Abstract
Objective
The human adaptive fasting response enables survival during periods of caloric deprivation. A crucial component of the fasting response is the shift from glucose metabolism to utilization of lipids, underscoring the importance of adipose tissue as the central lipid-storing organ. The objective of this study was to investigate the response of adipose tissue to a prolonged fast in humans.
Methods
We performed RNA sequencing of subcutaneous adipose tissue samples longitudinally collected during a 10-day, 0-calorie fast in humans. We further investigated observed transcriptional signatures utilizing cultured human monocytes and Thp1 cells. We examined the cellularity of adipose tissue biopsies with transmission electron microscopy and tested for associated changes in relevant inflammatory mediators in the systemic circulation by ELISA assays of longitudinally collected blood samples.
Results
Coincident with the expected shift away from glucose utilization and lipid storage, we demonstrated downregulation of pathways related to glycolysis, oxidative phosphorylation, and lipogenesis. The canonical lipolysis pathway was also downregulated, whereas fasting drove alternative lysosomal paths to lipid digestion. Unexpectedly, the dominant induced pathways were associated with immunity and inflammation, although this only became evident at the 10-day time point. Among the most augmented transcripts were those associated with macrophage identity and function, such as members of the erythroblast transformation-specific (ETS) transcription factor family. Key components of the macrophage transcriptional signal in fasting adipose tissue were recapitulated with induced expression of two of the ETS transcription factors via cultured macrophages, SPIC and SPI1. The inflammatory signal was further reflected by an increase in systemic inflammatory mediators.
Conclusions
Collectively, this study demonstrates an unexpected role of metabolic inflammation in the human adaptive fasting response.
Keywords: Adipose tissue, Fasting, Inflammation, Macrophage, Human, SPIC
Highlights
-
•
Prolonged fasting drives an inflammatory transcriptome in human subcutaneous fat.
-
•
Fasting induction of a macrophage signature includes the transcription factor SPIC.
-
•
SPIC expression in macrophages recapitulates aspects of the fasting transcriptome.
-
•
Prolonged fasting drives circulating markers of systemic inflammation.
-
•
Fasting drives lysosomal pathways, while canonical lipolysis pathways are attenuated.
1. Introduction
Humans can survive periods of fasting lasting well beyond a month, in contrast to rodent models that succumb to starvation within days. The adaptive responses that enable survival during fasting periods are relevant beyond the context of famine or psychiatric conditions associated with self-induced starvation [1]. On the one hand, evolutionary selection for efficiencies that have enabled humans to survive cycles of famine may explain human obesogenic tendencies, the so-called thrifty gene hypothesis [2]. Conversely, caloric restriction or fasting may drive beneficial pathways in disease states including hypertension, atherosclerosis, diabetes, and cancer and may prolong lifespans [3]. These collective factors underscore the ongoing interest in regulatory mechanisms that drive the fasting response.
Adapting to fasting involves a coordinated series of metabolic shifts [4]. The requirement for glucose is initially supported by glycogenolysis and utilization of muscle-derived amino acids as substrate for gluconeogenesis [4,5]. By 48–72 h into a fast, lipid metabolism facilitates sparing the vital protein backbone of the organism [4], resulting in catabolism of lipid-derived fatty acids via beta oxidation, or in the case of the central nervous system, utilization of lipid-derived ketone bodies.
As the dominant lipid storage depot, adipose tissue (AT) plays a central role in the adaptive transition from glucose metabolism to catabolism of stored lipids during fasting. Adipocytes mobilize lipids from intracellular protein-coated triglyceride droplets. Since triglycerides cannot directly cross the plasma membrane, the canonical view holds that they are released by lipolysis into the circulation in the form of free fatty acids, the coordinated release of which involves three enzymes, adipose triglyceride lipase (ATGL), hormone sensitive lipase (HSL), and monoacylglycerol lipase (MGL), which catalyze the sequential removal of the three fatty acid chains from the glycerol backbone [6]. However, several lines of evidence suggest that alternative mechanisms may also be involved in the mobilization of adipocyte lipid stores. First, loss of function of the gene encoding HSL, LIPE, is manifested by relatively small adipocytes, not the large adipocytes that might be expected from a pure defect in lipid catabolism [7]. Second, genetic deletion of critical lipase genes in mice attenuates, but does not neutralize, AT lipolysis [8,9]. Third, recent murine studies pointed to an alternative mechanism of adipocyte lipid mobilization, whereby resident macrophages may support lipolysis in AT by catabolizing triglycerides that have been released into the interstitial space as a core component of adipocyte-derived vesicles [10].
Challenges to the canonical view of lipolysis, coupled with the more general centrality of the role of AT in the adaptive fasting response, provided a rationale to revisit the AT response to fasting through unbiased, genome-scale transcriptional profiling. We previously performed a 10-day zero-calorie fast in healthy volunteers [11]. Here, we describe the transcriptional response to fasting in a subset of subjects from whom we also obtained serial subcutaneous AT biopsies at three prespecified time points, inclusive of the transition to lipid metabolism and contraction of AT mass. We unexpectedly discovered attenuation of transcripts for genes involved in the canonical lipolysis pathways, instead finding induction of transcriptional signatures associated with lysosomal function and inflammation, including SPIC, an ETS transcription factor and candidate regulator of macrophage specification. SPIC gain of function in cultured monocytes recapitulated key components of the inflammatory transcriptional signature evident in whole AT with fasting. The transcriptional inflammatory signal was further reflected by increased resident macrophages in AT biopsy specimens and an increase in systemic inflammatory mediators. Collectively, this study demonstrates an unexpected role for metabolic inflammation in the human adaptive fasting response.
2. Methods
2.1. Human fasting study
This study was approved by the Partners HealthCare Institutional Review Board (Boston, MA, USA) and complied with the Health Insurance Portability and Accountability Act guidelines. Written informed consent was obtained from all the subjects.
We previously published the protocol for the 10-day inpatient fasting study [11]. In this report, we present data for a subset of 7 subjects who underwent serial subcutaneous AT biopsies. Subjects were recruited through online advertisements. All had normal thyroid function and regular menstrual cycles (women). Subjects with a history of an eating disorder or any chronic illness, including diabetes mellitus, were excluded. The subjects were admitted to the Center for Clinical Investigation at Brigham and Women's Hospital for a 10-day fast in the morning after fasting overnight. During the inpatient fast, their only oral intake consisted of water ad libitum, a daily multivitamin, 20 mEq of potassium chloride daily to prevent hypokalemia, and 200 mg of allopurinol daily. Serial subcutaneous AT biopsies were collected from the periumbilical region on the morning of admission (baseline, time = 0), day 1 (morning after admission), and the morning of the final day of the fast (day 10). A 14-gauge Temno biopsy device was utilized, which enabled the directed collection of core samples such that different regions could be sampled at different time points. Human fat specimens were immediately placed in RNAlater (Life Technologies) and stored at −80 °C. One sample was also fixed in 4% paraformaldehyde.
