Abstract
We present manually curated transcriptomics data of psoriasis and atopic dermatitis patients retrieved from the NCBI Gene Expression Omnibus and EBI ArrayExpress repositories. We collected 39 transcriptomics datasets, deriving from DNA microarrays and RNA-Sequencing technologies, for a total of 1677 samples. We provide quality-checked, homogenised and preprocessed gene expression matrices and their corresponding metadata tables along with the estimated surrogate variables. These data represent a ready-made valuable source of knowledge for translational researchers in the dermatology field.
Subject terms: Psoriasis, Atopic dermatitis
| Measurement(s) | transcriptomic data • Psoriasiform dermatitis • atopic dermatitis |
| Technology Type(s) | digital curation |
| Factor Type(s) | dermatitis type • technology type |
| Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12967916
Background & Summary
Psoriasis (PSO) and Atopic dermatitis (AD) are among the most common inflammatory skin disorders associated with immunologic impairment. While the first signs of AD tend to appear in the early childhood, the manifestation of PSO is most common during the third decade of life1. Both the diseases have a substantial negative impact on the quality of life of affected patients. Although a number of therapeutic approaches have been developed in the last two decades to mitigate PSO and AD symptoms, their pathophysiology is still not completely understood2,3. AD is believed to be driven by epidermal barrier disruption, activation of specific T-cell subsets, and dysbiosis of the commensal skin microbiome2 while psoriatic inflammation is sustained by uncontrolled responses of the innate and adaptive cutaneous immune system, which lead to intense keratinocyte proliferation and dysfunctional differentiation4.
Transcriptomics technologies, such as DNA microarray and RNA Sequencing (RNA-Seq), have been used to characterise the molecular alterations of human diseases5, including PSO and AD. To date, only marginal efforts have been carried out in order to collect, quality-check and harmonize PSO- and AD-related transcriptomics data in order to make them easily reusable by the research community. Therefore, the motivation behind this study was to create a source of ready-to-use data of gene expression profiles of PSO and AD patients derived from both DNA microarray and RNA-Seq publicly available datasets.
The preprocessed and harmonized microarray data provided in this study were collected from the NCBI Gene Expression Omnibus (GEO) and EBI ArrayExpress public repositories, while the RNA-Seq datasets were retrieved from the European Nucleotide Archive (ENA). Overall, 26 microarrays datasets were collected, for a total of 991 samples, 632 of which from patients affected by psoriasis and 70 by atopic dermatitis. Some of the microarray datasets contain samples collected from patients affected by other skin diseases such as psoriatic arthritis, psoriasis sebaceous hyperplasia, palmoplantar pustulosis, lichen planus and discoid lupus. These datasets were generated with commercially available Affymetrix and Agilent platforms. All of the analytical steps performed in this work were carried out through the use of the eUtopia software6. We also retrieved 13 RNA-Seq datasets, for a total of 686 samples, 392 of which from patients affected by psoriasis and 94 by atopic dermatitis. RNA-seq data were mostly produced through Illumina platforms, while a minority of datasets were produced through other platforms. All the datasets underwent meta-data curation and harmonisation, data quality check and preprocessing with standardised procedures. The curation and harmonisation of the meta-data consisted in the definition and usage of a common data model for all of the collected datasets. The data models, to which the raw meta-data were mapped to, are reported in the data dictionary files (enclosed with the preprocessed data). The data dictionary describes all the variables reported in the final metadata tables. For each variable, the description, type and allowed values are reported. At the same time, this work is aimed at homogenising the preprocessing procedures in order to improve the comparability of the gene expression data across different studies and platforms. Therefore, in this work we provide meta-data tables, along with the inferred surrogate batch variables, as well as the preprocessed gene expression estimates.
Our analysis significantly increases the FAIRness7 of publicly available PSO and AD transcriptomics data and represents a valuable “ready-to-use” resource available to the scientific community.
