Abstract
Ubiquitin-conjugating enzyme 2C (UBE2C) involves in numerous cellular processes and the tumor progression in many cancers. However, its role in oral squamous cell carcinoma (OSCC) is unclear. We aimed to investigate the role and clinical significance of UBE2C in OSCC. The expression levels of UBE2C were examined by immunohistochemistry in 185 buccal mucosa squamous cell carcinomas, 247 tongue squamous cell carcinomas (TSCCs) and 75 lip squamous cell carcinomas. The roles of UBE2C in cell growth, invasion/migration and cancer stemness were also examined in OSCC cells. The expression levels of UBE2C protein were higher in tumor tissues than they were in the corresponding tumor adjacent normal tissues from OSCC patients. Higher UBE2C expression was associated with poor cell differentiation and lymph node invasion in OSCC patients. High UBE2C expression was also correlated with shorter disease-specific survival in TSCC patients having poor cell differentiation, advanced pathological stages, lymph node metastasis as well as receiving radiation therapy. Compared to control cells, OSCC cells in which UBE2C was silenced showed decreased cell proliferation, migration/invasion and colony formation and they exhibited lower expression levels of the following cancer stemness markers—ALDH1/A2, CD44, CD166 and EpCAM. High co-expression levels of UBE2C/CD44, UBE2C/CD166 and UBE2C/EpCAM were associated with poor prognosis in oral cancer patients from The Cancer Genome Atlas database. Our findings indicated that UBE2C might be a potential biomarker for tumorigenesis and prognosis in TSCC.
Keywords: UBE2C, tongue squamous cell carcinoma, tumorigenesis, prognosis, biomarker
1. Introduction
More than 90% of oral cancers are oral squamous cell carcinoma (OSCC) which is typically observed on the tongue, buccal mucosa and lips [1]. Cigarette smoking, alcohol drinking, betel quid chewing, chronic periodontitis [2] and viral infections [3] are major risk factors contributing to the incidence of OSCC. OSCC is a highly aggressive cancer with frequent local recurrences and lymph node metastases [4]. The overall 5-year survival rate for OSCC patients have not been improved and remained at approximately 50% for several decades [5]. Therefore, how a diagnostic/prognostic assessment of gene expression could be translated into better patient survival. Ultimately, the aim of biomarker tests is probably to identify/stratify patients for certain therapeutic intervention. Thus, identifying reliable diagnostic and prognostic biomarkers for OSCC is urgent.
Human ubiquitin-conjugating enzyme 2C (UBE2C) is a member of the E2 ubiquitin-conjugating enzyme family [6]. Ubiquitin-dependent protein degradation is involved in numerous cell processes, such as cell cycle progression, signal transduction and malignant transformation [7]. UBE2C is required for the destruction of mitotic cyclin and participates in the regulation of cell cycle progression [8]. UBE2C overexpression causes loss of mitotic spindle checkpoint activity and genomic stability [8]. The expression level of UBE2C is very high in various human cancers, such as breast cancer [9], ovarian cancer [10], head and neck squamous cell carcinoma (HNSCC) [11] and melanoma [12], suggesting that UBE2C is associated with tumorigenesis. Moreover, overexpression of UBE2C is correlated with tumor progression [13], so it acts as a potential prognostic/diagnostic biomarker in various types of tumors [11,14,15]. However, the role of UBE2C in OSCC is still unclear. Herein, we found that high expression level of UBE2C was associated with the tumorigenesis, poor clinicopathological outcomes and poor prognosis in OSCC patients. Moreover, UBE2C played roles in cell growth, migration/invasion and cancer stemness in OSCC cells. Furthermore, The Cancer Genome Atlas (TCGA) oral cancer cohorts with co-expression level of high UBE2C/cancer stemness markers had poor prognosis. Our results indicated the potential roles and clinical significance of UBE2C in TSCC.
2. Materials and Methods
2.1. Patients and Tissue Specimens
All paraffin-embedded tissues of buccal mucosa squamous cell carcinoma (BMSCC, n = 185), tongue squamous cell carcinoma (TSCC, n = 247) and lip squamous cell carcinoma (LSCC, n = 75) were obtained from patients at the Department of Pathology at KVGH between 1990 and 2013. This study was approved by the Institutional Review Board of the Kaohsiung Veterans General Hospital (IRB, approved on: 13-06-14, code number: VGHKS14-CT6-18). Informed consent was obtained from patients before collecting their tissue specimens. Clinicopathological data about TNM stages were determined according to the eight editions of the AJCC Cancer Staging Manual [16]. The survival time of patients was calculated from the date of the primary tumor operation to the date of death or the last follow-up until October 2013.
2.2. Tissue Microarray (TMA) Construction
All TMA blocks were constructed with a 1.5 mm diameter and each block consisted of 134 cores, including 43 sets of three (each set of three contained 2 cores from the tumor tissue, 1 core from the corresponding tumor adjacent normal (CTAN) of the same patient) and another 5 cores of normal uvula epithelium. To avoid false positives, noncancerous tissue of patients was excluded. After exclusion, a total of 12 TMA blocks were constructed and subsequently cut into 4-μm paraffin sections.
2.3. IHC Analysis and Scoring
Before IHC analysis, paraffin sections were first dewaxed in xylene then rehydrated in a graded alcohol series (30–100%). Then, IHC analysis was performed by using the Novolink Polymer Detection Systems (Leica Biosystem, RE7280-K, Richmond, IL, USA). First, these dewaxed and rehydrated sections were immersed in sodium citrate (pH 6.0) for 10 min at 125 °C in a pressure boiler for antigen retrieval. Then, they were incubated in methanol containing 3% hydrogen peroxide for 30 min to block endogenous peroxidase activity. Afterward, these sections were incubated with an anti-UBE2C monoclonal antibody (dilution 1:500; H00011065-M01, Abnova, Walnut, CA, USA) in 5% BSA/TBS at 4 °C overnight and washed with TBS. The sections were incubated with Poly-HRP-labeled secondary antibody (Novolink™ Polymer) for 30 min, developed with 0.03% diaminobenzidine for 5 min and counterstained with hematoxylin. The IHC staining was scored according intensity and percentage as described in our previous study [17,18]. For the association of UBE2C with patient survival, the ‘high’ and ‘low’ expression level of cytoplasmic UBE2C was defined based on the cutoff point, which was set at the 50th percentile according to the immunoreactive scores of UBE2C.
2.4. Cell Culture
Two OSCC cell lines from tongue squamous cell carcinoma (SAS) and gingival squamous cell carcinoma (Ca9-22) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen-GIBCO, Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine serum (Biological Industries, Cromwell, CT, USA), 100 U/mL penicillin (Invitrogen-GIBCO, Carlsbad, CA, USA), 1% MEM nonessential amino acids and 100 µg/mL streptomycin (Invitrogen-GIBCO, Carlsbad, CA, USA) at 37 °C in humidified 5% CO2 atmosphere.
