Abstract
Cynoglossus monopus, a small benthic fish, belongs to the Cynoglossidae, Pleuronectiformes. It was rarely studied due to its low abundance and cryptical lifestyle. In order to understand the mitochondrial genome and the phylogeny in Cynoglossidae, the complete mitogenome of C. monopus has been sequenced and analyzed for the first time. The total length is 16,425 bp, typically containing 37 genes with novel gene rearrangements. The tRNA-Gln gene is inverted from the light to the heavy strand and translocated from the downstream of tRNA-Ile gene to its upstream. The control region (CR) translocated downstream to the 3’-end of ND1 gene adjoining to inverted to tRNA-Gln and left a 24 bp trace fragment in the original position. The phylogenetic trees were reconstructed by Bayesian inference (BI) and maximum likelihood (ML) methods based on the mitogenomic data of 32 tonguefish species and two outgroups. The results support the idea that Cynoglossidae is a monophyletic group and indicate that C. monopus has the closest phylogenetic relationship with C. puncticeps. By combining fossil records and mitogenome data, the time-calibrated evolutionary tree of families Cynoglossidae and Soleidae was firstly presented, and it was indicated that Cynoglossidae and Soleidae were differentiated from each other during Paleogene, and the evolutionary process of family Cynoglossidae covered the Quaternary, Neogene and Paleogene periods.
Keywords: Cynoglossus monopus, mitochondrial genome, novel rearrangement, intramitochondrial recombination, phylogenetic analyses, divergence times
1. Introduction
Flatfishes (Pleuronectiformes) are unique animals with both eyes moved on one side of the body through asymmetrical development. Furtherly, according to the traditional morphological opinions, the tonguefishes (Cynoglossidae and Soleoidae) are the most specified and have an advanced classification in Pleuronectiformes [1,2]. Cynoglossus monopus belongs to Cynoglossidae in suborder Soleoidei, distributed from the Malay Archipelago to the Indian Ocean, including the west and northward along the South China Sea [1,2]. It is a small benthic fish with ctenoid scale covered on both-side, two lateral line on ocular-side and absent lateral line on blind-side. Especially, the small pedunculate eyes are the distinct diagnosis in C. monopus from other Cynoglossus species.
Traditionally, suborder Soleoidei includes three families: Achiridae, Cynoglossidae and Soleidae [3], among which Achiridae was thought to be separated from Soleidae and represented the primitive sister group to Soleidae and Cynoglossidae [4]. The Cynoglossidae was believed to be derived from the Soleidae as well, and have a closer relationship with Soleidae than Achiridae [5]. In morphology, both eyes of the species in Cynoglossidae and Soleidae turned to the left-side and the right-side, respectively, which is the principle basis to distinguish these two families [1]. Within Cynoglossidae, there are more than 140 species in three genera of two subfamilies. The genera Cynoglossus and Paraplagusia were sister groups belonging to subfamily Cynoglossinae, while Symphurus was the only genus in subfamily Symphurinae [3].
The mitochondrial DNA (mtDNA) is a double-stranded circular DNA molecule. Similar in composition and structure to most vertebrates, it is 15–20 kb in length, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA), a light-strand replication origin (OL), and a control region (CR) that possesses cis-regulatory elements [6,7]. Compared to the nuclear genome, the mitochondrial genome shares maternal inheritance, large copy number, stable gene composition, conserved gene arrangement, and high evolutionary rate [6,8]. Therefore, mitogenome has been widely used to infer phylogenetic relationships and population genetics in animal [9,10,11]. Furthermore, the gene rearrangements have been reported in various vertebrates [11,12,13,14,15,16], but the organization in most fish mitogenomes is generally considered quite conserved [17,18]. However, we analyzed the complete mitogenome sequence of C. monopus and found a novel gene order in this study. Although there are some phylogenetic studies focusing on genus Cynoglossus, the evolutionary histories of Cynoglossidae and Soleidae are still remain unclear since very few reports concentrated on the divergence time among them (Table 1). To fill the gap in genetic information, this study analyzes the phylogeny and evolutionary histories of Cynoglossidae and Soleidae based on the complete mitogenome of C. monopus and all available mitogenomic data of these two families.
Table 1.
Details of species and mitogenomes of Pleuronectiformes used in this study.
| Family | Genus | Species | Length (bp) | Accession ID | Reference |
|---|---|---|---|---|---|
| Cynoglossidae | Cynoglossus | Cynoglossus abbreviatus | 16,417 | JQ349004 | [23] |
| Cynoglossus bilineatus | 16,454 | JQ349000 | [24] | ||
| Cynoglossus gracilis | 16,565 | KT809367 | [25] | ||
| Cynoglossus interruptus | 17,262 | LC482306 | [20] | ||
| Cynoglossus itinus | 16,915 | JQ639062 | Unpublished | ||
| Cynoglossus joyneri | 16,428 | NC030256 | [27] | ||
| Cynoglossus monopus | 16,425 | MT798589 | This study | ||
| Cynoglossus nanhaiensis | 17,130 | MT117229 | [28] | ||
| Cynoglossus puncticeps | 17,142 | JQ349003 | [26] | ||
| Cynoglossus robustus | 16,720 | LC482305 | [21] | ||
| Cynoglossus roulei | 16,565 | MN966658 | [22] | ||
| Cynoglossus semilaevis | 16,731 | EU366230 | [19] | ||
| Cynoglossus senegalensis | 16,519 | MH709122 | [29] | ||