Methods
Microarray data
Data collection and homogenization
Transcriptomics data generated by DNA microarrays of psoriasis and atopic dermatitis patients were retrieved from NCBI GEO8 (GEO - https://www.ncbi.nlm.nih.gov/geo/) and EBI ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) repositories by using the R packages GEOquery9 and ArrayExpress10,11, respectively. For each dataset, a table specifying the disease (psoriasis/atopic dermatitis) and the origin of biopsy (lesional/non-lesional sample) in addition to other phenotypic information was also retrieved. Since the phenotypic information was heterogeneous across the datasets, rigorous harmonization procedure was performed. The GEO and Array Express identifiers of the retrieved datasets are reported in Tables 1–3.
Table 2.
DNA microarray and RNA-Sequencing datasets of Psoriasis and Atopic Dermatitis samples.
Table 1.
DNA microarray and RNA-Sequencing datasets of Atopic Dermatitis samples.
Table 3.
DNA microarray and RNA-Sequencing datasets of Psoriasis samples.
| Psoriasis | ||||
|---|---|---|---|---|
| Dataset ID | # of included samples | PMID | Technology | Platform |
| E-MTAB-3201 | 19 | 26086874 | Microarray | GPL571 |
| GSE2737 | 8 | 16283139 | Microarray | GPL91 |
| GSE6710 | 25 | 16858420 | Microarray | GPL96 |
| GSE13355 | 173 | 19169254 | Microarray | GPL570 |
| GSE14905 | 75 | 18648529 | Microarray | GPL570 |
| GSE30999 | 151 | 22763790 | Microarray | GPL570 |
| GSE34248 | 24 | 23308107 | Microarray | GPL570 |
| GSE41662 | 46 | 23308107 | Microarray | GPL570 |
| GSE50790 | 8 | 22479649 | Microarray | GPL570 |
| GSE52471 | 38 | 23771123 | Microarray | GPL571 |
| GSE58121 | 18 | 25058585 | Microarray | GPL14550 |
| GSE61281 | 52 | 25243786 | Microarray | GPL6480 |
| GSE67853 | 24 | 26763436 | Microarray | GPL570 |
| GSE68923 | 5 | 28570274 | Microarray | GPL13607 |
| GSE68924 | 5 | 28570274 | Microarray | GPL13607 |
| GSE68937 | 6 | 28570274 | Microarray | GPL13607 |
| GSE68939 | 5 | 28570274 | Microarray | GPL13607 |
| GSE78097 | 31 | 27185339 | Microarray | GPL570 |
| GSE80047 | 50 | 27152848 | Microarray | GPL13158 |
| GSE82140 | 8 | 27312025 | Microarray | GPL17692 |
| GSE83582 | 93 | 27448749 | Microarray | GPL19983 |
| GSE106087 | 6 | Unpublished | Microarray | GPL15207 |
| GSE41745 | 6 | 21850022 | RNA-Seq | GPL10999 |
| GSE47944 | 84 | 24909886 | RNA-Seq | GPL11154 |
| GSE54456 | 174 | 24441097 | RNA-Seq | GPL9052 |
| GSE63979 | 42 | 5723451 | RNA-Seq | GPL9052 |
| GSE67785 | 28 | 26251673 | RNA-Seq | GPL10999 |
| GSE74697 | 52 | 27793094 | RNA-Seq | GPL16791 |
| GSE83645 | 25 | 29031600 | RNA-Seq | GPL10999 |
| GSE107871 | 24 | 29273799 | RNA-Seq | GPL10999 |
| GSE117405 | 28 | 30054515 | RNA-Seq | GPL11154 |
| GSE123785 | 19 | 31539532 | RNA-Seq | GPL18573 |
| GSE123786 | 16 | 31539532 | RNA-Seq | GPL11154 |
Data quality check
The retrieved datasets were thoroughly quality checked. In particular, each sample was evaluated by visual inspection of the array pseudo-images, quality check reports and density plots of probe intensities by using the eUTOPIA software6. Further, outlier detection step, based on the sample distributions, was performed within each dataset by using ad hoc R scripts (see Code Availability section).
