Abstract
Background: Prognosis of oral squamous cell carcinoma (OSCC) is difficult to exactly assess on pre-operative biopsies. Since OSCC DNA methylation profile has proved to be a useful pre-operative diagnostic tool, the aim of the present study was to evaluate the prognostic impact of DNA methylation profile to discriminate OSCC with high and low aggressive potential. Methods: 36 OSCC cases underwent neoplastic cells collection by gentle brushing of the lesion, before performing a pre-operative biopsy. The CpG islands methylation status of 13 gene (ZAP70, ITGA4, KIF1A, PARP15, EPHX3, NTM, LRRTM1, FLI1, MiR193, LINC00599, MiR296, TERT, GP1BB) was studied by bisulfite Next Generation Sequencing (NGS). A Cox proportional hazards model via likelihood-based component-wise boosting was used to evaluate the prognostic power of the CpG sites. Results: The boosting estimation identified five CpGs with prognostic significance: EPHX3-24, EPHX3-26, ITGA4-3, ITGA4-4, and MiR193-3. The combination of significant CpGs provided promising results for adverse events prediction (Brier score = 0.080, C-index = 0.802 and AUC = 0.850). ITGA4 had a strong prognostic power in patients with early OSCC. Conclusions: These data confirm that the study of methylation profile provides new insights into the molecular mechanisms of OSCC and can allow a better OSCC prognostic stratification even before surgery.
Keywords: DNA methylation, bisulfite-SEQ, oral squamous cell carcinoma, pre-operative prognostic test, oral cancer, oral oncology
1. Introduction
Oral squamous cell carcinoma (OSCC) is the tenth most common cancer in the world. The annual estimated incidence is approximately 389,000 per year [1]. Despite the improvement in surgical techniques and radio-chemotherapy, the mortality rates have remained unchanged (around 50% within 5 years of diagnosis) [2].
Prevention, early detection, and adequate pre-operative diagnosis are needed to find out patients with high-risk lesions.
OSCC has a great variety of morphological patterns that can have an impact on prognosis, as the depth of invasion, infiltrative, and growth patterns.
Pre-operative biopsy of a suspected lesion in the oral cavity is necessary to confirm the diagnosis of OSCC but can give only limited information about pathological prognostic factors. Therefore, there is a great need for strategies that can allow an accurate prognostic evaluation of OSCC aggressiveness pre-operatively.
Recently, the interest in the epigenetic mechanisms that regulate tumor development has gained increasing attention [3].
DNA methylation is a common epigenetic mechanism leading to gene silencing in tumors. Specifically, DNA methylation refers to the covalent addition of a methyl group to the 5 carbon (C5) position of cytosine bases that are located 5′ to a guanosine base in a CpG dinucleotide. CpG dinucleotides are usually found clustered in specific regions, named CpG islands, which are often located in the promoter of several genes, including tumor suppressor genes and proto oncogenes [4]. Aberrant DNA methylation in these loci may contribute to cancer progression, leading to dysregulation of mRNA expression, an early and frequent event in tumors. On the other side, DNA hypomethylation promotes tumorigenesis via transcriptional activation of oncogenes and chromosomal instability [5,6].
The loss of function of tumor-suppressor genes, which often occurs in tumors, has been ascribed more frequently to epigenetic silencing through methylation than to genetic mutations, supporting the hypothesis that epigenetic alterations have a significant role in every step of carcinogenesis [7].
Attractive strategies to better characterize OSCC cases by analyzing the methylation status of a gene panel starting from saliva and/or brushing specimens have been proposed [8,9,10,11,12,13].
Previous studies, performed by our group, demonstrated the importance of epigenetic alterations and aberrant DNA methylation of specific genes to discriminate OSCC and its precursors lesions from benign oral mucosal lesions [14,15,16]. Specifically, 19 genes known to be altered in OSCC [14,17,18,19,20] were evaluated by bisulfite Next Generation Sequencing (NGS) with the aim of developing a noninvasive procedure for OSCC detection based on oral brushing. ROC analysis of all CpGs investigated allowed us to select the highest informative ones mapped within the following 13 genes: ZAP70, ITGA4, KIF1A, PARP15, EPHX3, NTM, LRRTM1, FLI1, MIR193, LINC00599, MIR296, TERT, and GP1BB. A linear discriminant analysis was used to develop an algorithm of choice that clearly discriminated benign oral lesions from potentially malignant or malignant oral lesions [13,14].
None of our previous studies analyzed the impact of an altered methylation pattern of these genes obtained from pre-operative oral brushing samples on post-surgical clinical outcome. Recently, few studies from other authors demonstrated that genetic and epigenetic alterations of some genes of our 13-gene panel may play a role in OSCC prognosis, presence of metastasis and response to treatment [18,21,22,23,24,25,26]. For example, Marsit et al., starting from a case series of 68 post-surgical Formalin Fixed Paraffin Embedded (FFPE) OSCC samples revealed that an altered methylation pattern of ZAP70 and GP1BB is associated with poor survival [18]. Shintani et al., starting from 7 OSCC lines, showed that an altered methylation pattern of FLI1 is a prediction marker gene for OSCC radiotherapy resistance [22].
The aim of the present study is to correlate the same epigenetic alterations with prognosis. To this purpose, samples were collected before surgery through oral brushing in a group of OSCC patients. The relationship between methylation profile of each gene, clinic-pathological features and follow-up was evaluated. Most informative CpGs were also used to generate a unique prognostic model in OSCC patients based on a Cox proportional hazards model via likelihood-based component-wise boosting.
2. Results
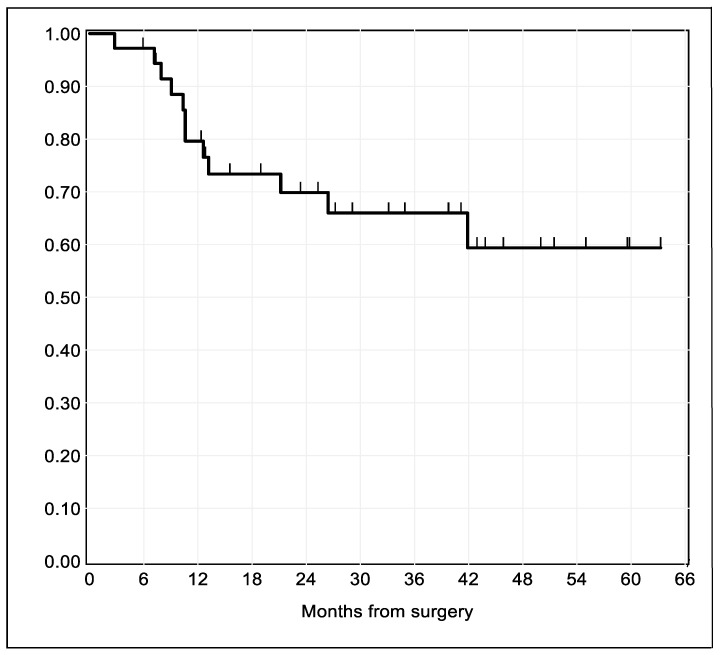
Thirty-six patients met the inclusion criteria. Median follow-up time was 25.8 months (2.1 years), ranging from 2.8–63.1 months (0.2–5.3 years). In total, one third of the patients (n = 12) experienced disease relapse during follow-up. The Kaplan–Meier estimate of recurrence-free survival for the entire study sample is illustrated in Figure 1.
