Abstract
Keratin intermediate filaments constitute the primary cytoskeletal component of epithelial cells. Numerous human disease phenotypes related to keratin mutation remain mechanistically elusive. Our recent crystal structures of the helix 1B heterotetramer from keratin 1/10 enabled further investigation of the effect of pathologic 1B domain mutations on keratin structure. We used our highest resolution keratin 1B structure as a template for homology-modeling the 1B heterotetramers of keratin 5/14 (associated with blistering skin disorders), keratin 8/18 (associated with liver disease), and keratin 74/28 (associated with hair disorder). Each structure was examined for the molecular alterations caused by incorporating pathogenic 1B keratin mutations. Structural modeling indicated keratin 1B mutations can harm the heterodimer interface (R265PK5, L311RK5, R211PK14, I150VK18), the tetramer interface (F231LK1, F274SK74), or higher-order interactions needed for mature filament formation (S233LK1, L311RK5, Q169EK8, H128LK18). The biochemical changes included altered hydrophobic and electrostatic interactions, and altered surface charge, hydrophobicity or contour. Together, these findings advance the genotype-structurotype-phenotype correlation for keratin-based human diseases.
Keywords: keratin, intermediate filament, structure, modeling, skin disease, liver disease, epidermolysis bullosa, cytoskeleton, mutation
1. Introduction
Intermediate filaments (IFs) work with actin microfilaments and microtubules to provide essential cytoskeleton functions within eukaryotic cells. IFs are aptly named because their diameter (~10 nm) is in between that of actin (~4–5 nm) and microtubules (~25 nm) [1]. There are six types of IFs [2], with type I and II representing keratins. The 54 keratin genes outnumber all the other IF types combined. Keratins are obligate heterodimers, with one type I keratin dimerizing in register and parallel with one type II keratin [3]. Parallel heterodimers then form anti-parallel tetramers, which serve as the building block for mature IFs.
Keratins are differentially expressed across human tissues, providing unique biological functions to support the host organism. For example, keratins 5 and 14 (K5/14) and K1/10 are expressed in stratified epithelia. K5/14 are the primary keratins in basal layer keratinocytes, whereas K1/10 are the primary suprabasal IF proteins in differentiating keratinocytes of the epidermis. Hard keratins (K25–86), found in hair and nails, utilize abundant cysteine cross-linking to form more rigid filaments than epithelial keratin IFs (K1–K24) [4].
Keratins support human health, as exemplified by the fact that more than 80 human IF-related diseases (termed IF-pathies [5]) exist. For example, different keratin mutations are responsible for blistering skin disorders (e.g., epidermolysis bullosa simplex, EBS), keratodermas (e.g., epidermolytic palmoplantar keratoderma, EPPK), hair and nail defects (e.g., pachyonychia congenita), and liver disease (e.g., cryptogenic cirrhosis) [6]. Keratins have also been linked to cancer, corneal dystrophy, and pancreatitis [7]. IF mutations vary in type: deletion, insertion, nonsense, and missense. Efforts have been made to correlate a patient’s keratin genotype with clinical phenotype for these diseases [7,8,9,10,11,12,13]. Understanding how keratin genotype impacts the molecular structure of keratin filaments is also important, and we have termed this concept genotype-structurotype-phenotype correlation [14,15].
Keratins share a common protein organization: flexible, glycine- and serine-rich head and tail domains bookend a highly-conserved central coiled-coil rod domain, itself comprised of 1A, 1B, 2A, and 2B subdomains. We recently determined two keratin 1/10 1B domain crystal structures [16]. The first was a 3.0 Å resolution structure of wild-type K1/K10-1B (Protein Data Bank (PDB) ID 6EC0), and the second was a 2.4 Å resolution structure of K1/K10-1B incorporating the S233LK1 mutation responsible for EPPK (PDB ID 6E2J). 1B domain structures of homomeric IF proteins have also been determined (vimentin, glial fibrillary acidic protein (GFAP), lamin A) [17,18,19,20,21]. The keratin structures represent the only heteromeric 1B structures to date, and as such they are experimentally determined quality templates for modeling 1B domains from other keratin pairs. Here, we used our highest resolution K1/K10 1B structure to model 1B domains of K5/K14, K8/K18, and K74/K28 in order to evaluate the structural alterations associated with pathogenic keratin missense mutations in this region. In particular, we wanted to ascertain whether the mutations would impact the dimer, tetramer, or high-order assembly of the keratins, and how the mutations would affect the molecular surface properties (charge, hydrophobicity, contour) of the keratins.
2. Results
2.1. Identification of Keratin 1B Mutations
The Human Intermediate Filament Database (HIFD) [6] was analyzed to find all missense mutations in the 1B region of keratins. We identified 15 mutations: 9 missense, 3 nonsense, 2 leading to frameshift, and 1 in-frame deletion (indel) (Table 1). To predict the potential impact of the missense mutations on keratin structure and function, we utilized Polyphen-2 to analyze the primary keratin sequences [22]. Six out of nine keratin missense mutations had a polyphen score classified as possibly or probably damaging (F231LK1, S233LK1, R265PK5, L311RK5, R211PK14, F274SK74) (Table 1). The other three mutations (Q169EK8, H128LK18, I150VK18) were predicted to be benign.
Table 1.
Keratin 1B domain mutations identified from the Human Intermediate Filament Database. Fifteen DNA and protein mutations are listed, with 9 missense mutations that were modeled in this study. The Polyphen-2 algorithm was used to generally predict a mutation effect on the keratin. “Disease Mechanism” depicts the hypothesized or experimentally validated filamentous structural changes believed to contribute to the disease phenotype. Abbreviations: (N)EPPK, (Non)-epidermal Palmoplantar Keratoderma; BCIE, Bullous Congenital Ichthyosiform Erythroderma; EHK, Epidermolytic Hyperkeratosis; (R)EBS-K/WC, (Recessive) Epidermolysis Bullosa Simplex: Koebner or Weber–Cockayne subtypes; CIEH, Cyclic Ichthyosis with Epidermolytic Hyperkeratosis; ADWH, Autosomal-Dominant Woolly Hair; LOF, Loss of Function.
