Abstract
Episodic ataxia type 2 (EA2) is an autosomal dominant neurological disorder characterized by paroxysmal attacks of ataxia, vertigo, and nausea that usually last hours to days. It is caused by loss-of-function mutations in CACNA1A, the gene encoding the pore-forming α1 subunit of P/Q-type voltage-gated Ca2+ channels. Although pharmacological treatments, such as acetazolamide and 4-aminopyridine, exist for EA2, they do not reduce or control the symptoms in all patients. CACNA1A is heavily spliced and some of the identified EA2 mutations are predicted to disrupt selective isoforms of this gene. Modulating splicing of CACNA1A may therefore represent a promising new strategy to develop improved EA2 therapies. Because RNA splicing is dysregulated in many other genetic diseases, several tools, such as antisense oligonucleotides, trans-splicing, and CRISPR-based strategies, have been developed for medical purposes. Here, we review splicing-based strategies used for genetic disorders, including those for Duchenne muscular dystrophy, spinal muscular dystrophy, and frontotemporal dementia with Parkinsonism linked to chromosome 17, and discuss their potential applicability to EA2.
Keywords: episodic ataxia type 2, P/Q-type Ca2+ channels, alternative splicing, antisense oligonucleotides, SMaRT, CRISPR/Cas9
1. Introduction
Episodic ataxia type 2 (EA2) is an autosomal dominant neurological disorder characterized by recurrent disabling attacks of imbalance, vertigo, nausea and ataxia, typically lasting hours to days [1,2]. Symptoms may also include fatigue, migraine headaches, and visual disturbances. Nystagmus commonly occurs between attacks and progressive cerebellar atrophy can also be observed. The frequency of attacks ranges from once a year to four times a week. Ataxic episodes can be triggered by emotional stress, physical exercise, alcohol intake, or fever. Although late-onset cases have been reported, the onset is typically in childhood or early adolescence [3,4]. Acetazolamide and 4-aminopyridine can reduce or control the symptoms in some patients. However, treatments are sometimes discontinued because the drugs are either no longer effective or because the patients develop adverse effects to them [5].
EA2 is caused by mutations in CACNA1A, the gene encoding the pore-forming α1 subunit of P/Q-type voltage-gated Ca2+ channels (VGCCs; CaV2.1) [6,7]. These channels are enriched in the cerebellum where they control neurotransmitter release in cooperation with N-type (CaV2.2) and R-type (CaV2.3) VGCCs. P/Q-type channels are however more efficient than the other two types of VGCCs in supporting synaptic transmission at mature synapses, partly because they are more tightly coupled to the neurotransmitter release machinery [8,9,10]. The CACNA1A gene, located on chromosome 19p13, contains 47 exons, many of which are subject to alternative splicing (AS). As a consequence, the total number of CACNA1A splice isoforms is estimated to be in the order of thousands. Moreover, functional studies indicate that some of them exhibit differential properties and expression patterns [7,11]. This creates a large molecular variability that is thought to optimize Ca2+ signaling to specific cellular tasks.
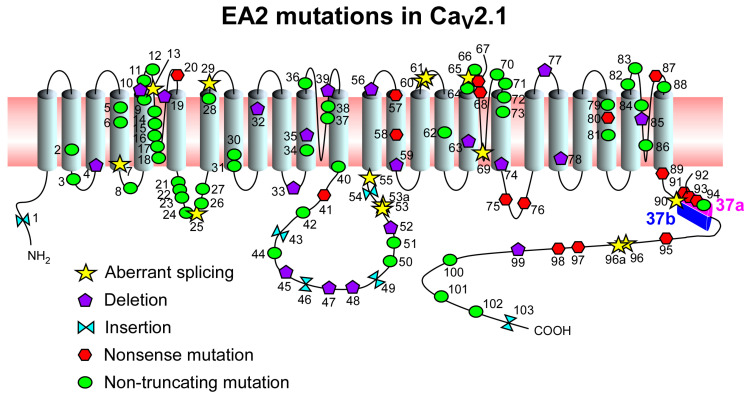
Over 100 different mutations can cause EA2 (Figure 1; Table 1). Some of them are nonsense loss-of-function mutations that disrupt the open reading frame of CACNA1A and result in truncated channels generating little or no current. EA2 mutations are found in all regions of the channel but most of them reside in the pore loop region (Figure 1) [4,6,12]. Interestingly, some mutations are predicted to induce aberrant splicing.
Figure 1.
Scheme of human CaV2.1 highlighting the position of 103 mutations causing EA2. Topology of CaV2.1 as for Uniprot entry O00555. The two mutually exclusive exons 37a and 37b are shown in pink and blue, respectively. The 103 mutations are depicted in the scheme and listed in Table 1.
Table 1.
List of CaV2.1 mutations causing EA2.
| No. | Amino Acid | DNA Mutation | References |
|---|---|---|---|
| 1 | p.(Ala56Serfs*20) | c.165dupA | [4] |
| 2 | p.Glu147Lys | c.439G>A | [27] |
| 3 | p.Gly162Val | c.485G>T | [28] |
| 4 | p.(Trp168Glyfs*10) | c.504delC | [4] |
| 5 | p.Arg192Trp | c.574C>T | [29] |
| 6 | p.Arg198Gln | c.593G>A | [30] |
| 7 | c.868+5G>A; possible aberrant splicing | [16] | |
| 8 | p.Ser218Leu | c.653C>T | [30] |
| 9 | p.Tyr248Asn/Cys | c.742T>A/c.743A>G | [31,32] |
| 10 | p.Gly250Glufs*60 | c.749delG | [12] |
| 11 | p.His253Tyr | c.1032C >T | [33] |
| 12 | p.(Cys256Arg) | c.1041T>C | [34] |
| 13 | c.983-1G>A; aberrant splicing | [4] | |
| 14 | p.Arg279Cys | c.835C>T | [28] |
| 15 | p.Cys287Tyr | c.1096G>A | [1] |
| 16 | p.Gly293Arg | c.1152G>A | [35] |
| 17 | p.Gly297Arg | c.889G>A | [36] |
| 18 | p.Asp302Asn | c.904G>A | [28] |
| 19 | p.Thr310 fs*5 | c.928_931delACTG | [28] |
| 20 | p.Trp320* | c.959G>A | [12] |
| 21 | p.Arg387Gly | c.1159C>G | [28] |
| 22 | p.Glu388Lys | c.1161G>A | [37] |
| 23 | p.(Leu389Phe) | c.1165C>T | [4] |
| 24 | p.Gly411Trp | c.1231G>T | [28] |
| 25 | c.1253+1G>A; probable aberrant splicing | [15] | |
| 26 | p.Ala454Thr | c.1360G>A | [38] |
| 27 | p.Arg455Gln | c.1364G>A | [39] |
| 28 | p.(Thr501Met) | c.1502C>T | [4] |
| 29 | c.1557+1G>A; aberrant splicing | [13] | |
| 30 | p.Glu533Lys | c.1597G>A | [40] |
| 31 | p.Gly540Arg | c.1618G>A | [41] |
| 32 | p.Val558Serfs*13 | c.1672-1_1675delGGTTA | [28] |
| 33 | p.Leu600 fs*41 | c.1799_1800delTC | [28] |
| 34 | p.Leu621Arg | c.2144T>G | [41] |
| 35 | p.Leu624Phe | c.1870-1873del | [13] |
| 36 | p.Gly638Asp | c.1913G>A | [42] |
| 37 | p.Trp670Cys | c.2010G>C | [30] |
| 38 | p.Gly677Glu | c.2030G>A | [31] |
| 39 | p.(Gln681Argfs*100) | c.2042_2043del | [33] |
| 40 | p.Ile712Val | c.2134A>G | [43] |
| 41 | p.Gln736* | c.2206C>T | [44] |
| 42 | p.(Met798Thr) | c.2393T>C | [4] |
| 43 | p.(Arg822Profs*246) | c.2464dupC | [4] |
| 44 | p.(Pro897Arg) | c.2690C>G | [4] |
| 45 | p.Gly939* | c.2816delG | [45] |
| 46 | p.Ser943Gln | c.2825+1insG | [46] |
| 47 | p.(Ala952Serfs*115) | c.2852_2861del | [4] |
| 48 | p.(Arg957Aspfs*113) | c.2867_2869del | [16] |
| 49 | p.Glu1004Argfs*66 | c.3244+1insG | [1] |
| 50 | p.Glu998Gln | c.2992G>C | [31] |
| 51 | p.Gln1154* | c.3460C>T | [45] |
| 52 | p.Cys1178Pro | c.3531delC | [13] |
| 53 | c.3089+2T; possible aberrant splicing | [17] | |
| 53a | c.3102+2T; possible aberrant splicing | [30] | |
| 54 | c.3603dupC | [30] | |
| 55 | c.3977+1G>A; aberrant splicing | [14] | |
| 56 | p.Pro1267Leu | c.4073delC | [6] |
| 57 | p.(Arg1278*) | c.3832C<T | [16] |
| 58 | p.Arg1281* | c.4077C>T | [1] |
| 59 | p.Glu1294Del | c.3871_3873delGAG | [31] |
| 60 | c.4270+1G>A; aberrant splicing |
[6] | |
| 61 | c.4001+3insT; cryptic splice donor site in exon 24 |
[18] | |
| 62 | p.Arg1350Gln | c.4049G>A | [47] |
| 63 | p.Phe1394Leu | c.4182delC | [33] |
| 64 | p.Phe1404Cys | c.4486T>G | [48] |
| 65 | c.4261+1G>A; aberrant splicing | [13] | |
| 66 | p.Arg1433Gln | c.4298G>A | [49] |
| 67 | p.Tyr1443* | c.4331C>G | [13] |
| 68 | p.Trp1451* | c.4588G>A | [1] |
| 69 | c.4636+1G>T; aberrant splicing | [1] | |
| 70 | p.(Gly1483Arg) | c.4722G>A | [34] |
| 71 | p.Met1488_Ser1489del | c.4739_4744del | [34] |
| 72 | p.Phe1491Ser | c.4747T>C | [50] |
| 73 | p.(Val1494Ile) | c.4755G>A | [34] |
| 74 | p.Phe1503Del | c.4509-11delCTT | [13] |
| 75 | p.Arg1549* | c.4645C>T | [51] |
| 76 | p.Gln1561* | c.4963C>T | [52] |
| 77 | p.Tyr1594Del | c.4778-80delCTT | [13] |
| 78 | p.Val1620Ser | c.4854delG | [46] |
| 79 | p.Arg1666His/Gln | c.5260G>A/c.4991G>A | [53,54] |
| 80 | p.Arg1669* | c.5005C>T | [31] |
| 81 | p.(Arg1680Cys) | c.5038C>T | [4] |
| 82 | p.His1737Leu | c.5211A>T | [55] |
| 83 | p.Leu1749Pro | c.5246T>C | [28] |
| 84 | p.Arg1751Trp | c.5251C>T | [45] |
| 85 | p.Ser1753Cysfs*2 | c.5253-2259_5403+1135del | [12] |
| 86 | p.Glu1757Lys | c.5271G>A | [56] |
| 87 | p.Arg1785* | c.5589C>T | [1] |
| 88 | p.Ser1799Leu | c.5396C>T | [57] |
| 89 | p.Arg1824* | c.5733C>T | [58] |
| 90 | c.IVS36+2T>C (c.4582+2T>C); possible aberrant splicing |
[19] | |
| 91 | p.Tyr1849* | c.5547T>A | [12] |
| 92 | p.Tyr1854* | c.5562C>G | [26] |
| 93 | p.Arg1858* | c.5571C>T | [26] |
| 94 | p.(Cys1870Arg) | c.5608T>C | [4] |
| 95 | p.(Glu1927*) | c.5779G>T | [4] |
| 96 | c.IVS41+(3–6)delGAGT (c.6068+(3-6)delGAGT); aberrant splicing | [18] | |
| 96a | c.6335+4delAGTG; aberrant splicing |
[14] | |
| 97 | p.Gln1986* | c.5956C>T | [31] |
| 98 | p.Gln2039* | c.6351C>T | [1] |
| 99 | p.Pro2058Leufs*69 | c.6404delC | [1] |
| 100 | p.Arg2090Gln | c.6269G>A | [59] |
| 101 | p.(Arg2136Cys) | c.6681C>T | [34] |
| 102 | p.Pro2222Leu | c.6665C>T | [12] |
| 103 | p.2319Gln[n] | c.7191CAG[n] | [60] |
Note: For most mutations, the numbering is as cited in the original publications. If not present in the original publication it refers to the translated region of the GeneBank access number AF004883.1. The asterisk (*) indicates a stop codon.