2.5. siRNA Knockdown and Transient Transfection
UBE2C siRNA pools were synthesized (5′-UCCUUUUUGUGAUUUCUGUTT-3′, Ambion, Austin, TX, USA) and cells were transfected for 48–96 h with 5 nM scrambled siRNA or an siRNA against UBE2C using RNAiMAX (Life Technologies, Carlsbad, CA, USA). The knockdown efficiency was determined by western blot.
2.6. Real-Time PCR (RT-PCR)
Total RNA was extracted with TRIzol reagent (Biocompare, South San Francisco, CA, USA). A total of 1 μg of RNA was reverse-transcribed with SuperScriptIII RNase Reverse Transcriptase (Invitrogen-GIBCO, Carlsbad, CA, USA) for cDNA synthesis. Real-time polymerase chain reaction (PCR) was carried out using a StepOnePlusTM system (Applied Biosystems, Foster City, CA, USA) with SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). All genes expression levels in cells was normalized with internal control GAPDH gene. The UBE2C primers sequences were shown as follows—the forward—5′-TGATGTCTGGCCATAAAGGGA-3′, the reverse—5′-AGCGAGAGCTTATACCTCAGG-3′.
2.7. Western Blot Analysis
Cells were initially washed with phosphate-buffered saline (PBS) and lysed with RIPA buffer (1% NP40, 50 mM Tris Cl pH 7.5, 150 mM NaCl, 0.25% sodium deoxycholate 1% sodium dodecyl sulphate (SDS) and a protease inhibitor cocktail). All proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with primary anti-UBE2C antibodies (Abnova, Walnut, CA, USA) at 4 °C overnight and then probed with an HRP-labeled secondary antibody (Santa Cruz Biotechnology sc-2004, Santa Cruz, CA, USA). Finally, a LI-COR Odyssey Imaging System (LI-COR Inc, Lincoln, NE, USA) was used to analyze protein expression levels on the probed membrane before incubation with an enhanced chemiluminescence (ECL) reagent (Thermo Fisher Scientific, 32106, Carlsbad, CA, USA).
2.8. Cell Viability
Cells were seeded into 96-well flat bottom plates (5 × 103 cells/well) containing 100 µL of medium. Cell viability was determined by Cell Titer-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA). The resultant luminescence signal was measured using a Fluoroskan Ascent FL reader (Thermo Fisher Scientific, Carlsbad, MA, USA).
2.9. Colony Formation
1 × 103. cells were seeded into each well of 6-well plates and cultured for 2–3 weeks until colony formation; fresh media was provided every 3 days. The colonies were fixed with paraformaldehyde (3.75% v/v), stained with crystal violet (0.25% w/v) and quantified.
2.10. Migration and Invasion
For the migration assay, UBE2C-knockdown cells (2 × 105 cells) were cultured for 24 h in dishes fitted with IBIDI Culture-Inserts (35 mm with high culture-insert coating). Afterward, the plastic inserts were removed, wound healing was observed for 7 h and the migration distance was measured. For the invasion assay, UBE2C-knockdown cells were seeded (1.5 × 105 cells/well) into the top chamber of transwell inserts with 8-μm pores (Greiner Bio-One, Monroe, NC, USA) coated with 0.5% Matrigel in 300 μL of DMEM containing 1% Fetal Bovine Serum (FBS). Twenty-four hours after seeding, the cells on the bottom of the inserts were fixed, stained with 0.1% crystal violet and quantified.
2.11. Statistical Analysis and TCGA Database
All statistical analyses were performed with SPSS software (version 20.0, SPSS Inc., Chicago, IL, USA). Wilcoxon signed-rank tests were used to compare the protein expression levels between tumor tissues and tumor-adjacent normal tissues. Student’s t-test, one-way ANOVA, Mann-Whitney U test and Kruskal-Wallis one-way ANOVA were used to evaluate the correlation of UNE2C expression levels at different subsites of OSCC with clinicopathologic outcomes. A Cox proportional hazard model and a log-rank test were used to evaluate the contribution of UBE2C expression to disease-specific survival (DSS) in OSCC patients. The association of co-expression level of UBE2C/cancer stemness markers with survival was analyzed using RNA-sequencing transcriptome profiles of 315 oral cancer patients from TCGA database (https://cancergenome.nih.gov). The gene expression levels of UBE2C and cancer stemness markers were dichotomized into ‘high’ and ‘low’ expression according to a receiver operating characteristic (ROC) curve.
3. Results
3.1. The Clinical Significance of UBE2C in OSCC Patients
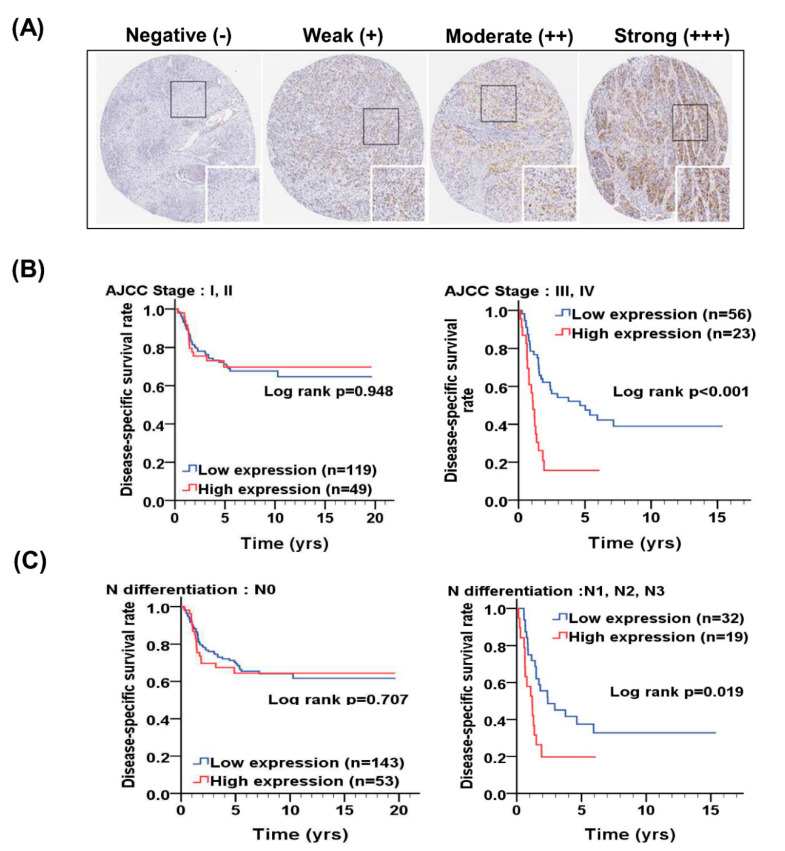
To investigate the clinical significance of UBE2C in OSCC patients, the expression level of UBE2C in tissues of OSCC patients was evaluated by scoring of IHC staining according intensity and percentage. For scoring intensity, the expression level of UBE2C in OSCC patients was determined using a numerical scale (−, negative; +, weak; ++, moderate; and +++, strong; Figure 1). After multiplying scores of intensity and percentage in positive cells of each tissues section, we found that the expression level of UBE2C was higher in tumor tissues than it was in CTAN tissues in all subsites of OSCC (p < 0.001; Table 1). Moreover, the high expression level of UBE2C was associated with lymph node invasion (p = 0.017; Supplementary Table S1) and poor cell differentiation (p < 0.001; Supplementary Table S1) in OSCC patients, especially in BMSCC (p = 0.002; Supplementary Table S1) and TSCC (p = 0.001; Supplementary Table S1). Furthermore, a high expression level of UBE2C was associated with shorter DSS in TSCC patients with poor cell differentiation (p = 0.042; Table 2), advanced pathological stage (p = 0.001; Table 2), lymph node metastasis (p = 0.041; Table 2) and postoperative radiation therapy (p = 0.002; Table 2). By log rank test, we found that OSCC patients with high UBE2C level had a significantly shorter DSS, especially for those with advanced pathological stage (Figure 1B, p < 0.001) and lymph node metastasis (Figure 1C, p = 0.019). Taken together, our results indicated that UBE2C expression was significantly associated with tumorigenesis, clinicopathological outcomes and prognosis in OSCC patients, especially in TSCC patients.
