Oomycetes cause hundreds of diseases in economically and environmentally significant plants. How these microbes acquire host nutrients is not well understood. Many oomycetes insert specialized hyphae called haustoria into plant cells, but unlike their fungal counterparts, a role in nutrition has remained unproven. The discovery that Phytophthora invertases localize to haustoria provides the first strong evidence that these structures participate in feeding. Since regions of proteins containing signal peptides targeted proteins to the haustorium-plant interface, haustoria appear to be the primary machinery for secreting proteins during biotrophic pathogenesis. Although oomycete invertases were acquired laterally from fungi, their expression patterns have adapted to the Phytophthora lifestyle by abandoning substrate-level regulation in favor of developmental control, allowing the enzymes to be produced in anticipation of plant colonization. This study highlights how a widely distributed hydrolytic enzyme has evolved new behaviors in oomycetes.
KEYWORDS: metabolism, nutrition, oomycetes, plant pathogens, transcriptional regulation
ABSTRACT
The oomycete Phytophthora infestans, the causal agent of potato and tomato blight, expresses two extracellular invertases. Unlike typical fungal invertases, the P. infestans genes are not sucrose induced or glucose repressed but instead appear to be under developmental control. Transcript levels of both genes were very low in mycelia harvested from artificial medium but high in preinfection stages (sporangia, zoospores, and germinated cysts), high during biotrophic growth in leaves and tubers, and low during necrotrophy. Genome-wide analyses of metabolic enzymes and effectors indicated that this expression profile was fairly unusual, matched only by a few other enzymes, such as carbonic anhydrases and a few RXLR effectors. Genes for other metabolic enzymes were typically downregulated in the preinfection stages. Overall metabolic gene expression during the necrotrophic stage of infection clustered with artificial medium, while the biotrophic phase formed a separate cluster. Confocal microscopy of transformants expressing green fluorescent protein (GFP) fusions indicated that invertase protein resided primarily in haustoria during infection. This localization was not attributable to haustorium-specific promoter activity. Instead, the N-terminal regions of proteins containing signal peptides were sufficient to deliver proteins to haustoria. Invertase expression during leaf infection was linked to a decline in apoplastic sucrose, consistent with a role of the enzymes in plant pathogenesis. This was also suggested by the discovery that invertase genes occur across multiple orders of oomycetes but not in most animal pathogens or a mycoparasite.
INTRODUCTION
A primary goal of pathogens is to acquire host nutrients to support growth and reproduction. Important to plants and pathogens is the disaccharide sucrose. During normal plant growth, sucrose is made from photosynthate in green source tissues, exported to the apoplast, loaded into phloem, and delivered to sink tissues such as roots, tubers, and fruits (1). In these sinks, sucrose enters plant cells through sucrose transporters or is taken up by hexose transporters after hydrolysis into glucose and fructose by cell wall invertases (β-fructofuranosidase; EC 3.2.1.26). Interestingly, these plant invertases are activated during pathogen attack, increasing the hexose/sucrose ratio in the apoplast (2, 3). While this may stimulate the plant defense response, it may also nourish apoplast-adapted pathogens, which include many fungi, bacteria, and oomycetes.
Many of these microbes encode their own invertases as well as sugar transporters, which cause the pathogen to acquire more sugar by becoming a sucrose sink (4, 5). For example, bacterial invertases, typically termed sucrose hydrolases, are considered pathogenicity factors (6). The Uromyces fabae invertase has been shown to generate hexoses that are taken up by that fungus’ own transporter (7). Oomycetes such as Phytophthora spp. also express invertases. These have been little studied but are thought to have been acquired from fungi by horizontal gene transfer (8).
The present study focused on invertases in the potato and tomato blight pathogen Phytophthora infestans. Infections by this hemibiotrophic oomycete usually begin when asexual sporangia release zoospores, which later encyst on host tissues and extend germ tubes that enter the host (9). Biotrophic growth then typically proceeds for several days, during which time the pathogen feeds from the apoplast using intercellular hyphae. These also produce haustoria, which establish membrane interfaces with plant cells. The transition between biotrophy and necrotrophy is associated with a decline in haustorium formation and alterations in metabolic enzyme production (10, 11). For example, only late in infection does P. infestans produce much enzyme for degrading nutrients, such as starch, that are normally sequestered within plant cells.
Unlike their fungal counterparts, oomycete haustoria have not been proved to play a role in nutrient uptake. Haustoria are known to deliver effectors into plants using conventional and nonconventional (brefeldin A-insensitive) secretion pathways (12). The RXLR motif, the signature of the major class of Phytophthora effectors, has been proposed to direct proteins to the plant-haustorium interface, although the role of that sequence is controversial (13).
How invertases are regulated in any pathogen, including oomycetes, is not well understood. Studies in nonpathogenic yeasts and filamentous fungi have shown that invertase transcription is usually induced by sucrose (14, 15). Glucose often inhibits expression, through either the carbon catabolite repression pathway or translational control (16).
In this study, we focused on the expression pattern and localization of invertases in P. infestans. Unlike most fungal invertases studied to date, the P. infestans genes do not appear to be sucrose induced but are under developmental control. Expression begins in spores, continues during biotrophic growth, and is suppressed during necrotrophy and growth on artificial medium. The invertases localize to haustoria, providing the first strong evidence that oomycete haustoria play a role in acquiring nutrients. We also propose that a signal peptide may be the only sequence needed to target proteins to haustoria, which helps to explain its role as an organ for polarized secretion.
RESULTS
Apoplastic sucrose declines during infection.
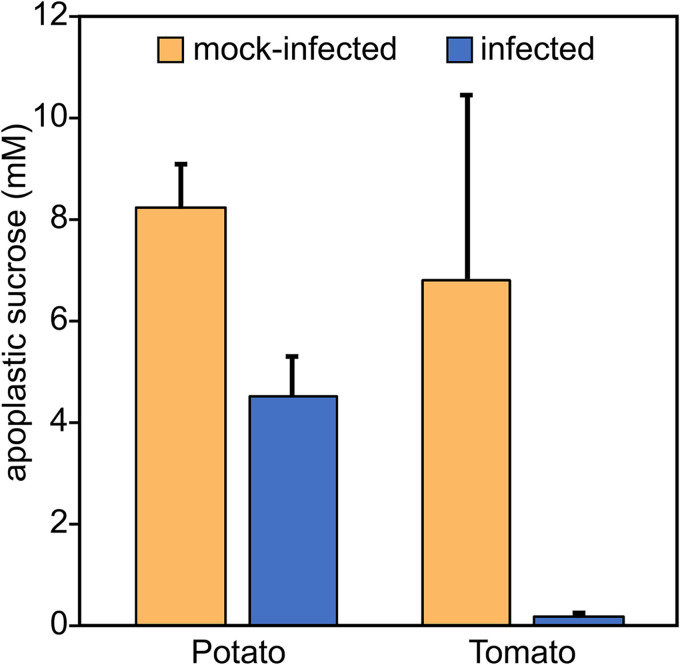
To help address which nutrients may be used by P. infestans during biotrophic growth, potato and tomato leaflets were infected with zoospores. After 24 h, apoplastic wash fluids were obtained by vacuum infiltration from infected and mock-infected samples. Disaccharide (mostly sucrose) levels were then quantified by liquid chromatography-mass spectrometry. Sucrose levels were found to decline sharply in infected leaflets compared to the controls (Fig. 1). This reduction may be sharper than would occur within 24 h in a natural field infection, since about 1,000 infection events occurred per leaflet in our experiment, compared to a single penetration in a normal agricultural setting.
FIG 1.
Apoplastic sucrose declines during P. infestans infection. Infected and mock-infected potato and tomato leaflets, after 24 h, were infiltrated with buffer. After the fluid was recovered by centrifugation, disaccharides (mostly sucrose) were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a standard curve. Values were adjusted to the true apoplastic concentration based on an indigo carmine dilution assay.
P. infestans expresses two invertases.
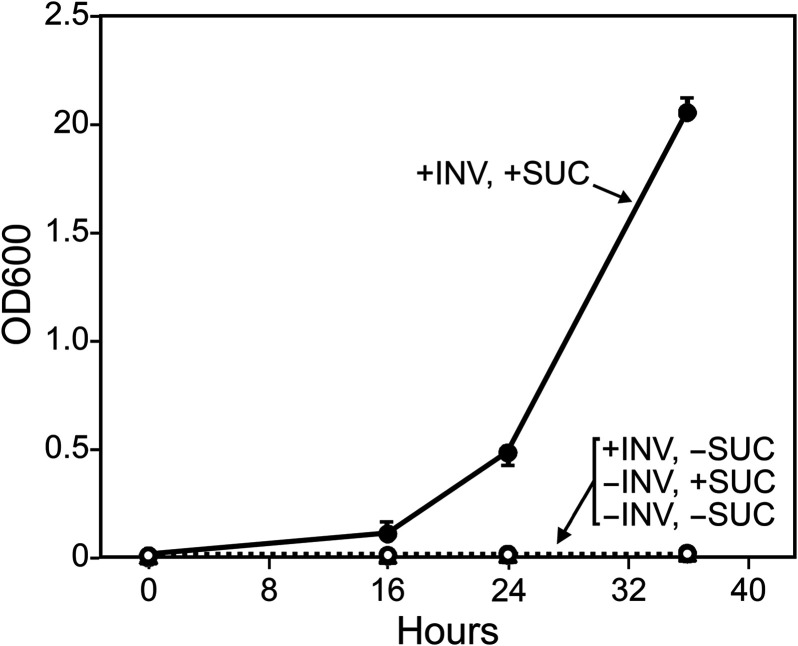
Factors contributing to the drop in apoplastic sucrose during infection may include reduced photosynthesis, reduced efflux or increased uptake of photosynthate by the plant, or the action of host or pathogen invertases. To investigate the latter, a search of the P. infestans reference genome (strain T30-4) detected three genes encoding putative invertases. PITG_14237 and PITG_14238 were predicted to encode 68.5-kDa (617-amino-acid [aa]) and 74.4-kDa (666-aa) proteins, respectively. Transcriptome sequencing (RNA-seq) reads supported both gene models, which reside 13.3 kb from each other. Both are closely related in amino acid sequence (86% similarity), contain N-terminal signal peptides, and bear domains associated with invertases, including PFAM 00251 and PFAM 08244, which represent the N- and C-terminal domains, respectively, of glycoside hydrolase family 32. Invertase activity was confirmed by heterologous expression in Pichia pastoris, a species that lacks invertase. As shown in Fig. 2, the P. infestans gene allowed P. pastoris to grow using sucrose as a carbon source.
FIG 2.