| Cynoglossus sinicus | 16,478 | JQ348998 | [30] | ||
| Cynoglossus trigrammus | 18,369 | KP057581 | [31] | ||
| Cynoglossus zanzibarensis | 16,569 | KJ433559 | [32] | ||
| Paraplagusia | Paraplagusia bilineata | 16,985 | NC023227 | Unpublished | |
| Paraplagusia blochii | 16,611 | JQ349002 | [33] | ||
| Paraplagusia japonica | 16,694 | JQ639066 | [34] | ||
| Soleidae | Aesopia | Aesopia cornuta | 16,737 | KF000065 | [35] |
| Aseraggodes | Aseraggodes kobensis | 16,944 | KJ601760 | [36] | |
| Brachirus | Brachirus orientalis | 16,600 | KJ513134 | [37] | |
| Heteromycteris | Heteromycteris japonicus | 17,111 | JQ639060 | [38] | |
| Liachirus | Liachirus melanospilos | 17,001 | KF573188 | [39] | |
| Pardachirus | Pardachirus pavoninus | 16,573 | KJ461620 | [40] | |
| Pseudaesopia | Pseudaesopia japonica | 16,789 | KJ433482 | [41] | |
| Solea | Solea ovata | 16,782 | KF142459 | [42] | |
| Solea senegalensis | 16,659 | AB270760 | [18] | ||
| Zebrias | Zebrias crossolepis | 16,734 | KJ433564 | [43] | |
| Zebrias quagga | 17,045 | NC023225 | [44] | ||
| Zebrias zebra | 16,758 | JQ700100 | [45] | ||
| Zebrias zebrinus | 16,762 | KC491209 | Unpublished | ||
| Paralichthyidae | Paralichthys | Paralichthys olivaceus | 17,090 | NC002386 | Unpublished |
| Psettodidae | Psettodes | Psettodes erumei | 17,315 | FJ606835 | Unpublished |
2. Results and Discussions
2.1. Genome Organization and Nucleotide Composition
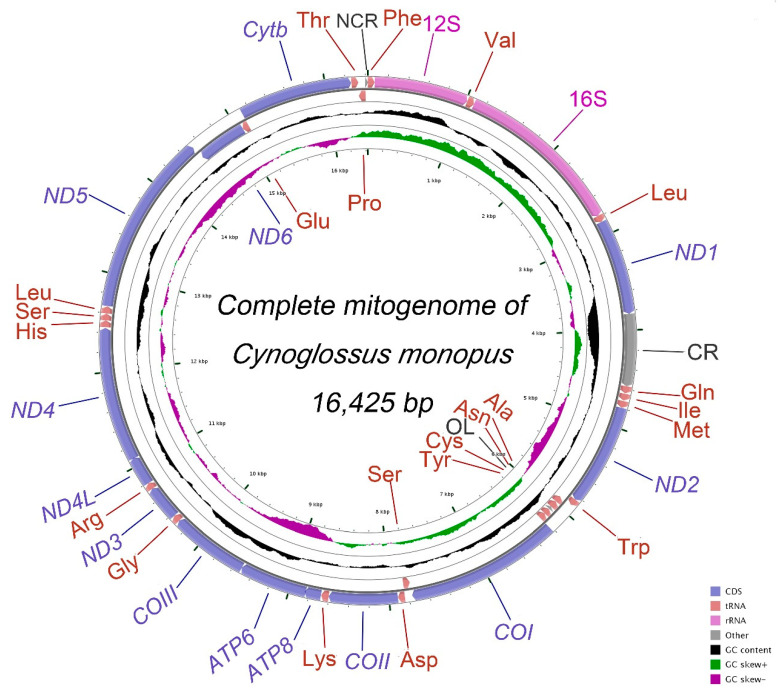
The complete mitochondrial genome of C. monopus is 16,425 bp in length (GenBank accession number MT798589), within the range of other reported Pleuronectiformes mitogenomes from 15,973 bp (Kareius bicoloratus) to 18,369 bp (Cynoglossus trigrammus). This mitogenome contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, the origin of light-strand replication (OL) and a control region (CR) that possesses cis-regulatory elements (Figure 1, Table 2). Except ND6 and eight tRNA genes encoded on the Light-strand, others are encoded on the Heavy-strand. The tRNA-Gln gene was inverted from the L-strand position to the H-strand in the other position. This result is consistent with the findings of previous studies where species in subfamily Cynoglossinae have been found to have large-scale gene rearrangements, and a unique gene order, CR-Gln-Ile-Met, which is different from the typical gene order of CR-Phe-12S-Val-16S-Leu1-ND1-Gln-Ile-Met [19,20,21,22,23,24,25,26,27].
Figure 1.
Graphical genome map of the mitogenome of C. monopus. The genes outside the outermost circle are transcribed clockwise, whereas the genes inside the outermost circle are transcribed counterclockwise. The inside circle shows the GC content and GC skews.
Table 2.
Features of the C. monopus mitochondrial genome.
| Gene | Strand | Position | Size (bp) | Amino Acids (aa) | Codon | Anti-Codon | Intergenic Nucleotides (bp) | ||
|---|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||||
| tRNA-Phe | H | 1 | 68 | 68 | GAA | 0 | |||
| 12S rRNA | H | 69 | 1014 | 946 | 0 | ||||
| tRNA-Val | H | 1015 | 1087 | 73 | TAC | 0 | |||
| 16S rRNA | H | 1088 | 2789 | 1702 | 0 | ||||
| tRNA-Leu | H | 2790 | 2859 | 70 | TAA | 0 | |||
| ND1 | H | 2860 | 3834 | 975 | 324 | ATG | TAA | 0 | |
| Control region | H | 3835 | 4570 | 736 | 0 | ||||
| tRNA-Gln | H | 4571 | 4643 | 73 | GAT | 0 | |||
| tRNA-Ile | H | 4649 | 4717 | 69 | TTG | 5 | |||
| tRNA-Met | H | 4724 | 4793 | 70 | CAT | 6 | |||
| ND2 | H | 4795 | 5838 | 1044 | 347 | ATG | TAA | 1 | |
| tRNA-Trp | H | 5838 | 5905 | 68 | TCA | −1 | |||
| tRNA-Ala | L | 5908 | 5976 | 69 | TGC | 2 | |||
| tRNA-Asn | L | 5979 | 6051 | 73 | GTT | 2 | |||
| OL | L | 6056 | 6079 | 24 | 4 | ||||
| tRNA-Cys | L | 6086 | 6150 | 65 | GCA | 6 | |||
| tRNA-Tyr | L | 6151 | 6218 | 68 | GTA | 0 | |||
| COI | H | 6220 | 7770 | 1551 | 516 | GTG | TAA | 1 | |
| tRNA-Ser | L | 7771 | 7841 | 71 | TGA | 0 | |||
| tRNA-Asp | H | 7844 | 7912 | 69 | GTC | 2 | |||
| COII | H | 7914 | 8604 | 691 | 230 | ATG | T | 1 | |
| tRNA-Lys | H | 8605 | 8680 | 76 | TTT | 0 | |||
| ATP8 | H | 8683 | 8847 | 165 | 54 | ATG | TAA | 2 | |
| ATP6 | H | 8838 | 9521 | 684 | 227 | ATG | TAA | −10 | |
| COIII | H | 9521 | 10,306 | 786 | 261 | ATG | TAA | −1 | |
| tRNA-Gly | H | 10,306 | 10,375 | 70 | TCC | −1 | |||
| ND3 | H | 10,376 | 10,726 | 351 | 116 | ATG | TAA | 0 | |
| tRNA-Arg | H | 10,725 | 10,793 | 69 | TCG | −2 | |||
| ND4L | H | 10,794 | 11,090 | 297 | 98 | ATG | TAA | 0 | |
| ND4 | H | 11,084 | 12,448 | 1365 | 454 | ATG | TAA | −7 | |
| tRNA-His | H | 12,456 | 12,524 | 69 | GTG | 9 | |||
| tRNA-Ser | H | 12,525 | 12,592 | 68 | GCT | 0 | |||
| tRNA-Leu | H | 12,595 | 12,667 | 73 | TAG | 2 | |||
| ND5 | H | 12,671 | 14,524 | 1854 | 617 | ATG | TAA | 3 | |
| ND6 | L | 14,530 | 15,051 | 522 | 183 | ATG | TAG | 5 | |
| tRNA-Glu | L | 15,052 | 15,120 | 69 | TTC | 0 | |||
| Cytb | H | 15,123 | 16,259 | 1137 | 378 | ATG | TAG | 2 | |
| tRNA-Thr | H | 16,263 | 16,331 | 69 | TGT | 3 | |||
| tRNA-Pro | L | 16,331 | 16,401 | 71 | TGG | −1 | |||
| Noncoding region | H | 16,402 | 16,425 | 24 | 0 | ||||
The overall base composition is 30.80% A, 24.04% C, 14.77% G, and 30.39% T, with a high AT content (61.19%). The AT-skew and GC-skew of the C. monopus mitogenome are 0.01 and −0.24, respectively (Table 3). C. monopus showed a higher AT content (63.19%) in 16S rRNA gene than the 12S rRNA (55.31%). The control region is 736 bp in length with the rich AT (70.92%) and poor G (11.41%) content. Moreover, the AT content of 13 PCGs ranged from 54.21% (ND4L) to 66.67% (ATP8). The AT content of tRNAs is 61.10% in average, while the CG content is 38.90%.
