Abstract
For the functional food applications, antioxidant properties and the bioactive compounds of the 23 Curcuma species commercially cultivated in Thailand were studied. Total phenolic content and DPPH radical scavenging activity were determined. The concentrations of eight bioactive compounds, including curcumin (1), demethoxycurcumin (2), bisdemethoxycurcumin (3), 1,7-diphenyl-(4E,6E)-4,6-heptadien-3-ol (4), germacrone (5), furanodienone (6), zederone (7), and ar-turmerone (8), were determined from the Curcuma by HPLC. While the total phenolic content of C. longa was highest (22.3 ± 2.4 mg GAE/g, mg of gallic acid equivalents), C. Wan Na-Natong exhibited the highest DPPH (2,2-diphenyl-1-picryl-hydrazyl-hydrate) radical scavenging activity. Twenty-three Curcuma species showed characteristic distributions of the bioactive compounds, which can be utilized for the identification and authentication of the cultivated Curcuma species. C. longa contained the highest content of curcumin (1) (304.9 ± 0.1 mg/g) and C. angustifolia contained the highest content of germacrone (5) (373.9 ± 1.1 mg/g). It was noteworthy that 1,7-diphenyl-(4E,6E)-4,6-heptadien-3-ol (4) was found only from C. comosa at a very high concentration (300.7 ± 1.4 mg/g). It was concluded that Thai Curcuma species have a great potential for the application of functional foods and ingredients.
Keywords: antioxidant, bioactive, Curcuma, curcuminoids, HPLC, sesquiterpenoids, total phenolic content
1. Introduction
Turmeric, Curcuma longa, is the only Curcuma species extensively cultivated and traded in the world [1]. Taxonomically, it belongs to the Curcuma genus of the Zingiberaceae family, and is mostly distributed in Asia [2,3]. Various beneficial health-promoting effects of C. longa, such as Alzheimer′s disease prevention, anti-inflammatory effects, and HIV-1 protease inhibition, were reported [4,5]. Curcuminoids and sesquiterpenoids were identified as the major bioactive constituents of C. longa [6]. More than a hundred species of Curcuma are reported worldwide, and about 30 species among them are cultivated and consumed in Thailand as food additives, cosmetics, and traditional medicines [7]. In particular, many of them are used for the treatment of various diseases due to the existence of bioactive compounds. However, bioactive compounds in other Curcuma species, other than C. longa, have never been studied extensively.
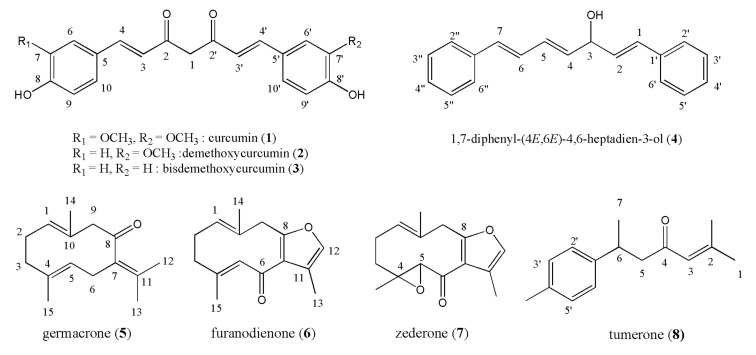
To expand the application of Curcuma species as functional foods, we have collected and cultivated 23 Thai Curcuma species widely consumed in Thailand. Total phenolic content and antioxidant activity were measured in the ethanol extracts of dried Curcuma rhizomes. For the bioactive compound study, four curcuminoids and four sesquiterpenoids were isolated and used as reference compounds after complete characterizations, because these are popularly studied representative phytochemicals in Curcuma species. In detail, the curcuminoids curcumin (1), demethoxycurcumin (2), and bisdemethoxycurcumin (3) were isolated from C. longa, and the other curcuminoid 1,7-diphenyl-(4E,6E)-4,6-heptadien-3-ol (4) was isolated from C. comosa. The three germacrane-type sesquiterpenoids germacrone (5), furanodienone (6), and zederone (7) were isolated from C. latifolia, and ar-turmerone (8) was isolated from C. zedoaria (Figure 1). The compositions of these representative bioactive compounds in the Curcuma species were also analyzed by HPLC.
Figure 1.
Structure of curcuminoids and sesquiterpenoids isolated from Curcuma species.
2. Materials and Methods
2.1. Curcuma Species
The rhizomes of 23 Curcuma species were collected in Kanchanaburi in 2013 and cultivated in Tapong, Meaung Rayong, Thailand in 2014 throughout the year. The plants that formed flowers were processed to prepare depository specimens at Bangkok Herbarium, Plant Variety Protection Division, Department of Agriculture, Bangkok, Thailand.
2.2. Plant Materials and Extraction
Fresh rhizomes of the 23 Curcuma species were rinsed several times with tap water to make them dust and debris free. The rhizomes were then cut into small pieces and dried at room temperature in the shade for 2–3 days. The dried samples were ground to a fine powder and kept in a deep freeze (−70 °C) for further experiments. For extraction, each sample (1 g) of Curcuma sp. was macerated in EtOH (20 mL) for 24 h. The ethanol extract was filtered and dried under reduced pressure. The residue was weighed for the extraction yield and dissolved in MeOH for the determination of total phenolic content and DPPH radical scavenging assay.