Figure 1.
Kaplan–Meier survival estimates of time to relapse after surgical resection for oral squamous cell carcinoma (OSCC). The spikes indicate censoring times.
2.1. Pathological Findings and Tumor Stage
Evaluation of TNM was assigned according to p-TNM Classification of Tumors (AJCC 8th Edition) [27]. Tumor size (T): 17 patients were T1 (47%), 10 patients were T2 (28%), 3 patients were T3 (8%), 6 patients were T4 (17%). Lymph nodes involvement (N): 32 patients were N0 (89%); N1: 2 patients (6%); N2: 1 patient (3%); N3: 1 patient (3%).
Sixteen patients were diagnosed as T1 N0, 1 as T1 N2, 10 as T2 N0, 1 as T3N0, 2 as T3 N1, 5 as T4 N0 and 1 as T4 N3 (see Table 1 for details).
Table 1.
Clinical-pathological features of the studied population: Asterisks (*) indicate significant prognostic variables of loco-regional secondary neoplastic manifestation at the 5%-level. p-values were obtained with the log-rank test.
| Clinical-Pathological Variables | ||||
|---|---|---|---|---|
| Patients | Relapses Observed | p-Value | ||
| Sex | Male | 17 (47%) | 6 (35%) | 0.82 |
| Female | 19 (53%) | 6 (31%) | ||
| Age | <65 | 15 (42%) | 8 (53%) | 0.35 |
| >65 | 21 (58%) | 4 (19%) | ||
| Smoke | Yes | 7 (19%) | 3 (43%) | 0.26 |
| No | 29 (81%) | 9 (31%) | ||
| Site | Tongue and floor of mouth | 13 (36%) | 4 (31%) | 0.96 |
| Buccal and labial mucosa | 7 (19%) | 3 (43%) | ||
| Gingiva, Hard Palate, Retromolar region | 16 (45%) | 5 (31%) | ||
| T stage | T1-T2 | 27 (75%) | 8 (29%) | 0.33 |
| T3-T4 | 9 (25%) | 4 (44%) | ||
| N stage | N− | 32 (89%) | 9 (28%) | 0.06 |
| N+ | 4 (11%) | 3 (75%) | ||
| Grading | G1 | 20 (56%) | 5 (25%) | 0.18 |
| G2 | 14 (39%) | 6 (43%) | ||
| G3 | 2 (5%) | 1(50%) | ||
| Surgical margins | Free | 29 (81%) | 9 (31%) | 0.83 |
| Close | 4 (11%) | 1 (25%) | ||
| Displasia | 3 (8%) | 2 (66%) | ||
| Involved | 0 (0%) | |||
| Presence of associated OPMD | None | 26 (72%) | 11 (42%) | 0.07 |
| Lichen | 6 (17%) | 0 (0%) | ||
| Leucoplakia | 4 (11%) | 1 (25%) | ||
| Depth of invasion (DOI) | <4 mm | 21 (58%) | 5 (24%) | 0.07 |
| >4 mm | 15 (42%) | 7 (47%) | ||
| Pattern of invasion | P1-P2 | 22 (61%) | 4 (18%) | 0.01 * |
| P3-P4 | 14 (39%) | 8 (57%) | ||
| Radiotherapy | Yes | 8 (22%) | 4 (50%) | 0.2 |
| No | 28 (78%) | 8 (29%) | ||
The log-rank test revealed that the only clinic-pathological factor significantly associated with worse prognosis was higher pattern of invasion (P3-P4 according to Chang et al. classification [28]).
2.2. Methylation Profile and Clinical-Pathological Characteristics
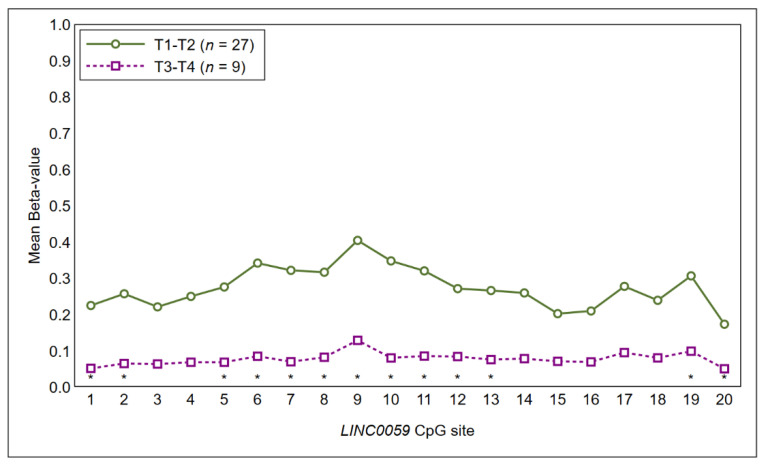
Tumor size: the Mann–Whitney U test showed higher methylation levels of most CpG sites of LINC00599 (coordinates hg38: Chr8: 9903242-9903378) in T1-T2 tumors as compared to T3-T4 tumors, although results failed to achieve statistical significance at the 5%-level (Figure 2).
Figure 2.
Methylation profile of the CpG sites of LINC00599 segregated between T1-T2 tumors and T3-T4 tumors. Statistical significance at 10% is marked with an asterisk.
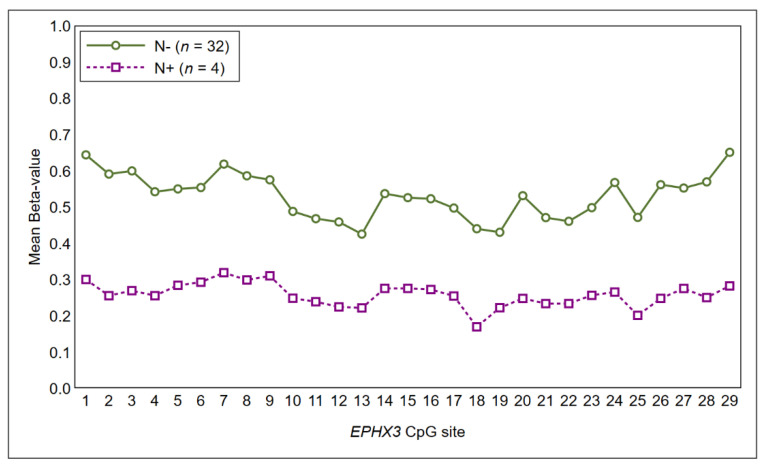
Lymph-node metastases: As shown in Figure 3, a higher methylation profile of CpG islands in EPHX3 was found in OSCC without lymph-node involvement as compared to OSCC with a positive lymph-node status at the diagnosis (Figure 3), although results failed to achieve statistical significance.
