], were considered here. Although data on previous tests in other laboratories were sought, we conducted confirmatory testing without awaiting this information.
Samples from patients without a preceding positive SARS-CoV-2 NAAT underwent repeat testing on both platforms (same- and cross-platform), starting from the original sample or aliquot. Aliquots were stored at −20°C for at most 24 hours, with at most one freeze–thaw cycle, before confirmatory testing. Samples with at least two positive results were reported as confirmed positive; samples with one of three positive results (i.e. that twice failed to confirm) were reported as negative, i.e. as a false positive. Where a sample was insufficient for two confirmatory tests, we aimed to prioritize cross-platform confirmation; we reported one of two positive results as indeterminate and requested a repeat sample.
For all positive tests, we recorded whether clinical details indicated symptoms or a previous positive SARS-CoV-2 NAAT, using data submitted with testing requests, or electronic patient records where available. We initially advised requesters of positive results awaiting confirmation, but following consultation with stakeholders during the study period we switched to reporting only after confirmation, as we judged risks from reporting false-positive results exceeded risks from short delays in reporting true positives in our population at this stage.
Effect of storage on confirmation
We investigated the effect of freezing/thawing on positive result repeatability: see Supplementary Material (Methods).
Data analysis
We prospectively recorded and analysed all samples with first test results issued between 22nd June and 21st July 2020. Descriptive statistics (patient sex and age, sample type) were collected and analysed by testing platform and initial result. Statistical analysis was undertaken in R, version 3.2.3 [13]. The Supplementary Material contains code to reproduce the analyses. Binomial conditional probabilities for RNA detection were calculated using the GenBinomApps package (version 1.1). The odds ratio of RNA detection by initial test given a previous positive testing sample from the individual versus no previous positive was calculated using the exact2x2 package (version 1.4.1). Additional calculations used the packages stats4 (base package), numDeriv (version 2016.8.1.1).
For samples from patients without a previous positive result, specificity, prevalence of samples containing virus, and positive predictive value (PPV) were estimated for both Aptima SARS-CoV-2 and in-house PCR assays using a maximum likelihood approach, described in the Supplementary Material (Methods). As these estimates depend upon test sensitivity, which is unknown but not expected to impact estimates heavily in a low-prevalence setting for plausible sensitivity values [14], estimates were generated for a range of sensitivities (50–100%) to ensure that results were not excessively sensitive to assumed sensitivity values. To obtain a full range of possible estimates, specificity, prevalence and PPV were estimated under extreme cases of all indeterminate results confirming, and no indeterminate result confirming, the initial positive result. There were few positive in-house assay results, so these were grouped together regardless of extraction platform or number of targets.
The impact of clinical information on predicting positive result confirmation was evaluated using Freeman–Halton tests. The impact of quantitative assay output—relative light units (RLUs) for the Panther system, threshold cycle (Ct) for RNA-dependent RNA polymerase detection by the in-house assay—on predicting positive result confirmation was evaluated using Mann–Whitney U tests (conducting a sensitivity analysis for indeterminate results by removing them from analysis, and grouping them with positive and with negative results).
Ethical approval
All testing was undertaken within routine care; analysis was undertaken within routine service evaluation. Ethical approval for this study was therefore not required. Our laboratory evaluates assays for validation and improves them as part of its role [15]. Testing and analysis were undertaken in accordance with the UK Human Tissue Act 2004, as amended [16].