| DNA Mutation | Protein Mutation | Protein | Hetero Pair |
Mutation Type | PolyphnScore | PolyphenPrediction | Disease Association |
Disease Mechanism |
References |
|---|---|---|---|---|---|---|---|---|---|
| c.693T>G | p.Phe231Leu | K1 | K1/10 | Missense | 0.971 | Probably Damaging | NEPPK | Disrupts 1B knob-pocket interaction | [23] |
| c.698C>T | p.Ser233Leu | K1 | K1/10 | Missense | 0.704 | Possibly Damaging | EPPK, NEPPK, BCIE/EHK | Tonotubular filament formation | [16,23,24,25] |
| c.794G>C | p.Arg265Pro | K5 | K5/14 | Missense | 0.975 | Probably Damaging | EBS-K | Unknown | [26] |
| c.932T>G | p.Leu311Arg | K5 | K5/14 | Missense | 0.908 | Possibly Damaging | EBS-WC | Unknown | [27] |
| c.475C>G | p.Gln169Glu | K8 | K8/18 | Missense | 0.171 | Benign | Cryptogenic cirrhosis | Unknown | [28] |
| c.846T>A | p.Tyr282X | K10 | K1/10 | Nonsense (truncation) | N/A | N/A | CIEH | LOF (K10 K/O), aggregated K1, compensatory upregulation of K14 & K17 | [29] |
| c.526-2A>C | p.[Ile176ValfsX2, Ile176ProfsX30] | K14 | K5/14 | Frame-shift (truncation) | N/A | N/A | REBS, REBS-K | Basal K14 IF loss, compensatory K15 protofilaments | [30,31,32] |
| c.[612T>A]+[612T>A] | p.[Tyr204X]+[Tyr204X] | K14 | K5/14 | Nonsense (truncation) | N/A | N/A | REBS-K | "Natural K14 K/O", Basal K14 IF loss, insoluble keratin aggregation | [33,34] |
| c.632G>C | p.Arg211Pro | K14 | K5/14 | Missense | 0.999 | Probably Damaging | EBS-WC | Unknown | [35] |
| c.740_748delCCTACCTGAinsGAA | p.Ala247_Lys250delinsGlu | K14 | K5/14 | Indel (in-frame) | N/A | N/A | EBS-WC | Unknown | [36] |
| c.744delCinsAG | p.Tyr248X | K14 | K5/14 | Nonsense (truncation) | N/A | N/A | REBS-K | "Natural K14 K/O", Basal K14 IF loss, insoluble keratin aggregation | [37] |
| c.749delA | p.Lys250ArgfsX8 | K14 | K5/14 | Deletion (frame-shift) | N/A | N/A | REBS | Absent K14 expression | [38] |
| c.383A>T | p.His128Leu | K18 | K8/18 | Missense | 0.014 | Benign | Cryptogenic cirrhosis | Abnormal IF assembly | [39] |
| c.448A>G | p.Ile150Val | K18 | K8/18 | Missense | 0.247 | Benign | Liver disease | Unknown | [40] |
| c.821T>C | p.Phe274Ser | K74 | K74/14 or K74/28 | Missense | 0.998 | Probably Damaging | Ectodermal Dysplasia, Pure Hair-Nail Type, ADWH | Disrupts 1B knob-pocket interaction | [16,41] |
2.2. K1/K10 1B Missense Mutations Associated with Keratodermas
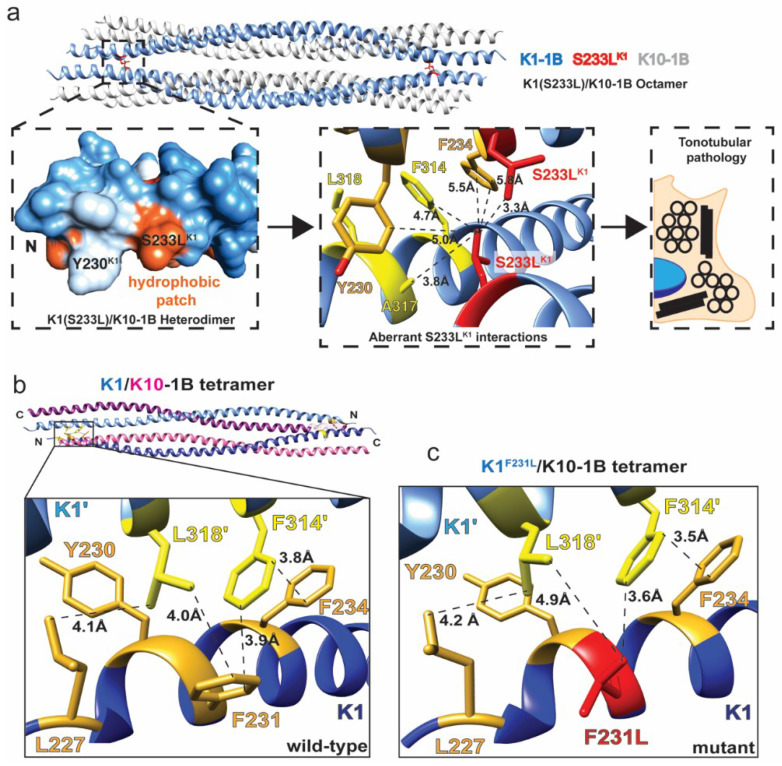
We previously determined the crystal structure of K1/K10 containing S233LK1 and also characterized the 1B domain harboring S233LK1 by multi-angle light scattering (MALS) [16]. This particular mutation is associated with epidermolytic palmoplantar keratoderma (EPPK) and histologically has the key finding of tonotubular keratin [24,42]. The structure identified that S233LK1 caused a new hydrophobic patch on the molecular surface of the K1/K10 dimer, and MALS demonstrated it also caused keratin aggregation in solution. The structure also showed that the aberrant leucine residue directly participated in inter-tetramer contacts promoting octameric assembly between K1/K10 tetramers (Figure 1a). How this excess hydrophobicity on the K1/K10 dimer surface leads to tubule formation for full-length K1/K10 remains unclear, but the tendency for hydrophobicity-driven aggregation is established.
Figure 1.