Figure 3.
Methylation profile of the CpG sites of EPHX3 segregated between N− tumors and N+ tumors.
The Mann–Whitney U test and Kruskal–Wallis test did not show other differences in the methylation profile of 13 genes in relationship with other clinical-pathological characteristics of the studied population.
2.3. Methylation Profile and Adverse Event (AE)
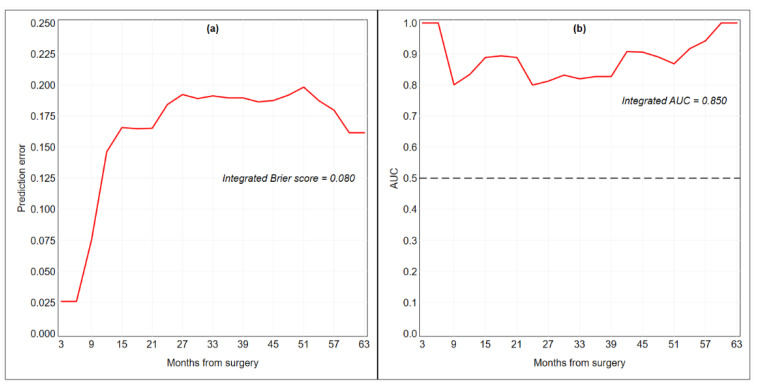
As possible molecular predictors, we analyzed the methylation data of 238 CpG sites from 13 genes (ZAP70, ITGA4, KIF1A, PARP15, EPHX3, NTM, LRRTM1, FLI1, MiR193, LINC00599 TERT, MiR296, and GP1BB). Permutation-based p-value estimates for each candidate covariate are presented in Table S1. The boosting estimation resulted in five non-zero regression coefficients associated with EPHX3-24, EPHX3-26, ITGA4-3, ITGA4-4, and MiR193-3. As shown in Table 2, EPHX3 was associated with worse prognostic outcomes in case of hypomethylation (HR < 1), while ITGA4 and MiR193 were associated with worse prognosis outcomes in case of hypermethylation (HR > 1). The coefficient estimates (ln HR) at each boosting step are illustrated in Figure S1. The integrated Brier score of the final risk prediction model was 0.080, the C-index was 0.802 and the integrated AUC was 0.850 (Table 2, Figure 4).
Table 2.
Cox proportional hazards model resulting from likelihood-based component-wise boosting after seven steps. The five non-zero regression coefficients (ln HR) are presented along with hazard ratios (HRs) and permutation-based p-values. Coefficients are scaled to be at the level of the original methylation Beta-values.
| CpG Site | Ln HR | HR | p-Value |
|---|---|---|---|
| EPHX3-24 (Chr19:15232040) | −0.0234 | 0.9769 | 0.0157 |
| EPHX3-26 (Chr19:15232034) | −0.0226 | 0.9777 | 0.0172 |
| ITGA4-3 (Chr2:181458175) | 0.0163 | 1.0165 | 0.0078 |
| ITGA4-4 (Chr2:181458181) | 0.0306 | 1.0310 | 0.0027 |
| MiR193-3 (Chr17:31559856) | 0.0089 | 1.0090 | 0.0099 |
| Integrated Brier score | 0.080 | ||
| C-index | 0.802 | ||
| Integrated AUC | 0.850 | ||
Figure 4.
Time-dependent Brier score (a) and area under the receiver operator characteristic curve (AUC) (b) for the final Cox proportional hazards model fitted by component-wise likelihood-based boosting. The dashed line represents a non-discriminatory model (random guessing). Performance and predictive accuracy of the model were estimated using the original training set of 36 patients.
Removing the connection information from the boosting algorithm, results were quite stable (Table S2): the CpG sites retained in the final Cox model were EPHX3-23, EPHX3-24, ITGA4-4 and MiR193-3; the discriminative ability was virtually identical (C-index = 0.805; integrated AUC = 0.847), as well as the overall performance (integrated Brier score = 0.089). As shown in Table S2 and Figure S2, re-running all analyses after converting Beta-values into M-values, the overall performance of the regression model was similar, but its calibration and discriminative ability were poorer in the last months and in the first months of follow-up, respectively.
In the subgroup analysis of patients with early tumor diagnosis (26 patients with T1-2N0), we found that ITGA4 had a strong prognostic power for future disease recurrence (Table 3).
Table 3.
Cox proportional hazards model resulting from likelihood-based component-wise boosting after nine steps. The study sample is made of patients with an early diagnosis of OSCC (n = 26). The four non-zero regression coefficients (ln HR) are presented along with hazard ratios (HRs) and permutation-based p-values. Coefficients are scaled to be at the level of the original methylation Beta-values.
| CpG Site | Ln HR | HR | p-Value |
|---|---|---|---|
| ITGA4-3(Chr2:181458175) | 0.0616 | 1.0636 | 0.0001 |
| ITGA4-4(Chr2:181458181) | 0.0467 | 1.0478 | 0.0003 |
| ITGA4-7(Chr2: 181458229) | 0.0176 | 1.0177 | 0.0046 |
| ITGA4-12(Chr2: 181458289) | 0.0152 | 1.0153 | 0.0052 |
| Integrated Brier score | 0.059 | ||
| C-index | 0.892 | ||
| Integrated AUC | 0.903 | ||
3. Discussion
Maxillo-facial surgeons and oncologists are called not only to surgically treat an OSCC but also to prevent AEs that could affect their patients. Therefore, pre-operative assessment plays a crucial role in OSCC work-up.
Clinical evaluation and radiological assessment (CT-scan and MRI) are essential for tumor staging.
A pre-operative biopsy is required to confirm the diagnosis of OSCC and to give information about pathological prognostic factors. It is now clear that clinic-pathological findings, as well as radiological evaluation, alone are not sufficient in predicting OSCC clinical behavior. Local recurrences, nodal metastases, or second primary tumors can hamper the prognosis particularly in early staged OSCC. If the poor prognosis for locally advanced tumors is well explainable, also T1-T2 stages can be occasionally unpredictable in terms of disease control [29]. Despite an apparent favorable condition, sometimes early staged OSCC can have a very aggressive biological behavior, even if resected with free surgical margins.
As in other body sites (as lung or colon), OSCC molecular profile can give interesting information about tumor aggressiveness: Specifically, gene silencing by promoter methylation seems to play a crucial role in determining tumor aggressive potential [30].