2.3. Determination of Total Phenolic Contents
Total phenolic contents of all plant extracts were determined using Folin–Ciocalteu reagent as described by Singleton and Rossi [8]. The plant extracts were dissolved in MeOH (1 mL, 0.5 mg/mL). Samples were added to microtiter plates and mixed with 100 µL of a 10-fold diluted Folin–Ciocalteu reagent and 80 µL of 7.5% sodium carbonate. The absorbance at 765 nm was measured using microplate reader spectrophotometers (Spectramax190, Molecular Device, CA, USA) after 30 min. Total phenolic content was expressed as mg GAE/g. The calibration curve of standard gallic acid solution was obtained as y = 0.005 x (A765) + 0.081, R2 = 0.998, where y = mg GAE/g, at the region between 5 and 200 µg/mL.
2.4. DPPH Radical Scavenging Assay
The antioxidant activity of the Curcuma extracts was evaluated by DPPH radical scavenging. The extract in MeOH (2 mg/mL) and ascorbic acid in MeOH (200 µg/mL) were prepared. The test solution (50 µL) was mixed with 950 µL of DPPH (0.1 mM in MeOH). After 30 min, the absorbance at 516 nm was measured using spectrophotometer (UV/visible detector) in triplicate. DPPH radical scavenging activity was calculated using the following formula: % scavenging = 100 × ((absorbance of control–absorbance of sample test)/absorbance of control).
2.5. Isolation of Compounds
The dried rhizomes (500 g) of C. longa were ground and extracted with EtOH (1 L) by maceration (24 h). The dried crude extracts (5 g) were suspended in water and extracted by EtOAc (500 mL × 3). The combined organic layer (500 mg) was isolated by vacuum liquid chromatography using hexanes and an acetone gradient. Out of the 36 fractions, fractions 25–34 (150 mg) were combined and rechromatographed by column chromatography (4% MeOH in chloroform) to afford 40 fractions. Curcumin (1) was isolated from fractions 9–14 (30 mg), demethoxycurcumin (2) was isolated from fractions 19–23 (15 mg), and bisdemethoxycurcumin (3) was isolated from fractions 25–35 (55 mg).
Curcumin (1). Yellow powder; ESI m/z 369 [M + H]+; m.p. 181–183 °C; Anal. calcd for C21H20O6: C 68.47, H 5.47, found: C 67.12, H 5.47. 1H NMR (600 MHz, DMSO-d6): δ 7.54 (2H, d, J = 15.8 Hz, H-4, 4′), 7.32 (2H, d, J = 2.0 Hz, H-6, 6′), 7.15 (2H, dd, J = 8.3 Hz, 2.0 Hz, H-10, 10′), 6.82 (2H, d, J = 8.1 Hz, H-9, 9′), 6.75 (2H, d, J = 15.8 Hz, H-3, 3′), 6.06 (1H, s, H-1), 3.84 (6H, s, 7-OCH3, 7′-OCH3). 13C NMR (151 MHz, DMSO-d6): δ 183.26 (C-2, 2′), 149.43 (C-8, 8′), 148.06 (C-7, 7′), 140.76 (C-4, 4′), 126.37 (C-5, 5′), 123.21 (C-10, 10′), 121.13 (C-3, 3′), 115.76 (C-9, 9′), 111.35 (C-6, 6′), 100.91 (C-1), 55.76 (7-OCH3, 7′-OCH3).
Demethoxycurcumin (2). Yellow orange powder; ESI m/z 339 [M + H]+; m.p. 170–171 °C; Anal. calcd for C20H18O5: C 70.99, H 5.36, found: C 68.53, H 5.17. 1H NMR (600 MHz, DMSO-d6): δ δ 10.0 (1H, br s, 8-OH), 7.56 (2H, d, J = 8.7 Hz, H-6, 10), 7.57 (2H, dd, J = 15.8 Hz, 5.3 Hz, H-4, 4′), 7.31 (1H, d, J = 2.0 Hz, H-6′), 7.14 (1H, dd, J = 8.2 Hz, 2.0 Hz, H-10′), 6.82 (3H, dm, H-7, 9, 9′), 6.75 (H, d, J = 16 Hz, H-3′), 6.69 (H, d, J = 16 Hz, H-3), 6.04 (1H, s, H-1), 3.51 (1H, s, H-1′), 3.83 (3H, s, 7′-OCH3). 13C NMR (125 MHz, DMSO-d6): δ 183.69 (C-2), 183.56 (C-2′), 160.25 (C-8), 149.81 (C-8′), 148.43 (C-7′), 141.13 (C-4′), 140.79 (C-4), 130.76 (C-6, C-10), 126.76 (C-5′), 126.24 (C-5), 123.63 (C-10′), 121.46 (C-3′), 121.24 (C-3), 116.34 (C-7, C-9), 116.12 (C-9′), 111.68 (C-6′), 101.32 (C-1), 56.13 (OMe).
Bisdemethoxycurcumin (3). Yellow orange crystals; ESI m/z 309 [M + H]+; m.p. 220–221 °C; Anal. calcd for C19H16O5: C 74.01, H 5.23, found: C 69.72, H 5.51. 1H NMR (600 MHz, DMSO-d6): δ 10.1 (2H, br s, 8,8′-OH), 7.57 (4H, dd, J = 6.6 Hz, 2.1 Hz, H-6, 6′, 10, 10′), 7.54 (2H, d, J = 16 Hz, H-4, 4′), 6.82 (4H, dm, J = 8.6 Hz, H-7, 7′, 9, 9′), 6.70 (2H, d, J = 16 Hz, H-3, 3′), 6.04 (1H, s, H-1), 3.51 (1H, s, H-1′). 13C NMR (151 MHz, DMSO-d6): δ 183.32 (C-2, 2′), 159.88 (C-8, 8′), 140.48 (C-4, 4′), 130.44 (C-6, 6′, 10, 10′), 125.93 (C-5, 5′), 120.90 (C-3, 3′), 116.04 (C-7, 7′, 9, 9′), 101.05 (C-1).