Results
Sample and test outcome characteristics
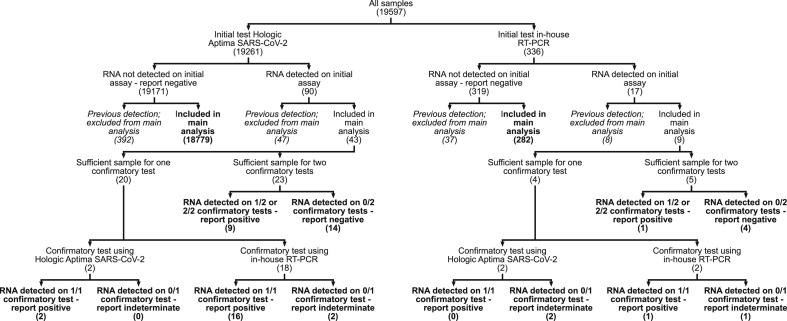
In total, 19 597 samples had valid test results during the 1-month period (Fig. 1 ); 19 261 were initially tested with the Aptima SARS-CoV-2 assay, and 336 by in-house RT-PCR (Table 1 gives patient demographics and sample characteristics). Four hundred and eighty-four samples were from people with a previous positive test, of which 55 (11.3%, 95% confidence interval (CI) (8.7%,14.5%)) tested positive; 19 113 samples were from people without a previous positive test, of which 52 (0.27%, 95%CI (0.20%,0.36%)) had RNA detected in the first test. The odds ratio of RNA detection by initial test, given a previous positive testing sample from the individual versus no previous positive, was 46.9 (95%CI (31.4,70.5), p < 0.001). There was substantial heterogeneity in specimen type and patient sex and age between samples tested on different platforms and between samples with different testing outcomes (Table 1). Notably, first positive initial tests from the Aptima SARS-CoV-2 assay came predominantly from upper-airway samples, whereas first positive initial tests from the in-house RT-PCR predominantly came from lower-airway samples.
Fig. 1.
Platforms on which samples were tested, and outcomes of initial and confirmatory testing. Numbers in parentheses correspond to the numbers of samples at each stage of analysis. Bold endpoints correspond to the outcomes analysed to derive the main specificity, prevalence and positive predictive value results. Italic endpoints correspond to the outcomes analysed to derive results relating to samples from patients with previous SARS-CoV-2 RNA detection.
Table 1.
Characteristics of patients and sample types
| All samples | Hologic Aptima SARS-CoV-2 TMA |
In-house PCR |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) All samples | People with previous positive test |
People without previous positive test |
(1) All samples | People with previous positive test |
People without previous positive test |
||||||||||
| (2) All | (3) Negative test | (3) Initial positive test | (2) All | (4) Negative test | (4) Initial positive test | (5) All | (6) Negative test | (6) Initial positive test | (5) All | (7) Negative test | (7) Initial positive test | ||||
| No. of samples | 19 597 | 19 261 | 439 | 392 | 47 | 18 822 | 18 779 | 43 | 336 | 45 | 37 | 8 | 291 | 282 | 9 |
| Female sex n (%) |
11 992 (61.2) | 11 808 (61.3)a | 249 (56.7)b | 231 (58.9)c | 18 (38.3)3 | 11 559 (61.4)b | 11 523 (61.3)d | 36 (83.7)d | 184 (54.8)a | 12 (26.7)e | 12 (32.4) | 0 (0) | 172 (59.1)e | 167 (59.2) | 5 (55.6) |
| Median age, y (IQR) | 58 (38–77) | 58 (38–77)f | 74 (57–86)g | 74 (56–87) | 74 (65–83) | 58 (38–77)g | 58 (38–77)h | 71 (56-88)h | 64 (47–77)f | 69 (52–75) | 63 (49–78)i | 74 (74–74)i | 63 (45–77) | 63 (46–77) | 61 (37–77) |
| URT specimens – no. (%) | 19 359 (98.8) | 19 148 (99.4)j | 431 (98.2)k | 384 (97.8) | 47 (100) | 18 717 (99.4)k | 18 677 (99.5)l | 40 (93.0)l | 211 (62.8)j | 33 (73.3) | 25 (67.6) | 8 (100) | 178 (61.2) | 177 (62.8)m | 1 (11.1)m |
TMA, transcription-mediated amplification; PCR, polymerase chain reaction; URT, upper respiratory tract.