In this study, the relationship between the methylation profile of a panel of 13 genes (ZAP70, ITGA4, KIF1A, PARP15, EPHX3, NTM, LRRTM1, FLI1, MIR193, LINC00599 TERT, MIR296, and GP1BB) and their role in predicting some factors (tumor size, lymph node involvement, AE), not otherwise identifiable from pathological pre-operative diagnosis was investigated. The same gene panel was recently validated as diagnostic marker of OSCC and proved to be accurate in the early detection of oral cancer [15].
The use of bisulfite NGS method allowed to test many CpG sites for a single gene and to align longer reads to the reference sequence more easily and with greater accuracy. Superiority of amplicon bisulfite sequencing in terms of accuracy, robustness, and throughput compared to other DNA methylation analysis methods such as methylation-sensitive PCR was recently reported by Bock et al. [31]. Furthermore, DNA methylation analysis was assessed starting from minimal invasive collection specimens obtained by oral brushing. This approach may be advantageous for its very limited invasiveness and because it avoids formalin fixation of the neoplastic cells. It is well known that DNA extraction and consequent DNA methylation analysis can be easier performed in fresh-unfixed material. Furthermore, brushing cell collection from the whole tumor area can provide a sample representative of the complex epigenetic landscape of each single OSCC lesion. In this setting intratumoral heterogeneity can be evaluated pre-operatively.
The comparative analysis of methylation profiles with clinical-pathological characteristics suggested the presence of an association, albeit not significant at the 5%-level, between LINC00599 and T stage. In particular, several CpG islands in exon 1 of LINC00599 segregated between early-OSCC (T1–2) and locally advanced-OSCC (T3–4). LINC00599 (Long Intergenic Non-Protein Coding RNA 599) is a long noncoding RNA Gene mapped close to the MIR124 gene. A number of studies have reported that LINC00599 exerts important regulatory functions in cell proliferation differentiation, and in atherosclerosis [32,33]. Little is known about the role of LINC0059 methylation in oral cancer. This finding supports the idea that changes in epigenetic landscape occur during tumor progression. However, further studies and larger studies are needed to elucidate contribution LINC00599 methylation in oral carcinogenesis. Similarly, there was a tendency toward hypomethylation for most CpG islands of EPHX3 in case of positive lymph-node status at the diagnosis.
In relation to post-surgical clinical outcomes, the analysis of methylation profiles identified 19/238 CpG sites significantly associated with the occurrence of loco regional adverse events (see Table S1). It also emerged that only 4 out of 13 studied genes in the panel harbored CpGs with prognostic relevance (9 CpG islands for EPHX3, 7 CpG islands for ITGA4, and only one CpG island respectively for MiR193, MiR296, and TERT). The boosting estimation resulted in five non-zero regression coefficients associated with EPHX3-24, EPHX3-26, ITGA4-3, ITGA4-4, and MiR193-3 significantly associated with the occurrence of loco regional adverse events. Boosting is a method for incrementally building linear combinations of “weak” models, to generate a “strong” predictive model. This process shows similarities to lasso-like approaches, with many of the estimated coefficients shrunk toward zero. Boosting algorithms have been shown to be particularly useful to handle models in which the number of candidate predictors exceeds the number of observations (high-dimensional settings).
Putative prognostic predictors methylation analysis showed that EPHX3 was associated with worse prognostic outcomes when hypomethylated (HR < 1), while ITGA4 and MiR193 were associated with worse prognosis outcomes in case of hypermethylation (HR > 1).
Prior studies have noted that the methylation of epoxide hydrolase 3 (EPHX3) is significantly associated with the prognosis of prostate cancer and its inclusion into clinical practice has been suggested as a tool to a more accurate prediction of which patients may experience prostate cancer recurrence. Curiously, it was found hypermethylated in human salivary gland Adenoid Cystic Carcinoma [34]. In OSCC its role is yet to be elucidated but emerging evidence from previous published research papers and the relationship with lymph-node involvement and occurrence of secondary neoplastic events suggest its promising use in risk predictive models.
ITGA4 is involved in gastric and colorectal cancer [35,36]. Previous reports indicate ITGA4 as a promising marker for oral squamous cell carcinoma and especially tongue oral cancer.
MiR193 was previously identified as associated with oral cancers in previous studies by Kozaki and our group [15,19].
These findings suggest that patients experiencing less favorable behavior of the disease may actually have a distinctive methylation profile. Noteworthy, ITGA4 showed a strong prognostic power for future disease recurrence in the subgroup analysis of patients with early stage tumor diagnosis. T1-T2 are commonly considered at lower risk of unfavorable outcome but they can pose a problem in the correct planning of surgical management. Despite the apparent ease to manage these early staged tumors, sometimes the surgeon faces to unpleasant surprises during the follow-up period. In fact, local recurrence can be experienced even if resection margins were clear and neck nodal metastases can appear in 10–30% of clinically negative necks (cN0) patients. Indeed, neck management for cT1-2 N0 patients remains debated. In these cases, known prognostic clinical-pathological factors (grading, depth of invasion, surgical margins, pattern of invasion) are usually less informative as compared to advanced tumors. The presence of a biomarker with the ability of recognition of early tumors that will eventually show aggressive behaviors before surgery starting from pre-operative oral brushing specimen is of primary importance in oral cancer management.
Nevertheless, the great amount of data deriving from several CpGs makes methylation analysis hardly applicable for models of single patient risk assessment. For this reason, in the last decade researchers have called in biostatistical techniques to translate complex results into clinical practice.
In the present study, the combination of significant CpGs (EPHX3-24, EPHX3-26, ITGA4-3, ITGA4-4, and MiR193-3) to build up a risk assessment prediction model provided promising results as shown by its performance and predictive accuracy. Interestingly, when connection information from the boosting algorithm was removed, EPHX3-24, ITGA4-4, and MiR193-3 retained a strong predictive prognostic ability in the final Cox model. More in detail, EPHX3 gene showed fluctuation and changes in significant CpGs depending on which model was used (EPHX3-18 to EPHX3-26). However, prognostic ability of CpGs in the area resulted virtually equivalent. In addition, the conversion of Beta-values into M-values resulted in similar but poorer overall performance of the regression model, suggesting the use of Beta values.
More in detail, EPHX3 gene showed fluctuation and changes in significant CpGs depending on which model was used (EPHX3-18 to EPHX3-26). However, prognostic ability of CpGs in the area resulted almost equivalent.
In addition, the conversion of Beta-values into M-values, resulted in similar but poorer overall performance and discriminative ability of the regression model suggesting a preferable use of Beta values.
Combination of epigenetic profiles of several genes as useful source of information is in agreement with emerging published literature in which several authors have tried to identify most relevant methylated genes to be used for prognostic purposes. Shen et al. in 2017 identified an OSCC DNA-methylation signature overall survival, based on 7 significant CpGs from genes AJAP1, SHANK2, FOXA2, MT1A, ZNF570, HOXC4, and HOXB4 [37].