Figure 1.
Representative immunohistochemical staining of UBE2C in tumor tissues and compared DSS of OSCC patients according to expression level of UBE2C. (A) The expression level of UBE2C was determined using a numerical scale (no signal (−), weak (+), moderate (++) or strong (+++) staining) for scoring intensity. (B) The comparison of DSS in OSCC patients with early pathological stage (I + II) and advanced pathological stage (III + IV) according to expression level of UBE2C. (C) The comparison of DSS in OSCC patients without lymph node metastasis (N0) and with lymph node metastasis (N1, N2, N3) according to expression level of UBE2C.
Table 1.
Comparison of UBE2C expression between corresponding tumor adjacent normal tissues and tumor tissues in three primary subsites of OSCC patients.
| Variables | No. | Tumor Adjacent Normal | Tumor | Z | p-Value * | ||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | Mean ± SD | Median | ||||
| Buccal mucosal SCC | 136 | 0.57 ± 0.92 | 0.00 | 2.56 ± 1.33 | 3.00 | 9.267 | <0.001 |
| Tongue SCC | 188 | 0.08 ± 0.34 | 0.00 | 1.69 ± 1.38 | 1.50 | 10.162 | <0.001 |
| Lip SCC | 63 | 0.38 ± 0.85 | 0.00 | 1.56 ± 1.12 | 2.00 | 5.177 | <0.001 |
| Total: Oral SCC | 387 | 0.30 ± 0.72 | 0.00 | 1.97 ± 1.39 | 2.00 | 14.717 | <0.001 |
Abbreviations: SCC, squamous cell carcinoma; SD, standard deviation. * p-values were estimated by Wilcoxon signed-rank test. Bold values denote statistically significant.
Table 2.
Association of UBE2C expression with disease-specific survival stratified by clinicopathologic factors in three primary subsites of OSCC patients.
| Buccal Mucosal SCC (n = 185) | Tongue SCC (n = 247) | Lip SCC (n = 75) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | UBE2C | No. (%) | AHR (95% CI) | p Value † | No. (%) | AHR (95% CI) | p Value † | No. (%) | AHR (95% CI) | p Value † |
| Sex | ||||||||||
| Female | Low | 1 (25.0) | 1.00 | 17 (58.6) | 1.00 | 4 (57.1) | 1.00 | |||
| High | 3 (75.0) | Incalculable | 12 (41.4) | 0.45 (0.08–2.59) | 0.370a | 3 (42.9) | Incalculable | |||
| Male | Low | 72 (39.8) | 1.00 | 158 (72.5) | 1.00 | 56 (82.4) | 1.00 | |||
| High | 109 (60.2) | 1.08 (0.67–1.74) | 0.751 a | 60 (27.5) | 1.86 (1.20–2.89) | 0.006 a | 12 (17.6) | 2.17 (0.57–8.24) | 0.254 a | |
| Age, yrs | ||||||||||
| ≤5 0 | Low | 36 (43.4) | 1.00 | 92 (71.9) | 1.00 | 14 (82.4) | 1.00 | |||
| High | 47 (56.6) | 0.76 (0.37–1.54) | 0.439 a | 36 (28.1) | 1.50 (0.86–2.61) | 0.156 a | 3 (17.6) | 1.64 (0.14–18.90) | 0.693 a | |
| >50 | Low | 37 (36.3) | 1.00 | 83 (69.7) | 1.00 | 46 (79.3) | 1.00 | |||
| High | 65 (63.7) | 1.54 (0.79–3.01) | 0.207 a | 36 (30.3) | 1.62 (0.84–3.14) | 0.149 a | 12 (20.7) | 2.06 (0.38–11.15) | 0.403 a | |
| Cell differentiation | ||||||||||
| Well | Low | 26 (52.0) | 1.00 | 22 (81.5) | 1.00 | 29 (82.9) | 1.00 | |||
| High | 24 (48.0) | 0.54 (0.18–1.67) | 0.287 b | 5 (18.5) | 1.48 (0.15–14.40) | 0.738 b | 6 (17.1) | Incalculable | ||
| Moderate, poor | Low | 47 (34.8) | 1.00 | 153 (69.5) | 1.00 | 31 (77.5) | 1.00 | |||
| High | 88 (65.2) | 1.27 (0.74–2.17) | 0.387 b | 67 (30.5) | 1.57 (1.02–2.41) | 0.042 b | 9 (22.5) | 0.59 (0.07–4.86) | 0.621 b | |
| AJCC pathological stage | ||||||||||
| I, II | Low | 48 (42.1) | 1.00 | 119 (70.8) | 1.00 | 47 (79.7) | 1.00 | |||
| High | 66 (57.9) | 1.18 (0.56–2.49) | 0.664 c | 49 (29.2) | 0.95 (0.51–1.75) | 0.859 c | 12 (20.3) | 0.88 (0.10–7.98) | 0.907 c | |
| III, IV | Low | 25 (35.2) | 1.00 | 56 (70.9) | 1.00 | 13 (81.3) | 1.00 | |||
| High | 46 (64.8) | 1.06 (0.58–1.95) | 0.842 c | 23 (29.1) | 2.67 (1.46–4.86) | 0.001 c | 3 (18.8) | 4.66 (0.55–39.46) | 0.158 c | |
| T classification | ||||||||||
| T1, T2 | Low | 57 (40.7) | 1.00 | 134 (69.1) | 1.00 | 49 (79.0) | 1.00 | |||
| High | 83 (59.3) | 1.23 (0.68–2.22) | 0.489 d | 60 (30.9) | 1.45 (0.88–2.40) | 0.144 d | 13 (21.0) | 0.87 (0.17–4.50) | 0.870 d | |
| T3, T4 | Low | 16 (35.6) | 1.00 | 41 (77.4) | 1.00 | 11 (84.6) | 1.00 | |||
| High | 29 (64.4) | 0.99 (0.45–2.18) | 0.982 d | 12 (22.6) | 1.62 (0.71–3.71) | 0.251 d | 2 (15.4) | 3.08 (0.13–74.18) | 0.488 d | |
| N classification | ||||||||||
| N0 | Low | 57 (41.0) | 1.00 | 143 (73.0) | 1.00 | 56 (80.0) | 1.00 | |||