The dried rhizome (100 g) of C. comosa was ground and macerated with MeOH (200 mL) for 24 h. The dried crude extracts (2 g) were partitioned by EtOAc and water (300 mL × 3). The EtOAc extract (300 mg) was further isolated by vacuum liquid chromatography using hexanes and acetone. Fractions 12–15 gave 4 (48 mg). 1,7-Diphenyl-(4E,6E)-4,6-heptadien-3-ol (4) Pale yellow solid; ESI m/z 247 [M + H]+; Anal. calcd for C19H18O: C 86.99, H 6.92, found: C 86.37, H 7.59. 1H NMR (300 MHz, CDCl3): δ 7.27 (2H, br, H-3′, H-5′), 6.75 (2H), 5.83 (H-4), 4.21 (1H, s, OH), 2.71 (H-7).
The sesquiterpenoids were isolated from the ethanol extract of C. latifolia by the same procedure as for curcuminoids. Germacrone (5) was isolated from fractions 9–10 (18 mg), furanodienone (6) was isolated from fraction 15 (10 mg), and zederone (7) was isolated from fractions 20–22 (38 mg).
Germacrone (5). White crystal; EI m/z 218 [M]+; m.p. 55–56 °C; Anal. calcd for C15H22O: C 82.52, H 10.16, found: C 80.81, H 9.83. 1H NMR (600 MHz, CDCl3): δ 4.99 (1H, br d, J = 12.2 Hz, H-1), 4.71 (1H, br d, J = 11.2 Hz, H-5), 3.41 (1H, d, J = 10.6 Hz, H-9a), 2.99–2.83 (3H, m, H-6a, H-6b, H-9b), 2.37 (1H, m, H-2a), 2.19–2.05 (3H, m, H-2b, H-3a, H-3b), 1.78 (3H, s, 13-CH3), 1.73 (3H, s, 12-CH3), 1.63 (3H, s, 14-CH3), 1.44 (3H, s, 15-CH3). 13C NMR (151 MHz, CDCl3): δ 207.92 (C-8), 137.25 (C-11), 135.00 (C-4), 132.68 (C-1), 129.48 (C-7), 126.68 (C-10), 125.38 (C-5), 55.91 (C-9), 38.09 (C-3), 29.23 (C-6), 24.09 (C-2), 22.34 (C-13), 19.90 (C-12), 16.72 (C-14), 15.59 (C-15).
Furanodienone (6). Yellow pale oil; EI m/z 230 [M]+; m.p. 84–86 °C; Anal. calcd for C15H18O2: C 78.23, H 7.88, found: C 64.84, H 6.91. 1H NMR (600 MHz, CDCl3): δ 7.08 (1H, m, H-12), 5.81 (1H, m, H-5), 5.18 (1H, br dd, J = 11.7 Hz, 4.3 Hz, H-1), 3.70 (2H, br d, J = 22 Hz, H-9), 2.47 (1H, dt, J = 11.4 Hz, 3.6 Hz, H-3a), 2.32 (1H, m, H-2a), 2.18 (1H, m, H-2b), 2.13 (3H, d, J = 1.2 Hz, 13-CH3), 2.00 (3H, d, J = 1.2 Hz, 15-CH3), 1.89 (1H, m, H-3b), 1.31 (3H, bd, J = 0.7 Hz, 14-CH3). 13C NMR (151 MHz, CDCl3): δ 189.81 (C-6), 156.52 (C-8), 145.79 (C-4), 138.07 (C-12), 135.39 (C-10), 132.44 (C-5), 130.51 (C-1), 123.71 (C-11), 122.18 (C-7), 41.70 (C-3), 40.66 (C-9), 26.44 (C-2), 18.98 (C-15), 15.78 (C-14), 9.55 (C-13).
Zederone (7). White crystal; EI m/z 246 [M]+; m.p. 149–151 °C; (69.13 %C, 7.11 %H) Anal. calcd for C15H18O3: C 73.15, H 7.37, found: C 69.13, H 7.11. 1H NMR (600 MHz, CDCl3): δ 7.08 (1H, m, H-12), 5.48 (1H, m, H-1), 3.81 (1H, s, H-5), 3.75 (1H, d, J = 16.3 Hz, H-9a), 3.69 (1H, d, J = 16.3 Hz, H-9b), 2.52 (1H, m, H-2a), 2.30 (1H, dt, J = 13.1 Hz, 3.5 Hz, H-3a), 2.26–2.20 (1H, m, H-2b), 2.11 (3H, d, J = 1.3 Hz, 13-CH3), 1.34 (3H, d, J = 0.8 Hz, 15-CH3), 1.32–1.25 (1H, m, H-3b). 13C NMR (151 MHz, CDCl3): δ 192.19 (C-6), 157.08 (C-8), 138.05 (C-12), 131.19 (C-1), 131.04 (C-10), 123.24 (C-11), 122.21 (C-7), 66.54 (C-5), 63.96 (C-4), 41.88 (C-9), 37.98 (C-3), 24.64 (C-2), 15.72 (C-14), 15.14 (C-15), 10.26 (C-13).
ar-Turmerone (8) was isolated from C. zedoaira, and 65 mg of 8 was obtained from 100 g of the dried rhizomes. ar-Turmerone (8). Colorless oil; EI m/z 216 [M]+; Anal. calcd for C15H20O: C 83.28, H 9.32, found: C 79.31, H 8.82. 1H NMR (600 MHz, CDCl3): δ 7.10 (4H, aromatic, H), 6.02 (1H, J = 1.3 Hz), 3.28 (1H, dd, J = 8.1 Hz), 2.70 (1H, dd, J = 15.6 Hz), 2.60 (1H, dd, J = 15.6 Hz), 2.31 (3H, s, -CH3), 2.10 (3H, d, J = 1.3 Hz, -CH3), 1.85 (3H, d, J = 1.3 Hz, -CH3), 1.25 (3H, d, J = 1.3 Hz, 12-CH3). 13C NMR (151 MHz, CDCl3): δ 199.85, 155.06, 143.68, 135.53, 129.10, 126.65, 124.08, 52.68, 35.28, 27.63, 21.98, 20.97, 20.70.