Zhu in 2019 found out that the combined analysis of the methylation status of 5 genes (CENPV, SYTL2, OCLN, CASD1, and TUB) helped in the construction of a risk prediction model of the prognosis of OSCC [38]
Pan et al. in 2019 reported that subsequent combined survival analysis on six genes (INA, LINC01354, TSPYL4, MAGEB2, EPHX3, and ZNF134) could be used as independent prognostic markers and potentially used as drug targets [39].
A limit of the present study is the low number of patients, especially the number of advanced OSCC tumors (T3-T4). However, component-wise boosting was specifically used to overcome the limits imposed by the small sample size. A second limit of the present study is the absence of a prospective test set of patients that we hope to undertake in future investigations in order to provide an unbiased predictive evaluation of the fitted model. A validated Cox model including molecular and clinical predictors has the potential to estimate for each patient the risk of disease relapse at any given time points following surgery.
4. Materials and Methods
4.1. Ethics Statement
All clinical investigations were conducted according to the principles of the Declaration of Helsinki. The study was approved by the local ethics committee (520/2018/Sper/AOUBo, 2018/Dec/12th). All information regarding the human material used in this study was managed using anonymous numerical codes.
4.2. Study Setting and Data Collection
Selection criteria were the following: oral brushing performed at the time of pre-operative biopsy; histological diagnosis of primary oral squamous cell carcinoma; no other neoplastic diseases reported on anamnesis; radical surgery with histologically negative resection margins; availability of follow-up information.
All these cases were treated at the Departments of Oral Sciences and Oral and Maxillofacial Surgery, University of Bologna, from March 2013 to December 2018. Histological diagnoses were performed at the Section of Anatomic Pathology at Bellaria Hospital, Department of Biomedical and Neuromotor Sciences, University of Bologna (VPF, SA) and Unit of Anatomic Pathology, S. Orsola Hospital, Bologna (TB).
All patients presenting with a suspected oral neoplastic lesion underwent oral brushing sampling before diagnostic incisional biopsy. Oral brushing was performed according to a previously described protocol [14,15].
After surgery, histological examination was performed blindly from the methylation profile. A multi-head microscope discussion was made on discordant cases to obtain a common diagnosis. Histological diagnoses and staging were established following currently accepted criteria [27,40].
4.3. Treatment Modality
After the diagnostic workup and multidisciplinary discussion, all 36 patients underwent surgical resection of OSCC in accordance with standard treatment practice [41]. Surgery consisted of composite resections, including excision of the primary tumor with ipsilateral or bilateral neck dissection, in accordance to the N-status. Both primary closure and local flaps were performed for early stages. Microvascular reconstruction was performed for patients with locally advanced disease. Post-operative radiation therapy was performed according to currently accepted criteria [42].
Follow-up was performed every two weeks for the first two months after surgery and then monthly during the first year after surgery, every three months during the second year after surgery, and finally every six months. A CT scan or MRI was requested every six months during the first three years after surgery and then once a year.
The disease-free survival endpoints, defined as the duration between oral brushing cell collection and the diagnosis of local recurrence, lymph node or distant metastasis, death, or the last follow-up visit, were evaluated in June 2020.
4.4. DNA Methylation Analysis
DNA methylation analysis was performed as previously described by Morandi et al. [15]. Shortly, DNA from exfoliating brush specimens was purified using The MasterPure™ Complete DNA Purification Kit (Lucigen, Middleton, WI, USA, cod. MC85200), and treated with sodium bisulfite using the EZDNA Methylation-Lightning™ Kit (ZymoResearch, Irvine, CA, USA, cod. D5031) according to the manufacturer’s instructions. Quantitative DNA methylation analysis was performed by next-generation sequencing for the following genes: ZAP70, ITGA4, KIF1A, PARP15, EPHX3, NTM, LRRTM1, FLI1, MIR193, LINC00599, MIR296, TERT, and GP1BB (Table 4).
Table 4.
List of 13 genes target.
| Gene | Map | Position | Amplicon Length | Position Respect to TSS | Number of Interrogated CpG | hg38 Coordinates |
|---|---|---|---|---|---|---|
| ZAP70 | 2q11.2 | Exon 3 | 180 | 10728 | 20 | Chr2: 97724265-97724445 |
| GP1BB | 22q11.21 | Exon 1 | 192 | 363 | 18 | Chr22: 19723282-19723460 |
| KIF1A | 2q37.3 | Exon 1 | 189 | −1 | 27 | Chr2: 240820168-240820310 |
| PARP15 | 3q21.1 | Exon 1 | 206 | 93 | 19 | Chr3:122577695-122577901 |
| ITGA4 | 2q31.3 | Exon 2 | 214 | 912 | 14 | Chr2:181457647-181457879 |
| NTM | 11q25 | Exon 1 | 190 | 62 | 15 | Chr11:131911126-131911314 |
| MIR193A | 17q11.2 | Promoter | 256 | −178 | 26 | Chr17:31559818-31560073 |
| EPHX3 | 19p13.12 | Exon 1 | 223 | 215 | 29 | Chr19:15231995-15232217 |
| LINC00599 | 8p23.1 | Exon 1 | 199 | 69 | 20 | Chr8:9903205-9903403 |
| FLI1 | 11q24.3 | Exon 1 | 186 | 187 | 12 | Chr11:128694103-128694288 |
| MIR296 | 20q13.32 | Exon 1 | 238 | 180 | 15 | Chr20:58817149-58817363 |
| LRRTM1 | 2p12 | Promoter | 179 | −431 | 24 | Chr2:80304527-80304705 |
| TERT | 5p15.33 | Intron4-5 | 109 | 14976 | 6 | Chr5:1279604-1279759 |
Libraries were prepared using the Nextera™ Index Kit (Illumina, San Diego, CA, USA, FC-121-1012) following a two steps approach with locus-specific bisulfite amplicon [15] and loaded onto MiSEQ (Illumina, San Diego, CA, USA, cod. 15027617). Each NGS experiment was designed to allocate at least 1000 reads/amplicon, with the aim to reach a depth of coverage of 1000×. FASTQ output files were processed for quality control (>Q30) and converted into FASTA format in a Galaxy Project environment [43]. The methylation ratio of each CpG was calculated in parallel by different tools: BSPAT (http://cbc.case.edu/BSPAT/index.jsp) [44] BWAmeth in a Galaxy Project environment (Europe) followed by the Methyl Dackel tool (https://github.com/dpryan79/MethylDackel), EPIC-TABSAT [45], and finally Kismeth [46]. Methylation plotter tool was used to compare DNA methylation level of 13 gene panel and clinic-pathological variables [47].