P. infestans invertase enables P. pastoris to use sucrose as a carbon source. P. pastoris strain KM71H and a version expressing PITG_14238 (INV) were grown on minimal medium with or without sucrose (SUC), plus methanol to induce the gene. Similar results were obtained with three independent transformants of KM71H, as well as with three independent transformants of strain X33, and when cells from methanol-induced cultures were pelleted and resuspended in methanol-free medium with or without sucrose.
A third gene, PITG_14243, was also detected in T30-4. This was predicted to encode a 54-kDa (487-aa) protein but with truncated PFAM 00251 and PFAM 08244 domains. RNA reads from infected plant and artificial-medium samples failed to map to PITG_14243, suggesting that it is a pseudogene. An identical truncated gene was detected in an assembly of isolate 1306 of P. infestans (17).
Not all oomycetes encode invertase.
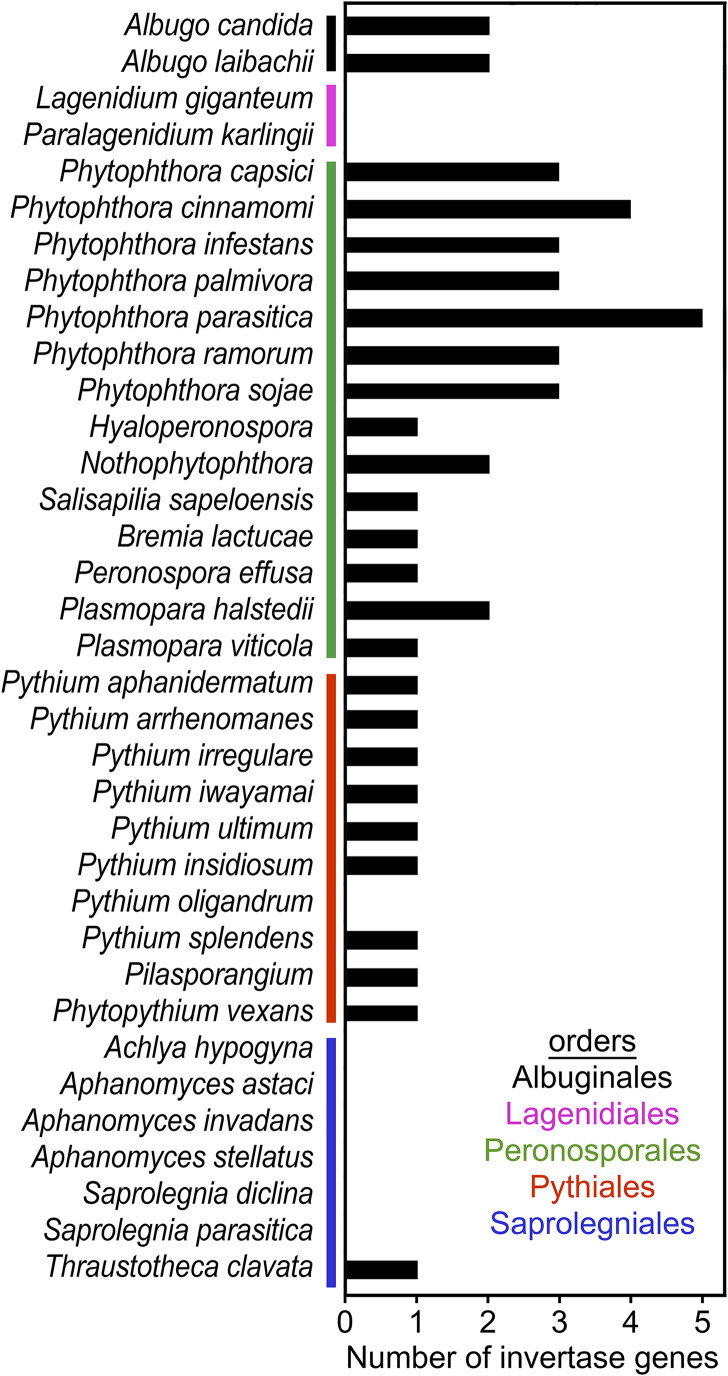
The distribution of invertases was assessed using genome data from 35 species representing most oomycete orders (Fig. 3). Invertases were detected throughout the order Peronosporales. Consistent with our findings in P. infestans, small gene families were observed in each Phytophthora sp. However, no truncated genes resembling PITG_14243 were identified in the other species. A single gene was detected in Salisapilia sapeloensis, a peronosporalean that grows on plant litter. Single genes were also present in most of the obligate plant pathogens known as downy mildews (Bremia, Hyaloperonospora, Peronospora, and Plasmopara). In contrast, obligate plant pathogens of the order Albuginales (e.g., Albugo candida and Albugo laibachii) were each predicted to express two invertases.
FIG 3.
Occurrence of invertase genes in oomycetes. Genome sequences and annotated proteins were searched using representative invertases as queries in TBLASTn and BLASTP searches, using an E value cutoff of 10−20. The number shown may include some nonfunctional proteins.
Most representatives of the order Pythiales were found to encode a single invertase. This group mostly includes saprophytes and opportunistic pathogens of plants, or animal pests in the case of Pythium insidiosum. Phytopythium vexans, which is intermediate between the Pythiales and Peronosporales, also encoded a single invertase. Interestingly, no invertases were detected in the mycoparasite Pythium oligandrum. In this and other species lacking invertase, when possible, the searches were performed using both predicted proteins and whole-genome data from multiple strains (e.g., strains Po37 and ATCC 38472_TT for Py. oligandrum) (Table S1).
Summed expression of enzymes in KEGG orthology groups. (A) Aggregate expression levels of each KEGG orthology group in hyphae (minimal medium), hyphae (rye-sucrose medium), sporangia, chilled sporangia, zoospores, germinated cysts, potato leaf at 2 and 6 dpi, potato leaf at 6 dpi, and tubers at 4 dpi, with the annotated enzyme and EC number for each KEGG group and the pathway classification. (B) P. infestans genes assigned to each KEGG group. Download Table S1, XLSX file, 0.2 MB (190.1KB, xlsx) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Most members of the order Saprolegniales lacked invertase. This included the three sequenced species of Aphanomyces, two of Saprolegnia, and Achlya hypogyna. These are animal or insect pathogens that also grow as saprophytes. Invertases were also absent from the order Lagenidiales, which are also insect pathogens. However, an invertase was detected in the nonpathogenic saprolegnian Thraustotheca clavata.
P. infestans invertases are induced in spores and infection stages.
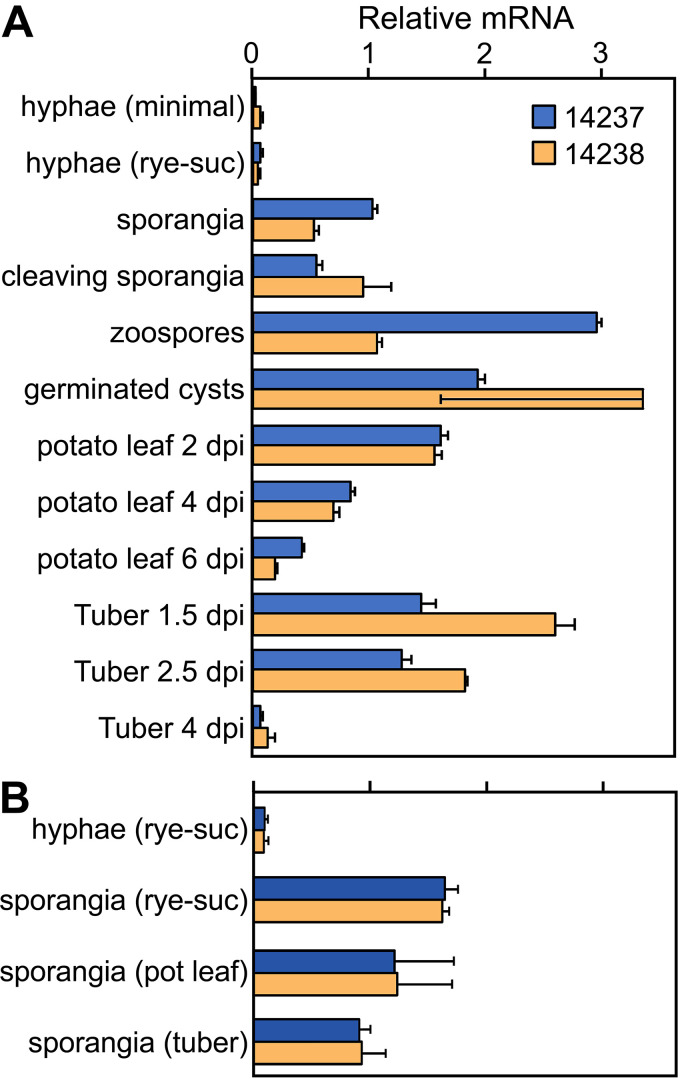
RNA-seq revealed that PITG_14237 and PITG_14238 had similar expression patterns (Fig. 4). Their mRNA levels were very low in mycelia grown in artificial medium and collected before the onset of visible sporulation. This included nonsporulating mycelia from both minimal medium with 20 g/liter glucose and rye grain medium with 20 g/liter sucrose (Fig. 4A). The relative transcript abundance of both genes rose >10-fold in sporangia purified from rye medium cultures compared to nonsporulating mycelia and stayed elevated through germination (zoospore release) and zoospore cyst germination. In biotrophic stages of potato leaf and potato tuber infection (2 and 1.5 days postinfection [dpi], respectively), expression of both genes was >20-fold higher than in the artificial medium. Transcripts of both genes then fell to low levels as the infections entered their necrotrophic stages (6 and 4 dpi, respectively). The expression of markers for biotrophic growth, necrotrophy, and sporulation in the plant samples are shown in Fig. S1.
FIG 4.
Expression of invertase genes. (A) mRNA levels of PITG_14237 and PITG_14238 were measured using RNA-seq data from young nonsporulating hyphae grown on glucose-containing minimal medium or rye-sucrose medium, sporangia from rye-sucrose (rye-suc) medium, sporangia chilled for 60 min to stimulate zoosporogenesis (cleaving sporangia), zoospores after 60 min of swimming, germinated zoospore cysts, potato leaves at 2, 4, and 6 dpi, and tubers at 1.5, 2.5, and 4 dpi. The first time point for each type of plant tissue represents biotrophic growth, and the last time point represents the necrotrophic stage. (B) mRNA levels in nonsporulating hyphae from rye-sucrose medium and sporangia isolated from rye-sucrose cultures, potato leaves, or tubers. Per-gene normalization was performed separately in each panel to a mean of 1.0.