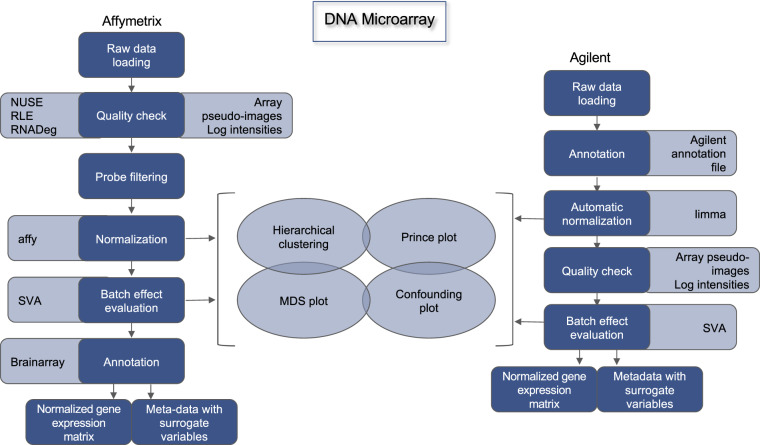
Moreover, for the Affymetrix datasets, outlier samples were detected by computing the Normalized Unscaled Standard Error (NUSE)12 and the Relative Log Expression (RLE)12 from the affyPLM v1.64.0 R package, and the RNA degradation curves (RNADeg)13 from the affy v1.64.0 R package (Fig. 1).
Fig. 1.
DNA microarray data preprocessing pipeline.
The distributions of the values of these three metrics were investigated by means of boxplots and the sample outlierness was evaluated for each measure based on the data distribution. Eventually, a concordance outlierness score was computed across the three metrics. In particular, a sample was removed from the analysis if considered an outlier in at least two out of three metrics, one of them being the RNA degradation curve.
Normalization
Data normalization was performed by using the eUTOPIA software. Affymetrixbased studies were normalized by using the justRMA from the R affy v1.66.0 package14. Agilent-based studies were quantile normalized with the normalizeQuantiles function from the limma v3.44.3 package15.
Surrogate variable analysis
In order to investigate the effect of unknown batches that might mask biological variability, Surrogate Variable Analysis (SVA) was performed with the eUtopia software, which implements the sva R package16. The analysis was performed by using origin of biopsy or diagnosis as variable of interest. The other biological variables (if present and if not confounded with the variable of interest) were used as covariates6. The estimated surrogate variables for each dataset are included in the meta-data tables.
Probe annotation
Custom annotation files (CDF files) were downloaded from Brainarray (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF\_download.asp) for Affymetrix-based microarrays. The latest version of Agilent probe annotation was retrieved from the Agilent website (https: //earray.chem.agilent.com/earray/). The probesets were mapped to the Ensembl gene IDs and the expression matrix was aggregated by computing the median of the expression of the Agilent probes mapping to the same Ensembl transcript ID. The entire DNA microarray data preprocessing is depicted in Fig. 1.
RNA Sequencing data
Data collection and homogenization
Raw files in “.fastq” format were retrieved from the European Nucleotide Archive (ENA). Along with the raw data files, the metadata tables reporting the samplewise clinical features for each dataset were also collected. As for the DNA microarray data, the metadata tables of RNASeq data were carefully harmonized to improve the across-datasets comparability. Phenotypic information for each dataset is reported along with the gene expression tables. GEO and ENA identifiers of the retrieved datasets are reported in Tables 1–3.
Quality control
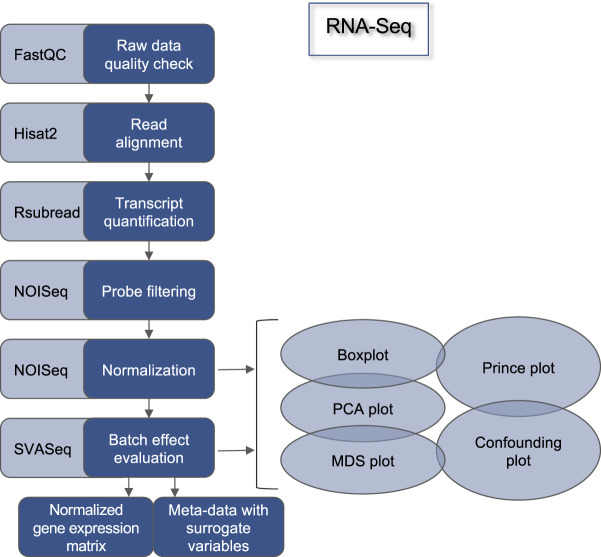
All the RNA-Seq datasets underwent quality check through the use of FastQC v0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastq c/). Reads were trimmed for low-quality ends in addition to adapters removal by TrimGalore v0.4.4_dev (http://www.bioinformatics.babraham.ac.uk/ projects/trim_galore/). In particular, the reads were trimmed if the Phred score was lower than 20 and discarded if the number of undetected nucleotides was greater than 50. The trimmed and adapter-clipped raw files were further quality checked with FastQC v0.11.7.
