NuMA disruption causes nuclear defects, and it was unclear if this is due to NuMA's essential spindle function. Serra-Marques et al. show that NuMA keeps chromosomes together at nuclear formation and promotes a mechanically robust nucleus independently of its spindle function.
Abstract
Eukaryotic cells typically form a single, round nucleus after mitosis, and failures to do so can compromise genomic integrity. How mammalian cells form such a nucleus remains incompletely understood. NuMA is a spindle protein whose disruption results in nuclear fragmentation. What role NuMA plays in nuclear integrity, and whether its perceived role stems from its spindle function, are unclear. Here, we use live imaging to demonstrate that NuMA plays a spindle-independent role in forming a single, round nucleus. NuMA keeps the decondensing chromosome mass compact at mitotic exit and promotes a mechanically robust nucleus. NuMA’s C terminus binds DNA in vitro and chromosomes in interphase, while its coiled-coil acts as a central regulatory and structural element: it prevents NuMA from binding chromosomes at mitosis, regulates its nuclear mobility, and is essential for nuclear formation. Thus, NuMA plays a structural role over the cell cycle, building and maintaining the spindle and nucleus, two of the cell’s largest structures.
Introduction
Most eukaryotic cells enclose their genome within a single, round nucleus. When chromosomes are not properly incorporated into the nucleus, they can form micronuclei, which can be vulnerable to DNA damage and chromothripsis, leading to genomic instability and disease (Andreassi et al., 2011; Crasta et al., 2012; Hatch et al., 2013; Ly et al., 2019; Zhang et al., 2015). Furthermore, while different cell types have nuclei of different shapes, within a given cell type, nuclear shape is regulated and stereotypical, and nuclear shape defects can correlate with altered gene expression, disease, and aging (Le Berre et al., 2012; Dahl et al., 2008; Skinner and Johnson, 2017; Thomas et al., 2002). While some of the mechanisms required for nuclear formation are known, how mammalian cells pack many chromosomes into a single nucleus with a simple rounded shape is not well understood. More broadly, understanding how nanometer-scale molecules give rise to higher-order micrometer-scale structures of a given number and shape remains a frontier.
Several mechanisms are known to promote the formation of a single nucleus and to give rise to its shape. For example, bringing anaphase chromosomes into a tight cluster at mitosis promotes the formation of a single nucleus. Indeed, erroneous chromosome-to-spindle attachments (Cimini et al., 2004; Liu et al., 2018; Liu and Pellman, 2020; Thompson and Compton, 2011), misregulated centrosome amplification (Ganem et al., 2009), and defective metaphase chromosome alignment and anaphase compaction (Fonseca et al., 2019) can all lead to micronucleation. At late mitosis, the protein Barrier-to-autointegration factor (BAF) builds a dense chromatin layer around the anaphase chromosome mass, limiting membrane penetration to prevent the formation of multiple nuclei (Samwer et al., 2017). Similarly, diverse factors are thought to influence nuclear shape. These range from the shape of the anaphase chromosome mass (Fonseca et al., 2019; Mora-Bermúdez et al., 2007) and chromatin organization and compaction (Furusawa et al., 2015; Stephens et al., 2017) to the mechanics of the nuclear lamina, cytoskeleton, and extracellular environment (Buxboim et al., 2010; Chu et al., 2017; Dahl et al., 2008; Lammerding et al., 2005).
Despite what we know about nuclear formation, understanding when, at mitosis versus at nuclear formation, and how directly certain molecules play a role in this process remains challenging. The protein nuclear mitotic apparatus (NuMA; Lydersen and Pettijohn, 1980), for example, is essential to spindle assembly (Compton and Luo, 1995; Kallajoki et al., 1991; Yang and Snyder, 1992) and chromosome segregation (Silk et al., 2009; Haren et al., 2009), as well as to the formation of a single nucleus (Compton and Cleveland, 1993; Kallajoki et al., 1991, 1993). NuMA contains two globular domains, its N and C termini, connected through seven coiled-coil domains (Compton et al., 1992; Yang et al., 1992). At mitosis, NuMA localizes to the spindle and cell cortex, where it drives spindle structural stability and orientation, respectively (Compton et al., 1992; Kallajoki et al., 1991; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2013). At interphase, NuMA localizes to the nucleus. It has been proposed to have a postmitotic role in nuclear formation (Compton and Cleveland, 1993; Kallajoki et al., 1993; Van Ness and Pettijohn, 1983), which could be a direct or indirect role (Cleveland, 1995; Kallajoki et al., 1991, 1993; Merdes and Cleveland, 1998), and to promote DNA repair and chromatin organization in the nucleus (Abad et al., 2007; Kivinen et al., 2010; Vidi et al., 2014). Furthermore, NuMA has been reported to self-assemble into a filamentous network in the nucleus (Gueth-Hallonet et al., 1998; Zeng et al., 1994), and to directly bind DNA in vitro (Ludérus et al., 1994). However, NuMA’s essential function at mitosis has made it difficult to determine whether it also acts at nuclear formation independently of its spindle function. If it does, we do not know what this nuclear formation function is, or how NuMA’s structure supports it and regulates it over the cell cycle.
Here, we show that NuMA is essential for the formation of a single, round nucleus in human cells independent of its spindle role. We find that in the absence of NuMA, nuclear defects appear at the moment of nuclear envelope reassembly, even without prior spindle defects. We also show that in cells lacking NuMA, the chromosome mass expands faster at mitotic exit, and that nuclei are mechanically compromised. We demonstrate that NuMA’s coiled-coil is essential to forming a single nucleus and to promoting the formation of stable nuclear NuMA assemblies. In addition, we find that NuMA’s coiled-coil prevents the premature binding of NuMA’s C terminus, which we show directly binds DNA in vitro and to chromosomes at mitosis. Thus, NuMA’s coiled-coil serves as a central element that controls both its localization and its function over the cell cycle. Together, these findings establish a spindle-independent role for NuMA in nuclear formation and mechanics, and suggest that NuMA’s long coiled-coil helps it organize chromosomes at interphase. More broadly, they highlight NuMA’s key structural role in building and maintaining two of the largest cellular structures throughout the cell cycle, the spindle and nucleus.
Results
NuMA plays a spindle-independent role in the formation of a single, round nucleus
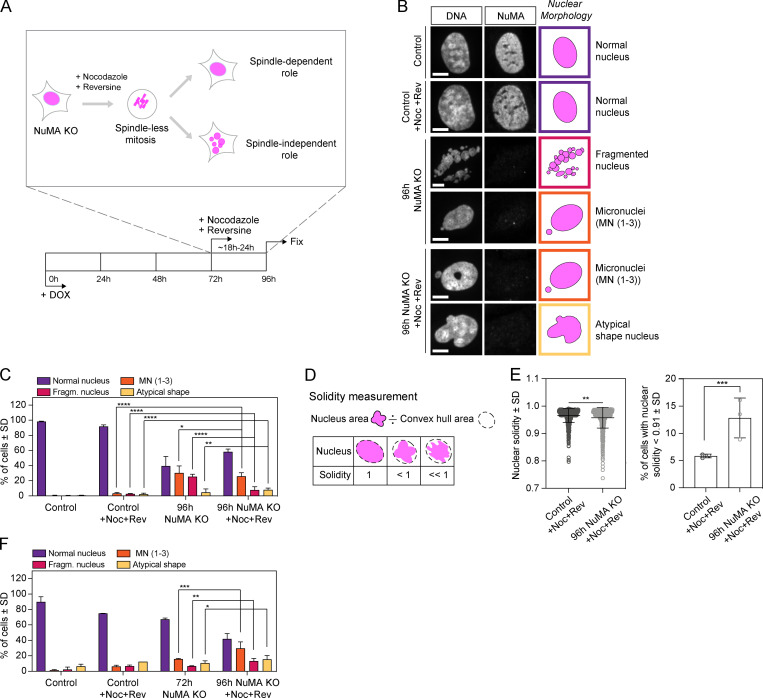
To investigate whether NuMA contributes to the formation of the mammalian nucleus independently of its spindle function, we used inducible CRISPR/Cas9 human retinal pigment epithelial (RPE1) cells to knock out (KO) NuMA (Hueschen et al., 2017), and looked at nuclear formation after a spindle-less mitosis. To do so, we treated cells with nocodazole to depolymerize spindle microtubules, and with reversine to bypass the spindle assembly checkpoint. With this approach, mitotic cells do not assemble a spindle, exit mitosis and form a single nucleus of unsegregated chromosomes (Samwer et al., 2017). We induced NuMA KO cells at time (t) = 0 with doxycycline (DOX) to deplete endogenous NuMA (Fig. S1 A), treated them with nocodazole and reversine after 72 h, and fixed and imaged them 18–24 h after drug addition (termed “96 h” onward for simplicity; Fig. 1 A). As expected, control (uninduced) drug-treated cells did not assemble a spindle (Fig. S1 B), and yet still formed a single, round (termed “normal”) nucleus after mitosis (Fig. 1, B and C; Samwer et al., 2017), as did Cas9-induced cells without a single guide RNA (sgRNA) for NuMA (Fig. S1 C).
Figure S1.
Validation of the RPE1 inducible Cas9 NuMA KO cell line and of the nocodazole/reversine treatment. (A) Western blot (top) and immunofluorescence measurement (bottom) of endogenous NuMA levels in RPE1 cells after DOX-induced NuMA depletion for the indicated times. n = 147 (0 h), 190 (24 h), 141 (48 h), 107 (72 h), and 64 (96 h) cells. (B) Representative immunofluorescence images showing spindle morphology (tubulin), NuMA, and chromosomes (Hoechst) in the indicated conditions. Cells were untreated or treated with nocodazole (664 nM) and reversine (320 nM) for 18-24 h. Scale bar, 5 µm. Control cells assemble a spindle, NuMA KO cells assemble a perturbed spindle, and nocodazole- and reversine-treated cells do not assemble a spindle. (C) Nuclear morphology in immunofluorescence images of DOX-induced Cas9 RPE1 cells not expressing sgRNAs. n = 209 (no DOX control) and 328 (DOX) cells. (D) Representative immunofluorescence images of nuclear shapes in control and NuMA KO cells after DOX-induced NuMA depletion for the indicated times. The green line outlines the nucleus. Scale bar, 10 µm. (E) Nuclear solidity of interphase cells with respect to endogenous NuMA levels after DOX-induced NuMA depletion for 0–96 h. Whiskers represent minimum to maximum. n = 61 (1.2–1.8), 183 (0.8–1.2), 191 (0.4–0.8), and 214 (0.0–0.4) cells. Two-sided Mann–Whitney test: ***, P = 0.0009; *, P = 0.01; ns, nonsignificant. MN, micronuclei; Noc, nocodazole; and Rev, reversine.
Figure 1.
NuMA plays a spindle-independent role in the formation of a single, round nucleus. (A) Experimental design for inducible CRISPR/Cas9 NuMA KO RPE1 cells to undergo and exit a spindle-less mitosis. Endogenous NuMA is depleted by DOX addition starting at 0 h, and cells treated with nocodazole (664 nM) and reversine (320 nM) for a spindle-less mitosis starting at 72 h, and subsequently fixed and stained for immunofluorescence at 96 h. (B) Representative immunofluorescence images of nuclear morphologies observed in uninduced NuMA KO RPE1 (control) cells, control cells treated with nocodazole and reversine (for 24 h), 96 h NuMA KO cells, and 96 h NuMA KO cells treated with nocodazole and reversine (for last 24 h). Cells were stained for tubulin (not shown), DNA (Hoechst), and NuMA. Scale bar, 10 µm. (C) Percentage of cells with different nuclear morphologies in the indicated conditions, from DNA images in B. Plot shows mean ± SD n = 1,385 (control), 1,123 (control +Noc+Rev), 638 (96 h NuMA KO), and 649 (96 h NuMA KO +Noc+Rev) cells, from three independent experiments. Two-sided Fisher’s exact test: ****, P < 0.0001; **, P = 0.003; *, P = 0.01. The same nuclear morphologies are compared within different conditions. (D) The solidity of nuclei corresponds to the ratio of the nucleus’ area to its convex hull area. (E) Nuclear solidity in nocodazole- and reversine-treated control and 96 h NuMA KO cells, and corresponding percentage of cells with nuclear solidity 2 SDs below the control mean. Plot shows mean ± SD n = 750 (control) and 656 (96 h NuMA KO) cells, from three independent experiments. Left plot, two-sided Mann–Whitney test: **, P = 0.001. Right plot, two-sided Fisher’s exact test: ***, P = 0.0001. (F) Percentage of cells with different nuclear morphologies in the indicated conditions, from DNA images. Plot shows mean ± SD n = 285 (control), 456 (control +Noc+Rev), 321 (72 h NuMA KO), and 347 (96h NuMA KO +Noc+Rev) cells, from two independent experiments. Two-sided Fisher’s exact test: ***, P = 0.0001; **, P = 0.009; *, P = 0.03. The same nuclear morphologies are compared within different conditions. Fragm., fragmented; MN, micronuclei; Noc, nocodazole; and Rev, reversine.
In turn, NuMA KO cells with a defective spindle (untreated with drugs; Fig. S1 B) presented nuclei with several fragments (Fig. 1, B and C), as previously observed (Compton and Cleveland, 1993; Kallajoki et al., 1991, 1993). Additionally, these cells also presented micronuclei (one to three small nuclei in addition to the primary nucleus) and atypically shaped nuclei (Fig. 1, B and C). In spindle-less NuMA-KO cells (i.e., treated with nocodazole and reversine drugs), the fraction of cells with fragmented nuclei was significantly reduced, while the fraction of cells with micronuclei and atypically shaped nuclei remained similar (Fig. 1, B and C). This suggests that fragmented nuclei result at least partly from spindle-related failures, and that micronucleation and atypically shaped nuclei result from spindle-independent failures. To quantitatively compare nuclear roundness, we measured the solidity of primary nuclei. Solidity significantly decreased in spindle-less NuMA KO cells compared with control cells (Fig. 1, D and E), and decreased as NuMA depletion extent increased (Fig. S1, D and E). Finally, because NuMA depletion occurred gradually over 4 d (Fig. S1 A), we sought to determine whether nuclear defects appeared when spindles were present (before drug addition) or absent (after drug addition). To do so, we repeated the same assay (Fig. 1 A) and compared cells fixed at 72 h, just before drug addition, and at 96 h, after drug treatment. The fraction of NuMA KO cells with fragmented nuclei, micronuclei, and atypically shaped nuclei all increased significantly in the 18–24 h spindle-less period (Fig. 1 F). Together, these findings indicate that NuMA plays a role in the formation of a single and round nucleus, independent of its spindle function.