Markers of developmental stages during colonization of potato tubers and leaves. Shown are per-gene normalized expression values from RNA-seq of genes encoding RXLR protein PITG_23226, NPP1 protein PITG_16866, and centrin PITG_21764, which are markers of the biotrophic, necrotrophic, and sporulation stages, respectively (52, 61). Error bars represent standard deviations based on three biological replicates. Download FIG S1, TIF file, 0.6 MB (712.1KB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The induction of the invertases in sporangia was interesting, since we showed previously that most metabolic genes were expressed at lower levels in sporangia than mycelia (18). To ensure that the higher level of invertase mRNA in sporangia was not an artifact of growth in artificial medium, sporangia from rye-sucrose medium, potato leaf infections, and tuber infections were compared. The two invertases had similar mRNA levels regardless of their source (Fig. 4B).
Spore, biotrophic, and necrotrophic stages form distinct clusters based on metabolic gene expression.
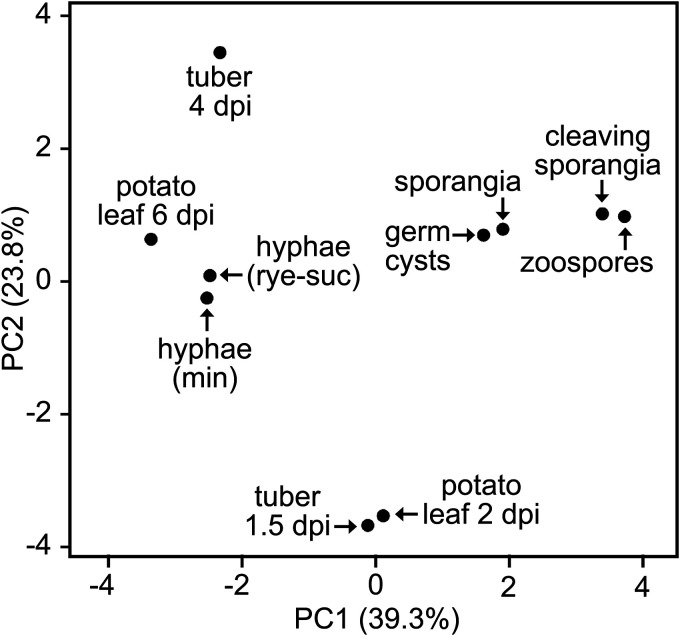
To help understand the regulation of the invertases in the context of other metabolic enzymes, we studied the total complement of genes encoding metabolic enzymes from P. infestans, excluding those participating directly in protein, DNA, or cell wall modification (10). This encompassed 1,507 genes representing 586 Enzyme Commission numbers and 634 KEGG orthology groups (19). Principal-component analysis showed tight clustering of the biotrophic samples (1.5-dpi tubers and 2-dpi leaves) (Fig. 5). A second cluster included rye-sucrose medium, minimal medium, and the late stages of potato leaf and tuber infection, which indicated that metabolism during necrotrophy resembles growth on artificial medium (Fig. 5). A third cluster included sporangia, cleaving sporangia (i.e., chilled to initiate zoosporogenesis), zoospores, and germinated cysts.
FIG 5.
Principal-component analysis (PCA) of metabolic gene expression. The analysis was performed based on the summed FPKM values of genes in each KEGG Orthology (KO) group. Tissue samples are described in the legend to Fig. 4, and the underlying data are shown in Fig. S2.
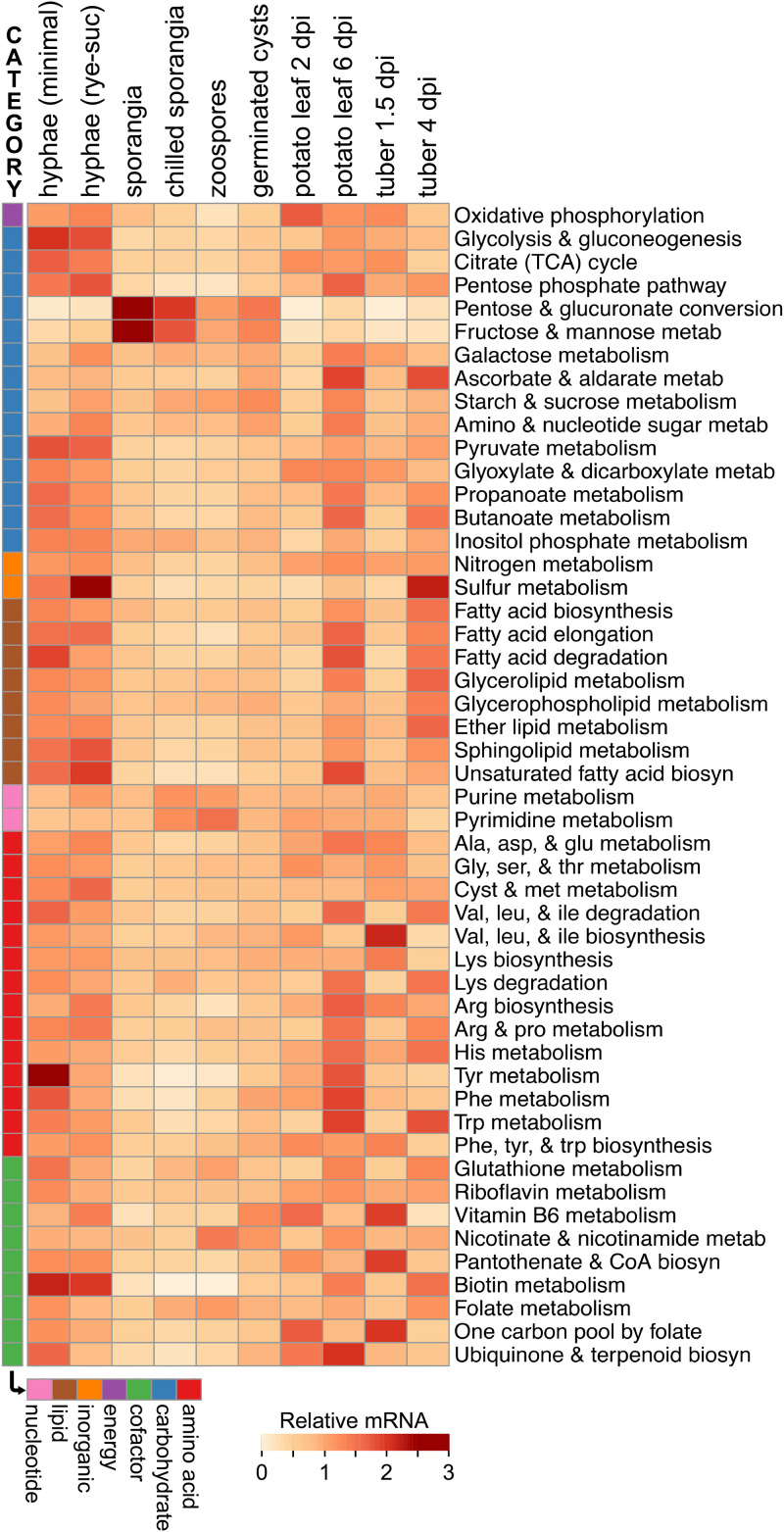
Expression of metabolic genes classified by KEGG orthology (KO) number. RNA levels in the samples described in the legend to Fig. 3 were calculated by averaging the summed FPKM values of genes in each KO group, followed by normalization across all samples to a mean of 1. Invertase (K01193) and other enzymes mentioned in the text are highlighted by a blue arrow and diamonds, respectively. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. The FPKM data used to make the heat map are shown in Table S1. Download FIG S2, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Invertase expression is distinct from that of most other metabolic genes.
Analyses of KEGG pathways (ko numbers) (Fig. 6) and KEGG orthology groups (K numbers) (Fig. S2) further demonstrated that the expression profile of the invertases was unusual. We first examined the summed FPKM (fragments per kilobase per million) values of genes in the 50 major metabolic pathways defined by KEGG. Invertases (K01193) are included, along with multiple other enzymes, in the KEGG pathways for galactose metabolism (ko00052) and starch and sucrose metabolism (ko00500). Transcript levels of most pathways were lower in the spore stages than during growth in plants or medium, which is opposite to that observed for the invertases (Fig. 6). Four pathways were upregulated in spores. The fructose/mannose metabolism and pentose/glucuronate conversion pathways (KEGG pathways ko00040 and ko00051) were higher in spores, primarily due to spore-specific sugar alcohol dehydrogenases in KEGG orthology group K00008. Pyrimidine and purine metabolism (ko00230 and ko00240) were higher in spores due to upregulated biosynthetic enzymes such as IMP dehydrogenase (K00088) and others. Also upregulated in spores were ribonucleoside-diphosphate reductase subunits (K10807 and K10808), which may be related to preparing for DNA synthesis after germination, and cyclic nucleotide phosphodiesterases (K01120), which may participate in signal transduction during zoosporogenesis (20). Unlike invertases, none of these were expressed more during biotrophic (early) than necrotrophic (late) stages of infection.
FIG 6.
Expression of genes in metabolic pathways. Genes were grouped based on KEGG pathways, with the addition of categories for assimilating inorganic nitrogen and sulfur. The aggregate (summed) FPKM value for genes in each pathway was calculated and normalized across the tissue samples to an average of 1.0.
About one-third of KEGG pathways were transcribed more during early than late infection. Examples include the oxidative phosphorylation (ko00750) and vitamin biosynthesis pathways (e.g., ko00190 and ko00770). However, unlike invertases, these were downregulated in spores.
Similar conclusions about the atypical nature of invertase expression were drawn by inspecting the 634 KEGG orthology groups encoding metabolic enzymes (Fig. S2; Table S1). When the average expression in spore stages was compared to the average in the mycelia from artificial medium, only 16 and 25 groups were higher in spores based on 10-fold and 4-fold cutoffs, respectively. Besides those mentioned above, these included tyrosinase (K00505), sugar epimerase (K08679), ceramidase (K04711), carnitine O-palmitoyltransferase (K08765), and two cofactor biosynthesis enzymes, nicotinamidase (K08281) and protoporphyrinogen oxidase (K00231). However, few were expressed more during early than late infection. Instead, most spore-induced genes had higher mRNA levels during late infection, probably since this is when sporulation initiates. Examples include ceramidase and carnitine O-palmitoyltransferase, which might mobilize carbon from the lipid reserves of spores.
About 69% of the 634 KEGG orthology groups showed higher expression during late versus early infection in leaves and tubers, with 16% showing the opposite pattern (Fig. S2). Nearly all of the latter, however, differed from invertases by not being upregulated in spores compared to medium. Only a few KEGG orthology groups (<1%) had patterns of expression that roughly matched the invertases. These included mostly cytoplasmic enzymes, including a threonine catabolic enzyme (K15789) and the ketogenic enzyme hydroxymethylglutaryl-CoA lyase (K01640).