2.2. Body composition
Body composition, including fat mass (kg) and body fat percentage, was measured in all the study subjects using DXA (Hologic Discovery A; Hologic Inc.). Coefficients of variation of DXA have been reported as less than 2.7% for fat mass [29].
2.3. RNA sequencing
Total RNA was extracted and purified from human fat specimens using a Qiagen RNeasy Micro kit (Qiagen), and residual genomic DNA was further removed by an on-column DNase digestion kit (Qiagen). Library construction, sequencing, and data analysis were performed at the Center for Cancer Computational Biology Core Facilities at Dana-Farber Cancer Institute (DFCI). Sequencing libraries were prepared using a SMART-Seq Ultra Low Input RNA kit (Clontech). The resulting library size distributions were analyzed using a Bioanalyzer (Agilent). The concentration of the library was determined using a DNA High-Sensitivity Qubit assay, and the final functional library concentration was determined through qPCR using Illumina adaptor-specific primers with a KAPA SYBR FAST Universal qPCR kit (Sigma–Aldrich).The library pools were loaded at final concentrations of 2 pM on single-read 75 flow cells and sequenced on an Illumina NextSeq 500 platform. Sequencing reads were aligned to the reference genome (Ensembl GRCh37.75) using the RNA-specific STAR aligner (v2.3.1z4).
2.4. Quantitative real-time PCR
RNA extraction, cDNA preparation, and qPCR were performed as previously described [25]. Briefly, cells were placed in TRIzol (Life Technologies) and RNA extracted according to the manufacturer's protocol. cDNA was synthesized using a High-Capacity cDNA Reverse Transcription kit (Life Technologies). qPCR was performed using Power SYBR Green Master (Applied Biosystems) and a QuantStudio 5 Real-Time PCR System (Applied Biosystems). The mix expression was normalized to beta-actin (ACTB), and the ΔΔCt method was used to calculate the fold change in transcript levels. Primers used in the qPCR reactions are listed in Table S3.
2.5. Transmission electron microscopy
Human AT samples that had been collected in 4% paraformaldehyde were post-fixed and embedded as previously described [30] with 2% osmium tetroxide (9 h) followed by overnight incubation with 1% osmium tetroxide and 1.5% potassium ferrocyanide prior to embedding in EPON. Thin sections (80 nm) were mounted on slot grids and imaged with a JEOL 1200EX scope. In adjacent sections that had been stained with toluidine blue, the total number of adipocytes were counted.
2.6. Serum and plasma analyses
CRP was measured by a solid-phase sandwich ELISA (R&D Systems) with an intra-assay CV of 3.8–8.3% and an inter-assay CV of 6.0–7.0%. CCL18 was measured by a solid-phase ELISA (R&D Systems) with an intra-assay CV of 3.2–3.7% and an inter-assay CV of 4.5–6.5%. Interleukin 10 was measured by a solid-phase sandwich ELISA (R&D Systems) with an intra-assay CV of 1.7–5.0% and an inter-assay CV of 5.9–7.3%. Interleukin 6 was measured by a solid-phase sandwich ELISA (R&D Systems) with an intra-assay CV of 1.6–4.2% and an inter-assay CV of 3.3–6.4%. TNFα was measured by a solid-phase ELISA (R&D Systems) with an intra-assay CV of 4.2–5.2% and an inter-assay CV of 4.6–7.4%. CCL2 was measured by a solid-phase sandwich ELISA (R&D Systems) with an intra-assay CV of 4.7–7.8% and an inter-assay CV of 4.6–6.7%. Serum analyses were performed on all the available samples from the subjects who participated in the fasting study (n = 12 study subjects and 9 study completers) [31].
2.7. Lentiviral constructs and lentivirus packing
The pCW-Cas9 (Addgene #50661) plasmid was used as the backbone to construct the doxycycline-inducible overexpression lentiviral vector. The CDSs of GFP, human SPIC, and human SPII (Origene) were respectively subcloned into the original vector whose Cas9 fragment was excised via BamHI/NheI sites. Lentiviral packaging was carried out by co-transfection of pCW lentiviral vector, psPAX2, and VSV-G envelope plasmids into 293T cells with lipofectamine-3000 (Invitrogen). Lentiviral supernatant was concentrated using Lenti-X concentrator (Takara) following the manufacturer's instructions to yield 100-fold titer viral stock.
2.8. Monocyte culture
Thp-1 monocytic cells (ATCC, TIB-202) were cultured in RPMI 1640 (Corning) supplemented with 10% fetal bovine serum (Corning), 0.05 mM 2-mercaptoethanol, and 100 U/ml penicillin/streptomycin (Life Technologies) at 5% CO2 and 37.0 °C. Monocyte suspension at a concentration of 500,000 cells/ml was differentiated with 100 ng/ml PMA (Sigma–Aldrich) for 48 h and maintained in normal medium for 24 h. For lentiviral transduction, 1 × 106 Thp-1 cells were resuspended in 0.5 ml of complete medium containing 50 μl of concentrated lentiviral stock (1 × 108 IU/ml) and transferred into the wells of a 12-well plate. Centrifuging transduction was performed in the presence of polybrene (5 μg/mL) at 1000 rpm for 2 h at 37.0 °C. Doxycycline at a final concentration of 1 μg/ml was added to the cells for 48 h to induce the gene overexpression. Cells demonstrating successful overexpression by q-PCR were used for differentiation experiments.
Human peripheral blood monocytes were isolated by density gradient centrifugation using Ficoll–Paque Premium 1.073 (GE Healthcare) and further enriched for CD14+ monocytes using CD14+ magnetic microbeads and MACS separation. The primary monocytes were subsequently treated with different chemicals or differentiation cocktails in complete M0 medium consisting of RPMI 1640 supplemented with 10% FBS, 10 ng/ml M-CSF, 100 U/ml penicillin/streptomycin, and 2 mM l-glutamine. For M1 macrophage differentiation, medium was supplemented with LPS (50 ng/ml, Sigma–Aldrich) and interferon-γ (INF-γ, 20 ng/ml, R&D Systems). For M2 macrophage differentiation, medium was supplemented with IL-4, IL-10, and TGF-β (20 ng/ml of each, R&D Systems).