For example, G to A substitutions at position +1 of the donor splice sites in exons 6, 11, 21, 24, 26, and 27 are all predicted to form aberrant mRNAs [6,13,14,15]. Likewise, mutations G to A at position +5 of the donor splice site in exon 4, G to A at position −1 in exon 7, T to C at position +2 of the donor splice site in exon 19 and a four-base pair deletion at position +4 of the donor splice site in exon 41 also impair splicing [4,14,16,17]. Further, the insertion of a T at position +3 of intron 24 creates a cryptic splice donor site within exon 24, leading to an aberrant splicing event [18]. While all the above mutations affect constitutive splicing, defects in AS are also important for EA2. Specifically, an A to G substitution at position −2 of the acceptor splice site in the mutually exclusive exon 37a has been found in EA2 patients [19]. This mutation likely impairs the insertion in the final transcript of exon37a while not affecting the inclusion of its mutually exclusive exon 37b.
The mutually exclusive exons 37a and 37b produce two major isoforms of the channel, CaV2.1[EFa] and CaV2.1[EFb], which diverge in an EF-hand-like domain located in the proximal intracellular C-terminus [20,21,22]. The ratio between the synaptic expression of CaV2.1[EFa] and CaV2.1[EFb] is key to controlling synaptic efficacy: CaV2.1[EFa] is tightly coupled to the neurotransmitter release machinery and support efficient vesicle release while CaV2.1[EFb] is loosely coupled to the release machinery and participates in vesicle release only upon repetitive stimulations [7,23,24]. The developmental expression pattern of the two isoforms is also markedly different. Whereas, for CaV2.1[EFb], the expression levels do not undergo major developmental changes, being high in most brain regions from the early stages of postnatal development, for CaV2.1[EFa], the expression levels build up during synapse maturation, presumably contributing to the developmental increase in neurotransmitter release efficiency. As a result, both splice isoforms are expressed at similar levels in most regions of the adult brain [20,21,22,23,25]. In addition to the aforementioned mutation impairing inclusion of exon 37a [19], four loss-of-function mutations have been identified within exon 37a in four unrelated families [4,26], while none has been found, to date, in exon 37b (Figure 1; Table 1). The presence of five mutations impairing selectively CaV2.1[EFa] may therefore suggest a major role of this splice isoform in EA2.
Altogether, these findings indicate that aberrant splicing of CACNA1A may be an important pathogenic mechanism for EA2. Targeting RNA splicing may therefore be a promising approach for developing new EA2 therapies. After reviewing RNA splicing, AS, and splicing-based therapies developed for other diseases, we will discuss the potential benefits of splicing-based therapies for EA2.
2. Alternative Splicing
During the maturation of a pre-mRNA into an mRNA, non-coding introns are removed and exons are joined together, a process known as RNA splicing. By modulating which exons and how they are included in the mature mRNA, it is possible to generate multiple transcripts from a single gene, thereby producing protein isoforms with potentially different functions. This mechanism, known as AS, is a critical determinant of protein diversity. Importantly, more than 90% of human genes are alternatively spliced [61,62].
The splicing reaction is catalyzed by a large complex, the spliceosome, which is composed of five small nuclear RNAs (snRNAs) and more than 200 proteins [63]. snRNAs and proteins combine to form small nuclear ribonucleoproteins (snRNPs) that bind cis-regulatory elements on the pre-mRNA to catalyze the splicing reaction. The core splicing elements include a donor site at the 5′ end of the intron (the exon/intron junction) and an acceptor site at the 3′ end of the intron defined by a polypyrimidine tract and the actual intron/exon junction. The 3′ splice site is functionally coupled to the branch point, which is usually within 40 nucleotides upstream of the 3′ splice site.
Additional cis-regulatory sequences, known as exonic and intronic splicing enhancers/silencers (ESE/S and ISE/S), recruit specific RNA-binding proteins, such as serine/arginine-rich (SR) family proteins and heterogeneous nuclear ribonucleoprotein particles (hnRNPs) [64]. These additional elements facilitate or inhibit the assembly of the spliceosome, thereby regulating RNA splicing and making AS possible [65,66,67].
The most common type of AS is exon skipping, also known as cassette exon, in which a whole exon can be either included in or spliced out of the mRNA. Other types of AS comprise intron retention, in which an intron is included in the mature mRNA, mutually exclusive exons, whereby the transcript contains only one of two (or more) possible exons, and alternative 3′ and 5′ splice site selection, leading to alternate inclusion or exclusion of part of an exon [68,69].
Aberrant splicing is associated with a growing number of diseases [70,71,72]. Mutations affecting the core splice sites generally result in exon skipping or intron retention, which disrupt the reading frame, thus generating truncated non-functional proteins with potentially pathological consequences. In addition, also defects in AS, by mutations in splicing enhancers/silencers or in trans-acting elements, can be pathological. Several therapeutic approaches aiming at correcting aberrant splicing have therefore been developed, as detailed below.
3. Splicing Modulating Therapies
3.1. Antisense Oligonucleotides (AONs)
The antisense oligonucleotide (AON) approach is based on short oligonucleotides that bind the pre-mRNA and modify gene expression by either promoting mRNA degradation or interfering with the splicing process [73,74,75,76]. RNA-degrading AONs typically contain a phosphorothioate (PS) backbone modification that increases their stability and promotes the recruitment of the RNase H to induce cleavage of the target RNA. Other modifications, in the phosphodiester backbone and at the 2′ position of the sugar moiety, are also possible for improved AON properties [76,77]. For example, the morpholino backbone modification promotes highly stable RNA complementarity without inducing degradation of the target sequence. Likewise, 2′-O-methyl (2′-O-Me) and 2′-O-methoxyethyl (2′MOE) modifications enhance AON binding to the mRNA while inhibiting RNase H cleavage. AONs with modifications that protect from RNA degradation can regulate splicing if they are designed to recognize intron/exon junctions or splicing regulatory elements [74,78,79,80] because they prevent binding of splicing factors, thereby inducing exon skipping or forcing the inclusion of alternatively spliced exons [74,81]. Several AON-based therapeutic approaches, some of which are currently in clinical trials, have been developed for pathologies in which splicing is defective [82], as further discussed below.
3.1.1. AON-Mediated Exon Inclusion
One disease for which AONs have been used to promote exon inclusion is spinal muscular atrophy (SMA; Table 2). SMA is a recessive, progressive, neuromuscular disorder caused by mutations in the survival motor neuron 1 (SMN1) gene [83]. It results in motor neuron degeneration and subsequent muscular atrophy leading to paralysis and respiratory defects, ultimately reducing the life expectancy of patients. Depending on the age of onset and level of motor function, SMA can be classified into four different subtypes SMA types I–IV [84,85,86,87]. Humans have a paralogous of SMN1, referred to as SMN2, which differs from SMN1 in only few bases. Interestingly, the severity of SMA is inversely correlated to SMN2 expression levels [88,89]. SMN2 fails however to fully compensate for the lack of SMN1 because of a C-to-T substitution at position 6 of exon 7, which disrupts an exonic splicing enhancer, thus promoting skipping of this exon with consequent formation of an unstable protein [86,90,91]. Instead of trying to correct the SMN mutations in SMN1, researchers have designed AONs targeting an intronic splicing silencer (ISS) in SMN2 to prevent the binding of negative splicing factors, thus favoring exon 7 inclusion in the SMN2 mRNA. This increases the amount of SMN protein in human fibroblasts and in the mouse brain [92,93,94]. Following these results, nusinersen, a 2′-O-methyl-modified ribose AON with a full-length phosphorothioate backbone (2′OMePS), which increases the inclusion of exon 7 in the SMN2 mRNA, has been delivered in patients using intrathecal injections. These clinical trials indicated that the drug is generally well tolerated and effective in alleviating SMA symptoms and improving motor functions [95,96,97,98,99]. In addition, investigations of neuronal tissue in treated deceased patients have revealed a two- to six-fold increase in SMN2 transcripts containing exon 7, as well as an increase in SMN protein, as compared to untreated patients [100]. Following these positive outcomes, nusinersen has been approved as SMA treatment by the US Food and Drug Administration (FDA) in 2016 and by the European Medicines Agency in 2017 [100]. Ongoing studies are evaluating the long-term safety and tolerability of nusinersen and its effects on presymptomatic patients [101].