Table 3.
Nucleotide composition and skewness levels calculated for sequenced majority strand of the C. monopus.
| Regions | Size (bp) | Nucleotides Composition (%) | AT-Skew | GC-Skew | |||||
|---|---|---|---|---|---|---|---|---|---|
| T | C | A | G | AT | GC | ||||
| Whole genome | 16,425 | 30.39 | 24.04 | 30.80 | 14.77 | 61.19 | 38.81 | 0.01 | −0.24 |
| PCGs | 11,422 | 32.38 | 25.03 | 28.38 | 14.21 | 60.76 | 39.24 | −0.07 | −0.28 |
| 1st codon position | 3808 | 24.91 | 24.52 | 27.10 | 23.46 | 52.02 | 47.98 | 0.04 | −0.02 |
| 2nd codon position | 3807 | 42.10 | 26.48 | 18.15 | 13.27 | 60.25 | 39.75 | −0.40 | −0.33 |
| 3rd codon position | 3807 | 30.94 | 24.06 | 38.92 | 6.08 | 69.86 | 30.14 | 0.11 | −0.60 |
| ATP6 | 684 | 30.85 | 31.43 | 27.49 | 10.23 | 58.33 | 41.67 | −0.06 | −0.51 |
| ATP8 | 165 | 32.73 | 25.45 | 33.94 | 7.88 | 66.67 | 33.33 | 0.02 | −0.53 |
| COI | 1551 | 31.72 | 23.86 | 26.69 | 17.73 | 58.41 | 41.59 | −0.09 | −0.15 |
| COII | 691 | 32.27 | 22.43 | 30.39 | 14.91 | 62.66 | 37.34 | −0.03 | −0.20 |
| COIII | 786 | 30.53 | 27.35 | 25.83 | 16.28 | 56.36 | 43.64 | −0.08 | −0.25 |
| Cytb | 1137 | 34.21 | 25.77 | 26.30 | 13.72 | 60.51 | 39.49 | −0.13 | −0.31 |
| ND1 | 975 | 32.51 | 24.62 | 28.41 | 14.46 | 60.92 | 39.08 | −0.07 | −0.26 |
| ND2 | 1044 | 31.90 | 26.44 | 31.32 | 10.34 | 63.22 | 36.78 | −0.01 | −0.44 |
| ND3 | 351 | 32.76 | 26.21 | 27.35 | 13.68 | 60.11 | 39.89 | −0.09 | −0.31 |
| ND4 | 1365 | 32.60 | 24.62 | 30.26 | 12.53 | 62.86 | 37.14 | −0.04 | −0.33 |
| ND4L | 297 | 29.97 | 29.29 | 24.24 | 16.50 | 54.21 | 45.79 | −0.11 | −0.28 |
| ND5 | 1854 | 31.01 | 25.57 | 30.91 | 12.51 | 61.92 | 38.08 | 0.00 | −0.34 |
| ND6 | 522 | 41.38 | 12.26 | 21.65 | 24.71 | 63.03 | 36.97 | −0.31 | 0.34 |
| tRNAs | 1540 | 30.19 | 17.92 | 30.91 | 20.97 | 61.10 | 38.90 | 0.01 | 0.08 |
| rRNAs | 2648 | 25.11 | 20.85 | 35.31 | 18.73 | 60.42 | 39.58 | 0.17 | −0.05 |
| Control region | 736 | 35.87 | 17.66 | 35.05 | 11.41 | 70.92 | 29.08 | −0.01 | −0.21 |
2.2. Protein-Coding Genes (PCGs), Transfer RNAs and Ribosomal RNAs
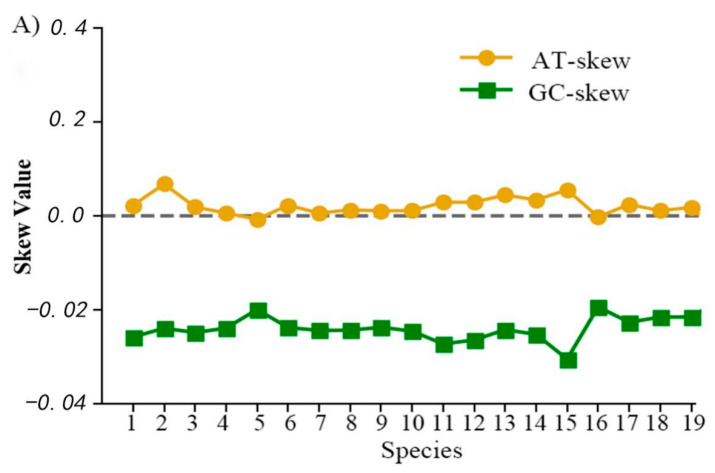
In the mitogenome of C. monopus, except for the ND3, started with ATT and COII terminated with T, most PCGs have typical initiation codons (ATG or GTG) and termination codons (TAA or TAG) (Table 2). The size of 13 PCGs ranged from 165 bp (ATP8) to 1854 bp (ND5), and the total length of PCGs is 11,422 bp, similar to other Cynoglossidae species (Figure 2B). The GC-skews of the 13 PCGs were all negative and the majority of the AT-skew values were negative, similarly. The AT skews of ND3 and ND6 were the lowest and the GC-skews of COI and COII were the highest (Table 3).
Figure 2.
Graphical illustration shown the usage bias of AT and GC (AT-skew and GC-skew values) (A), and the length of protein-coding genes (PCGs), tRNAs, rRNAs, and control regions (B) in the mitogenomes of 19 species in the subfamily Cynoglossinae. Note: Species 1–21: C. abbreviatus, C. bilineatus, C. gracilis, C. interruptus, C. itinus, C. joyneri, C. monopus, C. nanhaiensis, C. puncticeps, C. robustus, C. roulei, C. semilaevis, C. senegalensis, C. sinicus, C. trigrammus, C. zanzibarensis, P. bilineata, P. blochii and P. japonica.
The 22 tRNA genes of C. monopus ranged from 65 bp (tRNA-Cys) to 76 bp (tRNA-Lys), with a total of 1540 bp. The 12S rRNA (946 bp) and 16S rRNA genes (1702 bp) of the C. monopus mitogenome are located in the typical position between tRNA-Phe and tRNA-Leu (UUR), and separated by tRNA-Val, with a high AT content of 60.42%. Additionally, the origin of light-strand is 24 bp in length between tRNA-Asn and tRNA-Cys.
