Abstract
The serotonin 2A receptor (5‐HT2AR) is implicated in the pathophysiology and treatment of various psychiatric disorders. [18F]altanserin and [11C]Cimbi‐36 positron emission tomography (PET) allow for high‐resolution imaging of 5‐HT2AR in the living human brain. Cerebral 5‐HT2AR binding is strongly genetically determined, though the impact of specific variants is poorly understood. Candidate gene studies suggest that HTR2A single nucleotide polymorphisms including rs6311/rs6313, rs6314, and rs7997012 may influence risk for psychiatric disorders and mediate treatment response. Although known to impact in vitro expression of 5‐HT2AR or other serotonin (5‐HT) proteins, their effect on human in vivo brain 5‐HT2AR binding has as of yet been insufficiently studied. We thus assessed the extent to which these variants and the commonly studied 5‐HTTLPR predict neocortex in vivo 5‐HT2AR binding in healthy adult humans. We used linear regression analyses and likelihood ratio tests in 197 subjects scanned with [18F]altanserin or [11C]Cimbi‐36 PET. Although we observed genotype group differences in 5‐HT2AR binding of up to ~10%, no genetic variants were statistically significantly predictive of 5‐HT2AR binding in what is the largest human in vivo 5‐HT2AR imaging genetics study to date. Thus, in vitro and post mortem results suggesting effects on 5‐HT2AR expression did not carry over to the in vivo setting. To any extent these variants might affect clinical risk, our findings do not support that 5‐HT2AR binding mediates such effects. Our observations indicate that these individual variants do not significantly contribute to genetic load on human in vivo 5‐HT2AR binding.
Keywords: 5‐HTTLPR, positron emission tomography, serotonin 2A receptor, single nucleotide polymorphism
We evaluated whether common HTR2A single nucleotide polymorphisms rs6313, rs6314, rs7997012, and the 5‐HTTLPR predict 5‐HT2AR binding, assessed with [18F]altanserin and [11C]Cimbi‐36 positron emission tomography in healthy individuals. This is the largest study to date evaluating genetic predictors of a human in vivo serotonin protein (n = 197). Although relative differences of up to 10% were detected, no statistically significant effect was observed. Our results do not support a significant contribution of these individual variants to genetic load on 5‐HT2AR binding.
1. INTRODUCTION
[18F]altanserin and [11C]Cimbi‐36 positron emission tomography (PET) provide high‐resolution measures of serotonin 2A receptor (5‐HT2AR) levels in the living human brain (Ettrup et al., 2016; Pinborg et al., 2003). PET studies have shown altered cerebral 5‐HT2AR binding in depression (Mintun et al., 2004), schizophrenia (Rasmussen et al., 2010), and in patients at increased risk for these disorders (Frokjaer et al., 2010; Hurlemann et al., 2005). 5‐HT2AR function mediates antidepressant (Artigas, 2013) and antipsychotic (Meltzer & Massey, 2011) efficacy. A PET study from our group, performed in twins, indicates that cerebral 5‐HT2AR binding is strongly genetically determined (Pinborg et al., 2008). Identifying genetic sources of variation in brain 5‐HT2AR elucidates how variation emerges and highlights mechanisms mediating genetic effects on neuropsychiatric risk.
Candidate gene studies suggest that single nucleotide polymorphisms (SNPs) within the HTR2A gene might affect psychiatric risk and treatment effects. rs6311 (i.e., −1438G > A, promotor) and rs6313 (i.e., 102C > T, Exon 1, in perfect linkage disequilibrium) G/C alleles were linked to increased risk for depression (Petit et al., 2014) and schizophrenia (Williams et al., 1996) as well as superior antidepressant (Minov et al., 2001), but lesser antipsychotic response (Arranz et al., 1995). The rs6314 (i.e., 1354C > T, Exon 3) T allele was associated with poor response to antipsychotic treatment (Blasi et al., 2013) and rs7997012 (i.e., IVS2G > A, intronic) A‐carriers may show greater antidepressant response than noncarriers (McMahon et al., 2006). Although recent genome‐wide association studies (GWAS) for depression (Wray et al., 2018) and schizophrenia (Pardinas et al., 2018) do not support significant independent roles in mediating clinical risk, understanding these variants' effects on 5‐HT2AR expression supports elucidation of mechanisms of psychiatric pathophysiology.
In vitro studies are inconclusive regarding these SNP's impact on 5‐HT2AR protein expression. The concordant rs6311/rs6313 A/T alleles showed higher expression in some studies (Parsons, D'Souza, Arranz, Kerwin, & Makoff, 2004; Polesskaya & Sokolov, 2002), though this was contradicted by others (Bray, Buckland, Hall, Owen, & O'Donovan, 2004). rs6314 induces a histidine to tyrosine amino acid change, alters 5‐HT2AR intracellular signaling (Ozaki et al., 1997), and was associated with lower expression levels in vitro and post mortem (Blasi et al., 2013). Effects of rs7997012 on 5‐HT2AR protein levels or function have yet to be reported, though an association with in vivo cerebral serotonin transporter (SERT) binding was observed in humans (Laje et al., 2010). PET provides the unique opportunity to assess whether these SNPs affect 5‐HT2AR binding in the living human brain. The specific impact of individual HTR2A SNPs on 5‐HT2AR binding has been insufficiently elucidated. Two previous PET studies failed to demonstrate an effect of the HTR2A SNPs assessed here. However, these studies were in small samples (Hurlemann et al., 2008), particularly considering the small effect sizes attributed to singular variants in psychiatry (Bogdan et al., 2017), or only assessed one HTR2A variant (Erritzoe et al., 2009).
In addition, a study in marmosets linked a SERT gene (SLC6A4) promotor haplotype associated with low SERT expression and an anxiety‐prone phenotype (Santangelo et al., 2016) to reduced brain 5‐HT2AR binding (Santangelo et al., 2019). In humans, 5‐HTTLPR (rs4795541) and the associated rs25531 variant have been associated with depressive pathophysiology (Caspi et al., 2003) and may affect SERT binding (Praschak‐Rieder et al., 2007), though null findings have been observed, including from our lab (Fisher et al., 2017; Murthy et al., 2010). Although associations between 5‐HTTLPR and other serotonin receptors have been detected with PET in humans (Fisher et al., 2012; Lothe et al., 2009), the impact on 5‐HT2AR binding has yet to be investigated.
Here, we aimed to assess whether the aforementioned HTR2A SNPs (rs6313, rs6314, rs7997012) and the 5‐HTTLPR predict neocortex 5‐HT2AR binding in the living human brain. To do so, we probed their effects in a uniquely large (n = 197) [18F]altanserin and [11C]Cimbi‐36 PET data set available through the Center for Integrated Molecular Brain Imaging (CIMBI) database (Knudsen et al., 2016). Hereby, we explore whether these variants contribute to genetic load on 5‐HT2AR binding.
2. MATERIALS AND METHODS
2.1. Participants
All available [18F]altanserin and [11C]Cimbi‐36 PET data acquired in healthy subjects were extracted from the CIMBI database (Knudsen et al., 2016), providing 291 scans (169/122 [18F]altanserin/[11C]Cimbi‐36). Eleven scans (two [18F]altanserin/nine [11C]Cimbi‐36) performed in subjects with self‐reported non‐European ancestry were excluded to limit genetic background confounders and variation in allele frequencies. We restricted our analyses to baseline scans to eliminate pharmacologic intervention effects. Thus, 67 (5/62) rescans as well as eight [18F]altanserin scans performed in subjects also scanned with [11C]Cimbi‐36 were excluded from analyses. This step resulted in 205 scans (154/51), each performed in a unique individual. Finally, scans without corresponding genotype information were excluded; 5‐HTTLPR data were available in two subjects for whom HTR2A variants were not. Thus, our dataset included 197 (154/43) and 195 (153/42) scans, each from a unique individual, for 5‐HTTLPR and HTR2A analyses, respectively.
All sampling, including scanning and blood draw for genotyping, took place during participation in studies carried out at the Neurobiology Research Unit, Rigshospitalet, Denmark, that have been detailed previously (Adams et al., 2004; Adams et al., 2005; Erritzoe et al., 2008; Erritzoe et al., 2010; Erritzoe et al., 2011; Ettrup et al., 2014; Ettrup et al., 2016; Frokjaer et al., 2008; Haahr et al., 2015; Hasselbalch et al., 2008; Haugbol et al., 2007; Madsen et al., 2019; Marner et al., 2009; Marner et al., 2011; Marner et al., 2012; Pinborg et al., 2003; Pinborg et al., 2004; Pinborg et al., 2008; Rasmussen et al., 2010). All subjects provided written informed consent and the study protocols were approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark (KF‐11‐061‐03, KF‐01‐124‐04, KF‐01‐2006‐20, KF‐02‐058‐99, KF‐01‐001‐02, KF‐01‐156‐04, H‐4‐2012‐105, H‐16026898, H‐15004506). Healthy subjects included in the CIMBI database are deemed free from primary psychiatric disorders (according to the DSM‐IV axis 1 or WHO ICD‐10 diagnostic classifications), severe systemic or neurologic disease, pregnancy, head trauma, drug or alcohol abuse, and current or prior intake of psychopharmacologic substances through an interview with a trained clinician. Absence of psychopathologic symptoms is confirmed on the day of the PET‐scan using the symptom check list revised SCL‐90‐R (Derogatis & Savitz, 1999). Additional information on CIMBI database inclusion criteria are detailed in (Knudsen et al., 2016). A largely overlapping dataset was recently utilized by Stenbaek et al., 2019) to assess the impact of openness, a specific personality trait, on 5‐HT2AR binding. Erritzoe et al. (2009) previously assessed one HTR2A SNP (rs6311, in perfect LD with rs6313) in a subset (n = 95) of the data presented here.