| High | 82 (59.0) | 1.03 (0.55–1.93) | 0.938 e | 53 (27.0) | 1.21 (0.69–2.12) | 0.496 e | 14 (20.0) | 1.77 (0.36–8.84) | 0.486e | |
| N1, N2 | Low | 16 (34.8) | 1.00 | 32 (62.7) | 1.00 | 4 (80.0) | 1.00 | |||
| High | 30 (65.2) | 1.29 (0.62–2.66) | 0.493 e | 19 (37.3) | 2.06 (1.03–4.14) | 0.041 e | 1 (20.0) | 1.41 (0.09–23.57) | 0.809 e | |
| Postoperative RT | ||||||||||
| No | Low | 55 (41.7) | 1.00 | 125 (69.4) | 1.00 | 57 (79.2) | 1.00 | |||
| High | 77 (58.3) | 1.40 (0.75–2.61) | 0.290 a | 55 (30.6) | 1.18 (0.69–2.02) | 0.544 a | 15 (20.8) | 1.71 (0.45–6.57) | 0.432 a | |
| Yes | Low | 18 (34.0) | 1.00 | 50 (74.6) | 1.00 | 3 (100.0) | 1.00 | |||
| High | 35 (66.0) | 0.79 (0.38–1.63) | 0.522 a | 17 (25.4) | 3.01 (1.49–6.09) | 0.002 a | 0 (0.0) | Incalculable | ||
Abbreviations: CHR, crude hazard ratio; CI, confidence interval; AHR, adjusted hazard ratio; AJCC, American Joint Committee on Cancer; RT, radiotherapy. † p values were estimated by multivariate Cox’s regression. a Adjusted for cell differentiation (moderate + poor vs. well) and AJCC pathological stage (stage III + IV vs. stage I + II). b Adjusted for AJCC pathological stage (stage III + IV vs. stage I + II). c Adjusted for cell differentiation (moderate + poor vs. well). d Adjusted for cell differentiation (moderate + poor vs. well) and N classification (N1, N2 vs. N0). e Adjusted for cell differentiation (moderate+poor vs. well) and T classification (T3, T4 vs. T1, T2). Bold values denote statistically significant.
3.2. The role of UBE2C in the Growth of OSCC Cells
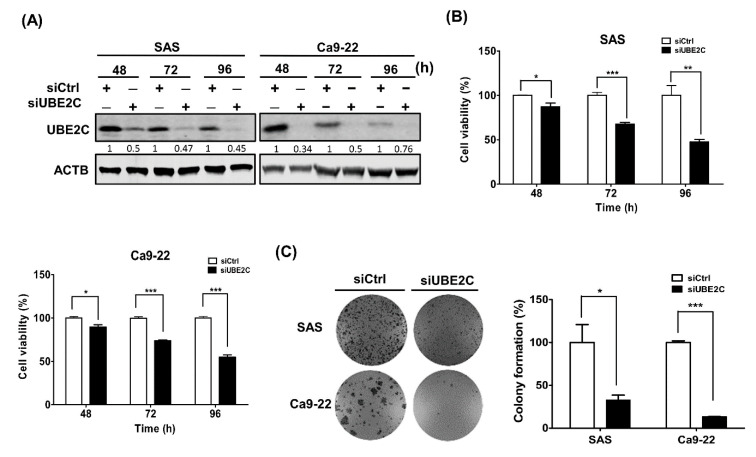
Our results indicated that UBE2C expression was associated with tumorigenesis and prognosis in TSCC patients. To further investigate the role of UBE2C in the tumorigenesis of TSCC, UBE2C was knocked down in two OSCC cell lines, SAS and Ca9-22 cells, for 48, 72 and 96 h and the knockdown efficiency was verified by western blot (Figure 2A). Moreover, we found that cell viability (Figure 2B) and colony formation (Figure 2C) were significantly decreased in UBE2C-knockdown OSCC cells compared to control cells. These results suggested that UBE2C might be involved in the growth of OSCC cells.
Figure 2.
Cell viability and colony formation in UBE2C-knockdown SAS and Ca9-22 cells. (A) UBE2C knockdown efficiency of cells transfected with scrambled siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) for 48, 72 and 96 h was analyzed by western blot analysis. (B) The viability of cells transfected with scrambled siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) for 48 h was measured by Cell Titer Glo. (C) The colony formation of cells transfected with scrambled s siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) was monitored for 1–3 weeks. The colonies of UBE2C-knockdown cells are shown in the left panel and their quantification is shown in the right panel. The quantitative results are expressed as the mean ± SD from three independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs. nontargeting control siRNA (siCtrl).
3.3. The Role of UBE2C in Invasion and Migration of OSCC Cells
To further investigate the possible role of UBE2C in the metastasis of TSCC, UBE2C was knocked down in SAS and Ca9-22 cells by transfecting with an siRNA against UBE2C for 48 h and their invasion and migration abilities were subsequently evaluated. The results showed that the migration of UBE2C-knockdown cells was significantly suppressed by 65% and 67% in SAS and Ca9-22 cells compared to that of control cells (Figure 3A), respectively. Moreover, UBE2C-knockdown SAS and Ca9-22 cells also showed decreased invasion by 70% and 44%, respectively (Figure 3B). The results indicated that UBE2C might be involved in OSCC metastasis.
Figure 3.
Cell migration and invasion in UBE2C-knockdown SAS and Ca9-22 cells. (A) Cells were transfected with scrambled siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) for 7 h and then their migration was assessed by wound–healing assay. (B) The invasion of cells transfected with scrambled siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) for 24 h was measured by transwell invasion assay. The quantitative results are expressed as the mean ± SD from three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. nontargeting control siRNA (siCtrl).