2.3. Mitochondrial Gene Codon Usage and Skewness
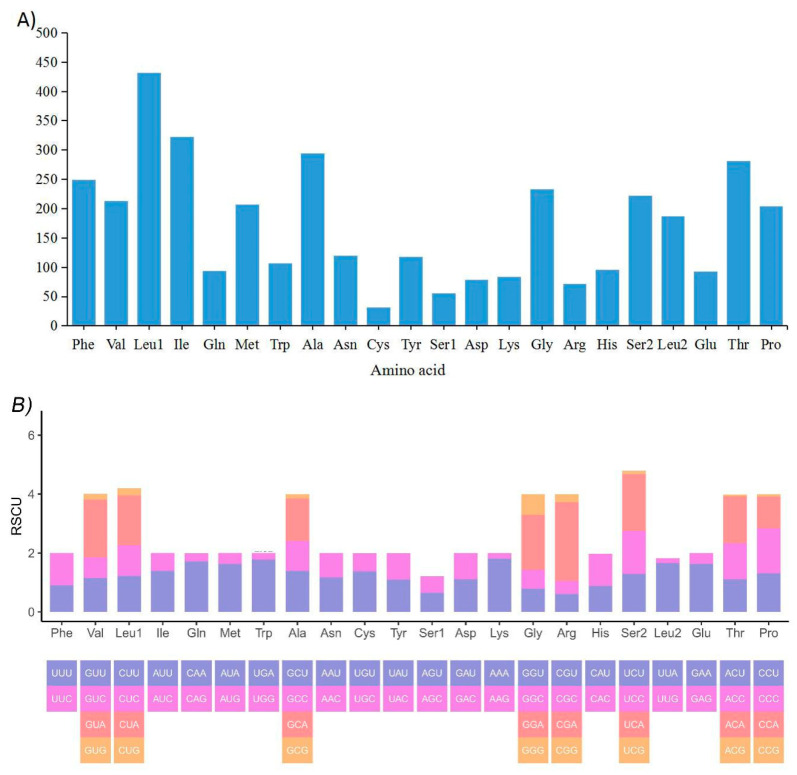
The amino acids were utilized by either two or four different codons, respectively. Ile was the most frequently used, while Met and Trp were the least frequently used (Figure 3A). In addition, the relative synonymous codon usage (RSCU) analysis indicated that Arg (AGG, AGA), Pro (CCC) and Ala (GCU) were the most frequent, and Ser (UCG), Thr (ACG), Pro (CCG) and Lys (AAG) were rare.
Figure 3.
Amino acid composition (A) and relative synonymous codon usage (B) in the mitogenome of C. monopus.
Moreover, the AT-skew and GC-skew in the mitogenome of the subfamily Cynoglossinae were reflected in the codon usage consistently (Figure 3B). The RSCU values indicated that codons with A or U in the third position were more frequent than C or G. Additionally, the GC-skew values are negative in the available mitogenomes of Cynoglossidae species (Figure 2A). In contrast, most of the AT skews are positive, except for C. itinus.
2.4. Gene Rearrangement
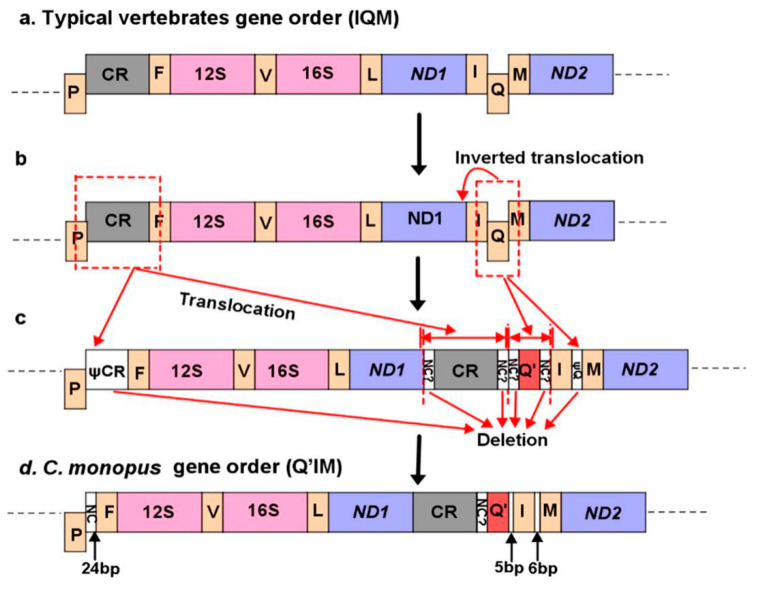
Similar to other Cynoglossus and Paraplagusia species which mitogenome available, the CR of C. monopus is translocated downstream to the place between ND1 and tRNA-Gln instead of the typical location between tRNA-Pro and tRNA-Phe (Figure 1). The translocation left a functionally undefined 24 bp trace fragment in the original CR position. Furthermore, the tRNA-Gln gene (Q) translocated from the downstream of tRNA-Ile gene (I) to its upstream with inverted encoding direction (Q’). The result was the formation of the Q’-I-M gene order, which is different from the typical I-Q-M gene order in most vertebrates (Figure 4a).
Figure 4.
Inferred gene rearrangement between the gene order of typical vertebrates and the mitogenome of C. monopus. The typical vertebrate gene order (a); inferred intermediate processes of gene rearrangement (b,c); the gene order in the mitogenome of C. monopus (d).
Among several mitochondrial gene rearrangement models [46,47,48], the intramitochondrial recombination is the most probable mechanism to explain the rearrangement events in the mitogenome of C. monopus based on the principle of parsimony. The hypothesized intermediate steps are as follows. Firstly, the whole Control region (likely carrying some neighbour sequences) translocated to the downstream of ND1 gene, and left a duplicated partial Control region in the original position. At the same time, the tRNA-Gln gene was inversely translocated to the upstream of tRNA-Ile with some neighbour sequences and left a duplicated partial tRNA-Gln fragment between tRNA-Ile and tRNA-Met. The rearrangement formed a new ND1-CR-Q’-I-M region in the mitogenome of C. monopus with unfunctional gene fragments connecting each gene (Figure 4b). Secondly, after a rapid deletion process under the strong selective pressure, the unfunctional gene fragments in each gene junction left a 24 bp trace fragment in the original CR position and 5 and 6 bp intergenic spacers between Q’-I and I-M gene junctions, respectively (Figure 4c,d).