Table 2.
Examples of Splicing-based Therapeutic Approaches.
| Disease | Gene | Aberrant Splicing | Therapy | References |
|---|---|---|---|---|
| SMA | Mutated: SMN1 Therapy: SMN2 |
Skipping of exon 7 due to C > T mutation at position 6 of exon 7 | 2′OMePS AONs targeting ISS-N1 to promote exon 7 inclusion in human fibroblasts and mouse brain | [92,93,94] |
| Intrathecal injection of nusinersen to increase exon 7 inclusion in patients | [95,96,97,98,99,100] | |||
| Splice switching and lncRNA-targeting AONs to promote inclusion of exon 7 in patient-derived cells and mouse brain | [102] | |||
| Trans-splicing-mediated insertion of exon 7 in fibroblasts and SMAΔ7 mice | [145,146,147,148] | |||
| CRISPR/Cas9-mediated disruption of intronic splicing silencers to enhance exon 7 inclusion in iPSC-derived motor neurons and mice | [153] | |||
| DMD | DMD | Formation of nonfunctional truncated dystrophin due to point mutations and frame-shifting deletions | 2′OMePS- or PMO-mediated skipping of exon 51 in patients | [109,110,111,112,113,114,115] |
| tcDNA-mediated exon skipping in mice | [116,117] | |||
| Restoration of open reading frame by dual Cas9-mediated exon skipping in mdx mice | [155,156,157] | |||
| dCas9-cytidine deaminase-mediated base editing to induce skipping of exon 50 and restore reading frame in iPSCs | [161] | |||
| FTDP-17 | MAPT | Increased exon 10 inclusion | Trans-splicing to reverse aberrant exon 10 inclusion in mice | [142,143,144] |
| dCas13 targeted to exon 10 splice sites on pre-mRNA to mediate exon exclusion in iPSCs-derived cortical neurons | [164] | |||
| Ataxia telangiectasia | ATM | Activation of cryptic splice sites | AMOs to mask cryptic splice sites and restore correct splicing in cell lines | [123,124] |
| Hutchinson-Gilford progeria syndrome | LMNA | Activation of an exonic cryptic donor splice site in exon 11 | AMOs to block the recognition of cryptic splice sites in fibroblasts and mice | [128,129] |
| CF | CFTR | Three base-pair deletion in exon 10 causing deletion of F508 | Correction of exon 10 by trans-splicing in epithelial cells and a xenograft model | [131,136] |
Abbreviations: 2′OMePS: 2’-O-methyl-phosphorothioate RNA; AMO: Antisense morpholino oligonucleotide; AON: Antisense oligonucleotide; CF: cystic fibrosis; DMD: Duchenne muscular dystrophy; FTDP-17: Frontotemporal dementia with parkinsonism linked to chromosome 17; PMO: Phosphorodiamidate antisense morpholino oligonucleotide; SMA: Spinal muscular atrophy.
Interestingly, combining different AONs can have synergic effect. For example, SMN-AS1, a long noncoding RNA (lncRNA) arising from the antisense strand of the SMN genes, represses SMN expression. AONs degrading SMN-AS1 increase SMN expression in patient-derived cells and in the mouse brain. When lncRNA-targeting and splice-switching AONs are combined there is an even larger increase in SMN levels containing exon 7 [102].
3.1.2. AON-Mediated Exon Skipping
AON-based approaches have been used as therapeutic strategy also in Duchenne muscular dystrophy (DMD; Table 2). This is a genetic X-linked recessive disease caused by mutations in the DMD gene that encode the cytoskeletal protein dystrophin. Patients manifest initial symptoms, such as muscular weakness and walking abnormalities, within the first years of life, and lose the ability to walk by the age of 12–14 years. Children with DMD eventually develop also cardiomyopathies and breathing problems due to weakness in respiratory muscles, which eventually lead to death in late adolescence [103,104]. Among inherited pediatric muscular dystrophies, DMD is the most common, with an incidence of 1/3500 male births [105]. Most DMD patients lack a functional dystrophin protein as a consequence of point mutations or frame-shifting deletions/insertions in the DMD gene [106,107]. By contrast, mutations leading to a partially functioning dystrophin protein are associated with the milder Becker muscular dystrophy (BMD) [108].
Over the last years, researchers have developed AON-mediated approaches that promote exon skipping of specific exons in order to restore the open reading frame of DMD. Two main types of compounds have been investigated: drisapersen, a 2′OMePS, and eteplirsen, a phosphorodiamidate antisense morpholino oligonucleotide (PMO), both of which promote skipping of exon 51 by binding to exon 51-splicing enhancers on the DMD pre-mRNA [109,110]. Skipping of this exon restores the open reading frame, thus promoting the translation of an internally truncated dystrophin protein that is capable of converting a severe DMD into a milder BMD [110,111,112]. Despite initial encouraging results with intramuscular injections of drisapersen in four DMD patients [110,111,112], phase II and phase III clinical trials have failed to demonstrate a significant clinical benefit or a clear increase in dystrophin expression [113,114]. Drisapersen has therefore not received FDA approval [73,114]. By contrast, eteplirsen was approved by the FDA in September 2016 for DMD patients harboring a mutation that can be compensated by exon 51 skipping (which occurs in about 13% of the DMD population) [113]. The extent of eteplirsen clinical benefits are however still debated and larger clinical trials are being conducted [115].
Another class of promising drugs for treating DMD are tricyclo-DNAs (tcDNA). These are conformationally constrained AONs specially designed to limit torsional flexibility of the sugar backbone in order to stabilize tcDNA/RNA heteroduplexes [115]. Their efficacy has recently been demonstrated in mouse models of DMD, where they induced consistently higher levels of exon skipping and rescue of dystrophin protein levels than 2′OMePSs and PMOs [116,117]. Toxicological and tolerance studies are currently being conducted to evaluate the possibility of clinical applications.
Although deletions flanking the exon 51 are the most common cause of DMD [118], multiple genomic defects affecting different regions of the DMD gene have been described in DMD. To circumvent this issue, ongoing studies aim at developing multiple exon skipping strategies [115].
3.1.3. AON-Mediated Masking of Cryptic Splice Sites
In addition to promoting inclusion or skipping of exons, AONs can block the recognition of cryptic splice sites. This approach has for example been used successfully to correct the genetic defects of ataxia-telangiecatasia and Hutchinson-Gilford progeria syndrome (Table 2).
Ataxia-telangiecatasia is a rare, neurodegenerative, autosomal recessive disease characterized by cerebellar degeneration, telangiectasia (spider veins), immunodeficiency, cancer susceptibility and radiation sensitivity. It is caused by mutations in the ATM gene, which encodes the ubiquitously expressed serine/threonine kinase ATM, involved in controlling cell cycle checkpoints and in repairing damaged DNA [119]. About 50% of identified mutations affect splicing and 30–40% of these splicing mutations create new cryptic sites or interfere with splice regulatory elements [120,121,122]. Customized antisense morpholino oligonucleotides (AMOs) were designed to target three different mutations in ATM: (i) 7865C3T (A2622V), which creates a new 5′ splice site within exon 55, resulting in a deletion of the last 64 nt; (ii) 513C3T (Y171Y), which activates a cryptic 3′ splice site within exon 8, inducing deletion of the first 22 nt of exon 8; (iii) IVS28-159 A3G in intron 28, which activates cryptic 5′ and 3′ splice sites, resulting in insertion of a 112 nt segment from intron 28. The AMOs were effective in masking the pathogenic cryptic splice sites, thus restoring correct splicing and increasing functional ATM protein levels [123]. Further, coupling ATM-targeting AMOs to arginine-rich cell-penetrating peptides improved their capacity to cross the blood brain barrier and hereby their efficiency in correcting splicing [124]. Similar AMO-mediated strategies were also used effectively to correct splicing defects induced by other ATM mutations [120,121], thus highlighting the general applicability of this approach for ataxia-telangiecatasia.
Hutchinson-Gilford progeria syndrome is a rare autosomal genetic disorder whose main feature is premature aging [125]. The disease is caused by mutations in the LMNA gene, which encodes two nucleophilic A-type lamins, lamin A and lamin C. The most common mutation is a de novo substitution c.1824C>T in exon 11, which has the double effect of creating a cryptic splice site and disrupting a probable exon splicing enhancer [126,127]. The use of the cryptic splice site produces a truncated mRNA lacking the last 150 nucleotides of exon 11, thereby resulting in a muted protein, which is called progerin. AMOs designed to prevent access of the splicing machinery to the cryptic splice site restored normal splicing and rescued nuclear morphology in fibroblasts from patients [128]. Likewise, treating knock-in mice harboring the c.1827C>T mutation (equivalent to the c.1824C>T mutation in humans) with a combination of AMOs targeting the cryptic splice site in exon 11 and a 5′ splice site in exon 10 reduced the accumulation of progerin, thereby extending the life span of knock-in mice [129].
3.2. Spliceosome-Mediated RNA Trans-Splicing (SMaRT)
A different approach for reprogramming RNA splicing is spliceosome-mediated RNA trans-splicing (SMaRT). It consists of a trans-splicing reaction between an exogenous pre-trans-splicing RNA molecule (PTM) and a target endogenous pre-mRNA that induces the formation of a chimeric mRNA [130]. A typical PTM is composed of a binding domain, allowing the recognition of the target mRNA, intronic splicing elements, necessary for the splicing reaction, and the coding sequence to be substituted [131]. Depending on the orientation of the PTM, it is possible to replace 5′-, 3′- or internal gene portions. Although not yet used in clinical applications, this strategy has been successful in in vitro and in vivo models of cystic fibrosis (CF), frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), and SMA (Table 2) [130,132].
