In this study, we report that the ability of the human pathogen Leishmania to sense and monitor the lack of arginine in the phagolysosome of the host macrophage is essential for disease development. Phagolysosomes of macrophages are the niche where Leishmania resides and causes human leishmaniasis. During infection, the arginine concentration in the phagolysosome decreases as part of the host innate immune response. An arginine sensor on the Leishmania cell surface activates an arginine deprivation response pathway that upregulates the expression of a parasite arginine transporter (AAP3). Here, we use CRISPR/Cas9-mediated disruption of the AAP3 locus to show that this response enables Leishmania parasites to successfully compete with the host macrophage in the “hunger games” for arginine.
KEYWORDS: host-pathogen interaction, Leishmania, amino acid sensing, amino acid transport, intracellular parasitism
ABSTRACT
Arginine homeostasis in lysosomes is critical for the growth and metabolism of mammalian cells. Phagolysosomes of macrophages are the niche where the parasitic protozoan Leishmania resides and causes human leishmaniasis. During infection, parasites encounter arginine deprivation, which is monitored by a sensor on the parasite cell surface. The sensor promptly activates a mitogen-activated protein kinase 2 (MAPK2)-mediated arginine deprivation response (ADR) pathway, resulting in upregulating the abundance and activity of the Leishmania arginine transporter (AAP3). Significantly, the ADR is also activated during macrophage infection, implying that arginine levels within the host phagolysosome are limiting for growth. We hypothesize that ADR-mediated upregulation of AAP3 activity is necessary to withstand arginine starvation, suggesting that the ADR is essential for parasite intracellular development. CRISPR/Cas9-mediated disruption of the AAP3 locus yielded mutants that retain a basal level of arginine transport but lack the ability to respond to arginine starvation. While these mutants grow normally in culture, they were impaired in their ability to develop inside THP-1 macrophages and were ∼70 to 80% less infective in BALB/c mice. Hence, inside the host macrophage, Leishmania must overcome the arginine “hunger games” by upregulating the transport of arginine via the ADR. We show that the ability to monitor and respond to changes in host metabolite levels is essential for pathogenesis.
INTRODUCTION
Protozoan parasites of the genus Leishmania are the causative agents of a wide spectrum of human and veterinary diseases. Leishmania species cause morbidity and mortality throughout the Old and New Worlds, with clinical manifestations ranging from lesions of the skin (cutaneous leishmaniasis [CL]) and mucous membranes (mucocutaneous leishmaniasis [MCL]) to lethal infection of the spleen and liver (visceral leishmaniasis [VL]) (1). Approximately 350 million people in 88 countries are at risk of VL, with as many as 500,000 new cases diagnosed every year, 10% of which are fatal (2).
Leishmania donovani, the causative agent of kala-azar (VL), exhibits a digenetic life cycle that includes both insect and mammalian forms. Extracellular promastigotes develop in the alimentary tract of sand flies. Following infection of the mammalian host, promastigotes differentiate into intracellular amastigotes within the phagolysosome of macrophages (3, 4). This differentiation process in the host can be mimicked in axenic cultures by shifting promastigotes from an insect-like (26°C and pH 7) to an intralysosomal (37°C, pH 5.5, and 5% CO2) environment (5–7).
During infection, Leishmania parasites encounter macrophage defense mechanisms designed to interdict parasite invasion and block their intracellular survival, including the release of reactive oxygen species (ROS) and the synthesis of cytotoxic nitric oxide (NO) by inducible nitric oxide synthase (iNOS). Yet for macrophages to produce effective amounts of NO, they must import arginine from the extracellular environment. Countering the host NO attack, parasitic invasion activates macrophage arginase 1, which converts arginine to ornithine, the first substrate of the polyamine pathway, thereby suppressing NO synthesis and promoting parasite survival (8). However, the reduction of the host arginine pool becomes a double-edged sword for Leishmania parasites in infected macrophages since they cannot synthesize arginine de novo. Instead, they must import exogenous arginine via a monospecific amino acid transporter (AAP3) (9), using it primarily in the polyamine pathway to provide precursors for trypanothione biosynthesis (10). Thus, they encounter an intriguing metabolic dilemma: on the one hand, emptying the host arginine pool provides an advantage (reduction of NO), but at the same time, it causes a potentially deadly disadvantage by blocking the supply of an essential amino acid (8). Therefore, to survive, Leishmania parasites need to sense and respond to changes in arginine availability.
A few years ago, we discovered that upon arginine starvation, Leishmania parasites promptly activate a mitogen-activated protein kinase 2 (MAPK2)-mediated arginine deprivation response (ADR) pathway, resulting in the upregulation of the expression and activity of the Leishmania arginine transporter (AAP3) (11). Significantly, the ADR is also activated during macrophage infection due to intracellular parasites actively depleting arginine within the host phagolysosome (12).
The L. donovani genome contains two (haploid) AAP3 gene copies that are tandemly arrayed on chromosome 31 (chr31), which is tetrasomic in L. donovani (and most other Leishmania species). While the coding sequences (CDSs) of the AAP3.1 and AAP3.2 genes are (almost) identical, their 3′ untranslated regions (UTRs) are quite different, and only (4.5-kb and 3.5-kb) mRNAs from the latter (AAP3.2) are upregulated under conditions of arginine deprivation (10). Genome-scale transcriptomic analysis (RNAseq) revealed only five other changes in gene expression (mostly other transporters) associated with arginine deprivation, indicating the presence of a coordinated ADR (11).
Nutrient sensing is an essential means for parasites to adapt to and successfully settle inside the vector and host (13). Studies by several laboratories have recently identified intriguing nutrient sense response mechanisms in Leishmania. Martin et al. (14) reported that intracellular sensing of purine starvation directs promastigotes into long-term cell cycle arrest at G1-G0 until a new supply of purine is provided. Starvation of Leishmania mexicana promastigotes for glucose induced a 50-fold increase in the abundance of LmGT1, a glucose transporter that localizes to the flagellum (15). In the case of the facultative intracellular parasite Leishmania, arginine availability is of particular interest since it is important for both host defense and parasite proliferation. Sensing of arginine levels in the lysosome lumen is a key mechanism that regulates mTORC1 activity in mammalian cells (16). In macrophages, mTORC1 activation induces a Th1 response (17). This pathway is the major means by which macrophages kill invading pathogenic microorganisms. Interestingly, Leishmania parasites are able to counteract this outcome by activating a Th2 response, directing arginine toward polyamine biosynthesis instead of NO production, thereby enabling the parasites to persist and cause long-term nonhealing infections (18, 19). However, both Th1 and Th2 responses result in arginine depletion in the macrophage phagolysosome, presenting an existential threat to parasite survival since they are unable to synthesize this essential amino acid.
In this study, we explore the mechanism that enables intracellular Leishmania to win this “hunger game.” Using CRISPR/Cas9, we created mutants that lack AAP3.2 and thereby are unable to upregulate AAP3 expression after arginine starvation. These mutants were unable to grow in either THP-1 macrophages or BALB/c mouse livers. This study shows that sensing host nutrients is essential for intracellular parasite development.