2.9. Statistical analyses
For the RNA-seq dataset, read quantifications were created using Subread featureCounts v1.4.4 with reads counted to exon feature (-t exon flag) [32]. Differential expression analysis was performed via DESeq2 [33] using a linear model including the time points as discrete levels (desigñ time). Differential expression was determined using the likelihood ratio test against a reduced model that did not include the time factor (∼1). Multiple-test correction was performed by DESeq2 using the Benjamini-Hochberg procedure. Additional statistical analyses were conducted using JMP version 14.0 (SAS Institute) and Prism 8 (GraphPad Software). A heat map was produced using Morpheus (Broad). Gene set enrichement analyses (GSEA) were performed using the pre-ranked method and publicly available software [34]. For CRP and CCL18, paired-sample Wilcoxon signed-rank tests were conducted comparing baseline values to values from the day on which the normalized-to-baseline median value peaked (day 3 for CRP and day 5 for CCL18).
3. Results
3.1. Transcriptional reprogramming of adipose tissue with prolonged fasting
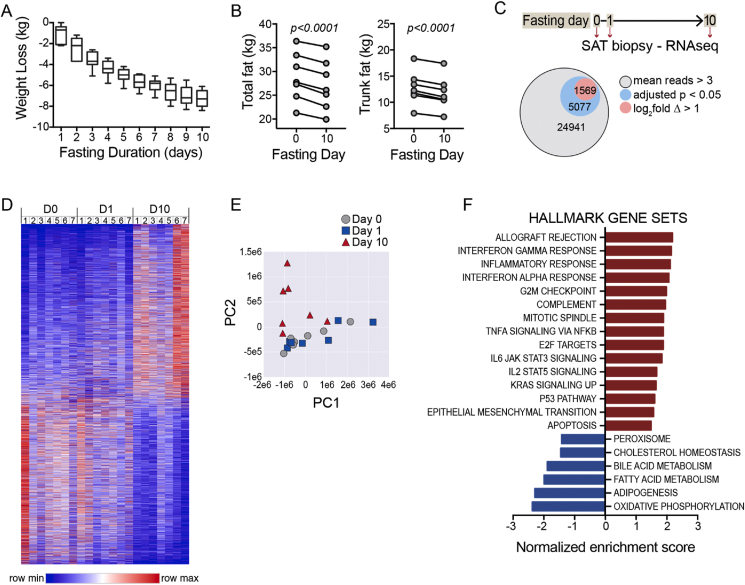
We performed a 10-day zero-calorie inpatient fast in humans [11]. In a subset of the subjects (n = 7, Table 1), we conducted serial subcutaneous AT biopsies from the periumbilical region. The participants uniformly lost weight over the course of the fast (Figure 1A, mean = 7.2 ± 0.8 kg, max = 8.4 kg, min = 6.1 kg) coincident with the contraction of fat mass as measured by dual energy X-ray absorptiometry (DXA) (Figure 1B). Our protocol employed a biopsy device that enabled directed collection of core biopsies from different sites in the periumbilical region at three different time points: on the morning of admission to the clinical research center after an overnight fast (time = 0), 24 h after admission, and then upon completion of the 10-day fast (Figure 1C). We performed RNA sequencing of these samples, revealing 24,941 transcripts with a mean normalized read frequency of >3 (Figure 1C). A subset of 5077 transcripts (20%) changed in a statistically significant manner as defined by a Benjamini-Hochberg (BH) adjusted p-value of <0.05. A smaller fraction of detectable transcripts (1569 or 6%) demonstrated both an adjusted p-value < 0.05 and a log2fold change of >1. The AT transcriptomes demonstrated the most dramatic changes at the 10-day time point (Figure 1D,E). These data demonstrate the dramatic effect of fasting on the AT transcriptome.
Table 1.
Baseline characteristics of the study participants.
| Mean ± SD (N = 7) | Range | |
|---|---|---|
| Age (years) | 29.1 ± 5.9 | 22.4–39.8 |
| Male/female | 2/5 | N/A |
| Baseline weight (kg) | 80.1 kg ± 11.3 | 67.5–98.0 |
| Baseline BMI (kg/m2) | 27.6 ± 1.9 | 24.7–29.3 |
| Total body fat mass by DXA (kg) | 28.8 ± 5.2 | 21.3–36.4 |
| % Body fat by DXA | 35.7 ± 7.6 | 26.4–46.8 |
Figure 1.
Fasting-induced adipose tissue contraction and transcriptional reprogramming. A) Weight loss during the 10-day fasting study. Mean weight loss = 7.2 kg (SD = 0.8, max/min = 8.4/6.1). B) Total fat mass (left) and trunk fat mass (right) measured by DXA at baseline day 0 and day 10 of fasting. Subjects lost a mean of 1.69 kg fat mass, SD: 0.37. Paired two-sided t-test. Subjects lost a mean of 1.00 kg trunk fat mass, SD: 0.20. Paired two-sided t-test. C) Schematic depicting serial subcutaneous adipose tissue (SAT) biopsies. Parts of whole chart showing total transcripts with detectable reads and the fractions with adjusted p-value < 0.05 and those reads with adjusted p-value < 0.5 and log2fold change >1. D) Heat map demonstrating global changes in gene expression in SAT during 10-day fast. E) Principal component analysis (PCA) showing transcriptomics shift with fasting, most evident at the 10-day time point. F) Gene set enrichment analysis (GSEA) using Hallmark gene sets. Gene sets that met false discovery rate (FDR) p-values < 0.05 are shown.
Quantitative PCR (qPCR) is often used as an orthogonal quality control method to validate transcriptional changes identified by RNA-seq. We did not have residual tissue for such analyses; however, in a prior manuscript [11], we performed qPCR on AT specimens obtained in parallel to those used for this analysis. Comparing the RNA-seq data to that prior qPCR data, we found strong directional consistency (Table S1), which supports the reproducibility of our findings.
We next applied GSEA to identify candidate pathways modulated by fasting in AT utilizing the Hallmark (Figure 1F) and Reactome (Fig. S1) gene sets. Metabolic processes were the unifying theme of downregulated gene sets that met a false discovery threshold of adjusted p < 0.05, including gene sets related to carbohydrate metabolism and energy expenditure such as glycolysis, oxidative phosphorylation, fatty acid metabolism, and triglyceride biosynthesis. The top enriched gene sets were allograft rejection, interferon gamma response, inflammatory response, and interferon alpha response.