Read alignment
RNA Sequencing reads were then aligned against the human reference genome assembly GRCh38. The alignment was performed through the use of the HISAT2 algorithm17,18 using the genome indexes built for usage with HISAT2 (retrieved from https://ccb.jhu.edu/software/hisat2/manual.shtml).
Conversions between.sam and.bam file formats, sorting and extraction of uniquely mapped reads were performed through the use of samtools version 1.8-27-g089626219.
Read counts extraction
Transcript abundance was computed by using the featurecounts function from the Rsubread v2.2.3 R package20. To accomplish this task, the Gencode version 31 annotation was downloaded from https://www.gencodegenes.org, and then utilized for read counts extraction.
Low counts filtering
In order to filter out the transcripts with low expression levels in all the samples of each dataset, the proportion test strategy was used as implemented in the function filtered.data of the R package NOISeq v2.31.021.
Normalization
RNASeq expression data were normalized using the upper quantile method22 implemented in the R package NOISeq v2.31.0.
Surrogate Variable Analysis
As for the DNA microarray data, in order to identify unknown sources of technical variability, a Surrogate Variable Analysis (SVA) was performed through the use of the svaseq function implemented in the sva v3.36.0 R/Bioconductor package16. The analysis was performed by using disease state or diagnosis as variable of interest. The other biological variables (if present and if not confounded with the variable of interest) were used as covariates6. The estimated surrogate variables for each dataset are included in the meta-data tables, along with the gene expression tables. The entire RNA-Seq data preprocessing is depicted in Fig. 2.
Fig. 2.
RNA-Sequencing data preprocessing pipeline.
Data Records
The complete list of DNA microarray and RNA Sequencing datasets discussed in this work is reported in Tables 1–3. All the preprocessed transcriptomics data, along with harmonised meta-data, were submitted to Zenodo23.
Technical Validation
DNA microarray and RNA-Seq data are linked to clinical meta-data, reporting multiple information such as gender, age or the treatment (including e.g. drug dose). Additionally, sample meta-data is recorded, such as the tissue type a sample was taken from, or whether the tissue derives from a lesional or nonlesional sample.
In order to ensure that the data is recorded in a consistent and well-formed way, we created data dictionaries describing each of these variables. The data dictionaries contain detailed information describing the content of a variable, the data type (numeric, categorical, text, date, etc), the allowed values of categorical data or ranges of numeric variables.
The data was validated by checking compliance with the rules encoded in the data dictionaries. Data that was found not to comply with the rules was manually curated by consulting the original data sources. In fact, a large proportion of the datasets were found not to meet the requirements encoded in the data dictionaries. For instance, big heterogeneity was found in the description of the skin status. “Involved skin”, “psoriatic skin” were reported in order to describe the “lesional” status of the skin. “Normal”, “ctrl”, “Non-involved skin of healthy individual” were used to describe the “healthy control” samples. Yet, to define the gender, “m”, “f”, “male” and “female” were used across the datasets. All of these variables were mapped to the allowed values reported in the data dictionaries to improve the comparability across the datasets.
Usage Notes
The transcriptomics data presented in this article is an unprecedented source of preprocessed, harmonized, “ready-to-use” and FAIR datasets, made available to the scientific community. Data derived from both DNA microarray and RNASeq technologies can be exploited in order to uncover the molecular mechanisms underlying psoriasis and atopic dermatitis. Differential expression analysis can be carried for instance by the limma package15 for the microarray data, and the edgeR24, DESeq 225 or NOISeq21 packages for RNA-Seq data, respectively. Functional analysis of differentially expressed genes can be performed by using FunMappOne26, the R/Bioconductor package ReactomePA27 or Ingenuity Pathway Analysis (Qiagen, http://www.ingenuity.com/products/ipa). The inference and analysis of co-expression networks can be performed, for instance, by using the INfORM tool28. Altogether, these analyses can aid the stratification of PSO and AD patients, the identification of relevant biomarkers and novel therapeutic targets.