3.4. The Role of UBE2C in Cancer Stemness of OSCC Cells
UBE2C plays roles in some cancer stemness properties, such as drug resistance [19,20]. To investigate whether UBE2C is also involved in cancer stemness in TSCC, the expression of several cancer stemness markers was evaluated by RT-PCR. The expression levels of aldehyde dehydrogenase A2 (ALDH1A2), CD44, CD166 and epithelial cell adhesion molecule (EpCAM) were significantly decreased in UBE2C-knockdown SAS (Figure 4A) and Ca9-22 cells (Figure 4B). Moreover, high coexpression of UBE2C/CD44, UBE2C/CD166 and UBE2C/EpCAM genes (Table 3) was associated with overall survival and recurrence in oral cancer patients including TSCC patients from TCGA database, indicating that UBE2C might be involved in controlling cancer stemness of OSCC.
Figure 4.
Expression of cancer stemness markers in UBE2C-knockdown OSCC cells. (A) SAS cells. (B) Ca9-22 cells. The expression levels of the ALDH1/A1, ALDH1/A2, CD44, ABCG2, CD166 and EpCAM in cells transfected with scrambled s siRNA (5 nM, siCtrl) or siRNAs against UBE2C (5 nM, siUBE2C) were detected by real time polymerase chain reaction (RT-PCR) and GAPDH was used as an internal control. The quantitative results are expressed as the mean ± SD from three independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs. nontargeting control siRNA (siCtrl).
Table 3.
Association of co-expression of UBE2C and cancer stemness markers with survival in oral cancer patients from TCGA database.
| Variable | No. (%) | CHR (95% CI) | p Value * | AHR (95% CI) | p Value † |
|---|---|---|---|---|---|
| Overall survival | |||||
| UBE2C (L) CD44(L) | 38 (14.4) | 1.00 | 1.00 | ||
| UBE2C (H) CD44(L) | 141 (53.4) | 1.30 (0.90–1.88) | 0.166 | 2.91 (1.39–6.08) | 0.004 |
| UBE2C (L) CD44(H) | 17 (6.4) | 0.74 (0.34–1.59) | 0.435 | 1.91 (0.69–5.30) | 0.212 |
| UBE2C (H) CD44(H) | 68 (25.8) | 1.38 (0.92–2.07) | 0.115 | 3.29 (1.51–7.16) | 0.003 |
| UBE2C (L) ALCAM(L) | 51 (19.3) | 1.00 | 1.00 | ||
| UBE2C (H) ALCAM(L) | 171 (64.8) | 1.12 (0.76–1.65) | 0.562 | 2.24 (1.23–4.07) | 0.008 |
| UBE2C (L) ALCAM(H) | 4 (1.5) | 0.83 (0.21–3.37) | 0.795 | 1.81 (0.41–8.07) | 0.438 |
| UBE2C (H) ALCAM(H) | 38 (14.4) | 2.05 (1.32–3.18) | 0.001 | 3.91 (1.99–7.68) | <0.001 |
| UBE2C (L) EPCAM(L) | 51 (19.3) | 1.00 | 1.00 | ||
| UBE2C (H) EPCAM(L) | 141 (53.4) | 1.21 (0.84–1.75) | 0.308 | 2.42 (1.31–4.50) | 0.005 |
| UBE2C (L) EPCAM(H) | 4 (1.5) | 0.90 (0.28–2.91) | 0.862 | 1.93 (0.53–6.98) | 0.319 |
| UBE2C (H) EPCAM(H) | 68 (25.8) | 2.50 (1.01–2.24) | 0.045 | 2.99 (1.55–5.78) | 0.001 |
| Disease-free survival | |||||
| UBE2C (L) CD44(L) | 37 (16.4) | 1.00 | 1.00 | ||
| UBE2C (H) CD44(L) | 10 (4.4) | 2.15 (0.77–5.95) | 0.143 | 3.90 (1.04–14.57) | 0.043 |
| UBE2C (L) CD44(H) | 149 (65.9) | 0.82 (0.47–1.40) | 0.461 | 1.75 (0.69–4.48) | 0.242 |
| UBE2C (H) CD44(H) | 30 (13.3) | 1.89 (1.00–3.60) | 0.051 | 3.21 (1.13–9.12) | 0.029 |
| UBE2C (L) ALCAM(L) | 130 (57.5) | 1.00 | 1.00 | ||
| UBE2C (H) ALCAM(L) | 18 (8.0) | 0.98 (0.35–2.70) | 0.961 | 1.15 (0.40–3.30) | 0.792 |
| UBE2C (L) ALCAM(H) | 56 (24.8) | 0.80 (0.42–1.52) | 0.492 | 0.99 (0.50–1.96) | 0.977 |
| UBE2C (H) ALCAM(H) | 22 (9.7) | 2.84 (1.50–5.40) | 0.001 | 2.87 (1.45–5.67) | 0.002 |
| UBE2C (L) EPCAM(L) | 158 (69.9) | 1.00 | 1.00 | ||
| UBE2C (H) EPCAM(L) | 32 (14.2) | 1.25 (0.61–2.56) | 0.537 | 1.42 (0.68–2.97) | 0.354 |
| UBE2C (L) EPCAM(H) | 28 (12.4) | 0.78 (0.33–1.82) | 0.565 | 0.93 (0.39–2.23) | 0.878 |
| UBE2C (H) EPCAM(H) | 8 (3.5) | 4.99 (2.24–11.12) | <0.001 | 5.24 (2.30–11.95) | <0.001 |
Abbreviations: OSCC, Oral squamous cell carcinoma; CHR, crude hazard ratio; CI, confidence interval; AHR, adjusted hazard ratio; AJCC, American Joint Committee on Cancer; RT, radiotherapy. * p values were estimated by Cox’s regression. † p values were adjusted for cell differentiation (moderate + poor vs. well) and AJCC pathological stage (stage III + IV vs. stage I + II) by multivariate Cox’s regression. Bold values denote statistically significant.
4. Discussion
UBE2C is involved in tumor progression and is considered a potential cancer biomarker [21,22]. However, its role has not been reported in OSCC, especially for TSCC. In the present study, we first indicated that (1) the expression level of UBE2C was higher in tumor tissues of OSCC patients than it was in CTAN tissues; (2) the high expression level of UBE2C in tumor tissues was associated with poor cell differentiation and lymph node invasion in OSCC patients, especially in BMSCC and TSCC patients; (3) the high expression level of UBE2C in tumor tissues was associated with shorter DSS in TSCC patients with poor cell differentiation, advanced pathological stage, lymph node metastasis and postoperative radiation therapy; (4) UBE2C was involved in cell proliferation, migration, invasion, colony formation and cancer stemness of OSCC cells; and (5) the high co-expression levels of UBE2C and cancer stemness markers such as CD44, CD166 and EpCAM were associated with poor prognosis in oral cancer patients including TSCC patients from TCGA database. These findings suggested the potential clinical significance and roles of UBE2C in OSCC.