Invertase expression resembles that of only a few other secreted proteins.
We predicted that approximately 40 types of metabolic enzymes (encoded by 111 genes with an FPKM value of >1) were secreted. Interestingly, mRNA levels of 28% of these genes were >10-fold higher in one or more spore stages (sporangia to germinated cysts) than mycelia from artificial medium. Also, 22% were >10-fold higher in one or more plant tissues than mycelia from medium (Fig. S3). Fifteen genes were expressed at >10-fold-higher levels both in a spore stage and during plant infection compared to artificial medium, including the invertases. These included three carbonic anhydrases, three acid phosphatases, an aldose epimerase, two phospholipases, one tyrosinase, a β-glucosidase, and a catalase. Hierarchical clustering indicated that the catalase and carbonic anhydrases were closest in expression pattern to the invertases.
Expression of secreted metabolic enzymes. Indicated are per-gene-normalized expression values based on the samples described in the legend to Fig. 3. Each gene is annotated with its function, Enzyme Commission (EC) number, and gene name (PITG_XXXXX). The two invertases are highlighted with arrows. Data were subjected to hierarchical clustering using the Pearson correlation method with complete linkage. Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also checked whether genes encoding nonmetabolic secreted proteins traditionally associated with plant-pathogen interactions had mRNA profiles similar to those of invertase genes. Only 4 of the 312 expressed genes for RXLR effectors resembled invertases in being upregulated in spores and in early compared to late infection in both leaves and tubers (e.g., PITG_02843) (Fig. S4). Interestingly, many of the RXLRs were differentially expressed between the two types of host tissues. In contrast, no cell wall-degrading enzymes (Fig. S4) or proteases (Fig. S5) had expression patterns that resembled those of the invertases. It was nevertheless interesting to observe that many of these were transcribed at much higher levels in tubers than in leaves.
Expression of secreted RXLR and protease effectors. Data are per-gene-normalized expression values based on the samples described in the legend to Fig. 3. Invertase is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S4, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of cell wall-degrading (CWDE) enzymes. Data are per-gene-normalized expression values based on the samples in Fig. 3. Appended to the name of each gene is its enzymatic function, as follows: CUT (cutinase); PE (pectin esterase, carbohydrate esterase family CE8); pectate lyase families PL1, PL2, and PL3; GH28 (polygalacturonase); GH53 (endo-β-1,4-galactanase); GH54 (β-xylosidase); GH78 (α-l-rhamnosidase); and GH105 (rhamnogalacturonyl hydrolase). Invertase expression is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S5, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Some sugar transporters are induced in spores and plants.
Since invertases are expected to liberate monosaccharides for use by P. infestans, we also examined sugar transporters (Fig. S6). Of 44 genes expressed with FPKM values of >1, none exhibited patterns similar to those of the invertases. A majority were expressed at higher levels during late infection than early infection. One gene encoding a SWEET/MtN3 transporter (PITG_04173) was expressed preferentially at early time points in both leaves and tubers, like the invertases. However, PITG_04173 was also expressed at similar levels in artificial medium and plants.
Expression of predicted sugar transporters. Data are per-gene-normalized expression values based on the samples described in Fig. 3. Genes belong to the MFS or MTN3 (SWEET) families, as noted. Invertase is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S6, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
P. infestans invertases are not induced by sucrose.
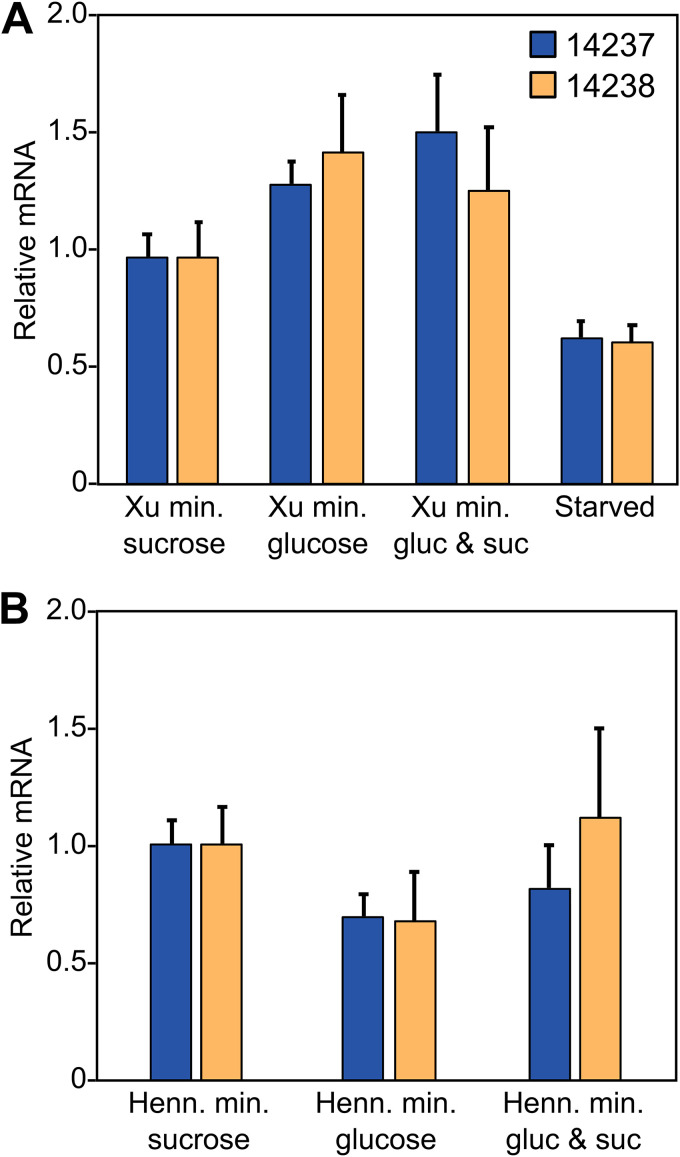
Fungal invertases are typically upregulated by sucrose (14, 15). Although such data come mostly from studies of saprophytic fungi, it was nevertheless surprising that the P. infestans invertases were transcribed at similarly low levels in both the glucose-based minimal medium and the sucrose-based rye grain medium (Fig. 4). To better ascertain whether sucrose induces the invertases, their expression was measured by RT-qPCR in two distinct types of minimal medium (21, 22), each modified to include glucose, sucrose, or both (Fig. 7). PITG_14237 and PITG_14238 exhibited similar mRNA levels regardless of the sugar, indicating that sucrose did not induce either invertase. We also considered whether nutrient limitation might upregulate the invertases, since sporangia are metabolically active but reliant on carbon reserves. However, placing mycelia for 6 h in medium lacking a carbon source did not induce either gene.
FIG 7.
Effect of sugars on expression of P. infestans invertase genes. (A) qRT-PCR analysis of the invertases in young (nonsporulating) hyphae from modified Xu’s minimal medium containing 20 g/liter sucrose, glucose, or both sugars. Also shown are values for mycelia grown in sucrose but then starved for 6 h in medium lacking a carbon source. Expression values were calculated by the ΔΔCT method, using two constitutive genes as a control, and normalized to the samples containing only sucrose. (B) Same as panel A except that modified Henninger’s medium was used.
We also attempted to test the effect of fructose, which is the other end product of the invertase reaction. This was not feasible, however. With Xu’s minimal medium (21), no growth was observed when fructose or a combination of fructose and sucrose was used. With Henninger’s medium (22), some growth occurred with fructose but very slow growth occurred when both sugars were present.
Invertase accumulates in haustoria.
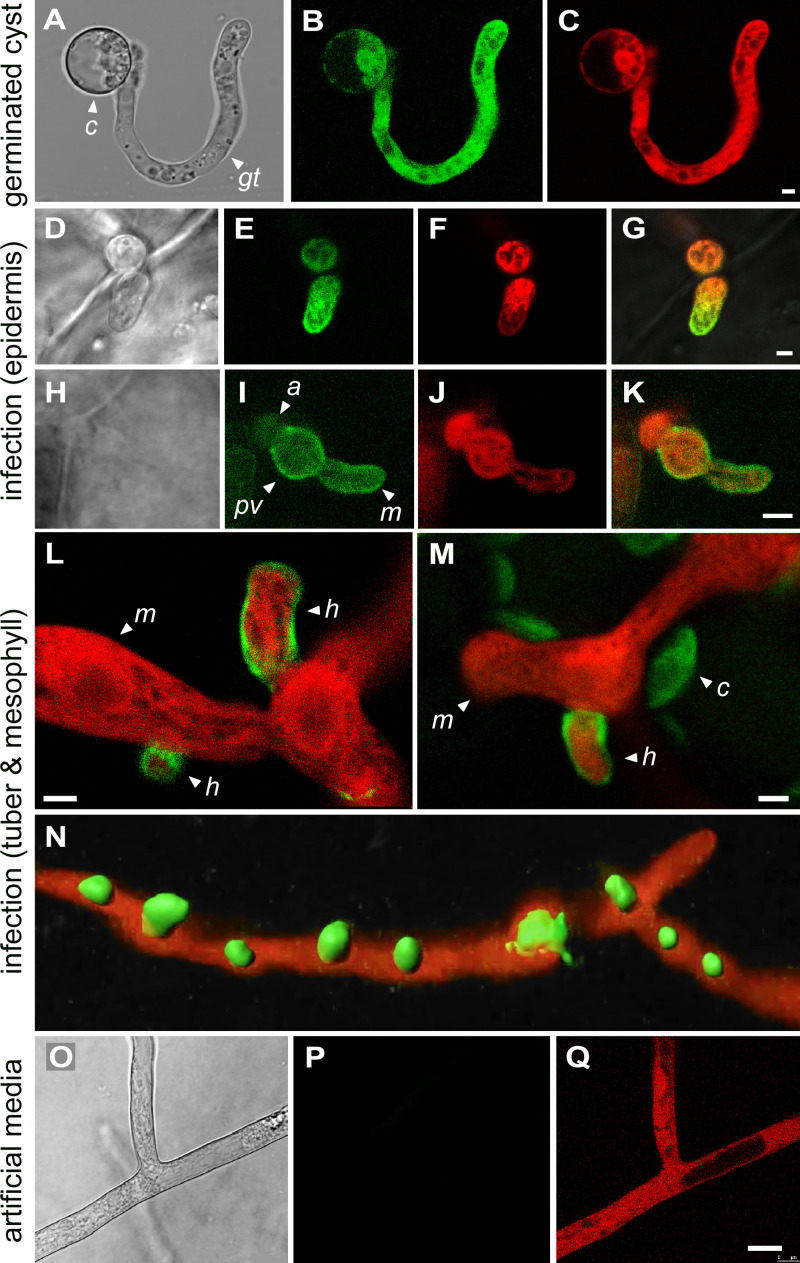
To study the location of the protein, we generated transformants of P. infestans expressing the invertases fused to green fluorescent protein (GFP), driven by the native promoters. Even though mRNA from both invertase genes was detected in sporangia, no invertase-GFP was detected by confocal microscopy in ungerminated sporangia ranging in age from 1 to 5 days. It is possible that not all mRNA in that dormant stage is efficiently translated. However, a GFP signal was detected in germinated cysts (Fig. 8A to C). This transformant, and those in the subsequent panels of Fig. 8, coexpress invertase-GFP from the native promoter along with cytoplasmic tdTomato driven by the constitutive ham34 promoter. Invertase-GFP was distributed throughout the cytoplasm of germinated cysts, even though our constructs included the N-terminal signal peptide. This might be explained by the fact that secreted proteins would be below detection levels due to dilution in the medium.
