Abstract
The MITF(E318K) variant confers moderate risk for cutaneous melanoma. While there are small studies suggesting that this risk is associated with other malignancies (e.g. renal cell carcinoma), little is known about the role of this variant in specifying risk for other cancers. In this study, we perform a systematic review and meta-analysis of the published data as a backdrop to a whole-exome sequence(WES)-based characterization of MITF(E318K) risk for various cancers in sporadic samples from the TCGA and several genetically-enriched patient cohorts. We found minimal evidence of MITF(E318K)’s contribution to non-melanoma cancer risk among individuals with low inherited risks of melanoma (OR 1.168; 95% CI 0.78–1.74; p = 0.454), suggesting that earlier reports of an association between this variant and other malignancies may be related to shared environmental or polygenic risk factors rather than MITF(E318K). Interestingly, an association was observed with uterine carcinosarcoma, (OR 9.24; 95% CI 2.08–37.17; p = 0.024), which was not previously described. While more research needs to be completed, this study will help update cancer screening recommendations for patients with the MITF(E318K) variant.
Subject terms: Cancer genetics, Skin cancer, Genetics
Introduction
The microphthalmia-associated transcription factor (MITF) is a known master regulator of melanocyte development with functions ranging from pigment production to differentiation, survival, and cell-cycle production of melanocytes. A rare functional variant of MITF(E318K) has been shown to disrupt a conserved SUMOylation site and to confer a two- to fourfold risk for cutaneous melanoma1,2. Moreover, there have been small studies suggesting that this variant is also associated with other cancers such as renal cell carcinoma (RCC)1–13. However, since epidemiologic observations have documented an association between cutaneous melanoma and many other malignancies, including RCC, it is possible that MITF(E318K) represents a germline “passenger” mutation in these other cancers without directly impacting the risk of these other cancers such as RCC. In this study we perform a systematic review and meta-analysis of the published data as a backdrop to a whole-exome sequence (WES)-based characterization of MITF(E318K) risk for various cancers in sporadic populations from the TCGA database and genetically-enriched patient populations.
Results
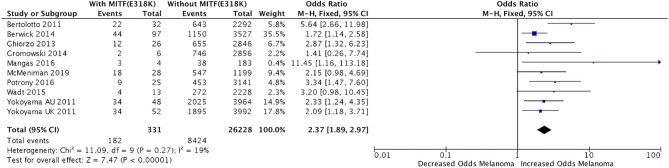
Meta-analysis of the data compiled from nine published studies (n = 331)1–10 demonstrated that the variant was significantly correlated with melanoma (Fig. 1; odds ratio (OR) 2.37; 95% confidence interval (CI) 1.89–2.97; p < 1E−5; I2=19%). Studies were conducted on a mixture of hereditary and sporadic melanoma populations. A few studies identified a relationship of this variant with renal cell carcinoma (RCC), pancreatic cancer, and pheochromocytoma/paraganglioma2,4,10. Bertolotto et al. identified over a fivefold increased risk for carriers to develop RCC, melanoma, or both cancers in 829 patients (OR 5.55; 95% CI 2.59–12.91; p = 1.2E−6) and identified a fivefold risk of developing RCC only in 164 “genetically-enriched” patients who were wild-type for RCC-predisposing genes (OR 5.19; 95% CI 1.37–16.87; p = 0.008)2. In contrast, other studies did not find an association with sporadic RCC5,12,13. Ghiorzo et al. identified a 31-fold increased risk of melanoma in carriers with a personal or family history of pancreatic cancer (OR 30.85; 95% CI 6.85–138.9; p = 0.0005), but did not find significant enrichment of the variant in sporadic pancreatic cancer patients (0/210 patients had the variant)4. Castro-Vega et al. screened 555 unrelated pheochromocytoma/paraganglioma patients, and found that the variant was enriched compared to controls (OR 3.19; 95% CI 1.34–7.59; p = 0.005)11.
Figure 1.
Meta-analysis Odds Ratio of the Association Between MITF(E318K) and Personal History of Melanoma. 1Ozola et al. also studied relationship of MITF(E318K) with melanoma, but found no variants in cases or controls and thus was not included in calculations10.