NuMA acts at the time of nuclear formation, compacting the chromosome mass at mitotic exit
To gain insight into NuMA’s nuclear function, we sought to determine whether it is required before, after, or at the time of nuclear formation. To do so, we performed live cell imaging of inducible NuMA KO RPE1 cells treated with nocodazole and reversine and stably expressing the histone marker mCherry-H2B and the nuclear envelope marker EGFP-Lap2β. As for fixed cells (Fig. 1), live imaging revealed a significant increase in the fraction of spindle-less NuMA KO cells with fragmented nuclei, micronuclei, and atypically shaped nuclei at 96 h compared with control cells (Fig. 2, A and B). To define when NuMA is needed, we followed the trajectories of individual cells that entered mitosis with a single, round nucleus (“mother cells”) until they exited spindle-less mitosis, formed a nucleus, and beyond (“daughter cells”; Fig. 2 C). From mother cells with normal nuclei, NuMA KO daughter cells exited mitosis with more fragmented nuclei, micronuclei, and atypically shaped nuclei than control cells (Fig. 2, D and E; and Videos 1 and 2). In these cell trajectories, we observed morphology defects appear as chromosomes decondensed and the nuclear envelope formed. We did not observe defects appear before or soon after nuclear formation. To test whether NuMA could help keep the decondensing chromosome mass together (either directly or indirectly) as the nuclear envelope formed, we measured the expansion of this mass with and without NuMA. We found that the chromosome mass expanded faster, and further, at nuclear formation in NUMA KO cells than in control cells (Fig. 2 F). Together, these findings indicate that NuMA’s spindle-independent role in building the nucleus takes place at the time of nuclear envelope formation, rather than before or after, and that NuMA keeps the chromosome mass compact during this time.
Figure 2.
NuMA acts at the time of nuclear formation, compacting the chromosome mass at mitotic exit. (A) Representative live images of observed nuclear phenotypes after 16-18 h of nocodazole and reversine treatment (spindle-less mitosis) in 96 h NuMA KO RPE1 cells stably expressing mCherry-H2B and EGFP-Lap2β. Arrow indicates a micronucleus. Scale bar, 5 µm. (B) Percentage of cells with different nuclear morphologies before and 16-18 h after nocodazole and reversine treatment (spindle-less mitosis). All cells, and not only those that went through mitosis, were analyzed. Plot shows mean ± SD n = 476 (control) and 820 (NuMA KO) cells from five and two independent experiments, respectively. Two-sided Fisher’s exact tests: ****, P < 0.0001; *, P < 0.04. (C) Experimental design as in Fig. 1 A, except that individual cell trajectories were followed by live imaging for the 16-18 h period in nocodazole (664 nM) and reversine (control 1 µM; NuMA KO 320 nM). Mother cell, cell before mitotic entry; daughter cell, cell after mitotic exit. (D) Representative time-lapse images of control and NuMA KO RPE1 cells stably expressing mCherry-H2B (magenta) and EGFP-Lap2β (green) that were followed live through spindle-less mitosis for 16-18 h, noting nuclear envelope breakdown (NEB) and nuclear envelope reformation (NER). White arrows indicate nuclear defects. Time in hours:minutes. Scale bar, 10 µm. See also Video 1 (control) and Video 2 (NuMA KO). (E) Percentage of cells with the indicated nuclear phenotypes in uninduced control and NuMA-KO cells treated with nocodazole and reversine just before mitotic entry (mother cells) into a spindle-less mitosis and just after mitotic exit (daughter cells). Mother cells include only cells that entered mitosis with a single, round nucleus. Plot shows mean ± SD n = 508 (control) and 308 (NuMA KO) cells from five and two independent experiments, respectively. Two-sided unpaired t test: ***, P < 0.001; *, P = 0.01. (F) Expansion of the chromosome mass from the time of nuclear envelope reformation (t = 0) in control and NuMA KO cells treated with nocodazole and reversine for a spindle-less mitosis. Plot shows mean ± SEM n = 5 cells in each condition. Two-sided Mann–Whitney test: **, P = 0.008 at 132 min. Fragm., fragmented; MN, micronuclei; Noc, nocodazole; and Rev, reversine.
Video 1.
Control cells exit mitosis with a single and round nucleus. Time-lapse spinning disk confocal imaging of a representative control RPE1 cell stably expressing mCherry-H2B (left, magenta) and EGFP-Lap2β (center, green) and entering and exiting a spindle-less mitosis (nocodazole/reversine). Cells enter and exit mitosis with a single and round nucleus. Time in minutes:seconds, with the frame corresponding to nuclear envelope breakdown set to 00:00. The start of NER becomes detectable at 00:12. Scale bar, 10 µm. Video corresponds to the still images in Fig. 2 D.
Video 2.
Nuclear defects arise at nuclear envelope formation as NuMA KO cells exit mitosis. Time-lapse spinning disk confocal imaging of a representative NuMA KO RPE1 cell stably expressing mCherry-H2B (left, magenta) and EGFP-Lap2β (center, green) and entering and exiting a spindle-less mitosis (nocodazole/reversine). Cells enter mitosis with a single and round nucleus, and exit with nuclear defects, which appear during nuclear envelope reformation. Time in minutes:seconds, with the frame corresponding to nuclear envelope breakdown set to 00:00. The start of nuclear envelope reformation becomes detectable at 0:24. Scale bar, 10 µm. Video corresponds to the still images in Fig. 2 D.
NuMA’s coiled-coil is required for the formation of a single nucleus and modulates its mobility in the nucleus
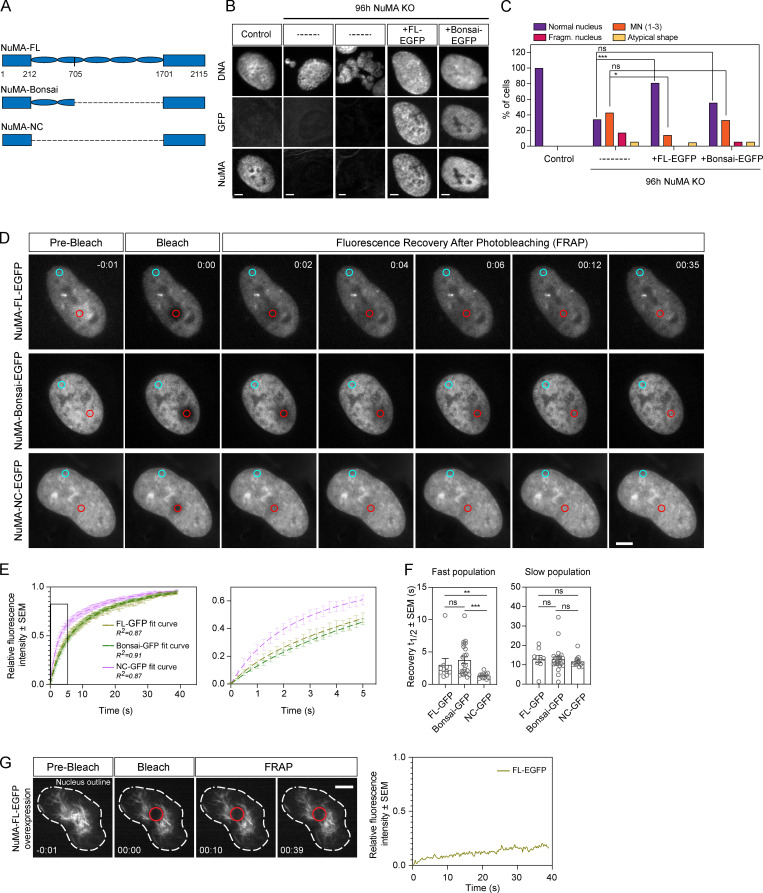
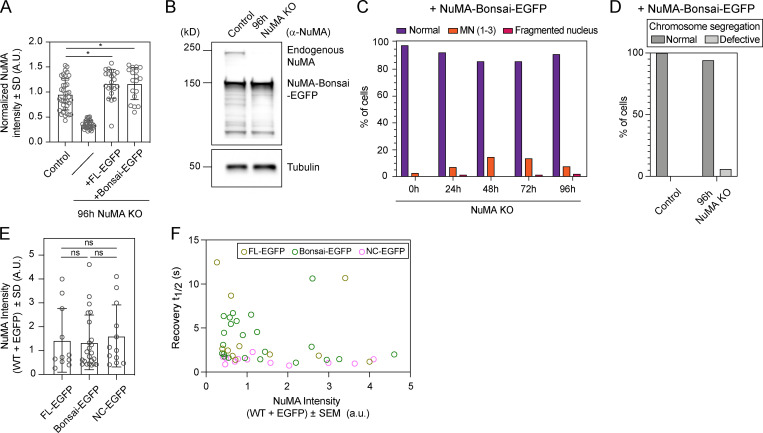
Electron microscopy and overexpression of NuMA coiled-coil truncations suggest that the coiled-coil helps form nuclear NuMA filaments, which could in principle contribute to nuclear stability (Gueth-Hallonet et al., 1998; Harborth et al., 1999; Zeng et al., 1994). To determine if NuMA’s coiled-coil is required for nuclear formation, we tested the ability of a truncation of NuMA that lacks most of the coiled-coil (NuMA-Bonsai; Fig. 3 A) to rescue the KO of endogenous NuMA, when expressed at near endogenous levels (Fig. 3, B and C; and Fig. S2 A). As a control, overexpression of similar levels of NuMA–full-length (FL)–EGFP (Fig. S2 A) in NuMA KO cells mostly rescued the nuclear defects observed in the absence of endogenous NuMA, even though some cells still presented micronuclei (Fig. 3, B and C). In turn, NuMA-Bonsai-EGFP, which contains only a short portion of the coiled-coil, failed to significantly rescue micronucleation (Fig. 3, B and C). This is consistent with NuMA-Bonsai being sufficient for NuMA’s role in the spindle body (Forth et al., 2014; Hueschen et al., 2017) but not at nuclear formation, with nuclear fragmentation coming largely from spindle defects, and micronucleation coming from spindle-independent defects, being revealed without spindle defects (Fig. 1 C). Consistently, inducible NuMA KO cells stably expressing NuMA-Bonsai-EGFP (at higher levels than endogenous NuMA; Fig. S2 B) had micronuclei (Fig. S2 C) but no detectable chromosome segregation errors (Fig. S2 D). Importantly, this indicates that NuMA’s nuclear formation role is essential not only after a drug-induced spindle-less mitosis but also after normal mitosis. Together, these findings indicate that either all or part of NuMA’s coiled-coil is necessary for formation of a single nucleus.
Figure 3.
NuMA’s coiled-coil is required for the formation of a single nucleus and modulates its mobility in the nucleus. (A) Schematic representations of FL NuMA and truncations NuMA-Bonsai and NuMA-NC, with amino acid numbers indicated. (B) Representative immunofluorescence images of nuclear morphologies observed in uninduced NuMA KO RPE1 cells (control), NuMA KO cells without exogenous constructs (–, two examples), and NuMA KO cells transiently expressing the rescue constructs NuMA-FL-EGFP or NuMA-Bonsai-EGFP. Cells were stained for tubulin (not shown), DNA (Hoechst), GFP, and NuMA. Scale bar, 5 µm. (C) Percentage of cells with different nuclear morphologies observed in experiment from B. n = 38 (control), 35 (NuMA KO), 21 (NuMA KO + NuMA-FL-EGFP), and 18 (NuMA KO + NuMA-Bonsai-EGFP) cells. These rescues were repeated one more time, with similar results (not shown). Two-sided Fisher’s exact test: ***, P < 0.0006; *, P = 0.04; ns, nonsignificant. (D and E) FRAP of NuMA-FL-EGFP, NuMA-Bonsai-EGFP, and NuMA-NC-EGFP in the nucleus of uninduced NuMA KO RPE1 cells. GFP intensity was measured in the bleached area (red circle) and in a nonbleached area (blue circle) to account for photobleaching. Scale bar, 10 µm. Time in minutes:seconds; 0:00 indicates the time of bleaching. n = 11 (NuMA-FL-EGFP), 23 (NuMA-Bonsai-EGFP), and 12 (NuMA-NC-EGFP) cells. (F) Distribution of the fast and slow recovery halftimes (t1/2) of the different GFP-tagged NuMA proteins during FRAP. Plot shows mean ± SEM n = 11 (NuMA-FL-EGFP), 23 (NuMA-Bonsai-EGFP), and 12 (NuMA-NC-EGFP) cells. Two-sided Mann–Whitney test: ***, P < 0.001; **, P = 0.001; ns, nonsignificant. (G) Time-lapse images showing the nucleus (dashed white line) of an uninduced NuMA KO RPE1 cell highly overexpressing NuMA-FL-EGFP and forming cable-like structures (left) and FRAP of the same cell (right). NuMA was bleached in the indicated red circle at 0:00, and only minimal GFP intensity recovered by 39 s. Time in minutes:seconds. Scale bar, 5 µm. See also Video 3. Fragm., fragmented; and MN, micronuclei.
Figure S2.