2.5. Phylogenetic Analyses
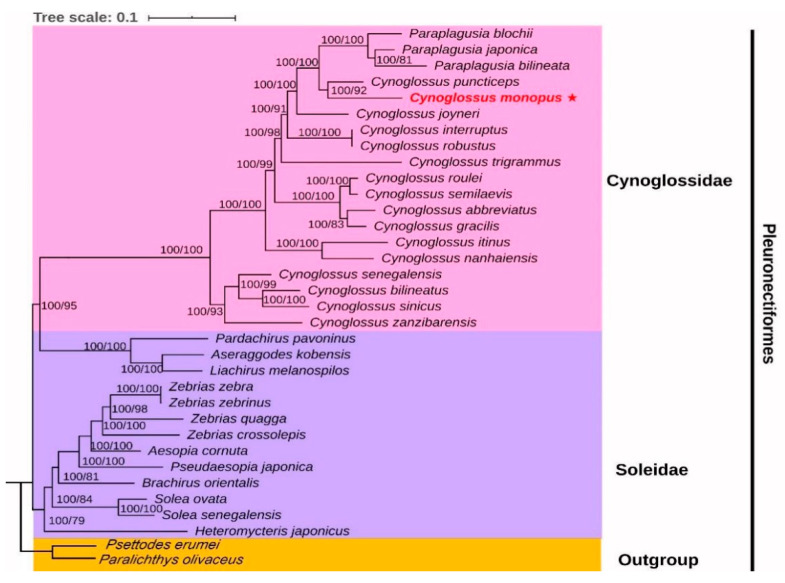
Both BI and ML analyses produced almost identical topologies with similar branch lengths. Most clades were strong supported by high bootstraps (ML) and posterior probabilities (Bayesian) values (Figure 5). Contrary to the traditional classification [36,39,40], the molecular phylogenetic tree showed a clade with three Soleidae species (P. pavoninus, A. kobensis and L. melanospilos) clustered to Cynoglussidae as a sister group rather than other Soleidae species (Figure 5). This suggests that Soleidae is not a monophyletic group. However, the insufficient species could cause questionable phylogeneic results [49], and as Soleidae is a large family including 32 genera and at least 180 species [50], more species will be necessarily needed for further investigation. Furthermore, the most terminal branch relationship within Cynoglossus is consistent with traditioanal taxon. This supports the opinion that lateral line number and scale characteristics are valuable morphological diagnoses in classification [1].
Figure 5.
Phylogenetic tree of C. monopus was performed using partial genomes of 32 species of Pleuronectiformes and Psettodes erumei and Paralichthys olivaceus were used as outgroups, with Bayesian analyses and Maximum likelihood analyses. Species in red indicates sequence generated in this study. Bootstrap support (right) and Bayesian posterior probability values (left) of each clade are displayed next to the nodes.
Among Cynoglussidae, compared to other Cynoglossus species, C. monopus had the closest phylogenetic relationship to C. puncticeps. Traditionally, they were also classified into the subgenus Cynoglossoides by sharing the most similarities with both sides covered with ctenoid scale, two lateral lines on the ocular-side and an absent lateral line on the blind-side [1]. However, the clade of C. monopus and C. puncticeps clustered to the Paraplagusia rather than other Cynoglossus species. This suggests that the relationship between Cynoglossus and Paraplagusia is more complex than expected and needs further study.
2.6. Divergence Time Analyses
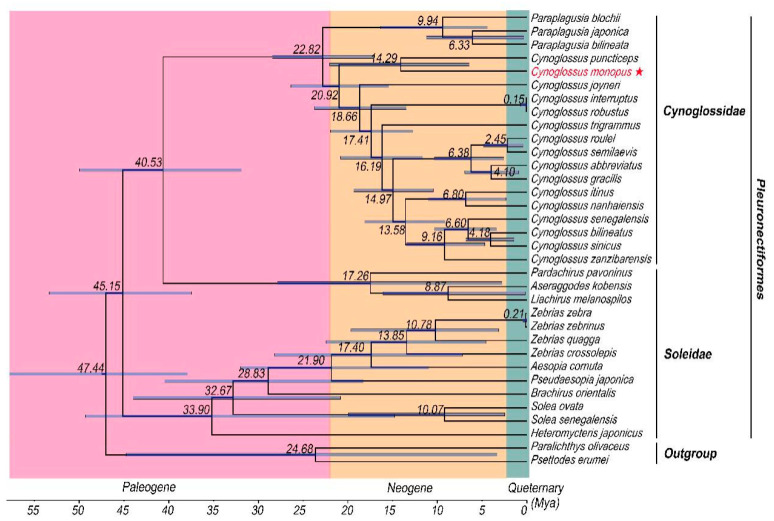
The advent of the phylogenomic era has significantly improved our understanding of the taxonomy and phylogenetic relationships of many animals [51]. Compared with other flatfish studies, the divergence time estimations within or between families Cynoglossidae and Soleidae were additionally conducted based on two calibration constraints in this study. The divergence time between Cynoglossidae and Soleidae could be occurred at about 45.15 Mya (37.47–49.99 Mya). Furthermore, the divergence time of the subfamily Cynoglossinae could be dated back to 20.92 Mya (17.11–28.44 Mya) earlier than the fossil record of C. leuchsi (lower and middle Miocene) [5].
The Chronogram for the 34 species of Pleuronectiformes covered three geological epochs, including the Quaternary, Neogene, and Paleogene periods (Figure 6). It was indicated that family Soleidae (33.90 Mya (14.79–49.30 Mya)) evolved earlier than family Cynoglossidae (22.82 Mya (17.11–28.44 Mya)). The fossil-calibrated divergence time estimated that genus Paraplagusia diverged at 22.82 Mya (17.11–28.44 Mya) within the Neogene as the most primitive species in family Cynoglossidae. During this period, C. monopus and C. puncticeps began to differentiate from other species at about 14.29 Mya (6.43–22.04 Mya). In particular, we found that most species of family Soleidae were divided during the Neogene period and the speciation of family Cynoglossidae was happening during the Quaternary period. More importantly, large-scale gene rearrangement was recently detected to occur in genera Cynoglossus and Paraplagusia, and they were differentiated from the clade of Soleidae (P. pavoninus, A. kobensis and L. melanospilos as a subgroup) about 40.53 Mya (31.87–49.99 Mya). However, another clade of family Soleidae found no evidence of gene rearrangement, and the two clades began to differentiate from each other at about 45.15 Mya (37.47–49.99 Mya).
Figure 6.
Chronogram for the 32 species of Pleuronectiformes with P. erumei and P. olivaceus as outgroups based on the concatenated nucleotide sequences of the 12 PCGs (except ND6 and the third codon positions) and 2 rRNA genes using BEAST analysis. Numbers near the nodes indicate the average estimated divergence time estimated in Mya, and the 95% confidence intervals for each node are shown in blue bars.
3. Materials and Methods
3.1. Specimen Collection and DNA Extraction
A single specimen of C. monopus was collected from Sanya (E 108°56′, N 18°09′), Hainan Province. The voucher specimen (Voucher No. HNSY2010060432) was deposited in the College of Marine Sciences, South China Agricultural University, Guangzhou, China (SCAU). Animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of SCAU. Genomic DNA was extracted from the muscle of C. monopus according to the standard phenol-chloroform procedure [52]. The data analysis method is based on the previous study [53].
3.2. PCR Amplification and Sequencing
To ensure a sufficient amount of DNA for the amplification and sequencing of these small fishes, the parameters of the LA-PCR reactions were mostly in accordance with the manufacturer’s recommendations. PCR products were purified using the gel purification kit (Invitrogen) after gel-cutting (1.5% TBE agarose). Purified PCR products were sequenced directly on an ABI 3730 automated sequencer (Life Technologies Holdings Pte Ltd, Tuas, Singapore) with ABI PRISM BigDye Terminators v3.0 Cycle Sequencing (ABI) using the primer-walking strategy. The eight fragments were separated from the complete mitogenome of C. monopus, with universal primer for Cynoglossidae mitogenome.