To better understand the risks of this rare functional polymorphism on other cancers, we then systematically evaluated prevalence of the MITF(E318K) variant in multiple cancer cohorts using germline whole-exome sequence data from the TCGA panel and from several genetically-enriched cohorts to validate these associations (Table 1). The TCGA cohort consists of patients mostly with late-onset cancer and unascertained family history, and therefore their risk of comorbid cancers is theoretically low. This set up enables evaluation of primary cancer risk conferred by MITF(E318K). Since the variant is mainly found in the European population (minor allele frequency (MAF) in gnomAD14 = 0.00245 for European; MAF = 6.01E−04 for African; MAF = 0 for Asian), we performed the analysis using only European patients identified through principal component analysis (PCA). We used a set of common (MAF > 0.01) LD-pruned variants to perform PCA and used k-means to identify European cluster of samples. The final dataset contained European 6143 cancer cases and 11,106 European non-cancer controls. Comparison of MITF(E318K) frequency in the control sample (MAF = 0.28%, Table 1) matched well with the frequency expectation for Europeans (MAF in gnomAD Europeans = 0.25%), suggesting that our control cohort provides a reference point for MAF free from ancestral bias.
Table 1.
Relationship of MITF(E318K) with different cancer types in TCGA and genetically-enriched patient cohorts.
| Cohort | # MITF(E318K) carriers | N total samples | MAF | Fisher OR (95% CI) | p-value |
|---|---|---|---|---|---|
| Control | 63 | 11,106 | 0.00284 | Ref | Ref |
| TCGA thyroid carcinoma | 1 | 363 | 0.00138 | 0.48 (0.05–2.57) | 0.724 |
| TCGA glioblastoma | 1 | 326 | 0.00153 | 0.54 (0.05–2.86) | 1.000 |
| TCGA breast carcinoma | 3 | 776 | 0.00193 | 0.68 (0.22–1.96) | 0.800 |
| TCGA lung adenocarcinoma | 2 | 501 | 0.00200 | 0.70 (0.17–2.63) | 1.000 |
| TCGA low grade glioma | 2 | 447 | 0.00224 | 0.79 (0.19–2.96) | 1.000 |
| TCGA bladder carcinoma | 1 | 209 | 0.00239 | 0.84 (0.08–4.50) | 1.000 |
| TCGA lung SCC | 2 | 391 | 0.00256 | 0.90 (0.22–3.39) | 1.000 |
| TCGA/enriched breast cancer | 6 | 976 | 0.00307 | 1.08 (0.50–2.39) | 0.823 |
| TCGA prostate adenocarcinoma | 1 | 226 | 0.00221 | 1.13(0.11–6.55) | 0.597 |
| TCGA pheochromocytoma/paraganglioma | 1 | 145 | 0.00345 | 1.22 (0.12–6.55) | 0.565 |
| TCGA liver carcinoma | 1 | 125 | 0.00400 | 1.41 (0.14–7.64) | 0.512 |
| TCGA kidney/papillary clear cell carcinoma | 1 | 123 | 0.00407 | 1.44 (0.14–7.77) | 0.507 |
| TCGA cervical SCC/endocervical adenocarcinoma | 1 | 114 | 0.00439 | 1.51 (0.14–8.24) | 0.496 |
| TCGA pancreatic adenocarcinoma | 1 | 116 | 0.00431 | 1.52 (0.15–8.26) | 0.487 |
| TCGA rectum adenocarcinoma | 1 | 113 | 0.00442 | 1.57 (0.15–8.49) | 0.478 |
| TCGA head/neck SCC | 4 | 436 | 0.00459 | 1.62 (0.63–4.23) | 0.322 |
| TCGA sarcoma | 1 | 102 | 0.00490 | 1.74 (0.17–9.44) | 0.444 |
| TCGA colon adenocarcinoma | 3 | 300 | 0.00500 | 1.77 (0.58–5.15) | 0.251 |
| TCGA uterine corpus endometrial carcinoma | 4 | 385 | 0.00519 | 1.79 (0.66–4.83) | 0.295 |
| MGH genetically-enriched cutaneous melanoma (GECM) | 3 | 250 | 0.00600 | 2.13 (0.69–6.22) | 0.178 |
| TCGA cutaneous melanoma | 5 | 409 | 0.00611 | 2.17 (0.93–5.29) | 0.094 |
| TCGA cutaneous melanoma + MGH GECM | 8 | 659 | 0.00607 | 2.15 (1.03–4.37) | 0.061 |
| MGH genetically-enriched breast cancer | 3 | 200 | 0.00750 | 2.67 (0.87–7.83) | 0.111 |
| TCGA ocular melanoma | 1 | 47 | 0.01064 | 3.81 (0.37–21.69) | 0.237 |
| TCGA uterine carcinosarcoma | 2 | 39 | 0.02564 | 9.24 (2.08–37.17) | 0.024 |
1Cancers whose lower boundary of 95% CI is greater than 1.0 are highlighted in Italics.