Validation of the RPE1 NuMA KO line stably expressing NuMA-Bonsai-EGFP and estimate of NuMA-EGFP intensity in cells used in the FRAP analysis. (A) Normalized NuMA intensity in uninduced NuMA KO RPE1 cells (control), NuMA KO cells, and NuMA KO cells overexpressing the rescue constructs NuMA-FL-EGFP and NuMA-Bonsai-EGFP. An antibody against the N terminus was used in order to detect both endogenous and overexpressed protein. Plot shows mean ± SD n = 38 (control), 35 (NuMA KO), 21 (NuMA KO + NuMA-FL-EGFP), and 18 (NuMA KO + NuMA-Bonsai-EGFP) cells. Two-sided unpaired t test: *, P < 0.02. (B) Western blot of endogenous NuMA and NuMA-Bonsai-EGFP in uninduced NuMA KO RPE1 cells (control) and 96 h NuMA KO cells stably overexpressing NuMA-Bonsai-EGFP. (C) Percentage of cells with different nuclear phenotypes at 0 h, 24 h, 48 h, 72 h, and 96 h of endogenous NuMA KO in cells stably overexpressing NuMA-Bonsai-EGFP and fixed for immunofluorescence as in Fig. 1 B. n = 205 (0 h), 177 (24 h), 181 (48 h), 181 (72 h), and 177 (96 h) cells. (D) Chromosome segregation defects in live control and NuMA KO cells stably overexpressing Bonsai-NuMA-EGFP. Normal segregation indicates anaphase progression without lagging chromosomes or anaphase bridges. n = 8 (control) and 17 (NuMA KO) cells. 1 out of 17 NuMA KO cells exited mitosis with a defective nucleus, and without completing anaphase, and was scored as segregation defective. (E) NuMA-FL-EGFP, NuMA-Bonsai-EGFP, and NuMA-NC-EGFP fluorescence intensity in the cells analyzed in the FRAP experiment presented in Fig. 3, D and E. Plot shows mean ± SD n = 11 (NuMA-FL-EGFP), 23 (NuMA-Bonsai-EGFP), and 12 (NuMA-NC-EGFP). Two-sided Mann–Whitney test: ns, nonsignificant. (F) Fast recovery halftime as a function of NuMA-FL-EGFP, NuMA-Bonsai-EGFP, and NuMA-NC-EGFP intensity in the cells from Fig. 3, D and E. n = 11 (NuMA-FL-EGFP), 23 (NuMA-Bonsai-EGFP), and 12 (NuMA-NC-EGFP). MN, micronuclei; and WT, wild-type.
Given the role of NuMA’s coiled-coil in nuclear formation (Fig. 3, B and C; and Fig. S2 C), and that NuMA can assemble into higher-order filament-like structures (Zeng et al., 1994) whose size depends on the coiled-coil’s length (Gueth-Hallonet et al., 1998; Harborth et al., 1999), we hypothesized that the coiled-coil contributes to NuMA’s self-assembly in the nucleus. To test this, we measured the turnover of NuMA-FL-GFP, NuMA-Bonsai-GFP, and NuMA-NC-GFP, a construct that has only N and C termini and lacks the entire coiled-coil, in the nucleus using FRAP (Fig. 3, D–F). NuMA-FL-GFP and NuMA-Bonsai-GFP displayed similar recovery kinetics, while NuMA-NC-GFP recovered faster. Using a double exponential model, the fast half-life was shorter for NuMA-NC-GFP than NuMA-FL-GFP and NuMA-Bonsai-GFP, while the slow half-life was indistinguishable for all three constructs, independent of GFP-tagged protein expression levels within the analyzed range (Fig. S2, E and F). Furthermore, we observed that cells expressing the highest NuMA-FL-EGFP levels, excluded from the above analysis, can form stable cable-like nuclear structures that show much slower recovery after photobleaching (Fig. 3 G and Video 3). This supports the notion that NuMA can self-assemble in the nucleus when highly overexpressed, though it may or may not do so when endogenously expressed. In turn, we never observed such cable-like structures in NuMA-NC-EGFP cells, even at high expression levels. Together, our findings indicate that NuMA’s coiled-coil is necessary for nuclear formation, and suggest that it modulates NuMA’s mobility in the nucleus, by modulating its interaction either with itself or with other players.
Video 3.
Overexpressed NuMA-FL-EGFP can form stable cable-like structures in the nucleus. Time-lapse imaging of a representative RPE1 cell transiently overexpressing NuMA-FL-EGFP imaged before and during FRAP. The red circle highlights the photobleached area. Fluorescence recovery in the bleached area is minimal, consistent with NuMA-FL being able to form higher-order structures in the nucleus. Time in minutes:seconds, with the frame corresponding to bleaching set to 00:00. Scale bar, 5 µm. Video corresponds to the still images in Fig. 3 G.
NuMA’s coiled-coil regulates chromatin binding by its C terminus over the cell cycle
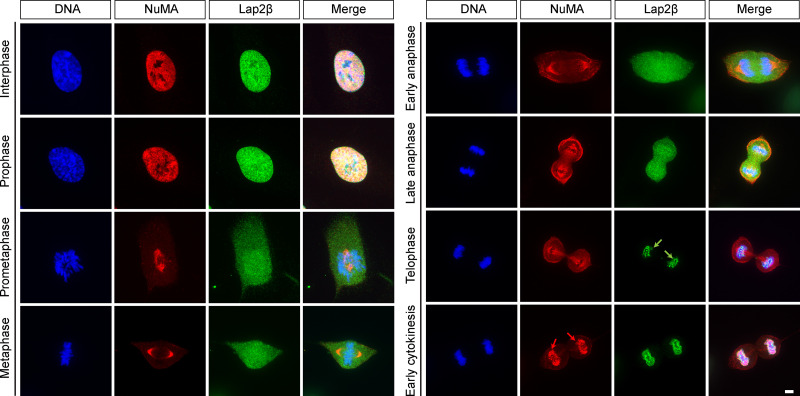
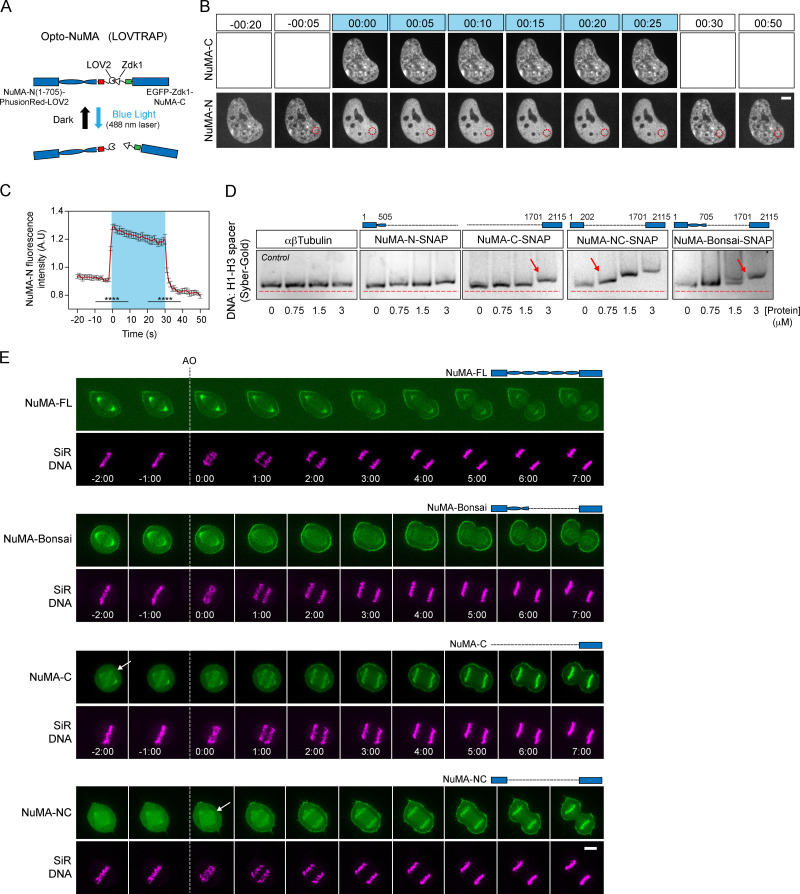
NuMA is only detected on chromosomes after initiation of nuclear envelope reassembly (Fig. S3), when it is imported into the nucleus (Compton et al., 1992). What mechanism ensures that NuMA only associates with chromosomes in interphase, when chromosomes are kept together, and not in mitosis, when chromosomes must move apart? To begin answering this question, we asked how NuMA binds chromosomes in interphase, testing whether NuMA’s globular N- and C-terminal domains associate with chromosomes once in the nucleus. NuMA’s N and C termini were suggested to bind DNA (Radulescu and Cleveland, 2010), though no specific and direct comparisons were made. Using a light-controlled dissociation system, we sought to disconnect NuMA’s N and C termini from each other once they were both imported together into the nucleus. We engineered an optocontrollable NuMA-Bonsai protein (Opto-NuMA) using the LOV2 trap and release of protein (LOVTRAP) system (van Haren et al., 2018; Wang et al., 2016; Fig. 4 A). In the dark, when LOV2 and Zdk1 bind, NuMA-N(1–705)-PhusionRed-LOV2 and EGFP-Zdk1-NuMA-C localized to the nucleus, as expected since NuMA’s C terminus contains a NLS (Tang et al., 1994). Upon blue light illumination, the nuclear localization of NuMA-C (imaged with the same wavelength that activates LOV2 and induces dissociation) remained unchanged. Furthermore, NuMA-C’s nuclear localization pattern resembles that of NuMA-N before and after blue light illumination, but not during illumination (Fig. S4 A), suggesting that NuMA-C drives NuMA-N localization. Meanwhile, NuMA-N became diffuse (Fig. 4, B and C; and Video 4), likely remaining in the nucleus due to its high molecular weight (123 kD). Thus, whether chromatin binding is direct or indirect, NuMA's C terminus has a higher affinity for chromatin than its N terminus.
Figure S3.
NuMA is only detectable on chromosomes after initial Lap2β recruitment. Representative immunofluorescence images of uninduced RPE1 NuMA KO cells showing localization of the nuclear envelope protein Lap2β (green), NuMA (red), and DNA (Hoechst, blue) over the cell cycle. Lap2β is detectable around the chromosome mass (green arrows) before NuMA appears on chromosomes (red arrows). Scale bar, 5 µm.
Figure 4.
NuMA’s coiled-coil regulates its C terminus’ chromatin binding over the cell cycle. (A) LOVTRAP light-induced dissociation system: in the dark, LOV2 (phototropin) binds to Zdk1, while blue light induces a conformational change in LOV2 that prevents binding to Zdk1. In the Opto-NuMA engineered protein NuMA-N(1–705)-PhusionRed-LOV2 + EGFP-Zdk1-NuMA-C, both NuMA ends are linked through LOVTRAP, and blue light dissociates them from each other. (B) Representative time-lapse of an uninduced NuMA KO RPE1 cell stably expressing Opto-NuMA in the nucleus before, during (0:00-0:25), and after illumination with blue (488 nm) light, showing NuMA-N but not NuMA-C becoming diffuse upon dissociation. EGFP-Zdk1-NuMA-C cannot be imagined without dissociating Opto-NuMA. Red circle corresponds to the area chosen for intensity analysis in C. Time in minutes:seconds. Scale bar, 5 µm. See also Video 4. (C) NuMA-N(1–705)-PhusionRed-LOV2 intensity in the nucleus before, during (blue box), and after illumination with blue light. Red trace = mean; black = SEM. n = 9 cells. Two-sided Mann–Whitney test: ****, P < 0.0001. (D) Top: The schematics represent the NuMA constructs used in each gel, with amino acid numbers indicated. Bottom: EMSA using 60 ng H1-H3 spacer DNA fragment and increasing concentrations of αβtubulin, NuMA-N-SNAP, NuMA-C-SNAP, NuMA-Bonsai-SNAP, and NuMA-NC-SNAP. DNA was labeled with Sybr-Gold. Arrows indicate the minimum protein concentration for each protein with a noticeable DNA band shift. (E) Representative time-lapse images of RPE1 cells transiently expressing NuMA-FL-EGFP and stably expressing NuMA-Bonsai-EGFP, EGFP-Zdk1-NuMA-C, or NuMA-NC-EGFP and labeled with SiR-Hoechst (DNA), followed from metaphase through cytokinesis. NuMA-C and NuMA-NC localize to chromosomes at mitosis, while NuMA-Bonsai does not. Time in minutes:seconds; 0:00 corresponds to anaphase onset (AO, vertical dashed line). Arrows indicate when we detect different NuMA truncations on chromosomes. Scale bar, 5 µm. See also Video 5.
Figure S4.
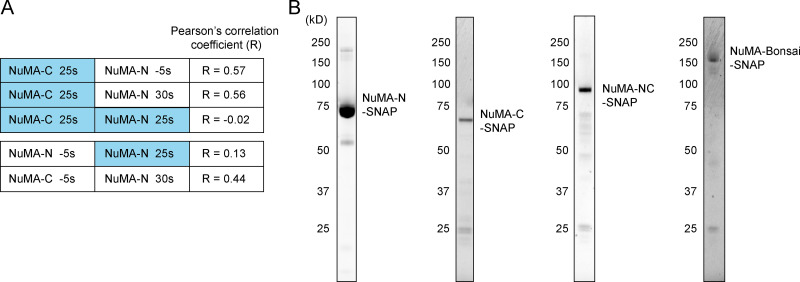
NuMA-N and -C responses in Opto-NuMA in vivo experiments, and purified NuMA truncation proteins used for in vitro EMSAs. (A) Pearson’s correlation coefficient between NuMA-N and/or NuMA-C images of nuclear regions in Opto-NuMA experiments at the indicated times (related to Fig. 4 B). Blue represents illumination with blue light. Lines 1 and 2 show that NuMA-C localization during illumination correlates with NuMA-N localization both before and after illumination, consistent with NuMA-N being recruited to the same NuMA-C localization before and after illumination; line 3 shows that NuMA-C localization does not correlate with NuMA-N localization during illumination, consistent with NuMA-N unbinding from NuMA-C during illumination; line 4 shows that NuMA-N localizations before and during illumination do not correlate; line 5 shows that NuMA-N localizations before and after illumination correlate, consistent with NuMA-N being recruited to the same NuMA-C localization before and after illumination; line 6 shows that NuMA-C localizations at the start and end of illumination correlate. Together these results suggest that NuMA-C drives NuMA-N localization, and that NuMA-C has a higher affinity for chromatin than NuMA-N. The representative cell shown in Fig. 4 B was analyzed. (B) Coomassie blue–stained gels with SNAP-tagged NuMA-N, NuMA-C, NuMA-Bonsai, and NuMA-NC proteins purified from insect Sf9 cells. These purified NuMA truncation proteins were used in EMSAs (Fig. 4 D).