FIG 8.
Localization of invertase expressed from native promoter. (A to C) Germinating zoospore cyst from strain expressing invertase fused with GFP and a separate gene encoding cytoplasmic tdTomato. Panels show bright-field, green, and red channels, from left to right. The cyst (c) and germ tube (gt) are marked. (D to G) Strain coexpressing invertase-GFP and cytoplasmic tdTomato in tomato leaf epidermal cell. The appressorium (a) and a young intracellular mycelium (m) are marked. Panels show bright-field, green, red, and merged images, from left to right. (H to K) Similar to panels D to G but at a later stage of growth. An appressorium (a), intracellular primary vesicle (pv), and mycelium (m) are marked. (L) Strain expressing invertase-GFP and cytoplasmic tdTomato in the medulla of an infected potato tuber. Emerging from the mycelium and penetrating host cells are several haustoria (h). (M) Same as panel L but showing mesophyll of a tomato leaf. The green channel detects both a GFP signal surrounding the haustorium (h) and autofluorescing chloroplasts (c). (N) Similar tissue as in panels L and M but showing a z-stack reconstruction of haustoria formed in the plant using Imaris software. (O to Q) Same strain as panel N but illustrating growth in rye-sucrose medium, showing an absence of invertase-GFP expression. Panels show bright-field, green, and red channels, from left to right. Each labeling pattern was observed in a minimum of three transformants. The images were obtained with PITG_14238, but similar results were obtained with PITG_14237. Bars, 2.5 μm.
During the initial stages of penetration of an epidermal cell of a tomato leaf, invertase-GFP continued to show a cytoplasmic signal (Fig. 8D to G). However, instead of being uniformly distributed, it accumulated preferentially at the hyphal tip. At a later stage of development in epidermal cells, soon after formation of the so-called primary vesicle, invertase-GFP accumulated in the pathogen cell wall in addition to the cytoplasm (Fig. 8H to K). During subsequent growth in the medulla (flesh) of potato tubers or tomato leaf mesophyll, invertase-GFP resided mostly in the haustorial cell wall (Fig. 8L and M).
While the invertase-GFP signal was not difficult to observe in cyst germ tubes or haustoria, the same transformants exhibited little signal in hyphae in artificial medium (Fig. 8O to Q). This is consistent with the expression patterns in Fig. 6. Patches of invertase-GFP were seen occasionally in older cultures, which we assume represent zones in which sporulation was occurring.
In contrast, when invertase-GFP was expressed from the constitutive ham34 promoter (23) in artificial medium, the protein was detected in hyphal walls (Fig. S7A and B). It also concentrated in sporangial papilla (Fig. S7C and D). Since we showed previously that fluorescent proteins lacking signal peptides are cytoplasmic in sporangia (24), we hypothesized that all secreted proteins might accumulate in papilla. As shown in Fig. S7, this appeared to be true in transformants in which the ham34 promoter drove the expression of tdTomato fused to 33 amino acids from the N-terminal region of the secreted P. infestans INF1 protein (25), which includes the 20-amino-acid signal peptide predicted by TargetP. Although the papillar location of secreted proteins does not appear to be relevant to the invertase story, papilla have been suggested to contain enzymes that facilitate germination (26).
Expression of invertase in rye-sucrose medium using ham34 promoter. (A) Hypha of P. infestans transformant expressing invertase-GFP (showing expression in the cell wall [w]) and mCherry-tagged histone H2b as a marker of nuclei (red channel [n]). (B) Sporangium from P. infestans strain expressing invertase-GFP, showing strong fluorescence at papilla (p). (C) Sporangium of strain expressing invertase-GFP and histone H2b-mCherry. (D) Strain expressing tdTomato fused to the Inf1 signal peptide. Download FIG S7, TIF file, 2.7 MB (2.8MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Haustoria function as secretion organelles.
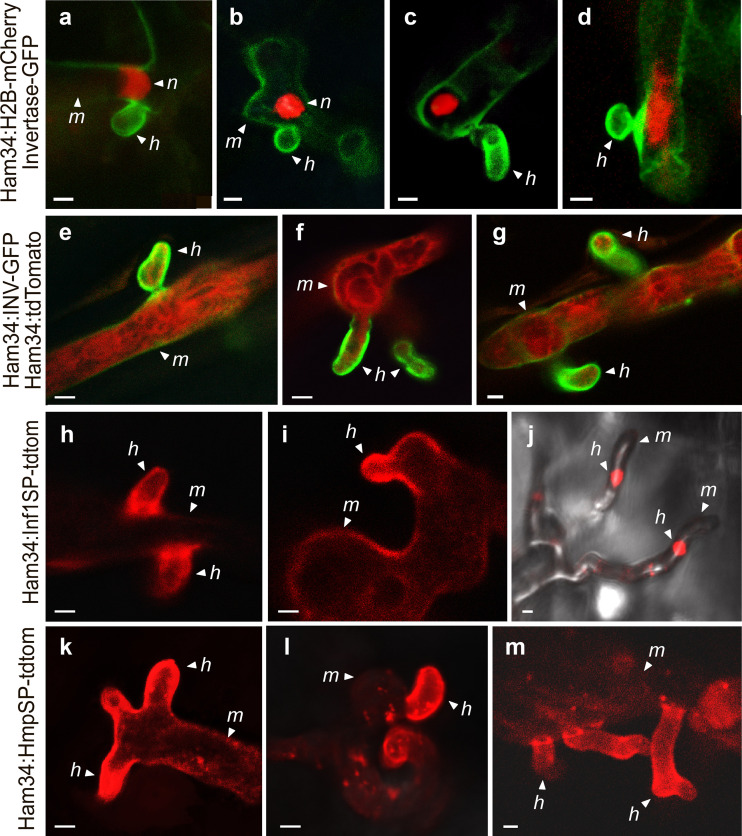
It was interesting that most invertase during infection resided in the haustorial wall, based on transformants expressing invertase-GFP from their native promoters (Fig. 8). Since we observed that nuclei typically reside adjacent to haustoria (Fig. 9a to d), we considered the possibility that invertase promoters are specifically active in those nuclei. However, preferential deposition of the protein in haustoria also occurred when invertase-GFP expression was driven by ham34 (Fig. 9e to g).
FIG 9.
Investigating basis of haustoria targeting. (a to d) Transformants coexpressing invertase-GFP with H2B-mCherry during tuber infection. Nuclei (n), haustoria (h), and intercellular mycelia (m) are marked. (e to g) Transformants coexpressing cytoplasmic tdTomato and invertase-GFP in tubers, but unlike in Fig. 8, data were obtained using the ham34 promoter. (h to j) Transformants expressing a fusion of the N-terminal region of INF1 containing its signal peptide with tdTomato, driven by the ham34 promoter, in tuber infection. Note that in panel j, the haustoria extend upward from the hyphae. (k to m) Transformants expressing a fusion of the N-terminal region of HMP1 containing its signal peptide and tdTomato in tubers. Bars, 2.5 μm.
An alternative theory to explain the haustorial accumulation of invertase is that all secreted proteins are delivered to that structure. This would be consistent with a recent study that showed that diverse extracellular infection-associated proteins also accumulated near haustoria (27). We tested this further by using the ham34 promoter to express tdTomato fused to 33 amino acids from the N terminus of the P. infestans INF1 protein, which is downregulated during biotrophic growth and thus not associated with haustoria (25), or 42 N-terminal amino acids from HMP1, which is a known haustorial protein (11). Within these regions, both INF1 and HMP1 contain canonical signal peptides based on TargetP (Fig. S8) (28). Both tdTomato fusions accumulated preferentially in haustoria (Fig. 9h to m).
N-terminal regions of proteins mentioned in this paper. The first 64 amino acids of the INF1 (PITG_12251), HMP1 (PITG_00375), and invertase (PITG_14238) proteins are shown. Grey shading indicates the signal peptides predicted by TargetP 2.0 (28). The regions used in tdTomato fusions involving INF1 and HMP1 are underlined. PITG_21410, PITG_22926, PITG_01029, and PITG_14371 are the secreted INF4, pectinesterase, and RXLR proteins, which were shown in other studies to concentrate near haustoria (27). Sequences C-terminal of the signal peptide are dissimilar in chemical features such as hydropathy, ranging from moderately hydrophilic to hydrophobic, and lack conserved motifs based on alignments and motif discovery programs. Download FIG S8, PDF file, 0.05 MB (49.7KB, pdf) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Unlike characterized fungal invertases, those of P. infestans are not sucrose inducible but instead are expressed primarily during preinfection stages (spores) and biotrophic growth (Fig. 4). That the substrate-induction paradigm does not extend to P. infestans might seem surprising, since oomycetes acquired invertases by lateral transfer from fungi (8). Expression neofunctionalization is understandable if lifestyle differences are considered, however. Most Phytophthora spp. depend on a living host for survival, as they survive poorly as saprophytes except for some clade 6 species (P. infestans is clade 1c) (29). In contrast, most fungal pathogens alternate between saprophytic and pathogenic growth in nature. Since the normal environment of P. infestans is a sucrose-bearing plant, a mechanism for regulation based on substrate induction would be superfluous. It is more logical to trigger invertase transcription in spores to allow sucrose to be used immediately upon invasion of the host. Similar reasoning justifies the expression of a subset of plant defense-suppressing RXLR proteins and other effectors in spores (Fig. S4 and S5).