2.2. PET imaging
As recently detailed by Stenbaek et al. (2019), [18F]altanserin and [11C]Cimbi‐36 were synthesized according to Lemaire, Cantineau, Guillaume, Plenevaux, and Christiaens (1991) and Ettrup et al. (2014), respectively. All PET scans were acquired using either an 18‐ring GE‐Advance scanner (General Electric, Milwaukee, WI, n = 142 [18F]altanserin) or a Siemens ECAT High Resolution Research Tomograph (HRRT, CTI/Siemens, Knoxville, TN, n = 12/43 [18F]altanserin/[11C]Cimbi‐36). Both were used in 3D acquisition mode and with in‐plane resolutions of approximately 6 mm and <2 mm, respectively (Knudsen et al., 2016). [18F]altanserin scans were performed as detailed by Pinborg et al. (2003), using a bolus plus infusion radioligand administration scheme. A 10 min transmission scan was followed by 40 min of dynamic PET scanning (frames: 5 × 8 min), which started 2 hr after initiation of [18F]altanserin administration (Pinborg et al., 2003). [11C]Cimbi‐36 scans were performed after bolus radioligand administration using a protocol established by Ettrup et al., 2014 and Ettrup et al. (2016). A 6 min transmission scan was followed by 120 min (frames: 6 × 10 s, 6 × 20 s, 6 × 60 s, 8 × 2 min, and 19 × 5 min) of dynamic PET scanning, which commenced with [11C]Cimbi‐36 injection. PET data from the GE‐Advance scanner were reconstructed using a filtered back projection algorithm followed by attenuation‐, dead time‐, and scatter correction. Scans performed on the HRRT scanner were reconstructed using a 3D‐OSEM‐PSF algorithm (Hong et al., 2007; Knudsen et al., 2016; Sureau et al., 2008).
2.3. MR imaging
Each subject underwent one magnetic resonance imaging (MRI) session for acquisition of a high‐resolution, T1‐weighted structural brain scan. MRI scans were utilized for coregistration, gray matter‐, white matter‐, and cerebrospinal fluid segmentation, as well as region of interest (ROI) delineation. MRI scans were acquired using either a Siemens 1.5 T Vision scanner (Erlangen, DE, n = 63 [18F]altanserin), a Siemens 3 T Magnetom Trio scanner (n = 91 [18F]altanserin), a Siemens 3 T Magnetom Verio scanner (n = 21 [11C]Cimbi‐36) or a Siemens 3 T Prisma scanner (n = 22 [11C]Cimbi‐36) (Knudsen et al., 2016). See previous publications for scan protocol details (Ettrup et al., 2014; Madsen et al., 2019; Stenbaek et al., 2019).
2.4. PET data processing
As previously described (Knudsen et al., 2016; Stenbaek et al., 2019), motion correction utilized the AIR algorithm based on Woods, Cherry, and Mazziotta (1992) and PET scans were smoothed using a 12 or 10 mm within‐frame Gaussian filter for the GE‐Advance and HRRT scanners, respectively. The AIR algorithm was also used for coregistration of scans acquired on the GE‐Advance scanner. For HRRT scans, Statistical Parametric Mapping (SPM8) was used for this purpose.
[18F]altanserin BPP and [11C]Cimbi‐36 BPND were calculated based on Pinborg et al. (2003) and (Ettrup et al. (2014) and Ettrup et al. (2016), respectively. BPP and BPND are considered indices of 5‐HT2AR density (Innis et al., 2007). The neocortex was defined as the primary ROI and delineated using corresponding subject‐level MRI data and PVElab as published by Svarer et al., 2005). PVElab defines the neocortex to include the middle/inferior frontal‐, middle/inferior temporal‐, superior frontal‐, superior temporal‐, and pre/post central gyri as well as occipital‐, orbitofrontal‐, and parietal cortices. Several biological and methodological characteristics motivated selection of this singular ROI for analyses. Neocortex has high 5‐HT2AR binding, whereas subcortical levels are low (Ettrup et al., 2014) and show lower reproducibility (Ettrup et al., 2016). [11C]Cimbi‐36 has serotonin 2C receptor affinity, relevant in hippocampus (Finnema et al., 2014), confounding genetic associations. In the neocortex, BPP and BPND values from [18F]altanserin and [11C]Cimbi‐36 scans are correlated (Ettrup et al., 2016), supporting pooling of data. Finally, 5‐HT2AR binding is strongly correlated between neocortical subregions (Figure 1). Thus, our focus on a neocortex ROI represents a statistically efficient strategy for evaluating genetic predictors of brain 5‐HT2AR binding.
FIGURE 1.
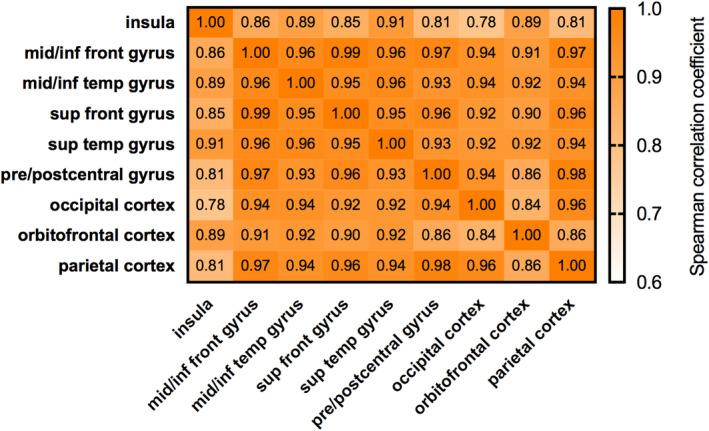
Heat map of correlation coefficients between neocortex ROI subregions. Heat map depicts strength of correlations of 5‐HT2AR binding (pooled [18F]altanserin BPP: n = 154; [11C]Cimbi‐36 BPND: n = 43) between subregions included in the neocortex ROI and insula. Insula is not included in the PVE lab neocortex ROI, but correlation with neocortex ROI subregions is strong. Each rectangle depicts one region‐to‐region correlation; a darker orange color depicts higher Spearman correlation coefficients. front, frontal; mid/inf, middle/inferior; ROI, region of interest; sup, superior; temp: temporal
2.5. Genotyping
Genotyping was done on material from either buffy coat lymphocytes or whole blood samples. Samples or extracted DNA were stored at −20°C in the CIMBI biobank until analyzed. DNA extractions were performed with Qiagen DNA extraction kits (Qiagen, Hilden, Germany, https://www.qiagen.com). Quality control of DNA samples utilized Thermo Scientific NanoDrop spectrophotometry (Thermo Fisher Scientific, Waltham, MA, https://www.thermofisher.com). In case of insufficient purity, DNA decontamination was carried out by sodium acetate precipitation, using 0.1 volume of 3 M sodium acetate and 3 volumes of ice‐cold 96–99% ethanol. To verify consistent genotype results, five previously genotyped rs6313, rs6314, and rs7997012 samples were re‐analyzed with perfect correspondence. rs6311 genotype information was available in 129 subjects and used to confirm perfect LD with rs6313. All other SNPs are in linkage equilibrium based on LDmatrix tool (https://ldlink.nci.nih.gov/?tab=ldmatrix).
All rs6311, rs6313, rs6314, and rs7997012 genotypes were determined using Applied Biosystems TaqMan 5′‐exonuclease allelic discrimination assays (Thermo Fisher Scientific). Previously available genotyping was performed using the ABI 7500 multiplex PCR machine (Thermo Fisher Scientific), while current genotyping utilized the Roche LightCycler 480 II (Roche, Penzeberg, Germany, https://www.lifescience.roche.com).
All 5‐HTTLPR (rs4795541, rs25531) genotyping was performed as described by Madsen et al., 2016). rs4795541 genotyping used TaqMan 5′‐exonuclease allelic discrimination assays and PCR. rs25531 genotyping comprised PCR amplification from the forward and reverse primers 5′‐GGCGTTGCCGCTCTGAATGC‐3′ and 5′‐GAGGGACTGAGCTGGACAACCAC‐3′ followed by MspI restriction enzyme degradation and separation by gel electrophoresis. All kits used in genotyping procedures were utilized according to the manufacturers' instructions.
2.6. Statistical analyses
Hardy–Weinberg equilibrium (HWE) was evaluated with a chi‐square test. HWE was not evaluated for 5‐HTTLPR because this variant was an inclusion criterion in some of the original studies.