2.6. Validation of HPLC Analysis
Validation of HPLC analysis was performed using the purified reference compounds. Calibration curves were constructed from the HPLC peak areas of the reference standards, 1–8, versus their concentrations. Linearity was calculated by measuring the eight points of the calibration curve in the range of 0.003125-0.4 mM (1–3) and 0.00625-0.8 mM (4–8) with triplicate measurements. The limits of detection (LOD) for 1–8 were found at the micromolar level. Demethoxycurcumin (2) and furanodienone (6) showed the lowest LOD (2.0 μM) and limits of quantification (LOQ) at 420 nm and 245 nm, respectively (Table 1).
Table 1.
Validation of HPLC assay of curcuminoids and sesquiterpenoids.
| Compounds | Accuracy 1 | Linearity | Correlation Coefficient (R²) | LOD (mM) | LOQ (mM) |
|---|---|---|---|---|---|
| Curcumin (1) | 104.8 ± 7.9 | y = 56,277,482x + 476,424 | 0.9951 | 0.0081 | 0.0245 |
| Demethoxycurcumin (2) | 101.0 ± 7.9 | y = 50,455,350x + 56,885 | 0.9997 | 0.0020 | 0.0061 |
| Bisdemethoxycurcumin (3) | 96.5 ± 16.2 | y = 54,929,237x + 369,859 | 0.9964 | 0.0069 | 0.0209 |
| 1,7-diphenyl-(4E,6E)-4,6-heptadien-3-ol (4) | 106.9 ± 11.6 | y = 28,237,427x + 612,834 | 0.9963 | 0.0125 | 0.0377 |
| Germacrone (5) | 97.5 ± 16.6 | y = 3,051,741x + 22,600 | 0.9977 | 0.0110 | 0.0333 |
| Furanodienone (6) | 101.9 ± 6.8 | y = 8,043,402x − 10,111 | 0.9999 | 0.0020 | 0.0060 |
| Zederone (7) | 99.9 ± 5.1 | y = 5,130,378x + 25,981 | 0.9999 | 0.0026 | 0.0080 |
| ar-Turmerone (8) | 98.5 ± 15.0 | y = 35,723,577x + 423,877 | 0.9962 | 0.0125 | 0.0379 |
1 All values are presented as the mean ± SD of triplicate determinations.
2.7. Compositional Analysis of Curcuma Species
Each sample (1 g) of the Curcuma species was macerated in EtOH (20 mL) for 24 h. The extract was filtered and dried under reduced pressure. The dried residue was dissolved in DMF (N,N-dimethylformamide, 1 mg/1 mL) for the analysis with a Finnigan Surveyor Plus HPLC system. The mobile phase comprised 0.1% acetic acid in water (solvent A) and 0.1% acetic acid in MeCN (solvent B). The injection volume for HPLC analysis was 10 μL and the flow was 1.0 mL/min. For the quantitative analysis, UV absorption at 420 nm was adopted for 1–3, and UV absorption at 245 nm was adopted for 4–8.
2.8. Statistical Analyses
All measurements were performed in triplicate. Data were processed by Microsoft Excel and reported as means ± standard deviation.
3. Results and Discussion
3.1. Taxonomy of Curcuma Species
Among the 23 Curcuma species, 11 Curcuma species, such as C. manga, C. aeruginosa, C. comosa, C. aurantiaca, C. aromatic, C. latifolia, C. zedoaria, C. longa, C. parviflora, C. angustifolia, and C. petiolate, were identified by the taxonomist at the Bangkok Herbarium. The other 12 species are new species, and designated by common names in this report (Table 2).
Table 2.
Total phenolic content and antioxidant activity of Curcuma species.