In contrast to invertase, most metabolic genes and pathways were downregulated in spores compared to artificial medium or infected plants (Fig. 6 and Fig. S2). By mining public proteomic data (30) for 35 randomly selected enzymes in such pathways (glycolysis, oxidative phosphorylation, pentose phosphate pathway, amino acid metabolism), we observed that their protein levels decline an average of 34% between mycelia and germinating cysts; this residual amount is apparently enough to allow successful pathogenesis. That study focused on cytoplasmic proteins and thus did not yield data on invertase.
The novelty of the pattern of invertase expression in P. infestans versus characterized nonoomycetes is supported by a survey of the literature and our mining of public RNA-seq data. For example, while little expression of the P. infestans invertases occurred in artificial medium compared to plant infection, no significant difference between medium and in planta growth was observed in the rust fungus Uromyces fabae and the bacterium Erwinia amylovora (31, 32). Moreover, while P. infestans invertases were expressed primarily during early infection, those of the biotroph Ustilago maydis and hemibiotroph Fusarium graminearum were expressed at higher levels during late infection (33, 34). Whether the U. maydis and F. graminearum invertases are developmentally regulated or induced in response to a rise in sucrose during late infection is unknown. This would be interesting to explore, since most research on fungal invertases has involved saprophytes, not host-adapted pathogens.
In P. infestans, extracellular carbonic anhydrases were among the few enzymes that exhibited transcriptional patterns resembling those of invertases, i.e., upregulation in spores and during biotrophy (Fig. S3). By converting carbon dioxide and water to bicarbonate and protons, carbonic anhydrases are candidate effectors that may acidify the haustorial interface to benefit the pathogen. It is interesting that the invertase of Phytophthora palmivora was shown to function optimally at a slightly acidic pH (35). Our analysis may underestimate the number of secreted enzyme activities that, like carbonic anhydrases, are coexpressed with invertases. For example, although distinct aldose epimerases were spore or biotrophy induced, their aggregate activity profile may match that of invertase. These secreted epimerases may work in concert with invertase by speeding the anomeric conversion of α- to β-d-glucose. P. infestans glucokinases are believed to be of bacterial origin, in which the enzymes more efficiently bind the β form (36).
The idea that oomycete haustoria participate in nutrient uptake is supported by our localization of invertases to that structure (Fig. 8). We hypothesize that invertase targeting does not require any motif other than its signal peptide; haustoria are simply the default destination for in planta-secreted proteins. Several data support this proposition. First, a prior study showed that three diverse extracellular infection-related proteins were all secreted from haustoria (27). Second, we found that tdTomato fused to the N-terminal regions of INF1 and HMP1 accumulated in haustoria (Fig. 9). Since our constructs included a few amino acids C-terminal to the signal peptide and since the N-terminal regions of only a few proteins were tested, additional studies could test if the requisite sequences are entirely within the signal peptide, if a second motif related to haustorial delivery resides within the signal peptide, and if N-terminal peptides from additional proteins provide the same result. We did examine the N-terminal regions of the haustoria-delivered proteins studied here and in a previous study (27) (Fig. S8) but found no shared characteristics (sequence, hydrophobicity, or secondary structure) other than those diagnostic of a signal peptide.
The concentration of P. infestans invertase in the haustorial wall is consistent with a prior study of Phytophthora sojae in medium that showed that its invertase is a glycoprotein, with >50% of the activity bound to the cell wall (37). We also observed invertase-GFP in the cell walls of P. infestans hyphae in medium when the transgene was expressed from the constitutive ham34 promoter (Fig. S7). Cell walls are known to trap many secreted proteins, especially glycoproteins, which include most fungal and plant invertases (38–40). PITG_14237 and PITG_14238 contain predicted N-glycosylation sites, particularly C-terminal to their GH32 (PFAM 00251) domains. However, using the native promoter, we observed that the invertase-GFP signal in germinating cysts was cytoplasmic. This might be explained if protein secretion by young germinated cysts is slow, if the wall signal becomes obvious only after a more extended time of secretion, or if protein glycosylation or cell wall structure in cysts differs from that of more mature hyphae. The latter is consistent with our observation that β-glucanases digest the walls of cyst germ tubes faster than those of more mature hyphae (41).
Although they are known to serve as a location for enzyme secretion, the full scope of involvement of oomycete haustoria in nutrition requires further investigation, since the site of glucose uptake is unknown. Unlike the case in fungi (7), no sugar transporter of P. infestans was transcriptionally upregulated in the haustoria-forming phases of potato leaf and tuber colonization (Fig. S5). Oomycete haustoria also lack a neckband, which encircles fungal haustoria and may help establish electrochemical gradients that aid nutrient transport (42). Instead, the extrahaustorial matrix of P. infestans is continuous with the rest of the apoplast (43). Consequently, hexoses generated by invertase may diffuse from the haustorial interface to transporters at other locations. Also, some invertase may escape the haustorial wall and become dispersed to other regions of the apoplast.
Regardless of their precise location, the combination of hexose transporters and invertases would improve the sink strength of P. infestans, providing carbohydrate to enhance pathogen growth. The relative contributions of the two types of proteins cannot be measured at present. Although ex planta studies suggest that glucose is a preferred carbon source (44), other apoplastic compounds may also supply carbon. If P. infestans has sugar transporters that can outcompete host transporters, its invertase may be dispensable, as suggested for Ustilago maydis (45). We have not yet succeeded at generating invertase knockdown strains of P. infestans using homology-based silencing methods that we have used successfully for other genes. Thus, evidence for a role of the P. infestans invertases in pathogenesis remains indirect. Nevertheless, the taxonomic distribution of invertases in oomycetes argues in favor of their contribution to plant pathogenicity: most oomycetes that infect animals also lack invertase (Fig. 3). Also, the P. infestans invertases likely contribute to the decline in apoplastic sucrose during leaf infection (Fig. 1), although enhanced activity of plant cell wall invertases or sugar transporters may also be responsible (46). It is well-known that some plants intensify sugar uptake during infection, which may reduce nutrients available to pathogens (47). In Arabidopsis, for example, sugar transporter STP13 was shown to contribute to resistance against both fungal and bacterial pathogens (48, 49). The movement of sugars into plant cells may also explain why we previously observed only small decreases in total, as opposed to apoplastic, leaf sucrose early in infection (50).
MATERIALS AND METHODS
P. infestans growth and development.
Strain 1306 was maintained in the dark in rye-sucrose agar (51) at 18°C. For RNA analysis, cultures were grown in rye-sucrose medium, minimal medium based on the recipe of Xu (21) using 0.11 M glucose and/or sucrose as the carbon source, or Henninger’s minimal medium (22) using a total of 0.055 M concentrations of the sugar(s). Nonsporulating mycelia were from 3-day cultures. Spore stages were obtained from plants as described below or rye medium (18). For the latter, sporangia were scraped from 10-day cultures using water and a glass rod and frozen pending RNA extraction or chilled at 10°C for 60 min to initiate zoosporogenesis (sporangial cleavage). Zoospores were obtained by incubating sporangia for an additional 90 min. Germinating cysts were made by adding 0.25 mM CaCl2 to zoospores, vortexing for 1 min, and incubating the resulting cysts for 6 h in water at 18°C. Starved hyphae were obtained by washing mycelia grown in rye medium twice in modified Petri’s solution (52) and then incubating the hyphae for a further 6 h in Petri’s solution.
Plant infection.
Tubers (cv. Russet Burbank) were washed in water, soaked in 10% (vol/vol) household bleach for 15 min, rinsed in water, and cut into 2-mm slices. After a second rinse, the slices were blotted dry and placed on a rack 8 mm above moist towels in a sealed box. Inoculations were performed by spreading about 0.2 ml of a 5 × 105/ml zoospore suspension per slice. These were kept at 18°C in the dark and harvested after 1.5, 2.5, and 4 days.
Leaf infections used potato cv. Russet Burbank or tomato cv. New Yorker grown using 14-h days (400 μmol m−2 s−1) at 25°C and 10-h nights at 18°C. Prior to infection, plants were equilibrated in a chamber using 12-h days (200 μmol m−2 s−1) and 12-h nights at 18°C. For RNA-seq, whole plants were sprayed with 105/ml zoospores to the point at which runoff occurred, placed in a plastic bag, and incubated in the 18°C chamber. For analyses of apoplastic sucrose, detached leaflets were inoculated with approximately 0.2 ml of water or zoospores at 2 × 104/ml. These were placed in the 18°C chamber in humidified boxes.
Sporangia for RNA-seq were washed from tubers or leaves and collected by centrifugation. Apoplastic wash fluids from leaves were prepared using modifications of existing methods (53, 54). This involved vacuum infiltration, wrapping in plastic film, and centrifugation at 1,750 × g at 4°C above a plastic support. Tissue integrity was assessed using propidium iodide (54) and by measuring β-glucose-6-phosphate. Metabolite concentrations were determined by the UCR Metabolomics Core Facility using external and internal standards, with a Waters TQ-XS triple quadrupole mass spectrometer coupled to a two-dimensional ultraperformance liquid chromatograph (UPLC).
P. infestans transformants.
Coding sequences plus 500 nucleotides (nt) of promoters were amplified using Platinum High Fidelity polymerase (Invitrogen), cloned into pGEMT-Easy (Promega), and subcloned in place of the ham34 promoter in pGFPH (hygromycin resistance) (24) to make invertase-GFP fusions. Other constructs had the coding sequences downstream of the ham34 promoter. In some experiments, these plasmids were cotransformed into P. infestans with pTdTomatoN or pH2B-TdTomato (G418 resistance). Other vectors were constructed by making translational fusions of the N-terminal regions of INF1 or HMP1 with TdTomato in pTdTomatoN (Fig. S8). Vectors were transformed into P. infestans by the protoplast method (24) using 12 μg/ml G418 and 50 μg/ml of hygromycin.
Confocal microscopy was performed using tissue from medium, tomato leaves (cv. New Yorker), or tuber slices (cv. Yukon Gold). Just before examination, tuber slices were cut into 5-mm cubes, while 5- by 5-mm squares were cut from leaves. These were inverted (infection side down) on 35-mm glass-bottom culture dishes (MatTak) under a cover glass. Other samples were mounted on a slide. Microscopy was performed using a Leica SP5 laser scanning inverted confocal microscope with a 40× water objective, using fluorescein isothiocyanate (FITC; 488 nm; for GFP) and tetramethyl rhodamine isocyanate (TRITC; 543 nm; for tdTomato) channels at 15% and 33% intensity, respectively. Two-color images were obtained using sequential scanning to prevent bleed-through.