RESULTS AND DISCUSSION
The Leishmania genome contains two genes (AAP3.1 and AAP3.2) in a tandem array on chromosome 31 that encode arginine transporters. They are highly conserved (only 3 amino acid differences) within their CDSs and 5′ UTRs but have very different 3′ UTRs. However, only AAP3.2 is responsive to arginine deprivation (10). To assess whether the AAP3.2 response to arginine deprivation is necessary for parasite intracellular development, we aimed to delete it from the L. donovani genome. Unfortunately, chromosome 31 is tetraploid in L. donovani (20), making classical homologous gene replacement approaches cumbersome and tedious, especially since the plasticity of the Leishmania genome enables parasites to retain an additional wild-type (WT) chromosome as well as those containing the selectable marker(s) (21). Therefore, we used the Leishmania-adapted CRISPR/Cas9 system (22) to expedite the disruption of the AAP3 genes.
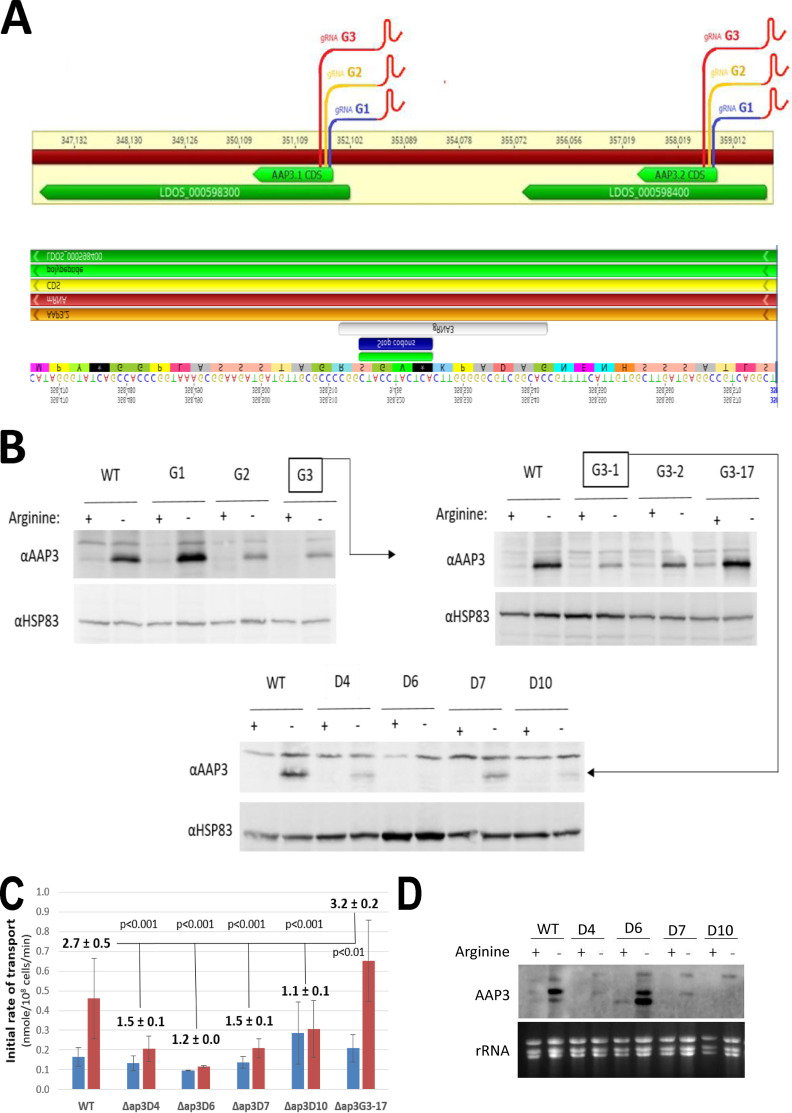
WT L. donovani was transfected (separately) with CRISPR plasmids containing 21-nucleotide (nt) guide RNAs (gRNAs) (G1, G2, and G3) targeting the 5′ ends (positions +19, 85, and 173) of both the AAP3.1 and AAP3.2 CDSs (Fig. 1A, top), and the cultures were examined 6 weeks later for AAP3 protein levels in the presence or absence of arginine. While the G1-transfected cultures showed enhanced ADR-mediated increases in AAP3 levels compared to the WT, the G2 and G3 cultures showed a diminished capacity to upregulate AAP3 levels after arginine starvation (Fig. 1B, top left). The G3 culture was seeded onto agar plates, and individual colonies were examined for an ADR, with different clones showing normal (Δap3G3-2), enhanced (Δap3G3-17), or reduced (Δap3G3-1) increases in AAP3 protein abundance compared to the WT (Fig. 1B, top right). In order to increase the efficiency of CRISPR/Cas9-mediated gene disruption, the Δap3G3-1 mutant was transfected (four times at 3-day intervals) with a 61-nt “donor” oligonucleotide consisting of an 11-nt insertion with a stop codon in all frames and 25-nt sequences flanking the Cas9 cleavage site (Fig. 1A, bottom). Single colonies were isolated 3 weeks after transfection and examined for AAP3 expression levels after arginine deprivation. Two clones (Δap3D6 and Δap3D10, D6 and D10, respectively) showed no increase in AAP3 protein abundance after arginine deprivation, while two others (Δap3D4 and -D7, D4 and D7, respectively) showed a small (less than WT) increase in AAP3 protein levels (Fig. 1B, bottom). Analyses of the initial rates of arginine transport confirmed that these mutants had lost all or some of their response to arginine deprivation (Fig. 1C; see also Fig. S1 in the supplemental material). While WT cells showed a 2.7-fold increase in the initial rate of arginine transport 2 h after arginine starvation, most of the mutants showed little or no increase in the transport rate after arginine starvation (1.5-, 1.2-, 1.5-, and 1.1-fold for Δap3D4, Δap3D6, Δap3D7, and Δap3D10, respectively). Interestingly, the Δap3G3-17 mutant showed a 3.2-fold (larger than WT) increase in transport after arginine deprivation, consistent with additional copies of the AAP3.2 gene (see below).
FIG 1.
Targeting CRISPR guides to the 5′ end of the AAP3 open reading frame (ORF) impairs their response to arginine deprivation. (A, top) Leishmania-adapted CRISPR/Cas9 targeted to the 5′ end of the AAP3 ORF at bp 19, 85, and 173 in the CDS with gRNAs designated G1, G2, and G3, respectively. gRNA-transfected promastigotes were grown in cultures supplemented with neomycin (50 μg/ml) for 6 weeks. (B, top left) Subsequently, proteins extracted from these cells were subjected to Western analysis using anti-LdAAP3 antiserum (and HSP83 antiserum for a loading control). (B, top right) G3 cells (highlighted in squares) were then seeded on agar plates, and three types of colonies were raised, Δap3G3-1, Δap3G3-2, and Δap3G3-17. (A, bottom) The Δap3G3-1 mutant (highlighted in a square) was then further transfected with a donor, i.e., the gRNA G3-based sequence containing an 11-bp insertion with stop codons. (B, bottom) These transfectants gave rise to several colonies of Δ(aa3-adr) mutants that we named Δap3D4, Δap3D6, Δap3D7, and Δap3D10. (C) Fold change of the initial rate of arginine transport (2 min) (see Fig. S1 in the supplemental material) before and after 2 h of arginine starvation (blue and red, respectively). (D) Northern analysis of AAP3 mRNA expression before and after 2 h of arginine starvation.