| Curcuma Species | Extraction Yield (%) | Total Phenolic Content (mg GAE/g Dry Weight) | Antioxidant Activity (%) 1 |
|---|---|---|---|
| Curcuma Wan Ma-Leung | 6.5 ± 0.4 | 0.9 ± 0.0 | 18.2 ± 0.3 |
| Curcuma mangga | 6.9 ± 0.3 | 0.6 ± 0.3 | 9.6 ± 0.5 |
| Curcuma Wan Ma-Hor | 8.9 ± 0.2 | 0.5 ± 0.1 | 13.2 ± 0.5 |
| Curcuma Wan Khamin-Dam | 13.5 ± 0.2 | 3.9 ± 0.4 | 20.4 ± 0.4 |
| Curcuma Wan Rang- Jud | 6.3 ± 0.3 | 0.4 ± 0.1 | 11.2 ± 0.2 |
| Curcuma aeruginosa | 8.6 ± 0.2 | 3.2 ± 0.3 | 21.5 ± 0.3 |
| Curcuma comosa | 16.8 ± 0.3 | 4.2 ± 0.1 | 90.0 ± 0.3 |
| Curcuma Wan Kanta-Mala | 12.0 ± 1.0 | 4.8 ± 0.1 | 12.7 ± 0.3 |
| Curcuma aurantiaca | 11.9 ± 0.1 | 1.5 ± 0.1 | 15.2 ± 0.4 |
| Curcuma aromatica | 18.5 ± 0.3 | 11.0 ± 0.2 | 20.8 ± 0.1 |
| Curcuma latifolia | 20.3 ± 0.2 | 12.9 ± 0.3 | 58.6 ± 0.0 |
| Curcuma zedoaria | 8.5 ± 0.6 | 9.3 ± 0.7 | 65.7 ± 0.2 |
| Curcuma longa | 21.6 ± 0.5 | 22.3 ± 2.4 | 73.9 ± 0.1 |
| Curcuma parviflora | 11.4 ± 0.5 | 15.3 ± 1.2 | 17.5 ± 0.5 |
| Curcuma angustifolia | 3.2 ± 0.3 | 2.6 ± 0.3 | 3.0 ± 0.2 |
| Curcuma Wan Khabitong | 11.7 ± 0.1 | 4.6 ± 0.1 | 17.0 ± 0.1 |
| Curcuma Wan Pataba | 5.8 ± 0.2 | 3.3 ± 0.1 | 44.5 ± 0.8 |
| Curcuma Wan Kortong | 8.4 ± 0.5 | 3.3 ± 0.1 | 10.4 ± 0.4 |
| Curcuma Wan Na-Natong | 3.7 ± 0.2 | 5.0 ± 0.1 | 91.8 ± 0.6 |
| Curcuma petiolata | 5.5 ± 0.1 | 2.8 ± 0.4 | 11.8 ± 0.4 |
| Curcuma Wan Khamin-Khao-Padtalod | 6.2 ± 0.3 | 1.9 ± 0.8 | 7.7 ± 0.2 |
| Curcuma Wan Chai-Dam | 3.9 ± 0.2 | 8.9 ± 0.2 | 89.8 ± 0.6 |
| Curcuma Wan Khamintong | 8.6 ± 0.2 | 7.7 ± 1.3 | 33.3 ± 0.7 |
1 Antioxidant activity (%): final concentration of sample = 100 µg/mL and ascorbic acid final concentration = 10 µg/mL (95.1 ± 0.2).
3.2. Total Phenolic Content and Antioxidant Activity
Total phenolic contents of the Curcuma species varied from 0.4 ± 0.1 to 22.3 ± 2.4 mg GAE/g. The rhizomes of C. longa contained the highest phenolic contents (22.3 ± 2.4 mg GAE/g), followed by C. parfivlora (15.3 ± 1.2 mg GAE/g) and C. latifolia (12.9 ± 0.3 mg GAE/g). The DPPH radical scavenging activity of Curcuma extracts is also presented in Table 2. Curcuma Wan Na-Natong showed the highest DPPH radical scavenging activity (91.8 ± 0.6%), followed by C. comosa (90.0 ± 0.3%) and Curcuma Wan Chai-Dam (89.8 ± 0.6%). These three species showed very strong antioxidant activity, even stronger than C. longa, and require further study.
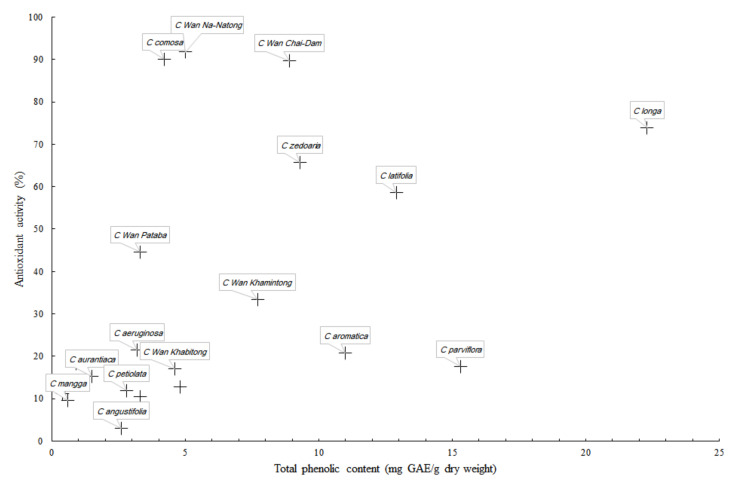
From the results, it was clear that total phenolic contents of Curcuma extracts are not necessarily correlated with the antioxidant activity (Figure 2). Among the three most antioxidant Curcuma rhizomes, C. Wan Na-Natong and C. Wan Chai-Dam are the new species that have never been extensively studied, regardless of their local popularity. The correlation chart (Figure 2) shows that the total phenolic content and DPPH radical scavenging antioxidant activity among the Curcuma species are not necessarily linearly correlated. This is believed to be due to different phytochemical compositions of Curcuma species, as discussed below.
Figure 2.
Correlation chart of total phenolic content and DPPH radical scavenging antioxidant activity. The data are from Table 2, and only notable data are labeled in the chart.
3.3. Isolation of Bioactive Curcuminoids and Sesquiterpenoids
Curcuminoids 1–3 were isolated from C. longa, and 4 was isolated from C. comosa. Sesquiterpenoids 5–7 were isolated from C. latifolia for the first time in this study. Compound 8 was isolated from C. zedoaria. The structures of the isolated reference compounds (Figure 1) were confirmed by the comparison of melting point, elemental analysis, 1H, and 13C NMR data (see the Supplementary Materials) [9,10,11,12,13,14,15,16].