Overexpression of UBE2C is associated with tumorigenesis and tumor progression in various types of cancer. For example, UBE2C is involved in the tumorigenesis of colorectal cancer [23] and non–small cell lung cancer (NSCLC) [24]. UBE2C is highly expressed in breast microcalcification lesions [25] and its overexpression is correlated with relapse in early HR+/HER2− breast cancer patients [9]. UBE2C expression is elevated and predicts poor prognosis in hepatocellular carcinoma, esophageal squamous cell carcinoma and intestinal-type gastric cancer [14,26,27]. UBE2C promotes the progression of HNSCC [11] and nasopharyngeal carcinoma progression [28]. High expression of UBE2C is associated with the tumor progression and unfavorable outcome in patients with malignant glioma [29]. UBE2C expression was positively correlated with unfavorable overall survival in ovarian and bladder cancers [30,31]. High expression levels of UBE2C were correlated with high rates of tumor recurrence in meningiomas [32]. Similarly, our results showed that UBE2C was highly expressed in the tumor tissues of OSCC patients and that a high expression level of UBE2C was associated with shorter DSS in TSCC patients having poor cell differentiation, lymph node metastasis and postoperative radiation therapy, indicating that UBE2C might be a potential diagnostic and prognostic biomarker in OSCC patients, especially in TSCC patients with certain clinicopathological outcomes.
UBE2C is a key regulator of cell cycle progression in various types of cancers, such as NSCLC, gastric cancer and breast cancer [33]. Silencing UBE2C arrests cell cycle progression at the G1/S phases, inhibits cell proliferation in pancreatic ductal adenocarcinoma [34] and blocks the G2/M transition in melanoma [12]. Moreover, the downregulation of UBE2C suppresses cell proliferation in osteosarcoma by decreasing the expression of the cell cycle-related protein Ki-67 [35]. UBE2C overexpression accelerates the proliferation of colon cancer by alternating the cell cycle profile [36]. On the other hand, knockdown of UBE2C arrests G2/M phase of cell cycle and enhances cell apoptosis through induction of Bax/p53 and downregulation of Bcl-2 [37]. Our results indicated that silencing UBE2C decreased the viability of OSCC cells (Figure 2) but further verification is needed to determine whether UBE2C is involved in cell proliferation or apoptosis.
UBE2C also modulates migration and invasion in various types of cancers, such as hepatocellular carcinoma [13], NSCLC [7,24], HNSCC [11] and pancreatic ductal adenocarcinoma [34]. The expression of UBE2C is correlated with lymphatic metastasis and serosa invasion [15]. Moreover, UBE2C induces Wnt/β-catenin and PI3K/Akt signaling to regulate phosphorylation levels of Aurora-A for epithelial–mesenchymal transition (EMT) in gastric adenocarcinoma [38]. UBE2C promotes EMT via p53 and p21 in endometrial cancer [39]. Our results indicated that UBE2C was involved in cell invasion and migration in OSCC cells (Figure 3); higher UBE2C expression was associated with lymph node metastasis in OSCC patients (Supplementary Table S1) and with poor DSS in TSCC patients having lymph node metastasis (Table 3). These results indicated that UBE2C was associated with TSCC metastasis but their molecular mechanisms regarding metastasis also need to be further investigated.
Knockdown of UBE2C sensitizes epirubicin- and docetaxel-resistant breast cancer cells to chemotherapeutic agents [40]. Moreover, UBE2C induces cisplatin resistance in NSCLC cells [41]. These findings suggested that UBE2C plays an important role in drug resistance. However, the role of UBE2C in the drug resistance of OSCC is not clear. Cancer stem cells are responsible for drug resistance and to contribute to incomplete therapeutic responses of tumors [20]. Cancer stemness markers such as ALDH1/A2, CD44, CD166 and EpCAM are associated with drug resistance in cancers [42]. Among them, CD44 is involved in drug resistance in oral cancer [43]. Our results indicated that knockdown of UBE2C decreased the expression levels of these cancer stemness markers including ALDH1A2, CD44, CD166 and EpCAM in both Ca9-22 and SAS cells (Figure 4). However, reduced expression levels of ADLH/A1 and ABCG2 stemness markers were only found in UBE2C-knockdown Ca9-22 cells but not in UBE2C-knockdown SAS cells, which indicating that UBE2C might play a more important role in cancer stemness in tongue carcinoma. Moreover, high co-expression of UBE2C/CD44, UBE2C/CD166 and UBE2C/EpCAM was associated with recurrence in oral cancer patients including TSCC patients (Table 3). These results indicated that UBE2C might be involved in stemness–related properties of drug resistance in TSCC.
Indeed, UBE2C expression is associated in worse survival in several cancer types except oral cancer (https://www.proteinatlas.org/ENSG00000175063-UBE2C), suggesting low discrimination of UBE2C expression between different forms of cancers. Nevertheless, we further found that high co-expression of UBE2C and cancer stemness markers, including CD44, D166 and EpCAM, have even worse poor prognosis in TSCC patients, which provided potential diagnostic and prognostic markers in oral cancer patients. Although our study indicated that UBE2C played the role in cancer stemness through regulating several cancer stemness markers including ALDH1A2, CD44, CD166 and EpCAM in oral cancer, their detailed molecular mechanisms for cancer stemness are still largely unresolved, which require further work to elucidate. More studies will be focused on investigating how UBE2C regulating these cancer markers including ALDH1A2, CD44, CD166 and EpCAM and the relationship between these cancer markers for cancer stemness in vitro. The regulation pathway of UBE2C-modulating cancer stemness will be further validated in oral cancer patient. Moreover, we found that UBE2C expression was associated with worse survival in TSCC patients. To further verify the clinical significance of UBE2C in tumor prognosis of oral cancer, the data from more independent cohorts or using external databases, such as The International Cancer Genome Consortium (ICGC) and the Gene Expression Omnibus (GEO), which will be applied for further analysis.
5. Conclusions
In conclusion, our study reported that UBE2C expression was associated with cell growth, invasion/migration and cancer stemness in TSCC. Moreover, a high level of UBE2C expression was associated with tumorigenesis, poor cell differentiation and lymph node invasion in OSCC patients and with poor DSS in TSCC patients having poor cell differentiation, advanced pathological stages and lymph node metastasis and receiving radiation therapy. These new findings indicated that UBE2C may be a potential diagnostic and prognostic biomarker for TSCC patients with certain clinicopathological outcomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4418/10/9/674/s1, Table S1. Association of UBE2C expression with clinicopathologic outcomes in three primary subsites of OSCC patients.