Ages are rounded to the nearest year. Heterogeneity comparisons are made within rows between the columns whose headers are marked with identical numbers in parentheses. Proportion female and proportion URT specimen comparisons are made using Fisher's exact test. Age comparisons are made using the Mann–Whitney U test. Values yielding p < 0.05 are marked and reported, together with odds ratios (ORs) reported to two significant figures or medians of difference between samples reported to one decimal place, as appropriate, followed by 95% confidence intervals. Given 21 separate independent tests, p < 0.05 would be expected to occur by chance on average 1.05 times. Note that the table's construction means that the tests are not independent of each other.
p 0.015, OR 1.3 (1.1,1.6).
p 0.047, OR 0.82 (0.68,1.00).
p 0.0081, OR 2.3 (1.2,4.5).
p 0.0025, OR 0.31 (0.13,0.68).
p < 0.001, OR 0.25 (0.12,0.52).
p 0.0043, median of difference –3.7 y (–6.3,–1.2).
p < 0.001, median of difference 12.6 y (10.3,14.9).
p 0.0023, median of difference –11.6 y (–19.5,–4.2).
p 0.045, median of difference –12.3 y (–24.9,0.0).
p < 0.001, OR 100 (75 130).
p 0.004, OR 0.30 (0.15,0.68).
p 0.0018, OR 14 (3.5,43).
p 0.0027, OR 13 (2.0, 300).
Effect of freeze–thaw cycle
Clinical sample storage did not significantly affect positive results: see Supplementary Material (Results).
Specificity, prevalence and positive predictive value
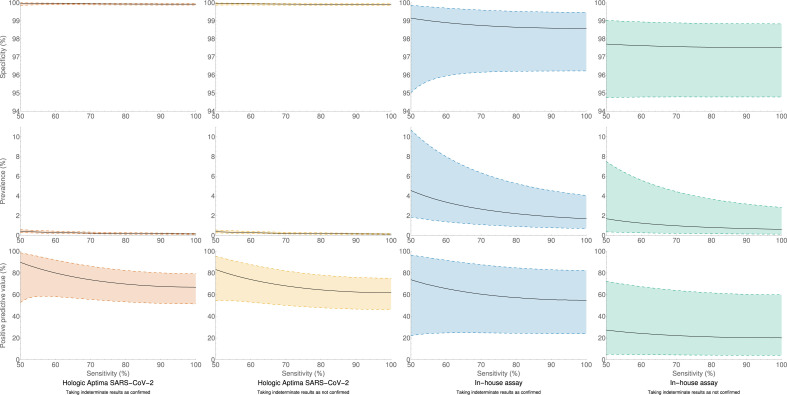
Depending upon the assumed sensitivity and status of indeterminate results, testing with the Aptima SARS-CoV-2 assay was found to have a specificity of 99.91–99.98%, with positive sample prevalence of 0.14–0.41% and PPV 61.8–89.8% (unions of 95%CIs (99.85%,100.00%), (0.10%,0.59%) and (46.3%,98.6%), respectively). Testing with the in-house RT-PCR had a specificity of 97.4–99.1%, with positive sample prevalence 0.62–4.6% and PPV 20.1–73.8% (unions of 95%CIs (94.6%,99.9%), (0.13%,10.7%) and (4.0%,96.5%), respectively). These results are summarized in Fig. 2 and Supplementary Material Fig. S1. Supplementary Material Tables S1–S4 contain output data.
Fig. 2.
Calculated specificity, prevalence of samples containing virus, and positive predictive value of the Hologic Aptima SARS-CoV-2 assay and the in-house assay. Calculated values are solid lines, with 95% confidence intervals (shaded regions enclosed by dashed lines), for assumed test sensitivities from 50% to 100%, and for the two extreme assumptions of all indeterminate samples being confirmed as positive, and all indeterminate samples not confirming as positive. A version of the Hologic Aptima SARS-CoV-2 plots with zoomed vertical scales may be found in web-only Supplementary Figure S1.
Demographic and analytical characteristics of confirming versus non-confirming results
Few specimens were submitted with sufficient clinical information to determine whether a patient was symptomatic or had previously had a positive test from another laboratory. Of the 43 specimens with initial first positive results from the Hologic Aptima SARS-CoV-2 assay, four had information indicating symptoms or a previous positive test (two both, one symptoms only, one previous positive only), of which one specimen with both symptoms and a previous positive test was ultimately reported as indeterminate and the remaining three were reported as positive (Freeman–Halton p 0.071). Of the nine specimens with initial first positive results from the in-house assay, five had information indicating symptoms (and none a previous positive test), of which two were ultimately reported as positive, two as indeterminate, and one as negative (Freeman–Halton p 0.38). Eight of these nine specimens, including all five with information of symptoms, were lower-airway specimens.