3.4. Chemical Composition of 23 Curcuma Species
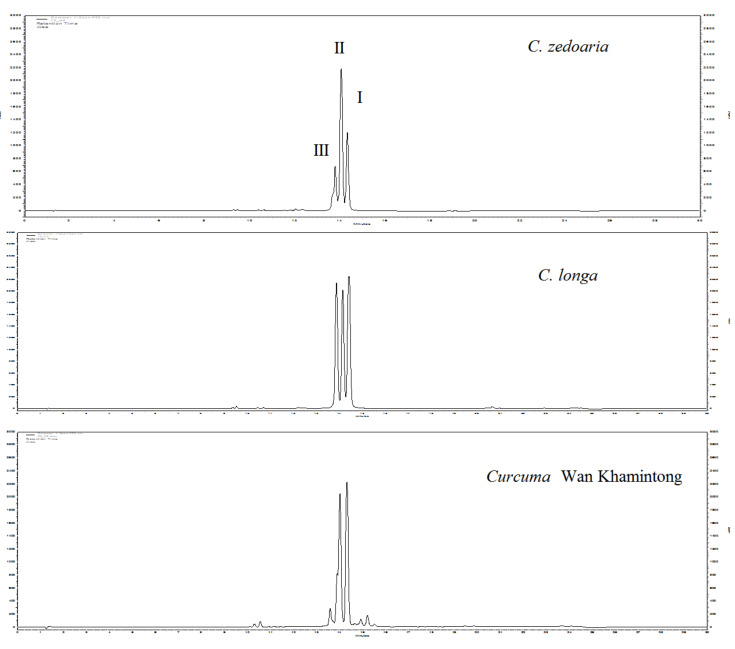
Curcuminoids 1–3 are considered as the most characteristic bioactive secondary metabolites found in turmeric. Therefore, we analyzed the collected Curcuma species to find out whether the rhizomes contain 1–3. The EtOH extracts (1 mg/1 mL) were analyzed with HPLC at 420 nm [17,18], and only ten species were found to contain curcuminoids (Table 3). Furthermore, only three Curcuma species, C. zedoaria, C. longa, and Curcuma Wan Khamintong, contained all three curcuminoids (Figure 3). Trace amounts of curcuminoids were identified from C. manga, Curcuma Wan Rang-Jud, C. comosa, C. latifolia, C. parviflora, Curcuma Wan Pataba, Curcuma Wan Na-Natong, and C. petiolate under LOQ, only when the higher concentrations of the analytes were analyzed by HPLC. It is noticeable that C. longa was found to contain the highest curcuminoid content (653 mg/g). Even though curcumin (1) is the representative bioactive compound of turmeric, only nine Curcuma species contained curcumin (1). Demethoxycurcumin (2) was the major curcuminoid of four other Curcuma species, C. aeruginosa, C. aurantiaca, C. aromatica, and C. zedoaria.
Table 3.
Analysis of curcuminoids from the Curcuma species.
| Curcuma Species | 1 | 2 | 3 |
|---|---|---|---|
| Curcuma Wan Ma-Leung | 5.2 ± 0.1 | 4.1 ± 0.0 | ND 1 |
| Curcuma Wan Ma-Hor | ND | 0.2 ± 0.0 | ND |
| Curcuma aeruginosa | 35.5 ± 0.7 | 107.2 ± 1.0 | ND |
| Curcuma aurantiaca | 3.2 ± 0.0 | 6.5 ± 0.0 | ND |
| Curcuma aromatica | 24.3 ± 0.1 | 112.5 ± 1.0 | ND |
| Curcuma zedoaria | 72.3 ± 0.6 | 201.5 ± 1.5 | 28.2 ± 0.8 |
| Curcuma longa | 304.9 ± 0.1 | 189.2 ± 0.4 | 158.8 ± 0.7 |
| Curcuma Wan Khabitong | 10.2 ± 0.1 | 6.6 ± 0.0 | ND |
| Curcuma Wan Kortong | 0.1 ± 0.0 | 1.8 ± 0.0 | ND |
| Curcuma Wan Khamintong | 47.2 ± 0.1 | 37.1 ± 0.1 | 8.0 ± 0.0 |
1 ND = in trace below detection limit.
Figure 3.
HPLC chromatogram (420 nm) for C. zedoaria, C. longa, and Curcuma Wan Khamintong; I: curcumin (1), II: demethoxycurcumin (2), III: bisdemethoxycurcumin (3).
From Indonesian Curcuma species, C. mangga, C. heyneana, C. aeruginosa, and C. soloensis were reported to contain curcuminoids [19]. Our analysis also confirmed that C. mangga and C. aeruginosa contained curcuminoids. C. aurantiaca contained high contents of 1–3, and this is the first report of the identification of 1–3 in the rhizomes of C. aurantiaca. Diarylheptanoid 1,7-diphenyl-(4E,6E)-4,6-heptadien-3-ol (4) belongs to the curcuminoids, but it was found only from C. comosa in a high content of 300.7 ± 1.4 mg/g. It is noteworthy that C. comosa does not contain 1–3.
There are more than 100 different sesquiterpenoids reported from Curcuma rhizomes [20], and these secondary bioactive metabolites are also responsible for some of the biological activities observed from the rhizomes of Curcuma species. Therefore, we isolated three germacrane-type sesquiterpenoids, germacrone (5), furanodienone (6), and zederone (7), which is the first report from C. latifolia. In addition, ar-turmerone (8) was isolated from C. zedoaria for the first time. Germacrone (5) is known to exhibit anti-inflammatory [21], antiviral [22], and anti-cancer activities [23], and was found in the various Curcuma species, such as C. aeruginosa, C. amanda, C. aromatica, C. xanthrrhiza, and C. zedoaria.
Among the 23 Curcuma species analyzed, 21 species were found to contain at least one of the four sesquiterpenoids, and only two Curcuma species, C. aurantiaca and C. zedoaria, contained all four sesquiterpenoids (Table 4). Generally, germacrone (5) was the most abundant sesquiterpenoid in the Curcuma species, and C. angustifolia contained only 5, out of the eight reference compounds. In particular, C. angustifolia showed a very high concentration of 5 (37% of extract), and Curcuma Wan Khamin-Khao-Padtalod contained only ar-turmerone (8).
Table 4.