Initial rates of arginine transport in wild-type and mutant L. donovani promastigotes. Two-minute arginine transport was determined using the rapid filtration technique (see Materials and Methods). Transport was assayed in the wild type before (diamond) and after (square) 2 h of arginine starvation and in the Δap3D6 mutant before (diamond) and 2 h after (square) arginine starvation. Download FIG S1, EPS file, 1.0 MB (1MB, eps) .
Copyright © 2020 Goldman-Pinkovich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
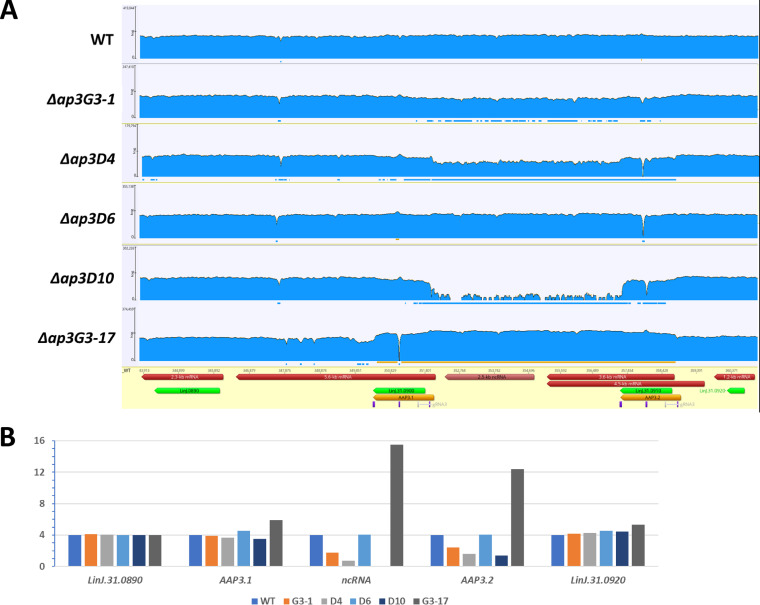
Whole-genome sequencing was carried out to map the precise location of the CRISPR/Cas9-induced mutations in each cell line. Illumina libraries were constructed and sequenced on a HiSeq 4000 instrument using paired-end 75-bp reads with TruSeq standard primers. Between 45.9 million (Δap3D4) and 96.3 million (Δap3G3-17) reads were obtained per library, and they were aligned against the L. donovani 1S genome sequence assembled from a combination of PacBio and Illumina reads (A. Sur, J. R. McDonald, G. Ramasamy, and P. J. Myler, unpublished data). Changes in coverage (read counts) were seen (Fig. 2A) in the AAP3 locus of all mutants (Δap3D7 is not shown since it was essentially identical to Δap3D4). As expected, the WT parent had four copies of both AAP3.1 and AAP3.2 as well as the genes flanking this locus (Fig. 2B). However, the Δap3G3-1 mutant and most of its progeny (D4 and D10) had substantially lower coverage in the sequence between the 5′ UTR of AAP3.1 and the 3′ end of AAP3.2, consistent with recombination near the double-strand break(s) introduced by Cas9 at the site of the gRNA G3 sequence to create an AAP3.1/AAP3.2 fusion that contains the 5′ UTR of AAP3.2 and the 3′ UTR of AAP3.1 along with a deletion of the 3′ UTR of AAP3.2 (and the non-coding RNA [ncRNA] located in the intergenic region between the two AAP3 genes). Quantitation of the read counts (Fig. 2B) indicated that the Δap3G3-1 mutant retained two intact copies of AAP3.1 and AAP3.2, along with two copies of the AAP3.1/AAP3.2 fusion, and Δap3G3-1/D4 (and Δap3G3-1/D7) retained one copy of the intact AAP3 locus (containing both AAP3.1 and AAP3.2) and three copies of the AAP3.1/AAP3.2 fusion, while in Δap3D10, all four copies of the AAP3 locus contained the AAP3.1/AAP3.2 fusion. A small number of reads, 102, from this region are present in the Δap3D10 mutant, suggesting that some cells in this population may retain an intact copy of chr31 (or the sample was contaminated with another cell line). In contrast, the Δap3G3-17 mutant showed higher coverage in this region, consistent with recombination between the two copies of the AAP3 gene resulting in additional (2 to 3 for each copy of chr31) copies of AAP3.2 and the intergenic sequence (including the ncRNA). Interestingly, the Δap3D6 mutant shows slightly higher than WT levels of read coverage for the AAP3.1 gene, suggesting that it retained one copy of chr31 with the AAP3.1/AAP3.2 fusion along with three copies containing both the AAP3.1 and AAP3.2 genes (one of which may contain an additional AAP3.2 gene). Examination of the read coverage in the gRNA G3 region indicated that 78% (418/536) contain the 11-bp insertion with a stop codon in all frames, consistent with the hypothesis that the Δap3D6 mutant contained seven copies of the AAP3.1 and AAP3.2 genes with stop codons that render them nonfunctional at the protein level and only one (or two) functional version(s). However, the inability of this mutant to present with even a partial ADR on the protein level (such as that of Δap3D4 or Δap3D7) suggests that the only functional copies without stop codons are AAP3.1/AAP3.2 fusions that lack ADR capacity.
FIG 2.
Whole-genome sequencing of the AAP3 region in CRISPR/Cas9-treated L. donovani promastigotes. Whole-genome sequencing was carried out for the WT and each of the Δ(aap3-adr) mutants. Illumina libraries were constructed and sequenced on a HiSeq 4000 instrument using paired-end 75-bp reads with TruSeq standard primers. (A) Reads were aligned against the L. donovani 1S reference genome and revealed that mutants Δap3G3-1, Δap3D4, and Δap3D10 harbor a large deletion essentially fusing the AAP3.1 CDS with that of AAP3.2 and eliminating the AAP3.2 3′ UTR and all intergenic ncRNA sequences. Quantification of reads indicates that a gene fusion event has occurred on two copies of chromosome 31 in the Δap3G3-1 mutant, on three copies in Δap3D4, and on four copies in Δap3D10. The Δap3G3-17 mutant harbors 16 copies of the region. Δap3D6 alignment to a version of LdoS_31 revealed an 11-bp insertion (with a stop codon in each frame) in the AAP3.2 gene. The read counts for each gene represent only those that can be unambiguously assigned to the WT or mutant version of the gene. These results are consistent with the hypothesis that 3 copies of AAP3.2 and 3 copies of the 4 AAP3.1 genes contain stop codons in the Δap3D6 mutant. (B) Read analysis indicates that neighboring genes such as LinJ.31.0890 and LinJ.31.0920 were not affected by the AAP3 mutation.