Figure 3 shows the RLU output of the Panther analysis of Aptima SARS-CoV-2 assay results initially determined to be first positives, and Supplementary Material Fig. S2 shows output by total number of positive assays. There is a statistically significant difference between RLU output of initial positive results ultimately reported negative versus those ultimately reported positive (p < 0.001 in all cases, difference in RLU and 95%CIs 389 (271,456), 397 (311,461) and 378 (199,444) as indeterminate results are removed, considered negative, and considered positive, respectively). However, there is overlap in RLU outputs for samples ultimately reported positive and samples ultimately reported negative.
Fig. 3.
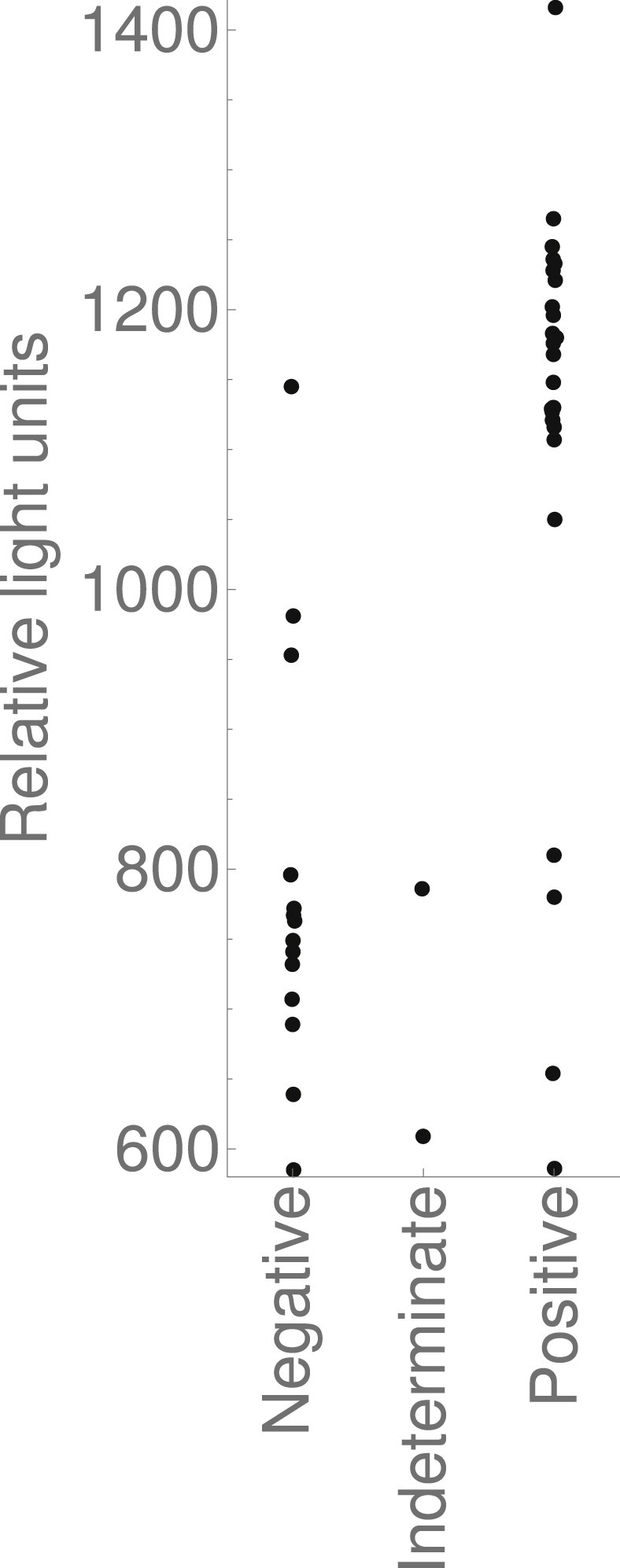
Relative light unit output of the Hologic Panther analysis of the Aptima SARS-CoV-2 assay of samples initially determined to be first positives. Grouped by the reporting categories detailed in the flowchart in Fig. 1.