3.3. Sequene Analysis
Sequence data were analyzed and compiled to create the complete genome using the SeqMan program from Lasergene soft package (DNASTAR, Madison, WI, USA), and manually adjusted in a few cases. The complete mitogenome was annotated using the software of Sequin v16.0 (National Library of Medicine, Bethesda, MD, USA). Mitochondrial tRNA genes and their secondary structures were obtained by ARWEN v1.2 [54], and identified by tRNAscan-SE Search Server v2.0 (Washington University School of Medicine, St Louis, MO, USA) [55] using default search mode, then anticodons were further confirmed. Annotation and accurate boundary determination of protein-coding and ribosomal RNA genes were first performed by NCBI-BLAST searches, and then by alignment and manual comparisons with the other released reference mitogenomes of Cynoglossidae species using DNAMAN v6.0 (Lynnon Biosoft, San Ramon, CA, USA).
The complete mitogenome of C. monopus was uploaded to GenBank with accession number MT798589. The graphical genome map of the C. monopus mitogenome was drawn using CGView Server v1.0 [56]. The base composition, codon usage and RSCU values were obtained using MEGA 7.0 (Tokyo Metropolitan University, Tokyo, Japan) [57]. Strand asymmetry was estimated using the following formulas by Perna and Kocher (1995) [58]: AT skew = [A − T]/[A + T] and GC skew = [G − C]/[G + C].
3.4. Phylogenetic and Divergence Time Analyses
Phylogenetic analyses were conducted using 19 Cynoglossidae species and 13 Soleidae species with P. erumei and P. olivaceus as outgroups. All sequences were available in GenBank (27/7/2020). We aligned DNA sequences of 12 protein-coding genes (except ND6) and two rRNA genes in the 34 species using the MAFFT program with the default parameters [59]. The alignments of PCGs (except ND6) excluded the start codon and the stop codon. Then, ambiguously aligned fragments of two alignments were removed using Gblocks [60], and exported two gblocks alignments. We used a dataset comprised of concatenated the gblocks of 12 protein-coding genes (the first and second codon positions, except ND6) and 2 rRNA genes.
Then, the data were divided into three pre-defined partitions for the best partitioning scheme using PartitionFinder 2.0 (Macquarie University Genes to Geosciences Centre, North Ryde, NSW, Australia) [61], with the greedy algorithm. ModelFinder [62] lugin integrated into PhyloSuite v1.2.1 (Bio-Transduction Lab, Wuhan, China) [63] was used to select the best-fit partition model (Edge-unlinked). The best-fit model according to BIC: GTR + F + G4 was selected as the optimal model for the glocks of first codons of PCGs (except ND6) and rRNAs, respectively, whereas TVM + F + G4 was chosen for the gblocks of two rRNA genes. Additionally, phylogenetic analyses were performed using the Bayesian analyses (BI) and Maximum Likelihood (ML) methods [64,65]. ML analysis was inferred using IQ-TREE v1.6.2 [66] plugin integrated into PhyloSuite v1.2.1 under Edge-linked partition model for 10,000 ultrafast [67] bootstraps, approximate Bayes test [64], as well as the Shimodaira–Hasegawa-like approximate likelihood-ratio test [68]. Additionally, Bayesian inference with partition model was conducted in MrBayes 3.2.6 (University of California, La Jolla, San Diego, CA, USA) [65] under the partition model (two parallel runs, 2,000,001 generations), in which the initial 25% of sampled data were discarded as burn-in with default settings and 5 × 106 metropolis-coupled Markov chain Monte Carlo (MCMC) generations. The iTOL dataset files produced by PhyloSuite were then used to visualize and annotate the phylograms and gene orders in iTOL [69].
The evolutionary analysis was inferred by BEAST v1.10.4 (open source under the GNU lesser general public license) using Bayesian Information Criterion based on the two gblocks (the first and second codon positions of PCGs, except ND6) and two rRNA genes of 34 Pleuronectiformes species [70]. The divergence times were presented in the Time Tree database (http://www.timetree.org/) [71] and fossil-based comparative analyses [72]. The time tree was computed using two calibration constraints: the most recent common ancestor (MRCA) of genus Cynoglossus was estimated to be lower and middle Miocene period based on fossilized C. leuchsi and the MRCA of both Cynoglossidae and Soleidae was estimated to be at the lower Eocene at least 45 Mya [5]. Molecular dating involved a Birth-Death process as the tree prior, and an uncorrelated relaxed clock as the best model. The chains of 1 × 108 samples were run for the MCMC analysis, and the 10% of all samples was burn-in using TreeAnnotator. Tracer v1.7.1 was used to confirm the output [73]. FigTree v1.4.3 was used to edit the results.
4. Conclusions
This study suggests that mitochondrial gene rearrangement in Cynoglossidae is a single originated evolutionary event, which occurred in the common ancestor of Cynoglossus and Paraplagusia before at least 22.82 Mya (17.11–28.44 Mya). The highly similar rearranged gene order in all available Cynoglossus and Paraplagusia species [18,35,36,37,38,39,40,41,42,43,44,45] inferred that novel gene order possesses some selective advantage in this group, then kept it conserved in the whole lineage. Even the details of the molecular mechanism are still unclear; the intramitochondrial recombination is the most probable model to explain the process of gene rearrangement in this group based on the principle of parsimony.
The phylogenetic relationships constructed by ML and BI method are consistent. The time tree covers three geological epochs, including the Quaternary, Neogene, and Paleogene periods. The divergence times show that C. monopus and C. puncticeps begin to differentiate from other species at about 14.29 Mya (6.43–22.04 Mya) within the Neogene period. Tonguefish is a highly specialized body. The traditional morphological classifications of tonguefish are often controversial due to the unstable diagnosis caused by its asymmetrical development. The mitochondrial rearrangement is a helpful gene marker to reconstruct the phylogenetic relationship in this group with mitogenome and other molecular data.