Author Contributions
Wrote manuscript, P.-F.L., conducted experiments, C.-F.C., H.-M.C., contributed materials and reagents, C.-W.S., C.-L.C., L.-P.G., performed the data analysis, C.-H.L., performed IHC and IHC scoring, H.-H.L., designed the project and experiments, P.-F.L., B.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Technology (MOST 108-2320-B-037-038, MOST 109-2320-B-037 -015 -MY3), Kaohsiung Medical University Research Foundation (KMU-Q109008), NSYSU-KMU Joint Research Project (NSYSUKMU 109-I007) and Kaohsiung Veterans General Hospital (VGHKS107-134).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Liu P.-F., Chen H.-C., Cheng J.-S., Tsai W.-L., Lee H.-P., Wang S.-C., Peng W.-H., Lee C.-H., Ger L.-P., Shu C.-W. Association of ATG4B and Phosphorylated ATG4B Proteins with Tumorigenesis and Prognosis in Oral Squamous Cell Carcinoma. Cancers. 2019;11:1854. doi: 10.3390/cancers11121854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y., Choung H., Lee J., Rhyu I., Kim H.-D. Association of Periodontitis with Oral Cancer: A Case-Control Study. J. Dent. Res. 2019;98:526–533. doi: 10.1177/0022034519827565. [DOI] [PubMed] [Google Scholar]
- 3.Rahman R., Poomsawat S., Juengsomjit R., Buajeeb W. Overexpression of Epstein-Barr virus-encoded latent membrane protein-1 (LMP-1) in oral squamous cell carcinoma. BMC Oral Health. 2019;19:142. doi: 10.1186/s12903-019-0832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding D., Stokes W., Eguchi M., Hararah M., Sumner W., Amini A., Goddard J., Somerset H., Bradley C., McDermott J., et al. Association Between Lymph Node Ratio and Recurrence and Survival Outcomes in Patients With Oral Cavity Cancer. JAMA Otolaryngol. Neck Surg. 2019;145:53. doi: 10.1001/jamaoto.2018.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimura H., Yoshida H., Matsuda S., Ryoke T., Ohta K., Ohmori M., Yamamoto S., Kiyoshima T., Kobayashi M., Sano K. The therapeutic potential of epigallocatechin-3-gallate against human oral squamous cell carcinoma through inhibition of cell proliferation and induction of apoptosis: In vitro and in vivo murine xenograft study. Mol. Med. Rep. 2019;20:1139–1148. doi: 10.3892/mmr.2019.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L., Ding Z., Huang N., Huang Z., Zhang N., Xia Z. Forkhead Box M1 positively regulates UBE2C and protects glioma cells from autophagic death. Cell Cycle. 2017;16:1705–1718. doi: 10.1080/15384101.2017.1356507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo J., Wu Y., Du J., Yang L., Chen W., Gong K., Dai J., Miao S., Jin D., Xi S. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenisis. 2018;7:49. doi: 10.1038/s41389-018-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj S., Alam S.K., Roy K.S., Datta A., Nath S., Roychoudhury S. E2 Ubiquitin-conjugating Enzyme, UBE2C Gene, Is Reciprocally Regulated by Wild-type and Gain-of-Function Mutant p53*. J. Boil. Chem. 2016;291:14231–14247. doi: 10.1074/jbc.M116.731398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y.-J., Lee G., Han J., Song K., Choi J.-S., Choi Y.-L., Shin Y.K. UBE2C Overexpression Aggravates Patient Outcome by Promoting Estrogen-Dependent/Independent Cell Proliferation in Early Hormone Receptor-Positive and HER2-Negative Breast Cancer. Front. Oncol. 2020;9:9. doi: 10.3389/fonc.2019.01574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Zhi X., Shen X., Chen C., Yuan L., Dong X., Zhu C., Yao L., Chen M. Depletion of UBE2C reduces ovarian cancer malignancy and reverses cisplatin resistance via downregulating CDK1. Biochem. Biophys. Res. Commun. 2020;523:434–440. doi: 10.1016/j.bbrc.2019.12.058. [DOI] [PubMed] [Google Scholar]
- 11.Jin Z., Zhao X., Cui L., Xu X., Zhao Y., Younai F., Messadi D., Hu S. UBE2C promotes the progression of head and neck squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2020;523:389–397. doi: 10.1016/j.bbrc.2019.12.064. [DOI] [PubMed] [Google Scholar]
- 12.Liu G., Zhao J., Pan B., Ma G., Liu L. UBE2C overexpression in melanoma and its essential role in G2/M transition. J. Cancer. 2019;10:2176–2184. doi: 10.7150/jca.32731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y., Lu J., Fang Q., Lu Y., Xie C., Wu H., Yin Z. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci. Rep. 2019;39:39. doi: 10.1042/BSR20182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Liu X., Yu G., Liu L., Wang J., Chen X., Bian Y., Ji Y., Zhou X., Chen Y., et al. UBE2C Is a Potential Biomarker of Intestinal-Type Gastric Cancer With Chromosomal Instability. Front. Pharmacol. 2018;9:847. doi: 10.3389/fphar.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H.-Q., Zhao G., Ke B., Ma G., Liu G.-L., Liang H., Liu L.-R., Hao X.-S. Overexpression of UBE2C correlates with poor prognosis in gastric cancer patients. Eur. Rev. Med. Pharmacol. Sci. 2018;22:1665–1671. doi: 10.26355/eurrev_201803_14578. [DOI] [PubMed] [Google Scholar]
- 16.Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA A Cancer J. Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 17.Chen H., Tseng Y., Shu C., Weng T., Liou H., Yen L., Hsieh I., Wang C., Wu P., Shiue Y., et al. Differential clinical significance of COL 5A1 and COL 5A2 in tongue squamous cell carcinoma. J. Oral Pathol. Med. 2019;48:468–476. doi: 10.1111/jop.12861. [DOI] [PubMed] [Google Scholar]
- 18.Chen H.C., Tseng Y.K., Shu C.W., Fu T.Y., Liou H.H., Huang C.H., Chen C.C., Wang J.S., Wu P.C., Ger L.P., et al. Prognostic role of RECK in pathological outcome-dependent buccal mucosa squamous cell carcinoma. Oral Dis. 2020;26:62–71. doi: 10.1111/odi.13214. [DOI] [PubMed] [Google Scholar]