Keratin 1 1B mutations affect the oligomeric state of the K1/K10-1B heterocomplex. (a) The crystal structure of the K1S233L/K10-1B domain (PDB ID 6E2J) is an octamer composed of two tetramers (top). A surface-exposed hydrophobic patch is created by the serine to leucine mutation (left), resulting in the leucine making multiple aberrant hydrophobic interactions between tetramers (center). Histologically, this mutation results in tonotubular keratin, a more aggregated form than wild-type tonofilaments (right). (b) The crystal structure of the wild-type K1/K10-1B domain (PDB ID 6E2J) is a tetramer composed of anti-parallel dimers. A key interaction occurs between heterodimers: K1 residues comprised of an anchoring knob from one dimer (F314, L318, yellow) bind into a hydrophobic pocket on the surface of the other dimer (L227, Y230, F231, F234, gold). (c) The F231L mutation occurs in one of the key hydrophobic pocket residues in K1 (red). Modeling of this mutation reveals disruption of the knob-pocket interaction, likely leading to tetramer destabilization.
In contrast to S233LK1, F231LK1 alters the tetrameric interface between two K1/K10 dimers at the anchoring knob-hydrophobic pocket assembly mechanism [16]. F231LK1 is associated with non-epidermolytic palmoplantar keratoderma (NEPPK) [23]. In the wild-type K1/K10 complex, F231K1 makes molecular contacts with F314 and L318 from the partner K1 molecule to help form the K1/K10 tetramer (Figure 1b). Modeling of F231LK1 suggests there is an increase in the contact distance between the mutant L231 and L318’ as well as a loss of the aromatic–aromatic interactions by F314’ (Figure 1c). These molecular changes caused by F231LK1 likely destabilize the keratin 1B tetrameric interface, which in turn affects mature filament formation and function.
2.3. K5/K14 1B Missense Mutations Cause Epidermolysis Bullosa Simplex
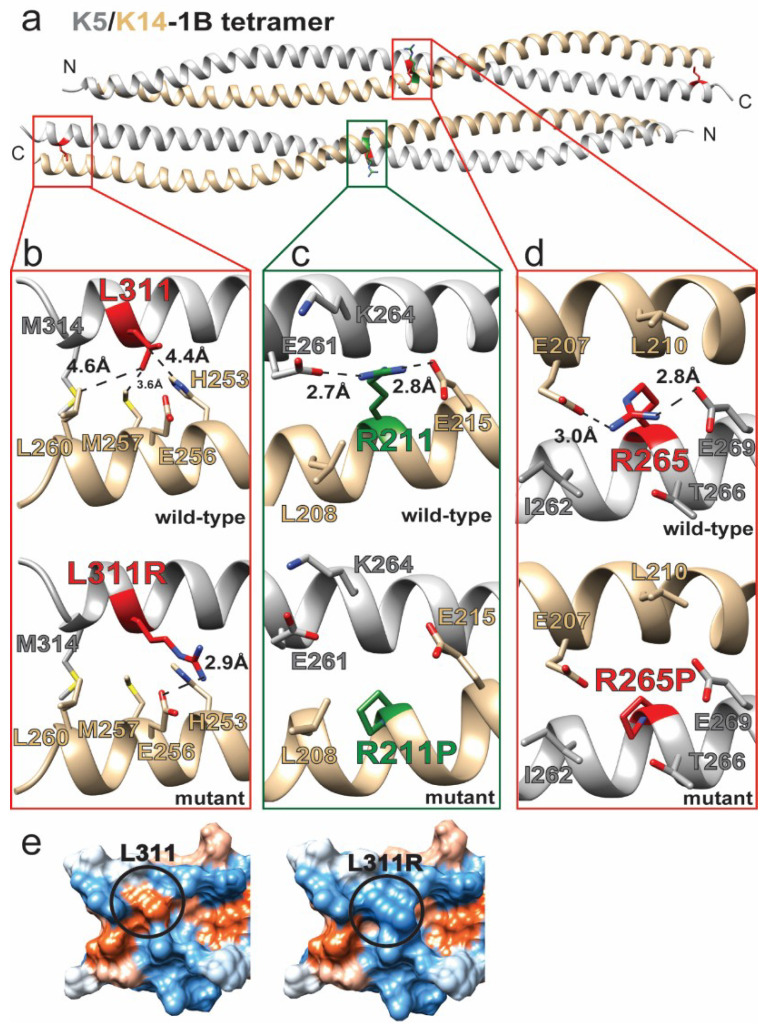
Mutations in keratins 5 and 14 are commonly associated with EBS, but the vast majority of these mutations lie within the 1A or 2B subdomains [13]. Of the 9 unique mutations we identified in the 1B subdomain (Table 1), three were in K5/K14: two K5 (R265P, L311R) and one K14 (R211P) missense mutations that are documented to cause EBS (Figure 2).
Figure 2.
Keratin 5 and keratin 14 1B mutations destabilize the K5/K14 heterodimer. (a) The K5/K14-1B tetramer was generated by homology modeling using the crystal structure of K1/K10-1B (PDB ID 6E2J) as a template. Two of the three identified missense mutations occur in the middle of the domain and the other at the carboxy-terminal end. (b) The L311RK5 mutation disrupts intra-dimeric hydrophobic interactions with nearby K14 residues and may cause an aberrant electrostatic interaction (with E256K14). (c) R211PK14 disrupts two electrostatic interactions (with E261K5 and E215K14) at the dimer interface. (d) R265PK5 disrupts two electrostatic interactions (with E269K5 and E207K14) at the dimer interface. (e) The comparison of hydrophobic surfaces (orange = hydrophobic, white = neutral, blue = polar) of the wild-type (left) and L311RK5 (right) K5/K14-1B C-terminus demonstrates that the mutation eliminates a surface-exposed hydrophobic patch.
R211PK14 was identified during screening of 27 German EBS patients; the individual from which the mutation was identified also suffered from palmoplantar symptoms [35]. The authors hypothesized that proline incorporation due to R211PK14 led to helix kinking, which is a known structural property of proline [43]. R211K14 lies in the center of the K5/14 1B domain at the heterodimer interface (Figure 2a). Modeling of wild-type K5/14 1B shows R211 forms electrostatic interactions or salt bridges with E215K14 and E261K5 (Figure 2c). By having intra-K14 and inter-K5 interactions, R211K14 provides stabilization to the K5/14 1B dimer interface. Mutation to proline eliminates the dual electrostatic/salt bridge interactions, in addition to kinking the K14 helix, thereby destabilizing the K5/14 heterodimer.
R265PK5 is a mutation identified in Korean EBS patients [26] and located in the middle of the 1B domain (Figure 2a). The structural model of K5/14 1B demonstrates that R265K5, like R211K14, forms electrostatic interactions or salt bridges with residues that are intra-strand (E269K5) as well as opposite dimer strand (E207K14) (Figure 2d). Mutation R265PK5 eliminates these important dual interactions that stabilize the K5/14 heterodimer, in addition to the expected helix kinking.