The risk of developing any cancer in aggregate with the MITF(E318K) mutation was slightly increased, but this did not reach significance (OR 1.294; 95% CI 0.88–1.88; p = 0.19). Similarly, the risk for all non-melanoma cancers was also increased but not significantly (OR 1.168; 95% CI 0.78–1.74; p = 0.454). Among the 25 cancers tested, uterine carcinosarcoma (OR 9.24; 95% CI 2.08–37.17; p = 0.024) and melanoma (OR 2.15; 95% CI 1.03–4.37; p = 0.061) exhibited the strongest associations with the variant. We did not find any significant association of MITF(E318K) with renal cell carcinoma (RCC) (p = 0.5068), pancreatic cancer (p = 0.487), or pheochromocytoma/paraganglioma (p = 0.565) in the TCGA cohort.
Further, we tested whether differences in ascertainment between cancer cohorts could explain the lack of the previously reported association with RCC. Specifically, it is expected that the burden of germline risk variants should be lower in cohorts with unascertained family history (TCGA) than in genetically-enriched samples. We compared allele frequencies of MITF (E318K) for the familial (MAF = 0.6%) and sporadic (MAF = 0.6%) melanoma cohorts and did not observe any difference, suggesting that the sporadic cohort should be powerful enough to detect primary cancer risks associated with MITF(E318K).
Therefore, in our analysis of RCC, pancreatic cancer, pheochromocytoma/paraganglioma cohorts, there does not appear to be a strong risk of primary cancer conferred by MITF(E318K). Importantly, TCGA non-melanoma cohorts are unlikely to have substantial inherited risk of cutaneous melanoma due to the relatively high age of participants and no ascertainment, resulting in the lack of an association signal for MITF(E318K). Previous studies reporting the non-melanoma cancer association of MITF(E318K) have not yet assessed inherited melanoma risks for the RCC and other cancer cohorts.
Discussion
This study is an important addition to the published literature as it represents the largest MITF(E318K) variant analysis to date with more samples collectively between cases and controls than found in previously published studies. In addition, with WES, we utilize principal component analysis (PCA) to match for European ancestry which allows for a much cleaner calculation of risk compared to most other studies that relied solely on MITF(E318K) counts without regard for population mix. Although we corroborated the association of the variant with melanoma, we found minimal evidence of MITF(E318K)’s contribution to the risk of non-melanoma cancers among individuals with low inherited risks of melanoma.
These results suggest that earlier reports of an association between this variant and other malignancies may be related to shared environmental or polygenic risk factors rather than this specific MITF polymorphism. For example, multiple studies have shown an association between RCC and melanoma. In a 2018 analysis of the Surveillance, Epidemiology, and End Results (SEER) database, the Standardized Incidence Ratio (SIR) for developing secondary primary melanoma among RCC patients was 2.31, and the SIR for developing secondary primary RCC among melanoma patients was 2.8715. In previous MITF(E318K) studies, inclusion of high melanoma risk patients with a primary manifestation of RCC or other cancers could have driven increased frequency of MITF(E318K) in observed non-melanoma cohorts. Common putative risk genes for RCC and melanoma – BAP1, MITF, CDKN2B – suggest that similar pathways are disrupted in both disorders; however, individual variant risks should be evaluated when shared genetic background is taken into account. For example, Christensen et al. investigated 48 families with early onset RCC, a family history of RCC, a family history of RCC and melanoma, or both RCC and melanoma diagnosis in the same individual. MITF(E318K) was found only in a RCC-affected member of a family with multiple melanomas. Consequently, authors concluded that BAP1, MITF or CDKN2B are not frequent causes of hereditary renal cancer12.