Northern analyses (Fig. 1D) confirmed the reduction in the AAP3.2 gene copy number in the D4, D7, and D10 cell lines by showing that its mRNA levels after arginine deprivation were significantly lower than those of the WT. These changes paralleled the changes in protein abundance observed in Fig. 1B, confirming that AAP3.2 accounts for most (if not all) of the increase in arginine transport after arginine starvation. Importantly, even though the AAP3.2 mRNA levels in the Δap3D6 mutant were similar to WT levels in both the presence and absence of arginine, AAP3 protein levels were not upregulated in response to arginine deprivation because of the stop codons in the AAP3.2 gene. Two other ADR-responsive genes (LinJ.10.1450 and LinJ.36.2900, which encode pteridine and MFS family transporters, respectively) (11) responded normally to arginine deprivation in all mutants (Fig. S2), indicating that the CRISPR/Cas9 mutations affected only the response of AAP3.2 to arginine deprivation, not the entire ADR pathway.
Other members of the ADR retained sensitivity to arginine deprivation. Real-time PCR analysis was carried out to determine mRNA levels of pteridine (top) and MFS (bottom) transporters before and 2 h after arginine starvation. The analyses were carried out in Δap3D4 (D4), Δap3D6 (D6), Δap3D7 (D7), and Δap3D10 (D10) mutants. Results are relative to 2-h arginine-deprived WT cells. Download FIG S2, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2020 Goldman-Pinkovich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
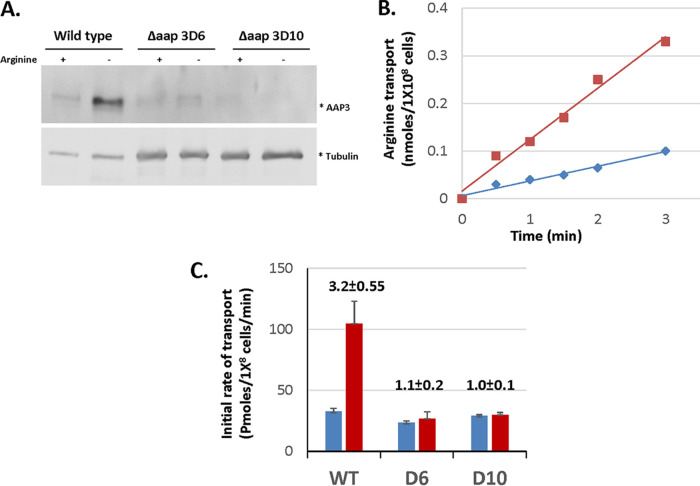
The analyses described above indicated that the Δap3D6 and Δap3D10 mutants are the most informative AAP3 mutants, retaining near-WT basal levels of arginine transport (when grown in normal medium), but are unable to upregulate transporter activity after arginine deprivation. Therefore, we decided to conduct further studies with these two mutants to determine whether the upregulation of AAP3 expression is necessary for intracellular growth. First, we assessed whether amastigotes retain the phenotypes observed for promastigotes. Axenic promastigotes, WT and mutants, differentiated into amastigotes in culture (7). The rates of differentiation were identical in both the WT and the Δap3D6 and Δap3D10 mutants; i.e., they reached maturation within 5 days (6). Mature amastigotes were then subjected to 2 h of arginine starvation. As shown in Fig. 3A, WT axenic amastigotes responded to arginine deprivation by increasing the AAP3 protein abundance 2.3-fold. In contrast, Δap3D6 and Δap3D10 amastigotes were insensitive to arginine deprivation, as were mutant promastigotes.
FIG 3.
Wild-type axenic amastigotes, but not AAP3.2-null mutants, respond to arginine deprivation. Wild-type, Δap3D6 (D6), and Δap3D10 (D10) L. donovani amastigotes were subjected to 2 h of arginine deprivation as described in Materials and Methods. (A) A portion of each culture was subjected to SDS-PAGE, transferred to nitrocellulose, and then subjected to anti-AAP3 antibodies. (B) The other portion was subjected to analysis of the initial rate of arginine transport. (C) The slopes of each transport assay were used to calculate the rate of transport.
In correlation with ADR-driven AAP3 expression in amastigotes, the initial rate of arginine transport in the WT increased by 3-fold following 2 h of arginine deprivation (Fig. 1B and C). Both Δap3D6 and Δap3D10 amastigotes transported arginine at a rate similar to that of unstarved WT amastigotes but remained unchanged after arginine deprivation. Hence, the results indicate that wild-type and mutant amastigote responses to arginine deprivation were identical to those of promastigotes. This further indicates that the rate of arginine transport in L. donovani is regulated by the abundance of the AAP3 protein.
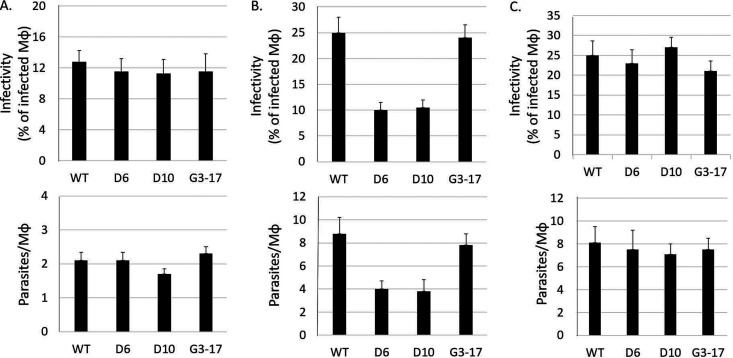
To determine whether the ADR is necessary for intracellular growth, we infected THP-1 macrophages with late-log-phase L. donovani promastigotes of the WT and Δap3D6, Δap3D10, and Δap3G3-17 mutants in medium containing a physiologically relevant (0.1 mM) concentration of arginine (12, 23). As shown in Fig. 4A, the initial level of infection (i.e., the percentage of macrophages infected after 4 h of coincubation) (Fig. 4A, top) and parasite burden (i.e., the average number of parasites per infected macrophage) (bottom) with all three mutants were similar to those of the WT. However, by 48 h postinfection (Fig. 4B), the infectivity and parasite burden for both the Δap3D6 and Δap3D10 mutants were reduced by 2-fold or more compared to the WT, while the levels for Δap3G3-17 were comparable to those of the WT. Hence, our results indicate that while the mutants can infect macrophages as well as the WT, they fail to proliferate normally thereafter. Indeed, it appears that a significant number of macrophages completely cleared their internalized parasites (since the percentage of infected macrophages was lower after 48 h than that after 4 h), while the parasites in the remaining infected macrophages underwent only 1 or 2 rounds of replication (since the parasites per macrophage increased only ∼2-fold), compared to the normal 3 to 4 rounds of replication and reinfection of new macrophages seen with WT parasites.
FIG 4.