2.6.1. Genotype grouping
Subjects were grouped for each HTR2A variant based on genotype, resulting in three groups for each SNP with the exception of the rs6314, for which we identified only one YY homozygote (i.e., rs6313 TT vs. CT vs. CC; rs6314 HH vs. Y‐carriers; rs7997012 AA vs. AG vs. GG). For the 5‐HTTLPR, LG or S alleles were coded together as an S′ allele and genotypes were grouped accordingly (i.e., S′S′, LAS′, LALA). This grouping is based on evidence that the LG and S alleles are associated with similarly low SERT expression levels (Hu et al., 2006). See Table 1 for an overview of genotype grouping used in statistical analyses.
TABLE 1.
Genotype grouping for statistical analyses
| Variant | Alleles | ||
|---|---|---|---|
| rs6313 | TT | CT | CC |
| 27 | 88 | 80 | |
| rs6314 | HH | Y‐carriers | |
| 165 | 30 (one homozygote) | ||
| rs7997012 | AA | AG | GG |
| 45 | 92 | 58 | |
| 5‐HTTLPR a | LALA | LAS′ | S′S′ |
| 55 | 95 | 47 | |
Note: HTR2A variants: n = 195. 5‐HTTLPR: n = 197.
5‐HTTLPR comprises rs4795541 and rs25531 with S′ denoting either composite LG allele or S allele, in accordance with Hu et al. (2006).
2.6.2. Analyses of genotype effects on neocortex 5‐HT2AR binding
[18F]altanserin BPP and [11C]Cimbi‐36 BPND data were pooled for analyses and will subsequently be referred to as 5‐HT2AR binding. Continuous variables were mean‐centered prior to model fit. First, the effects of the covariates age, sex, radioligand ([18F]altanserin, [11C]Cimbi‐36), PET scanner (HRRT, GE), MR scanner field strength (1.5 T, 3 T), and BMI were assessed in a linear regression model. Age, sex (Moses‐Kolko et al., 2011), and BMI (Erritzoe et al., 2009) were previously shown to affect 5‐HT2AR binding. MR scanner field strength, radioligand, and PET scanner were included to address methodological heterogeneity. Interaction effects between radioligand and the covariates age, sex, and BMI were tested for, but were nonsignificant (all p unc > .05).
Next, likelihood ratio testing was used to test for a main effect of each variant (rs6313, rs6314, rs7997012, 5‐HTTLPR) by comparing the covariate model with and without each variant. Each variant was evaluated separately. Genotype effects were not moderated by PET ligand (all p unc > .05), which is supportive of pooling [18F]altanserin BPP and [11C]Cimbi‐36 BPND. Likelihood ratio testing was also performed to test for the combined effect of all variants on neocortex 5‐HT2AR. Specific genotype effects were then estimated by linear regression analyses.
Visual and graphical (e.g., QQ plot) evaluation of our model including only covariates indicated nonnormally distributed residuals. This was likely due to inclusion criteria of original studies, some of which specifically recruited overweight (Erritzoe et al., 2009) or elderly individuals (Marner et al., 2009; Marner et al., 2011). To derive estimates robust to this deviation, parameter estimates, 95% confidence intervals (95% CI) and p‐values for linear regression models were estimated with 1,000 bootstrap resamples. For all linear regression analyses, only bootstrapped results are reported. Statistical significance of likelihood ratio testing was assessed with a permutation test (10,000 permutations).
2.6.3. Radioligand subgroup models
Linear regression analyses and likelihood ratio testing was also performed within the [18F]altanserin (HTR2A: n = 153; 5‐HTTLPR: n = 154) and [11C]Cimbi‐36 (HTR2A: n = 42; 5‐HTTLPR: n = 43) subgroups in order to address potential radioligand differences in genotype effects. Analyses were performed as described above for the pooled sample with the exception that covariates lacking more than one level were dropped (i.e., radioligand for both subsamples, PET scanner, and MR field strength for [11C]Cimbi‐36).
2.6.4. Exploratory analyses of genotype effects on insula 5‐HT2AR binding
In addition, exploratory analysis probing for effects of 5‐HTTLPR genotype on insula 5‐HT2AR binding was performed in the n = 197 pooled sample based on animal studies reporting effects of SLC6A4 genotype on 5‐HT2AR binding (Santangelo et al., 2019). Again, linear regression analyses and likelihood ratio testing were implemented using the same procedures as for the neocortex ROI.
Statistical analyses were performed using IBM SPSS Software 26.0.0.0 and R (v3.5.0, https://cran.r-project.org/). p < .05 was considered statistically significant.
3. RESULTS
3.1.1. Descriptive results
See Table 2 for descriptive information. HTR2A variants were in HWE (df = degrees of freedom, rs6313: X 2 = 0.13, df = 1, p = .72; rs6314: X 2 = 0.05, df = 1, p = .82; rs7997012: X 2 = 0.53, df = 1, p = .47). As expected (Spurlock et al., 1998), rs6311 and rs6313 were in perfect LD, validating the consideration of only rs6313.
TABLE 2.
Descriptive information
| [18F]altanserin (n = 154) | [11C]Cimbi‐36 (n = 43) | |||||
|---|---|---|---|---|---|---|
| Mean ± SD | Median | Range | Mean ± SD | Median | Range | |
| Age (years) | 40.76 ± 17.97 | 35.10 | 18.47–81.73 | 26.84 ± 5.81 | 26.42 | 18.43–49.82 |
| BMI (kg/m2) | 25.51 ± 4.70 | 24.44 | 18.38–45.91 | 23.86 ± 2.84 | 23.08 | 19.32–33.20 |
| Inj. dose a (MBq) | 275.19 ± 64.83 | 278.00 | 143.00–447.00 | 497.74 ± 100.62 | 511.12 | 213.00–604.04 |
| SA a (GBq/μmol) | 76.14 ± 62.32 | 55.71 | 10.30–357.78 | 345.13 ± 243.01 | 273.14 | 73.36–1,039.63 |
| Inj. mass a (μg) | 2.84 ± 2.22 | 2.12 | 0.26–10.99 | 0.84 ± 0.48 | 0.75 | 0.11–1.50 |
| Neocortical binding b | 1.46 ± 0.56 | 1.47 | 0.32–4.66 | 1.30 ± 0.21 | 1.28 | 0.99–2.02 |
| PET scanner (GE/HRRT) | 142/12 | 0/43 | ||||
| MR field strength (1.5 T/3 T) | 63/91 (1.5 T: Siemens Vision 3 T: Siemens Magnetom Trio) | 0/43 (3 T: Siemens Prisma, Siemens Magnetom Verio) | ||||
| Sex (m/f) | 94/60 | 25/18 | ||||
Abbreviations: BMI, body mass index; HRRT, High Resolution Research Tomograph; Inj., injected (dose/mass); MR, magnetic resonance; PET, Positron emission tomography; SA, specific activity; SD, standard deviation.
Inj. dose: n = 129/43; SA: n = 81/43; Inj. mass: n = 77/43 ([18F]altanserin/[11C]Cimbi‐36).
[18F]altanserin: BPP; [11C]Cimbi‐36: BPND.
3.1.2. Covariate model
In the covariate model, age (parameter estimate [95% CI], p value: −.02 [−0.02, −0.01], p < .001, units: [11C]Cimbi‐36 BPND change per year), radioligand (0.81 [0.31, 1.31], p < .001, units: change in 5‐HT2AR binding from [11C]Cimbi‐36 BPND to [18F]altanserin BPP), and BMI (0.03 [0.002, 0.06], p = .04, units: [11C]Cimbi‐36 BPND change per kg/m2) had significant effects on neocortex 5‐HT2AR binding. Effects of PET scanner (−0.45 [−0.98, 0.06], p = .09, units: [11C]Cimbi‐36 BPND change from HRRT to GE‐Advance scanner), MR field strength (−0.12 [−0.23, 0.001], p = .05, units: [11C]Cimbi‐36 BPND change from 3 to 1.5 T) and sex (−0.07 [−0.16, 0.02], p = .13, units: [11C]Cimbi‐36 BPND change from female to male) were not statistically significant predictors of 5‐HT2AR binding but nevertheless included when estimating genetic effects.
3.1.3. Genotype effects on neocortex 5‐HT2AR binding
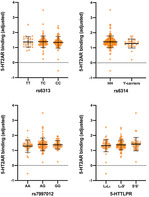
We did not observe evidence that any of the genetic variants considered were statistically significant predictors of 5‐HT2AR binding (likelihood ratio estimation effects, rs6313: X 2 = 0.38, p = .84; rs6314: X 2 = 2.15, p = .15; rs7997012: X 2 = 2.70, p = .28; 5‐HTTLPR: X 2 = 1.80, p = .43; all variants combined: X 2 = 7.56, p = .42). See Figure 2 for depiction of specific genotype effects and Table 3 for respective parameter estimates, 95% CI, and % differences in 5‐HT2AR between genotype groups.
FIGURE 2.