4.5. Statistical Analysis
Summary statistics for the study sample were presented as frequencies and percentages. Recurrence-free survival was calculated using the date of surgery as the time origin. The endpoints that we considered were local recurrence, second primary tumor, lymph-node metastasis, and disease-related death, whichever occurred first. Lost-to-follow-up were right-censored at the time of the status last known. The survival function was estimated with the Kaplan–Meier method.
Differences in methylation levels across clinical-pathological groups were evaluated with the Mann–Whitney U test. Permutation-based p-values were corrected using Simes’ method for false discovery rate control.
To evaluate the prognostic power of the CpG sites, we fit a Cox proportional hazards model via likelihood-based component-wise boosting [48,49]. This forward procedure, which is rooted in the field of machine learning [50], starts with an empty model and updates only one regression coefficient in each step of the algorithm. Updates are done by adding small fractions of the estimated regression coefficients to the estimates obtained in previous steps; these fractions are governed by a penalty term attached to the log-likelihood (usually 0.1). This process results in sparse fits similar to lasso-like approaches, with many of the estimated coefficients shrunk toward zero. Boosting algorithms have been shown to be particularly useful to handle models in which the number of candidate predictors exceeds the number of observations (high-dimensional settings) [51,52].
The number of boosting steps, which is the main tuning parameter of the procedure and determines both shrinkage of effect estimates and variable selection, was optimized by cross-validation [52]. To deal with the high variability of the results due to small sample sizes, we averaged over 100 repetitions of cross-validation, then repeated the same procedure by changing the number of partitions (leave-one-out, 3-, 6-, and 9-fold) and selected the median value among the four penalty parameters [53]. This approach led to an optimal boosting step number of seven. All methylation Beta-values specified in the Cox proportional hazards model were standardized in order to meet the homoscedasticity requirement of the likelihood-based boosting algorithm [52]. Permutation-based p-value estimates were calculated for each candidate CpG site [54]. By including pathway information in boosting estimation, CpG sites were more likely to be chosen in subsequent boosting steps if a connected CpG site from the same gene had been chosen in an earlier boosting step [55].
The overall performance of the risk prediction model resulting from seven boosting steps was evaluated with the Brier score, which is a weighted average of the squared distances between the observed survival status and the survival probability predicted by the model. A prediction error curve was obtained by following the Brier score over time, and the integral under the curve was calculated as a summary measure for model performance; the lower the curve, the better is the performance of the model [56,57]. The discriminative ability of the model was evaluated with the C-index for survival data, a measure that quantifies the proportion of all patient pairs for whom the predicted and observed survival outcomes are concordant. A C-index = 0.5 indicates a non-informative prediction rule, while a C-index = 1 indicates perfect association. We used Uno’s estimate of the index, which is based on inverse-probability-of-censoring weights and has been shown to be more robust than Harrell’s estimate to the presence of many censored observations [58]. We also estimated the time-dependent area under the receiver operator characteristic (ROC) curve (AUC) as an additional measure of discriminative ability [59], because it is more robust than the C-index to deviations from the proportional hazards assumptions [60]. The summary measure for time-dependent AUC was the integrated AUC, which averages all available statistics over time. It should be recognized that the performance of the risk prediction model was evaluated using the original training set of 36 patients, therefore all our estimates have to be considered as overly optimistic.
In a sensitivity analysis, we removed connection information from the algorithm in order to treat all CpG sites as unconnected covariates. In another sensitivity analysis, Beta-values were converted into M-values, i.e., the binary logarithm of the intensities of methylated probes versus non-methylated probes [61]. In a subgroup analysis, the prognostic power of the CpG sites was reevaluated on the 26 study patients with an early diagnosis of OSCC. All analyses were conducted using R version 4.0.1 [62]; likelihood-based boosting was performed with the R package CoxBoost [63].
5. Conclusions
The present preliminary study, even if performed in a small population of patients affected by Oral Cancer, confirms the attractive use of the preoperative evaluation of methylation profile also for the prognostic assessment of patients with OSCC. Adding more details than the simple biopsy, molecular findings from oral brushing could help clinicians to stratify patients at high- versus low-risk of recurrences, metastases and second tumors, and to plan the adequate treatment. Further studies with a larger and homogeneous cohort are needed to elucidate the intrinsic prognostic potential of our assay.
Acknowledgments
We are grateful to Andrea Ravaioli and Cristina Bettini from Knowledge Transfer Office, University of Bologna, for their outstanding support. We also thank the Galaxy server that was used for some calculations which is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR (de.NBI)).
Abbreviations
| AE | Adverse Event |
| NGS | Next Generation Sequencing |
| OSCC | Oral Squamous Cell Carcinoma |
| PNI | Perineural invasion |
| POI | Pattern of infiltration |
| WPOI | Worst pattern of infiltration |
| DOI | Depth of Invasion |
| HR | Hazard Ratio |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/18/6691/s1.
Author Contributions
Conceptualization: L.M. (Luca Morandi), D.B.G., J.L.; Data curation: D.B.G., A.G., S.M., A.T., J.L.; Formal analysis: J.L., L.M. (Luca Morandi), L.M. (Lucio Montebugnoli); Funding acquisition: L.M. (Luca Morandi), C.M.; Investigation: S.M., S.A., A.T., V.P.F.; Methodology: L.M. (Lucio Montebugnoli), J.L., D.B.G., L.M. (Luca Morandi).; Project administration: M.P.F., L.M. (Lucio Montebugnoli), C.M.; Resources: S.A., S.M., A.T., T.B.; Supervision: C.M., L.M. (Lucio Montebugnoli); Validation: S.A., L.M. (Luca Morandi); Writing—original draft: A.G., D.B.G., J.L., V.P.F.; Writing, review and editing: L.M. (Luca Morandi), D.B.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by an academic grant (Proof of Concept—PoC 2019 assigned to Luca Morandi) from the University of Bologna.