Mutants lacking the AAP3 ADR lost infectivity in THP-1 macrophages. Human THP-1 differentiated macrophages grown in RPMI 1640 medium containing 0.1 mM arginine were subjected to infection by mid-log-phase L. donovani promastigotes of the WT and Δap3D6 (D6), Δap3D10 (D10), and Δap3G3-17 (G3-17) mutants at an MOI of 10 for 4 h, as described in Materials and Methods. Following 4 h of coincubation, extracellular parasites were washed away, and this point is referred to as time zero for the purpose of the infection start point. At time zero and 48 h after infection, macrophages were fixed, Giemsa stained, and subsequently subjected to counting. (A and B) The intracellular growth capacity was determined by calculating infectivity (top) and parasitemia (bottom). Infectivity was calculated as the percentage of infected macrophages (Mϕ) of the total counted macrophages (n = ≥100 cells). Parasitemia is calculated as the average number of parasites per infected macrophage. Macrophage infections were carried out in the presence of 0.1 mM (A and B) or 1.5 mM (C) arginine. For panel B, one-way ANOVA indicated that the Δap3D10 and Δap3D6 mutants were 57% less infective than the WT (P < 0.05; n = 16), and parasitemia was 50% lower than for the WT (P < 0.05; n = 4). (C) When infection was performed at a 1.5 mM external arginine concentration, one-way ANOVA was insignificant for mutant impairment in either infectivity (top) or parasitic burden (bottom) parameters (P < 0.1).
To assess whether the lower AAP3 expression levels (and consequently less arginine import) were responsible for the reduction in the infectivity and parasite burden of the Δap3D6 and Δap3D10 mutants, we infected THP-1 macrophages grown in medium containing 1.5 mM arginine, a concentration that prevented the ADR in intracellular WT amastigotes (12). Under these conditions, both the Δap3D6 and Δap3D10 mutants developed normally into amastigotes, with infectivity and parasitemia at 48 h similar to those of the WT (Fig. 4C). The results indicate that the inability to develop inside macrophages is due to arginine deprivation in macrophage phagolysosomes during infection and that the ability to respond to this deprivation by upregulating AAP3.2 protein levels is essential for successful intracellular Leishmania development.
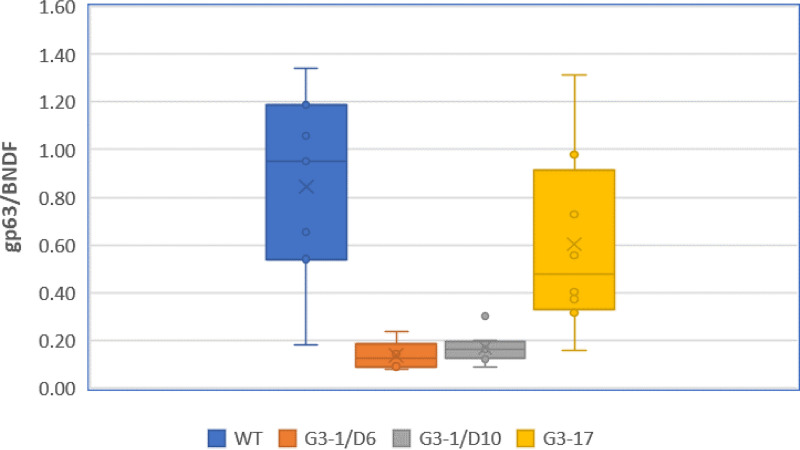
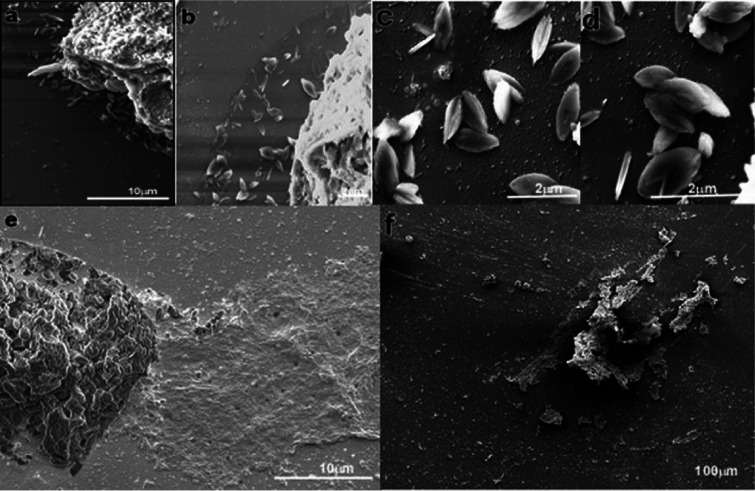
To further assess the role of arginine transport in vivo, we infected BALB/c mice (8 per group) with Δap3D6, Δap3D10, Δap3G3-17, and WT parasites. On day 21 postinfection, the mice were sacrificed, and tissue parasite burdens were determined by quantitative PCR (qPCR) on the DNA extracted from the liver of each mouse. As shown in Fig. 5, the parasite burdens of the Δap3D6 and Δap3D10 mutants averaged only 20 and 24%, respectively, of that of the WT. One-way analysis of variance (ANOVA) and Tukey post hoc honestly significant difference (HSD) testing showed a significant difference (P < 0.001) from the WT for both the Δap3D6 and Δap3D10 mutants, while the Δap3G3-17 mutant was not significantly different from the WT (P = 0.387). To further evaluate the level of infection, dissected mouse liver samples (n = 6 for each group) were crushed in the presence of Karnovsky fixative to prepare crude liver homogenates. These were fixed and subsequently subjected to scanning electron microscopy (Fig. 6). As shown, livers from mice infected with WT parasites were highly infected with amastigotes (Fig. 6a to d), while livers infected with the Δap3D6 mutant were almost clear of parasites (Fig. 6e and f), supporting the qPCR data that mutant parasites that lack AAP3.2 are unable to develop colonies in their host.
FIG 5.
Parasites that lack AAP3.2 are unable to develop in mice. Late-stationary-phase L. donovani promastigotes of the WT and Δap3D6, Δap3D10, and Δap3G3-17 mutants were injected intravenously (i.v.) into 8-week-old female BALB/c mice (1 × 108 cells per injection; eight mice per mutant). On day 21 postinfection, mice were sacrificed, and liver DNA was extracted using the proteinase K method (26). Analyses were carried out as described in Materials and Methods.
FIG 6.
Scanning electron microscopy of crude WT and mutant L. donovani-infected liver homogenates. Dissected mouse liver samples were crushed in the presence of Karnovsky fixative to prepare a crude liver homogenate, which was processed further with tannic acid fixation and 4% osmium tetroxide. The samples were serially dehydrated in ethanol, coated with gold, and visualized using an FEI Quanta 200 FEG electron microscope at 20 kV. (a) WT-infected liver tissue piece with amastigotes seen underneath the tissue piece. (b to d) Enlarged images from the wild-type-infected liver tissue piece. (e and f) D6-infected liver tissue piece with amastigotes found negative.
These results show that the inability of the mutants to express higher levels of AAP3 in order to compensate for the reduced level of arginine in the phagolysosomes of infected macrophages severely compromised their ability to develop in the liver. Since spleens were not included in this analysis, it is possible that differential expression of arginine metabolism-related enzymes in the different organs may influence the role and significance of the AAP3-mediated ADR in parasite survival. Nevertheless, we show here that the liver as a major target of this pathogen presents with significantly lower parasite burdens for mutant parasites than for both WT and AAP3.2-overexpressing parasites (G3-17), which serve as an alternative to an add-back control. Therefore, it appears that the ADR is a crucial mechanism for enabling intracellular Leishmania parasites to overcome the arginine bottleneck.