Our initial analyses involved longitudinal repeated measures testing inclusive of all three time points. Given that the global analyses (Figure 1D,E) suggested that the most dramatic transcriptional effects were evident at the 10-day time point, we considered the extent to which the enriched gene sets were detectable at the early time point. A GSEA analysis restricted to a comparison of the day 1 time point to baseline demonstrated several downregulated gene sets that had a false discovery threshold of 0.05 and were similar to the full longitudinal analysis, including peroxisome, fatty acid metabolism, adipogenesis, and oxidative phosphorylation. However, no upregulated gene sets met the false discovery threshold (Fig. S2A). When we performed a similar paired analysis comparing the day 10 time point to baseline, we again noted negatively regulated gene sets related to metabolic processes; however, we also detected the emergence of numerous upregulated gene sets related to inflammation and immunity, similar to the full longitudinal analysis (Figs. S2B and 1F). Therefore, the unexpected and dominant theme of the positively regulated gene sets was inflammation and immunity; however, this signature was only evident after the prolonged 10-day fast.
3.2. Fasting drives an alternative transcriptional program of lysosomal lipolysis
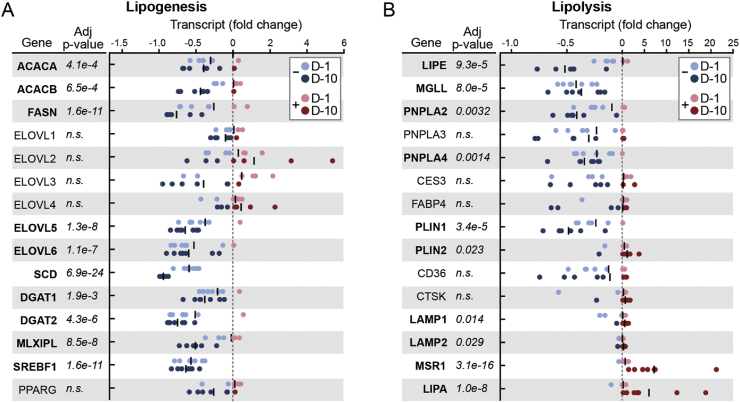
Given the central role played by AT in mobilization of lipid stores during fasting, we hypothesized that transcriptomic analysis would reveal a shift away from anabolic pathways in favor of augmented transcription of lipid catabolism regulators. In the context of the GSEA, which demonstrated negative regulation of the fatty acid metabolism gene set (Figure 1F), we specifically considered decreased transcription of genes related to de novo lipogenesis. Indeed, the canonical regulators of lipogenesis in AT were uniformly downregulated by fasting, including the transcriptional regulators, carbohydrate-responsive element-binding protein (MLXIPL), sterol regulatory element-binding protein 1 (SREBF1), and downstream genes coding for key enzymatic nodes (Figure 2A).
Figure 2.
Fasting drives an alternative lysosomal lipolysis program. A) Lipogenesis gene transcript expressed as fold change. Each dot represents an individual subject time point. B) Lipolysis gene transcript expressed as fold change. Each dot represents an individual subject time point. A and B: Black line = mean. Gene transcripts that reached significance with an adjusted p-value < 0.05 are bold.
Conversely, transcription of the canonical lipolysis pathway genes was not augmented as we hypothesized (Figure 2B). Instead, transcript levels of the 3 key enzymes responsible for the sequential release of the three fatty acid chains from the glycerol backbone were significantly attenuated, including hormone-sensitive lipase (LIPE), adipose triglyceride lipase (PNPLA2), and monoacylglycerol lipase (MGLL). Additional genes involved in the coordinated mobilization of triglycerides from adipocyte lipid droplets were also either significantly downregulated or unchanged. In contrast to the temporal dynamics of the inflammatory signature, which emerged at the 10-day time point, many of the lipolysis pathway genes were trending down at the day 1 time point (for example, MGLL, PNPLA2, and PLIN1; Figure 2B). These data in isolation cannot distinguish a regulated response from non-specific exhaustion of lipolytic pathways. However, this provided a rationale to examine transcripts related to an alternative path to triglyceride catabolism involving digestion of lipids in lysosomes, where the activity of lysosomal acid lipase (LIPA) is critical. Indeed, transcript levels for LIPA and related genes were markedly increased during fasting. Although we cannot conclude that downregulation of canonical lipolysis genes is concordantly reflected by their respective protein levels or enzymatic activities, these data raise the possibility that an alternative lipolytic pathway involving lysosomes is operative in the subcutaneous AT of fasting humans.
3.3. Prolonged fasting is associated with a macrophage signature in subcutaneous AT
Adipose tissue contains macrophages with diverse phenotypes, including those that may play a role in metabolism [12]. Given that GSEA revealed pathways related to immunity and inflammation, we examined the dataset for transcripts linked to macrophage recruitment, differentiation, and activation (for example, cytokines, chemokines, cell surface markers, and transcription factors). Among the list of the most significantly modulated transcripts (ranked by ascending adjusted p-value), numerous monocyte/macrophage-related genes were present and in each case their expression was augmented by fasting (Table S2), including the two most significantly induced transcripts (HO1 and SPIC).