Author Contributions
C.W. analyzed the data, prepared the figures and tables, and wrote the paper. H.C. collected the samples and revised the drafts of the paper. S.T. and C.Y. performed the experiments, analyzed the data. X.C. conceived and designed the experiments, supervised the work, and polished the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Special National Project on investigation of basic resources of China (Grant SQ2019FY010009), the National Natural Science Foundation of China (41666008) and China-ASEAN Maritime Cooperation Fund (CAMC-2018F).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Li S.Z., Wang H.M. Fauna Sinica (Osteichthyes): Pleuronectiformes. Science Press; Beijing, China: 1995. [Google Scholar]
- 2.Menon A. A systematic monograph of the tongue soles of the genus Cynoglossus Hamilton-Buchanan (Pisces: Cynoglossidae) Smithson. Contrib. Zool. 1977;238:1–129. doi: 10.5479/si.00810282.238. [DOI] [Google Scholar]
- 3.Chapleau F. A phylogenetic reassessment of the monophyletic status of the family Soleidae, with notes on the suborder Soleoidei. Can. J. Zool. 1988;66:2797–2810. doi: 10.1139/z88-408. [DOI] [Google Scholar]
- 4.Chapleau F. Pleuronectiform relationships: A cladistic reassessment. Bull. Mar. Sci. 1993;52:516–540. [Google Scholar]
- 5.Schwarzhans W.A. comparative morphological treatise of recent and fossil otoliths of the order Pleuronectiformes/Werner Schwarzhans. München Pfeil. 1999;2:1–396. [Google Scholar]
- 6.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzeng C.S., Hui C.F., Shen S.C., Huang P.C. The complete nucleotide sequence of the Crossostoma lacustre mitochondrial genome: Conservation and variations among vertebrates. Nucleic Acids Res. 1992;20:4853–4858. doi: 10.1093/nar/20.18.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolstenholme D.R. Animal Mitochondrial DNA: Structure and Evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y., Liu H., Qi L., Kong L., Li Q. Complete Mitochondrial Genomes of Two Toxin-Accumulated Nassariids (Neogastropoda: Nassariidae: Nassarius) and Their Implication for Phylogeny. Int. J. Mol. Sci. 2020;21:10. doi: 10.3390/ijms21103545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X., He H., Shen H., Tang J., Dong W., Shi Y., Wu S., Zhang F., Liang G. Comparative mitochondrial genome analysis of Dendrolimus houi (Lepidoptera: Lasiocampidae) and phylogenetic relationship among Lasiocampidae species. PLoS ONE. 2020;15:e0232527. doi: 10.1371/journal.pone.0232527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C., Huang X., Deng J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020;158:854–864. doi: 10.1016/j.ijbiomac.2020.04.257. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Yu J., Zhou M., Yang J. Complete mitochondrial genome of Japalura flaviceps: Deep insights into the phylogeny and gene rearrangements of Agamidae species. Int. J. Biol. Macromol. 2019;125:423–431. doi: 10.1016/j.ijbiomac.2018.12.068. [DOI] [PubMed] [Google Scholar]
- 13.Eberhard J.R., Wright T.F. Rearrangement and evolution of mitochondrial genomes in parrots. Mol. Phylogenet. Evol. 2016;94:34–46. doi: 10.1016/j.ympev.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macey J.R., Larson A., Ananjeva N.B., Fang Z., Papenfuss T.J. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- 15.Yuan M.L., Wang D.D., Wang B.J., Dou W., Wang J.J. The complete mitochondrial genome of the citrus red mite Panonychus citri (Acari: Tetranychidae): High genome rearrangement and extremely truncated tRNAs. BMC Genom. 2010;11:597. doi: 10.1186/1471-2164-11-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong L., Lu X., Luo H., Zhang Y., Shi W., Liu L., Lu Z., Liu B., Jiang L. Novel gene rearrangement pattern in Cynoglossus melampetalus mitochondrial genome: New gene order in genus Cynoglossus (Pleuronectiformes: Cynoglossidae) Int. J. Biol. Macromol. 2020;149:1232–1240. doi: 10.1016/j.ijbiomac.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Clayton D.A. Transcription and Replication of Animal Mitochondrial DNAs. Int. Rev. Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- 18.Manchado M., Catanese G., Ponce M., Funes V., Infante C. The complete mitochondrial genome of the Senegal sole, Solea senegalensis Kaup. Comparative analysis of tandem repeats in the control region among soles. DNA Seq. 2007;18:169–175. doi: 10.1080/10425170701308956. [DOI] [PubMed] [Google Scholar]
- 19.Kong X., Dong X., Zhang Y., Shi W., Wang Z., Yu Z. A novel rearrangement in the mitochondrial genome of tongue sole, Cynoglossus semilaevis: Control region translocation and a tRNA gene inversion. Genome. 2009;52:975–984. doi: 10.1139/G09-069. [DOI] [PubMed] [Google Scholar]
- 20.Song H.Y., Kim J.K., Jo S., Jung S.H., Kim B., Choi Y.J., Yoo J.S., Lee D.S. The complete mitochondrial genome of Cynoglossus interruptus and its novel rearrangement (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA Part B. 2019;4:2924–2925. doi: 10.1080/23802359.2019.1660262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song H.Y., Kim J.K., Jo S., Jung S.H., Lee D.S., Kim B., Choi Y.J., Yoo J.S. Gene rearrangements in the mitochondrial genome of robust tonguefish, Cynoglossus robustus (Pleuronectiformes: Cynoglossidae) and a comparative analysis with other Cynoglossus fishes. Mitochondrial DNA Part B. 2019;5:353–354. doi: 10.1080/23802359.2019.1637297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C., Liu M., Tian S., Yang C., Chen X. The complete mitochondrial genome of Cynoglossus roulei (Pleuronectiformes: Cynoglossidae): Novel rearrangement and phylogenetic position analysis. Mitochondrial DNA Part B. 2020;5:1439–1440. doi: 10.1080/23802359.2020.1734501. [DOI] [Google Scholar]
- 23.Shi W., Gong L., Kong X.Y. The complete mitochondrial genome sequence of Cynoglossus abbreviatus (Pleuronectiformes: Cynoglossidae) with control region translocation and tRNA-Gln gene inversion. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:2157–2158. doi: 10.3109/19401736.2014.982604. [DOI] [PubMed] [Google Scholar]
- 24.Shi W., Li D.H., Kong X.Y. Gene rearrangements in the mitochondrial genome of Cynoglossus bilineatus (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:2161–2162. doi: 10.3109/19401736.2014.982606. [DOI] [PubMed] [Google Scholar]
- 25.Wei M., Liu Y., Guo H., Zhao F., Chen S. Characterization of the complete mitochondrial genome of Cynoglossus gracilis and a comparative analysis with other Cynoglossinae fishes. Gene. 2016;591:369–375. doi: 10.1016/j.gene.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Yang M., Shi W., Miao X.G., Kong X.Y. The complete mitochondrial genome of Cynoglossus puncticeps (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:1233–1234. doi: 10.3109/19401736.2014.945533. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B., Jia L., Liu K., Sun J. The complete mitochondrial genome of Cynoglossus joyneri and its novel rearrangenment. Mitochondrial DNA Part B. 2017;2:581–582. doi: 10.1080/23802359.2017.1365642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian S.L., Wang C., Yan Y.R., Chen X. Characterization of the complete mitochondrial genome of Cynoglossus nanhaiensis (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA Part B. 2020;5:2613–2614. doi: 10.1080/23802359.2020.1781582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zealous G.F., Kim N., Andriyono S., Kim H.W. Characterization of the mitogenome of Cynoglossus Senegalensis (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA Part B. 2018;3:1191–1192. doi: 10.1080/23802359.2018.1524728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi W., Jiang J.X., Miao X.G., Kong X.Y. The complete mitochondrial genome sequence of Cynoglossus sinicus (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA. 2015;26:865–866. doi: 10.3109/19401736.2013.861433. [DOI] [PubMed] [Google Scholar]