In summary, this study demonstrates that the ability to monitor metabolic deprivation and subsequently induce a specific response at the level of gene expression is essential for the pathogenesis of a protozoan parasite. Furthermore, we have shown that for pathogenic microorganisms to respond to host-inflicted environmental changes and survive, they must employ external sensing and response mechanisms to serve as their monitoring device.
MATERIALS AND METHODS
Leishmania strains and culture.
L. donovani MHOM/SD/00/1S (24) promastigotes were grown in medium M-199, Earle’s salts (Biological Industries Ltd.) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco Ltd.) and a 1% penicillin-streptomycin solution (Biological Industries Ltd.). Axenic differentiation of L. donovani promastigotes to amastigotes was carried out as described previously (7). Briefly, mid-log-phase promastigotes were washed twice in Earle’s salt solution and finally suspended in amastigote medium containing M-199 at pH 5.5 supplemented with 0.5 mM sodium succinate, 25% bovine serum, and 1% penicillin-streptomycin solution. Mature amastigotes developed at 5 days after exposure of promastigotes to the differentiation medium.
Arginine deprivation was carried out as described previously by Pawar et al. (12). Briefly, mid-log-phase promastigotes (1 × 107 cells/ml) were washed with Earle’s balanced salt solution twice and resuspended in arginine-deficient medium M-199 (Biological Industries Ltd.) at 26°C for the specified period before being transferred to ice. Arginine-deprived cells were washed twice with ice-cold Earle’s balanced salt solution before being used for transport assays and Northern and Western blot analyses.
CRISPR/Cas9 guide and donor transfections.
gRNA sequences were designed using the Eukaryotic Pathogen CRISPR Guide RNA/DNA Design Tool (EuPaGDT) (http://grna.ctegd.uga.edu/) and cloned into Leishmania-adapted vector pLdCN using the single-step digestion-ligation cloning protocol previously described (22), and the constructs were transfected into mid-log-phase promastigotes. Following gRNA-pLdCN transfections, cells were grown for 4 weeks with G418 at 50 μg/ml and subsequently screened for their ability to increase LdAAP3 protein abundance after arginine deprivation. Next, the G3 donor sequence was introduced into a gRNA G3-originated clone exhibiting the desired phenotype. The G3 donor is a single-strand oligonucleotide donor (sense) containing 25-nt sequences flanking the Cas9 cleavage site (shown in boldface type below) and an 11-nt sequence with a stop codon in all three frames (underlined and italicized below). Three transfections with 10 μl (100 μm) of this oligonucleotide were performed on promastigotes at 3-day intervals as previously described (22). gRNA sequences are as follows: GTCTATTCCAGCACAGGCGG for gRNA G1, GCCGTCGATAAACACCCGAG for gRNA G2, GTGCCGACGCCGCCAAGCCG for gRNA G3, and ATGAAAACGGTGCCGACGCCGCCAAGTGAGTAGGTAGCCGGGGCGCAACATCATCTTCCG for the G3-based donor.
Arginine deprivation.
Arginine deprivation of axenic promastigotes and amastigotes was carried out as described previously by Goldman-Pinkovich et al. (11). Briefly, logarithmic-phase promastigotes or amastigotes (1 × 107 cells/ml) were washed twice in ice-cold Earle’s salt solution at pH 7 (promastigotes) or pH 5.5 (amastigotes) and subsequently suspended to a final density of 1 × 107 cells/ml in arginine-free Earle’s salt-based medium 199 (Biological Industries Ltd., Beit Haemek, Israel). This deprivation medium was supplemented with heat-inactivated dialyzed (10,000-kDa cutoff) fetal calf serum at 1% for promastigotes and 2.5% for amastigotes. Deprivation was carried out at either 26°C or 37°C for 2 h and terminated by washing twice in ice-cold Earle’s salt solution at the respective pHs. Final suspension was done according to the analyses required.
Western blot analyses.
Western blot analysis was done as described previously (10), using a 1:1,000 dilution of rabbit anti-AAP3 N terminus antisera (11).
Transport assays.
The uptake of 25 μM l-[3H]arginine (600 mCi/mmol) into axenic L. donovani promastigotes and amastigotes was determined using the rapid filtration technique as described previously (11). Briefly, the transport reaction mixture contained 1 × 108 cells/ml in ice-cold Earle’s balanced salt solution at pH 7 (promastigotes) or pH 5.5 (amastigotes) supplemented with 5 mM glucose. Cells were prewarmed at either 30°C for 10 min (promastigotes) or 37°C for 5 min (amastigotes) prior to the addition of radiolabeled arginine. At 0, 0.5, 1, 1.5, 2, and 3 min, the cell suspensions were filtered through glass fiber GF/C filters that were then washed twice with ice-cold Earle’s solution.
The amount of radiolabel associated with the cells was linear with time over the 2-min time course of the assay, so the initial rate of transport was calculated from the slope of the line fitted by linear regression (see Fig. S1 in the supplemental material for promastigotes and Fig. 3B for amastigotes).
RNA isolation and real-time quantitative reverse transcription-PCR.
RNA was isolated using Tri reagent (Sigma-Aldrich Ltd.) and a Direct-zol RNA MiniPrep kit (Zymo Research), according to the manufacturers’ instructions. Eluted RNA samples were quantified using a NanoDrop One spectrophotometer (Thermo Scientific). Two micrograms of the extracted RNA was subjected to DNase treatment using RQ1 (Promega). Successful DNase treatment was verified by PCR to make sure that no residual DNA could be responsible for amplification. cDNA was synthesized from 2 μg of DNase-treated RNA using a qScript cDNA synthesis kit (Quanta Biosciences) in a 40-μl total volume. Real-time quantitative reverse transcription-PCR (qRT-PCR) was carried out with the reagents of SsoAdvanced universal SYBR green supermix (Bio-Rad Ltd.) in a 10-μl reaction volume (5 μl SYBR green, 0.5 nM forward primers, 0.5 nM reverse primers, and 2.5 μl cDNA template) on a CFX96 Touch real-time PCR system (Bio-Rad). The AAP3 primers matched both AAP3.1 and AAP3.2. Primers specific for a regulatory subunit of protein kinase A (PKAR′) were used as a control. All the samples were run in triplicates, including a no-template (negative) control for all primers used. Also, RNAseq data of the arginine deprivation response indicated that PKAR′ is not affected by the ADR, thus serving as a good control for testing LdAAP3 behavior under ADR and related conditions (Table 1).
TABLE 1.