Figure 4 shows the Ct output of in-house assay results initially determined to be first positives, and Supplementary Material Fig. S3 shows output by total number of positive assays. There is no statistically significant difference between Ct values of initial positive results ultimately reported negative versus those ultimately reported positive (p 0.53, 0.38 and 1.00 as indeterminate results are removed, considered negative, and considered positive, respectively).
Fig. 4.
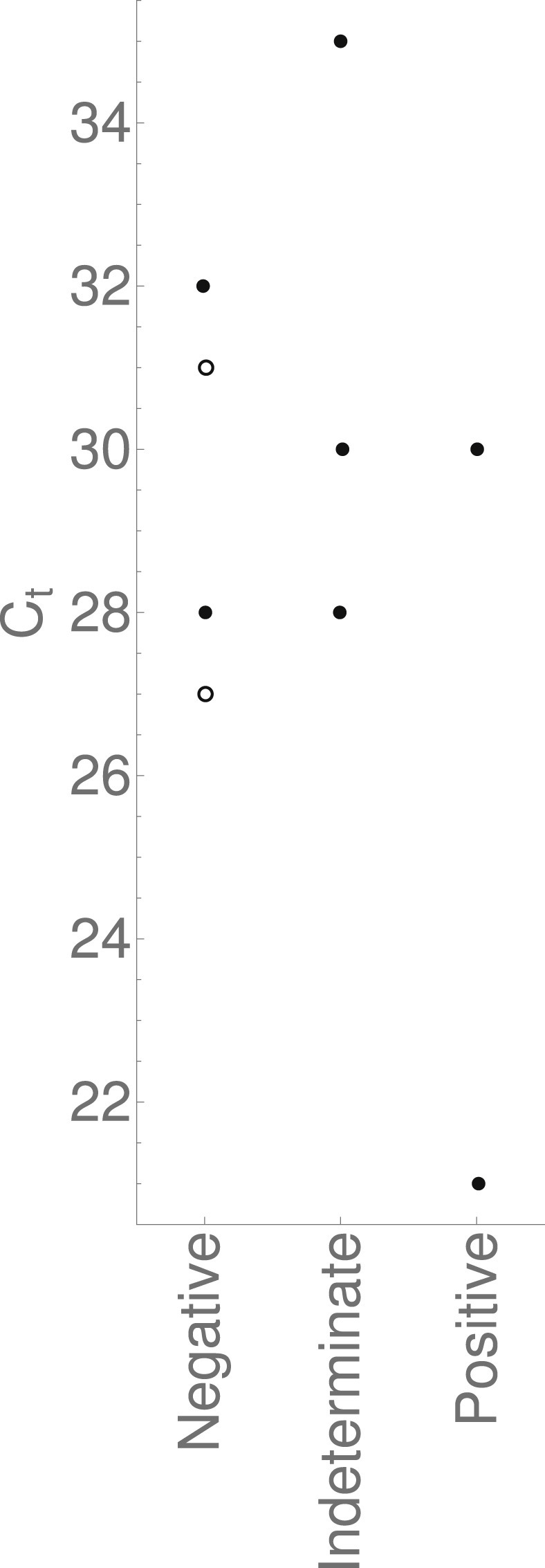
Threshold cycles of the samples initially determined by the in-house assay to be first positives. Grouped by the reporting categories detailed in the flowchart in Fig. 1. The solid data points correspond to samples assayed when the in-house assay had a single target; the hollow points correspond to samples assayed when the in-house assay had two targets. In both cases only the RdRp target, and not the S target, was detected.
Discussion
Our results show that, regardless of testing sensitivity, both platforms have high specificity, and single positive results from either are reliable for diagnosis in high-risk populations. However, despite a small absolute difference in false-positive rates, the relative difference between testing platforms is marked. This requires consideration when screening low-risk populations, to avoid adverse effects to individuals from overdiagnosis and to avoid heterogeneity in estimates of infection rates attributable to different testing platforms. We have shown that when prevalence is low, a clinically significant proportion of first positive results are unrepeatable on the same testing platform, meaning confirmatory testing has utility for smaller laboratories using single platforms. Conducting confirmatory tests on all platforms, including the first used, ensures that the most sensitive platform is used for confirmation, even with unknown sensitivities. We have shown that clinically significant positive results are repeatable, so confirmatory testing helps to discriminate true-from false-positive results.