CF is a common autosomal recessive disease due to loss-of-function mutations in a gene for a chloride channel, cystic fibrosis trans-membrane conductance regulator (CFTR). Deficiencies in CTFR cause the production of mucus secretions with higher viscosity, eventually obstructing pancreatic and respiratory tracts [133]. The most common mutation is ΔF508, a three base-pair deletion at codon 508 in exon 10, which results in deletion of a phenylalanine residue inducing misfolding and mislocalization of the chloride channel [134,135]. Trans-splicing with a PTM containing the coding sequence of exons 10–24 was effective in correcting this mutation at the mRNA level in both cultured human epithelial cells and a xenograft model of the disease [131,136].
SMaRT has also been used successfully to regulate AS in the MAPT gene, which encodes the microtubule associated protein tau, and whose mutation causes several pathologies including FTDP-17. MAPT contains 16 exons. AS of exons 2, 3, and 10 generates six different isoforms. In particular, exclusion or inclusion of exon 10 gives rise to isoforms containing three (3R) or four (4R) microtubule-binding repeats, respectively [137,138]. Normal function of the adult brain requires 3R and 4R isofoms to be expressed in similar amounts. Many FTDP-17-causing mutations promote inclusion of exon 10, thus increasing the levels of 4R tau, which alters axonal transport and hereby triggers FTDP-17 [139,140,141]. In a mouse model of tauopathies, it was possible to regulate the ratio between 4R and 3R tau isoforms by delivering, into the mouse brain, a PTM containing a binding domain complementary to the 3′ end of intron 9 followed by either exons 10–13 or exons 11–13 to produce full-length tau chimeric proteins with or without exon 10, respectively [142,143,144].
Likewise, a SMaRT strategy has been developed to compensate for the deficiency of functional SMN proteins in SMA [145,146,147,148]. Researchers have used a PTM containing the SMN1 exon 7 sequence, an optimized splice site and a sequence annealing to the SMN intron 6 to increase the expression of exon 7-containing SMN2 mRNAs in SMA fibroblasts [147]. Combining this PTM with an AON designed to inhibit the downstream splice site at exon 8 further increased the efficiency of trans-splicing [145] and could ameliorate the SMA phenotype of SMAΔ7 mice [146,148].
3.3. CRISPR-Based Approaches
In recent years, the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) system has emerged as a powerful tool for developing new gene editing therapies [149,150]. It relies on short guide RNA sequences (gRNAs) that bring the endonuclease Cas9 to specific genomic loci to induce double strand breaks (DSBs). Upon DSB, endogenous cell mechanisms mediate non-homologous end joining (NHEJ) DNA repair, resulting in insertion/deletion (InDels) mutations near the DSB site. Alternatively, in the presence of a homologous DNA donor template, CRISPR/Cas9 can be used to introduce precise mutations through homology-directed repair (HDR) [151]. HDR is however not as efficient as NHEJ and it is thought to occur primarily in the S and G2 phases of the cell cycle, making its use in post-mitotic cells, such as neurons, challenging [152]. Recently, CRISPR approaches have been adopted also to modulate RNA splicing.
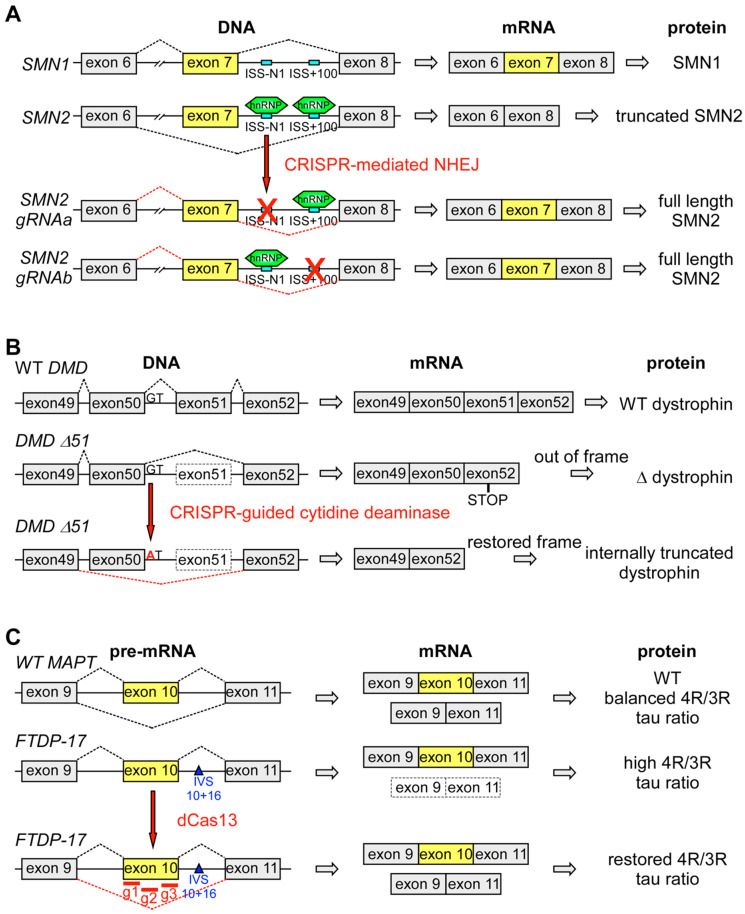
An NHEJ-based CRISPR strategy has for example been developed to promote exon 7 inclusion in SMN2, thus compensating for SMN1 deficiency in SMA (Table 2) [153]. As previously done using AONs [93], the authors did not try to correct the causative mutation in SMN1, rather they aimed at inactivating by NHEJ two intronic splicing silencers in intron 7 of SMN2 (ISS-N1 and ISS + 100; Figure 2A). To this end, they expressed, in human iPSC-derived motor neurons, Cas9 and a gRNA targeting one or the other ISS. The inactivation of either ISS effectively promoted inclusion of exon 7 in SMN2, thus increasing the expression of full length SMN protein. Importantly, this was sufficient to inhibit degeneration of iPSCs-derived motor neurons carrying a SMA mutation. Likewise, co-injecting Cas9 mRNA and a gRNA targeting ISS-N1 into the zygote of a SMA mouse model was effective in improving motor functions and extending the lifespan of treated mice [153].
Figure 2.
CRISPR-based splicing therapies (A) CRISPR-mediated non-homologous end joining (NHEJ) disrupts intronic splicing silencer (ISS) sites in intron 7 of SMN2 preventing binding of hnRNPs, thus favoring inclusion of exon 7, normally not present in SMN2. This helps compensate for mutated SMN1. (B) Deletion of exon 51 of DMD leads to a frameshift resulting in a truncated nonfunctional dystrophin. CRISPR-guided cytidine deaminase mutates the 5′ splice site of exon 50, leading to skipping of exon 50, which restores the reading frame. The resulting internally truncated dystrophin is able to restore partially WT function. (C) AS of MAPT exon 10 leads to formation of 4R and 3R tau isoforms. The FTDP-17-associated IVS 10 + 16 mutation results in increased exon 10 inclusion and higher 4R tau levels. Expression of dCas13 together with three gRNAs targeting the exon 10 splice acceptor site and two putative exonic splicing enhancers (ESEs) promotes skipping of exon 10, thus restoring a balanced 4R/3R tau ratio. Schemes adapted from [149,161,164].
NHEJ-based approaches have also been used successfully to skip mutated exons in the Dmd gene of the mdx mouse model of DMD (Table 2). The mdx mouse harbors a nonsense mutation in exon 23 of Dmd, which generates a premature stop codon leading to the formation of a truncated non-functional dystrophin protein [154]. Expression of Cas9 together with two gRNAs targeting the mutated region and the 3′ or 5′ end of exon 23 induced skipping of this exon by NHEJ-mediated genome editing, thus restoring the open reading frame of Dmd [155]. Exon 23 could also be skipped by targeting Cas9 to the regions flanking exon 23 [156,157]. Both approaches were effective in increasing the expression of internally truncated dystrophin, thereby partially rescuing cardiac muscle deficiencies and improving muscle force in mdx mice [155,156,157].
A different but equally effective CRISPR-based strategy to regulate splicing and restore the open reading frame of DMD consists of fusing a cytidine deaminase to a mutated nuclease dead (d)Cas9. The chimeric protein can be targeted to specific DNA sequences by gRNAs where it mediates a C to T conversion without inducing a DSB [158,159]. Likewise, adenine base editors can mediate an A to G conversion in specific DNA regions when fused to dCas9 [160]. A CRISPR-guided cytidine deaminase was used to mutate the 5′ splice site of exon 50 of DMD in iPSCs derived from a DMD patient who lacked dystrophin because of a deletion of exon 51 (Table 2) [161]. Following this mutation, exon 50 was skipped, which restored the open reading frame, thus generating an internally truncated dystrophin capable of partially rescuing the phenotype of iPSC-derived cardiomyocytes (Figure 2B) [161]. By targeting specific splice sites, we believe that CRISPR-mediated base editing technologies may become a powerful tool also for regulating the usage of mutually exclusive exons, for favoring the utilization of alternative splice sites and for promoting intron retention.
Finally, the discovery of Cas nucleases that target and cleave RNA rather than DNA, such as those of the Cas13 superfamily, has led to the development of CRISPR therapeutic strategies based on RNA manipulations [162,163]. Targeting RNA presents the advantage of correcting disease-relevant transcripts without permanently altering the genome. A recent study has shown that this strategy is feasible and has therapeutic potential in FTDP-17. Expression of a nuclease dead (d)Cas13 together with three gRNAs targeting the splice acceptor and two putative ESEs in exon 10 of MAPT increased skipping of exon 10 by preventing binding of splicing factors. This effectively restored a normal 4R/3R tau ratio in iPSCs-derived cortical neurons derived from FTDP-17 patients (Table 2; Figure 2C) [164].