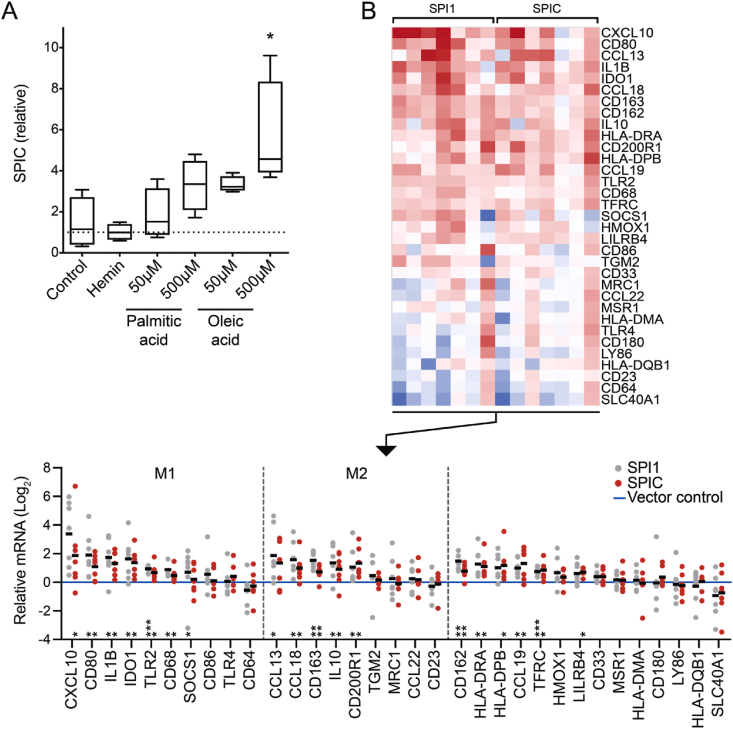
SPIC, which increased by 5.6-fold (adjusted p = 8.16e-48), encodes a member of the ETS family of transcription factors. Another member of this family, the better studied PU.1 (SPI1), ranked lower on the list, but its transcripts also increased 1.6-fold with fasting (adjusted p = 0.0012). SPIC has also been shown to be induced in murine bone marrow-derived monocytes by heme [13]. In the murine spleen, heme-induced SPIC is critical for the specification of splenic red pulp macrophages [14]. We hypothesized that SPIC might also play a role in driving the specification of macrophages in human AT during starvation. Given that lipid flux is a critical metabolic event in AT with fasting, we first tested whether exposure to fatty acids would induce SPIC expression in primary human peripheral blood monocytes in the same manner as the metabolic activator, heme, which we included as a putative positive control. We observed an increase in SPIC expression that was particularly evident with oleic acid, but we did not detect an augmentation of expression in response to hemin (Figure 3A). Other markers of macrophage specification were augmented by hemin exposure, however, suggesting that the absence of a heme-mediated SPIC effect was not due to inactivity of the reagent (Fig. S3A). We performed a similar assessment after 24 h of treatment with fatty acids or other physiologically relevant stimuli inclusive of classic inflammatory stimuli and components of M1/M2 differentiation cocktails, again finding that the most robust signal was from fatty acids or lipids (Fig. S3B). Given that there was a trend toward augmentation of SPIC with both M1 and M2 differentiation reagents, we also retested whether SPIC expression was specific to one pathway or the other, examining a time course including a later 48-hour time point. We found significant induction of SPIC after 48 h of directed differentiation to both M1 and M2 stimuli, although there was a trend toward greater induction with exposure to the M2 differentiation cocktail (Fig. S3C). Collectively, these data suggest that SPIC is transcriptionally induced in human peripheral blood monocytes in the process of macrophage specification and/or activation, including with exposure to a fatty acid stimulus of contextual relevance to the fasted state in AT where lipid and fatty acid flux is operative.
Figure 3.
SPIC as a candidate transcriptional regulator of macrophage specification in fasting adipose tissue. A) Tukey box plot of SPIC expression in human peripheral blood monocytes treated for 24 h with hemin or fatty acids. ∗p < 0.05, ANOVA with Dunnett's multiple comparisons test (n = 4 technical replicates, similar induction to fatty acids in 3 independent experiments). B) Transcriptional pattern resulting from SPIC gain of function compared to SPI1 gain of function. Thp1 cells were transduced with lentiviral vectors delivering doxycycline-inducible SPIC. Control and experimental cells were treated with doxycycline for 48 h after viral transduction, then differentiated via 48-hour PMA stimulation and 24-hour rest to induce adherence to the culture dish prior to qPCR analysis. Results from 7 independent experiments are shown as a heat map (top), demonstrating directional concordance of SPIC induction relative to SPI1. Bottom: dot plots, where each dot represents the mean of 4 technical replicates. To compare the scope of transcriptional changes across experiments, each biological replicate was normalized to vector control (blue line at log2 = 0). Genes associated with M1 or M2 phenotypes are grouped together. Significance assessed by one sample testing: two-sided t test for normally distributed data or Wilcoxon signed-rank test for non-normal datasets (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The identification of SPIC as a marker and candidate transcriptional regulator of AT macrophages in the fasted state provided a rationale to test whether SPIC gain of function in monocytes would replicate some aspects of the macrophage gene signature evident in fasted AT. To test this, we used Thp1 cells, which are amenable to viral transduction and express low levels of SPIC in their basal state. We transduced Thp1 cells with constructs that enabled doxycycline (TET-ON)-inducible expression of SPIC compared to a control transgene (GFP) or the better studied SPI1 (Fig. S3D). We tested the transcriptional effect of SPIC gain of function in the context of stimulation with phorbol 12-myristate 13-acetate (PMA), a standard method of activating Thp1 cells to transition from suspension to adherence culture and macrophage specification [15]. We assessed a panel of macrophage genes (Figure 3B), drawing on standard markers and with a particular focus on a number of genes enriched in AT during fasting. Of the 33 genes tested, SPIC gain of function resulted in induction of 16 genes, 13 of which were significantly and directionally concordant with Thp1 cells induced with SPI1. These data in cultured monocytes collectively demonstrate that (1) a fatty acid stimulus, which has physiological relevance in fasting AT, drives the expression of the transcription factor SPIC, the most significantly modified transcription factor in fasting AT and (2) SPIC gain of function in Thp1 cells drives a gene program that partially overlaps with the macrophage signature elucidated in human AT with fasting.
3.4. Fasting increases adipose tissue macrophages
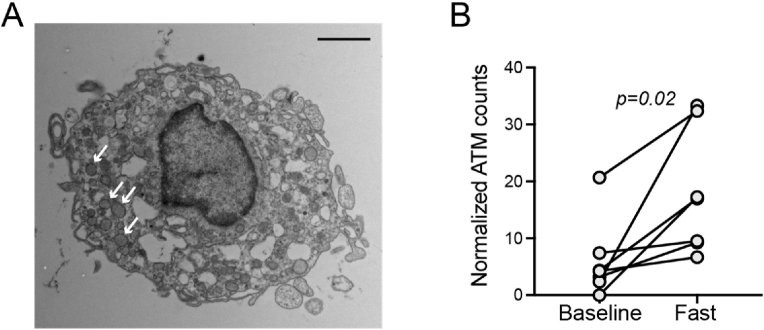
Given evidence of a macrophage gene signature in AT with fasting, we considered the possibility that this might be partially attributable to increased numbers of monocytes/macrophages. To test this, we performed a histological analysis. As the amount of available tissue was limited for standard histology and immunostaining, we imaged ultrathin sections by transmission electron microscopy (TEM). We scanned the entire section, capturing TEM images of any observable cells interspersed between adipocytes, and then a blinded observer reviewed the images and identified cells that exhibited stereotypical features consistent with monocytes or macrophages. Because there was stochastic variability in the total area of the AT sections, we also counted the total number of adipocytes in the section to which we normalized the number of monocytes/macrophages. After 10 days of fasting, the AT biopsy specimens revealed a significant increase (p = 0.02) in the frequency of interstitial cells exhibiting a monocytic or macrophage morphology (Figure 4). In addition, some of the cells demonstrated intracellular vesicles that stained darkly with osmium tetroxide consistent with lipid containing lysosomes or lipid droplets (Figure 4A). These data suggest that the increase in macrophage-specific transcripts could be partially attributable to an increase in the number of macrophages with fasting either due to increased monocyte recruitment or local proliferation.