We have further demonstrated that, in a large clinical laboratory, appropriately stratifying patients by prior test probability is key to achieving accurate and timely results whilst minimizing workload. The most important subpopulation to stratify is those with prior evidence of SARS-CoV-2 shedding, where both the risk and the impact of a false-positive test are much lower. Because the biochemical properties of assaying SARS-CoV-2 in samples from those with late acute immune responses might differ from assaying in other samples, it is especially important to analyse this population separately when inferring prevalences and test properties.
In clinical laboratory settings, much focus is on minimizing and detecting false-positive results from contamination or non-specific probe breakdown, as opposed to other causes. The quantitative difference between confirming and non-confirming results for one of our assays supports the idea that many false-positive results could be attributed to non-specific reactions or low-level sample contamination. (Another explanation is a low-level true-positive but non-repeatable result, which would seldom be clinically significant.) When confirming all initial first positive results is impossible, this observation allows stratification to decide which tests to repeat. However, the substantial overlap in quantitative results between those confirming and those not confirming highlights the presence of other causes of false-positive results. Restricting confirmatory testing to an intermediate category will incorrectly categorize samples giving apparently strongly positive, but non-repeatable, results. Considering sources of false-positive results also yields this study's largest limitation: errors before the confirmatory testing process branches from the initial process will not be detected, and confirmatory testing may falsely reassure.
When analytical test sensitivity is low, failure to confirm may represent a false-negative confirmatory test. However, our data on repeatability, coupled with validation literature indicating that SARS-CoV-2 tests have high analytical sensitivity [17,18], suggest that lower reported overall clinical sensitivity predominantly arises during sampling; confirmation tests of an initially positive sample would not reflect such insensitivity. Any clinical sensitivity issues must be considered when making prevalence estimates in a population of people, rather than the population of samples they have produced.
We observed substantial heterogeneity between samples following different testing pathways through our laboratory. We highlight the different extraction methods used for our in-house assay and the additional primer/probe set in some tests, meaning the reported assay specificity represents a composite; for real-world clinical and epidemiological applications, this remains useful. Another issue is overrepresentation of more complex (lower respiratory) samples in the in-house assay positive results. Processing these samples involves additional steps, yielding a potential confounder when comparing our assays' performances. Nonetheless, it is perhaps unsurprising that the commercial assay, with fewer manual steps, shows a lower false-positive rate. Multiple possible factors may explain other aspects of observed heterogeneity, e.g. whether there are differences in age and sex representation in screened residential facilities, in symptomatic individuals, or in those shedding viral RNA over a prolonged period.
Prevalence and PPV estimates reported refer to the populations tested, rather than the general populations from which they are drawn. These estimates are appropriate when considering clinical interpretation of assay results, but differences between these populations and the underlying general populations must be accounted for when making epidemiological estimates in underlying populations.
This study gives quantitative estimates of SARS-CoV-2 NAAT specificity, useful to guide clinical interpretation of test results, in planning clinical services, and for power calculations for vaccine and therapeutic trials. The methods and code can calculate testing parameters in other laboratories. We have shown the utility and limitations of confirmatory testing, reminding us to focus on managing patients and not treat test results in isolation.
Author contributions
JPS: conceptualization, methodology, software, formal analysis, investigation, data curation, writing—original draft, visualization, project administration. MW: investigation, writing—review and editing. AAS: investigation, writing—review and editing. SP: methodology, validation, resources, writing—review and editing. MDF: conceptualization, methodology, formal analysis, writing—review and editing. DS: investigation, writing—review and editing. MDC: methodology, resources, writing—review and editing. HZ: conceptualization, writing—review and editing. HJ: conceptualization, methodology, writing—review and editing, supervision.
Transparency declaration
The authors declare that they have no conflicts of interest. JPS is funded by a fellowship from the Mason Medical Research Foundation. No other author has received external funding for this study.