This common polygenic background and shared environmental factors likely contributed, at least in part, to Bertolotto et al.’s observation of an enrichment of the variant in RCC and melanoma cases2. While this study did report an association with RCC-only, the cohort was young, had family history of RCC, and had rare histological subtypes, unlike the TCGA patients. Thus, MITF(E318K) may play a role in driving cancer formation in a subset of “genetically-enriched” RCC patients by a still undisclosed biological mechanism. Of note, several other researchers have also failed to identify an association of sporadic RCC with MITF(E318K)5,12,13. This may also explain Ghiorzo et al.’s findings, as their association was only noted in melanoma patients with a personal or family of pancreatic cancer, and not in patients with sporadic pancreatic cancer. The underpinnings of variance regarding pheochromocytoma/paraganglioma is unclear, but may be due to the difference in age between our and Castro-Vega et al.’s cohorts.
Beyond cutaneous melanoma, our study also found a putative risk association between the germline MITF(E318K) and uterine carcinosarcoma (malignant mixed Müllerian tumors) although the low frequency of the variant and, expectedly, low numbers of variant carriers, preclude us from rigorously adjusting for multiple hypotheses. Interestingly, uterine carcinosarcomas (mixed Mullerian tumors) have been reported to exhibit melanocytic differentiation, and increased MITF expression16,17. While more research needs to be completed, this study will help update cancer screening recommendations for patients with the MITF(E318K) variant.
A limitation of this study is the relatively small sample size of some of the TCGA cohorts. This, along with the rarity of the MITF variant, contributes to the limited power of analysis. Nevertheless, it may be worthwhile to better characterize the relationship of MITF(E318K) with different cancer types in a larger study with explicit evaluation of inherited polygenic predisposition to melanoma, and to look for biology of how MITF(E318K) mediates risks for co-occurrence of other cancers with melanoma.
Methods
Literature review and meta-analysis
We searched MEDLINE and Harvard HOLLIS from database inception to February 25, 2020 by using various combinations of the terms “melanoma”, “MITF”, “E318K”, and “cancer”. Studies without non-cancer controls were excluded. No pertinent non-English articles were found. Meta-analysis was conducted using the Review Manager 5.3 software, published by the Cochrane community. All variables were considered dichotomous, and analyzed using the Mantel–Haenszel statistical method and fixed effect analytic method. Heterogeneity was assessed using the Chi2 test and I2 statistic. See Supplementary File S1 for full methods and excluded studies.
TCGA and enriched cohorts
Datasets of germline cancer (TCGA sporadic and MGH genetically-enriched cases) and control exome sequences are described in previous publications from our lab18,19. Details of enriched patient cohorts can be found in Supplementary Methods S1. All datasets can be accessed through dbGAP using the following accession numbers: phs000178.v1.p1, phs000823.v1, phs000822.v1.p1, phs000806.v1.p1, and phs000814.v1.p1.
Aggregated set of samples was used for joint variant calling using Picard/GATK pipeline. Principal component analysis (PCA) using common LD-pruned autosomal variants was used to identify a cluster of European samples (k-means). Individual genotypes for the MITF(E318K) variant were filtered. Only genotypes with depth of coverage more than 10X and genotype quality more than 20 × were used for analysis and all cohorts had the target variant called in more than 95% of the samples. Fisher exact test (two-sided) was used to test association with a specific cancer type. Odds ratio and 95% confidence interval was calculated using Graph Prism using the Baptista-Pike method. MAF was calculated by MITF(E318K) # Minor Alleles/# Total Samples/2.
Supplementary information
Acknowledgements
We thank the staff of the Wellman Center for Photomedicine at Massachusetts General Hospital and the Broad Institute. We also appreciate the generosity and support of the patients who made this project possible.
Author contributions
S.G. and H.T. conducted the systematic review and meta-analysis. M.A., S.G., and H.T. conducted the TCGA analysis. A.S., K.S., L.E., M.A., S.M., C.N., S.G., and H.T. prepared and analyzed the genetically-enriched data. All authors prepared and edited the manuscript. S.G. submitted the manuscript. S.G. and H.T. serve as guarantors.
Data availability
The TCGA datasets were generated using the data publically available at https://portal.gdc.cancer.gov. The genetically-enriched datasets can be accessed through dbGAP using the following accession numbers: phs000178.v1.p1, phs000823.v1, phs000822.v1.p1, phs000806.v1.p1, and phs000814.v1.p1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74237-z.