4. Conclusions: Potential Advantages of Splicing Therapies for EA2
Given the number of mutations predicted to disrupt the splicing of CACNA1A in EA2 [4,6,13,14,15,16,17,18,19,26], splicing-targeting approaches, as described in the previous paragraphs, may hold great promise for developing new EA2 therapies. Furthermore, skipping of mutated exons could be used in some cases to restore the open reading frame of CACNA1A, thereby making it possible to generate functional CaV2.1 channels. These strategies would, however, require a specific design and an experimental validation for each targeted mutation.
As for SMA, where therapies target SMN2 to compensate for a deficiency in SMN1, EA2 therapies could regulate AS in presynaptic VGCCs other than CaV2.1 to compensate for a deficiency in this channel. At most central synapses, CaV2.1 is aided by CaV2.2 (aka: N-type channel; gene for the α1 subunit: CACNA1B) and CaV2.3 (aka: R-type channel; gene for the α1 subunit: CACNA1E). CACNA1B and CACNA1E are homologous to CACNA1A (59.4% and 54.0% identity at the amino acidic level, respectively) and, like CACNA1A, are extensively spliced, with the splicing pattern often conserved across the three mammalian CaV2 channels [7]. Notably, in mouse models of EA2, lack, reduced expression or functional impairment of CaV2.1 induces a compensatory up-regulation in the expression of the other two synaptic VGCCs [165,166]. Despite this compensation, synaptic transmission remains severely compromised at many synapses, including those in the cerebellum, mostly because CaV2.2 and CaV2.3 channels maintain a loose and inefficient coupling configuration to the neurotransmitter release machinery [167,168,169,170,171]. In principle, this functional diversity could mainly be due to intrinsic differences between the three CaV2 genes. Some interesting data indicate however that it is also extensively influenced by alternative splicing. For example, the mutually exclusive exons 37a (EFa) and 37b (EFb) are conserved across CaV2.1, CaV2.2 and CaV2.3 channels [23,172]. While little is known about the functional relevance of this splicing event in CaV2.3, the EFa isoforms of both CaV2.1 and CaV2.2 are more efficient than the respective EFb isoforms in supporting synaptic transmission [23,173,174]. The EFa isoform of CaV2.2 is, however, enriched only in capsaicin-responsive nociceptors of dorsal root ganglia [175], where it mediates efficient synaptic transmission [173,174], whereas EFb is the predominant isoform of CaV2.2 in most brain regions [175]. Molecular approaches designed to increase the relative abundance of the EFa isoform of CaV2.2 (and possibly of CaV2.3) in the brain could therefore help compensate for a CaV2.1 deficiency. Importantly, such strategies that leverage a generalized up-regulation of ‘secondary’ presynaptic Ca2+ channels (CaV2.2 and CaV2.3) in EA2 rather than targeting individual EA2 mutations in the ‘primary’ presynaptic Ca2+ channel (CaV2.1), of which there are more than 100, would have the advantage of being suitable for most EA2 patients.
Abbreviations
| 2′MOE | 2′-O-methoxyethyl |
| 2′OMePS | 2’-O-methyl-phosphorothioate RNA |
| AMO | Antisense morpholino oligonucleotide |
| AON | Antisense oligonucleotide |
| AS | Alternative splicing |
| BMD | Becker muscular dystrophy |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| DMD | Duchenne muscular dystrophy |
| DSB | Double strand break |
| EA2 | Episodic ataxia type 2 |
| ESE/S | Exonic splicing enhancer/silencer |
| FDA | Food and Drug Administration |
| FTDP-17 | Frontotemporal dementia with parkinsonism linked to chromosome 17 |
| gRNA | Guide RNA |
| HDR | Homology-directed repair |
| hnRNP | Heterogeneous nuclear ribonucleoprotein particle |
| ISE/S | Intronic splicing enhancer/silencer |
| NHEJ | Non-homologous end joining |
| PMO | Phosphorodiamidate antisense morpholino oligonucleotide |
| PS | Phosphorothioate |
| PTM | Pre-trans-splicing RNA molecule |
| SMA | Spinal muscular atrophy |
| SMaRT | Spliceosome-Mediated RNA Trans-Splicing |
| SMN | Survival of motor neuron |
| tcDNA | tricyclo-DNA |
| VGCC | Voltage-gated calcium channel |
Author Contributions
Conceptualization, L.A.C. and F.Z.; original draft preparation, F.J., S.B., and I.M.; writing editing, L.A.C.; visualization, A.T.; funding acquisition, L.A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by Ataxia UK (to L.A.C.) and the Telethon foundation (grant number: GGP19181 to L.A.C. and F.Z.).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Jen J., Kim G.W., Baloh R.W. Clinical spectrum of episodic ataxia type 2. Neurology. 2004;62:17–22. doi: 10.1212/01.WNL.0000101675.61074.50. [DOI] [PubMed] [Google Scholar]
- 2.Strupp M., Zwergal A., Brandt T. Episodic ataxia type 2. Neurother. J. Am. Soc. Exp. Neurother. 2007;4:267–273. doi: 10.1016/j.nurt.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Imbrici P., Eunson L.H., Graves T.D., Bhatia K.P., Wadia N.H., Kullmann D.M., Hanna M.G. Late-onset episodic ataxia type 2 due to an in-frame insertion in CACNA1A. Neurology. 2005;65:944–946. doi: 10.1212/01.wnl.0000176069.64200.28. [DOI] [PubMed] [Google Scholar]
- 4.Mantuano E., Romano S., Veneziano L., Gellera C., Castellotti B., Caimi S., Testa D., Estienne M., Zorzi G., Bugiani M., et al. Identification of novel and recurrent CACNA1A gene mutations in fifteen patients with episodic ataxia type 2. J. Neurol. Sci. 2010;291:30–36. doi: 10.1016/j.jns.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Jen J.C., Graves T.D., Hess E.J., Hanna M.G., Griggs R.C., Baloh R.W., The CINCH Investigators Primary episodic ataxias: Diagnosis, pathogenesis and treatment. Brain J. Neurol. 2007;130:2484–2493. doi: 10.1093/brain/awm126. [DOI] [PubMed] [Google Scholar]
- 6.Ophoff R.A., Terwindt G.M., Vergouwe M.N., van Eijk R., Oefner P.J., Hoffman S.M., Lamerdin J.E., Mohrenweiser H.W., Bulman D.E., Ferrari M., et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/S0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 7.Thalhammer A., Jaudon F., Cingolani L.A. Emerging Roles of Activity-Dependent Alternative Splicing in Homeostatic Plasticity. Front. Cell Neurosci. 2020;14:104. doi: 10.3389/fncel.2020.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y.Q., Piedras-Renteria E.S., Smith G.B., Chen G., Harata N.C., Tsien R.W. Presynaptic Ca2+ channels compete for channel type-preferring slots in altered neurotransmission arising from Ca2+ channelopathy. Neuron. 2004;43:387–400. doi: 10.1016/j.neuron.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Mintz I.M., Sabatini B.L., Regehr W.G. Calcium control of transmitter release at a cerebellar synapse. Neuron. 1995;15:675–688. doi: 10.1016/0896-6273(95)90155-8. [DOI] [PubMed] [Google Scholar]
- 10.Wu L.G., Westenbroek R.E., Borst J.G., Catterall W.A., Sakmann B. Calcium channel types with distinct presynaptic localization couple differentially to transmitter release in single calyx-type synapses. J. Neurosci. 1999;19:726–736. doi: 10.1523/JNEUROSCI.19-02-00726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipscombe D., Allen S.E., Toro C.P. Control of neuronal voltage-gated calcium ion channels from RNA to protein. Trends Neurosci. 2013;36:598–609. doi: 10.1016/j.tins.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sintas C., Carreno O., Fernandez-Castillo N., Corominas R., Vila-Pueyo M., Toma C., Cuenca-Leon E., Barroeta I., Roig C., Volpini V., et al. Mutation Spectrum in the CACNA1A Gene in 49 Patients with Episodic Ataxia. Sci. Rep. 2017;7:2514. doi: 10.1038/s41598-017-02554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denier C., Ducros A., Vahedi K., Joutel A., Thierry P., Ritz A., Castelnovo G., Deonna T., Gerard P., Devoize J.L., et al. High prevalence of CACNA1A truncations and broader clinical spectrum in episodic ataxia type 2. Neurology. 1999;52:1816–1821. doi: 10.1212/WNL.52.9.1816. [DOI] [PubMed] [Google Scholar]
- 14.Eunson L.H., Graves T.D., Hanna M.G. New calcium channel mutations predict aberrant RNA splicing in episodic ataxia. Neurology. 2005;65:308–310. doi: 10.1212/01.wnl.0000169020.82223.dd. [DOI] [PubMed] [Google Scholar]
- 15.Subramony S.H., Schott K., Raike R.S., Callahan J., Langford L.R., Christova P.S., Anderson J.H., Gomez C.M. Novel CACNA1A mutation causes febrile episodic ataxia with interictal cerebellar deficits. Ann. Neurol. 2003;54:725–731. doi: 10.1002/ana.10756. [DOI] [PubMed] [Google Scholar]
- 16.Damaj L., Lupien-Meilleur A., Lortie A., Riou E., Ospina L.H., Gagnon L., Vanasse C., Rossignol E. CACNA1A haploinsufficiency causes cognitive impairment, autism and epileptic encephalopathy with mild cerebellar symptoms. Eur. J. Hum. Genet. 2015;23:1505–1512. doi: 10.1038/ejhg.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachbauer W., Nocker M., Karner E., Stankovic I., Unterberger I., Eigentler A., Schneider R., Poewe W., Delazer M., Boesch S. Episodic ataxia type 2: Phenotype characteristics of a novel CACNA1A mutation and review of the literature. J. Neurol. 2014;261:983–991. doi: 10.1007/s00415-014-7310-2. [DOI] [PubMed] [Google Scholar]
- 18.Wan J., Carr J.R., Baloh R.W., Jen J.C. Nonconsensus intronic mutations cause episodic ataxia. Ann. Neurol. 2005;57:131–135. doi: 10.1002/ana.20343. [DOI] [PubMed] [Google Scholar]
- 19.Kaunisto M.A., Harno H., Kallela M., Somer H., Sallinen R., Hamalainen E., Miettinen P.J., Vesa J., Orpana A., Palotie A., et al. Novel splice site CACNA1A mutation causing episodic ataxia type 2. Neurogenetics. 2004;5:69–73. doi: 10.1007/s10048-003-0161-0. [DOI] [PubMed] [Google Scholar]