Video 4.
NuMA-C has a higher affinity than NuMA-N for chromosomes in the nucleus. Time-lapse spinning disk confocal imaging of a representative RPE1 cell stably expressing EGFP-Zdk1-NuMA-C (left) and NuMA-N(1–705)-mCherry-LOV2 (right), before (<0:15), during (0:15-0:44, “+Light”), and after (>0:45) exposure to blue light (488-nm laser). Upon blue light illumination, NuMA-C remains associated with chromosomes, while NuMA-N becomes diffusive. This indicates the NuMA-C binds to chromosomes in the nucleus with higher affinity than NuMA-N. Time in minutes:seconds. Scale bar, 5 µm. Video corresponds to the still images in Fig. 4 B.
Previous work suggested that FL NuMA can directly bind DNA (Ludérus et al., 1994), but whether binding occurs through the C or N terminus or both, and the relative affinity of each for DNA, are not known. To address these questions and understand how NuMA binds DNA, we purified different SNAP-tagged NuMA truncation constructs (Fig. S4 B) and tested their ability to bind a DNA fragment in vitro using an electrophoretic mobility shift assay (EMSA) under stringent binding conditions (Fig. 4 D). We observed that the construct that lacks the coiled-coil (NuMA-NC) bound DNA with the highest affinity, followed by NuMA-Bonsai (NuMA-NC with an additional 493–amino acid–long coiled-coil) and finally NuMA-C with the lowest detectable affinity. NuMA-N did not have detectable DNA affinity in our conditions, and negative control tubulin had no detectable affinity. Thus, NuMA directly binds DNA primarily through its C terminus, and this binding could in principle underlie NuMA-C’s chromatin binding in vivo (Fig. 4, A–C). Furthermore, these in vitro observations suggest that NuMA’s DNA binding may be intramolecularly regulated, as NuMA’s N terminus increases its DNA binding, and the coiled-coil appears to decrease it.
Given that NuMA-C binds chromatin in interphase (Fig. 4, A–C) and DNA in vitro (Fig. 4 D), we asked what prevents NuMA-C from binding chromosomes in mitosis. To answer this question, we mapped the localization of NuMA-FL, NuMA-Bonsai, NuMA-NC, and NuMA-C through mitosis (Fig. 4 E and Video 5), testing whether other parts of NuMA regulate NuMA-C’s chromatin binding, as suggested in vitro (Fig. 4 D). Similar to endogenous NuMA (Fig. S3) and NuMA-FL, NuMA-Bonsai did not localize to chromosomes in mitosis. In contrast, NuMA-NC and NuMA-C were recruited to mitotic chromosomes as early as anaphase onset. Therefore, NuMA’s coiled-coil actively regulates NuMA’s localization during mitosis by preventing the premature binding of NuMA-C to chromosomes. NuMA is heavily phosphorylated by the mitotic kinase CDK1 at mitotic entry (Compton and Luo, 1995; Kotak et al., 2013; Yang et al., 1992). CDK1 activity negatively controls NuMA levels at the cell cortex during metaphase, and NuMA’s dephosphorylation at anaphase promotes the protein’s translocation to the cortex (Kiyomitsu and Cheeseman, 2013; Kotak et al., 2013). Premature RO3366-mediated CDK1 inhibition at metaphase did not change NuMA-Bonsai’s localization, while NuMA-C and NuMA-NC still bound to chromosomes (Fig. S5). This indicates that the coiled-coil regulates NuMA localization during mitosis, apparently doing so independently of CDK1 activity, and may depend on other regulatory processes. Together, these findings indicate that the coiled-coil of NuMA acts as a central element that drives not only the protein’s nuclear specific function and the dynamics of its assemblies (Fig. 3), but also when NuMA is allowed to bind chromosomes and which structures it acts on over the cell cycle.
Video 5.
NuMA’s coiled-coil prevents its C terminus from binding chromosomes at mitosis. Time-lapse spinning disk confocal imaging of a representative noninduced NuMA KO RPE1 cell transiently expressing NuMA-FL-EGFP, or stably expressing NuMA-Bonsai-EGFP, EGFP-Zdk1-NuMA-C, or NuMA-NC-EGFP (in this order, green, left) and labeled with SiR-Hoechst (magenta, center) transitioning from metaphase to anaphase. NuMA-FL-EGFP and NuMA-Bonsai-EGFP, which contains part of the coiled-coil, do not bind chromosomes at mitosis, while NuMA-C and NuMA-NC, which lacks the entire coiled-coil, do. These results indicate that NuMA’s coiled-coil regulates NuMA-C’s binding to chromosomes in mitosis. Time in minutes:seconds, with the frame corresponding to anaphase onset set to 00:00. Scale bar, 10 µm. Video corresponds to the still images in Fig. 4 E.
Figure S5.
CDK1 activity does not affect NuMA binding to chromosomes during mitosis. Representative time-lapse images of uninduced NuMA KO RPE1 cells stably expressing NuMA-Bonsai-EGFP, EGFP-Zdk1-NuMA-C, or NuMA-NC-EGFP, treated with SiR-Hoechst (DNA), and treated with the CDK1 inhibitor RO-3366 ∼10 s before 0:00. Time in minutes:seconds. Cells were imaged during metaphase to monitor the localization of the different EGFP-tagged NuMA truncation proteins on chromosomes. Arrows indicate the time at which we detect the tested EGFP-tagged NuMA truncations on chromosomes upon CDK1 inhibition. Scale bars, 5 µm.
NuMA provides mechanical stability to the nucleus
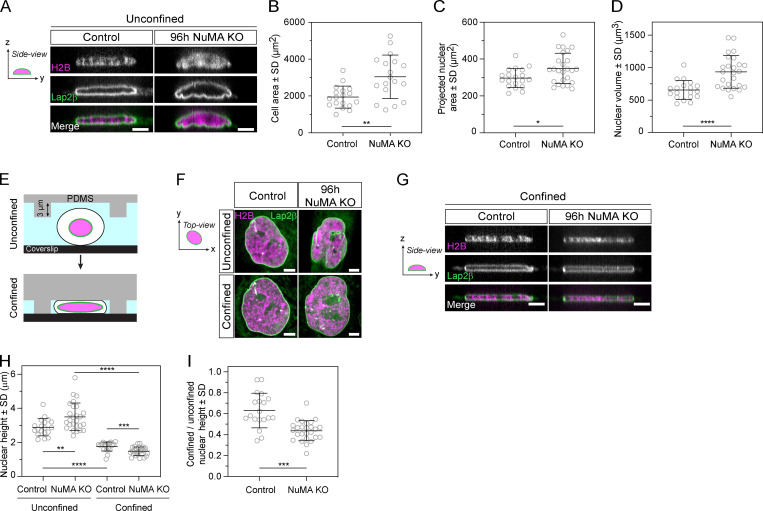
So far, we have shown that NuMA is essential for forming a single, round nucleus (Figs. 1 and 2), keeps the chromosome mass compact at mitotic exit (Fig. 2 F), can form higher-order nuclear assemblies (Fig. 3 G; Zeng et al., 1994), and binds chromatin in a cell cycle–regulated manner (Fig. 4). Together, these findings indicate that NuMA plays a structural role in forming the nucleus, and suggest how it may do so. This led us to hypothesize that NuMA also plays a structural role in maintaining the nucleus. To test for such a role, we compared nuclear morphologies and then mechanically perturbed the nuclei of cells stably expressing EGFP-Lap2β and mCherry-H2B with and without NuMA. If nuclei were more deformable without NuMA, this would indicate that NuMA plays a structural role in nuclear stability. Imaging in 3D, and at higher resolution than above experiments, revealed invaginations in the nuclear surface of NuMA KO cells that were undetectable in control cells (Fig. 5 A). While reduced cell spreading can cause a reduced nuclear area (Buxboim et al., 2017) and nuclear wrinkling (Li et al., 2015), this is unlikely the reason NuMA KO nuclei have wrinkles as NuMA KO cells spread more (Fig. 5 B) and their nuclear area is larger (Fig. 5 C) than control cells. The presence of wrinkles suggested that nuclei are more deformable in NuMA KO cells. Furthermore, nuclei had a larger volume in NuMA KO than control cells (Fig. 5 D), suggesting that NuMA may keep chromosomes together not only at nuclear formation (Fig. 2 F) but also during nuclear maintenance. To compare how nuclear shape responds to mechanical force with and without NuMA, we confined and thereby compressed cells using a polydimethylsiloxane (PDMS) device (Guild et al., 2017; Le Berre et al., 2012) with 3-µm-high pillars (Fig. 5 E), and measured resulting changes in nuclear morphology. Under confinement, the nuclei of NuMA KO cells lost their nuclear invaginations as these “unfolded” into a smooth nuclear surface similar to control cells (Fig. 5 F). In addition, nuclei without NuMA were more deformable under external mechanical force than control nuclei: while nuclei were thicker (i.e., higher in the confinement axis) before confinement in NuMA KO cells, and had a larger volume in NuMA KO cells (Fig. 5 C), they became thinner than in control cells under confinement (Fig. 5, G–I). Thus, NuMA either directly or indirectly provides mechanical stability to the nucleus, which could help maintain nuclear structure and function. Altogether, our work indicates that NuMA plays a spindle-independent, structural role in both nuclear formation and mechanics in human epithelial cells.
Figure 5.
NuMA provides mechanical robustness to the nucleus. (A) Representative side (y-z) view images of a live control and 96 h NuMA KO RPE1 cell expressing mCherry-H2B (magenta) and EGFP-Lap2β (green), with images acquired every 0.25 µm over 8 µm. Scale bar, 5 µm. (B) Cell area of unconfined control and NuMA KO cells. Plot shows mean ± SD n = 18 cells in both conditions. Two-sided unpaired t test: **, P < 0.002. (C) Projected nuclear area of unconfined control and NuMA KO cells. Plot shows mean ± SD n = 19 (control) and 25 (NuMA KO) cells. Two-sided unpaired t test: *, P = 0.019. (D) 3D nuclear volume of unconfined control and NuMA KO cells. Plot shows mean ± SD n = 19 (control) and 25 (NuMA KO) cells. Two-sided unpaired t test: ****, P < 0.0001. (E) Schematic representation of cell confinement using a PDMS device (gray) with 3-µm-high pillars. Nuclear (pink) compression occurs when pillars are brought down to contact the coverslip (black). (F) Top (x-y) view images of a live control and 96 h NuMA KO RPE1 cells from A, shown before (unconfined) and during confinement by the PDMS device depicted in B. Scale bar, 5 µm. (G) Representative side (y-z) view images of a confined live control and NuMA KO RPE1 cell from A and C. Scale bar, 5 µm. (H and I) Nuclear height of unconfined and confined control and NuMA KO cells (E), calculated based on EGFP-Lap2β localization, and unconfined nuclear height ratio in control and NuMA KO cells (F). Plot shows mean ± SD n = 19 (control) and 25 (NuMA KO) cells. Two-sided Mann–Whitney test: ****, P < 0.0001; ***, P = 0.007; **, P < 0.008. Same dataset as in Fig. 5 B.
Discussion
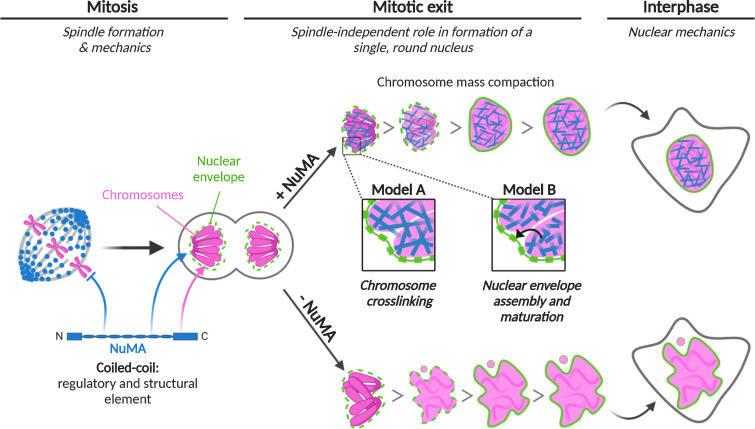
Although many of the molecules required for nuclear formation are known, we still do not understand how they together give rise to a single, round, and robust mammalian nucleus. Because mitotic errors can lead to abnormal nuclei, one challenge is to define when, at mitosis or nuclear formation, a given molecule acts, and what roles it plays at different times. A long-standing example has been NuMA, an ∼200-nm-long coiled-coil protein (Harborth et al., 1995, 1999; Yang et al., 1992) essential for both mitosis (Compton and Luo, 1995; Kallajoki et al., 1991; Yang and Snyder, 1992) and nuclear integrity (Compton and Cleveland, 1993; Kallajoki et al., 1991, 1993). Here, we uncouple NuMA’s mitotic and nuclear functions. We show that NuMA has a spindle-independent role in building a single, round nucleus (Fig. 1), keeping the decondensing chromosome mass compact at nuclear formation (Fig. 2) and promoting the nucleus’ mechanical robustness (Fig. 5). Thus, NuMA plays a structural role in both building and maintaining the nucleus, as it does in the spindle (Compton and Luo, 1995; Hueschen et al., 2019; Silk et al., 2009; Yang and Snyder, 1992). We find that NuMA’s coiled-coil acts as a central element: it is required for nuclear function, and regulates NuMA’s mobility in the nucleus (Fig. 3) and its binding to chromosomes throughout the cell cycle (Fig. 4). As such, the coiled-coil compartmentalizes NuMA’s function to where and when it is needed: on chromosomes at interphase (when these are kept together) and off chromosomes at mitosis (when these are pulled apart). We propose a model whereby NuMA drives cellular organization throughout the cell cycle, building and maintaining the nucleus at interphase and the spindle at mitosis (Fig. 6).