L311RK5 was one of several K5/14 mutations detected from sequencing of 10 Israeli patients suffering from EBS [27]. L311K5 is located at the C-terminus of the K5/14 1B heterodimer, where it makes hydrophobic interactions in the dimer interface with K14 residues H253, M257, and L260 (Figure 2b). L311RK5 mutation leads to the substitution of a bulkier basic residue that disrupts hydrophobic interactions and may generate new electrostatic interactions with E256K14. The molecular surface properties of K5/14 1B are also changed, notably the loss of a surface-exposed hydrophobic patch at the C-terminus of the K5/14 1B heterodimer (Figure 2e). The loss of surface hydrophobicity likely disrupts higher-order filament interactions needed for mature IF formation.
2.4. Structural Alterations in K8/K18 1B Are Associated with Liver Disease
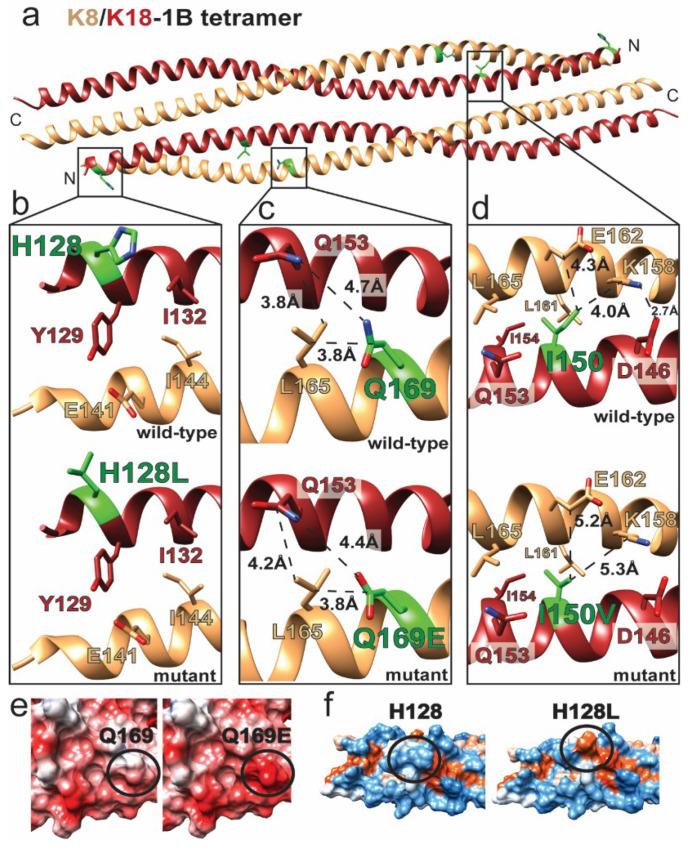
Keratins 8 and 18 (K8/18) are predominantly expressed in “simple” epithelia, and mutations in them are frequently associated with liver disease (Table 1 and Figure 3) [44]. Q169EK8 was identified during sequencing of 162 hemochromatosis patients as part of an analysis of K8/18 intron mutations associated with hereditary hemochromatosis or liver fibrosis [28]. Q169K8 is located in the central aspect of the K8/18 1B domain as part of the heterodimer interface (Figure 3a). It makes local van der Waals interactions with L165K8 and Q153K18 (Figure 3c). Q169EK8 does not appear to significantly alter the local heterodimer packing, although the OE2 atom on Q169EK8 might form a hydrogen bond with the Q153K18 NE2 atom. The larger effect of Q169EK8 is on the molecular surface, where the acidic nature of the K8/K18 1B dimer and tetramer is enhanced by the mutation (Figure 3e). This acidic patch created by Q169EK8 may disturb higher-order K8/K18 filament assembly.
Figure 3.
Keratin 8 and keratin 18 mutations alter intra-dimeric interactions and molecular surface properties. (a) The K8/K18-1B tetramer was generated by homology modeling using K1/K10-1B (PDB ID 6E2J) as a template. Missense mutated residues are identified in green. (b) The H128LK18 mutation does not disrupt intra-dimeric or tetrameric interactions. (c) Q169EK8 has limited impact on packing at the dimer interface. (d) I150VK18 preserves most hydrophobic interactions at the dimer interface but may destabilize a salt bridge at the dimer interface (between K158K8 and D146K18) by eliminating a contact with K158K8. (e) Comparison of electrostatic surfaces (red = acidic, white = neutral, blue = basic) of the wild-type (left) and Q169EK8 (right) K8/K18-1B tetramer demonstrates that the mutation enhances the surface acidity of the molecule. (f) The comparison of the hydrophobic surfaces (orange = hydrophobic, white = neutral, blue = polar) of the wild-type (left) and H128LK18 (right) K8/K18-1B tetramer demonstrates that the mutation introduces a surface-exposed hydrophobic patch.
H128K18 is located at the N-terminus of the K8/18 1B heterodimer (Figure 3a). It is solvent-exposed and does not contribute to the hydrophobic dimeric interface (Figure 3b). It also faces solvent in the K8/18 1B tetramer, suggesting that H128LK18 mutation does not destabilize the K8/18 dimer or tetramer, but rather alters the higher-order assembly of mature filaments. H128LK18 correlated with liver cirrhosis in a study involving patients with K8 and K18 mutations [45]. Mapping of hydrophobic potential onto the molecular surface of the K8/18 1B tetramer demonstrated that H128LK18 creates a bulky surface-exposed hydrophobic patch (Figure 3f). This patch would occur at both ends of the K8/18 1B tetramer due to the antiparallel alignment between dimers.
I150VK18 is another K18 mutation associated with liver disease [40]. I150K18 is located in the center of K8/18 1B at the heterodimer interface (Figure 3a). It makes several hydrophobic contacts to stabilize the dimer, including interactions with I154K18, L161K8, and L165K8. The CD1 atom of I150K18 also interacts with the aliphatic portions of K158K8 and E162K8, which stabilizes K158K8 so that it can make a salt bridge with D146K18 (Figure 3d). I150VK18 preserves the core hydrophobic interactions with I154K18, L161K8, and L165K8, but loss of the CD1 atom likely destabilizes contacts with K158K8 and E162K8 and potentially compromises the E162K8-D146K18 salt bridge.