Primers used
| Primer | Target | Amplicon size (bp) | Direction | Sequence |
|---|---|---|---|---|
| LinJ.31.0910 | AAP3 | 137 | Forward | 5′-GGCTTCATCTTCCCTGCGTA-3′ |
| LdBPK_3109010.1 | Reverse | 5′-CGGTCGAAATGGTGCCAAAC-3′ | ||
| LinJ.34.2680 | PKAR′ | 239 | Forward | 5′-AGACCGCGAAGCTCAGTC-3′ |
| LDBPK_342680.1 | Reverse | 5′-TTGTCGCAGAGAGTACCG-3′ | ||
| LinJ.10.1450 | Pteridine transporter | 171 | Forward | 5′-TGCATGTGCGGCATCTTTGGA-3′ |
| LdBPK_101450.1 | Reverse | 5′-GCACAGGATAACCGAGATGC-3′ | ||
| LinJ.36.2900 | Nodulin-like/MFS transporter | 160 | Forward | 5′-GTCATCGGGCTGGCCAAG-3′ |
| LdBPK_362900.1 | Reverse | 5′-GAGGCAGTGCAATGAGAAGC-3′ | ||
PCR was performed at 95°C for 30 s, followed by 39 cycles of 95°C for 10 s and 60°C for 30 s; melt curve analysis was carried out at 65°C to 95°C with 0.5°C increments for 5 s/step; and data analysis was carried out as described previously (25). Briefly, CP values were obtained from all samples in triplicate (except negative controls). Primers were calibrated on pooled samples, and primer efficiencies (Etarget and Eref) were calculated and incorporated into the equation below, where LdAAP3, pteridine, or MFS transporters were the target genes and PKAR′ served as the reference gene. Ratios were calculated with the CP (crosspoint) of 0-h or 48-h infected THP-1 macrophages. WT L. donovani 48-h infected macrophages served as the control.
Northern blot analyses.
Total RNAs from 5 × 108 L. donovani promastigotes (before or after 2 h of arginine deprivation) or infected THP-1 cell cultures were extracted using Tri reagent (catalog no. T9424; Sigma Ltd.) and a previously described extraction protocol (9). Blots were hybridized to the CDS of LdAAP3 as described previously (9), using PCR-amplified L. donovani genomic DNA. The probe was amplified with the following primers to yield a 538-bp amplicon (LinJ.31.0900/0910 and LdBPK_310900.1/09010.1): forward primer 5′-GGCTTCATCTTCCCTGCGTA-3′ and reverse primer 5′-GTACGTCGCCAGCCAGTG-3′.
Whole-genome sequencing.
Genomic DNA was prepared from Leishmania promastigotes using proteinase K digestion followed by phenol-chloroform extraction, ethanol precipitation, and fragmentation to 200 bp using a Covaris S2 sonicator according to the manufacturer’s protocol. Next-generation sequencing (NGS) libraries were prepared using a New England BioLabs (NEB) NextUltra II kit, quantified on Agilent bioanalyzer and Qubit instruments, and sequenced on an Illumina HiSeq 4000 instrument to generate 46 million to 96 million paired-end 75-bp reads, depending on the sample. Reads were aligned to the L. donovani 1S genome sequence assembled from a combination of PacBio and Illumina reads (Sur et al., unpublished) using Bowtie2 within Geneious, and the gene copy number was estimated by normalizing the read counts per gene to account for library size and assuming 4 copies of chr31.
Infectivity assays.
THP-1 cells grown in RPMI 1640 medium with 10% FBS (catalog no. R8758) and a 1% penicillin-streptomycin solution (catalog no. 03-031-1B; Biological Industries ) in a humidified 37°C, 5% CO2 air atmosphere were seeded onto glass coverslips (1 × 106 cells/well) in a 6-well plate and treated with 50 ng/ml of phorbol myristate acetate (PMA) (catalog no. P8139; Sigma-Aldrich, USA) for 48 h. The cells were infected as described previously (12), and the intracellular parasite load was visualized by Giemsa staining. Briefly, differentiated THP-1 macrophages were infected with log-phase L. donovani promastigotes at a multiplicity of infection (MOI) of 10 for 4 h. Parasites were washed three times in ice-cold Earle’s solution (X1) prior to infection to wash out arginine in the growth medium. Infection and subsequent incubation were performed with either 0.1 or 1.5 mM arginine in RPMI 1640 medium. Totals of 0.1 and 1.5 mM arginine in RPMI 1640 medium were prepared from no-arginine, no-leucine, and no-lysine RPMI 1640 medium (catalog no. R1780; Sigma) supplemented with 0.1 or 1.5 mM arginine, leucine (120 g/liter), lysine (70 g/liter), 10% FBS, and a 1% penicillin-streptomycin solution (catalog no. 03-031-1B; Biological Industries). Following 4 h of coincubation, the medium was aspirated from the wells. Cells were washed a total of five times with warm phosphate-buffered saline (PBS) and either collected (0-h time point) or incubated for 48 h (48-h time point) in RPMI 1640 medium containing either 0.1 mM or 1.5 mM arginine until coverslips were collected for Giemsa staining (catalog no. 32884; Sigma), and RNA was collected by direct resuspension of well contents in 600 μl Tri reagent (catalog no. T9424; Sigma).
In vivo BALB/c mouse infections and analysis.
L. donovani infections were done by tail intravenous (i.v.) injection of 108 stationary-phase promastigotes per mouse. On day 21 of infection, the mice were sacrificed, and mouse liver DNA was isolated using the proteinase K method (26). Quantitative PCR was performed on mouse liver DNA using mouse BDNF as the reference gene and parasite gp63 as the target gene with previously reported primer sequences (27, 28).
qPCR was carried out with the reagents of SYBR green (catalog no. A25776; Thermo Fisher Scientific) in a 10-μl reaction volume (5 μl SYBR green, final primer concentrations of 300 nM forward primer and 300 nM reverse primer, and 125 ng DNA template) on a CFX96 real-time PCR system (Bio-Rad). All samples were run in triplicate separately for the 2 primer sets, and the PCR cycle included a 30-s incubation step at 95°C and then 40 cycles of 5 s at 95°C and 30 s at 60°C. The output of normalized expression was determined using the Bio-Rad software of the instrument.
Scanning electron microscopy images from crude liver homogenates.
Crude liver homogenates were prepared from dissected mouse livers (n = 6) of different cohorts of infection. The dissected mouse liver samples were crushed in the presence of Karnowsky fixative to prepare a crude liver homogenate. The homogenate was processed further with tannic acid fixation followed by 4% osmium tetroxide. The samples were serially dehydrated in ethanol and freon dried. The grids were coated with gold and visualized using an FEI Quanta 200 field emission gun (FEG) electron microscope. The samples were screened for the presence of amastigotes in 200 fields per grid.
ACKNOWLEDGMENTS
We thank Ido Izhaki of Haifa University for his help with the statistical analyses. We also thank Michal Almoznino for experimental analyses.
Funding for this work was provided by University Grant Commission (UGC)-Israel Science Foundation (Indo-Israel) research grant no. 2316/15 to D.Z., A.G.-P., and R.M. and USA-Israel Binational Foundation grant no. 2017030 to D.Z. and P.J.M.
Footnotes
Citation Goldman-Pinkovich A, Kannan S, Nitzan-Koren R, Puri M, Pawar H, Bar-Avraham Y, McDonald J, Sur A, Zhang W-W, Matlashewski G, Madhubala R, Michaeli S, Myler PJ, Zilberstein D. 2020. Sensing host arginine is essential for Leishmania parasites’ intracellular development. mBio 11:e02023-20. https://doi.org/10.1128/mBio.02023-20.