Figure 6.
Model for NuMA’s role in nuclear formation and mechanics, and its structural role over the cell cycle. NuMA (blue) plays a spindle-independent role in nuclear formation (“Mitotic exit,” center) and mechanics (“Interphase,” right). It keeps the chromosome (pink) mass compact at nuclear formation, and is essential to building a single, round and mechanically robust nucleus (“+NuMA”, top). Without NuMA (“-NuMA”, bottom), micronucleation and nuclear shape defects occur. We propose two models for how NuMA, whose C terminus binds interphase chromosomes (pink arrow), performs its nuclear function. To promote nuclear formation and mechanics, NuMA could cross-link chromosomes (Model A, blue filaments) or regulate nuclear envelope (green) assembly and maturation (Model B, black arrow), either directly or indirectly. At “Mitosis” (left), NuMA plays a critical role in spindle formation and mechanics, and its coiled-coil prevents it from binding chromosomes (blue inhibitory arrow), when these must but segregated instead of kept together. At mitotic exit and interphase, the coiled-coil drives NuMA’s nuclear dynamics and function (blue arrow). As such, we propose that NuMA’s coiled-coil acts as a central regulatory and structural element to control its function in space and time. Altogether, NuMA is essential to the formation and stability of two of the cell’s largest structures, the spindle and the nucleus. Figure created with Biorender.com.
NuMA’s structure, regulation, and function in the nucleus
Our current understanding of NuMA’s spindle function and its underlying basis much surpasses that of its nuclear function. Here, our work raises the question of how NuMA’s structure can give rise to its function in nuclear formation (Figs. 1 and 2) and mechanics (Fig. 5). While most of NuMA’s coiled-coil is dispensable for building the spindle (Forth et al., 2014; Hueschen et al., 2017), it is essential for building a single, round nucleus (Fig. 3, B and C; and Fig. S2 C). Additionally, the coiled-coil can stabilize nuclear NuMA (Fig. 3, D–F), and nonphysiological overexpression of FL NuMA, but not coiled-coil truncations, induces the formation of stable filament-like nuclear structures (Fig. 3 G). Furthermore, we know that NuMA can, when ectopically expressed in the cytoplasm, assemble into an organized network (Saredi et al., 1996) and, when overexpressed in the nucleus, assemble into a 3D network whose lattice size is determined by its coiled-coil’s length (Gueth-Hallonet et al., 1998; Harborth et al., 1999). We also know that NuMA self-assembles at the cell cortex to position the spindle using its coiled-coil and C terminus (Okumura et al., 2018). Together, these observations are consistent with a model where NuMA assembles into a network that shapes nuclear morphology and mechanics (Figs. 1, 2, and 5), with its coiled-coiled key to function and possibly assembly.
NuMA’s mobility in the nucleus (Fig. 3, D–G) suggests that it assembles into a dynamic structure. We do not know whether and how NuMA’s mobility on chromosomes changes over nuclear formation and maintenance, and how NuMA performs its functions given its mobility. One possibility is that NuMA’s dynamic structure enables its diverse nuclear functions. Indeed, NuMA regulates the localization and mobility of proteins involved in DNA repair (Salvador Moreno et al., 2019; Vidi et al., 2014) and in the nucleolar stress response (Jayaraman et al., 2017). Since NuMA-C binds DNA and chromosomes (Fig. 4) and altered NuMA expression impacts higher chromatin organization (Abad et al., 2007), such a network could in principle help cross-link DNA within and between chromosomes. Alternatively, NuMA may also establish connections with other nuclear networks that determine higher-order nuclear organization, such as RNA and ribonucleoproteins, as previously suggested (Barboro et al., 2002, 2003). Independently of the mechanism by which NuMA acts in the nucleus, its long structure and its self-assembling and DNA-binding properties make it well-suited to bridge large distances and impact higher-order nuclear organization. Determining to what extent nuclear NuMA self-assembles, and the structure of this assembly, will be essential to understanding how NuMA’s structure gives rise to its dynamics (Fig. 3, D–F) and architectural (Figs. 1, 2, and 5) and regulatory functions in the nucleus. Uncovering NuMA’s nuclear functions may in turn help understand why its localization varies depending on cell phenotype (Knowles et al., 2006; Vega et al., 2017). Similarly, it will shed light on why NuMA’s expression level and impact on nuclear architecture vary dramatically between epithelial cells and highly specialized or terminally differentiated cells (Merdes and Cleveland, 1998).
While NuMA-C binds DNA in vitro (Fig. 4 D) and appears to bind chromosomes in the nucleus (Fig. 4, A and B), FL NuMA does not bind chromosomes at mitosis. As such, its binding must be regulated. Here, we show that a small portion of NuMA’s coiled-coil is sufficient to prevent NuMA-C from binding chromosomes at mitosis (Fig. 4 E), and sufficient to decrease NuMA’s DNA binding affinity in vitro (Fig. 4 D). Preventing NuMA from binding chromosomes at mitosis could prevent it from keeping chromosomes together when they must segregate or, since NuMA recruits dynein (Hueschen et al., 2017), from recruiting dynein-based forces there. However, we find that prematurely localizing NuMA-NC on chromosomes at mitosis does not lead to detectable mitotic defects (Fig. 4 E). This could be because NuMA-NC lacks the coiled-coil needed for NuMA to keep chromosomes together (Fig. 3, B and C; and Fig. S2 C) and that is likely necessary to activate dynein motility (Hueschen et al., 2017; Reck-Peterson et al., 2018), or because its function on chromosomes is cell cycle–regulated. We propose NuMA’s coiled-coil plays a central role throughout the cell cycle, acting as a central structural (Fig. 3, B and C; and Fig. S2 C) and regulatory (Fig. 3, D–F; and Fig. 4 E) element for NuMA’s function on chromosomes. How the cell regulates this hub, such that NuMA binds chromosomes at mitotic exit and dissociates at mitotic entry, remains an open question. Answering it will require defining how NuMA’s C terminus binds DNA and chromosomes and will be key to understanding how the same protein performs such different physical roles in interphase and mitosis. Notably, NuMA’s C terminus also targets NuMA in the spindle, specifically to microtubule minus-ends (Hueschen et al., 2017).
Models for NuMA’s role in nuclear formation and mechanics
What role NuMA plays in nuclear formation (Figs. 1 and 2) and mechanics (Fig. 5) is unclear. One model is that NuMA directly contributes to the organization and mechanics of the chromosome mass at mitotic exit (Fig. 6, model A). Indeed, NuMA-C binds DNA in vitro (Fig. 4 D) and chromosomes in vivo (Fig. 4, A–C), NuMA keeps the mass of decondensing chromosomes compact at mitotic exit (Fig. 2 F), and it localizes throughout the entire nucleus (Compton et al., 1992; Yang et al., 1992; Fig. S3), where it can interact with chromosomes. RNA-dependent NuMA localization and tethering to lamins has been suggested (Barboro et al., 2002), and NuMA influences higher-order chromatin organization (Abad et al., 2007; Kivinen et al., 2010) and binds chromatin remodelers (Vidi et al., 2014). Furthermore, we know that chromatin compaction impacts the shape and robustness of the nucleus (Stephens et al., 2019). If NuMA cross-linked DNA and chromosomes, or established connections with other nuclear structures, its import into the nucleus as it is forming and chromatin starts decondensing (Schooley et al., 2012) could keep the chromosome mass more compact at nuclear formation (Fig. 2 F) and maintenance (Fig. 5 D), and make it more rigid (Fig. 5). This could make it harder for the assembling nuclear envelope to penetrate into the chromosome mass, thus decreasing multinucleation and abnormally shaped nuclei (Figs. 1 and 2), and provide mechanical robustness to the nucleus (Fig. 5). In this model, NuMA plays a nuclear formation role with similarities to BAF’s, with BAF localizing at the chromosome mass’s rim (Samwer et al., 2017) and NuMA through the entire volume of the nucleus. Finally, we note that NuMA could also organize chromosomes at mitotic exit by keeping them in a tight focus around spindle poles (Cleveland, 1995; Compton and Cleveland, 1993), given NuMA’s pole-focusing role. However, such a role cannot explain our observations that NuMA plays a role in nuclear formation after a spindle-less mitosis (Figs. 1 and 2).
Alternatively, and not mutually exclusively, NuMA’s binding to chromosomes during nuclear formation could promote assembly and maturation of the nuclear envelope and its components (LaJoie and Ullman, 2017; Ungricht and Kutay, 2017) or mediate the tethering of chromatin to the nuclear envelope (Schreiner et al., 2015; Fig. 6, model B). These functions, when compromised, could lead to defects in nuclear morphology (Figs. 1 and 2) and mechanics (Fig. 5; Davidson and Lammerding, 2014; Webster et al., 2009). Incidentally, NuMA-C–coated beads mixed with Xenopus extracts recruit membranes, suggesting that NuMA could contribute to nuclear envelope assembly (Lu et al., 2012). Though NuMA is not enriched at the nuclear rim (Figs. 1 A and S3), we cannot exclude that it regulates the nuclear envelope and as such indirectly impacts chromosome organization. While in the two models we discuss that NuMA plays a direct role in nuclear formation and mechanics (Fig. 6), we cannot exclude the possibility that it plays an indirect role. For example, as a nuclear protein, it could in principle change the expression of genes that contribute to nuclear formation or to nuclear deformation under force, for example by changing the mechanics of nonnuclear elements such as the cytoplasm or cell cortex.
Looking forward, testing these and other models will be critical to understanding NuMA’s physical role in building and maintaining the nucleus. Toward this goal, it will be important to quantitatively measure NuMA’s physical contributions and to determine whether NuMA can cross-link chromatin in vitro and in vivo, and whether and how it influences the nuclear envelope and regulates gene expression. Tools to acutely inhibit NuMA will be essential to separating its roles in nuclear formation versus maintenance. Defining NuMA’s physical role in the nucleus, and with which molecular players it acts, will be key to understanding its relationship to other mechanisms regulating nuclear formation and maintenance, and to understanding its role in other, nonepithelial cell types and contexts. More broadly, these efforts will provide insight into how NuMA is repurposed and regulated across the cell cycle to build and maintain both the nucleus and the spindle.
Materials and methods
Cell culture and transfection
Tet-on inducible CRISPR/Cas9 NuMA KO RPE1 cells (hereafter “NuMA KO RPE1 cells”; Hueschen et al., 2017) were cultured in DMEM/F12 with GlutaMAX (10565018; Thermo Fisher Scientific) supplemented with 10% tetracycline-screened fetal bovine serum (SH30070.03T; Hyclone Labs). Cells were maintained at 37°C and 5% CO2. NuMA was depleted by inducing spCas9 expression with 1 µg/ml DOX hyclate in Tet-on inducible CRISPR/Cas9 RPE1 sgNuMA cells for 96 h before each experiment, unless other induction times are indicated. For transient expression experiments (Fig. 3, B and D–G, NuMA-FL-EGFP only; Fig. 4 E, NuMA-FL-EGFP only; Fig. S2 A; Fig. S2, E and F, NuMA-FL-EGFP only), inducible NuMA KO RPE1 cells were transfected with DNA using ViaFect (E4981; Promega) according to the manufacturer’s instructions 48 h before imaging. For rescue experiments (Fig. 3 B; and Fig. S2, A), cells were transfected 72 h before imaging. For long-term live imaging in Fig. 2, cells were treated with nocodazole and reversine at 72 h after KO induction, and imaged for 16–18 h. NuMA depletion at the end of the live imaging has occurred for 88–90 h, but for simplicity, we refer to it as “96 h.” At 88–90 h of induction, NuMA is expected to be nearly fully depleted (Fig. S1 A). Cell lines were not short-tandem repeat (STR)-profiled for authentication. All cell lines tested negative for mycoplasma.
Cell line generation and lentiviral production
To generate a cell line stably expressing mCherry-H2B and EGFP-Lap2β, NuMA KO RPE1 cells were infected with mCherry-H2B and EGFP-Lap2β lentiviruses (Figs. 2 and 5). Viruses were made in HEK293T cells from a pLenti6/V5-DEST plasmid (Invitrogen) containing mCherry-H2B (gift from T. Wittmann, University of California, San Francisco, San Francisco, CA) or EGFP-Lap2β and lentiviral packaging plasmids using calcium phosphate transfection. The medium of transfected HEK293T cells was replaced after 24 h, and virus-containing medium was collected 48 h after transfection. NuMA KO RPE1 cells were infected with virus-containing medium supplemented with 10 µg/ml polybrene and selected with 2.5 µg/ml blasticidin. A polyclonal stable line with low expression of mCherry-H2B and EGFP-Lap2β was further selected by FACS sorting and used herein.
To generate the Opto-NuMA cell line (Fig. 4, B and C), NuMA KO RPE1 cells were infected with NuMA-N-PhusionRed-LOV2 and EGFP-Zdk1-NuMA-C using retroviruses made in HEK293T-GP cells expressing the retroviral packaging genes gag and pol and transfected with calcium phosphate, as described above. A polyclonal stable line was established after FACS sorting.