2.5. Anchoring Knob Mutation in the Hard Keratin K74
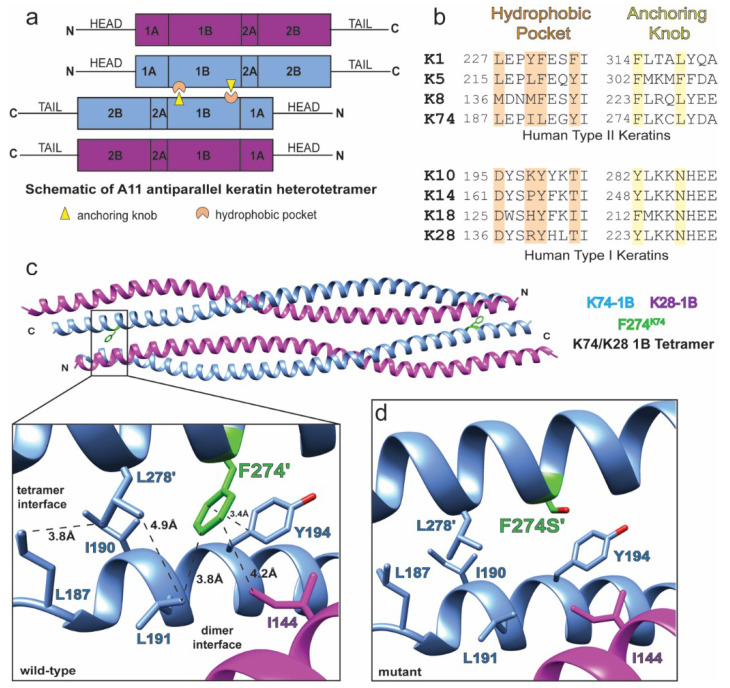
Keratin 74 (K74) is a type II IF protein whose obligate IF partner has yet to be unambiguously identified. Prior work showed that K74 can pair with K14 and K18 [46]. However, this is inconsistent with the fact that K74 is largely expressed in the inner root sheath (IRS) of hair, while K14 and K18 are expressed in basal keratinocytes and simple epithelia, respectively. There is evidence for K74 co-expression with K28 [47], which more closely aligns with the physiology of hair keratin IF formation (both are inner root sheath keratins). Therefore, we modeled the K74/28 1B heterotetramer (Figure 4).
Figure 4.
The F274SK74 mutation occurs in the anchoring knob and disrupts the tetramer interface. (a) The antiparallel keratin heterotetramer is stabilized by an anchoring knob-hydrophobic pocket interaction at both ends of the 1B domain. The “A11” alignment refers to the 1B domain of each heterodimer being in phase with each other in the tetramer state. (b) Knob and pocket residues are largely conserved in the type II keratins. These residues are not conserved in the type I keratins. (c,d) Modeling of the K74/K28-1B tetramer demonstrates loss of hydrophobic interactions in the knob-pocket mechanism as a result of the F274SK74 mutation.
F274SK74 was identified during sequencing of a consanguineous Pakistani family which had homozygous autosomal recessive pure hair and nail ectodermal dysplasia (PHNED) [41]. Patients with PHNED displayed loss of K74 when staining for expression in nail matrix, nail bed, and IRS of hair follicle, in addition to mouse hyponychium. With the available information at the time, it was hypothesized that F274SK74 would interfere with keratin hetero-dimerization. However, our recent crystal structures of K1/K10 1B tetramers (wild-type and S233LK1) demonstrated that the sequence position of F274K74 forms part of the anchoring knob involved in the anchoring knob–hydrophobic pocket tetramer assembly mechanism (Figure 4a,b). This mechanism is important for A11 tetramer formation (where A11 means the 1B domains are antiparallel and aligned in-phase in the tetramer) and mature filament assembly [16].
In the structural model, K74 residues F274’ and L278’ form the anchoring knob at the K74/28 tetramer interface (Figure 4c). These residues pack into a hydrophobic pocket formed by the partner heterodimer (K74 residues L187, I190, L191, and Y194, and K28 residue I144). F274SK74 mutation dramatically alters the knob-pocket interaction site, eliminating hydrophobic interactions and aromatic–aromatic interactions (Figure 4d). This will destabilize the K74/28 1B tetramer at both ends. Thus, F274SK74 is likely to severely alter mature filament assembly based on prior electron microscopy data showing keratins 1/10 and 8/18 and vimentin 1B knob mutations damage IF assembly [16].
3. Discussion
With 54 genes and differential expression by time and tissue type, keratins play a significant, but complex role in human physiology. Their medical relevance is exemplified by the high numbers of IF-related diseases associated with keratin mutation [5,7]. Their role in establishing and maintaining skin, hair, and nail aesthetics is also important. How keratins promote structural integrity within cells, facilitate protein interactions and signaling, and regulate tissue properties remains poorly understood at a molecular level. Their long, filamentous and insoluble nature contributes greatly to the difficulty in studying these proteins. Despite all the keratins known, only four out of 54 (K1, K10, K5, K14) have experimentally determined atomic resolution structures [12,13,14,16,48]. In total, there are six crystal structures of human keratins. Only one of these contains a mutation (S233LK1) known to cause human disease [16]. Therefore, a better understanding of how keratin mutations alter protein structure is needed to help correlate patient genotype with phenotype: a paradigm we call genotype-structurotype-phenotype correlation [14,15].
In the spirit of this paradigm, the work here examines nine pathologic keratin mutations that occur within the 1B subdomain of the keratin coiled-coil rod domain. Most keratin mutations associated with clinical disease are located in the 1A and 2B subdomains, with only a small number to date occurring in 1B. The molecular explanation for this difference is not clear, but evidence suggests the 1B domain is critical for establishing the tetramer building block of IFs, whereas the 1A and 2B domains may play a greater role in the higher-order packing interactions needed to form mature IFs [14,16,48,49]. In our analysis of the keratin 1B mutants, we considered: (1) What interface do the mutations affect, and (2) what do the mutations do to the molecular surface chemistry of the keratin?