Acknowledgements
We are extremely grateful to the staff of the Cambridge Clinical Microbiology and Public Health Laboratory, and particularly to the laboratory managers, senior virology biomedical scientists, staff involved in SARS-CoV-2 testing, volunteers from other Public Health England (PHE) sites, and data analysts, whose tireless work has been crucial in serving our patient population. We are particularly grateful to our clinical colleagues Mir Mubariz Hussain, Chloe Myers, and Kathryn Rolfe for their involvement in clinical case management and discussion as routine procedures were developed. The discussions of diagnostic assays and extraction platforms in this report should not be taken as a recommendation by PHE or any of the other institutions involved.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.10.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinnathamby M.A., Whitaker H., Coughlan L., Lopez Bernal J., Ramsay M., Andrews N. All-cause excess mortality observed by age group and regions in the first wave of the COVID-19 pandemic in England. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.28.2001239. pii=2001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelozzi P., De’Donato F., Scortichini M., de Sario M., Noccioli F., Rossi P. Mortality impacts of the coronavirus disease (COVID-19) outbreak by sex and age: rapid mortality surveillance system, Italy. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.19.2000620. 1 February to 18 April 2020. pii=2000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nogueirai P.J., De Araújo Nobre M., Nicola P.J., Furtado C., Vaz Carneiro A. Excess mortality estimation during the COVID-19 pandemic: preliminary data from Portugal. Acta Med Port. 2020;33:376–383. doi: 10.20344/amp.13928. [DOI] [PubMed] [Google Scholar]
- 6.Menachemi N., Yiannoutsos C.T., Dixon B.E., Duszynski T.J., Fadel W.F., Wools-Kaloustian K.K. Population point prevalence of SARS-CoV-2 infection based on a statewide random sample—Indiana, April 25–29, 2020. Morb Mortal Wkly Rep. 2020;69:960–964. doi: 10.15585/mmwr.mm6929e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pouwels K.B., House T., Robotham J.V., Birrell P.J., Gelman A., Bowers N. Community prevalence of SARS-CoV-2 in England: results from the ONS coronavirus infection survey pilot. medRxiv. 2020 doi: 10.1101/2020.07.06.20147348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reusken C.B.E.M., Broberg E.K., Haagmans B., Meijer A., Corman V.M., Papa A. Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.6.2000082. pii=2000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matheeussen V., Loens K., Lammens C., Vilken T., Koopmans M., Goossens H. Preparedness of European diagnostic microbiology labs for detection of SARS-CoV-2, March 2020. J Clin Virol. 2020;128:104432. doi: 10.1016/j.jcv.2020.104432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matheeussen V., Corman V.M., Donoso Mantke O., McCulloch E., Lammens C., Goossens H. International external quality assessment for SARS-CoV-2 molecular detection and survey on clinical laboratory preparedness during the COVID-19 pandemic, April/May 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.27.2001223. pii=2001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Public Health England COVID-19: laboratory investigations and sample requirements for diagnosis. https://www.gov.uk/government/publications/wuhan-novel-coronavirus-guidance-for-clinical-diagnostic-laboratories/laboratory-investigations-and-sample-requirements-for-diagnosing-and-monitoring-wn-cov-infection [Internet]. Available from:
- 12.Collier D., Assennato S., Sithole N., Sharrocks K., Ritchie A., Ravji P. Rapid point of care nucleic acid testing for SARS-CoV-2 in hospitalised patients: a clinical trial and implementation study. Cell Rep Med. 2020;1:100062. doi: 10.1016/j.xcrm.2020.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Core Team R: a Language and Environment for statistical computing. 2015. https://www.r-project.org/ [Internet]. Vienna, Austria. Available from:
- 14.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 15.Public Health England . UK Standards for Microbiology Investigations; 2017. UK Standards for Microbiology Investigations: Q1. Evaluation, validation and verifications of diagnostic tests. Report No Q1. [Google Scholar]
- 16.The National Archives. Human Tissue Act 2004.
- 17.Gorzalski A.J., Tian H., Laverdure C., Morzunov S., Verma S.C., VanHooser S. High-throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J Clin Virol. 2020;129:104501. doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.