P. pastoris experiments.
Strains KM71H and X33 were used to express P. infestans invertase using the EasySelect Pichia expression kit (Thermo). This involved cloning the mature peptide region of PITG_14238 behind the yeast signal peptide in pPICZα. Growth assays were performed at 30°C using shaking cultures in minimal medium (0.1 M potassium phosphate [pH 6.0], 13.4 g/liter yeast nitrogen base without amino acids, 40 pg/liter biotin, 5 ml/liter methanol, 40 mg/ml arginine) supplemented as appropriate with 10 g/liter sucrose.
RNA analysis.
For RNA-seq, experiments with plant samples used three biological replicates, while ex planta studies of development had two replicates. RNA was isolated using the Sigma Plant Total RNA kit. Single-end reads (75 nt) were aligned and mapped using HiSat2, and expression calls were made with edgeR, using systemPipeR (55). Low-expression-level genes were eliminated using a FPKM threshold of 1.0. Hierarchical clustering, heat map generation, and PCA used ClustVis (56). A minimum of 25 million reads were obtained per replicate of pure (nonplant) P. infestans samples, while 250, 160, and 80 million reads were obtained for early, middle, and late leaf samples and 1,000, 820, and 920 million reads were obtained for tuber samples.
RT-qPCR was performed as described elsewhere (52) using primers specific for PITG_14237 (5′-GTGCCAGCCAGACGAAGTC and 5′-GTGCTTAGGCTCCGTCCAGA) and PITG_14238 (5′-ACGGCATCAGGTAAGACCAC and 5′-CGACCTTCCCAGTCTTCTTG). cDNA made using the Maxima First-Strand kit (Thermo) was analyzed using a CFX Connect system (Bio-Rad) using the Dynamo HS SYBR green kit (Thermo). Assays employed two biological replicates, each with three technical replicates. Reverse transcriptase-lacking controls and melt curves confirmed the fidelity of amplification. Gene expression was determined by the ΔΔCT method using experimentally determined primer efficiencies with two constitutive genes as a reference. These were PITG_11766, which encodes ribosomal protein S3A, and PITG_09862, which encodes a Kelch repeat protein (57).
Sequence analysis.
Functions were assigned to metabolic proteins as described elsewhere (10). In brief, protein sequences were used as inputs for the KEGG-based KAAS and BlastKoala annotation servers. Annotations were checked for consistency with results from the Conserved Domain Database (CDD) and BLASTP searches of GenBank. Enzymes were placed into pathways based on the KEGG classification. Secreted proteins were identified using SignalP 4.1 and TargetP (28); genes were excluded if they contained a KDEL motif or a transmembrane domain based on TMHMM (58).
Oomycetes other than P. infestans (Table S2) were searched for invertases using BLASTP and TBLASTN for protein and genome searches, respectively. Glycosylation was predicted using N-GlyDE and GlycoEP (59, 60).
Sources of genome data. Download Table S2, PDF file, 0.05 MB (50.6KB, pdf) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
RNA-seq data have been deposited in NCBI GEO under BioProject nos. PRJNA361417 and PRJNA407960.
ACKNOWLEDGMENTS
This work was supported by awards from the U.S. Department of Agriculture National Institute of Food and Agriculture and National Science Foundation to H.S.J. and from UC-MEXUS and CONACYT to D.M.-S.
Footnotes
Citation Kagda MS, Martínez-Soto D, Ah-Fong AMV, Judelson HS. 2020. Invertases in Phytophthora infestans localize to haustoria and are programmed for infection-specific expression. mBio 11:e01251-20. https://doi.org/10.1128/mBio.01251-20.
REFERENCES
- 1.Griffiths CA, Paul MJ, Foyer CH. 2016. Metabolite transport and associated sugar signalling systems underpinning source/sink interactions. Biochim Biophys Acta 1857:1715–1725. doi: 10.1016/j.bbabio.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonfig KB, Gabler A, Simon UK, Luschin-Ebengreuth N, Hatz M, Berger S, Muhammad N, Zeier J, Sinha AK, Roitsch T. 2010. Post-translational derepression of invertase activity in source leaves via down-regulation of invertase inhibitor expression is part of the plant defense response. Mol Plant 3:1037–1048. doi: 10.1093/mp/ssq053. [DOI] [PubMed] [Google Scholar]
- 3.Moghaddam MRB, Van den Ende W. 2012. Sugars and plant innate immunity. J Exp Bot 63:3989–3998. doi: 10.1093/jxb/ers129. [DOI] [PubMed] [Google Scholar]
- 4.Parrent JL, Peay K, Arnold AE, Comas LH, Avis P, Tuininga A. 2010. Moving from pattern to process in fungal symbioses: linking functional traits, community ecology and phylogenetics. New Phytol 185:882–886. doi: 10.1111/j.1469-8137.2010.03190.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang Q, Liu J, Lin X, Hu S, Yang Y, Li D, Chen L, Huai B, Huang L, Voegele RT, Kang Z. 2017. A unique invertase is important for sugar absorption of an obligate biotrophic pathogen during infection. New Phytol 215:1548–1561. doi: 10.1111/nph.14666. [DOI] [PubMed] [Google Scholar]
- 6.Buttner D, Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 7.Voegele RT, Mendgen KW. 2011. Nutrient uptake in rust fungi: how sweet is parasitic life? Euphytica 179:41–55. doi: 10.1007/s10681-011-0358-5. [DOI] [Google Scholar]
- 8.Savory F, Leonard G, Richards TA. 2015. The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog 11:e1004805. doi: 10.1371/journal.ppat.1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leesutthiphonchai W, Vu AL, Ah-Fong AMV, Judelson HS. 2018. How does Phytophthora infestans evade control efforts? Modern insight into the late blight disease. Phytopathology 108:916–924. doi: 10.1094/PHYTO-04-18-0130-IA. [DOI] [PubMed] [Google Scholar]
- 10.Ah-Fong AMV, Kagda MS, Abrahamian M, Judelson HS. 2019. Niche-specific metabolic adaptation in biotrophic and necrotrophic oomycetes is manifested in differential use of nutrients, variation in gene content, and enzyme evolution. PLoS Pathog 15:e1007729. doi: 10.1371/journal.ppat.1007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PR, Whisson SC. 2008. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol 10:2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Boevink PC, Welsh L, Zhang R, Whisson SC, Birch PRJ. 2017. Delivery of cytoplasmic and apoplast effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol 216:205–215. doi: 10.1111/nph.14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawra S, Trusch F, Matena A, Apostolakis K, Linne U, Zhukov I, Stanek J, Koźmiński W, Davidson I, Secombes CJ, Bayer P, van West P. 2017. The rxlr motif of the host targeting effector avr3a of Phytophthora infestans is cleaved before secretion. Plant Cell 29:1184–1195. doi: 10.1105/tpc.16.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mormeneo S, Sentandreu R. 1982. Regulation of invertase synthesis by glucose in Saccharomyces cerevisiae. J Bacteriol 152:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vainstein MH, Peberdy JF. 1991. Regulation of invertase in Aspergillus nidulans effect of different carbon sources. J Gen Microbiol 137:315–322. doi: 10.1099/00221287-137-2-315. [DOI] [PubMed] [Google Scholar]
- 16.Neigeborn L, Carlson M. 1984. Genes affecting the regulation of suc-2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan W, Wanamaker SI, Ah-Fong AMV, Judelson HS, Lonardi S. 2018. Novo&stitch: accurate reconciliation of genome assemblies via optical maps. Bioinformatics 34:i43–i51. doi: 10.1093/bioinformatics/bty255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ah-Fong AM, Kim KS, Judelson HS. 2017. RNA-seq of life stages of the oomycete phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genomics 18:198. doi: 10.1186/s12864-017-3585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki-Kinoshita KF, Kanehisa M. 2007. Gene annotation and pathway mapping in kegg. Methods Mol Biol 396:71–91. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- 20.Yoshikawa M, Masago H. 1977. Effect of cyclic-AMP on catabolite-repressed zoosporangial formation in Phytophthora capsici. Can J Bot 55:840–843. doi: 10.1139/b77-099. [DOI] [Google Scholar]
- 21.Xu R. 1982. A defined medium for Phytophthora. Acta Mycol Sin 1:40–47. [Google Scholar]
- 22.Henninger H. 1963. Zur Kultur von Phytophthora infestans auf vollsynthetischen Nährsubstraten. Z Allg Mikrobiol 3:126–135. doi: 10.1002/jobm.3630030204. [DOI] [Google Scholar]
- 23.Judelson HS, Tyler BM, Michelmore RW. 1992. Regulatory sequences for expressing genes in oomycete fungi. Mol Gen Genet 234:138–146. doi: 10.1007/BF00272355. [DOI] [PubMed] [Google Scholar]
- 24.Ah-Fong AM, Judelson HS. 2011. Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biol 115:882–890. doi: 10.1016/j.funbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Kamoun S, Van West P, De Jong AJ, De Groot KE, Vleeshouwers VGAA, Govers F. 1997. A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol Plant Microbe Interact 10:13–20. doi: 10.1094/MPMI.1997.10.1.13. [DOI] [PubMed] [Google Scholar]
- 26.King JE, Colhoun J, Butler RD. 1968. Changes in the ultrastructure of sporangia of Phytophthora infestans associated with indirect germination and aging. Trans Br Mycol Soc 154:1794–1804. [Google Scholar]
- 27.Wang S, Welsh L, Thorpe P, Whisson SC, Boevink PC, Birch PRJ. 2018. The Phytophthora infestans haustorium is a site for secretion of diverse classes of infection-associated proteins. mBio 9:e01216-18. doi: 10.1128/mBio.01216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 29.Aram K, Rizzo DM. 2018. Distinct trophic specializations affect how Phytophthora ramorum and clade 6 Phytophthora spp. colonize and persist on Umbellularia californica leaves in streams. Phytopathology 108:858–869. doi: 10.1094/PHYTO-06-17-0196-R. [DOI] [PubMed] [Google Scholar]
- 30.Resjo S, Ali A, Meijer HJG, Seidl MF, Snel B, Sandin M, Levander F, Govers F, Andreasson E. 2014. Quantitative label-free phosphoproteomics of six different life stages of the late blight pathogen Phytophthora infestans reveals abundant phosphorylation of members of the CRN effector family. J Proteome Res 13:1848–1859. doi: 10.1021/pr4009095. [DOI] [PubMed] [Google Scholar]
- 31.Voegele RT, Wirsel S, Möll U, Lechner M, Mendgen K. 2006. Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol Plant Microbe Interact 19:625–634. doi: 10.1094/MPMI-19-0625. [DOI] [PubMed] [Google Scholar]
- 32.Bogs J, Geider K. 2000. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J Bacteriol 182:5351–5358. doi: 10.1128/jb.182.19.5351-5358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanver D, Müller AN, Happel P, Schweizer G, Haas FB, Franitza M, Pellegrin C, Reissmann S, Altmüller J, Rensing SA, Kahmann R. 2018. The biotrophic development of Ustilago maydis studied by RNA-seq analysis. Plant Cell 30:300–323. doi: 10.1105/tpc.17.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri KD, Yan C, Leng Y, Zhong S. 2016. RNA-seq revealed differences in transcriptomes between 3adon and 15adon populations of Fusarium graminearum in vitro and in planta. PLoS One 11:e0163803. doi: 10.1371/journal.pone.0163803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheard J, Farrar JF. 1987. Transport of sugar in Phytophthora palmivora butl. New Phytol 105:265–272. doi: 10.1111/j.1469-8137.1987.tb00863.x. [DOI] [Google Scholar]