Figure 4.
Fasting increases adipose tissue macrophages. A) Representative transmission electron micrograph capturing adipose interstitial cell with macrophage features after 10 days of fasting. Numerous darkly stained (osmium tetroxide) lysosomal structures are evident (arrows), consistent with lipids. Scale bar = 2 μm. B) Adipose tissue macrophages/monocytes were normalized to the number of adipocytes expressed as #ATM per 100 adipocytes. Significance assessed with paired t-test.
3.5. Evidence of a systemic inflammatory surge with prolonged fasting
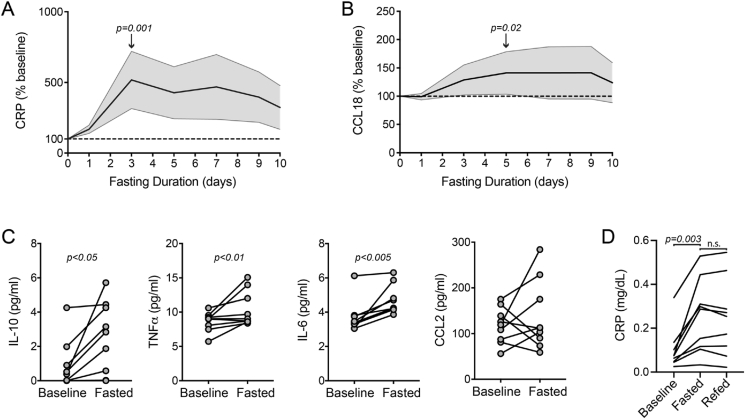
We next considered the possibility that the AT inflammatory signal would also be associated with evidence of systemic inflammation. To address this, we first performed serial analyses (ELISA) of two factors in the serum samples of the fasting study subjects: the general inflammatory biomarker C-reactive protein (CRP) and the chemokine CCL18, which we selected because it was the most significantly upregulated secreted cytokine/chemokine in our RNA-seq dataset (Table S2). Normalized to baseline values, the CRP levels peaked at day 3 and the CCL18 levels peaked at day 5. The levels of CRP and CCL18 were significantly higher at these time points compared to the baseline values (paired-sample Wilcoxon signed-rank test, p ≤ 0.02 for both) (Figure 5A,B).
Figure 5.
Fasting drives circulating inflammatory markers. A) Serial serum C-reactive protein (CRP) levels expressed as % of baseline. Center line = mean, shading = 95% CI, and dashed line = baseline. B) Serial serum CCL18 levels expressed as % of baseline. Center line = mean, shading = 95% CI, and dashed line = baseline. C) Serum IL-10, TNFα, IL-6, and CCL2 levels measured by ELISA at baseline and after completion of the 10-day fast. Significance assessed by paired Wilcoxon test. All of the values were significant except CCL2. D) Plasma CRP levels at baseline, the final day of prolonged fasting, and the day after refeeding. Note: plasma was used to assess CRP in this analysis as opposed to the serum analyses in A. Non-parametric Friedman's ANOVA with Dunn's correction for the two comparisons are shown in the graph.
We next examined additional systemic inflammatory mediators by measuring the levels of select canonical macrophage-derived cytokines by ELISA, including IL-10, IL-6, TNFα, and CCL2/MCP-1. Transcripts for each of these cytokines in AT also either increased or trended upward with fasting (IL-10: log2fold = 2.1, p = 3.64e-17, adjusted p = 1.08e-14; TNFα: log2fold = 0.81, p = 0.03, adjusted p = 0.11; IL-6: log2fold = 0.65, p = 0.09, adjusted p = 0.25; and CCL2: log2fold = 1.17, p = 0.051, adjusted p = 0.16). We performed ELISA on serum samples from baseline and day 10 of fasting, as serum was limited from the intermediate fasting time points. IL-10, IL-6, and TNFα significantly increased in the circulation at the end of the fast (Figure 5C), whereas we detected no difference in CCL2. Therefore, these analyses of circulating inflammatory (CRP, TNFα, IL-6, and CCL18) and immunomodulatory (IL-10) factors demonstrate that local inflammatory changes in AT are associated with directionally similar effects in the systemic circulation. In contrast to the adipose tissue analyses, where we did not collect samples between days 1 and 10 of fasting, the increased frequency of blood sampling enabled the detection of a surge in inflammatory markers between 1 and 3 days of fasting.
To test whether there was rapid resolution of systemic inflammatory markers with refeeding, we measured CRP in samples collected the day after refeeding. We did not have sufficient serum available to measure CRP in all of the subjects at the refeeding time point as had been done for the serial fasting time points in Figure 5A; however, we did have plasma samples available and measured the CRP levels after refeeding, with paired plasma at baseline and at completion of the prolonged fast. ELISA analysis confirmed the increase in CRP with prolonged fasting with no significant reduction at the one-day refeeding time point (Figure 5D). Collectively, these data support the concept that prolonged fasting drives metabolic inflammation.
4. Discussion
In this study, serial transcriptomics of subcutaneous AT over a 10-day fast demonstrated an unexpected dominant signal of inflammation, including augmentation of transcripts related to macrophage identity and function. Orthogonal analyses demonstrating macrophage influx in AT biopsy specimens and a corollary surge in systemically circulating inflammatory markers further established an inflammatory response to prolonged fasting.