- 31.Mu X., Wang X., Liu Y., Song H., Liu C., Gu D., Wei H., Luo J., Hu Y. An unusual mitochondrial genome structure of the tonguefish, Cynoglossus trigrammus: Control region translocation and a long additional non-coding region inversion. Gene. 2015;573:216–224. doi: 10.1016/j.gene.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 32.Shi W., Yang M., Si L., Li D., Kong X.Y. The complete mitochondrial genome of Cynoglossus zanzibarensis (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:2172–2173. doi: 10.3109/19401736.2014.982611. [DOI] [PubMed] [Google Scholar]
- 33.Li D.H., Shi W., Gong L., Yang M., Kong X.Y. The complete mitochondrial genome of Paraplagusia blochii (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:92–93. doi: 10.3109/19401736.2013.873918. [DOI] [PubMed] [Google Scholar]
- 34.Gong L., Shi W., Wang Z.M., Miao X.G., Kong X.Y. Control region translocation and a tRNA gene inversion in the mitogenome of Paraplagusia japonica (Pleuronectiformes: Cynoglossidae) Mitochondrial DNA. 2013;24:671–673. doi: 10.3109/19401736.2013.773984. [DOI] [PubMed] [Google Scholar]
- 35.Wang S.Y., Shi W., Wang Z.M., Gong L., Kong X.Y. The complete mitochondrial genome sequence of Aesopia cornuta (Pleuronectiformes: Soleidae) Mitochondrial DNA. 2015;26:114–115. doi: 10.3109/19401736.2013.803544. [DOI] [PubMed] [Google Scholar]
- 36.Chen X., Ai W., Peng X., Xiang D., Huang X. Complete mitochondrial genome of milk sole fish Aseraggodes kobensis (Pleuronectiformes: Soleidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:626–627. doi: 10.3109/19401736.2014.908376. [DOI] [PubMed] [Google Scholar]
- 37.Chen X., Ai W., Shi X., Pan L. Complete mitogenome of the oriental sole Brachirus orientalis. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:527–528. doi: 10.3109/19401736.2014.905840. [DOI] [PubMed] [Google Scholar]
- 38.Shi W., Jiang J.X., Miao X.G., Kong X.Y. The complete mitochondrial genome sequence of Heteromycteris japonicus (Pleuronectiformes: Soleidae) Mitochondrial DNA. 2014;25:257–258. doi: 10.3109/19401736.2013.800493. [DOI] [PubMed] [Google Scholar]
- 39.Gong L., Shi W., Wang S.Y., Kong X.Y. The complete mitochondrial genome of Liachirus melanospilos (Pleuronectiformes: Soleidae) Mitochondrial DNA. 2015;26:732–733. doi: 10.3109/19401736.2013.845763. [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Ai W., Xiang D., Shi X., Pan L. Complete mitogenome of the peacock sole Pardachirus pavoninus (Pleuronectiformes: Soleidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:463–464. doi: 10.3109/19401736.2014.900668. [DOI] [PubMed] [Google Scholar]
- 41.Chen X., Ai W., Xiang D., Shi X., Pan L. The complete mitochondrial genome of the wavyband sole Pseudaesopia japonica (Pleuronectiformes: Soleidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:420–421. doi: 10.3109/19401736.2014.898286. [DOI] [PubMed] [Google Scholar]
- 42.Shi W., Gong L., Wang S.Y., Kong X.Y. The complete mitochondrial genome of Solea ovata (Pleuronectiformes: Soleidae) Mitochondrial DNA. 2014;25:454–455. doi: 10.3109/19401736.2013.814117. [DOI] [PubMed] [Google Scholar]
- 43.Gong L., Si L.Z., Wei S., Kong X.Y. The complete mitochondrial genome of Zebrias crossolepis (Pleuronectiformes: Soleidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:1235–1236. doi: 10.3109/19401736.2014.945534. [DOI] [PubMed] [Google Scholar]
- 44.Li D.H., Shi W., Miao X.G., Kong X.Y. The complete mitochondrial genome of Zebrias quagga (Pleuronectiformes: Soleidae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:1125–1126. doi: 10.3109/19401736.2014.933331. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z.M., Shi W., Jiang J.X., Wang S.Y., Miao X.G., Huang L.M., Kong X.Y. The complete mitochondrial genome of a striped sole Zebrias zebrinus (Pleuronectiformes: Soleidae) Mitochondrial DNA. 2013;24:633–635. doi: 10.3109/19401736.2013.772160. [DOI] [PubMed] [Google Scholar]
- 46.Moritz C., Dowling T.E., Brown W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987;18:269–292. doi: 10.1146/annurev.es.18.110187.001413. [DOI] [Google Scholar]
- 47.Lavrov D.V., Boore J.L., Brown W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and non random loss. Mol. Biol. Evol. 2002;19:163–169. doi: 10.1093/oxfordjournals.molbev.a004068. [DOI] [PubMed] [Google Scholar]
- 48.Dowton M., Campbell N.J. Intramitochondrial recombination-is it why some mitochondrial genes sleep around? Trends Ecol. Evol. 2001;16:269–271. doi: 10.1016/S0169-5347(01)02182-6. [DOI] [PubMed] [Google Scholar]
- 49.Campbell M.A., Chen W.J., Lopez J.A. Are flatfishes (Pleuronectiformes) monophyletic? Mol. Phylogenet. Evol. 2013;69:664–673. doi: 10.1016/j.ympev.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Der Laan R., Eschmeyer W.N., Fricke R. Family-group names of Recent fishes. Zootaxa. 2014;3882:1–230. doi: 10.11646/zootaxa.3882.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Chen D., Liu Y., Davison G., Li Y.D., Gao S., Hu J., Li S.H., Zhang Z. Disentangling the evolutionary history and biogeography of hill partridges (Phasianidae, Arborophila) from low coverage shotgun sequences. Mol. Phylogenet. Evol. 2020;151:106895. doi: 10.1016/j.ympev.2020.106895. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning. A Laboratorymanual. Cold Spring Harbor Laboratory Press; New York, NY, USA: 1989. [Google Scholar]
- 53.Chen X., Ai W., Ye L., Wang X., Lin C., Yang S. The complete mitochondrial genome of the grey bamboo shark (Chiloscyllium griseum) (Orectolobiformes: Hemiscylliidae): Genomic characterization and phylogenetic application. Acta Oceanol. Sin. 2013;32:59–65. doi: 10.1007/s13131-013-0298-0. [DOI] [Google Scholar]
- 54.Laslett D., Canback B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 55.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucl Acids Res. 2016;44:54–57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant J.R., Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perna N.T., Kocher T.D. Patterns of Nueleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 59.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 61.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution formolecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 62.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang D., Gao F., Jakovlic I., Zhou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 64.Anisimova M., Gil M., Dufayard J.F., Dessimoz C., Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minh B.Q., Nguyen M.A., von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2010;32:268–274. [Google Scholar]
- 69.Letunic I., Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: A resource for timelines, timetrees, and divergence time. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 72.Betancur R., Orti G., Pyron R.A. Fossil-based comparative analyses reveal ancient marine ancestry erased by extinction in ray-finned fishes. Ecol. Lett. 2015;18:441–450. doi: 10.1111/ele.12423. [DOI] [PubMed] [Google Scholar]
- 73.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]