Structural modeling indicates that some keratin 1B mutations harm the heterodimer interface (R265PK5, L311RK5, R211PK14, I150VK18), some harm the tetramer interface (F231LK1, F274SK74), and some harm higher-order interactions needed for mature IF formation (S233LK1, L311RK5, Q169EK8, H128LK18). Some mutations, like L311RK5, alter multiple interfaces. Three of the 1B mutations have a prominent change in molecular surface properties: Q169EK8 generates an acidic surface patch, L311RK5 eliminates a hydrophobic patch, and H128LK18 creates a new hydrophobic patch. Solvent accessible hydrophobic surface has been shown to cause aberrant aggregation for S233LK1 [16], leading to dramatic morphological change from tonofilaments to tonotubules [24]. H128LK18 altered K8/K18 IFs in vitro by electron microscopy [39], likely due to aberrant hydrophobic interactions. Importantly, our analysis demonstrated structural changes conducive to IF pathology even for the three missense mutations (Q169EK8, H128LK18, I150VK18) allocated a “benign” prediction by Polyphen-2 analysis (Table 1). This indicates that primary sequence analysis alone is not sufficient to capture the pathogenicity of a keratin mutation, and that analysis of keratin structure plays an essential role in understanding the molecular basis of disease.
The molecular modeling presented here establishes a foundation for designing future biochemical studies to validate the predicted amino acid properties of these keratin mutations. While the modeling is in silico, this computational structural biology is based on an experimentally derived keratin 1/10 tetrameric crystal structure, and thus is more reliable than ab initio structure prediction. The analysis of pathogenic K1 mutations mapped onto the 1B structure advances our knowledge of how disease-causing keratin mutations alter the various stages of mature filament assembly.
Ultimately, each pathologic keratin mutation will exhibit its own “structurotype” or set of biochemical and structural changes driving pathogenesis. Identifying the molecular parameters of a keratin mutation is the initial step toward the goal of being able to manipulate keratin IFs for pharmacological purposes [50]. As the number of experimental keratin structures grows, so will the insights into the molecular mechanisms driving mature filament assembly. This knowledge is essential for understanding IF-related diseases and developing new approaches to treat keratinopathies.
4. Materials and Methods
Keratin primary sequences, mutations, and corresponding literature were identified using the HIFD (interfil.org) [6] and cross-referenced using the UniProt [51] and National Center for Biotechnology Information (NCBI) variant databases: dbSNP [52] and ClinVar [53]. NCBI Protein Accession Numbers for each keratin analyzed were: K1: NP_006112.3, K10: NP_000412.3, K5: NP_000415.2, K14: NP_000517.2, K8: NP_002264.1, K18: NP_000215.1, K74: NP_778223.2, K28: NP_853513.2. Polyphen-2 was used to assess mutation severity [22]. Clustal Omega was used to perform multiple sequence alignments [54]. Homology modeling was performed with SWISS-MODEL [55] using the K1(S233L)/K10-1B tetramer structure (PDB ID 6E2J) as a template [16]. Coot [56] and UCSF Chimera [57] were used for energy minimization, structure analysis, and figure preparation. Final figures were made using Adobe Illustrator.
Abbreviations
| IF | Intermediate filament |
| K | Keratin |
| EBS | Epidermolysis bullosa simplex |
| EPPK | Epidermolytic palmoplantar keratoderma |
| PDB | Protein Data Bank |
| HIFD | Human Intermediate Filament Database |
| MALS | Multi-angle light scattering |
| PHNED | Pure hair and nail ectodermal dysplasia |
| NEPPK | Non-epidermolytic palmoplantar keratoderma |
| GFAP | Glial fibrillary acidic protein |
| NCBI | National Center for Biotechnology Information |
Author Contributions
Conceptualization, C.G.B.; Data curation, A.J.H.; Formal analysis, C.G.B.; Funding acquisition, C.G.B.; Investigation, C.G.B. and A.J.H.; Validation, S.A.E. and M.H.; Writing—original draft, A.J.H.; Writing—review & editing, S.A.E., M.H. and C.G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIH/NIAMS Awards K08AR070290 and R03AR076484 (to CGB).
Conflicts of Interest
S.A.E., M.H., and C.G.B. are inventors on pending patent PCT/US19/55115.
References
- 1.Ku N.O., Zhou X., Toivola D.M., Omary M.B. The cytoskeleton of digestive epithelia in health and disease. Am. J. Physiol. 1999;277:1108–1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann H., Bär H., Kreplak L., Strelkov S.V., Aebi U. Intermediate filaments: From cell architecture to nanomechanics. Nat. Rev. Mol. Cell. Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 3.Coulombe P.A., Fuchs E. Elucidating the early stages of keratin filament assembly. J. Cell. Biol. 1990;111:153–169. doi: 10.1083/jcb.111.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacob J.T., Coulombe P.A., Kwan R., Omary M.B. Types I and II Keratin Intermediate Filaments. Cold Spring Harb. Perspect. Biol. 2018;10:a018275. doi: 10.1101/cshperspect.a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omary M.B. “IF-pathies”: A broad spectrum of intermediate filament-associated diseases. J. Clin. Investig. 2009;119:1756–1762. doi: 10.1172/JCI39894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szeverenyi I., Cassidy A.J., Chung C.W., Lee B.T., Common J.E., Ogg S.C., Chen H., Sim S.Y., Goh W.L., Ng K.W., et al. The Human Intermediate Filament Database: Comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 2008;29:351–360. doi: 10.1002/humu.20652. [DOI] [PubMed] [Google Scholar]