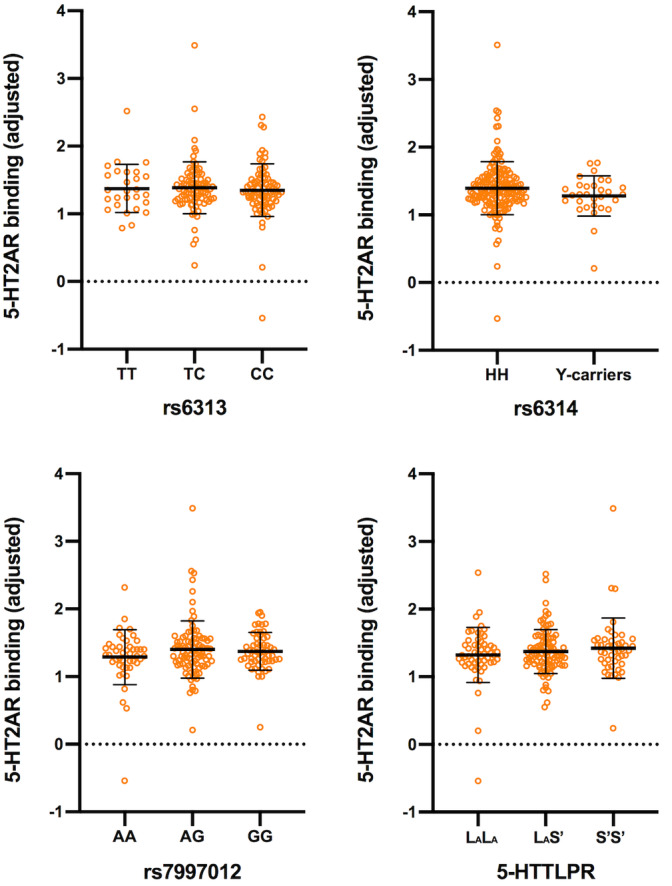
Genotype effects on covariate‐adjusted 5‐HT2AR binding. Orange circles represent covariate‐adjusted (i.e., age, sex, radioligand, positron emission tomography (PET) scanner, MR scanner field strength, and BMI) 5‐HT2AR binding from 195 (HTR2A variants) and 197 (5‐HTTLPR) healthy subjects. Negative binding values result from this adjustment, all observed BP values were greater than 0. Black lines illustrate mean ± SD. No statistically significant effects of rs6313, rs6314, rs7997012, nor 5‐HTTLPR on covariate‐adjusted 5‐HT2AR binding were observed. See Table 3 for parameter estimates and 95% CI. 5‐HT2AR: serotonin 2A receptor. 95% CI: 95% confidence intervals
TABLE 3.
Genotype effects on neocortical 5‐HT2AR binding
| Genotype/comparison | Estimate a | 95% CI |
|---|---|---|
| rs6313 | ||
| TT | 1.21 | 1.06, 1.38 |
| TC | 1.22 | 1.09, 1.36 |
| CC | 1.19 | 1.09, 1.31 |
| TT vs. TC | −0.01 (1.06%) | −0.17, 0.16 |
| TT vs. CC | 0.02 (1.42%) | −0.14, 0.18 |
| TC vs. CC | 0.03 (2.45%) | −0.09, 0.15 |
| rs6314 | ||
| Y‐carriers | 1.09 | 0.93, 1.25 |
| HH | 1.21 | 1.11, 1.32 |
| Y‐carriers vs. HH | −0.11 (10.24%) | −0.24, 0.01 |
| rs7997012 | ||
| GG | 1.23 | 1.10, 1.36 |
| AG | 1.25 | 1.13, 1.39 |
| AA | 1.14 | 1.02, 1.27 |
| GG vs. AG | −0.02 (1.97%) | −0.15, 0.10 |
| GG vs. AA | 0.09 (7.10%) | −0.06, 0.24 |
| AG vs. AA | 0.11 (8.90%) | −0.04, 0.28 |
| 5‐HTTLPR b | ||
| S′S′ | 1.24 | 1.09, 1.40 |
| LAS′ | 1.20 | 1.10, 1.31 |
| LALA | 1.15 | 1.01, 1.28 |
| S′S′ vs. LAS′ | 0.04 (3.34%) | −0.09, 0.18 |
| S′S′ vs. LALA | 0.09 (7.43%) | −0.07, 0.27 |
| LAS′ vs. LALA | 0.05 (4.23%) | −0.06, 0.17 |
Note: 5‐HT2AR: serotonin 2A receptor. 95% CI: 95% confidence intervals.
Genotype differences (%) noted where relevant.
5‐HTTLPR comprises rs4795541 and rs25531 with S′ denoting either composite LG allele or S allele, in accordance with Hu et al. (2006).
3.1.4. Radioligand subgroup models
We did not observe evidence that any of the genetic variants considered were statistically significant predictors of 5‐HT2AR binding in the [18F]altanserin (rs6313: X 2 = 0.37, p = .84; rs6314: X 2 = 2.56, p = .11; rs7997012: X 2 = 1.32, p = .54; 5‐HTTLPR: X 2 = 1.06, p = .61) nor [11C]Cimbi‐36 (rs6313: X 2 = 0.37, p = .86; rs6314: X 2 = .00003, p = .99; rs7997012: X 2 = 5.66, p = .08; 5‐HTTLPR: X 2 = 4.32, p = .15) subsamples.
3.1.5. Genotype effects on insula 5‐HT2AR binding
Linear regression analysis and likelihood ratio testing did not reveal an effect of 5‐HTTLPR genotype on insula 5‐HT2AR binding (5‐HTTLPR: X 2 = 3.00, p = .24). All reported results are unadjusted for multiple comparisons.
4. DISCUSSION
We did not find evidence for a statistically significant effect of three common HTR2A SNPs (rs6313, rs6314, rs7997012), nor the commonly studied 5‐HTTLPR, on 5‐HT2AR binding assessed with [18F]altanserin and [11C]Cimbi‐36 PET. This represents the largest evaluation of genetic predictors of a human in vivo 5‐HT protein marker to date. Our findings indicate that effects previously shown on gene expression and protein function in vitro (Parsons et al., 2004), post mortem (Blasi et al., 2013; Polesskaya, Aston, & Sokolov, 2006; Polesskaya & Sokolov, 2002; Turecki et al., 1999) and in animal studies (Santangelo et al., 2019) demonstrate only very limited translation to 5‐HT2AR binding in healthy adult humans. Although nominal effects detected for rs6313 and rs6314 were consistent with related in vitro reports (Blasi et al., 2013; Polesskaya & Sokolov, 2002), all observations were not statistically significant. Thus, despite evidence from twin studies for a substantial impact of genetic load on 5‐HT2AR binding (Pinborg et al., 2008), our findings suggest only marginal penetrance of the individual variants considered here. The potential explanations for this disparity differ between the individual SNPs due to their diverging effects on the HTR2A gene and how it is regulated.
The rs6311/rs6313 A/T‐alleles have been linked to increased promotor activity (Parsons et al., 2004), mRNA levels (Polesskaya & Sokolov, 2002) and post mortem 5‐HT2AR binding (Turecki et al., 1999). In our sample, TC and TT individuals showed nominally increased 5‐HT2AR binding relative to CC individuals, but observed group differences were ≤2.5% and not statistically significant. Thus, although we did not reject our null hypothesis, we observed effects nominally similar to previous in vitro and post mortem studies (Parsons et al., 2004; Polesskaya & Sokolov, 2002; Turecki et al., 1999). Importantly, rs6311/rs6313 may regulate transcription in a manner moderated by epigenetic mechanisms, including gene methylation (Polesskaya et al., 2006) and interaction with transcription factors (Falkenberg, Gurbaxani, Unger, & Rajeevan, 2011; Smith et al., 2013). Thus, we cannot rule out that epigenetic processes obscured main effects of this variant on 5‐HT2AR binding. Although epigenetic markers can be assayed from peripheral blood samples, this information is not available for the current data and the correspondence between peripheral blood and relevant neuronal populations is unclear. The above mentioned positive post mortem studies suggest that genotype effects override epigenetic‐induced variance. However, negative post mortem studies have also been published (Blasi et al., 2013; Bray et al., 2004). The susceptibility of these variants to epigenetic influence may explain these discrepancies, particularly as the corresponding studies were performed in small samples. In our uniquely large healthy adult sample, we estimate a small, nonsignificant main effect of rs6313 on 5‐HT2AR binding.
Previous studies linked rs6314 HY genotype to lower post mortem prefrontal mRNA levels (Blasi et al., 2013). Although not statistically significant (p = .15), we estimated a nominally consistent 10.2% lower 5‐HT2AR binding in Y‐carriers compared to HH homozygotes. rs6314 results in an amino acid change (histidine to tyrosine) (Ozaki et al., 1997). rs6313 is also exonic and could thus potentially also alter protein structure, though it has not been demonstrated in in vitro studies. Such conformational changes could influence binding of [18F]altanserin or [11C]Cimbi‐36 to 5‐HT2AR. For example, they may influence the affinity of these radioligands for 5‐HT2AR, KD, which is inversely proportional to binding potential. To our knowledge, rs6313 or rs6314 effects on radioligand affinity have not been evaluated. However, if these variants were to proportionally affect both available 5‐HT2AR (i.e., Bavail) and radioligand KD, their effects might not be elucidated in our study. Generally, conformational or functional changes to 5‐HT2AR that do not affect [18F]altanserin or [11C]Cimbi‐36 binding would not be detected with PET. Our binding outcome is thus likely insensitive to rs6314‐induced changes in intracellular signaling (Ozaki et al., 1997). Both [18F]altanserin and [11C]Cimbi‐36 likely bind intracellularly localized 5‐HT2AR, which comprises ~90% of 5‐HT2AR (Cornea‐Hebert, Riad, Wu, Singh, & Descarries, 1999); it is unknown whether internalization alters [18F]altanserin and [11C]Cimbi‐36 binding. If unaffected, variant effects on subcellular localization would not be detected in our analyses. In summary, the numerically lower 5‐HT2AR binding observed in the HY genotype is consistent with previous studies (Blasi et al., 2013), yet not statistically significant.