Prior cross-sectional studies of the AT transcriptome revealed associations between inflammatory pathways and obesity or clinical metrics of insulin resistance and diabetes [16]. Prospective longitudinal studies have demonstrated transcriptional attenuation of such inflammatory pathways in AT with weight loss achieved over weeks to months [[17], [18], [19]]. In murine AT, fasting leads to an acute reduction in macrophages found in close association with blood vessels [20]. In contrast, our observation of increased AT inflammation with fasting is perhaps most consistent with the augmentation of macrophage numbers observed in the AT of obese mice subjected to caloric restriction [21]. An important difference between our study and many prior human AT transcriptional analyses is that our study population was (1) normal to overweight and not obese and (2) relatively young and healthy without known metabolic disease. It is possible that the effects of fasting are quite different in the AT of obese/diabetic individuals with baseline inflammation. Differences in timing may also be important. When reconciling murine-human differences, for example, it may be that the mechanisms underpinning the mobilization of AT lipid concurrent with 10–15% weight loss in just 24 h are different than what is a slower process in humans who lose approximately 9.2% over 10 days [11]. In addition, while we did not have interval transcriptional data between the day 1 and day 10 time points, circulating inflammatory markers appear to peak between days 3 and 5 before trending downward. By day 10, we also observed an increase in local transcription and circulating levels of the anti-inflammatory cytokine IL-10. Whether this suggestion of an ongoing programmatic shift to immunomodulatory activity would continue in a sustained fashion weeks after refeeding is an important question for future study.
There also may be important differences between more subtle degrees of negative caloric balance and the response to a zero-calorie fast in which the shift to lipid metabolism and ketogenesis is imperative. One potential role for inflammatory cells, and particularly macrophages, in AT is to scavenge and metabolize lipids or lipid byproducts. When adipocytes undergo cell death, for example, macrophages surround and phagocytose the remnants including the lipid droplet(s) [22]. In mice, contraction of AT mass over several weeks of caloric restriction drives a macrophage population that supports lipolysis [21]. Recent data also support a role for macrophages in homeostatic lipid turnover in AT [10]. Therefore, we speculate that macrophages may also play a functional role in lipid catabolism during fasting; however, the analyses conducted in this study cannot exclude alternative mechanisms such as the elucidation of inflammatory pathways as a non-specific stress response to prolonged fasting.
Our observation of an AT inflammatory response to fasting is notable given the transcriptional downregulation of genes encoding the canonical lipases thought to effect lipolysis during fasting in adipocytes inclusive of MGLL, PNPLA2, and LIPE. Delayed transcriptional induction of LIPE with fasting was previously demonstrated and considered to represent a possible disconnect between gene transcription and the level and/or activity of the enzyme [23]. While our study certainly cannot exclude such a disconnect, our data point to the possibility of additional lysosomal-dependent mechanisms of lipolysis as also being operative during fasting. One challenge in interpreting the lysosomal signal, however, is that it was observed in analyses of unfractionated AT and therefore the signal could indicate lipophagic activity by either adipocytes or inflammatory cells. A unifying explanation to the dual signals of an inflammatory surge and lysosomal lipolysis pathways is that both arise from macrophages and that macrophages directly contribute to the mobilization and catabolic digestion of triglycerides. For this to be true, there would have to be a mechanism for transport of triglycerides from adipocyte lipid droplets to interstitial macrophages, as only fatty acids, not intact triglycerides, can freely diffuse through plasma membranes. Importantly, a recent study identified a new lipase-independent mechanism of adipocyte lipid droplet remodeling in which undigested triglycerides are directly released from adipocytes within extracellular vesicles [10]. We speculate that this process could be operative as a complementary mechanism for mobilizing stored lipids during fasting.
Our study also identified the transcription factor SPIC as a candidate marker and mediator of a fasting metabolic phenotype in AT macrophages. Ex vivo, the expression of SPIC was augmented in monocytes exposed to a fatty acid stimulus and SPIC gain of function drove a transcriptional signature that overlapped with that observed in AT with fasting, providing conceptual support for SPIC as a mediator of macrophage specification in AT. This potential role of SPIC deviates from murine studies in which SPIC lineage tracing and myeloid loss of function demonstrate a role of SPIC as a master regulator of red pulp macrophage specification in the spleen [13,14]. Our data may indicate a broader repertoire of SPIC functions or alternatively reflect interspecies differences in the transcriptional mechanisms of macrophage specification. An additional question is the underlying degree of macrophage heterogeneity and whether AT macrophages converge on a common phenotype with fasting. While each induced transcription factor (for example, SPIC and SPI1) could control distinct macrophage phenotypes, it is perhaps more likely that they represent a circuit of collaborating transcription factors that are increasingly recognized to establish specialized cell states [24,25]. However, our study cannot definitively answer this question, in part due to the challenge of deciphering the relative degrees to which transcriptional changes in AT arise from transcriptional reprogramming vs changes in cellularity. In the future, some of these questions may be addressed by applying single-cell sequencing methods to human AT with fasting, as has recently been performed in calorically restricted rats [26].
In recent years, there has been increasing interest in the use of fasting protocols to improve metabolic health and longevity [27]. One proposed mechanism of benefit from fasting has been modulation of inflammation and specifically that fasting may induce anti-inflammatory pathways. Our data suggest that the effects of fasting on the immune/inflammatory system may be more complex, as the transcriptional and systemic signals elicited by fasting in our study cannot be easily categorized as pro- or anti-inflammatory. Whether the reprogramming of AT with fasting would be beneficial if sustained cannot be addressed by this study. However, it is possible that the mechanisms that have evolved to enable humans to undertake the metabolic shifts required to survive starvation may include both beneficial and harmful factors. It is also possible that the net benefit of pathways activated by fasting are context dependent. For example, the psychiatric disorder anorexia nervosa, which is characterized by a state of self-induced chronic caloric restriction and low body weight, is associated with maladaptive pathology such as significant bone fragility and is among the psychiatric diseases with the highest mortality rate [1,28]. In contrast, individuals who are overweight or obese may incur additional metabolic benefits from fasting due to decreased adiposity. Therefore, an important open question, underscored but not answered by our study, is whether fasting in humans induces beneficial pathways that promote longevity independent of effects on adiposity. Nonetheless, our study provides a direct link between fasting physiology and reprogramming of metabolic inflammation in AT, the underlying mechanisms of which may hold one key to understanding the beneficial effects of fasting in humans.
Funding
KL2/Catalyst Medical Research Investigator Training award from the Harvard Catalyst/Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, NIH award KL2 TR002542); the Claflin Distinguished Scholar Award, Massachusetts General Hospital; and departmental funds, Division of Genetics, Department of Medicine, Brigham and Women's Hospital.
Author contributions
PKF: Conceptualization, investigation, funding acquisition, supervision, and wrote the original draft
YZ: Investigation and methodology
JO: Investigation
TP: Investigation
ML: Investigation
BL: Formal analysis
MLS: Conceptualization, investigation, funding acquisition, supervision, and wrote the original draft
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101082.
Contributor Information
Pouneh K. Fazeli, Email: pkfazeli@pitt.edu.