Conflicts of Interest
Luca Morandi, Davide B. Gissi, and Achille Tarsitano submitted a patent (the applicant is the University of Bologna) in November 2016 to the National Institute of Industrial Property; however, we believe that this is a natural step of translational research (bench-to-bedside) and guarantee that the scientific results are true. The other authors declare no conflict of interest
References
- 1.Rivera C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015;8:11884–11894. [PMC free article] [PubMed] [Google Scholar]
- 2.Amin R.W., Rivera B. A spatial study of oral & pharynx cancer mortality and incidence in the U.S.A.: 2000–2015. Sci. Total Environ. 2020;713:136688. doi: 10.1016/j.scitotenv.2020.136688. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Eden A., Gaudet F., Waghmare A., Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 6.Van Tongelen A., Loriot A., De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017;396:130–137. doi: 10.1016/j.canlet.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Mascolo M., Siano M., Ilardi G., Russo D., Merolla F., De Rosa G., Staibano S. Epigenetic disregulation in oral cancer. Int. J. Mol. Sci. 2012;13:2331–2353. doi: 10.3390/ijms13022331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demokan S., Chang X., Chuang A., Mydlarz W.K., Kaur J., Huang P., Khan Z., Khan T., Ostrow K.L., Brait M., et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int. J. Cancer. 2010;127:2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langevin S.M., Stone R.A., Bunker C.H., Grandis J.R., Sobol R.W., Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31:864–870. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattani K.M., Zhang Z., Demokan S., Glazer C., Loyo M., Goodman S., Sidransky D., Bermudez F., Jean-Charles G., McCaffrey T., et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev. Res. 2010;3:1093–1103. doi: 10.1158/1940-6207.CAPR-10-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schussel J., Zhou X.C., Zhang Z., Pattani K., Bermudez F., Jean-Charles G., McCaffrey T., Padhya T., Phelan J., Spivakovsky S., et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin. Cancer Res. 2013;19:3268–3275. doi: 10.1158/1078-0432.CCR-12-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagata S., Hamada T., Yamada N., Yokoyama S., Kitamoto S., Kanmura Y., Nomura M., Kamikawa Y., Yonezawa S., Sugihara K. Aberrant DNA methylation of tumor-related genes in oral rinse: A noninvasive method for detection of oral squamous cell carcinoma. Cancer. 2012;118:4298–4308. doi: 10.1002/cncr.27417. [DOI] [PubMed] [Google Scholar]
- 13.Sailer V., Holmes E.E., Gevensleben H., Goltz D., Dröge F., de Vos L., Franzen A., Schröck F., Bootz F., Kristiansen G., et al. PITX2 and PANCR DNA methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:75827–75838. doi: 10.18632/oncotarget.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morandi L., Gissi D., Tarsitano A., Asioli S., Monti V., Del Corso G., Marchetti C., Montebugnoli L., Foschini M.P. DNA methylation analysis by bisulfite next-generation sequencing for early detection of oral squamous cell carcinoma and high-grade squamous intraepithelial lesion from oral brushing. J. Cranio-Maxillofac. Surg. 2015;43:1494–1500. doi: 10.1016/j.jcms.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 15.Morandi L., Gissi D., Tarsitano A., Asioli S., Gabusi A., Marchetti C., Montebugnoli L., Foschini M.P. CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel. Clin. Epigenetics. 2017;9:85. doi: 10.1186/s13148-017-0386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gissi D.B., Gabusi A., Tarsitano A., Asioli S., Rossi R., Marchetti C., Montebugnoli L., Foschini M.P., Morandi L. Application of a non-invasive oral brushing procedure based on bisulfite sequencing of a 13-gene panel to study high-risk OSCC patients. Cancer Biomark. 2020 doi: 10.3233/CBM-190422. [DOI] [PubMed] [Google Scholar]
- 17.Guerrero-Preston R., Michailidi C., Marchionni L., Pickering C.R., Frederick M.J., Myers J.N., Yegnasubramanian S., Hadar T., Noordhuis M.G., Zizkova V., et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031–1046. doi: 10.4161/epi.29025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsit C.J., Christensen B.C., Houseman E.A., Karagas M.R., Wrensch M.R., Yeh R.-F., Nelson H.H., Wiemels J.L., Zheng S., Posner M.R., et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis. 2009;30:416–422. doi: 10.1093/carcin/bgp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 20.Li Y.-F., Hsiao Y.-H., Lai Y.-H., Chen Y.-C., Chen Y.-J., Chou J.-L., Chan M.W.Y., Lin Y.-H., Tsou Y.-A., Tsai M.-H., et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics. 2015;10:229–236. doi: 10.1080/15592294.2015.1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheriyan V.T., Thomas C., Balaram P. Augmentation of T-cell immune responses and signal transduction proteins in oral cancer patients: Potential for IL-2-mediated immunotherapy. J. Cancer Res. Clin. Oncol. 2011;137:1435–1444. doi: 10.1007/s00432-011-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shintani S., Hamakawa H., Nakashiro K., Shirota T., Hatori M., Tanaka M., Kuroshita Y., Kurokawa Y. Friend leukaemia insertion (Fli)-1 is a prediction marker candidate for radiotherapy resistant oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2010;39:1115–1119. doi: 10.1016/j.ijom.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 23.Yasukawa M., Fujihara H., Fujimori H., Kawaguchi K., Yamada H., Nakayama R., Yamamoto N., Kishi Y., Hamada Y., Masutani M. Synergetic Effects of PARP Inhibitor AZD2281 and Cisplatin in Oral Squamous Cell Carcinoma in Vitro and in Vivo. Int. J. Mol. Sci. 2016;17:272. doi: 10.3390/ijms17030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severino P., Oliveira L.S., Andreghetto F.M., Torres N., Curioni O., Cury P.M., Toporcov T.N., Paschoal A.R., Durham A.M. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med. Genom. 2015;8:31. doi: 10.1186/s12920-015-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghantous Y., Bahouth Z., Abu El-Naaj I. Clinical and genetic signatures of local recurrence in oral squamous cell carcinoma. Arch. Oral Biol. 2018;95:141–148. doi: 10.1016/j.archoralbio.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Boscolo-Rizzo P., Rampazzo E., Perissinotto E., Piano M.A., Giunco S., Baboci L., Spinato G., Spinato R., Tirelli G., Da Mosto M.C., et al. Telomere shortening in mucosa surrounding the tumor: Biosensor of field cancerization and prognostic marker of mucosal failure in head and neck squamous cell carcinoma. Oral Oncol. 2015;51:500–507. doi: 10.1016/j.oraloncology.2015.02.100. [DOI] [PubMed] [Google Scholar]
- 27.Amin M.B., Edge S., Greene F., Byrd D.R., Brookland R.K., Washington M.K., Gershenwald J.E., Compton C.C., Hess K.R., Sullivan D.C., et al., editors. AJCC Cancer Staging Manual. 8th ed. Springer International Publishing; Chicago, IL, USA: 2017. [Google Scholar]
- 28.Chang Y.-C., Nieh S., Chen S.-F., Jao S.-W., Lin Y.-L., Fu E. Invasive pattern grading score designed as an independent prognostic indicator in oral squamous cell carcinoma. Histopathology. 2010;57:295–303. doi: 10.1111/j.1365-2559.2010.03616.x. [DOI] [PubMed] [Google Scholar]