We did not observe a statistically significant impact of rs7997012 on 5‐HT2AR binding. rs7997012 A allele was previously associated with improved antidepressant treatment response (McMahon et al., 2006). Neither in vitro nor in vivo effects of rs7997012 on HTR2A gene expression have been reported previously, but an earlier PET study reported a marginal effect on SERT binding (Laje et al., 2010). We estimated 8.9 and 7.1% increases in 5‐HT2AR binding in AG and GG groups compared to AA individuals, respectively; AG and GG individuals showed similar 5‐HT2AR binding (~2% difference). In two cohorts, our lab has reported a nonlinear relation between 5‐HT2AR and SERT binding (Erritzoe et al., 2010; Haahr et al., 2015). Thus, it is plausible that small rs7997012 effects on 5‐HT2AR binding mediate effects on SERT, a model that requires further evaluation in independent samples. in vitro expression studies would support inference of the nominal differences that we observed, resolving rs7997012 functional effects.
We did not observe a statistically significant effect of 5‐HTTLPR on 5‐HT2AR binding. Our data nominally indicate that LAS′ and S′S′ status are associated with 4.2 and 7.4% higher 5‐HT2AR binding compared to LALA individuals, a somewhat linear effect. Notably, this remains not statistically significant even when considering LALA versus S′‐carriers (p = .27, data not shown), emphasizing a large degree of statistical uncertainty. We performed an exploratory analysis probing for an effect of 5‐HTTLPR on insula 5‐HT2AR binding, which was also nonsignificant (p = .24). This was motivated by a recent marmoset study showing an SLC6A4 haplotype associated with heightened anxiety and reduced insula 5‐HT2AR binding (Santangelo et al., 2016; Santangelo et al., 2019). The convergence with an anxiety phenotype is intriguing considering similar 5‐HTTLPR effects (Lesch et al., 1996). We observed nominally higher 5‐HT2AR with the S allele, as was observed within our neocortex ROI, inconsistent with what would be expected. However, the 5‐HTTLPR polymorphism does not occur in marmosets and is thus not included in the studies by Santangelo et al., who assess a different SLC6A4 variant. Thus, although we express caution in drawing a direct parallel to (Santangelo et al., 2016), our human in vivo results do not reflect their observations.
The variants included in this study were chosen based on their impact on 5‐HT protein expression (Blasi et al., 2013; Hu et al., 2006; Laje et al., 2010; Polesskaya et al., 2006), allele frequencies, as well as reported associations with disease risk (Caspi et al., 2003; Choi et al., 2004; Petit et al., 2014; Williams et al., 1996) and treatment outcomes of affective and psychotic disorders from candidate gene studies (Anttila et al., 2007; Arranz et al., 1995; Arranz et al., 1998; Blasi et al., 2013; Chen, Shen, & Chen, 2009; Choi, Kang, Ham, Jeong, & Lee, 2005; Kato et al., 2006; McMahon et al., 2006; Minov et al., 2001; Peters et al., 2009). This was done to allow for cautious interpretation of genotype effects vis‐à‐vis clinical findings. 5‐HT2AR binding appears altered in depression (Mintun et al., 2004) and schizophrenia (Rasmussen et al., 2010), and receptor function is modulated by antidepressant and antipsychotic drugs (Artigas, 2013; Meltzer & Massey, 2011). Thus, genetic influence on 5‐HT2AR binding could potentially moderate clinical effects. The relative effects we observed between genotype groups were <10% and not statistically significant, which does not support this assumption. It should be kept in mind that previous clinical effects were observed in diseased populations, whereas we assessed healthy subjects here. It is unclear whether our findings carry over to clinical populations, based on potential genotype by diagnosis interaction effects.
Our study is not without limitations. [18F]altanserin and [11C]Cimbi‐36 PET data were pooled, introducing data heterogeneity. Radioligand differences in KD as well as differences between [18F]altanserin fP and [11C]Cimbi‐36 fND may confound this data pooling, although such confounds are approximated by the inclusion of PET tracer as a covariate. Notably, we observed nonsignificant effects considering each radioligand separately. However, such a split‐sample strategy also negatively affects statistical power. Our healthy population varied in, for example, age and BMI. Although this can introduce measurement error and decrease statistical power, we note that collecting 200 datasets at only two PET scanners is a particularly homogenous sample than is otherwise available. Variation in ancestry was addressed by excluding from analyses persons self‐reported to be of non‐European ancestry. A limitation of the current study is that genetic relatedness and ethnic heterogeneity is not available and not directly modeled. The limitation of evaluating candidate genetic variants as predictors of brain imaging measures has been considered at length (Bogdan et al., 2017). In particular, recent GWAS do not support significant independent roles for the variants assessed here in depression (Wray et al., 2018) or schizophrenia (Pardinas et al., 2018). However, the sample sizes necessary for GWAS are currently not feasible in conjunction with human in vivo PET data. In the absence of available genome‐wide data, focusing on variants selected based on in vitro evidence and clinical candidate gene studies is a potentially useful alternative strategy. In addition, we point out that our outcome measure (5‐HT2AR binding) is more proximal to a genetic effect on protein levels vis‐à‐vis functional and structural brain MRI markers.
In conclusion, this study assessed the effect of three common HTR2A SNPs and the 5‐HTTLPR on 5‐HT2AR binding in the largest sample size to date (n = 197). Although we do not find evidence for statistically significant differences, we note effects nominally consistent with previous studies linking these variants to 5‐HT2AR levels and the 5‐HT system more broadly. Our study suggests not more than a limited direct effect of these variants on 5‐HT2AR binding in the adult human brain.
CONFLICT OF INTEREST
Within the last 3 years, M. S. has received travel grants and speaker honoraria from Janssen and Austroplant. All other authors declare no potential conflict of interest.
ACKNOWLEDGMENTS
The authors thank L. Freyr for assistance in scheduling and data collection at the MR and PET centers. The authors also thank S. Haugbøl and H. Eiberg for genotyping analyses. Then, they would like to thank the John and Birthe Meyer Foundation for the donation of the Cyclotron and PET scanner as well as the Simon Spies Foundation for the donation of the Siemens Trio MRI scanner. This study was supported by a grant from the Augustinusfonden to P. M. F. (19‐0489) and by a grant from the Lundbeck Foundation (R90‐A7722).