- 36.Lunin VV, Li Y, Schrag JD, Iannuzzi P, Cygler M, Matte A. 2004. Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose. J Bacteriol 186:6915–6927. doi: 10.1128/JB.186.20.6915-6927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegler E, Albersheim P. 1977. Host pathogen interactions. XIII. Extracellular invertases secreted by 3 races of a plant pathogen are glycoproteins which possess different carbohydrate structures. Plant Physiol 59:1104–1110. doi: 10.1104/pp.59.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-May E, Kim S-J, Brandizzi F, Rose JKC. 2012. The secreted plant N-glycoproteome and associated secretory pathways. Front Plant Sci 3:e117. doi: 10.3389/fpls.2012.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proels RK, Huckelhoven R. 2014. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol Plant Pathol 15:858–864. doi: 10.1111/mpp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzenberg RL. 1963. The localization of beta-fructofuranosidase in Neurospora. Biochim Biophys Acta 77:455–465. doi: 10.1016/0006-3002(63)90521-3. [DOI] [PubMed] [Google Scholar]
- 41.Judelson HS, Coffey MD, Arredondo FR, Tyler BM. 1993. Transformation of the oomycete pathogen Phytophthora megasperma f. sp. glycinea occurs by DNA integration into single or multiple chromosomes. Curr Genet 23:211–218. doi: 10.1007/BF00351498. [DOI] [PubMed] [Google Scholar]
- 42.Mims CW, Richardson EA, Holt BF, Dangl JL. 2004. Ultrastructure of the host-pathogen interface in Arabidopsis thaliana leaves infected by the downy mildew Hyaloperonospora parasitica. Can J Bot 82:1001–1008. doi: 10.1139/b04-073. [DOI] [Google Scholar]
- 43.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PR. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 44.Hodgson WA. 1958. Growth of four races of Phytophthora infestans (Mont.) de Bary in synthetic medium. Can J Plant Sci 38:145–154. doi: 10.4141/cjps58-026. [DOI] [Google Scholar]
- 45.Wahl R, Wippel K, Goos S, Kamper J, Sauer N. 2010. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol 8:e1000303. doi: 10.1371/journal.pbio.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch C, Noga G, Strittmatter G. 1994. Photosynthetic electron transport is differentially affected during early stages of cultivar/race-specific interactions between potato and Phytophthora infestans. Planta 193:551–557. doi: 10.1007/BF02411561. [DOI] [Google Scholar]
- 47.Sutton PN, Gilbert MJ, Williams LE, Hall JL. 2007. Powdery mildew infection of wheat leaves changes host solute transport and invertase activity. Physiol Plant 129:787–795. doi: 10.1111/j.1399-3054.2007.00863.x. [DOI] [Google Scholar]
- 48.Lemonnier P, Gaillard C, Veillet F, Verbeke J, Lemoine R, Coutos-Thevenot P, La Camera S. 2014. Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant Mol Biol 85:473–484. doi: 10.1007/s11103-014-0198-5. [DOI] [PubMed] [Google Scholar]
- 49.Yamada K, Saijo Y, Nakagami H, Takano Y. 2016. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 354:1427–1430. doi: 10.1126/science.aah5692. [DOI] [PubMed] [Google Scholar]
- 50.Judelson HS, Narayan RD, Tani S. 2009. Metabolic adaptation of Phytophthora infestans during growth on leaves, tubers, and artificial medium. Mol Plant Pathol 10:843–855. doi: 10.1111/j.1364-3703.2009.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caten CE, Jinks JL. 1968. Spontaneous variability in isolates of Phytophthora infestans. I. Cultural variation. Can J Bot 46:329–348. doi: 10.1139/b68-055. [DOI] [Google Scholar]
- 52.Ah-Fong AM, Shrivastava J, Judelson HS. 2017. Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization. BMC Genomics 18:764. doi: 10.1186/s12864-017-4151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joosten MH. 2012. Isolation of apoplastic fluid from leaf tissue by the vacuum infiltration-centrifugation technique. Meth Mol Biol 835:603–610. doi: 10.1007/978-1-61779-501-5_38. [DOI] [PubMed] [Google Scholar]
- 54.Gentzel I, Giese L, Zhao W, Alonso AP, Mackey D. 2019. A simple method for measuring apoplast hydration and collecting apoplast contents. Plant Physiol 179:1265–1272. doi: 10.1104/pp.18.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backman TWH, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metsalu T, Vilo J. 2015. Clustvis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu X, Ah-Fong AMV, Lopez LA, Judelson HS. 2018. Transcriptomic and proteomic analysis reveals wall-associated and glucan-degrading proteins with potential roles in Phytophthora infestans sexual spore development. PLoS One 13:e0198186. doi: 10.1371/journal.pone.0198186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Yu P, Luo J, Jiang Y. 2003. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14:859–865. doi: 10.1007/s00335-003-2296-6. [DOI] [PubMed] [Google Scholar]
- 59.Itti TP, Chen C-T, Lin H-N, Choong W-K, Hsu W-L, Sung T-Y. 2019. N-GlyDE: a two-stage N-linked glycosylation site prediction incorporating gapped dipeptides and pattern-based encoding. Sci Rep 9:15975. doi: 10.1038/s41598-019-52341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chauhan JS, Rao A, Raghava GPS. 2013. In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One 8:e67008. doi: 10.1371/journal.pone.0067008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vetukuri RR, Asman AK, Tellgren-Roth C, Jahan SN, Reimegard J, Fogelqvist J, Savenkov E, Soderbom F, Avrova AO, Whisson SC, Dixelius C. 2012. Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans. PLoS One 7:e51399. doi: 10.1371/journal.pone.0051399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summed expression of enzymes in KEGG orthology groups. (A) Aggregate expression levels of each KEGG orthology group in hyphae (minimal medium), hyphae (rye-sucrose medium), sporangia, chilled sporangia, zoospores, germinated cysts, potato leaf at 2 and 6 dpi, potato leaf at 6 dpi, and tubers at 4 dpi, with the annotated enzyme and EC number for each KEGG group and the pathway classification. (B) P. infestans genes assigned to each KEGG group. Download Table S1, XLSX file, 0.2 MB (190.1KB, xlsx) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Markers of developmental stages during colonization of potato tubers and leaves. Shown are per-gene normalized expression values from RNA-seq of genes encoding RXLR protein PITG_23226, NPP1 protein PITG_16866, and centrin PITG_21764, which are markers of the biotrophic, necrotrophic, and sporulation stages, respectively (52, 61). Error bars represent standard deviations based on three biological replicates. Download FIG S1, TIF file, 0.6 MB (712.1KB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of metabolic genes classified by KEGG orthology (KO) number. RNA levels in the samples described in the legend to Fig. 3 were calculated by averaging the summed FPKM values of genes in each KO group, followed by normalization across all samples to a mean of 1. Invertase (K01193) and other enzymes mentioned in the text are highlighted by a blue arrow and diamonds, respectively. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. The FPKM data used to make the heat map are shown in Table S1. Download FIG S2, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of secreted metabolic enzymes. Indicated are per-gene-normalized expression values based on the samples described in the legend to Fig. 3. Each gene is annotated with its function, Enzyme Commission (EC) number, and gene name (PITG_XXXXX). The two invertases are highlighted with arrows. Data were subjected to hierarchical clustering using the Pearson correlation method with complete linkage. Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of secreted RXLR and protease effectors. Data are per-gene-normalized expression values based on the samples described in the legend to Fig. 3. Invertase is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S4, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of cell wall-degrading (CWDE) enzymes. Data are per-gene-normalized expression values based on the samples in Fig. 3. Appended to the name of each gene is its enzymatic function, as follows: CUT (cutinase); PE (pectin esterase, carbohydrate esterase family CE8); pectate lyase families PL1, PL2, and PL3; GH28 (polygalacturonase); GH53 (endo-β-1,4-galactanase); GH54 (β-xylosidase); GH78 (α-l-rhamnosidase); and GH105 (rhamnogalacturonyl hydrolase). Invertase expression is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S5, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of predicted sugar transporters. Data are per-gene-normalized expression values based on the samples described in Fig. 3. Genes belong to the MFS or MTN3 (SWEET) families, as noted. Invertase is marked by the arrow. Data were subjected to hierarchical clustering using the Pearson correlation method with average linkage. Download FIG S6, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of invertase in rye-sucrose medium using ham34 promoter. (A) Hypha of P. infestans transformant expressing invertase-GFP (showing expression in the cell wall [w]) and mCherry-tagged histone H2b as a marker of nuclei (red channel [n]). (B) Sporangium from P. infestans strain expressing invertase-GFP, showing strong fluorescence at papilla (p). (C) Sporangium of strain expressing invertase-GFP and histone H2b-mCherry. (D) Strain expressing tdTomato fused to the Inf1 signal peptide. Download FIG S7, TIF file, 2.7 MB (2.8MB, tif) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N-terminal regions of proteins mentioned in this paper. The first 64 amino acids of the INF1 (PITG_12251), HMP1 (PITG_00375), and invertase (PITG_14238) proteins are shown. Grey shading indicates the signal peptides predicted by TargetP 2.0 (28). The regions used in tdTomato fusions involving INF1 and HMP1 are underlined. PITG_21410, PITG_22926, PITG_01029, and PITG_14371 are the secreted INF4, pectinesterase, and RXLR proteins, which were shown in other studies to concentrate near haustoria (27). Sequences C-terminal of the signal peptide are dissimilar in chemical features such as hydropathy, ranging from moderately hydrophilic to hydrophobic, and lack conserved motifs based on alignments and motif discovery programs. Download FIG S8, PDF file, 0.05 MB (49.7KB, pdf) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sources of genome data. Download Table S2, PDF file, 0.05 MB (50.6KB, pdf) .
Copyright © 2020 Kagda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq data have been deposited in NCBI GEO under BioProject nos. PRJNA361417 and PRJNA407960.