To generate cell lines stably expressing NuMA truncations, the above protocol was used to infect NuMAKO RPE1 cells. We made the following cell lines: monoclonal NuMA-Bonsai-EGFP (Fig. 3, D–F; Fig. 4 E; Fig. S2, B–D; and Fig. S5), EGFP-Zdk1-NuMA-C (Figs. 4 E and S5), and NuMA-NC-EGFP (Fig. 3, D–F; Fig. 4 E; Fig. S2, E and F; and Fig. S5). In the NuMA-Bonsai-EGFP line, exogenous NuMA is expressed at higher levels than endogenous NuMA (Fig. S2 B).
Drug and dye treatments
Cells were treated with 664 nM nocodazole (M1404; Sigma-Aldrich) to depolymerize spindle microtubules and with 320 nM reversine (R3904; Sigma-Aldrich) to inhibit MPS1 and silence the spindle assembly checkpoint (Fig. 1; and Fig. S1, A–C). To avoid the confounding negative effect of a too-short mitosis on successful nuclear formation, we adjusted reversine concentrations in control cells (1 µM) so that NuMA KO cells spent at least as much time in spindle-less mitosis as control cells (Fig. 2; and Fig. S1, D and E). Nocodazole and reversine were added 10–30 min before live imaging. Cells were treated with 9 µM RO-3306 to acutely inhibit CDK1. To visualize DNA by live imaging in cells not expressing mCherry-H2B (Figs. 4 E and S5), 500 nM silicon rhodamine (SiR)-DNA dye (CY-SC007; Cytoskeleton) was added 30 min before imaging.
Plasmids
NuMA-FL-EGFP (FL human NuMA; NM_006185.3), used for transient transfection, is Cas9-resistant and was made by creating silent mutations (5′-GTGTCAGAGAGACTGGACTTT-3′ mutated to 5′-GTTAGTGAACGCTTGGATTTT-3′, preserving amino acids 57–62 of NP_006176.2 [“VSERLD”]). The modified sequence was subsequently cloned into a pEGFP-N1 vector (Clonetech, Takara Bio). These silent mutations made the sequence resistant to the sgRNA with the sequence 5′-AAGTCCAGTCTCTCTGACAC-3′ (Hueschen et al., 2017). NuMA-Bonsai-EGFP used for transient transfection was made by amplifying NuMA1-705+1701-2115 from NuMA-FL-EGFP (Hueschen et al., 2017); for reference, the NuMA-Bonsai-EGFP herein is called NuMA-N-C-EGFP in Hueschen et al. (2017).
To make the mCherry-H2B and EGFP-Lap2β cell line, the EGFP-Lap2β coding sequence was amplified from a EGFP-Lap2β lentiviral plasmid (gift from D. Gerlich, Institute of Molecular Biotechnology of the Austrian Academy of Sciences [IMBA)] Vienna, Austria; plasmid laboratory ID: 1282; Samwer et al., 2017), and inserted into the lentiviral vector pLenti6/V5-DEST plasmid (Invitrogen, gift from T. Wittman). The lentiviral mCherry-H2B plasmid was subcloned from pH2B_mCherry_IRES_neo (Addgene plasmid 21044, gift from T. Wittman; Pemble et al., 2017).
To generate the Opto-NuMA cell line, NuMA-N-PhusionRed-LOV2 was cloned into a human cytomegalovirus (CMV) promoter plasmid as follows: NuMA-N1-705 was amplified from NuMA-Bonsai1-705+1701-2115-EGFP (Hueschen et al., 2017), and PhusionRed was amplified from pmKate2.7C-N1 (the Michael Davidson Fluorescent Protein Collection, University of California, San Francisco, Nikon Imaging Center). These were subsequently cloned into a CMV promoter plasmid containing the fast dissociation LOV2 domain (EB1-N-LZ-LOV2 described in van Haren et al., 2018) by replacing the EB1-N-LZ sequence (LZ stands for GCN4 leucine zipper) with the NuMA-N-PhusionRed sequence. EGFP-Zdk1-NuMA-C was cloned into the CMV promoter plasmid mCherry-Zdk1-EB1-C (van Haren et al., 2018) as follows: mCherry was replaced by EGFP amplified from pEGFP-C1 (Clonetech, Takara Bio), and EB1-C was replaced by NuMA-C1701-2115, which was amplified from NuMA-Bonsai-EGFP (Hueschen et al., 2017). As a final step, NuMA-N-PhusionRed-LOV2 and EGFP-Zdk1-NuMA-C were subcloned into a Moloney murine leukemia retroviral plasmid (Addgene plasmid 51403, gift from I. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, MA; Welburn et al., 2009). For this, EGFP-TEV-S-Ska1 (contaning a tobacco etch virus [TEV] protease cleavage site) was removed from the Addgene plasmid 51403, and NuMA-N-PhusionRed-LOV2 and EGFP-Zdk1-NuMA-C were amplified and inserted in that site.
To generate cell lines stably expressing NuMA truncations, NuMA-Bonsai1-705+1701-2115-EGFP was amplified from NuMA-Bonsai1-705+1701-2115-EGFP (Hueschen et al., 2017), EGFP-Zdk1-NuMA-C was made as described above, and NuMA-NC1-212+1701-2115-EGFP was made from assembling both NuMA-N1-212 and NuMA-C1701-2115 from NuMA-Bonsai1-705+1701-2115-EGFP (Hueschen et al., 2017). Then these were each inserted into the lentiviral vector pLenti6/V5-DEST plasmid (Invitrogen, gift from T. Wittmann). Notably, NuMA-FL-EGFP, NuMA-Bonsai-EGFP, and NuMA-NC-EGFP plasmids used in this study are resistant to the NuMA sgRNA, as described above and in Hueschen et al. (2017).
To express and purify proteins for EMSA, the SNAP-tagged NuMA proteins were cloned as follows: NuMA-N1-505, NuMA-C1701-2115, NuMA-Bonsai1-705+1701-2115, and NuMA-NC1-212+1701-2115 were amplified from NuMA-FL-EGFP and inserted into an Sf9 expression vector (pACEBac1). αβTubulin was purified from porcine brains (Hyman et al., 1991).
Western blotting
Cells were plated in six-well plates (08–772-1B; Thermo Fisher Scientific) in parallel with setting up an imaging experiment. The medium was removed, and cells were collected at the indicated time after induction of NuMA KO. Cells were subsequently lysed in cold radioimmunoprecipitation assay (RIPA) buffer, supplemented with protease inhibitor cocktail (11836153001; Roche), and incubated on ice for 30 min, and protein extracts were collected after 30 min of centrifugation at 4°C. Protein concentrations were measured using a Bradford assay kit (Bio-Rad) based on absorbance (A595 nm), and samples were prepared at equal concentrations in NuPage Sample buffer and NuPage Sample reduction agent (Invitrogen). Proteins were denatured for 10 min at 95°C, separated on a 3–8% Tris-Acetate gel (Invitrogen) by SDS-PAGE, and transferred to a nitrocellulose membrane by Western blotting using a Bio-Rad electrophoresis system. Membranes were blocked with 4% milk in TBS-Triton for 1 h at room temperature, probed with rabbit anti-NuMA (1:1,000; ab97585; Abcam; Fig. S2 B), rabbit anti-NuMA (1:300; NB500-174; Novus Biologicals; Fig. S1 A), and mouse anti-α-tubulin (1:5,000; T6199; Sigma-Aldrich) primary antibodies in 1% milk in TBS-T at 4°C overnight, washed with TBS-T, and incubated with anti-rabbit and anti-mouse HRP-conjugated secondary antibodies (0.01 µg/ml; sc-2357 and sc-2005; Santa Cruz Biotechnologies) for 1 h at room temperature. Proteins were detected with the ImageQuant LAS4000 using ECL (Thermo Fisher Scientific)–based chemiluminescence.
Immunofluorescence
Cells were plated on 25-mm glass coverslips coated with 1 mg/ml poly-L-lysine. Cells were fixed with 95% (vol/vol) methanol containing 5 mM EGTA for 2 min at −20°C, washed with 0.05% Triton X-100 in TBS-T, and blocked with 2% BSA in TBS-T for 1 h at room temperature. Primary and secondary antibodies were diluted in 2% BSA in TBS-T and incubated with cells for 1 h at room temperature. DNA was labeled with Hoechst 33342 (B2261; Sigma-Aldrich). Cells were mounted in ProLongGold Antifade (P36934; Thermo Fisher Scientific). Cells were imaged using a spinning disk confocal inverted or an epifluorescence microscope as described in the following section. The following antibodies were used: rabbit anti-NuMA (1:300; NB500-174; Novus Biologicals; Fig. 1; Fig. S1, A–E; and Fig. S3), rabbit anti-NuMA (1:130; ab97585; Abcam; Fig. 3, B and C; and Fig. S2, A and C), anti-GFP conjugated to Atto488 (1:100; ChromoTek gba-488; Fig. 3, B and C; and Fig. S2, A and C), anti-mouse secondary antibodies conjugated to Alexa Fluor 488 (5 µg/ml; A11001; Invitrogen), and anti-rabbit secondary antibodies conjugated to Alexa Fluor 647 (5 µg/ml; A21244; Invitrogen).
Microscopy
NuMA KO RPE1 cells were plated on no. 1.5 glass-bottom 35-mm MatTek dishes coated with poly-D-lysine (MatTek Corporation) and imaged in DMEM/F12 with GlutaMAX (10565018; Thermo Fisher Scientific). Imaging was performed on a spinning disk confocal (CSU-X1; Yokogawa) inverted microscope (Eclipse Ti-E, Nikon) with a perfect focus system (Nikon). Cells were imaged in a stage-top incubation chamber (Tokai Hit) and maintained at 37°C with 5% CO2. Experiments were performed with a Di01-T405/488/568/647 head dichroic (Semrock), along with 405-nm (100 mW), 488-nm (150 mW), 561-nm (150 mW), and 642-nm (100 mW) diode lasers, and emission filters ET455/50M, ET525/50M, ET630/75M, and ET705/72 (Chroma). Cells were imaged with a Zyla 4.2 sCMOS camera (Andor Technology) or an iXon3 camera (Andor Technology) operated by MetaMorph 7.7.8.0 (Molecular Devices; 50–100 ms exposures) with a 60× 1.4 Ph3 oil objective or a 100× 1.45 Ph3 oil objective (Fig. 4 B) in a single plane every 1–60 s. For the confinement experiments (Fig. 5), images were acquired with a 100× 1.45 Ph3 oil objective over 32 z planes spaced 0.25 µm apart. For long-term live imaging (Fig. 2), a 20× 0.5 Ph1 air objective was used, and images were acquired every 4 min over 11 z planes spaced 2 µm apart to account for focal plane changes over long periods. For fixed cell imaging (Figs. 1, 3, and S1), images were acquired over 30 z planes spaced 0.3 µm apart using the same microscope and objectives setting. Fixed cell imaging in Fig. S3 was performed on a Zeiss AxioPlan2 epifluorescence microscope. FRAP experiments were performed on a OMX-SR microscope (GE Health Care), equipped with a PCO Edge 5.5 cMOS camera (PCO), a four-line laser launch, 405/488/568/640 nm (Toptica), and a live cell chamber (GE Health Care) at 37°C with 5% CO2. All images were acquired through a Plan ApoN 60× 1.42 oil objective (Olympus) with Laser liquid 1.518 (Cargile). GFP was imaged with the 488-nm laser, with a 528/48M emission filter in place. Photobleaching was performed on a 2-µm-diameter circular region in the nucleus. After defining the region of interest, a 405-nm laser was used to bleach the region. Images were acquired at 250-ms intervals for a total of 39 s; five baseline images were captured 1 s before bleaching and a time-lapse of 39 s after the bleaching event. Cells were plated on glass-bottom 35-mm dishes coated with poly-D-lysine (MatTek Corporation) for all live imaging experiments or on 25 mm no. 1.5 glass coverslips acid-cleaned and coated with poly-L-lysine (0117650; Marienfeld) for all immunofluorescence experiments and imaged in DMEM/F12 with GlutaMAX (10565018; Thermo Fisher Scientific) supplemented as for NuMA KO RPE1 cell culture as described above.
SNAP-tagged protein expression and purification
NuMA-N1-505-SNAP, NuMA-Bonsai-SNAP, and NuMA-C1701-2115-SNAP were purified from baculovirus-infected Sf9 cells. Briefly, cell pellets were resuspended in lysis buffer (50 mM Hepes, pH 7.4, 300 mM NaCl, 10% glycerol, 1 mM DTT, and 2 mM PMSF) supplemented with 1× protease inhibitor cocktail (Roche cOmplete Protease Inhibitor Cocktail) and lysed using a Dounce homogenizer (20 strokes with a loose plunger followed by 20 strokes with a tight plunger). The lysate was clarified by centrifugation (65,000 rpm for 45 min) and incubated with IgG Sepharose (GE Healthcare) for 2 h at 12°C, applied to a gravity flow column, and washed extensively with wash buffer (same as lysis buffer without PMSF or protease inhibitor cocktail). Protein labeling was performed by concentrating bead slurry to 5 ml, followed by the addition of 5 nmol of BG-LD555 dye (Lumidyne) and then incubation (1 h at 12°C). The slurry was readded to a gravity flow column and washed extensively in wash buffer. The protein–bead complexes were then treated with TEV protease at 12°C overnight. The mixture was then centrifuged (4,000 rpm for 5 min), and the supernatant was retrieved. The mixture was then concentrated using an Amicon Ultra-0.5 ml 50k spin column (EMD Millipore). The protein concentration was determined using the Nanodrop 1000.
EMSA
Binding reactions were performed by mixing NuMA truncation proteins with 60 ng H1-H3 spacer DNA (Ludérus et al., 1994) followed by incubation on ice for 1 h. Binding buffer consisted of 20 mM Hepes, pH 7.9, 100 mM NaCl, 50 nM KCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 0.1 mg/ml BSA, and 0.1% NP-40. After incubation, binding reactions were then loaded onto a 2% Tris-glycine agarose gel, which was run for 90 min at 90 V. The gel was then poststained with 1× Sybr-Gold (S11494; Thermo Fisher Scientific) for 20 min followed by UV imaging to visualize DNA.