Spies M, Nasser A, Ozenne B, Jensen PS, Knudsen GM, Fisher PM. Common HTR2A variants and 5‐HTTLPR are not associated with human in vivo serotonin 2A receptor levels. Hum Brain Mapp. 2020;41:4518–4528. 10.1002/hbm.25138
Funding information Lundbeck Foundation, Grant/Award Number: R90‐A7722; Augustinusfonden, Grant/Award Number: 19‐0489
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author and CIMBI database (http://www.cimbi.dk/db). The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- Adams, K. H. , Hansen, E. S. , Pinborg, L. H. , Hasselbalch, S. G. , Svarer, C. , Holm, S. , … Knudsen, G. M. (2005). Patients with obsessive‐compulsive disorder have increased 5‐HT2A receptor binding in the caudate nuclei. The International Journal of Neuropsychopharmacology, 8(3), 391–401. 10.1017/S1461145705005055 [DOI] [PubMed] [Google Scholar]
- Adams, K. H. , Pinborg, L. H. , Svarer, C. , Hasselbalch, S. G. , Holm, S. , Haugbol, S. , … Knudsen, G. M. (2004). A database of [(18)F]‐altanserin binding to 5‐HT(2A) receptors in normal volunteers: Normative data and relationship to physiological and demographic variables. NeuroImage, 21(3), 1105–1113. 10.1016/j.neuroimage.2003.10.046 [DOI] [PubMed] [Google Scholar]
- Anttila, S. , Kampman, O. , Illi, A. , Rontu, R. , Lehtimaki, T. , & Leinonen, E. (2007). Association between 5‐HT2A, TPH1 and GNB3 genotypes and response to typical neuroleptics: A serotonergic approach. BMC Psychiatry, 7, 22 10.1186/1471-244X-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz, M. , Collier, D. , Sodhi, M. , Ball, D. , Roberts, G. , Price, J. , … Kerwin, R. (1995). Association between clozapine response and allelic variation in 5‐HT2A receptor gene. Lancet, 346(8970), 281–282 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7630250 [DOI] [PubMed] [Google Scholar]
- Arranz, M. J. , Munro, J. , Owen, M. J. , Spurlock, G. , Sham, P. C. , Zhao, J. , … Kerwin, R. W. (1998). Evidence for association between polymorphisms in the promoter and coding regions of the 5‐HT2A receptor gene and response to clozapine. Molecular Psychiatry, 3(1), 61–66 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9491814 [DOI] [PubMed] [Google Scholar]
- Artigas, F. (2013). Serotonin receptors involved in antidepressant effects. Pharmacology & Therapeutics, 137(1), 119–131. 10.1016/j.pharmthera.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Blasi, G. , de Virgilio, C. , Papazacharias, A. , Taurisano, P. , Gelao, B. , Fazio, L. , … Bertolino, A. (2013). Converging evidence for the association of functional genetic variation in the serotonin receptor 2a gene with prefrontal function and olanzapine treatment. JAMA Psychiatry, 70(9), 921–930. 10.1001/jamapsychiatry.2013.1378 [DOI] [PubMed] [Google Scholar]
- Bogdan, R. , Salmeron, B. J. , Carey, C. E. , Agrawal, A. , Calhoun, V. D. , Garavan, H. , … Goldman, D. (2017). Imaging genetics and genomics in psychiatry: A critical review of progress and potential. Biological Psychiatry, 82(3), 165–175. 10.1016/j.biopsych.2016.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N. J. , Buckland, P. R. , Hall, H. , Owen, M. J. , & O'Donovan, M. C. (2004). The serotonin‐2A receptor gene locus does not contain common polymorphism affecting mRNA levels in adult brain. Molecular Psychiatry, 9(1), 109–114. 10.1038/sj.mp.4001366 [DOI] [PubMed] [Google Scholar]
- Caspi, A. , Sugden, K. , Moffitt, T. E. , Taylor, A. , Craig, I. W. , Harrington, H. , … Poulton, R. (2003). Influence of life stress on depression: Moderation by a polymorphism in the 5‐HTT gene. Science, 301(5631), 386–389. 10.1126/science.1083968 [DOI] [PubMed] [Google Scholar]
- Chen, S. F. , Shen, Y. C. , & Chen, C. H. (2009). HTR2A A‐1438G/T102C polymorphisms predict negative symptoms performance upon aripiprazole treatment in schizophrenic patients. Psychopharmacology, 205(2), 285–292. 10.1007/s00213-009-1538-z [DOI] [PubMed] [Google Scholar]
- Choi, M. J. , Kang, R. H. , Ham, B. J. , Jeong, H. Y. , & Lee, M. S. (2005). Serotonin receptor 2A gene polymorphism (‐1438A/G) and short‐term treatment response to citalopram. Neuropsychobiology, 52(3), 155–162. 10.1159/000087847 [DOI] [PubMed] [Google Scholar]
- Choi, M. J. , Lee, H. J. , Lee, H. J. , Ham, B. J. , Cha, J. H. , Ryu, S. H. , & Lee, M. S. (2004). Association between major depressive disorder and the ‐1438A/G polymorphism of the serotonin 2A receptor gene. Neuropsychobiology, 49(1), 38–41. 10.1159/000075337 [DOI] [PubMed] [Google Scholar]
- Cornea‐Hebert, V. , Riad, M. , Wu, C. , Singh, S. K. , & Descarries, L. (1999). Cellular and subcellular distribution of the serotonin 5‐HT2A receptor in the central nervous system of adult rat. The Journal of Comparative Neurology, 409(2), 187–209 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10379914 [DOI] [PubMed] [Google Scholar]
- Derogatis, L. R. , & Savitz, K. L. (1999). The SCL‐90‐R, Brief Symptom Inventory, and Matching Clinical Rating Scales In Maruish M. E. (Ed.), The use of psychological testing for treatment planning and outcomes assessment (pp. 679–724). London, England: Lawrence Erlbaum Associates Publishers. [Google Scholar]
- Erritzoe, D. , Frokjaer, V. G. , Haugbol, S. , Marner, L. , Svarer, C. , Holst, K. , … Knudsen, G. M. (2009). Brain serotonin 2A receptor binding: Relations to body mass index, tobacco and alcohol use. NeuroImage, 46(1), 23–30. 10.1016/j.neuroimage.2009.01.050 [DOI] [PubMed] [Google Scholar]
- Erritzoe, D. , Frokjaer, V. G. , Holst, K. K. , Christoffersen, M. , Johansen, S. S. , Svarer, C. , … Knudsen, G. M. (2011). In vivo imaging of cerebral serotonin transporter and serotonin(2A) receptor binding in 3,4‐methylenedioxymethamphetamine (MDMA or "ecstasy") and hallucinogen users. Archives of General Psychiatry, 68(6), 562–576. 10.1001/archgenpsychiatry.2011.56 [DOI] [PubMed] [Google Scholar]
- Erritzoe, D. , Holst, K. , Frokjaer, V. G. , Licht, C. L. , Kalbitzer, J. , Nielsen, F. A. , … Knudsen, G. (2010). A nonlinear relationship between cerebral serotonin transporter and 5‐HT(2A) receptor binding: An in vivo molecular imaging study in humans. The Journal of Neuroscience, 30(9), 3391–3397. 10.1523/JNEUROSCI.2852-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erritzoe, D. , Rasmussen, H. , Kristiansen, K. T. , Frokjaer, V. G. , Haugbol, S. , Pinborg, L. , … Glenthoj, B. Y. (2008). Cortical and subcortical 5‐HT2A receptor binding in neuroleptic‐naive first‐episode schizophrenic patients. Neuropsychopharmacology, 33(10), 2435–2441. 10.1038/sj.npp.1301656 [DOI] [PubMed] [Google Scholar]
- Ettrup, A. , da Cunha‐Bang, S. , McMahon, B. , Lehel, S. , Dyssegaard, A. , Skibsted, A. W. , … Knudsen, G. M. (2014). Serotonin 2A receptor agonist binding in the human brain with [(1) (1)C]Cimbi‐36. Journal of Cerebral Blood Flow and Metabolism, 34(7), 1188–1196. 10.1038/jcbfm.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettrup, A. , Svarer, C. , McMahon, B. , da Cunha‐Bang, S. , Lehel, S. , Moller, K. , … Knudsen, G. M. (2016). Serotonin 2A receptor agonist binding in the human brain with [(11)C]Cimbi‐36: Test‐retest reproducibility and head‐to‐head comparison with the antagonist [(18)F]altanserin. NeuroImage, 130, 167–174. 10.1016/j.neuroimage.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Falkenberg, V. R. , Gurbaxani, B. M. , Unger, E. R. , & Rajeevan, M. S. (2011). Functional genomics of serotonin receptor 2A (HTR2A): Interaction of polymorphism, methylation, expression and disease association. Neuromolecular Medicine, 13(1), 66–76. 10.1007/s12017-010-8138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnema, S. J. , Stepanov, V. , Ettrup, A. , Nakao, R. , Amini, N. , Svedberg, M. , … Halldin, C. (2014). Characterization of [(11)C]Cimbi‐36 as an agonist PET radioligand for the 5‐HT(2A) and 5‐HT(2C) receptors in the nonhuman primate brain. NeuroImage, 84, 342–353. 10.1016/j.neuroimage.2013.08.035 [DOI] [PubMed] [Google Scholar]