Analysis of sesquiterpenoids from the ethanol extraction residues of Curcuma species.
| Curcuma Species | Sesquiterpenoids (mg/g) 1 | |||
|---|---|---|---|---|
| 5 | 6 | 7 | 8 | |
| Curcuma Wan Ma-Leung | 42.6 ± 0.9 | ND 2 | ND | ND |
| Curcuma Wan Ma-Hor | 47.5 ± 0.7 | 4.0 ± 0.0 | 6.6 ± 0.2 | ND |
| Curcuma Wan Khamin-Dam | 126.5 ± 0.3 | 18.5 ± 0.1 | ND | ND |
| Curcuma aeruginosa | ND | 3.1 ± 0.0 | ND | ND |
| Curcuma comosa | 31.1 ± 0.2 | 81.0 ± 0.9 | 37.6 ± 0.2 | ND |
| Curcuma Wan Kanta-Mala | 106.9 ± 0.4 | 15.7 ± 0.1 | ND | ND |
| Curcuma aurantiaca | 36.5 ± 0.4 | 15.4 ± 0.5 | 8.6 ± 0.1 | 4.9 ± 0.0 |
| Curcuma aromatica | 73.7 ± 2.8 | 4.7 ± 0.0 | 33.2 ± 0.6 | ND |
| Curcuma latifolia | 56.2 ± 0.3 | 73.3 ± 1.1 | 61.3 ± 0.1 | ND |
| Curcuma zedoaria | 7.5 ± 0.1 | 15.5 ± 0.4 | 3.3 ± 0.0 | 53.9 ± 0.2 |
| Curcuma longa | ND | ND | ND | 78.4 ± 0.2 |
| Curcuma parviflora | ND | 3.6 ± 0.1 | ND | ND |
| Curcuma angustifolia | 285.3 ± 0.6 | ND | ND | ND |
| Curcuma Wan Khabitong | 6.8 ± 0.1 | ND | ND | 4.4 ± 0.2 |
| Curcuma Wan Patab | 31.2 ± 0.3 | 2.5 ± 0.0 | ND | 0.2 ± 0.0 |
| Curcuma Wan Kortong | ND | ND | ND | 3.5 ± 0.2 |
| Curcuma Wan Na-Natong | 144.3 ± 2.8 | 47.1 ± 0.4 | 13.2 ± 0.1 | ND |
| Curcuma petiolata | ND | ND | ND | 0.4 ± 0.0 |
| Curcuma Wan Khamin-Khao-Padtalod | ND | ND | ND | 17.6 ± 0.1 |
| Curcuma Wan Chai-Dam | ND | 65.1 ± 0.2 | 13.5 ± 0.3 | ND |
| Curcuma Wan Khamintong | ND | ND | ND | 4.6 ± 0.0 |
1 Based on the weight of ethanol extraction residue from dried Curcuma species (wt%). 2 ND = not detected.
4. Conclusions
The phytochemical property of 23 Curcuma species was studied by means of total phenolic contents and antioxidant activity assays, as well as compositional analysis. Besides, eight bioactive curcuminoids and sesquiterpenoids were isolated from the rhizomes of C. longa, C. comosa, C. latifolia, and C. zedroaria. In particular, the isolation of germacrone (5), furanodienone (6), and zederone (7) is the first report from C. latifolia. From the compositional analysis, C. longa was distinct in its highest curcumin (1) concentration. C. angustifolia was found to contain the highest content of germacrone (5). Out of 23 Curcuma species, five Curcuma species did not contain any curcuminoids 1–4. None of the analyzed Curcuma species contained all eight bioactive compounds. Therefore, the data of phytochemical profiling can be used for the identification and authentication of cultivated Curcuma species. In addition, there is a tremendous potential for the utilization of less-studied Curcuma species as functional foods and ingredients.
Acknowledgments
The authors thank the staff at the Bangkok Herbarium, Thailand for the technical support on the taxonomic study of Curcuma species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2304-8158/9/9/1219/s1, Table S1: List of 23 Curcuma species, Figure S1: Spectral data for the isolated compounds, Table S2: Taxonomy of 23 Curcuma species.
Author Contributions
Conceptualization, J.H. and Y.P.; methodology, S.B. and M.K.; validation, M.K., B.E.E., and J.H.; investigation, S.B. and M.K.; resources, S.B. and Y.P.; data curation, S.B.; writing—original draft preparation, J.H.; writing—review and editing, B.E.E. and J.H.; visualization, X.X.; supervision, J.H.; funding acquisition, J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Chung-Ang University Research Grants in 2019 and the framework of the international cooperation program managed by the National Research Foundation of Korea (NRF-2017K2A9A1A01092924).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ravindran P.N., Nirmal B.K., Sivaraman K., editors. Tumeric—The Genus Curcuma. CRC Press; Boca Raton, FL, USA: 2007. p. 504. [Google Scholar]
- 2.Leong-Skornickova J., Sida O., Jarolimova V., Sabu M., Fer T., Travnicek P., Suda J. Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae) Ann. Bot. Lond. 2007;100:505–526. doi: 10.1093/aob/mcm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., Xia N., Zhao J., Chen J., Henny R. Chromosome numbers and ploidy levels of Chinese Curcuma species. Hortic. Sci. 2013;48:525–530. doi: 10.21273/HORTSCI.48.5.525. [DOI] [Google Scholar]
- 4.Burapan S., Kim M., Han J. Curcuminoid demethylation as an alternative metabolism by human Intestinal microbiota. J. Agric. Food Chem. 2017;65:3305–3310. doi: 10.1021/acs.jafc.7b00943. [DOI] [PubMed] [Google Scholar]