- 20.Bourinet E., Soong T.W., Sutton K., Slaymaker S., Mathews E., Monteil A., Zamponi G.W., Nargeot J., Snutch T.P. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat. Neurosci. 1999;2:407–415. doi: 10.1038/8070. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhuri D., Chang S.Y., DeMaria C.D., Alvania R.S., Soong T.W., Yue D.T. Alternative splicing as a molecular switch for Ca2+/calmodulin-dependent facilitation of P/Q-type Ca2+ channels. J. Neurosci. 2004;24:6334–6342. doi: 10.1523/JNEUROSCI.1712-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soong T.W., DeMaria C.D., Alvania R.S., Zweifel L.S., Liang M.C., Mittman S., Agnew W.S., Yue D.T. Systematic identification of splice variants in human P/Q-type channel alpha1(2.1) subunits: Implications for current density and Ca2+-dependent inactivation. J. Neurosci. 2002;22:10142–10152. doi: 10.1523/JNEUROSCI.22-23-10142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thalhammer A., Contestabile A., Ermolyuk Y.S., Ng T., Volynski K.E., Soong T.W., Goda Y., Cingolani L.A. Alternative Splicing of P/Q-Type Ca2+ Channels Shapes Presynaptic Plasticity. Cell Rep. 2017;20:333–343. doi: 10.1016/j.celrep.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 24.Thalhammer A., Jaudon F., Cingolani L.A. Combining Optogenetics with Artificial microRNAs to Characterize the Effects of Gene Knockdown on Presynaptic Function within Intact Neuronal Circuits. J. Vis. Exp. 2018 doi: 10.3791/57223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vigues S., Gastaldi M., Massacrier A., Cau P., Valmier J. The alpha(1A) subunits of rat brain calcium channels are developmentally regulated by alternative RNA splicing. Neuroscience. 2002;113:509–517. doi: 10.1016/S0306-4522(02)00213-0. [DOI] [PubMed] [Google Scholar]
- 26.Graves T.D., Imbrici P., Kors E.E., Terwindt G.M., Eunson L.H., Frants R.R., Haan J., Ferrari M.D., Goadsby P.J., Hanna M.G., et al. Premature stop codons in a facilitating EF-hand splice variant of CaV2.1 cause episodic ataxia type 2. Neurobiol. Dis. 2008;32:10–15. doi: 10.1016/j.nbd.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Imbrici P., Jaffe S.L., Eunson L.H., Davies N.P., Herd C., Robertson R., Kullmann D.M., Hanna M.G. Dysfunction of the brain calcium channel CaV2.1 in absence epilepsy and episodic ataxia. Brain J. Neurol. 2004;127:2682–2692. doi: 10.1093/brain/awh301. [DOI] [PubMed] [Google Scholar]
- 28.Maksemous N., Roy B., Smith R.A., Griffiths L.R. Next-generation sequencing identifies novel CACNA1A gene mutations in episodic ataxia type 2. Mol. Genet. Genom. Med. 2016;4:211–222. doi: 10.1002/mgg3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L., et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Indelicato E., Nachbauer W., Karner E., Eigentler A., Wagner M., Unterberger I., Poewe W., Delazer M., Boesch S. The neuropsychiatric phenotype in CACNA1A mutations: A retrospective single center study and review of the literature. Eur. J. Neurol. 2019;26:66–67. doi: 10.1111/ene.13765. [DOI] [PubMed] [Google Scholar]
- 31.Choi K.D., Kim J.S., Kim H.J., Jung I., Jeong S.H., Lee S.H., Kim D.U., Kim S.H., Choi S.Y., Shin J.H., et al. Genetic Variants Associated with Episodic Ataxia in Korea. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-14254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zafeiriou D.I., Lehmann-Horn F., Vargiami E., Teflioudi E., Ververi A., Jurkat-Rott K. Episodic ataxia type 2 showing ictal hyperhidrosis with hypothermia and interictal chronic diarrhea due to a novel CACNA1A mutation. Eur. J. Paediatr. Neurol. 2009;13:191–193. doi: 10.1016/j.ejpn.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Van den Maagdenberg A.M., Kors E.E., Brunt E.R., van Paesschen W., Pascual J., Ravine D., Keeling S., Vanmolkot K.R., Vermeulen F.L., Terwindt G.M., et al. Episodic ataxia type 2. Three novel truncating mutations and one novel missense mutation in the CACNA1A gene. J. Neurol. 2002;249:1515–1519. doi: 10.1007/s00415-002-0860-8. [DOI] [PubMed] [Google Scholar]
- 34.Mantuano E., Veneziano L., Spadaro M., Giunti P., Guida S., Leggio M.G., Verriello L., Wood N., Jodice C., Frontali M. Clusters of non-truncating mutations of P/Q type Ca2+ channel subunit Ca(v)2.1 causing episodic ataxia 2. J. Med. Genet. 2004;41:e82. doi: 10.1136/jmg.2003.015396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue Q., Jen J.C., Nelson S.F., Baloh R.W. Progressive ataxia due to a missense mutation in a calcium-channel gene. Am. J. Hum. Genet. 1997;61:1078–1087. doi: 10.1086/301613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tantsis E.M., Gill D., Griffiths L., Gupta S., Lawson J., Maksemous N., Ouvrier R., Riant F., Smith R., Troedson C., et al. Eye movement disorders are an early manifestation of CACNA1A mutations in children. Dev. Med. Child Neurol. 2016;58:639–644. doi: 10.1111/dmcn.13033. [DOI] [PubMed] [Google Scholar]
- 37.Nikaido K., Tachi N., Ohya K., Wada T., Tsutsumi H. New mutation of CACNA1A gene in episodic ataxia type 2. Pediatr. Int. 2011;53:415–416. doi: 10.1111/j.1442-200X.2011.03390.x. [DOI] [PubMed] [Google Scholar]
- 38.Cricchi F., Di Lorenzo C., Grieco G.S., Rengo C., Cardinale A., Racaniello M., Santorelli F.M., Nappi G., Pierelli F., Casali C. Early-onset progressive ataxia associated with the first CACNA1A mutation identified within the I-II loop. J. Neurol. Sci. 2007;254:69–71. doi: 10.1016/j.jns.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Isaacs D.A., Bradshaw M.J., Brown K., Hedera P. Case report of novel CACNA1A gene mutation causing episodic ataxia type 2. SAGE Open Med. Case Rep. 2017;5:2050313X17706044. doi: 10.1177/2050313X17706044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scoggan K.A., Friedman J.H., Bulman D.E. CACNA1A mutation in a EA-2 patient responsive to acetazolamide and valproic acid. Can. J. Neurol. Sci. 2006;33:68–72. doi: 10.1017/S0317167100004728. [DOI] [PubMed] [Google Scholar]
- 41.Rajakulendran S., Graves T.D., Labrum R.W., Kotzadimitriou D., Eunson L., Davis M.B., Davies R., Wood N.W., Kullmann D.M., Hanna M.G., et al. Genetic and functional characterisation of the P/Q calcium channel in episodic ataxia with epilepsy. J. Physiol. 2010;588:1905–1913. doi: 10.1113/jphysiol.2009.186437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuenca-Leon E., Banchs I., Serra S.A., Latorre P., Fernandez-Castillo N., Corominas R., Valverde M.A., Volpini V., Fernandez-Fernandez J.M., Macaya A., et al. Late-onset episodic ataxia type 2 associated with a novel loss-of-function mutation in the CACNA1A gene. J. Neurol. Sci. 2009;280:10–14. doi: 10.1016/j.jns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Guerin A.A., Feigenbaum A., Donner E.J., Yoon G. Stepwise developmental regression associated with novel CACNA1A mutation. Pediatr. Neurol. 2008;39:363–364. doi: 10.1016/j.pediatrneurol.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 44.Roubertie A., Echenne B., Leydet J., Soete S., Krams B., Rivier F., Riant F., Tournier-Lasserve E. Benign paroxysmal tonic upgaze, benign paroxysmal torticollis, episodic ataxia and CACNA1A mutation in a family. J. Neurol. 2008;255:1600–1602. doi: 10.1007/s00415-008-0982-8. [DOI] [PubMed] [Google Scholar]
- 45.Bertholon P., Chabrier S., Riant F., Tournier-Lasserve E., Peyron R. Episodic ataxia type 2: Unusual aspects in clinical and genetic presentation. Special emphasis in childhood. J. Neurol. Neurosurg. Psychiatry. 2009;80:1289–1292. doi: 10.1136/jnnp.2008.159103. [DOI] [PubMed] [Google Scholar]
- 46.Scoggan K.A., Chandra T., Nelson R., Hahn A.F., Bulman D.E. Identification of two novel mutations in the CACNA1A gene responsible for episodic ataxia type 2. J. Med. Genet. 2001;38:249–253. doi: 10.1136/jmg.38.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blumkin L., Michelson M., Leshinsky-Silver E., Kivity S., Lev D., Lerman-Sagie T. Congenital ataxia, mental retardation, and dyskinesia associated with a novel CACNA1A mutation. J. Child Neurol. 2010;25:892–897. doi: 10.1177/0883073809351316. [DOI] [PubMed] [Google Scholar]
- 48.Jen J., Wan J., Graves M., Yu H., Mock A.F., Coulin C.J., Kim G., Yue Q., Papazian D.M., Baloh R.W. Loss-of-function EA2 mutations are associated with impaired neuromuscular transmission. Neurology. 2001;57:1843–1848. doi: 10.1212/WNL.57.10.1843. [DOI] [PubMed] [Google Scholar]
- 49.Pietrobon D. CaV2.1 channelopathies. Pflug. Arch. Eur. J. Physiol. 2010;460:375–393. doi: 10.1007/s00424-010-0802-8. [DOI] [PubMed] [Google Scholar]
- 50.Guida S., Trettel F., Pagnutti S., Mantuano E., Tottene A., Veneziano L., Fellin T., Spadaro M., Stauderman K., Williams M., et al. Complete loss of P/Q calcium channel activity caused by a CACNA1A missense mutation carried by patients with episodic ataxia type 2. Am. J. Hum. Genet. 2001;68:759–764. doi: 10.1086/318804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jen J., Yue Q., Nelson S.F., Yu H., Litt M., Nutt J., Baloh R.W. A novel nonsense mutation in CACNA1A causes episodic ataxia and hemiplegia. Neurology. 1999;53:34–37. doi: 10.1212/WNL.53.1.34. [DOI] [PubMed] [Google Scholar]
- 52.Spacey S.D., Materek L.A., Szczygielski B.I., Bird T.D. Two novel CACNA1A gene mutations associated with episodic ataxia type 2 and interictal dystonia. Arch. Neurol. 2005;62:314–316. doi: 10.1001/archneur.62.2.314. [DOI] [PubMed] [Google Scholar]
- 53.Friend K.L., Crimmins D., Phan T.G., Sue C.M., Colley A., Fung V.S., Morris J.G., Sutherland G.R., Richards R.I. Detection of a novel missense mutation and second recurrent mutation in the CACNA1A gene in individuals with EA-2 and FHM. Hum. Genet. 1999;105:261–265. doi: 10.1007/s004399900101. [DOI] [PubMed] [Google Scholar]
- 54.Tonelli A., D’Angelo M.G., Salati R., Villa L., Germinasi C., Frattini T., Meola G., Turconi A.C., Bresolin N., Bassi M.T. Early onset, non fluctuating spinocerebellar ataxia and a novel missense mutation in CACNA1A gene. J. Neurol. Sci. 2006;241:13–17. doi: 10.1016/j.jns.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 55.Spacey S.D., Hildebrand M.E., Materek L.A., Bird T.D., Snutch T.P. Functional implications of a novel EA2 mutation in the P/Q-type calcium channel. Ann. Neurol. 2004;56:213–220. doi: 10.1002/ana.20169. [DOI] [PubMed] [Google Scholar]