Matthew L. Steinhauser, Email: msteinhauser@pitt.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Fazeli P.K., Klibanski A. Effects of anorexia nervosa on bone metabolism. Endocrine Reviews. 2018;39(6):895–910. doi: 10.1210/er.2018-00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neel J.V. Diabetes mellitus: a "thrifty" genotype rendered detrimental by "progress"? The American Journal of Human Genetics. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 3.Fontana L., Partridge L., Longo V.D. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahill G.F. Starvation in man. New England Journal of Medicine. 1970;282(12):668–675. doi: 10.1056/NEJM197003192821209. [DOI] [PubMed] [Google Scholar]
- 5.Rothman D.L., Magnusson I., Katz L.D., Shulman R.G., Shulman G.I. Quantitation of hepatic glycogenolysis and gluconeogenesis in fasting humans with 13C NMR. Science. 1991;254(5031):573–576. doi: 10.1126/science.1948033. [DOI] [PubMed] [Google Scholar]
- 6.Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metabolism. 2012;15(3):279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert J.S., Yerges-Armstrong L.M., Horenstein R.B., Pollin T.I., Sreenivasan U.T., Chai S. Null mutation in hormone-sensitive lipase gene and risk of type 2 diabetes. New England Journal of Medicine. 2014;370(24):2307–2315. doi: 10.1056/NEJMoa1315496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoiswohl G., Stefanovic-Racic M., Menke M.N., Wills R.C., Surlow B.A., Basantani M.K. Impact of reduced ATGL-mediated adipocyte lipolysis on obesity-associated insulin resistance and inflammation in male mice. Endocrinology. 2015;156(10):3610–3624. doi: 10.1210/en.2015-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proceedings of the National Academy of Sciences of the U S A. 2000;97(2):787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty S.E., Grijalva A., Xu X., Ables E., Nomani A., Ferrante A.W. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science. 2019;363(6430):989–993. doi: 10.1126/science.aaw2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fazeli P.K., Lun M., Kim S.M., Bredella M.A., Wright S., Zhang Y. FGF21 and the late adaptive response to starvation in humans. Journal of Clinical Investigation. 2015;125(12):4601–4611. doi: 10.1172/JCI83349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. Journal of Clinical Investigation. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haldar M., Kohyama M., So A.Y., Kc W., Wu X., Briseño C.G. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156(6):1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohyama M., Ise W., Edelson B.T., Wilker P.R., Hildner K., Mejia C. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457(7227):318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwende H., Fitzke E., Ambs P., Dieter P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. Journal of Leukocyte Biology. 1996;59(4):555–561. [PubMed] [Google Scholar]
- 16.Qatanani M., Tan Y., Dobrin R., Greenawalt D.M., Hu G., Zhao W. Inverse regulation of inflammation and mitochondrial function in adipose tissue defines extreme insulin sensitivity in morbidly obese patients. Diabetes. 2013;62(3):855–863. doi: 10.2337/db12-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clément K., Viguerie N., Poitou C., Carette C., Pelloux V., Curat C.A. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. The FASEB Journal. 2004;18(14):1657–1669. doi: 10.1096/fj.04-2204com. [DOI] [PubMed] [Google Scholar]
- 18.Kwok K.H.M., Rydén M., Andersson D.P., Beauchef G., Guere C., Vie K. Prospective analyses of white adipose tissue gene expression in relation to long-term body weight changes. International Journal of Obesity. 2020;44(2):377–387. doi: 10.1038/s41366-019-0385-1. [DOI] [PubMed] [Google Scholar]
- 19.Alemán J.O., Iyengar N.M., Walker J.M., Milne G.L., Da Rosa J.C., Liang Y. Effects of rapid weight loss on systemic and adipose tissue inflammation and metabolism in obese postmenopausal women. J Endocr Soc. 2017;1(6):625–637. doi: 10.1210/js.2017-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva H.M., Báfica A., Rodrigues-Luiz G.F., Chi J., Santos P.D.A., Reis B.S. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. Journal of Experimental Medicine. 2019;216(4):786–806. doi: 10.1084/jem.20181049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosteli A., Sugaru E., Haemmerle G., Martin J.F., Lei J., Zechner R. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. Journal of Clinical Investigation. 2010;120(10):3466–3479. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murano I., Barbatelli G., Parisani V., Latini C., Muzzonigro G., Castellucci M. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. The Journal of Lipid Research. 2008;49(7):1562–1568. doi: 10.1194/jlr.M800019-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Sztalryd C., Kraemer F.B. Regulation of hormone-sensitive lipase during fasting. American Journal of Physiology. 1994;266(2 Pt 1):E179–E185. doi: 10.1152/ajpendo.1994.266.2.E179. [DOI] [PubMed] [Google Scholar]
- 24.Saint-Andre V., Federation A.J., Lin C.Y., Abraham B.J., Reddy J., Lee T.I. Models of human core transcriptional regulatory circuitries. Genome Research. 2016;26(3):385–396. doi: 10.1101/gr.197590.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Federation A.J., Kim S., O'Keefe J.P., Lun M., Xiang D. Targeting nuclear receptor NR4A1-dependent adipocyte progenitor quiescence promotes metabolic adaptation to obesity. Journal of Clinical Investigation. 2018;128(11):4898–4911. doi: 10.1172/JCI98353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S., Sun S., Geng L., Song M., Wang W., Ye Y. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus norvegicus aging. Cell. 2020;180(5):984–1001. doi: 10.1016/j.cell.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Di Francesco A., Di Germanio C., Bernier M., de Cabo R. A time to fast. Science. 2018;362(6416):770–775. doi: 10.1126/science.aau2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshaviah A., Edkins K., Hastings E.R., Krishna M., Franko D.L., Herzog D.B. Re-examining premature mortality in anorexia nervosa: a meta-analysis redux. Comprehensive Psychiatry. 2014;55(8):1773–1784. doi: 10.1016/j.comppsych.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Johnson J., Dawson-Hughes B. Precision and stability of dual-energy X-ray absorptiometry measurements. Calcified Tissue International. 1991;49(3):174–178. doi: 10.1007/BF02556113. [DOI] [PubMed] [Google Scholar]
- 30.Guillermier C., Fazeli P.K., Kim S., Lun M., Zuflacht J.P., Milian J. Imaging mass spectrometry demonstrates age-related decline in human adipose plasticity. JCI Insight. 2017;2(5) doi: 10.1172/jci.insight.90349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinhauser M.L., Olenchock B.A., O'Keefe J., Lun M., Pierce K.A., Lee H. The circulating metabolome of human starvation. JCI Insight. 2018;3(16) doi: 10.1172/jci.insight.121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.