- 7.Toivola D.M., Boor P., Alam C., Strnad P. Keratins in health and disease. Curr. Opin. Cell. Biol. 2015;32:73–81. doi: 10.1016/j.ceb.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Steinert P.M., Bale S.J. Genetic skin diseases caused by mutations in keratin intermediate filaments. Trends Genet. 1993;9:280–284. doi: 10.1016/0168-9525(93)90014-9. [DOI] [PubMed] [Google Scholar]
- 9.Jones L.N., Steinert P.M. Hair keratinization in health and disease. Dermatol. Clin. 1996;14:633–650. doi: 10.1016/S0733-8635(05)70390-9. [DOI] [PubMed] [Google Scholar]
- 10.Chamcheu J.C., Siddiqui I.A., Syed D.N., Adhami V.M., Liovic M., Mukhtar H. Keratin gene mutations in disorders of human skin and its appendages. Arch. Biochem. Biophys. 2011;508:123–137. doi: 10.1016/j.abb.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karantza V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunick C.G., Milstone L.M. The X-ray Crystal Structure of the Keratin 1-Keratin 10 Helix 2B Heterodimer Reveals Molecular Surface Properties and Biochemical Insights into Human Skin Disease. J. Investig. Dermatol. 2017;137:142–150. doi: 10.1016/j.jid.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.H., Kim M.S., Chung B.M., Leahy D.J., Coulombe P.A. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 2012;19:707–715. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomakin I.B., Hinbest A.J., Ho M., Eldirany S.A., Bunick C.G. Crystal structure of keratin 1/10(C401A) 2B heterodimer demonstrates a proclivity for the C-terminus of helix 2B to form higher order molecular contacts. Yale J. Biol. Med. 2020;93:3–17. [PMC free article] [PubMed] [Google Scholar]
- 15.Eldirany S.A., Ho M., Bunick C.G. The Interface between Keratin Structurotype and Human Disease. Structure. 2020;28:271–273. doi: 10.1016/j.str.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eldirany S.A., Ho M., Hinbest A.J., Lomakin I.B., Bunick C.G. Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J. 2019;38:e100741. doi: 10.15252/embj.2018100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim B., Kim S., Jin M.S. Crystal structure of the human glial fibrillary acidic protein 1B domain. Biochem. Biophys. Res. Commun. 2018;503:2899–2905. doi: 10.1016/j.bbrc.2018.08.066. [DOI] [PubMed] [Google Scholar]
- 18.Aziz A., Hess J.F., Budamagunta M.S., Voss J.C., Kuzin A.P., Huang Y.J., Xiao R., Montelione G.T., FitzGerald P.G., Hunt J.F. The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography. J. Biol. Chem. 2012;287:28349–28361. doi: 10.1074/jbc.M111.334011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang A.H., Obiero J.M., Kulczyk A.W., Sviripa V.M., Tsodikov O.V. A crystal structure of coil 1B of vimentin in the filamentous form provides a model of a high-order assembly of a vimentin filament. FEBS J. 2018;285:2888–2899. doi: 10.1111/febs.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lilina A.V., Chernyatina A.A., Guzenko D., Strelkov S.V. Lateral A. J. Struct. Biol. 2020;209:107404. doi: 10.1016/j.jsb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Ahn J., Jo I., Kang S.M., Hong S., Kim S., Jeong S., Kim Y.H., Park B.J., Ha N.C. Structural basis for lamin assembly at the molecular level. Nat. Commun. 2019;10:3757. doi: 10.1038/s41467-019-11684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimberg G., Hausser I., Müller F.B., Wodecki K., Schaffrath C., Krieg T., Oji V., Traupe H., Arin M.J. Novel and recurrent mutations in the 1B domain of keratin 1 in palmoplantar keratoderma with tonotubules. Br. J. Dermatol. 2009;160:446–449. doi: 10.1111/j.1365-2133.2008.08831.x. [DOI] [PubMed] [Google Scholar]
- 24.Terron-Kwiatkowski A., van Steensel M.A., van Geel M., Lane E.B., McLean W.H., Steijlen P.M. Mutation S233L in the 1B domain of keratin 1 causes epidermolytic palmoplantar keratoderma with "tonotubular" keratin. J. Investig. Dermatol. 2006;126:607–613. doi: 10.1038/sj.jid.5700152. [DOI] [PubMed] [Google Scholar]
- 25.Hotz A., Oji V., Bourrat E., Jonca N., Mazereeuw-Hautier J., Betz R.C., Blume-Peytavi U., Stieler K., Morice-Picard F., Schonbuchner I., et al. Expanding the Clinical and Genetic Spectrum of KRT1, KRT2 and KRT10 Mutations in Keratinopathic Ichthyosis. Acta Derm. Venereol. 2016;96:473–478. doi: 10.2340/00015555-2299. [DOI] [PubMed] [Google Scholar]
- 26.Kang T.W., Lee J.S., Kim S.E., Oh S.W., Kim S.C. Novel and recurrent mutations in Keratin 5 and 14 in Korean patients with Epidermolysis bullosa simplex. J. Dermatol. Sci. 2010;57:90–94. doi: 10.1016/j.jdermsci.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Ciubotaru D., Bergman R., Baty D., Indelman M., Pfendner E., Petronius D., Moualem H., Kanaan M., Ben Amitai D., McLean W.H., et al. Epidermolysis bullosa simplex in Israel: Clinical and genetic features. Arch. Dermatol. 2003;139:498–505. doi: 10.1001/archderm.139.4.498. [DOI] [PubMed] [Google Scholar]
- 28.Strnad P., Kucukoglu O., Lunova M., Guldiken N., Lienau T.C., Stickel F., Omary M.B. Non-coding keratin variants associate with liver fibrosis progression in patients with hemochromatosis. PLoS ONE. 2012;7:e32669. doi: 10.1371/journal.pone.0032669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez J.A., Hannoush Z.C., Vargas L.G., Momany A., Garcia C.C., Murray J.C., Dunnwald M. A Novel non-sense Mutation in Keratin 10 Causes a Familial Case of Recessive Epidermolytic Ichthyosis. Mol. Genet. Genom. Med. 2013;1:108–112. doi: 10.1002/mgg3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonkman M.F., Heeres K., Pas H.H., van Luyn M.J., Elema J.D., Corden L.D., Smith F.J., McLean W.H., Ramaekers F.C., Burton M., et al. Effects of keratin 14 ablation on the clinical and cellular phenotype in a kindred with recessive epidermolysis bullosa simplex. J. Investig. Dermatol. 1996;107:764–769. doi: 10.1111/1523-1747.ep12365805. [DOI] [PubMed] [Google Scholar]