- 56.Denier C., Ducros A., Durr A., Eymard B., Chassande B., Tournier-Lasserve E. Missense CACNA1A mutation causing episodic ataxia type 2. Arch. Neurol. 2001;58:292–295. doi: 10.1001/archneur.58.2.292. [DOI] [PubMed] [Google Scholar]
- 57.Ohba C., Osaka H., Iai M., Yamashita S., Suzuki Y., Aida N., Shimozawa N., Takamura A., Doi H., Tomita-Katsumoto A., et al. Diagnostic utility of whole exome sequencing in patients showing cerebellar and/or vermis atrophy in childhood. Neurogenetics. 2013;14:225–232. doi: 10.1007/s10048-013-0375-8. [DOI] [PubMed] [Google Scholar]
- 58.Jouvenceau A., Eunson L.H., Spauschus A., Ramesh V., Zuberi S.M., Kullmann D.M., Hanna M.G. Human epilepsy associated with dysfunction of the brain P/Q-type calcium channel. Lancet. 2001;358:801–807. doi: 10.1016/S0140-6736(01)05971-2. [DOI] [PubMed] [Google Scholar]
- 59.Melzer N., Classen J., Reiners K., Buttmann M. Fluctuating neuromuscular transmission defects and inverse acetazolamide response in episodic ataxia type 2 associated with the novel CaV2.1 single amino acid substitution R2090Q. J. Neurol. Sci. 2010;296:104–106. doi: 10.1016/j.jns.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 60.Zhuchenko O., Bailey J., Bonnen P., Ashizawa T., Stockton D.W., Amos C., Dobyns W.B., Subramony S.H., Zoghbi H.Y., Lee C.C. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 61.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 62.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 64.De Conti L., Baralle M., Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 65.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh R., Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 67.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 68.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 69.Wang Q., Zhou T. Alternative-splicing-mediated gene expression. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2014;89:012713. doi: 10.1103/PhysRevE.89.012713. [DOI] [PubMed] [Google Scholar]
- 70.Havens M.A., Duelli D.M., Hastings M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montes M., Sanford B.L., Comiskey D.F., Chandler D.S. RNA Splicing and Disease: Animal Models to Therapies. Trends Genet. TIG. 2019;35:68–87. doi: 10.1016/j.tig.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tazi J., Bakkour N., Stamm S. Alternative splicing and disease. Biochim. Biophys. Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S. Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 74.DeVos S.L., Miller T.M. Antisense oligonucleotides: Treating neurodegeneration at the level of RNA. Neurother. J. Am. Soc. Exp. Neurother. 2013;10:486–497. doi: 10.1007/s13311-013-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mozaffari-Jovin S., Wandersleben T., Santos K.F., Will C.L., Luhrmann R., Wahl M.C. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341:80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 76.Schoch K.M., Miller T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron. 2017;94:1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cerritelli S.M., Crouch R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roberts J., Palma E., Sazani P., Orum H., Cho M., Kole R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. J. Am. Soc. Gene Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 80.Sazani P., Gemignani F., Kang S.H., Maier M.A., Manoharan M., Persmark M., Bortner D., Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 81.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kole R., Krainer A.R., Altman S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 84.Brzustowicz L.M., Lehner T., Castilla L.H., Penchaszadeh G.K., Wilhelmsen K.C., Daniels R., Davies K.E., Leppert M., Ziter F., Wood D., et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- 85.Farrar M.A., Kiernan M.C. The Genetics of Spinal Muscular Atrophy: Progress and Challenges. Neurother. J. Am. Soc. Exp. Neurother. 2015;12:290–302. doi: 10.1007/s13311-014-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lunn M.R., Wang C.H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 87.Melki J., Abdelhak S., Sheth P., Bachelot M.F., Burlet P., Marcadet A., Aicardi J., Barois A., Carriere J.P., Fardeau M., et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature. 1990;344:767–768. doi: 10.1038/344767a0. [DOI] [PubMed] [Google Scholar]
- 88.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wirth B., Brichta L., Schrank B., Lochmuller H., Blick S., Baasner A., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 90.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pao P.W., Wee K.B., Yee W.C., Pramono Z.A. Dual masking of specific negative splicing regulatory elements resulted in maximal exon 7 inclusion of SMN2 gene. Mol. Ther. J. Am. Soc. Gene Ther. 2014;22:854–861. doi: 10.1038/mt.2013.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Darras B.T., Farrar M.A., Mercuri E., Finkel R.S., Foster R., Hughes S.G., Bhan I., Farwell W., Gheuens S. An Integrated Safety Analysis of Infants and Children with Symptomatic Spinal Muscular Atrophy (SMA) Treated with Nusinersen in Seven Clinical Trials. CNS Drugs. 2019;33:919–932. doi: 10.1007/s40263-019-00656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 98.Hache M., Swoboda K.J., Sethna N., Farrow-Gillespie A., Khandji A., Xia S., Bishop K.M. Intrathecal Injections in Children With Spinal Muscular Atrophy: Nusinersen Clinical Trial Experience. J. Child Neurol. 2016;31:899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mercuri E., Finkel R.S., Muntoni F., Wirth B., Montes J., Main M., Mazzone E.S., Vitale M., Snyder B., Quijano-Roy S., et al. Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscul. Disord. NMD. 2018;28:103–115. doi: 10.1016/j.nmd.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 100.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E., et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 101.Wurster C.D., Ludolph A.C. Nusinersen for spinal muscular atrophy. Ther. Adv. Neurol. Disord. 2018;11:1756285618754459. doi: 10.1177/1756285618754459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D’Ydewalle C., Ramos D.M., Pyles N.J., Ng S.Y., Gorz M., Pilato C.M., Ling K., Kong L., Ward A.J., Rubin L.L., et al. The Antisense Transcript SMN-AS1 Regulates SMN Expression and is a Novel Therapeutic Target for Spinal Muscular Atrophy. Neuron. 2017;93:66–79. doi: 10.1016/j.neuron.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: Implementation of multidisciplinary care. Lancet Neurol. 2010;9:177–189. doi: 10.1016/S1474-4422(09)70272-8. [DOI] [PubMed] [Google Scholar]
- 104.Mendell J.R., Shilling C., Leslie N.D., Flanigan K.M., al-Dahhak R., Gastier-Foster J., Kneile K., Dunn D.M., Duval B., Aoyagi A., et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 105.Ryder S., Leadley R.M., Armstrong N., Westwood M., de Kock S., Butt T., Jain M., Kleijnen J. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: An evidence review. Orphanet J. Rare Dis. 2017;12:79. doi: 10.1186/s13023-017-0631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoffman E.P., Dressman D. Molecular pathophysiology and targeted therapeutics for muscular dystrophy. Trends Pharmacol. Sci. 2001;22:465–470. doi: 10.1016/S0165-6147(00)01770-3. [DOI] [PubMed] [Google Scholar]
- 107.Nowak K.J., Davies K.E. Duchenne muscular dystrophy and dystrophin: Pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Helderman-van den Enden A.T., Straathof C.S., Aartsma-Rus A., den Dunnen J.T., Verbist B.M., Bakker E., Verschuuren J.J., Ginjaar H.B. Becker muscular dystrophy patients with deletions around exon 51; a promising outlook for exon skipping therapy in Duchenne patients. Neuromuscul. Disord. NMD. 2010;20:251–254. doi: 10.1016/j.nmd.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 109.Arechavala-Gomeza V., Graham I.R., Popplewell L.J., Adams A.M., Aartsma-Rus A., Kinali M., Morgan J.E., van Deutekom J.C., Wilton S.D., Dickson G., et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- 110.Van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M., et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 111.Aartsma-Rus A., Janson A.A., Kaman W.E., Bremmer-Bout M., den Dunnen J.T., Baas F., van Ommen G.J., van Deutekom J.C. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- 112.Alter J., Lou F., Rabinowitz A., Yin H., Rosenfeld J., Wilton S.D., Partridge T.A., Lu Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 113.Aartsma-Rus A., Krieg A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kole R., Krieg A.M. Exon skipping therapy for Duchenne muscular dystrophy. Adv. Drug Deliv. Rev. 2015;87:104–107. doi: 10.1016/j.addr.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 115.Echevarria L., Aupy P., Goyenvalle A. Exon-skipping advances for Duchenne muscular dystrophy. Hum. Mol. Genet. 2018;27:R163–R172. doi: 10.1093/hmg/ddy171. [DOI] [PubMed] [Google Scholar]