- Fisher, P. M. , Holst, K. K. , Mc Mahon, B. , Haahr, M. E. , Madsen, K. , Gillings, N. , … Knudsen, G. M. (2012). 5‐HTTLPR status predictive of neocortical 5‐HT4 binding assessed with [(11)C]SB207145 PET in humans. NeuroImage, 62(1), 130–136. 10.1016/j.neuroimage.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Fisher, P. M. , Ozenne, B. , Svarer, C. , Adamsen, D. , Lehel, S. , Baare, W. F. , … Knudsen, G. M. (2017). BDNF val66met association with serotonin transporter binding in healthy humans. Translational Psychiatry, 7(2), e1029 10.1038/tp.2016.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer, V. G. , Mortensen, E. L. , Nielsen, F. A. , Haugbol, S. , Pinborg, L. H. , Adams, K. H. , … Knudsen, G. M. (2008). Frontolimbic serotonin 2A receptor binding in healthy subjects is associated with personality risk factors for affective disorder. Biological Psychiatry, 63(6), 569–576. 10.1016/j.biopsych.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Frokjaer, V. G. , Vinberg, M. , Erritzoe, D. , Baare, W. , Holst, K. K. , Mortensen, E. L. , … Knudsen, G. M. (2010). Familial risk for mood disorder and the personality risk factor, neuroticism, interact in their association with frontolimbic serotonin 2A receptor binding. Neuropsychopharmacology, 35(5), 1129–1137. 10.1038/npp.2009.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr, M. E. , Hansen, D. L. , Fisher, P. M. , Svarer, C. , Stenbaek, D. S. , Madsen, K. , … Knudsen, G. M. (2015). Central 5‐HT neurotransmission modulates weight loss following gastric bypass surgery in obese individuals. The Journal of Neuroscience, 35(14), 5884–5889. 10.1523/JNEUROSCI.3348-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbalch, S. G. , Madsen, K. , Svarer, C. , Pinborg, L. H. , Holm, S. , Paulson, O. B. , … Knudsen, G. M. (2008). Reduced 5‐HT2A receptor binding in patients with mild cognitive impairment. Neurobiology of Aging, 29(12), 1830–1838. 10.1016/j.neurobiolaging.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Haugbol, S. , Pinborg, L. H. , Regeur, L. , Hansen, E. S. , Bolwig, T. G. , Nielsen, F. A. , … Knudsen, G. M. (2007). Cerebral 5‐HT2A receptor binding is increased in patients with Tourette's syndrome. The International Journal of Neuropsychopharmacology, 10(2), 245–252. 10.1017/S1461145706006559 [DOI] [PubMed] [Google Scholar]
- Hong, I. K. , Chung, S. T. , Kim, H. K. , Kim, Y. B. , Son, Y. D. , & Cho, Z. H. (2007). Ultra fast symmetry and SIMD‐based projection‐backprojection (SSP) algorithm for 3‐D PET image reconstruction. IEEE Transactions on Medical Imaging, 26(6), 789–803. 10.1109/TMI.2002.808360 [DOI] [PubMed] [Google Scholar]
- Hu, X. Z. , Lipsky, R. H. , Zhu, G. , Akhtar, L. A. , Taubman, J. , Greenberg, B. D. , … Goldman, D. (2006). Serotonin transporter promoter gain‐of‐function genotypes are linked to obsessive‐compulsive disorder. American Journal of Human Genetics, 78(5), 815–826. 10.1086/503850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlemann, R. , Boy, C. , Meyer, P. T. , Scherk, H. , Wagner, M. , Herzog, H. , … Bauer, A. (2005). Decreased prefrontal 5‐HT2A receptor binding in subjects at enhanced risk for schizophrenia. Anatomy and Embryology, 210(5–6), 519–523. 10.1007/s00429-005-0036-2 [DOI] [PubMed] [Google Scholar]
- Hurlemann, R. , Matusch, A. , Kuhn, K. U. , Berning, J. , Elmenhorst, D. , Winz, O. , … Bauer, A. (2008). 5‐HT2A receptor density is decreased in the at‐risk mental state. Psychopharmacology, 195(4), 579–590. 10.1007/s00213-007-0921-x [DOI] [PubMed] [Google Scholar]
- Innis, R. B. , Cunningham, V. J. , Delforge, J. , Fujita, M. , Gjedde, A. , Gunn, R. N. , … Carson, R. E. (2007). Consensus nomenclature for in vivo imaging of reversibly binding radioligands. Journal of Cerebral Blood Flow and Metabolism, 27(9), 1533–1539. 10.1038/sj.jcbfm.9600493 [DOI] [PubMed] [Google Scholar]
- Kato, M. , Fukuda, T. , Wakeno, M. , Fukuda, K. , Okugawa, G. , Ikenaga, Y. , … Kinoshita, T. (2006). Effects of the serotonin type 2A, 3A and 3B receptor and the serotonin transporter genes on paroxetine and fluvoxamine efficacy and adverse drug reactions in depressed Japanese patients. Neuropsychobiology, 53(4), 186–195. 10.1159/000094727 [DOI] [PubMed] [Google Scholar]
- Knudsen, G. M. , Jensen, P. S. , Erritzoe, D. , Baare, W. F. C. , Ettrup, A. , Fisher, P. M. , … Frokjaer, V. G. (2016). The Center for Integrated Molecular Brain Imaging (CIMBI) database. NeuroImage, 124(Pt B), 1213–1219. 10.1016/j.neuroimage.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Laje, G. , Cannon, D. M. , Allen, A. S. , Klaver, J. M. , Peck, S. A. , Liu, X. , … McMahon, F. J. (2010). Genetic variation in HTR2A influences serotonin transporter binding potential as measured using PET and [11C]DASB. The International Journal of Neuropsychopharmacology, 13(6), 715–724. 10.1017/S1461145709991027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, C. , Cantineau, R. , Guillaume, M. , Plenevaux, A. , & Christiaens, L. (1991). Fluorine‐18‐altanserin: A radioligand for the study of serotonin receptors with PET: Radiolabeling and in vivo biologic behavior in rats. Journal of Nuclear Medicine, 32(12), 2266–2272 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1744713 [PubMed] [Google Scholar]
- Lesch, K. P. , Bengel, D. , Heils, A. , Sabol, S. Z. , Greenberg, B. D. , Petri, S. , … Murphy, D. L. (1996). Association of anxiety‐related traits with a polymorphism in the serotonin transporter gene regulatory region. Science, 274(5292), 1527–1531. 10.1126/science.274.5292.1527 [DOI] [PubMed] [Google Scholar]
- Lothe, A. , Boni, C. , Costes, N. , Gorwood, P. , Bouvard, S. , Le Bars, D. , … Ryvlin, P. (2009). Association between triallelic polymorphism of the serotonin transporter and [18F]MPPF binding potential at 5‐HT1A receptors in healthy subjects. NeuroImage, 47(2), 482–492. 10.1016/j.neuroimage.2009.04.067 [DOI] [PubMed] [Google Scholar]
- Madsen, M. K. , Fisher, P. M. , Burmester, D. , Dyssegaard, A. , Stenbaek, D. S. , Kristiansen, S. , … Knudsen, G. M. (2019). Psychedelic effects of psilocybin correlate with serotonin 2A receptor occupancy and plasma psilocin levels. Neuropsychopharmacology, 44, 1328–1334. 10.1038/s41386-019-0324-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, M. K. , Mc Mahon, B. , Andersen, S. B. , Siebner, H. R. , Knudsen, G. M. , & Fisher, P. M. (2016). Threat‐related amygdala functional connectivity is associated with 5‐HTTLPR genotype and neuroticism. Social Cognitive and Affective Neuroscience, 11(1), 140–149. 10.1093/scan/nsv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marner, L. , Frokjaer, V. G. , Kalbitzer, J. , Lehel, S. , Madsen, K. , Baare, W. F. , … Hasselbalch, S. G. (2012). Loss of serotonin 2A receptors exceeds loss of serotonergic projections in early Alzheimer's disease: A combined [11C]DASB and [18F]altanserin‐PET study. Neurobiology of Aging, 33(3), 479–487. 10.1016/j.neurobiolaging.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Marner, L. , Knudsen, G. M. , Haugbol, S. , Holm, S. , Baare, W. , & Hasselbalch, S. G. (2009). Longitudinal assessment of cerebral 5‐HT2A receptors in healthy elderly volunteers: An [18F]‐altanserin PET study. European Journal of Nuclear Medicine and Molecular Imaging, 36(2), 287–293. 10.1007/s00259-008-0945-4 [DOI] [PubMed] [Google Scholar]
- Marner, L. , Knudsen, G. M. , Madsen, K. , Holm, S. , Baare, W. , & Hasselbalch, S. G. (2011). The reduction of baseline serotonin 2A receptors in mild cognitive impairment is stable at two‐year follow‐up. Journal of Alzheimer's Disease, 23(3), 453–459. 10.3233/JAD-2010-100903 [DOI] [PubMed] [Google Scholar]
- McMahon, F. J. , Buervenich, S. , Charney, D. , Lipsky, R. , Rush, A. J. , Wilson, A. F. , … Manji, H. (2006). Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. American Journal of Human Genetics, 78(5), 804–814. 10.1086/503820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer, H. Y. , & Massey, B. W. (2011). The role of serotonin receptors in the action of atypical antipsychotic drugs. Current Opinion in Pharmacology, 11(1), 59–67. 10.1016/j.coph.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Minov, C. , Baghai, T. C. , Schule, C. , Zwanzger, P. , Schwarz, M. J. , Zill, P. , … Bondy, B. (2001). Serotonin‐2A‐receptor and ‐transporter polymorphisms: Lack of association in patients with major depression. Neuroscience Letters, 303(2), 119–122 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11311507 [DOI] [PubMed] [Google Scholar]
- Mintun, M. A. , Sheline, Y. I. , Moerlein, S. M. , Vlassenko, A. G. , Huang, Y. , & Snyder, A. Z. (2004). Decreased hippocampal 5‐HT2A receptor binding in major depressive disorder: in vivo measurement with [18F]altanserin positron emission tomography. Biological Psychiatry, 55(3), 217–224 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/14744461 [DOI] [PubMed] [Google Scholar]