- 31.Schuilenga-Hut P.H., Scheffer H., Pas H.H., Nijenhuis M., Buys C.H., Jonkman M.F. Partial revertant mosaicism of keratin 14 in a patient with recessive epidermolysis bullosa simplex. J. Investig. Dermatol. 2002;118:626–630. doi: 10.1046/j.1523-1747.2002.01715.x. [DOI] [PubMed] [Google Scholar]
- 32.Hut P.H., Vlies P.v.d., Jonkman M.F., Verlind E., Shimizu H., Buys C.H., Scheffer H. Exempting homologous pseudogene sequences from polymerase chain reaction amplification allows genomic keratin 14 hotspot mutation analysis. J. Investig. Dermatol. 2000;114:616–619. doi: 10.1046/j.1523-1747.2000.00928.x. [DOI] [PubMed] [Google Scholar]
- 33.Chan Y., Anton-Lamprecht I., Yu Q.C., Jackel A., Zabel B., Ernst J.P., Fuchs E. A human keratin 14 "knockout": The absence of K14 leads to severe epidermolysis bullosa simplex and a function for an intermediate filament protein. Genes Dev. 1994;8:2574–2587. doi: 10.1101/gad.8.21.2574. [DOI] [PubMed] [Google Scholar]
- 34.Yiasemides E., Trisnowati N., Su J., Dang N., Klingberg S., Marr P., Melbourne W., Tran K., Chow C.W., Orchard D., et al. Clinical heterogeneity in recessive epidermolysis bullosa due to mutations in the keratin 14 gene, KRT14. Clin. Exp. Dermatol. 2008;33:689–697. doi: 10.1111/j.1365-2230.2008.02858.x. [DOI] [PubMed] [Google Scholar]
- 35.Müller F.B., Küster W., Wodecki K., Almeida H., Bruckner-Tuderman L., Krieg T., Korge B.P., Arin M.J. Novel and recurrent mutations in keratin KRT5 and KRT14 genes in epidermolysis bullosa simplex: Implications for disease phenotype and keratin filament assembly. Hum. Mutat. 2006;27:719–720. doi: 10.1002/humu.9437. [DOI] [PubMed] [Google Scholar]
- 36.Bolling M.C., Lemmink H.H., Jansen G.H.L., Jonkman M.F. Mutations in KRT5 and KRT14 cause epidermolysis bullosa simplex in 75% of the patients. Br. J. Dermatol. 2011;164:637–644. doi: 10.1111/j.1365-2133.2010.10146.x. [DOI] [PubMed] [Google Scholar]
- 37.Lanschuetzer C.M., Klausegger A., Pohla-Gubo G., Hametner R., Richard G., Uitto J., Hintner H., Bauer J.W. A novel homozygous nonsense deletion/insertion mutation in the keratin 14 gene (Y248X; 744delC/insAG) causes recessive epidermolysis bullosa simplex type Köbner. Clin. Exp. Dermatol. 2003;28:77–79. doi: 10.1046/j.1365-2230.2003.01218.x. [DOI] [PubMed] [Google Scholar]
- 38.García M., Santiago J.L., Terron A., Hernandez-Martin A., Vicente A., Fortuny C., De Lucas R., Lopez J.C., Cuadrado-Corrales N., Holguin A., et al. Two novel recessive mutations in KRT14 identified in a cohort of 21 Spanish families with epidermolysis bullosa simplex. Br. J. Dermatol. 2011;165:683–692. doi: 10.1111/j.1365-2133.2011.10428.x. [DOI] [PubMed] [Google Scholar]
- 39.Ku N.O., Wright T.L., Terrault N.A., Gish R., Omary M.B. Mutation of human keratin 18 in association with cryptogenic cirrhosis. J. Clin. Investig. 1997;99:19–23. doi: 10.1172/JCI119127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ku N.O., Lim J.K., Krams S.M., Esquivel C.O., Keeffe E.B., Wright T.L., Parry D.A., Omary M.B. Keratins as susceptibility genes for end-stage liver disease. Gastroenterology. 2005;129:885–893. doi: 10.1053/j.gastro.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 41.Raykova D., Klar J., Azhar A., Khan T.N., Malik N.A., Iqbal M., Tariq M., Baig S.M., Dahl N. Autosomal recessive transmission of a rare KRT74 variant causes hair and nail ectodermal dysplasia: Allelism with dominant woolly hair/hypotrichosis. PLoS ONE. 2014;9:e93607. doi: 10.1371/journal.pone.0093607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wevers A., Kuhn A., Mahrle G. Palmoplantar keratoderma with tonotubular keratin. J. Am. Acad. Dermatol. 1991;24:638–642. doi: 10.1016/0190-9622(91)70099-N. [DOI] [PubMed] [Google Scholar]
- 43.Woolfson D.N., Williams D.H. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277:185–188. doi: 10.1016/0014-5793(90)80839-B. [DOI] [PubMed] [Google Scholar]
- 44.Strnad P., Paschke S., Jang K.H., Ku N.O. Keratins: Markers and modulators of liver disease. Curr. Opin. Gastroenterol. 2012;28:209–216. doi: 10.1097/MOG.0b013e3283525cb8. [DOI] [PubMed] [Google Scholar]
- 45.Ku N.O., Darling J.M., Krams S.M., Esquivel C.O., Keeffe E.B., Sibley R.K., Lee Y.M., Wright T.L., Omary M.B. Keratin 8 and 18 mutations are risk factors for developing liver disease of multiple etiologies. Proc. Natl. Acad. Sci. USA. 2003;100:6063–6068. doi: 10.1073/pnas.0936165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimomura Y., Wajid M., Petukhova L., Kurban M., Christiano A.M. Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am. J. Hum. Genet. 2010;86:632–638. doi: 10.1016/j.ajhg.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langbein L., Rogers M.A., Praetzel-Wunder S., Helmke B., Schirmacher P., Schweizer J. K25 (K25irs1), K26 (K25irs2), K27 (K25irs3), and K28 (K25irs4) represent the type I inner root sheath keratins of the human hair follicle. J. Investig. Dermatol. 2006;126:2377–2386. doi: 10.1038/sj.jid.5700494. [DOI] [PubMed] [Google Scholar]
- 48.Lee C.H., Kim M.S., Li S., Leahy D.J., Coulombe P.A. Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly. Structure. 2020;28:355–362. doi: 10.1016/j.str.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernot K.M., Lee C.H., Coulombe P.A. A small surface hydrophobic stripe in the coiled-coil domain of type I keratins mediates tetramer stability. J. Cell. Biol. 2005;168:965–974. doi: 10.1083/jcb.200408116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., Groppi V.E., Gui H., Chen L., Xie Q., Liu L., Omary M.B. High-Throughput Screening for Drugs that Modulate Intermediate Filament Proteins. Methods Enzymol. 2016;568:163–185. doi: 10.1016/bs.mie.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 53.Landrum M.J., Kattman B.L. ClinVar at five years: Delivering on the promise. Hum. Mutat. 2018;39:1623–1630. doi: 10.1002/humu.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]