References
- 1.Yokoyama S, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotto C, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 3.Berwick M, et al. MITF E318K's effect on melanoma risk independent of, but modified by, other risk factors. Pigment Cell Melanoma Res. 2014;27:485–488. doi: 10.1111/pcmr.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghiorzo P, et al. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res. 2013;26:259–262. doi: 10.1111/pcmr.12047. [DOI] [PubMed] [Google Scholar]
- 5.Gromowski T, et al. Prevalence of the E318K and V320I MITF germline mutations in Polish cancer patients and multiorgan cancer risk: a population-based study. Cancer Genet. 2014;207:128–132. doi: 10.1016/j.cancergen.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Mangas C, et al. Genetic susceptibility to cutaneous melanoma in southern Switzerland: role of CDKN2A, MC1R and MITF. Br. J. Dermatol. 2016;175:1030–1037. doi: 10.1111/bjd.14897. [DOI] [PubMed] [Google Scholar]
- 7.McMeniman EK, et al. The interplay of sun damage and genetic risk in Australian multiple and single primary melanoma cases and controls. Br. J. Dermatol. 2019 doi: 10.1111/bjd.18777. [DOI] [PubMed] [Google Scholar]
- 8.Potrony M, et al. Prevalence of MITF p. E318K in patients with melanoma independent of the presence of CDKN2A causative mutations. JAMA Dermatol. 2016;152:405–412. doi: 10.1001/jamadermatol.2015.4356. [DOI] [PubMed] [Google Scholar]
- 9.Wadt KA, et al. Molecular characterization of melanoma cases in Denmark suspected of genetic predisposition. PLoS ONE. 2015;10:e0122662. doi: 10.1371/journal.pone.0122662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozola A, Pjanova D. The lack of E318K MITF germline mutation in Latvian melanoma patients. Cancer Genet. 2015;208:355–356. doi: 10.1016/j.cancergen.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Castro-Vega LJ, et al. The MITF, p. E318K variant, as a risk factor for pheochromocytoma and paraganglioma. J. Clin. Endocrinol. Metab. 2016;101:4764–4768. doi: 10.1210/jc.2016-2103. [DOI] [PubMed] [Google Scholar]
- 12.Christensen MB, et al. Exploring the hereditary background of renal cancer in Denmark. PLoS ONE. 2019;14:e0215725. doi: 10.1371/journal.pone.0215725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoehr CG, et al. The microphthalmia-associated transcription factor p. E318K mutation does not play a major role in sporadic renal cell tumors from caucasian patients. Pathobiology. 2016;83:165–169. doi: 10.1159/000443311. [DOI] [PubMed] [Google Scholar]
- 14.Karczewski KJ, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia K, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England E, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, The Genome Aggregation Database Consortium, Neale BM, Daly MJ, MacArthur DJ Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019;1:1. doi: 10.1101/531210. [DOI] [Google Scholar]
- 15.Kim K, et al. Association between melanoma and renal-cell carcinoma for sequential diagnoses: a single-center retrospective study. Cancer Epidemiol. 2018;57:80–84. doi: 10.1016/j.canep.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Granter SR, Weilbaecher KN, Quigley C, Fisher DE. Role for microphthalmia transcription factor in the diagnosis of metastatic malignant melanoma. Appl. Immunohistochem. Mol. Morphol. 2002;10:47–51. doi: 10.1097/00129039-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Roma AA, Malpica A, Deavers MT. Malignant melanoma arising in an ovarian carcinosarcoma: case report and review of the literature. Int. J. Gynecol. Pathol. 2011;30:158–162. doi: 10.1097/PGP.0b013e3181f9eba5. [DOI] [PubMed] [Google Scholar]
- 18.Artomov M, et al. Case-control analysis identifies shared properties of rare germline variation in cancer predisposing genes. Eur. J. Hum. Genet. 2019;27:824–828. doi: 10.1038/s41431-019-0346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Artomov M, et al. Rare variant, gene-based association study of hereditary melanoma using whole-exome sequencing. J. Natl. Cancer Inst. 2017;109:1. doi: 10.1093/jnci/djx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA datasets were generated using the data publically available at https://portal.gdc.cancer.gov. The genetically-enriched datasets can be accessed through dbGAP using the following accession numbers: phs000178.v1.p1, phs000823.v1, phs000822.v1.p1, phs000806.v1.p1, and phs000814.v1.p1.