FRAP
NuMA KO RPE1 cells transiently overexpressing NuMA-FL-EGFP, or stably expressing NuMA-Bonsai-EGFP, NuMA-NC-EGFP, or EGFP-Zdk1-NuMA-C, were plated on no. 1.5 glass-bottom 35-mm MatTek dishes coated with poly-L-lysine (MatTek Corporation) for 48 h. Cells were subsequently imaged, and FRAP experiments were performed as described above in the Microscopy section.
NuMA optogenetic control
NuMA KO RPE1 cells, noninduced and stably expressing NuMA-N-PhusionRed-LOV2 and EGFP-Zdk1-NuMA-C, were imaged with a 561-nm laser (1 s interval) to visualize NuMA-N-PhusionRed-LOV2. The 488-nm laser was subsequently turned on (1 s interval) to simultaneously activate the LOV2 domain and visualize EGFP-Zdk1-NuMA-C. The 488-nm laser was turned off after 30 s to end activation of the LOV2 domain, while the 561-nm laser was used to continue imaging the behavior of NuMA-N-PhusionRed-LOV2.
Cell confinement
Cells were plated on no. 1.5 glass-bottom 35-mm MatTek dishes coated with poly-D-lysine (MatTek Corporation). A PDMS-based suction cup device that contains a 10-mm-diameter glass coverslip with PDMS-based pillar structures (3-µm height; 200-µm diameter; spacing 700 µm to center) was generated (Guild et al., 2017; Le Berre et al., 2012). The device was attached to a 1-ml syringe, placed on top of a glass-bottom 35-mm dish, and sealed by pulling the syringe to apply negative pressure. After imaging 0.3-µm-spaced z planes of unconfined cells and initiating live acquisition, additional negative pressure was created to lower the pillared coverslips onto cells. While the pillars are 3 µm high, the “roof” of the device may be <3 µm from the glass bottom as it can deform under negative pressure (Guild et al., 2017). The negative pressure was maintained while z-stacks of compressed cells were acquired. Confinement was applied gradually over a period of ∼1 min and sustained for 5 min.
Data analysis
Nuclear phenotype classification (Figs. 1, 2, and 3)
Observed nuclear phenotypes were qualitatively categorized into (1) single, round (shaped approximately like a sphere, without clear lobules or deep invaginations) nucleus, (2) nucleus with one to three micronuclei, (3) fragmented nucleus (one main nucleus and more than three micronuclei or two or more larger nuclei), and (4) atypical shape nucleus (cells with a single, abnormally shaped nucleus) based on DNA signal (Hoechst or mCherry-H2B). To determine percentages of these phenotypes before and after spindle-less mitosis (Fig. 2 B), the phenotype was analyzed based on mCherry-H2B signal for all cells at t = 0 (∼10 min after nocodazole and reversine addition) and at about t = 16 h.
Nuclear solidity and nuclear fluorescence intensity analysis (Figs. 1, 2, and 3)
A threshold of Hoechst or mCherry-H2B intensity was calculated and used to extract the outline of the nucleus using a particle analysis plugin from ImageJ. Based on this outline, the area and convex hull area were extracted. Solidity was calculated as the ratio of the area of the nucleus to the area of a convex hull (Fig. 1, D and E; and Fig. S1, D and E). NuMA and EGFP-tagged NuMA proteins mean intensities were measured in the nucleus based on a threshold of mCherry-H2B signal, as described above. The mean background intensity from three independent regions was subtracted. For each experiment, all values were normalized to the mean value of control (noninduced) cells (Fig. S1, A and E).
Mother–daughter cell analysis (Fig. 2 E)
Individual cells were followed, and only cells that entered and exited spindle-less mitosis during image acquisition were included in the analysis. Phenotypes were determined before mitotic entry (mother cells) and after mitotic exit (daughter cells) for all cells. Only cells that had a single and round nucleus at mitotic entry were analyzed.
Nuclear expansion analysis (Fig. 2 F)
Control and NuMA KO cells stably expressing mCherry-H2B and EGFP-Lap2β treated with nocodazole and reversine were randomly selected for analysis. Nuclear envelope reformation (NER; t = 0) was determined by the first appearance of EGFP-Lap2β around condensed mitotic chromosomes, and the area of the chromosome mass was calculated for the subsequent 132 min. Area was calculated based on a threshold of mCherry-H2B intensity using a particle analysis plugin from ImageJ. All thresholds were checked and manually corrected when the automated threshold was not accurate before areas were calculated.
FRAP analysis (Fig. 3, D–G; and Fig. S2, E and F)
The mean fluorescence intensity value in the bleached and unbleached area, as well as background, were measured using a custom macro in ImageJ written by Arthur Molines. The signal was corrected for background and photobleaching and normalized 0 to 1, with 0 corresponding to the lowest signal and 1 to the highest signal after photobleaching. Recovery measurements were quantified by fitting normalized fluorescence intensities of bleached areas to a double exponential for EGFP-tagged NuMA constructs using GraphPad Prism. Cells with an average intensity 3.5 SDs above the NuMA-FL-EGFP mean intensity were excluded. The recovery halftimes were calculated from double exponential fits (FL, R2= 0.87; Bonsai, R2 = 0.91; NC, R2 = 0.87).
NuMA-N photo-dissociation analysis (Fig. 4, B and C)
NuMA-N intensity in the nucleus was measured on a 2.2-µm-diameter circular region before, during, and after illumination with a 488-nm laser. The signal was corrected for background and normalized to average intensity.
Colocalization correlation analysis (Fig. S4 A)
The colocalization Pearson’s correlation coefficient was calculated by comparing images of nuclear regions with an area of 33.4 µm2 using the ImageJ plugin Coloc 2.
Cell area analysis (Fig. 5 B)
The area of the cell was estimated by calculating the cell area in the plane where it was largest, based on the transmitted light signal. Because the data in the rest of Fig. 5 did not allow us to view all cells in their entirety, we acquired a separate dataset of unconfined control and NuMA KO cells to image the entire cells, now all fully in the field of view.
Nuclear area analysis (Fig. 5 C)
To calculate the area of the nucleus, a sum projection from a z-stack was generated and the area of the projected nucleus was extracted, based on H2B signal and EGFP-Lap2β signal at the nuclear rim.
Nuclear volume analysis (Fig. 5 D)
3D volumes were measured using ImageJ and the 3D image segmentation plugin LimeSeg (Machado et al., 2019), selecting a region of interest corresponding to the nucleus. The segmentation plugin was used on z-stack images of EGFP-Lap2β with the following parameters: D_0 = 3.0, F_pressure = 0.025, Z_Scale = 3.8, Range_in_D0_units = 2.0, NumberOfIntegrationStep = −1, and RealXYsize = 0.066. The smallest feature size the segmentation could detect with these parameters was 3 pixels large. After each 3D analysis converged, we used the 2D result viewer to verify whether or not the contour found by LimeSeg overlapped with the EGFP-Lap2β outline. When there was an occasional discrepancy, we performed other analyses with multiple circular regions of interest.
Nuclear confinement analysis (Fig. 5, H and I)
To determine nuclear height before and after compression, orthogonal xz and yz views were generated from z-stacks of live cells (0.25-µm step size; 8-µm range), a line was drawn from top to bottom of the nucleus (based on EGFP-Lap2β signal), and its length was determined in ImageJ.
Statistics
All data are expressed as average ± SD or SEM, as indicated in the figures. All statistical analyses were performed using Prism 8 software (GraphPad). Data were tested for normality using the D’Agostino–Pearson normality test when n was not too small. P values were calculated using unpaired t tests and Mann–Whitney U tests as indicated in the figure legends. For the contingency tables of Fig. 1, C, E, and F; Fig. 2, B and C; and Fig. 3 C, the Fisher's exact test was applied. All P values were two-tailed, and we used P < 0.05 as the threshold for statistical significance. Quoted n’s are described in detail in the figure legends, and in general refer to individual measurements (individual cells, nuclei).
Image presentation
Time-lapse images show single planes from spinning disk confocal imaging, except Fig. 2, A and D, which show maximum intensity projections (20 µm total z distance) of spinning disk confocal z-stacks. Immunofluorescence images of nuclei and mitotic spindles (Fig. 1 B; Fig. 3 B; and Fig. S1 B and D) show maximum intensity projections (20–30 µm total z distance) of spinning disk confocal z-stacks or single planes (Fig. S3). All images and videos were formatted using ImageJ.
Video preparation
Videos 1 and 2 show a maximum intensity projection from a spinning disk confocal z-stack. Video 3 shows a single z-slice from an OMX-SR microscope. Videos 4 and 5 show a single z-slice from a spinning disk confocal microscope. Videos were prepared for publication using ImageJ and set to play at 10 (Videos 1, 2, 3, and 5) or 20 frames per second (Video 4).
Online supplemental material
Fig. S1 shows the validation of the RPE1-inducible Cas9 NuMA KO cell line and of the nocodazole/reversine treatment (related to Fig. 1). Fig. S2 shows the validation of the RPE1 NuMA KO line stably expressing NuMA-Bonsai-EGFP and the estimate of NuMA-EGFP intensity in cells used in the FRAP analysis (related to Fig. 3). Fig. S3 shows that NuMA is only detectable on chromosomes after initial Lap2β recruitment (related to Fig. 4). Fig. S4 shows NuMA-N and -C responses in Opto-NuMA in vivo experiments, and purified NuMA truncation proteins used for in vitro EMSAs (related to Fig. 4). Fig. S5 shows that CDK1 inhibition does not affect NuMA binding to chromosomes during mitosis (related to Fig. 4). Video 1 shows that control RPE1 cells enter and exit mitosis with a single and round nucleus (related to Fig. 2). Video 2 shows that nuclear defects arise at nuclear envelope formation as NuMA KO cells exit mitosis (related to Fig. 2). Video 3 shows that overexpressed NuMA-FL-EGFP can form stable cable-like structures in the nucleus (related to Fig. 3). Video 4 shows that NuMA-C has a higher affinity than NuMA-N for chromosomes in the nucleus (related to Fig. 4). Video 5 shows that NuMA’s coiled-coil prevents its C terminus from binding chromosomes at mitosis (related to Fig. 4).
Acknowledgments
We thank Christina Hueschen (Stanford University, Stanford, CA) for inducible RPE1 NuMA KO cells; Daniel Gerlich (Institute of Molecular Biotechnology of the Austrian Academy of Sciences [IMBA], Vienna, Austria) for reagents; Jeffrey van Haren (Erasmus University Medical Center Rotterdam, Netherlands) and Torsten Wittmann (University of California, San Francisco, San Francisco, CA) for LOVTRAP plasmids and discussions; Joël Lemière for help with FRAP and nuclear volume analyses and discussions; Arthur Molines for help with FRAP analysis and discussions; Amanda Jack for advice on DNA binding experiments; Delaine Larson and Kari Herrington at the University of California, San Francisco (UCSF), Nikon Imaging Center for assistance with FRAP experiments; Abby Buchwalter, Duane Compton, Daniel Gerlich, and Megan King for discussions; Abby Buchwalter and Christina Hueschen for critical reading of the manuscript; the Fred Chang laboratory for discussions; and the Dumont laboratory for discussions and critical reading of the manuscript. We thank Sachin Kotak for generously sharing findings prior to publication and for insightful discussions.
The FRAP data for this study were acquired at the UCSF Nikon Imaging Center with instruments obtained by using funding from the National Institutes of Health (5R35GM118119); the UCSF Program for Breakthrough Biomedical Research, funded in part by the Sandler Foundation; the UCSF Research Resource Fund Award; and the Howard Hughes Medical Institute. We acknowledge the UCSF Parnassus Flow Cytometry Core supported in part by National Institutes of Health grant P30 DK063720 and National Institutes of Health S10 Instrumentation Grant S10 1S10OD021822-01 for assistance. This work was funded by National Institutes of Health grants DP2GM119177, R01GM134132 and R35GM136420 (to S. Dumont), the Rita Allen Foundation (to S. Dumont), the National Science Foundation Center for Cellular Construction grant 1548297 (to S. Dumont), National Science Foundation CAREER grant 1554139 (to S. Dumont), National Science Foundation grant MCB-1617028 (to A. Yildiz), and a Human Frontier Science Program Long Term Postdoctoral Fellowship LT001061/2016-L (to A. Serra-Marques).
The authors declare no competing financial interests.
Author contributions: A. Serra-Marques, Conceptualization, Methodology, Software, Validation, Formal Analysis, Investigation, Resources, Data Curation, Writing – Original Draft Preparation, Writing – Review & Editing, Visualization, Supervision, and Funding Acquisition; R. Houtekamer, Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Data Curation, Writing – Review & Editing, and Visualization; D. Hintzen, Conceptualization, Methodology, Validation, Investigation, and Writing – Review & Editing; J.T. Canty, Methodology, Validation, Investigation, Data Curation, Writing – Review & Editing, and Visualization; A. Yildiz, Methodology, Writing – Review & Editing, Supervision, and Funding Acquisition; and S. Dumont, Conceptualization, Methodology, Resources, Writing – Review & Editing, Supervision, and Funding Acquisition.