- Moses‐Kolko, E. L. , Price, J. C. , Shah, N. , Berga, S. , Sereika, S. M. , Fisher, P. M. , … Meltzer, C. C. (2011). Age, sex, and reproductive hormone effects on brain serotonin‐1A and serotonin‐2A receptor binding in a healthy population. Neuropsychopharmacology, 36(13), 2729–2740. 10.1038/npp.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, N. V. , Selvaraj, S. , Cowen, P. J. , Bhagwagar, Z. , Riedel, W. J. , Peers, P. , … Grasby, P. M. (2010). Serotonin transporter polymorphisms (SLC6A4 insertion/deletion and rs25531) do not affect the availability of 5‐HTT to [11C] DASB binding in the living human brain. NeuroImage, 52(1), 50–54. 10.1016/j.neuroimage.2010.04.032 [DOI] [PubMed] [Google Scholar]
- Ozaki, N. , Manji, H. , Lubierman, V. , Lu, S. J. , Lappalainen, J. , Rosenthal, N. E. , & Goldman, D. (1997). A naturally occurring amino acid substitution of the human serotonin 5‐HT2A receptor influences amplitude and timing of intracellular calcium mobilization. Journal of Neurochemistry, 68(5), 2186–2193 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9109547 [DOI] [PubMed] [Google Scholar]
- Pardinas, A. F. , Holmans, P. , Pocklington, A. J. , Escott‐Price, V. , Ripke, S. , Carrera, N. , … Walters, J. T. R. (2018). Common schizophrenia alleles are enriched in mutation‐intolerant genes and in regions under strong background selection. Nature Genetics, 50(3), 381–389. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, M. J. , D'Souza, U. M. , Arranz, M. J. , Kerwin, R. W. , & Makoff, A. J. (2004). The ‐1438A/G polymorphism in the 5‐hydroxytryptamine type 2A receptor gene affects promoter activity. Biological Psychiatry, 56(6), 406–410. 10.1016/j.biopsych.2004.06.020 [DOI] [PubMed] [Google Scholar]
- Peters, E. J. , Slager, S. L. , Jenkins, G. D. , Reinalda, M. S. , Garriock, H. A. , Shyn, S. I. , … Hamilton, S. P. (2009). Resequencing of serotonin‐related genes and association of tagging SNPs to citalopram response. Pharmacogenetics and Genomics, 19(1), 1–10. 10.1097/FPC.0b013e3283163ecd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, A. C. , Quesseveur, G. , Gressier, F. , Colle, R. , David, D. J. , Gardier, A. M. , … Corruble, E. (2014). Converging translational evidence for the involvement of the serotonin 2A receptor gene in major depressive disorder. Progress in Neuro‐Psychopharmacology & Biological Psychiatry, 54, 76–82. 10.1016/j.pnpbp.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Pinborg, L. H. , Adams, K. H. , Svarer, C. , Holm, S. , Hasselbalch, S. G. , Haugbol, S. , … Knudsen, G. M. (2003). Quantification of 5‐HT2A receptors in the human brain using [18F]altanserin‐PET and the bolus/infusion approach. Journal of Cerebral Blood Flow and Metabolism, 23(8), 985–996. 10.1097/01.WCB.0000074092.59115.23 [DOI] [PubMed] [Google Scholar]
- Pinborg, L. H. , Adams, K. H. , Yndgaard, S. , Hasselbalch, S. G. , Holm, S. , Kristiansen, H. , … Knudsen, G. M. (2004). [18F]altanserin binding to human 5HT2A receptors is unaltered after citalopram and pindolol challenge. Journal of Cerebral Blood Flow and Metabolism, 24(9), 1037–1045. 10.1097/01.WCB.0000126233.08565.E7 [DOI] [PubMed] [Google Scholar]
- Pinborg, L. H. , Arfan, H. , Haugbol, S. , Kyvik, K. O. , Hjelmborg, J. V. , Svarer, C. , … Knudsen, G. M. (2008). The 5‐HT2A receptor binding pattern in the human brain is strongly genetically determined. NeuroImage, 40(3), 1175–1180. 10.1016/j.neuroimage.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Polesskaya, O. O. , Aston, C. , & Sokolov, B. P. (2006). Allele C‐specific methylation of the 5‐HT2A receptor gene: Evidence for correlation with its expression and expression of DNA methylase DNMT1. Journal of Neuroscience Research, 83(3), 362–373. 10.1002/jnr.20732 [DOI] [PubMed] [Google Scholar]
- Polesskaya, O. O. , & Sokolov, B. P. (2002). Differential expression of the "C" and "T" alleles of the 5‐HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. Journal of Neuroscience Research, 67(6), 812–822. 10.1002/jnr.10173 [DOI] [PubMed] [Google Scholar]
- Praschak‐Rieder, N. , Kennedy, J. , Wilson, A. A. , Hussey, D. , Boovariwala, A. , Willeit, M. , … Meyer, J. H. (2007). Novel 5‐HTTLPR allele associates with higher serotonin transporter binding in putamen: A [(11)C] DASB positron emission tomography study. Biological Psychiatry, 62(4), 327–331. 10.1016/j.biopsych.2006.09.022 [DOI] [PubMed] [Google Scholar]
- Rasmussen, H. , Erritzoe, D. , Andersen, R. , Ebdrup, B. H. , Aggernaes, B. , Oranje, B. , … Glenthoj, B. (2010). Decreased frontal serotonin 2A receptor binding in antipsychotic‐naive patients with first‐episode schizophrenia. Archives of General Psychiatry, 67(1), 9–16. 10.1001/archgenpsychiatry.2009.176 [DOI] [PubMed] [Google Scholar]
- Santangelo, A. M. , Ito, M. , Shiba, Y. , Clarke, H. F. , Schut, E. H. , Cockcroft, G. , … Roberts, A. C. (2016). Novel primate model of serotonin transporter genetic polymorphisms associated with gene expression, anxiety and sensitivity to antidepressants. Neuropsychopharmacology, 41(9), 2366–2376. 10.1038/npp.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, A. M. , Sawiak, S. J. , Fryer, T. , Hong, Y. , Shiba, Y. , Clarke, H. F. , … Roberts, A. C. (2019). Insula serotonin 2A receptor binding and gene expression contribute to serotonin transporter polymorphism anxious phenotype in primates. Proceedings of the National Academy of Sciences of the United States of America, 116(29), 14761–14768. 10.1073/pnas.1902087116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. M. , Papp, A. C. , Webb, A. , Ruble, C. L. , Munsie, L. M. , Nisenbaum, L. K. , … Sadee, W. (2013). Multiple regulatory variants modulate expression of 5‐hydroxytryptamine 2A receptors in human cortex. Biological Psychiatry, 73(6), 546–554. 10.1016/j.biopsych.2012.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock, G. , Heils, A. , Holmans, P. , Williams, J. , D'Souza, U. M. , Cardno, A. , … Owen, M. J. (1998). A family based association study of T102C polymorphism in 5HT2A and schizophrenia plus identification of new polymorphisms in the promoter. Molecular Psychiatry, 3(1), 42–49 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9491812 [DOI] [PubMed] [Google Scholar]
- Stenbaek, D. S. , Kristiansen, S. , Burmester, D. , Madsen, M. K. , Frokjaer, V. G. , Knudsen, G. M. , & Fisher, P. M. (2019). Trait openness and serotonin 2A receptors in healthy volunteers: A positron emission tomography study. Human Brain Mapping, 40(7), 2117–2124. 10.1002/hbm.24511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau, F. C. , Reader, A. J. , Comtat, C. , Leroy, C. , Ribeiro, M. J. , Buvat, I. , & Trebossen, R. (2008). Impact of image‐space resolution modeling for studies with the high‐resolution research tomograph. Journal of Nuclear Medicine, 49(6), 1000–1008. 10.2967/jnumed.107.045351 [DOI] [PubMed] [Google Scholar]
- Svarer, C. , Madsen, K. , Hasselbalch, S. G. , Pinborg, L. H. , Haugbol, S. , Frokjaer, V. G. , … Knudsen, G. M. (2005). MR‐based automatic delineation of volumes of interest in human brain PET images using probability maps. NeuroImage, 24(4), 969–979. 10.1016/j.neuroimage.2004.10.017 [DOI] [PubMed] [Google Scholar]
- Turecki, G. , Briere, R. , Dewar, K. , Antonetti, T. , Lesage, A. D. , Seguin, M. , … Rouleau, G. A. (1999). Prediction of level of serotonin 2A receptor binding by serotonin receptor 2A genetic variation in postmortem brain samples from subjects who did or did not commit suicide. The American Journal of Psychiatry, 156(9), 1456–1458. 10.1176/ajp.156.9.1456 [DOI] [PubMed] [Google Scholar]
- Williams, J. , Spurlock, G. , McGuffin, P. , Mallet, J. , Nothen, M. M. , Gill, M. , … Owen, M. J. (1996). Association between schizophrenia and T102C polymorphism of the 5‐hydroxytryptamine type 2a‐receptor gene. European Multicentre Association Study of Schizophrenia (EMASS) group. Lancet, 347(9011), 1294–1296 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8622505 [DOI] [PubMed] [Google Scholar]
- Woods, R. P. , Cherry, S. R. , & Mazziotta, J. C. (1992). Rapid automated algorithm for aligning and reslicing PET images. Journal of Computer Assisted Tomography, 16(4), 620–633 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1629424 [DOI] [PubMed] [Google Scholar]
- Wray, N. R. , Ripke, S. , Mattheisen, M. , Trzaskowski, M. , Byrne, E. M. , Abdellaoui, A. , … Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . (2018). Genome‐wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50(5), 668–681. 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author and CIMBI database (http://www.cimbi.dk/db). The data are not publicly available due to privacy or ethical restrictions.