- 5.Pinkaew D., Changtam C., Tocharus C., Thummayot S., Suksamrarn A., Tocharus J. Di-O-demethylcurcumin protects SK-N-SH cells against mitochondrial and endoplasmic reticulum-mediated apoptotic cell death induced by Aβ25–35. Neurochem. Int. 2015;80:110–119. doi: 10.1016/j.neuint.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal N.B., Jain S., Nagpal D., Agarwal N.K., Mediratta P.K., Sharma K.K. Liposomal formulation of curcumin attenuates seizures in different experimental models of epilepsy in mice. Fundam. Clin. Pharmacol. 2013;27:169–172. doi: 10.1111/j.1472-8206.2011.01002.x. [DOI] [PubMed] [Google Scholar]
- 7.Habsah M., Amran M., Mackeen M.M., Lajis N.H., Kikuzaki H., Nakatani N., Rahman A.A., Ali A.M. Screening of Zingiberaceae extracts for antimicrobial and antioxidant activities. J. Ethnopharmacol. 2000;72:403–410. doi: 10.1016/S0378-8741(00)00223-3. [DOI] [PubMed] [Google Scholar]
- 8.Singleton V.L., Rossi J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Viticult. 1965;16:144–158. [Google Scholar]
- 9.Uehara S.I., Yasua I., Akiyama K., Morita H., Takeya K., Itokawa H. Diarylheptanoids from the rhizomes of Curcuma xanthorrhiza and Alpinia officinarum. Chem. Pharm. Bull. 1987;35:3298–3304. doi: 10.1248/cpb.35.3298. [DOI] [Google Scholar]
- 10.Shibuya H., Hamamoto Y., Cai Y., Kitagawa I. A reinvestigation of the structure of zederone, a furanogermacrane-type sesquiterpene from zedoary. Chem. Pharm. Bull. 1987;35:924–927. doi: 10.1248/cpb.35.924. [DOI] [Google Scholar]
- 11.Firman K., Kinoshita T., Itai A., Sankawa U. Terpenoids from Curcuma heyneana. Phytochemistry. 1988;27:3887–3891. doi: 10.1016/0031-9422(88)83038-3. [DOI] [Google Scholar]
- 12.Dekebo A., Dagne E., Hansen L.K., Gautun O.R., Aasen A.J. Crystal structures of two furanosesquiterpenes from Commiphora sphaerocarpa. Tetrahed. Lett. 2000;41:9875–9878. doi: 10.1016/S0040-4039(00)01752-4. [DOI] [Google Scholar]
- 13.Barrero A.F., Herrador M.M., Lopez-Perez J., Arteaga J.F., Catalan J. New pathways in transannular cyclization of germacrone [germacra-1(10),4,7(11)-trien-8-one]: Evidence regarding a concerted mechanism. Org. Lett. 2009;11:4782–4785. doi: 10.1021/ol901066x. [DOI] [PubMed] [Google Scholar]
- 14.Sukari M.A., Wah T.S., Saad S.M., Rashid N.Y., Rahmani M., Lajis N.H., Hin T.Y.Y. Bioactive sesquiterpenes from Curcuma ochrorhiza and Curcuma heyneana. Nat. Prod. Res. 2010;24:838–845. doi: 10.1080/14786410903052951. [DOI] [PubMed] [Google Scholar]
- 15.Kader M.G., Habib M.R., Nikkon F., Yeasmin T., Rashid M.A., Rahman M.M., Gibbons S. Zederone from the rhizomes of Zingiber zerumbet and its anti-Staphylococcal activity. Lat. Am. Caribb. Bull. Med. Arom. Plants. 2010;9:63–68. [Google Scholar]
- 16.Lee J.H., Choung M.G. Determination of curcuminoid colouring principles in commercial foods by HPLC. Food Chem. 2011;124:1217–1222. doi: 10.1016/j.foodchem.2010.07.049. [DOI] [Google Scholar]
- 17.Paramasivam M., Poi R., Banerjee H., Bandyopadhyay A. High-performancethin layer chromatographic method for quantitative determination of curcuminoids in Curcuma longa germplasm. Food Chem. 2009;113:640–644. doi: 10.1016/j.foodchem.2008.07.051. [DOI] [Google Scholar]
- 18.Péret-Almeida L., Cherubino A.P.F., Alves R.J., Dufossé L., Glória M.B.A. Separation and determination of the physico-chemical characteristics of curcumin, demethoxycurcumin and bisdemethoxycurcumin. Food Res. Int. 2005;38:1039–1044. doi: 10.1016/j.foodres.2005.02.021. [DOI] [Google Scholar]
- 19.Bos R., Windono T., Woerdenbag H.J., Boersma Y.L., Koulman A., Kayser O. HPLC-photodiode array detection analysis of curcuminoids in Curcuma species indigenous to Indonesia. Phytochem. Anal. 2007;18:118–122. doi: 10.1002/pca.959. [DOI] [PubMed] [Google Scholar]
- 20.Balaji S., Chempakam B. Toxicity prediction of compounds from turmeric (Curcuma longa L) Food Chem. Toxicol. 2010;48:2951–2959. doi: 10.1016/j.fct.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 21.Makabe H., Maru N., Kuwabara A., Kamo T., Hirota M. Anti-inflammatory sesquiterpenes from Curcuma zedoaria. Nat. Prod. Res. 2006;20:680–685. doi: 10.1080/14786410500462900. [DOI] [PubMed] [Google Scholar]
- 22.Liao Q., Qian Z., Liu R., An L., Chen X. Germacrone inhibits early stages of influenza virus infection. Antivir. Res. 2013;100:578–588. doi: 10.1016/j.antiviral.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Lu J.J., Dang Y.Y., Huang M., Xu W.S., Chen X.P., Wang Y.T. Anti-cancer properties of terpenoids isolated from Rhizoma curcumae—A review. J. Ethnopharmacol. 2012;143:406–411. doi: 10.1016/j.jep.2012.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.