REFERENCES
- 1.Herwaldt BL. 1999. Leishmaniasis. Lancet 354:1191–1199. doi: 10.1016/S0140-6736(98)10178-2. [DOI] [PubMed] [Google Scholar]
- 2.Murray HW, Berman JD, Davies CR, Saravia NG. 2005. Advances in leishmaniasis. Lancet 366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 3.Chang KP, Dwyer DM. 1976. Multiplication of a human parasite (Leishmania donovani) in phagolysosomes of hamster macrophages in vitro. Science 193:678–680. doi: 10.1126/science.948742. [DOI] [PubMed] [Google Scholar]
- 4.McConville MJ, de Souza D, Saunders E, Likic VA, Naderer T. 2007. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol 23:368–375. doi: 10.1016/j.pt.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Bates PA, Robertson CD, Tetley L, Coombs GH. 1992. Axenic cultivation and characterization of Leishmania mexicana amastigote-like forms. Parasitology 105(Part 2):193–202. doi: 10.1017/S0031182000074102. [DOI] [PubMed] [Google Scholar]
- 6.Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. 2005. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol Biochem Parasitol 141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Zilberstein D, Nitzan Koren R. 2019. Host-free systems for differentiation of axenic Leishmania. Methods Mol Biol 1971:1–8. doi: 10.1007/978-1-4939-9210-2_1. [DOI] [PubMed] [Google Scholar]
- 8.Aoki JI, Laranjeira-Silva MF, Muxel SM, Floeter-Winter LM. 2019. The impact of arginase activity on virulence factors of Leishmania amazonensis. Curr Opin Microbiol 52:110–115. doi: 10.1016/j.mib.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Shaked-Mishan P, Suter-Grotemeyer M, Yoel-Almagor T, Holland N, Zilberstein D, Rentsch D. 2006. A novel high-affinity arginine transporter from the human parasitic protozoan Leishmania donovani. Mol Microbiol 60:30–38. doi: 10.1111/j.1365-2958.2006.05060.x. [DOI] [PubMed] [Google Scholar]
- 10.Darlyuk I, Goldman A, Roberts SC, Ullman B, Rentsch D, Zilberstein D. 2009. Arginine homeostasis and transport in the human pathogen Leishmania donovani. J Biol Chem 284:19800–19807. doi: 10.1074/jbc.M901066200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman-Pinkovich A, Balno C, Strasser R, Zeituni-Molad M, Bendelak K, Rentsch D, Ephros M, Wiese M, Jardim A, Myler PJ, Zilberstein D. 2016. An arginine deprivation response pathway is induced in Leishmania during macrophage invasion. PLoS Pathog 12:e1005494. doi: 10.1371/journal.ppat.1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pawar H, Puri M, Fischer Weinberger R, Madhubala R, Zilberstein D. 2019. The arginine sensing and transport binding sites are distinct in the human pathogen Leishmania. PLoS Negl Trop Dis 13:e0007304. doi: 10.1371/journal.pntd.0007304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landfear SM, Zilberstein D. 2019. Sensing what’s out there—kinetoplastid parasites. Trends Parasitol 35:274–277. doi: 10.1016/j.pt.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin JL, Yates PA, Boitz JM, Koop DR, Fulwiler AL, Cassera MB, Ullman B, Carter NS. 2016. A role for adenine nucleotides in the sensing mechanism to purine starvation in Leishmania donovani. Mol Microbiol 101:299–313. doi: 10.1111/mmi.13390. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Contreras D, Aslan H, Feng X, Tran K, Yates PA, Kamhawi S, Landfear SM. 2015. Regulation and biological function of a flagellar glucose transporter in Leishmania mexicana: a potential glucose sensor. FASEB J 29:11–24. doi: 10.1096/fj.14-251991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstein SL, Finn AJ, Dave SH, Meng F, Lowell CA, Sanghera JS, DeFranco AL. 2000. Phosphatidylinositol 3-kinase and mTOR mediate lipopolysaccharide-stimulated nitric oxide production in macrophages via interferon-beta. J Leukoc Biol 67:405–414. doi: 10.1002/jlb.67.3.405. [DOI] [PubMed] [Google Scholar]
- 18.Shahi M, Mohajery M, Shamsian SAA, Nahrevanian H, Yazdanpanah SMJ. 2013. Comparison of Th1 and Th2 responses in non-healing and healing patients with cutaneous leishmaniasis. Rep Biochem Mol Biol 1:43–48. [PMC free article] [PubMed] [Google Scholar]
- 19.Rath M, Muller I, Kropf P, Closs EI, Munder M. 2014. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol 5:532. doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schonian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laffitte MN, Leprohon P, Papadopoulou B, Ouellette M. 2016. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Res 5:2350. doi: 10.12688/f1000research.9218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang WW, Matlashewski G. 2015. CRISPR-Cas9-mediated genome editing in Leishmania donovani. mBio 6:e00861-15. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown CM, Becker JO, Wise PM, Hoofnagle AN. 2011. Simultaneous determination of six L-arginine metabolites in human and mouse plasma using hydrophilic-interaction chromatography and electrospray-tandem mass spectrometry. Clin Chem 57:701–709. doi: 10.1373/clinchem.2010.155895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saar Y, Ransford A, Waldman E, Mazareb S, Amin-Spector S, Plumblee J, Turco SJ, Zilberstein D. 1998. Characterization of developmentally-regulated activities in axenic amastigotes of Leishmania donovani. Mol Biochem Parasitol 95:9–20. doi: 10.1016/s0166-6851(98)00062-0. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofstetter JR, Zhang A, Mayeda AR, Guscar T, Nurnberger JI, Lahiri DK. 1997. Genomic DNA from mice: a comparison of recovery methods and tissue sources. Biochem Mol Med 62:197–202. doi: 10.1006/bmme.1997.2637. [DOI] [PubMed] [Google Scholar]
- 27.Tiwananthagorn S, Iwabuchi K, Ato M, Sakurai T, Kato H, Katakura K. 2012. Involvement of CD4+ Foxp3+ regulatory T cells in persistence of Leishmania donovani in the liver of alymphoplastic aly/aly mice. PLoS Negl Trop Dis 6:e1798. doi: 10.1371/journal.pntd.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tupperwar N, Vineeth V, Rath S, Vaidya T. 2008. Development of a real-time polymerase chain reaction assay for the quantification of Leishmania species and the monitoring of systemic distribution of the pathogen. Diagn Microbiol Infect Dis 61:23–30. doi: 10.1016/j.diagmicrobio.2007.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Initial rates of arginine transport in wild-type and mutant L. donovani promastigotes. Two-minute arginine transport was determined using the rapid filtration technique (see Materials and Methods). Transport was assayed in the wild type before (diamond) and after (square) 2 h of arginine starvation and in the Δap3D6 mutant before (diamond) and 2 h after (square) arginine starvation. Download FIG S1, EPS file, 1.0 MB (1MB, eps) .
Copyright © 2020 Goldman-Pinkovich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Other members of the ADR retained sensitivity to arginine deprivation. Real-time PCR analysis was carried out to determine mRNA levels of pteridine (top) and MFS (bottom) transporters before and 2 h after arginine starvation. The analyses were carried out in Δap3D4 (D4), Δap3D6 (D6), Δap3D7 (D7), and Δap3D10 (D10) mutants. Results are relative to 2-h arginine-deprived WT cells. Download FIG S2, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2020 Goldman-Pinkovich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.