Acknowledgements
This study was supported by the EU IMI2 Biomap Project (Grant agreement 821511).
Author contributions
A.F. and D.G. conceived and designed the study; A.F. and V.H. retrieved the data and performed the data preprocessing; A.F. and N.C. quality checked and harmonised the meta-data; A.K. supervised the quality check and the data harmonisation. A.F., V.H. and A.S. drafted the manuscript. A.S. and D.G. supervised the activities and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Code availability
R scripts for the analysis of DNA microarray and RNA-Seq transcriptomics data are available for download at: https://github.com/Greco-Lab/psoriasis-dermatitis-analysis.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Antonio Federico, Veera Hautanen.
References
- 1.Bowcock AM, Cookson WO. The genetics of psoriasis, psoriatic arthritis and atopic dermatitis. Hum. Mol. Genet. 2004;13:43–55. doi: 10.1093/hmg/ddh094. [DOI] [PubMed] [Google Scholar]
- 2.Tsoi LC, et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J. Allergy Clin. Immunol. 2019;5:1406–1415. doi: 10.1016/j.jaci.2019.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoi LC, et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rendon A, Schäkel K. Psoriasis Pathogenesis and Treatment. Int. J. Mol. Sci. 2019;6:1475. doi: 10.3390/ijms20061475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A. Transcriptome profiling in human diseases: new advances and perspectives. Int. J. Mol. Sci. 2017;8:1652. doi: 10.3390/ijms18081652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marwah, V. S. et al. eUTOPIA: solUTion for Omics data PreprocessIng and Analysis. Source Code Biol. Med. 14 (2019). [DOI] [PMC free article] [PubMed]
- 7.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;1:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, et al. NCBI GEO: archive for functional genomics data sets update. Nucleic Acids Res. 2012;1:991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;14:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 10.Kauffmann A, et al. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics. 2009;16:2092–2094. doi: 10.1093/bioinformatics/btp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athar A, et al. ArrayExpress - update from bulk to single-cell expression data. Nucleic Acids Res. 2018;1:711–715. doi: 10.1093/nar/gky964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality assessment for short oligonucleotide microarray data. Technometrics. 2008;3:241264. [Google Scholar]
- 13.Fasold M, Binder H. AffyRNADegradation: control and correction of RNA quality effects in GeneChip expression data. Bioinformatics. 2013;1:129–131. doi: 10.1093/bioinformatics/bts629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;3:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;7:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leek, J. T. et al. sva: Surrogate Variable Analysis. R package version 3.32.1. https://bioconductor.org/packages/release/bioc/html/sva.html (2019).
- 17.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;9:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;4:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;16:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;8:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;21:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNASeq experiments. BMC Bioinf. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Federico A. 2020. Preprocessed and Harmonised Transcriptomics Datasets for Psoriasis and Atopic Dermatitis. Zenodo. [DOI] [PMC free article] [PubMed]
- 24.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;3:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol. 2014;12:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scala G, Serra A, Marwah VS, Saarimäki LA, Greco D. FunMappOne: a tool to hierarchically organize and visually navigate functional gene annotations in multiple experiments. BMC Bioinf. 2019;1:79. doi: 10.1186/s12859-019-2639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;2:477–479. doi: 10.1039/C5MB00663E. [DOI] [PubMed] [Google Scholar]
- 28.Marwah VS, et al. INfORM: Inference of NetwOrk Response Modules. Bioinformatics. 2018;12:2136–2138. doi: 10.1093/bioinformatics/bty063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Federico A. 2020. Preprocessed and Harmonised Transcriptomics Datasets for Psoriasis and Atopic Dermatitis. Zenodo. [DOI] [PMC free article] [PubMed]
Data Availability Statement
R scripts for the analysis of DNA microarray and RNA-Seq transcriptomics data are available for download at: https://github.com/Greco-Lab/psoriasis-dermatitis-analysis.