- 19.Guo J., Jin D., Wu Y., Yang L., Du J., Gong K., Chen W., Dai J., Miao S., Xi S. The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells. EBioMedicine. 2018;35:204–221. doi: 10.1016/j.ebiom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Ling M., Yang X. Correlation between cancer stem cells (CSCs) and tumor- infiltrating lymphocytes (TILs): Do TILs interact with CSCs in non- small cell lung cancer? Ann. Transl. Med. 2020;8:914. doi: 10.21037/atm-20-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen R., Wu T., Huang P., Shao Q., Chen M. The clinicopathological significance of ubiquitin-conjugating enzyme E2C, leucine-rich repeated-containing G protein-coupled receptor, WW domain-containing oxidoreductase, and vasculogenic mimicry in invasive breast carcinoma. Medicine. 2019;98:e15232. doi: 10.1097/MD.0000000000015232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie C., Powell C., Yao M., Wu J., Dong Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int. J. Biochem. Cell Boil. 2014;47:113–117. doi: 10.1016/j.biocel.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Li S.-Z., Song Y., Zhang H.-H., Jin B.-X., Liu Y., Liu W.-B., Zhang X.-D., Du R.-L. UbcH10 overexpression increases carcinogenesis and blocks ALLN susceptibility in colorectal cancer. Sci. Rep. 2014;4:6910. doi: 10.1038/srep06910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallante P., Malapelle U., Berlingieri M.T., Bellevicine C., Sepe R., Federico A., Rocco D., Galgani M., Chiariotti L., Sanchez-Cespedes M., et al. UbcH10 overexpression in human lung carcinomas and its correlation with EGFR and p53 mutational status. Eur. J. Cancer. 2013;49:1117–1126. doi: 10.1016/j.ejca.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 25.Chou C.-P., Huang N.-C., Jhuang S.-J., Pan H.-B., Peng N.-J., Cheng J.-T., Chen C.-F., Chen J.-J., Chang T.-H. Ubiquitin-Conjugating Enzyme UBE2C Is Highly Expressed in Breast Microcalcification Lesions. PLoS ONE. 2014;9:e93934. doi: 10.1371/journal.pone.0093934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Zhao Y., Shi X., Fan C. Ube2s expression is elevated in hepatocellular carcinoma and predicts poor prognosis of the patients. Int. J. Clin. Exp. Pathol. 2018;11:781–787. [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto A., Ishibashi Y., Urashima M., Omura N., Nakada K., Nishikawa K., Shida A., Takada K., Kashiwagi H., Yanaga K. High UBCH10 protein expression as a marker of poor prognosis in esophageal squamous cell carcinoma. Anticancer. Res. 2014;34:955–961. [PubMed] [Google Scholar]
- 28.Shen Z., Jiang X., Zeng C., Zheng S., Luo B., Zeng Y., Ding R., Jiang H., He Q., Guo J., et al. High expression of ubiquitin-conjugating enzyme 2C (UBE2C) correlates with nasopharyngeal carcinoma progression. BMC Cancer. 2013;13:192. doi: 10.1186/1471-2407-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma R., Kang X., Zhang G., Fang F., Du Y., Lv H. High expression of UBE2C is associated with the aggressive progression and poor outcome of malignant glioma. Oncol. Lett. 2016;11:2300–2304. doi: 10.3892/ol.2016.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Y., Wang D., Lin L., Dai J., Yu L. The expression of ubiquitin-conjugating enzyme E2C and KAI1 in ovarian carcinoma and their clinical significance. Medicine. 2019;98:e17896. doi: 10.1097/MD.0000000000017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morikawa T., Kawai T., Abe H., Kume H., Homma Y., Fukayama M. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int. J. Clin. Exp. Pathol. 2013;6:1367–1374. [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang L., Wang T., Bao Y., Qian J., Wu X.-J., Hu G., Lu Y. A study of UbcH10 expression and its association with recurrence of meningiomas. J. Surg. Oncol. 2011;106:327–331. doi: 10.1002/jso.22141. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Gao L., Wang C., Wang S., Sun D., Li X., Liu M., Qi Y., Liu J., Lin B. Combining Bioinformatics and Experiments to Identify and Verify Key Genes with Prognostic Values in Endometrial Carcinoma. J. Cancer. 2020;11:716–732. doi: 10.7150/jca.35854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Yin L., Yang L., Zheng Y., Liu S., Yang J., Cui H., Wang H. Silencing ubiquitin-conjugating enzyme 2C inhibits proliferation and epithelial–mesenchymal transition in pancreatic ductal adenocarcinoma. FEBS J. 2019;286:4889–4909. doi: 10.1111/febs.15134. [DOI] [PubMed] [Google Scholar]
- 35.Wang S., Li D., Li J., Fang J., Li H., Tong P., Liu F. Lentivirus-mediated RNA interference targeting UbcH10 reduces cell growth and invasion of human osteosarcoma cells via inhibition of Ki-67 and matrix metalloproteinases. Oncol. Lett. 2015;9:2171–2176. doi: 10.3892/ol.2015.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujita T., Ikeda H., Taira N., Hatoh S., Naito M., Doihara H. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer. 2009;9:87. doi: 10.1186/1471-2407-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang L., Bao Y., Luo C., Hu G., Huang C., Ding X., Sun K., Lu Y.-C. Knockdown of ubiquitin-conjugating enzyme E2C/UbcH10 expression by RNA interference inhibits glioma cell proliferation and enhances cell apoptosis in vitro. J. Cancer Res. Clin. Oncol. 2009;136:211–217. doi: 10.1007/s00432-009-0651-z. [DOI] [PubMed] [Google Scholar]
- 38.Wang R., Song Y., Liu X., Wang Q., Wang Y., Li L., Kang C., Zhang Q. UBE2C induces EMT through Wnt/betacatenin and PI3K/Akt signaling pathways by regulating phosphorylation levels of Aurora-A. Int. J. Oncol. 2017;50:1116–1126. doi: 10.3892/ijo.2017.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Zhao R., Chi S., Zhang W., Xiao C., Zhou X., Zhao Y., Wang H. UBE2C Is Upregulated by Estrogen and Promotes Epithelial–Mesenchymal Transition via p53 in Endometrial Cancer. Mol. Cancer Res. 2019;18:204–215. doi: 10.1158/1541-7786.MCR-19-0561. [DOI] [PubMed] [Google Scholar]
- 40.Wang C., Pan Y.-H., Shan M., Xu M., Bao J.-L., Zhao L.-M. Knockdown of UbcH10 Enhances the Chemosensitivity of Dual Drug Resistant Breast Cancer Cells to Epirubicin and Docetaxel. Int. J. Mol. Sci. 2015;16:4698–4712. doi: 10.3390/ijms16034698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., Jin D., Wang X., Du J., Di W., An J., Shao C., Guo J. UBE2C Induces Cisplatin Resistance via ZEB1/2-Dependent Upregulation of ABCG2 and ERCC1 in NSCLC Cells. J. Oncol. 2019;2019:8607859. doi: 10.1155/2019/8607859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satar N.A., Fakiruddin K.S., Lim M.N., Mok P.L., Zakaria N., Fakharuzi N.A., Rahman A.Z.A., Zakaria Z., Yahaya B.H., Baharuddin P. Novel triple-positive markers identified in human non-small cell lung cancer cell line with chemotherapy-resistant and putative cancer stem cell characteristics. Oncol. Rep. 2018;40:669–681. doi: 10.3892/or.2018.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naik P.P., Mukhopadhyay S., Panda P.K., Sinha N., Das C.K., Mishra R., Patil S., Bhutia S.K. Autophagy regulates cisplatin-induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 2018:51. doi: 10.1111/cpr.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.