- 116.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T., et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 117.Relizani K., Griffith G., Echevarria L., Zarrouki F., Facchinetti P., Vaillend C., Leumann C., Garcia L., Goyenvalle A. Efficacy and Safety Profile of Tricyclo-DNA Antisense Oligonucleotides in Duchenne Muscular Dystrophy Mouse Model. Mol. Ther. Nucleic Acids. 2017;8:144–157. doi: 10.1016/j.omtn.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aartsma-Rus A., Fokkema I., Verschuuren J., Ginjaar I., van Deutekom J., van Ommen G.J., den Dunnen J.T. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 119.Perlman S., Becker-Catania S., Gatti R.A. Ataxia-telangiectasia: Diagnosis and treatment. Semin. Pediatr. Neurol. 2003;10:173–182. doi: 10.1016/S1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 120.Cavalieri S., Pozzi E., Gatti R.A., Brusco A. Deep-intronic ATM mutation detected by genomic resequencing and corrected in vitro by antisense morpholino oligonucleotide (AMO) Eur. J. Hum. Genet. EJHG. 2013;21:774–778. doi: 10.1038/ejhg.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakamura K., Du L., Tunuguntla R., Fike F., Cavalieri S., Morio T., Mizutani S., Brusco A., Gatti R.A. Functional characterization and targeted correction of ATM mutations identified in Japanese patients with ataxia-telangiectasia. Hum. Mutat. 2012;33:198–208. doi: 10.1002/humu.21632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: Underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Du L., Pollard J.M., Gatti R.A. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc. Natl. Acad. Sci. USA. 2007;104:6007–6012. doi: 10.1073/pnas.0608616104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Du L., Kayali R., Bertoni C., Fike F., Hu H., Iversen P.L., Gatti R.A. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum. Mol. Genet. 2011;20:3151–3160. doi: 10.1093/hmg/ddr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ullrich N.J., Gordon L.B. Hutchinson-Gilford progeria syndrome. Handb. Clin. Neurol. 2015;132:249–264. doi: 10.1016/B978-0-444-62702-5.00018-4. [DOI] [PubMed] [Google Scholar]
- 126.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 127.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scaffidi P., Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Osorio F.G., Navarro C.L., Cadinanos J., Lopez-Mejia I.C., Quiros P.M., Bartoli C., Rivera J., Tazi J., Guzman G., Varela I., et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 2011;3:106ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 130.Berger A., Maire S., Gaillard M.C., Sahel J.A., Hantraye P., Bemelmans A.P. mRNA trans-splicing in gene therapy for genetic diseases. Wiley Interdiscip. Rev. RNA. 2016;7:487–498. doi: 10.1002/wrna.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mansfield S.G., Chao H., Walsh C.E. RNA repair using spliceosome-mediated RNA trans-splicing. Trends Mol. Med. 2004;10:263–268. doi: 10.1016/j.molmed.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 132.Wally V., Murauer E.M., Bauer J.W. Spliceosome-mediated trans-splicing: The therapeutic cut and paste. J. Investig. Dermatol. 2012;132:1959–1966. doi: 10.1038/jid.2012.101. [DOI] [PubMed] [Google Scholar]
- 133.O’Sullivan B.P., Freedman S.D. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 134.Cheng S.H., Gregory R.J., Marshall J., Paul S., Souza D.W., White G.A., O’Riordan C.R., Smith A.E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 135.Welsh M.J., Smith A.E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-R. [DOI] [PubMed] [Google Scholar]
- 136.Liu X., Jiang Q., Mansfield S.G., Puttaraju M., Zhang Y., Zhou W., Cohn J.A., Garcia-Blanco M.A., Mitchell L.G., Engelhardt J.F. Partial correction of endogenous DeltaF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- 137.Andreadis A., Brown W.M., Kosik K.S. Structure and novel exons of the human tau gene. Biochemistry. 1992;31:10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- 138.Goedert M., Spillantini M.G., Jakes R., Rutherford D., Crowther R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 139.D’Souza I., Schellenberg G.D. Regulation of tau isoform expression and dementia. Biochim. Biophys. Acta. 2005;1739:104–115. doi: 10.1016/j.bbadis.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 140.Kar A., Kuo D., He R., Zhou J., Wu J.Y. Tau alternative splicing and frontotemporal dementia. Alzheimer Dis. Assoc. Disord. 2005;19(Suppl. 1):29–36. doi: 10.1097/01.wad.0000183082.76820.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qian W., Liu F. Regulation of alternative splicing of tau exon 10. Neurosci. Bull. 2014;30:367–377. doi: 10.1007/s12264-013-1411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Avale M.E., Rodriguez-Martin T., Gallo J.M. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum. Mol. Genet. 2013;22:2603–2611. doi: 10.1093/hmg/ddt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rodriguez-Martin T., Anthony K., Garcia-Blanco M.A., Mansfield S.G., Anderton B.H., Gallo J.M. Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing. Hum. Mol. Genet. 2009;18:3266–3273. doi: 10.1093/hmg/ddp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rodriguez-Martin T., Garcia-Blanco M.A., Mansfield S.G., Grover A.C., Hutton M., Yu Q., Zhou J., Anderton B.H., Gallo J.M. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: Implications for tauopathies. Proc. Natl. Acad. Sci. USA. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Coady T.H., Lorson C.L. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J. Neurosci. 2010;30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: Delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol. Ther. J. Am. Soc. Gene Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- 148.Shababi M., Glascock J., Lorson C.L. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum. Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- 149.Li B., Niu Y., Ji W., Dong Y. Strategies for the CRISPR-Based Therapeutics. Trends Pharmacol. Sci. 2020;41:55–65. doi: 10.1016/j.tips.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang D., Zhang F., Gao G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 152.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 153.Jin-Jing L., Xiang L., Cheng T., Ying-Qian L., Xinde H., Erwei Z., He L., Wenqin Y., Yidi S., Lu-Lu L., et al. Disruption of splicing-regulatory elements using CRISPR/Cas9 to rescue spinal muscular atrophy in human iPSCs and mice. Natl. Sci. Rev. 2020;7:92. doi: 10.1093/nsr/nwz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 155.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X., et al. In Vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A., et al. In Vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y., et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 160.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan J., Ma Y., Huang T., Chen Y., Peng Y., Li B., Li J., Zhang Y., Song B., Sun X., et al. Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol. Cell. 2018;72:380–394. doi: 10.1016/j.molcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 162.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jun K., Piedras-Renteria E.S., Smith S.M., Wheeler D.B., Lee S.B., Lee T.G., Chin H., Adams M.E., Scheller R.H., Tsien R.W., et al. Ablation of P/Q-type Ca2+ channel currents, altered synaptic transmission, and progressive ataxia in mice lacking the alpha(1A)-subunit. Proc. Natl. Acad. Sci. USA. 1999;96:15245–15250. doi: 10.1073/pnas.96.26.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Qian J., Noebels J.L. Presynaptic Ca2+ influx at a mouse central synapse with Ca2+ channel subunit mutations. J. Neurosci. 2000;20:163–170. doi: 10.1523/JNEUROSCI.20-01-00163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Inchauspe C.G., Forsythe I.D., Uchitel O.D. Changes in synaptic transmission properties due to the expression of N-type calcium channels at the calyx of Held synapse of mice lacking P/Q-type calcium channels. J. Physiol. 2007;584:835–851. doi: 10.1113/jphysiol.2007.139683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Inchauspe C.G., Martini F.J., Forsythe I.D., Uchitel O.D. Functional compensation of P/Q by N-type channels blocks short-term plasticity at the calyx of Held presynaptic terminal. J. Neurosci. 2004;24:10379–10383. doi: 10.1523/JNEUROSCI.2104-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ishikawa T., Kaneko M., Shin H.S., Takahashi T. Presynaptic N-type and P/Q-type Ca2+ channels mediating synaptic transmission at the calyx of Held of mice. J. Physiol. 2005;568:199–209. doi: 10.1113/jphysiol.2005.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Maejima T., Wollenweber P., Teusner L.U., Noebels J.L., Herlitze S., Mark M.D. Postnatal loss of P/Q-type channels confined to rhombic-lip-derived neurons alters synaptic transmission at the parallel fiber to purkinje cell synapse and replicates genomic Cacna1a mutation phenotype of ataxia and seizures in mice. J. Neurosci. 2013;33:5162–5174. doi: 10.1523/JNEUROSCI.5442-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mark M.D., Maejima T., Kuckelsberg D., Yoo J.W., Hyde R.A., Shah V., Gutierrez D., Moreno R.L., Kruse W., Noebels J.L., et al. Delayed postnatal loss of P/Q-type calcium channels recapitulates the absence epilepsy, dyskinesia, and ataxia phenotypes of genomic Cacna1a mutations. J. Neurosci. 2011;31:4311–4326. doi: 10.1523/JNEUROSCI.5342-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gray A.C., Raingo J., Lipscombe D. Neuronal calcium channels: Splicing for optimal performance. Cell Calcium. 2007;42:409–417. doi: 10.1016/j.ceca.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Altier C., Dale C.S., Kisilevsky A.E., Chapman K., Castiglioni A.J., Matthews E.A., Evans R.M., Dickenson A.H., Lipscombe D., Vergnolle N., et al. Differential role of N-type calcium channel splice isoforms in pain. J. Neurosci. 2007;27:6363–6373. doi: 10.1523/JNEUROSCI.0307-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Andrade A., Denome S., Jiang Y.Q., Marangoudakis S., Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat. Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bell T.J., Thaler C., Castiglioni A.J., Helton T.D., Lipscombe D. Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron. 2004;41:127–138. doi: 10.1016/S0896-6273(03)00801-8. [DOI] [PubMed] [Google Scholar]