- 29.Carsuzaa F., Thariat J., Gorphe P., Righini C., Cosmidis A., Thureau S., Roge M., De Mones E., Servagi-Vernat S., Tonnerre D., et al. Surgery or Radiotherapy of the Primary Tumor in T1-2 Head and Neck Squamous Cell Carcinoma with Resectable N3 Nodes: A Multicenter GETTEC Study. Ann. Surg. Oncol. 2019;26:3673–3680. doi: 10.1245/s10434-019-07589-0. [DOI] [PubMed] [Google Scholar]
- 30.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 31.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 32.Mercer T.R., Qureshi I.A., Gokhan S., Dinger M.E., Li G., Mattick J.S., Mehler M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan K., Jiang Q., Wang X.-Q., Wang Y.-N.-Z., Yang H., Yao M.-D., Liu C., Li X.-M., Yao J., Liu B., et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248. doi: 10.1038/cddis.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell A., Bell D., Weber R.S., El-Naggar A.K. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898–2909. doi: 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu Y., Dang S., Hou P. Gene methylation in gastric cancer. Clin. Chim. Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Lam K., Pan K., Linnekamp J.F., Medema J.P., Kandimalla R. DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta. 2016;1866:106–120. doi: 10.1016/j.bbcan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Shen S., Wang G., Shi Q., Zhang R., Zhao Y., Wei Y., Chen F., Christiani D.C. Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin. Epigenet. 2017;9:88. doi: 10.1186/s13148-017-0392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q., Tian G., Gao J. Construction of prognostic risk prediction model of oral squamous cell carcinoma based on co-methylated genes. Int. J. Mol. Med. 2019;44:787–796. doi: 10.3892/ijmm.2019.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y., Song Y., Cheng L., Xu H., Liu J. Analysis of methylation-driven genes for predicting the prognosis of patients with head and neck squamous cell carcinoma. J. Cell. Biochem. 2019;120:19482–19495. doi: 10.1002/jcb.29252. [DOI] [PubMed] [Google Scholar]
- 40.The International Agency for Research on Cancer. Eveson J.W., Reichart P., Sidransky D., World Health Organization Classification of Tumours . Pathology and Genetics of Head and Neck Tumours. 1st ed. IARC Press; Lyon, France: 2017. [Google Scholar]
- 41.Kademani D., Bell R.B., Bagheri S., Holmgren E., Dierks E., Potter B., Homer L. Prognostic factors in intraoral squamous cell carcinoma: The influence of histologic grade. J. Oral Maxillofac. Surg. 2005;63:1599–1605. doi: 10.1016/j.joms.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Adelstein D., Gillison M.L., Pfister D.G., Spencer S., Adkins D., Brizel D.M., Burtness B., Busse P.M., Caudell J.J., Cmelak A.J., et al. NCCN Guidelines Insights: Head and Neck Cancers, Version 2.2017. J. Natl. Compr. Cancer Netw. 2017;15:761–770. doi: 10.6004/jnccn.2017.0101. [DOI] [PubMed] [Google Scholar]
- 43.Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu K., Ting A.H., Li J. BSPAT: A fast online tool for DNA methylation co-occurrence pattern analysis based on high-throughput bisulfite sequencing data. BMC Bioinform. 2015;16:220. doi: 10.1186/s12859-015-0649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krainer J., Weinhäusel A., Hanak K., Pulverer W., Özen S., Vierlinger K., Pabinger S. EPIC-TABSAT: Analysis tool for targeted bisulfite sequencing experiments and array-based methylation studies. Nucleic Acids Res. 2019;47:W166–W170. doi: 10.1093/nar/gkz398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinform. 2008;9:371. doi: 10.1186/1471-2105-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallona I., Díez-Villanueva A., Peinado M.A. Methylation plotter: A web tool for dynamic visualization of DNA methylation data. Source Code Biol. Med. 2014;9:11. doi: 10.1186/1751-0473-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tutz G., Binder H. Generalized additive modeling with implicit variable selection by likelihood-based boosting. Biometrics. 2006;62:961–971. doi: 10.1111/j.1541-0420.2006.00578.x. [DOI] [PubMed] [Google Scholar]
- 49.Binder H., Schumacher M. Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models. BMC Bioinform. 2008;9:14. doi: 10.1186/1471-2105-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayr A., Binder H., Gefeller O., Schmid M. The evolution of boosting algorithms. From machine learning to statistical modelling. Methods Inf. Med. 2014;53:419–427. doi: 10.3414/ME13-01-0122. [DOI] [PubMed] [Google Scholar]
- 51.Sauerbrei W., Perperoglou A., Schmid M., Abrahamowicz M., Becher H., Binder H., Dunkler D., Harrell F.E., Royston P., Heinze G., et al. State of the art in selection of variables and functional forms in multivariable analysis-outstanding issues. Diagn. Progn. Res. 2020;4:3. doi: 10.1186/s41512-020-00074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Bin R. Boosting in Cox regression: A Comparison between the Likelihood-Based and the Model-Based Approaches with Focus on the R-Packages CoxBoost and Mboost. [(accessed on 3 July 2020)]; Available online: https://epub.ub.uni-muenchen.de/24466/
- 53.Seibold H., Bernau C., Boulesteix A.-L., De Bin R. On the choice and influence of the number of boosting steps for high-dimensional linear Cox-models. Comput. Stat. 2018 doi: 10.1007/s00180-017-0773-8. [DOI] [Google Scholar]
- 54.Binder H., Porzelius C., Schumacher M. Rank-Based p-Values for Sparse High-Dimensional Risk Prediction Models Fitted by Componentwise Boosting. University of Freiburg; Breisgau, Germany: 2009. FDM-Preprint Nr. 101. [Google Scholar]
- 55.Binder H., Schumacher M. Incorporating pathway information into boosting estimation of high-dimensional risk prediction models. BMC Bioinform. 2009;10:18. doi: 10.1186/1471-2105-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerds T.A., Schumacher M. Consistent estimation of the expected Brier score in general survival models with right-censored event times. Biom. J. 2006;48:1029–1040. doi: 10.1002/bimj.200610301. [DOI] [PubMed] [Google Scholar]
- 57.Steyerberg E.W., Vickers A.J., Cook N.R., Gerds T., Gonen M., Obuchowski N., Pencina M.J., Kattan M.W. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uno H., Cai T., Pencina M.J., D’Agostino R.B., Wei L.J. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uno H., Cai T., Tian L., Wei L.J. Evaluating Prediction Rules for t-Year Survivors with Censored Regression Models. J. Am. Stat. Assoc. 2007;102:527–537. doi: 10.1198/016214507000000149. [DOI] [Google Scholar]
- 60.Blanche P., Kattan M.W., Gerds T.A. The c-index is not proper for the evaluation of $t$-year predicted risks. Biostatistics. 2019;20:347–357. doi: 10.1093/biostatistics/kxy006. [DOI] [PubMed] [Google Scholar]
- 61.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [(accessed on 3 July 2020)]. Available online: https://www.R-project.org/ [Google Scholar]
- 63.Binder H. CoxBoost: Cox Models by Likelihood Based Boosting for a Single Survival Endpoint or Competing Risks. [(accessed on 3 July 2020)];2013 Available online: https://CRAN.R-project.org/package=CoxBoost.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.