References
- Abad, P.C., Lewis J., Mian I.S., Knowles D.W., Sturgis J., Badve S., Xie J., and Lelièvre S.A.. 2007. NuMA influences higher order chromatin organization in human mammary epithelium. Mol. Biol. Cell. 18:348–361. 10.1091/mbc.e06-06-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi, M.G., Barale R., Iozzo P., and Picano E.. 2011. The association of micronucleus frequency with obesity, diabetes and cardiovascular disease. Mutagenesis. 26:77–83. 10.1093/mutage/geq077 [DOI] [PubMed] [Google Scholar]
- Barboro, P., D’arrigo C., Diaspro A., Mormino M., Alberti I., Parodi S., Patrone E., and Balbi C.. 2002. Unraveling the Organization of the Internal Nuclear Matrix: RNA-Dependent Anchoring of NuMA to a Lamin Scaffold. Exp. Cell Res. 279:202–218. doi:. 10.1006/excr.2002.5605 [DOI] [PubMed]
- Barboro, P., D’Arrigo C., Mormino M., Coradeghini R., Parodi S., Patrone E., and Balbi C.. 2003. An intranuclear frame for chromatin compartmentalization and higher-order folding. J. Cell. Biochem. 88:113–120. 10.1002/jcb.10378 [DOI] [PubMed] [Google Scholar]
- Buxboim, A., Ivanovska I.L., and Discher D.E.. 2010. Matrix elasticity, cytoskeletal forces and physics of the nucleus: how deeply do cells ‘feel’ outside and in? J. Cell Sci. 123:297–308. 10.1242/jcs.041186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim, A., Irianto J., Swift J., Athirasala A., Shin J.W., Rehfeldt F., and Discher D.E.. 2017. Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Mol. Biol. Cell. 28:3333–3348. 10.1091/mbc.e17-06-0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F.Y., Haley S.C., and Zidovska A.. 2017. On the origin of shape fluctuations of the cell nucleus. Proc. Natl. Acad. Sci. USA. 114:10338–10343. 10.1073/pnas.1702226114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., Cameron L.A., and Salmon E.D.. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155. 10.1016/j.cub.2004.11.029 [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W. 1995. NuMA: a protein involved in nuclear structure, spindle assembly, and nuclear re-formation. Trends Cell Biol. 5:60–64. 10.1016/S0962-8924(00)88947-3 [DOI] [PubMed] [Google Scholar]
- Compton, D.A., and Cleveland D.W.. 1993. NuMA is required for the proper completion of mitosis. J. Cell Biol. 120:947–957. 10.1083/jcb.120.4.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A., and Luo C.. 1995. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J. Cell Sci. 108:621–633. [DOI] [PubMed] [Google Scholar]
- Compton, D.A., Szilak I., and Cleveland D.W.. 1992. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 116:1395–1408. 10.1083/jcb.116.6.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta, K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., and Pellman D.. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, K.N., Ribeiro A.J.S., and Lammerding J.. 2008. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102:1307–1318. 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, P.M., and Lammerding J.. 2014. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol. 24:247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, C.L., Malaby H.L.H., Sepaniac L.A., Martin W., Byers C., Czechanski A., Messinger D., Tang M., Ohi R., Reinholdt L.G., et al. 2019. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J. Cell Biol. 218:1148–1163. 10.1083/jcb.201807228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth, S., Hsia K.C., Shimamoto Y., and Kapoor T.M.. 2014. Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell. 157:420–432. 10.1016/j.cell.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa, T., Rochman M., Taher L., Dimitriadis E.K., Nagashima K., Anderson S., and Bustin M.. 2015. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 6:6138. 10.1038/ncomms7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet, C., Wang J., Harborth J., Weber K., and Osborn M.. 1998. Induction of a regular nuclear lattice by overexpression of NuMA. Exp. Cell Res. 243:434–452. 10.1006/excr.1998.4178 [DOI] [PubMed] [Google Scholar]
- Guild, J., Ginzberg M.B., Hueschen C.L., Mitchison T.J., and Dumont S.. 2017. Increased lateral microtubule contact at the cell cortex is sufficient to drive mammalian spindle elongation. Mol. Biol. Cell. 28:1975–1983. 10.1091/mbc.e17-03-0171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth, J., Weber K., and Osborn M.. 1995. Epitope mapping and direct visualization of the parallel, in-register arrangement of the double-stranded coiled-coil in the NuMA protein. EMBO J. 14:2447–2460. 10.1002/j.1460-2075.1995.tb07242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth, J., Wang J., Gueth-Hallonet C., Weber K., and Osborn M.. 1999. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18:1689–1700. 10.1093/emboj/18.6.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Gnadt N., Wright M., and Merdes A.. 2009. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res. Notes. 2:64. 10.1186/1756-0500-2-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, E.M., Fischer A.H., Deerinck T.J., and Hetzer M.W.. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154:47–60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueschen, C.L., Kenny S.J., Xu K., and Dumont S.. 2017. NuMA Targets Dynein to Microtubule Minus-Ends at Mitosis. eLife. 6. e29328. 10.1101/148692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueschen, C.L., Galstyan V., Amouzgar M., Phillips R., and Dumont S.. 2019. Microtubule End-Clustering Maintains a Steady-State Spindle Shape. Curr. Biol. 29:700–708.e5. 10.1016/j.cub.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., and Mitchison T.. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485. 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Jayaraman, S., Chittiboyina S., Bai Y., Abad P.C., Vidi P.-A., Stauffacher C.V., and Lelièvre S.A.. 2017. The nuclear mitotic apparatus protein NuMA controls rDNA transcription and mediates the nucleolar stress response in a p53-independent manner. Nucleic Acids Res. 45:11725–11742. 10.1093/nar/gkx782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki, M., Weber K., and Osborn M.. 1991. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO J. 10:3351–3362. 10.1002/j.1460-2075.1991.tb04899.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki, M., Harborth J., Weber K., and Osborn M.. 1993. Microinjection of a monoclonal antibody against SPN antigen, now identified by peptide sequences as the NuMA protein, induces micronuclei in PtK2 cells. J. Cell Sci. 104:139–150. [DOI] [PubMed] [Google Scholar]
- Kivinen, K., Taimen P., and Kallajoki M.. 2010. Silencing of Nuclear Mitotic Apparatus protein (NuMA) accelerates the apoptotic disintegration of the nucleus. Apoptosis. 15:936–945. 10.1007/s10495-010-0506-8 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu, T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu, T., and Cheeseman I.M.. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 154:391–402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, D.W., Sudar D., Bator-Kelly C., Bissell M.J., and Lelièvre S.A.. 2006. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proc. Natl. Acad. Sci. USA. 103:4445–4450. 10.1073/pnas.0509944102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak, S., Busso C., and Gönczy P.. 2013. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32:2517–2529. 10.1038/emboj.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJoie, D., and Ullman K.S.. 2017. Coordinated events of nuclear assembly. Curr. Opin. Cell Biol. 46:39–45. 10.1016/j.ceb.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Lammerding, J., Hsiao J., Schulze P.C., Kozlov S., Stewart C.L., and Lee R.T.. 2005. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 170:781–791. 10.1083/jcb.200502148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre, M., Aubertin J., and Piel M.. 2012. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol. 4:1406–1414. 10.1039/c2ib20056b [DOI] [PubMed] [Google Scholar]
- Li, Y., Lovett D., Zhang Q., Neelam S., Kuchibhotla R.A., Zhu R., Gundersen G.G., Lele T.P., and Dickinson R.B.. 2015. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys. J. 109:670–686. 10.1016/j.bpj.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., and Pellman D.. 2020. The coordination of nuclear envelope assembly and chromosome segregation in metazoans. Nucleus. 11:35–52. 10.1080/19491034.2020.1742064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Kwon M., Mannino M., Yang N., Renda F., Khodjakov A., and Pellman D.. 2018. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 561:551–555. 10.1038/s41586-018-0534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Lu Z., Liu Q., Guo L., Ren H., Fu J., Jiang Q., Clarke P.R., and Zhang C.. 2012. Chromatin-bound NLS proteins recruit membrane vesicles and nucleoporins for nuclear envelope assembly via importin-α/β. Cell Res. 22:1562–1575. 10.1038/cr.2012.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus, M.E., den Blaauwen J.L., de Smit O.J., Compton D.A., and van Driel R.. 1994. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 14:6297–6305. 10.1128/MCB.14.9.6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly, P., Brunner S.F., Shoshani O., Kim D.H., Lan W., Pyntikova T., Flanagan A.M., Behjati S., Page D.C., Campbell P.J., et al. 2019. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 51:705–715. 10.1038/s41588-019-0360-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydersen, B.K., and Pettijohn D.E.. 1980. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 22:489–499. 10.1016/0092-8674(80)90359-1 [DOI] [PubMed] [Google Scholar]
- Machado, S., Mercier V., and Chiaruttini N.. 2019. LimeSeg: a coarse-grained lipid membrane simulation for 3D image segmentation. BMC Bioinformatics. 20:2. 10.1186/s12859-018-2471-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., and Cleveland D.W.. 1998. The role of NuMA in the interphase nucleus. J. Cell Sci. 111:71–79. [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez, F., Gerlich D., and Ellenberg J.. 2007. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9:822–831. 10.1038/ncb1606 [DOI] [PubMed] [Google Scholar]
- Okumura, M., Natsume T., Kanemaki M.T., and Kiyomitsu T.. 2018. Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife. 7. e36559. 10.7554/eLife.36559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemble, H., Kumar P., van Haren J., and Wittmann T.. 2017. GSK3-mediated CLASP2 phosphorylation modulates kinetochore dynamics. J. Cell Sci. 130:1404–1412. 10.1242/jcs.194662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu, A.E., and Cleveland D.W.. 2010. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20:214–222. 10.1016/j.tcb.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Redwine W.B., Vale R.D., and Carter A.P.. 2018. The microtubule cytoskeleton is responsible for long-distance movements and spatial organization of intra-cellular vesicles, organelles and large protein-containing and RNA-containing complexes in many eukaryotic cells. Nat. Rev. Mol. Cell Biol. 19:1. 10.1038/s41580-018-0004-3 [DOI] [PubMed] [Google Scholar]
- Salvador Moreno, N., Liu J., Haas K.M., Parker L.L., Chakraborty C., Kron S.J., Hodges K., Miller L.D., Langefeld C., Robinson P.J., et al. 2019. The nuclear structural protein NuMA is a negative regulator of 53BP1 in DNA double-strand break repair. Nucleic Acids Res. 47:2703–2715. 10.1093/nar/gkz138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer, M., Schneider M.W.G., Hoefler R., Schmalhorst P.S., Jude J.G., Zuber J., and Gerlich D.W.. 2017. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell. 170:956–972.e23. 10.1016/j.cell.2017.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saredi, A., Howard L., and Compton D.A.. 1996. NuMA assembles into an extensive filamentous structure when expressed in the cell cytoplasm. J. Cell Sci. 109:619–630. [DOI] [PubMed] [Google Scholar]
- Schooley, A., Vollmer B., and Antonin W.. 2012. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma. 121:539–554. 10.1007/s00412-012-0388-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner, S.M., Koo P.K., Zhao Y., Mochrie S.G.J., and King M.C.. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6:7159. 10.1038/ncomms8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk, A.D., Holland A.J., and Cleveland D.W.. 2009. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184:677–690. 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, B.M., and Johnson E.E.P.. 2017. Nuclear morphologies: their diversity and functional relevance. Chromosoma. 126:195–212. 10.1007/s00412-016-0614-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A.D., Banigan E.J., Adam S.A., Goldman R.D., and Marko J.F.. 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 28:1984–1996. 10.1091/mbc.e16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A.D., Banigan E.J., and Marko J.F.. 2019. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 58:76–84. 10.1016/j.ceb.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, T.K., Tang C.J., Chao Y.J., and Wu C.W.. 1994. Nuclear mitotic apparatus protein (NuMA): spindle association, nuclear targeting and differential subcellular localization of various NuMA isoforms. J. Cell Sci. 107:1389–1402. [DOI] [PubMed] [Google Scholar]
- Thomas, C.H., Collier J.H., Sfeir C.S., and Healy K.E.. 2002. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA. 99:1972–1977. 10.1073/pnas.032668799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.L., and Compton D.A.. 2011. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. USA. 108:17974–17978. 10.1073/pnas.1109720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht, R., and Kutay U.. 2017. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18:229–245. 10.1038/nrm.2016.153 [DOI] [PubMed] [Google Scholar]
- van Haren, J., Charafeddine R.A., Ettinger A., Wang H., Hahn K.M., and Wittmann T.. 2018. Local control of intracellular microtubule dynamics by EB1 photodissociation. Nat. Cell Biol. 20:252–261. 10.1038/s41556-017-0028-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness, J., and Pettijohn D.E.. 1983. Specific attachment of nuclear-mitotic apparatus protein to metaphase chromosomes and mitotic spindle poles: possible function in nuclear reassembly. J. Mol. Biol. 171:175–205. 10.1016/S0022-2836(83)80352-0 [DOI] [PubMed] [Google Scholar]
- Vega, S.L., Liu E., Arvind V., Bushman J., Sung H.-J., Becker M.L., Lelièvre S., Kohn J., Vidi P.-A., and Moghe P.V.. 2017. High-content image informatics of the structural nuclear protein NuMA parses trajectories for stem/progenitor cell lineages and oncogenic transformation. Exp. Cell Res. 351:11–23. 10.1016/j.yexcr.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidi, P.-A., Liu J., Salles D., Jayaraman S., Dorfman G., Gray M., Abad P., Moghe P.V., Irudayaraj J.M., Wiesmüller L., et al. 2014. NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 42:6365–6379. 10.1093/nar/gku296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Vilela M., Winkler A., Tarnawski M., Schlichting I., Yumerefendi H., Kuhlman B., Liu R., Danuser G., and Hahn K.M.. 2016. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat. Methods. 13:755–758. 10.1038/nmeth.3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, M., Witkin K.L., and Cohen-Fix O.. 2009. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122:1477–1486. 10.1242/jcs.037333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, J.P.I., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., J.R. Yates, III, and Cheeseman I.M.. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.H., and Snyder M.. 1992. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell. 3:1259–1267. 10.1091/mbc.3.11.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.H., Lambie E.J., and Snyder M.. 1992. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 116:1303–1317. 10.1083/jcb.116.6.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C., He D., and Brinkley B.R.. 1994. Localization of NuMA protein isoforms in the nuclear matrix of mammalian cells. Cell Motil. Cytoskeleton. 29:167–176. 10.1002/cm.970290208 [DOI] [PubMed] [Google Scholar]
- Zhang, C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., and Pellman D.. 2015. Chromothripsis from DNA damage in micronuclei. Nature. 522:179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]